Abstract
Ovarian cancer is a female malignancy with high fatality-to-case ratio, which is due to late detection of cancer. Understanding the molecular mechanisms participating in these processes would facilitate design of therapeutic modalities and identification of novel tumor markers. Recent investigations have shown contribution of circular RNAs (circRNAs) in the evolution of ovarian cancer. These transcripts are produced through a back-splicing mechanism. The enclosed configuration of circRNAs protects them from degradation and potentiates them as biomarkers. Several circRNAs such as circMUC16, circRNA_MYLK, circRNA-UBAP2, circWHSC1, hsa_circ_0013958, circFGFR3, hsa_circRNA_102958 and circ_0072995 have been found to be up-regulated in this cancer, acting as oncogenes. On the other hand, circ-ITCH, circPLEKHM3, circ_100395, circ_0078607, circATRNL1, circHIPK3, circRHOBTB3, circEXOC6B, circ9119 and CDR1as are among down-regulated circRNAs in ovarian cancer. Expression levels of circCELSR1, circ_CELSR1, circATL2, circNRIP1, circTNPO3 and hsa_circ_0000714 have been shown to affect resistance of ovarian cancer cells to chemotherapy. Moreover, circ_100395, circFGFR3, circ_0000554, circCELSR1, circ-PTK2, circLNPEP, circ-CSPP1, circ_0000745, circ_100395 and circPLEKHM3 have been shown to regulate epithelial-mesenchymal transition and metastatic ability of ovarian cancer cells. In the current review, we explain the roles of circRNAs in the evolution and progression of ovarian cancer.
Keywords: circRNA, Ovarian cancer, Biomarker, Expression
Introduction
Epithelial ovarian cancer is the most fatal kind of malignancy among females [1]. Early detection of ovarian cancer is hindered by the lack of suitable tumor biomarkers, thus disease is usually diagnosed in advanced stages. Due to late diagnosis, this malignancy has the highest fatality-to-case ratio among gynecological cancers [2]. Malignant progression and prompt development of drug resistance are other problems encountered in clinical management of ovarian cancer [3]. The vast majority of ovarian tumors originate from the epithelial surface of the ovary. Others arise from germ cells or stromal cells. The main subclasses of epithelial cancers are serous, endometrioid, mucinous, clear cell, and undifferentiated cancers. These subclasses have different risk factors, clinical behaviors, and treatment responses [3]. From a molecular point of view, both genetic alterations in epithelial cells and reprogramming of the tumor microenvironment contribute in the evolution of ovarian cancer [3]. Understanding the molecular mechanisms participating in these processes would facilitate design of therapeutic modalities and identification of novel tumor markers [4, 5].
Circular RNAs (circRNAs) are a group of non-coding RNAs with a covalently closed configuration [6]. These transcripts have been initially regarded as a splicing error. However, their roles in the regulation of gene expression have been recognized during recent years. These transcripts are produced through back-splicing or exon skipping of precursor mRNAs [7]. These evolutionarily conserved transcripts have a high abundance in the cytoplasm and are more stable than linear transcripts. They can regulate expression of parental genes, modulate alternative splicing events or mRNA translation and act as molecular sponges for miRNAs or RNA-binding proteins. Moreover, they can occasionally produce peptides or proteins [7]. Recent studies have shown contribution of circRNAs in the pathogenesis of cancers [8]. In the current review, we explain the roles of circRNAs in the evolution and progression of ovarian cancer.
Up-regulated circRNAs in ovarian cancer
CircMUC16 is among up-regulated circRNAs in ovarian cancer tissues whose up-regulation in these tissues has been correlated with higher stage and grade. Down-regulation of circMUC16 in ovarian cancer cells has inhibited autophagy flux, while its forced over-expression has increased autophagy flux of cells. The impact of circMUC16 on autophagy has been shown to enhance invasion and metastasis of ovarian cancer cells. This effect has been exerted through binding to miR-199a-5p and releasing Beclin1 and RUNX1 from its suppressive roles. Moreover, RUNX1 has been found to elevate circMUC16 levels through increasing its transcription. Notably, circMUC16 can also directly bind to ATG13 and enhance its expression [9].
circRNA_MYLK is another up-regulated circRNAs in ovarian cancer tissues. Patients with over-expression of circRNA_MYLK have been found to have a more advanced stage and a lower overall survival time. In vitro studies have shown that circRNA_MYLK silencing attenuates proliferation ability of cells. Functionally, circRNA_MYLK can enhance the malignant progression of ovarian cancer cells through regulation of miR-652 levels [10].
Besides, circRNA-UBAP2 has been shown to be up-regulated this type of cancer. CircRNA-UBAP2 silencing has suppressed proliferation of ovarian cancer cells and induced their apoptosis. Mechanistically, circRNA-UBAP2 can target miR-382-5p and down-regulate its expression to release PRPF8 from its inhibitory effects [11].
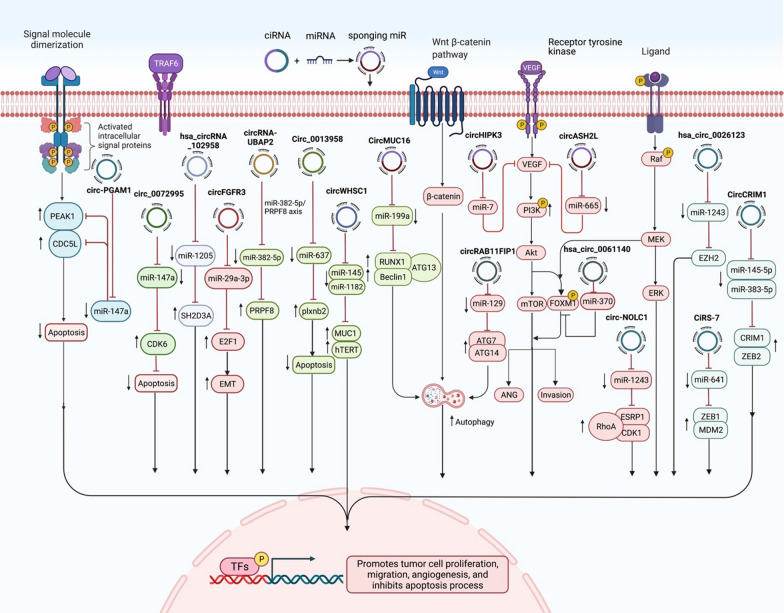
CircUBAP2 is another circRNA whose over-expression in ovarian cancer tissues has been correlated with clinical stage and survival of patients. This circRNA is mainly located in the cytoplasm. Up-regulation of circUBAP2 could enhance proliferative and migratory capacities of ovarian cancer cells. This circRNA acts as a sponge for miR-144 to release CHD2 from its inhibitory effects [12]. Figure 1 shows the effects of some oncogenic circRNAs in the progression of ovarian cancer.
Fig. 1.
A schematic representation of the effects of some oncogenic circRNAs in the progression of ovarian cancer. These circRNAs can sponge tumor suppressor miRNAs such as miR-147a, miR-1205, miR-29a, miR-382, miR-637, miR-145, miR-1182, miR-129 and miR-1243, thus increasing expression of certain oncogenes that affect activity of cancer-related signaling pathways
In order to find the impact of circRNAs in autophagy, Zhang et al. have assessed expression profile of circRNAs, miRNAs, and mRNAs in ovarian cancer cells after induction with Torin 1. They have reported up-regulation of 504 circRNAs and down-regulation of 478 ones. CircRAB11FIP1 has been among differentially expressed circRNAs. Expression of this circRNA has been found to be higher in epithelial ovarian cancer samples compared with normal ovarian tissues. Its silencing has suppressed the autophagic flux of SKOV3 cells. CircRAB11FIP1 has been shown to directly bind to miR-129 and regulate expression of miR-129 targets ATG7 and ATG14. CircRAB11FIP1 could also bind with DSC1 to assist its interaction with ATG101 [13]. Table 1 summarizes the results of studies that reported up-regulation of circRNAs in ovarian cancer.
Table 1.
Up-regulated circRNAs and Ovarian cancer
| circRNA | Clinical samples/animal model | Assessed cell lines | Targets/regulators/signaling pathways | Description | References |
|---|---|---|---|---|---|
| CircMUC16 | 3 EOC tissues and 4 healthy ovarian tissues/6-week-old BALB/c nude mice | SKOV3, ES-2, A2780 and CAOV-3 | miR-199a-5p, Beclin1, RUNX1 and ATG13 |
↑↑ circMUC16: ↑ autophagy flux of SKOV3 cells ↑ autophagy flux of SKOV3 cells: ↑ EOC invasion and metastasis |
[9] |
| circRNA_MYLK | 46 pairs of tumor tissues and ANCTs | SKOV3, OVCAR3, PEO1, 3AO, A2780, CAOV3 and HOSEPiCs | miR-652 |
Patients with high levels of circRNA_MYLK showed a higher pathological staging and a lower OS rate ∆ circRNA_MYLK: ↓ cell proliferation |
[10] |
| circRNA-UBAP2 | 20 pairs of tumor tissues and ANCTs | SKOV3, No. KL; OVCAR-3, No. H-OVCAR-3; ES-2, No. H-ES-2 and A2780, No. H-A2780 and IOSE80 | miR-382-5p/PRPF8 axis | ↑↑ circRNA-UBAP2: ↑ proliferation and ↓ apoptosis via sponging miR-382 | [11] |
| circRNA-UBAP2 | 24 pairs of tumor tissues and ANCTs | A2780, HEY, OVCAR3, HO8910, SKOV3 and IOSE | miR-144 |
Expression of circRNA-UBAP2 was negatively associated with TMN stage and five-year survival of OC patients ↑↑ circRNA-UBAP2: ↑ proliferation and migration via sponging miR-144 |
[12] |
| circWHSC1 | 79 patients and 13 healthy controls/4-week-old female BALB/c nude mice | CAOV3 and OVCAR3 | miR-145, miR-1182, MUC1 and hTERT | ↑↑ circWHSC1: ↑ cell proliferation, migration and invasion, and ↓ cell apoptosis via sponging miR-145 and miR-1182 | [14] |
| Hsa_circ_0013958 | 45 pairs of tumor tissues and ANCTs | A2780, OVCAR‐3 and HOSEpiC | – |
High levels of Hsa_circ_0013958 were related to patient FIGO stage and lymph node metastasis ∆ hsa_circ_0013958: ↓ proliferation, migration, and invasion and ↑ apoptosis |
[15] |
| Circ_0013958 | 30 pairs of tumor tissues and ANCTs/5-week-old female BALB/c nude mice | HOSE, SKOV3 and CAOV3 | miR-637/PLXNB2 axis | ∆ circ_0013958: ↓ proliferation, migration, invasion, and ↑ apoptosis | [16] |
| circFGFR3 | 35 pairs of tumor tissues and ANCTs | OSE, SKOV3, A2780, OV2008 and IGROV1 | miR-29a-3p/E2F1 axis | ↑↑ circFGFR3: ↑ EMT process | [17] |
| hsa_circRNA_102958 | 41 pairs of tumor tissues and ANCTs | OC cell lines and IOSE80 cells | miR-1205/SH2D3A axis | ∆ hsa_circRNA_102958: ↓ proliferation, migration and invasion | [18] |
| circ_0072995 | 40 pairs of tumor tissues and ANCTs/4 to 6-week-old BALB/c nude mice | HO8910, A2780, and IOSE80 | miR-147a/CDK6 axis | ↑↑ circ_0072995: ↑ cell proliferation, migration and ↓ apoptosis | [19] |
| circ_0072995 | 47 pairs of tumor tissues and ANCTs/6-week-old female BALB/c nude mice | IOSE-80, OVCAR-3 and SK-OV-3 | miR-122-5p/SLC1A5 axis | ∆ circ_0072995: ↓ cell proliferation, migration, invasion and ↑ cell apoptosis | [20] |
| circEPSTI1 | 50 pairs of tumor tissues and ANCTs/4‐week‐old BALB/c nude mice | – | miR-942/EPSTI1 axis | ∆ circEPSTI1: ↓ cell proliferation, invasion and ↑ apoptosis | [21] |
| circ-LOPD2 | normal ovarian tissue, benign tumor, borderline tumor and ovarian cancer tissues | CAOV3, A2780 and OVCAR3 | miR-378 | ∆ circ-LOPD2: ↓ cell growth | [22] |
| circGFRA1 | 50 pairs of tumor tissues and ANCTs/4-week old BALB/c nude mice | OV119 and A2780 | miR-449a/ GFRA1 axis | ∆ circGFRA1: ↓ cell proliferation and invasion and ↑ apoptosis | [23] |
| circ0004390 | 10 ovarian cancer tissues and 6 normal ovarian tissues | KOV3, HeyA8, OVCAR429 and HEK- 293 T | miR-198/MET axis | ∆ circ0004390: ↓ proliferation | [24] |
| circKIF4A | 50 pairs of tumor tissues and ANCTs/4-week-old female BALB/c nude mice | CAOV3 and SKOV3 | miR-127/ JAM3 axis | ∆ circKIF4A: ↓ cell proliferation and migration | [25] |
| circ_0000554 | – | HO8910 | miR-567 | ↑↑ circ_0000554: ↑ cell growth, invasion, and EMT process | [26] |
| circKRT7 | 5-week-old BALB/c mice | SKOV3, ES-2, CoC1, Caov-3, and Caov-4 | miR-29a-3p/COL1A1 axis | ∆ circKRT7: ↓ cell proliferation, migration and invasion via sponging miR-29a-3p | [27] |
| circCELSR1 | 4-week-old female athymic nude mice | SKOV3, A2780, IGROV1, CAOV3, and IOSE80 | miR-598/ BRD4 axis | ∆ circCELSR1: ↓ proliferation, migration, invasion and EMT process and ↑ apoptosis | [28] |
| circCELSR1 | 36 pairs of tumor tissues and ANCTs/6-week-old female BALB/c athymic nude mice | SKOV3, HeyA-8 and IOSE-80 | miR-1252/FOXR2 axis | ∆ circCELSR1: ↓ cell growth, ↑ G0/G1 arrest and apoptosis | [29] |
| circCELSR1 | – | – | miR-149-5p/SIK2 axis | ∆ circCELSR1: ↓ viability, colony formation and cell cycle process, ↑ paclitaxel sensitivity and cell apoptosis | [30] |
| circHIPK3 | 69 pairs of tumor tissues and ANCTs | A2780, HO8910, SKOV3, CAOV3 and HOEC | – | Patients with higher levels of circHIPK3 showed lymph node invasion, FIGO stage, and worse DFS and OS | [31] |
| circHIPK3 | 66 pairs of tumor tissues and ANCTs | SKOV3 | miR-7/VEGF axis | ∆ circHIPK3: ↓ tumorigenicity of ovarian cancer cells, proliferation and ↑ apoptosis | [32] |
| circRAB11FIP1 | ovarian cancer tissues and serum samples from 70 EOC and 30 matched non-carcinoma tissue samples/6-week-old BALB/c nude mice | SKOV3 | miR-129/ ATG7 and ATG14 |
∆ circRAB11FIP1: ↓ autophagic flux of ovarian cancer SKOV3 cells ↑↑ circRAB11FIP1: ↑ autophagy, proliferation and invasion |
[13] |
| Hsa_circ_0009910 | 50 pairs of tumor tissues and ANCTs | SKOV3 | miR-145 | Hsa_circ_0009910 induces proliferative and motile phenotypes via sponging miR-145 in ovarian cancer cells | [33] |
| circASH2L | 50 pairs of tumor tissues and ANCTs/4-week-old female athymic BALB/c nude mice | A2780, TOV112D, OVCAR-3, SKOV3 and ISOE80 | miR-665/VEGFA axis | ∆ circASH2L: ↓ invasion and cell growth in vitro, angiogenesis and lymphangiogenesis in vivo | [34] |
| circ-PGAM1 | 15 EOC tissues and 15 normal ovary tissues/nude mice | CAOV3, SKOV3, OVCAR3, ES‐2 and 293T cells | miR-542-3p/CDC5L/PEAK1 pathway | ∆ circ-PAGM1: ↓ proliferation, migration, and invasion of ovarian cancer cells and ↑ apoptosis | [35] |
| circRhoC | 127 ovarian cancer tissues and 24 normal ovarian tissues | A2780 cells | miR-302e/ VEGFA axis | ↑↑ circRhoC: ↑cell viability, migration and invasion via sponging miR-302e | [36] |
| circPUM1 | 62 EOC and 13 normal ovarian tissues/5-week-old female BALB/c nude mice | A2780, CAOV3 and HMrSV5 | miR-615-5p, miR-6753-5p, NF-κB and MMP2 |
↑↑ circPUM1: ↑ proliferation, migration, and invasion and ↓ apoptosis CircPUM1 showed to act on the peritoneum and increase metastasis of cancer in the form of cancer-derived exosomes |
[37] |
| circ_0007841 | 43 pairs of tumor tissues and ANCTs/6-week-old BALB/c nude mice | SKOV3, OVCAR3 and IOSE80 | miR-151-3p/MEX3C axis | ∆ circ_0007841: ↓ proliferation, migration and invasion | [38] |
| hsa_circ_0026123 | 20 pairs of tumor tissues and ANCTs/4 weeks old female BALB/c nude mice | A2780, TOV112D, SKOV3, OVCAR-3 and ISOE80 | miR-124-3p/EZH2 axis | ∆ hsa_circ_0026123: ↓ proliferation and metastasis | [39] |
| circFoxp1 | 112 EOC patients and 82 healthy controls/nude mice | COC1, OVCAR3, SKOV3, SKOV3/DDP and IOSE-80 | miR-22, miR-150-3p, CEBPG and FMNL3 |
↑↑ circFoxp1: ↑ proliferation and DDP resistance High levels of circFoxp1 were correlated with lymphatic metastasis, distant metastasis, FIGO stage, primary tumor size, residual tumor diameter, and clinical response |
[40] |
| CircCRIM1 | 130 ovarian cancer tissues and 24 normal ovarian tissues/4-week-old female BALB/c nude mice | OVCAR3 and CAOV3 | miR-145-5p, miR-383-5p, CRIM1 and ZEB2 | ↑↑ CircCRIM1: ↑ cancer progression in vitro and tumor growth in vivo | [41] |
| hsa_circ_0061140 | 4-week-old male BALB/c nude mice | SKOV3 and A2780 | miR-370/FOXM1 axis | ∆ hsa_circ_0061140: ↓ proliferation and migration | [42] |
| circ_0061140 | 20 PTX-resistant human ovarian cancer tissues and 19 PTX-sensitive human ovarian cancer tissues/5-week-old female BALB/c nude mice | SKOV3, HeyA8 and IOSE-80 | miR-136/CBX2 axis | ∆ circ_0061140: ↓ proliferation, migration and invasion, and ↑ apoptosis and PTX sensitivity | [43] |
| circ-PVT1 | GTEx database | SKOV3 and A2780 | miR-149-5p/FOXM1 axis |
∆ circ-PVT1: ↓ proliferation, migration and invasion High levels of PVT1 were correlated with shorter OS in OV patients |
[44] |
| circ-PVT1 | – | CAOV3, SKOV3, SNU119, OVCAR3 and HOSEpiC | miR-149 |
∆ circ-PVT1: ↓ proliferation and ↑ apoptosis ↑↑ circ-PVT1: ↑ proliferation and ↓ apoptosis |
[45] |
| circ_0015756 | 55 pairs of tumor tissues and ANCTs/5-week-old female BALB/c nude mice | OV90, SKOV3 and IOSE80 | miR-942-5p/CUL4B axis | ∆ circ_0015756: ↓ proliferation, migration and invasion and ↑ apoptosis | [46] |
| circ_0025033 | 39 pairs of tumor tissues and ANCTs/6–8 weeks old female BALB/c mice | KOV3 and A2780 and IOSE80 | miR-184/LSM4 axis | ∆ circ_0025033: ↓ colony formation, migration, invasion and glycolysis metabolism | [47] |
| circ_0005276 | 49 pairs of tumor tissues and ANCTs | CAOV3 and SKOV3 | ADAM9 |
∆ circ_0005276: ↓ migration High levels of circ_0005276 were associated with lymphatic metastasis and distant metastasis in EOC patients |
[48] |
| circ-NOLC1 | 118 ovarian cancer tissues, 11 borderline tumor tissues, 11 benign ovarian tissues, 15 normal ovarian tissues/5-week-old female BALB/c nude mice | A2780, CAOV3, ES-2, HO8910, OVCAR3, and SKOV3 | ESRP1, CDK1 and RhoA | ↑↑ circ-NOLC1: ↑ proliferation, migration, and invasion tumor growth by binding ESRP1 and modulating CDK1 and RhoA expression | [49] |
| circBIRC6 | – | SKOV3, SKOV3/DDP | miR-367-3p | ∆ circBIRC6: ↓ proliferation of ovarian cancer cisplatin-resistant cells and ↑ apoptosis | [50] |
| circ-0001068 | 20 pairs of tumor tissues and ANCTs 95 OC patients and 53 healthy controls | – | miR-28-5p | Circ-0001068 was found to be delivered into T cells and induced PD1 expression by sponging miR-28-5p | [51] |
| circRNA051239 | 30 EOC patients and 10 healthy controls | SKOV3.ip, SKOV3, A2780, CAOV3 and OVCAR3 | miR-509-5p/PRSS3 axis | ∆ circRNA051239: ↓ proliferation and migration | [52] |
| circVPS13C | 40 pairs of tumor tissues and ANCTs | A2780, SKOV3 and IOSE-80 | miR-145 and MEK/ERK signaling | Propofol treatment: ↓ circVPS13C levels and ↑ miR-145 levels, thus ↓ viability, cell cycle and motility and ↑ apoptosis | [53] |
| circANKRD12 | – | PA-1, SKOV3, Caov3, NIH:OVCAR-3 and APOCC | – | Downregulation of circANKRD12 compelled a strong phenotypic change in cell cycle, invasion and migration and metabolism in cancer cells | [54] |
| VPS13C-has-circ-001567 | 20 pairs of tumor tissues and ANCTs | SKOV3 and OV-1063 | – |
∆ VPS13C-has-circ-001567: ↓ proliferation, tumorigenicity and ↑ apoptosis High levels of VPS13C-has-circ-001567 were associated with tumor node metastasis stage and lymph node metastasis |
[55] |
| circPIP5K1A | 25 pairs of tumor tissues and ANCTs/4-week-old BALB/c nude mice | VCAR5, SKOV3, A2780, OV2008 and HCerEpiC | miR-661/IGFBP5 axis | ∆ circPIP5K1A: ↓ proliferation, migration and invasion | [56] |
| circATL2 | – | PTX-resistant OC tissues and cells | miR-506-3p/NFIB axis | ∆ circATL2: ↓ colony formation, resistance of OC to PTX and ↑ cell cycle arrest and apoptosis in PTX-resistant OC cells | [57] |
| hsa_circ_0004712 | 30 pairs of tumor tissues and ANCTs/4–6 weeks old female BALB/c mice | OVCAR-3, SKOV-3 and IOSE-80 | miR-331-3p/FZD4 axis | ∆ hsa_circ_0004712: ↓ proliferation, colony formation, invasion and migration, and ↑ apoptosis | [58] |
| CiRS-7 | 40 pairs of tumor tissues and ANCTs/4-week-old male BALB/c nude mice | SKOV3, A2780, OV2008, IGROV1, ES-2 and HOSE | miR-641/ZEB1 or miR-641/MDM2 axis |
∆ CiRS-7: ↓ cell growth and metastasis High levels of CiRS-7 were correlated with the TNM stages, lymph node metastasis status and overall survival rate in OC patients |
[59] |
| circ-PTK2 | 26 ovarian cancer tissues and 11 normal ovary tissues/4–5-week-old female nude mice | SK-OV-3 and OVCAR-3 | miR-639/FOXC1 axis | ↑↑ circ-PTK2: ↑tumor formation, migration and invasion and EMT process | [60] |
| circLNPEP | 40 pairs of tumor tissues and ANCTs/4-week-old male BALB/c nude mice | A2780, SKOV-3, OVCAR3, SK-BR-3, OV-56 and TOV-21 G | miR-876-3p/WNT5A axis | ∆ circLNPEP: ↓ cell viability, proliferation, migration, invasion, angiogenesis, and EMT process and ↑ apoptosis | [61] |
| circNRIP1 | 56 pairs of tumor tissues and ANCTs/BALB/c nude mice | HOEC, A2780, SKOV3 and A2780/PTX and SKOV3/PTX | miR-211-5p/HOXC8 axis | ∆ circNRIP1: ↓ PTX resistance of OC cells in vitro and OC tumor in vivo | [62] |
| circTNPO3 | 48 pairs of tumor tissues and ANCTs/nude mice | SKOV3, HeyA-8 and IOSE-80 | miR-1299/NEK2 axis | ∆ circTNPO3: ↑ sensitivity to PTX via promoting PTX-induced apoptosis in vitro and in vivo | [63] |
| hsa_circ_0051240 | 10 pairs of tumor tissues and ANCTs/6-week-old male nude mice | CAOV-3, SKOV-3, OVCAR-3 and H8910 and HOSE | miR-637/KLK4 axis | ∆ hsa_circ_0051240: ↓ cell proliferation, migration and invasion in vitro, and tumor formation in vivo | [64] |
| circSETDB1 | 73 pairs of tumor tissues and ANCTs/4–6-week-old female BALB/c nude mice | A2780, SKOV3, IOSE-80 and 293 T cells | miR-129-3p/MAP3K3 pathway | ∆ circSETDB1: ↓ cell proliferation, migration, invasion and ↑ apoptosis | [65] |
| circSETDB1 | 60 SOC patients [18 primary chemoresistance, 42 primary chemosensitive] and 60 healthy controls | – | – |
High levels of circSETDB1 Were correlated with advanced clinical stage, lymph node metastasis and a shorter PFS time of SOC patients |
[66] |
| hsa_circ_0000714 | – | SKOV3, A2780, SKOV3/PTX and A2780/PTX | miR-370-3p/RAB17 axis and CDK6/RB signaling pathway | Hsa_circ_0000714 was found to regulate RAB17 expression via sponging miR-370-3p, and through the CDK6/RB signaling pathway, so it showed to play a role in the malignant progression of the paclitaxel-resistant ovarian cancer cell A2780/PTX | [67] |
| circ_MUC16 | 30 pairs of tumor tissues and ANCTs/female BALB/c nude mice | A2780, SK-OV-3 and IOSE-80 | miR-1182/S100B axis |
∆ circ_MUC16: ↓ proliferation, glycolysis metabolism, migration and invasion ↑↑ circ_MUC16: ↓ effects of Propofol to promote the aggressive behaviors of ovarian cancer via sponging miR-1182 |
[68] |
| circ-FAM53B | 54 pairs of tumor tissues and ANCTs | HO8910, SKOV3, OVCAR3, A2780 and IOSE80 | miR-647, VAMP2 and MDM2 |
↑↑ circ-FAM53B: ↑ proliferation, migration, and invasion High levels of circ-FAM53B were correlated with clinical severity and poor prognosis of OC patients |
[69] |
| circ-ABCB10 | 103 EOC tumor tissues and 53 EOC adjacent tissues as control | OVCAR3, UWB1.289, SKOV3, CAOV3 and IOSE80 | miR-1271, miR-1252 and miR-203 |
High levels of circ-ABCB10 were associated with poor differentiation, large tumor size and advanced FIGO stage and worse OS in EOC patients ↑↑ circ-ABCB10: ↑ proliferation and ↓ apoptosis |
[70] |
| circ-ABCB10 | – | OVCAR3, UWB1.289, SKOV3, CAOV3 and IOSE80 | miR-1271, Capn4/Wnt/β-catenin signaling | ↑↑ circ-ABCB10: ↑ proliferation, invasion, Capn4/Wnt/β-catenin signaling pathway, ↓ apoptosis via sponging miR-1271 | [71] |
| circ-CSPP1 | 12 borderline tumors, 117 ovarian carcinomas tissues, 12 benign ovarian tissues and 15 normal ovarian tissues | AOV3, A2780, OVCAR3 | miR-1236-3p/ZEB1 axis |
∆ circ-CSPP1: ↓ cell growth, migration, invasion, and EMT process High levels of circ-CSPP1 were associated with correlated with FIGO staging and differentiation |
[72] |
| circ_0002711 | 54 pairs of tumor tissues and ANCTs/nude mice | SKOV3, OV90 and IOSE80 | miR-1244/ROCK1 axis | ∆ circ_0002711: ↓ cell viability, colony formation ability and aerobic glycolysis | [73] |
| circE2F2 | – | OC tissues and cell lines | – |
↑↑ circE2F2: ↑ proliferation, cell growth, metastasis, and glucose metabolism by stabilizing the E2F2 mRNA High levels of circE2F2 could strengthen the stability of the E2F2 mRNA through binding to the HuR protein High levels of circE2F2 were correlated with poor OS in OC patients |
[74] |
| circHIPK2 | 46 pairs of tumor tissues and ANCTs/4–5-week-old female BALB/c-nude mice | SKOV3, A2780, SKOV3/DDP and A2780/DDP and IOSE80 | miR-338-3p/CHTOP axis | ∆ circHIPK2: ↓ cell proliferation, cell cycle entrance, migration and invasion in SKOV3/DDP and A2780/DDP cells and ↑ apoptosis and suppresses the 50% inhibitory concentration of DDP | [75] |
| circ_0000745 | 50 pairs of tumor tissues and ANCTs/4-week-old BALB/c nude mice | CoC1, ES-2, SW626, SK-OV-3 and IOSE-80 | miR-3187-3p/ERBB4 axis and PI3K/AKT signaling pathway | ∆ circ_0000745: ↓ proliferation, aggressiveness, EMT process, and stemness of SK-OV-3 cells | [76] |
∆ knock-down or deletion, ANCTs adjacent non-cancerous tissues, EOC epithelial ovarian cancer, OS overall survival, EMT epithelial-mesenchymal transition, PFS progression-free survival, DFS disease-free survival, PTX paclitaxel, DDP cisplatin, HGSOC high-grade serous ovarian cancer, GTEx genotype-tissue expression, FIGO International Federation of Gynecology and Obstetrics, SOC high-grade serous ovarian cancer
Down-regulated circRNAs in ovarian cancer
A number of studies have reported down-regulation of certain circRNAs in ovarian cancer. For instance, circular RNA-ITCH has been shown to exert tumor suppressor role in this cancer. Down-regulation of circRNA-ITCH in this type of cancer has been associated with up-regulation of lncRNA HULC. Up-regulation of circRNA-ITCH has led to inhibition of cell proliferation, while up-regulation of HULC has resulted in opposite effects. Moreover, up-regulation of circRNA-ITCH has suppressed expression of HULC in these cells. While up-regulation of HULC has not affected expression of circRNA-ITCH, it has decraesed the inhibitory effect of circRNA-ITCH overexpression. Taken together, circRNA-ITCH can suppress proliferation of ovarian cancer cells through down-regulating HULC [77]. Moreover, circRNA-ITCH has been shown to suppress proliferation, invasiveness, and glycolysis of ovarian cancer cells through enhancing expression of CDH1 due to its sponging effect on miR-106a [78].
An RNA sequencing experiment has identified circPLEKHM3 as one of the utmost considerably down-regulated circRNAs in ovarian cancer samples versus normal tissues. Moreover, this circRNA has been found to be down-regulated in peritoneal metastatic ovarian cancers compared with primary cancers. Down-regulation of circPLEKHM3 has also been associated with poor prognosis. Mechanistically, up-regulation of circPLEKHM3 can inhibit cell growth, migration and epithelial-mesenchymal transition, while its silencing has led to opposite consequences. This circRNA acts through sponging miR-9 and regulation expressions of BRCA1, DNAJB6 and KLF4, and activity of AKT1 signaling. Moreover, the tumor-promoting effects of circPLEKHM3 silencing could be blocked by AKT inhibitor MK-2206 [79]. Another study has shown that the tumor suppressor role of curcumin in ovarian cancer is exerted through regulation of circ-PLEKHM3/miR-320a/SMG1 axis [80].
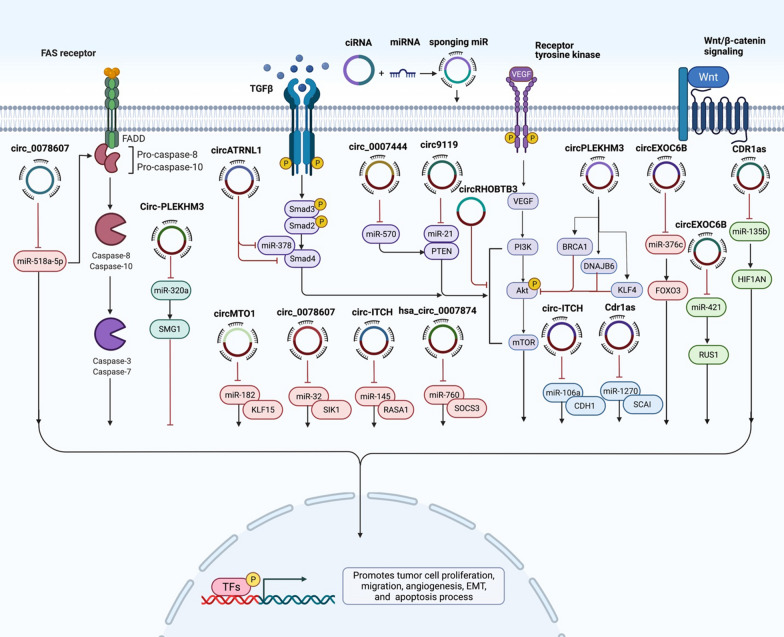
Hsa_circ_0078607 is another tumor suppressor circRNA whose inhibitory roles in ovarian cancer have been verified by different studies. This circRNA has been found to suppress progression of ovarian cancer through regulation of miR-518a-5p/Fas [81] and miR-32-5p/SIK1 [82] pathways. Moreover, down-regulation of this circRNA has predicted poor clinical outcome in high-grade serous ovarian cancer [83]. Figure 2 shows a number of tumor suppressor circRNAs in ovarian cancer.
Fig. 2.
A schematic representation of the effects of some tumor suppressor circRNAs in the progression of ovarian cancer. The sponging effects of tumor suppressor circRNAs on oncogenic miRNAs such as miR-182, miR-32, miR-145 and miR-740 decrease proliferation and induce apoptosis of ovarian cancer cells. Thus, down-regulation of these circRNAs promotes progression of ovarian cancer
CircEXOC6B is another tumor suppressor circRNA that inhibits proliferation and migratory potential of ovarian cancer cells and enhances their sensitivity to paclitaxel via modulation of miR-376c-3p/FOXO3 axis [84]. Moreover, it could progression of this cancer through influencing miR-421/RUS1 axis [85]. Notably, the tumor suppressor circRNA-9119 has been shown to affect miR-21-5p/PTEN/Akt axis [86]. Finally, circ-CDR1as could sequester miR-135b-5p to inhibit progression of ovarian cancer [87]. Moreover, it could up-regulate expression of SCAI to attenuate resistance of ovarian cancer cells to cisplatin through suppression of miR-1270 levels [88].
CircBNC2 is another tumor suppressor circRNA with potential biomarker role. It has been shown to perform better than HE4 and CA125 in differentiating patients with ovarian cancer from those with benign lesions or healthy subjects. Most notably, it could also separate early stage ovarian cancer from benign and healthy conditions. The performance of circBNC2 levels has been similar among pre- and postmenopausal subjects [89].
Table 2 shows the list of down-regulated circRNAs in ovarian cancer.
Table 2.
Down-regulated circRNAs in ovarian cancer
| circRNA | Clinical samples/animal model | Assessed cell lines | Targets/regulators/signaling pathways | Description | References |
|---|---|---|---|---|---|
| circPLEKHM3 | 5 tumor tissues and 5 normal ovarian tissues from patients with benign gynaecological diseases/4-week-old female athymic BALB/c nude mice | A2780, OV90 and MDAH2274 | miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis |
↑↑ circPLEKHM3: ↓ cell growth, migration and EMT process Patients with low levels of circPLEKHM3 showed a worse prognosis |
[79] |
| circPLEKHM3 | 35 pairs of tumor tissues and ANCTs/5-week-old female BALB/c athymic mice | SKOV3, A2780 and 293T cells | miR-320a/SMG1 axis | Curcumin treatment: ↑ circPLEKHM3 levels: ↓ cell proliferation and ↑ apoptosis | [80] |
| circ_100395 | 60 pairs of tumor tissues and ANCTs | A2780, OV2008, SKOV3, IGROV1 and ES-2 | miR-1228 | ↑↑ circ_100395: ↓ tumor growth, metastasis and EMT process | [90] |
| circ_0078607 | 20 pairs of tumor tissues and ANCTs | SKOV3 and A2780 | miR-518a-5p/Fas axis | ↑↑ circ_0078607: ↓ proliferation and ↑ apoptosis via sponging miR-518a-5p | [81] |
| circ_0078607 | 43 pairs of tumor tissues and ANCTs/female BALB/c nude mice | HEY, ES-2 and IOSE80 | miR-32-5p/SIK1 axis | ↑↑ circ_0078607: ↓ proliferation, migration, invasion, and ↑ apoptosis via sponging miR-32-5p | [82] |
| circ_0078607 | 49 pairs of tumor tissues and ANCTs | – | – | Patients with low levels of circ_0078607 had advanced FIGO stage, higher serum CA125 level, shorter PFS and OS | [83] |
| circATRNL1 | 56 pairs of tumor tissues and ANCTs/6-week-old nude mice | A2780, SKOV3, CAOV‐3, SNU119 and IOSE80 | miR-378/Smad4 axis | ↑↑ circATRNL1: ↓ proliferation, invasion, migration, angiogenesis and ↑ apoptosis | [91] |
| circHIPK3 | 21 high grade EOC and 21 normal ovarian tissues | A2780, SKOV3 and IOSE80 | – | ∆ circHIPK3: ↑ proliferation, migration, and invasion and ↓ apoptosis | [92] |
| circRHOBTB3 | – | ovarian cancer cells | PI3K/AKT signaling pathway | ↑↑ circRHOBTB3: ↓ cell proliferation, metastasis and glycolysis via inactivating PI3K/AKT signaling pathway | [93] |
| circEXOC6B | 60 ovarian cancer patients and 60 healthy controls/BALB/c mic | A2780, SKOV3 and IOSE-80 | miR-376c-3p/FOXO3 axis |
↑↑ circEXOC6B: ↓ proliferation, motility and chemoresistance of ovarian cancer cells to PTX via sponging miR-376c-3p Low levels of circEXOC6B were correlated with malignant pathological characteristics in ovarian cancer patients |
[84] |
| circEXOC6B | – | A2870, SKOV3, OVCAR3 and IOSE80 | miR-421/RUS1 axis | ↑↑ circEXOC6B: ↓ proliferation, invasion and ↑ apoptosis via sponging miR-421 | [85] |
| circ9119 | 40 ovarian cancer patients and 10 normal controls/5-week old female BALB/c nude mice | SKOV-3, HO-8910, A2780, ES-2, CAOV3, and OVCAR3 and FTE187 | miR-21 and PTEN/Akt pathway | ↑↑ circ9119: ↓ proliferation, viability and ↑ apoptosis | [86] |
| CDR1as | 65 ovarian cancer patients and 37 normal controls | HO8910 and A2780 | miR-135b-5p/HIF1AN axis | ↑↑ CDR1as: ↓ proliferation, invasion and migration | [87] |
| Cdr1as | 66 pairs of tumor tissues and ANCTs/4-week-old BALB/c female athymic mice | A2780, SKOV-3 and IOSE-80 | miR-1270/SCAI axis | ↑↑ CDR1as: ↓ proliferation and ↑ cisplatin-induced cell apoptosis in ovarian cancer cells via sponging miR-1270 | [88] |
| circRNA1656 | 60 HGSOC tissues and 60 benign ovarian tissues | SKOV-3, HO 8910, A2780 and OVCAR-3 | – | Downregulation of circRNA1656 was correlated with the FIGO stage of HGSOC | [94] |
| circ-ITCH | 45 pairs of tumor tissues and ANCTs/5-week-old BALB/c nude mice | A2780, OVCAR3 and ISOE80 | miR-106a/CDH1 axis |
↑↑ circ-ITCH: ↓ proliferation, invasion, glycolysis and ↑ apoptosis via sponging miR-106a Levels of circ-ITCH were positively associated with 5-year OS of patients |
[78] |
| circ-ITCH | 75 pairs of tumor tissues and ANCTs | UWB1.289 and UWB1.289 + BRCA1 | lncRNA HULC | ↑↑ circ-ITCH: ↓ proliferation via downregulating HULC | [77] |
| circ-ITCH | – | SKOV3, A-2780, OVCAR-3, HO-8910 and IOSE80 | miR-10a | ↑↑ circ-ITCH: ↓ proliferation and ↑ apoptosis via sponging miR-10a | [95] |
| circ-ITCH | 20 pairs of tumor tissues and ANCTs/6-weeks-old female BALB/c nude mice | SK-OV-3 and Caov-3 | miR-145/RASA1 axis | ↑↑ circ-ITCH: ↓ viability and motility by CCK8, cell cycle, wound healing assay and invasion via sponging miR-145 | [96] |
| circ-ITCH | 77 pairs of tumor tissues and ANCTs | SKOV3 and OVCAR-3 | – |
High levels of circ-ITCH were associated with small tumor size, decreased FIGO stage and prolonged OS ↑↑ circ-ITCH: ↓ proliferation and ↑ apoptosis |
[97] |
| circBNC2 | 83 EOC patients, 83 benign ovarian cysts, and 83 healthy controls | – | – | CircBNC2 was found to be downregulated in EOC and could be promising novel biomarker for EOC | [89] |
| circMTO1 | 48 pairs of tumor tissues and ANCTs | SKOV3 and OVCAR3 and IOSE80 | miR-182-5p/KLF15 axis | ↑↑ circMTO1: ↓ proliferation and invasion | [98] |
| circ_0007444 | 87 pairs of tumor tissues and ANCTs/5-week-old female nude mice | SKOV3, OV420, A2780, CAOV3, OVCAR3 and HOSEpiC | miR-570-3p/PTEN axis |
↑↑ circ_0007444: ↓ proliferation, migration, and invasion, and ↑ apoptosis via sponging miR-570-3p Low levels of circ_0007444 were correlated with advanced tumor stage and grade, large tumor size, and low 60-month percent survival |
[99] |
| circLARP4 | 78 pairs of tumor tissues and ANCTs | – | – | Low levels of circLARP4 were correlated with FIGO stage, lymph node metastases and poor prognosis of OC patients | [100] |
| circLARP4 | – | SKOV3, A2780, SW626, OVCAR3, OVCAR4 and HOSEpiC | miR-513b-5p/LARP4 axis | ↑↑ circLARP4: ↑proliferation, invasion and migration | [101] |
| hsa_circ_0007874 | 4-week-old BALB/c nude mice | IGROV1, A2780, ES‐2, OV2008, and SKOV3 and ISOE80 | miR-760/SOCS3 axis | ↑↑ hsa_circ_0007874: ↓ proliferation and migration | [102] |
| circN4BP2L2 | 126 EOC patients, 126 benign ovarian cyst, and 126 healthy controls | SKOV3, OVCAR3, CAOV3, HO8910, TOV-112D, and IOSE80 | – |
↑↑ circN4BP2L2: ↓ migration and invasion Low levels of circN4BP2L2 were correlated with advanced tumor stage, worse histological grade, lymph node metastasis and distant metastasis in EOC |
[103] |
∆ knock-down or deletion, ANCTs adjacent non-cancerous tissues, EOC epithelial ovarian cancer, OS overall survival, EMT epithelial-mesenchymal transition, PFS progression-free survival, DFS disease-free survival, PTX paclitaxel, DDP cisplatin, HGSOC high-grade serous ovarian cancer, GTEx genotype-tissue expression, FIGO International Federation of Gynecology and Obstetrics, SOC high-grade serous ovarian cancer
Discussion
Ovarian cancer is a malignancy with highly variable clinical behavior ranging from good prognosis and high chance of cure to fast progression and poor clinical outcome [3]. This variable clinical manifestation most probably reflects dissimilarity in the biological characteristics of tumors [3]. Recent studies have used bioinformatics tools for identification of dysregulated genes in this kind of cancer to find the most important pathways, targets for treatment and candidate drugs [104].
CircRNAs with prominent roles in determination of cancer cells malignant behavior [105] and response to therapeutic options can explain at least some parts of this variability. These transcripts have critical roles in the regulation of expression of known tumor suppressor genes or oncogenes, since they can sequester miRNAs that suppress expression of these genes [106, 107].
CircRNAs have been shown to participate in the pathogenesis ovarian cancer through sponging miRNAs. CircMUC16/miR-199a-5p, circRNA_MYLK/miR-652, circRNA-UBAP2/miR-382-5p, circRNA-UBAP2/miR-144, circWHSC1/miR-145, circ_0013958/miR-637, circFGFR3/miR-29a-3p, hsa_circRNA_102958/miR-1205, circ_0072995/miR-147a, circ_0072995/miR-122-5p and circEPSTI1/miR-942 are examples circRNAs/miRNA axes in which an oncogenic circRNA acts as a sponge for a tumor suppressor miRNA. On the other hand, circPLEKHM3/miR-9, circPLEKHM3/miR-320a, circ_100395/miR-1228, circ_0078607/miR-518a-5p, circ_0078607/ miR-32-5p, circATRNL1/miR-378 and circEXOC6B/miR-376c-3p are examples of tumor suppressor circRNAs/oncogenic miRNA axes.
Since expression of circRNAs is influenced in the process of carcinogenesis and they are stable in the circulation of patients, circRNAs can act as diagnostic and prognostic markers in ovarian cancer. The former application is highlighted by the stability of these transcripts in the circulation of affected individuals which potentiates them as candidates for non-invasive methods of cancer detection. It is expected that therapeutic modalities affect expression of circRNAs, thus evaluation of expression of these transcripts in the peripheral blood might reveal response to therapy or tumor recurrence. Thus, they might replace the conventional nonspecific ovarian cancer biomarkers. Application of circRNAs as prognostic markers is supported by the studies that reported correlations between their levels and clinical as well as pathological parameters related to cancer prognosis. Future studies are needed to elaborate the association between expression levels of circRNAs and standard staging and grading systems of ovarian cancer. High throughput sequencing techniques would pave the way for identification of stage-/grade-specific panels of dysregulated circRNAs in ovarian cancer.
Moreover, circRNAs can affect response of ovarian cancer cells to paclitaxel. CircCELSR1, circ_CELSR1, circATL2, circNRIP1, circTNPO3 and hsa_circ_0000714 are examples of circRNAs that have important roles in either determination or modulation of chemoresistant phenotype. Since expression levels of these circRNAs affect responses of ovarian cancer cells to chemotherapy, they are putative markers that could be useful for monitoring molecular responses. Epithelial-mesenchymal transition of ovarian cancer cells has also been shown to be affected by a number of oncogenic circRNAs such as circ_100395, circFGFR3, circ_0000554, circCELSR1, circ-PTK2, circLNPEP, circ-CSPP1 and circ_0000745 as well as tumor suppressor ones such as circ_100395 and circPLEKHM3. The impact of non-coding RNAs on activity of cancer-related signaling is a crucial element in the carcinogenesis [108].
Conclusion
Taken together, circRNAs can represent suitable candidate tumor markers in ovarian cancer and therapeutic targets to enhance response of cancer cells to conventional therapies. Moreover, results of in vitro and animal studies have proposed that targeting circRNAs can decrease malignant phenotype of ovarian cancer cells. A prominent limitation of studies conducted in this field is lack of verification of the obtained results in the clinical settings. Future studies are needed to verify these results in the clinical settings. Moreover, the importance of circRNAs in the determination of chemoresistance and possible targeted therapies for combating this phenotype should be assessed in future studies.
Acknowledgements
This study was financially supported by Grant from Medical School of Shahid Beheshti University of Medical Sciences.
Abbreviations
- circRNA
Circular RNA
- miRNA
MicroRNA
- ANCTs
Adjacent non-cancerous tissues
- EOC
Epithelial ovarian cancer
- OS
Overall survival
- EMT
Epithelial-mesenchymal transition
- PFS
Progression-free survival
- DFS
Disease-free survival
- PTX
Paclitaxel
- DDP
Cisplatin
- HGSOC
High-grade serous ovarian cancer
- GTEx
Genotype-tissue expression
- FIGO
International Federation of Gynecology and Obstetrics
- SOC
High-grade serous ovarian cancer
Author contributions
SGF wrote the manuscript and revised it. MT supervised and designed the study. TK, MS and BMH collected the data and designed the figures and tables. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participation
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Taheri, Email: Mohammad.taheri@uni-jena.de.
Majid Samsami, Email: samsamimd@gmail.com.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Ozols RF. Chemotherapy for ovarian cancer. Berlin: Springer; 1998. [Google Scholar]
- 3.Rosen DG, Yang G, Liu G, Mercado-Uribe I, Chang B, Xiao XS, et al. Ovarian cancer: pathology, biology, and disease models. Front Biosci. 2009;14:2089–2102. doi: 10.2741/3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghafouri-Fard S, Taheri M, Hussen BM, Vafaeimanesh J, Abak A, Vafaee R. Function of circular RNAs in the pathogenesis of colorectal cancer. Biomed Pharmacother. 2021;140:111721. doi: 10.1016/j.biopha.2021.111721. [DOI] [PubMed] [Google Scholar]
- 5.Hussen BM, Honarmand Tamizkar K, Hidayat HJ, Taheri M, Ghafouri-Fard S. The role of circular RNAs in the development of hepatocellular carcinoma. Pathol Res Pract. 2021;223:153495. doi: 10.1016/j.prp.2021.153495. [DOI] [PubMed] [Google Scholar]
- 6.Ghafouri-Fard S, Hussen BM, Taheri M, Ayatollahi SA. Emerging role of circular RNAs in breast cancer. Pathol Res Pract. 2021;223:153496. doi: 10.1016/j.prp.2021.153496. [DOI] [PubMed] [Google Scholar]
- 7.Geng X, Jia Y, Zhang Y, Shi L, Li Q, Zang A, et al. Circular RNA: biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics. 2020;12(3):267–283. doi: 10.2217/epi-2019-0295. [DOI] [PubMed] [Google Scholar]
- 8.Cheng D, Wang J, Dong Z, Li X. Cancer-related circular RNA: diverse biological functions. Cancer Cell Int. 2021;21(1):1–16. doi: 10.1186/s12935-020-01703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. 2020;19(1):1–13. doi: 10.1186/s12943-020-01163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Hu Y, Shen Q, Chen Q, Zhu X, Jiang S, et al. CircRNA_MYLK promotes malignant progression of ovarian cancer through regulating microRNA-652. Eur Rev Med Pharmacol Sci. 2020;24(10):5281–5291. doi: 10.26355/eurrev_202005_21310. [DOI] [PubMed] [Google Scholar]
- 11.Xu Q, Deng B, Li M, Chen Y, Zhuan L. circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of ovarian cancer though miR-382-5p/PRPF8 axis. J Ovarian Res. 2020;13(1):1–10. doi: 10.1186/s13048-020-00685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng M, Wei N, Yang H, Yan M, Zhao Q, Jing L. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA-144. Eur Rev Med Pharmacol Sci. 2019;23(17):7283–7294. doi: 10.26355/eurrev_201909_18833. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021;12(2):1–12. doi: 10.1038/s41419-021-03486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zong Z-H, Du Y-P, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):1–10. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei C, Wang H, Shi C, Zhang C, Wang M. CircRNA hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial-mesenchymal transition and apoptotic signaling pathways. J Clin Lab Anal. 2020;34(7):e23292. doi: 10.1002/jcla.23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Meng K, Qiu R. Circular RNA circ_0013958 functions as a tumor promoter in ovarian cancer by regulating miR-637/PLXNB2 axis. Front Genet. 2021 doi: 10.3389/fgene.2021.644451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Dong Z-N, Qiu B-Q, Hu M, Liang X-Q, Dai X, et al. CircRNA FGFR3 induces epithelial-mesenchymal transition of ovarian cancer by regulating miR-29a-3p/E2F1 axis. Aging. 2020;12(14):14080. doi: 10.18632/aging.103388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Zhang H, Li P. Upregulation of hsa_circRNA_102958 indicates poor prognosis and promotes ovarian cancer progression through miR-1205/SH2D3A axis. Cancer Manag Res. 2020;12:4045. doi: 10.2147/CMAR.S248560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding J, Wang Q, Guo N, Wang H, Chen H, Ni G, et al. CircRNA circ_0072995 promotes the progression of epithelial ovarian cancer by modulating miR-147a/CDK6 axis. Aging. 2020;12(17):17209. doi: 10.18632/aging.103668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Luo Y, Li X. Circ_0072995 promotes ovarian cancer progression through regulating miR-122-5p/SLC1A5 axis. Biochemical Genetics. 2021 doi: 10.1007/s10528-021-10092-5. [DOI] [PubMed] [Google Scholar]
- 21.Xie J, Wang S, Li G, Zhao X, Jiang F, Liu J, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi: 10.1111/jcmm.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei X, Lv H, Yang S, Yang X. CircRNA PLOD2 enhances ovarian cancer propagation by controlling miR-378. Saudi J Biol Sci. 2021;28(11):6260–6265. doi: 10.1016/j.sjbs.2021.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Yu F, Wang S, Zhao X, Jiang F, Xie J, et al. circGFRA1 promotes ovarian cancer progression by sponging miR-449a. J Cancer. 2019;10(17):3908. doi: 10.7150/jca.31615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu F, Ni M, Li J, Cheng J, Zhao H, Zhao J, et al. Circ0004390 promotes cell proliferation through sponging miR-198 in ovarian cancer. Biochem Biophys Res Commun. 2020;526(1):14–20. doi: 10.1016/j.bbrc.2020.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Sheng S, Hu Y, Yu F, Tong W, Wang S, Cai Y, et al. circKIF4A sponges miR-127 to promote ovarian cancer progression. Aging. 2020;12(18):17921. doi: 10.18632/aging.103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Zhang X, Qiao L, Wang H. CircRNA circ_0000554 promotes ovarian cancer invasion and proliferation by regulating miR-567. Environ Sci Pollut Res. 2021 doi: 10.1007/s11356-021-13710-2. [DOI] [PubMed] [Google Scholar]
- 27.An Q, Liu T, Wang MY, Yang YJ, Zhang ZD, Lin ZJ, et al. circKRT7-miR-29a-3p-COL1A1 axis promotes ovarian cancer cell progression. Onco Targets Ther. 2020;13:8963–8976. doi: 10.2147/OTT.S259033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng X-Y, Yuan J, Wang C, Zeng D, Yong J-H, Jiang X-Y, et al. circCELSR1 facilitates ovarian cancer proliferation and metastasis by sponging miR-598 to activate BRD4 signals. Mol Med. 2020;26(1):1–14. doi: 10.1186/s10020-020-00194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Cheng J, Quan C, Wen H, Feng Z, Hu Q, et al. circCELSR1 (hsa_circ_0063809) contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids. 2020;19:718–730. doi: 10.1016/j.omtn.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei S, Qi L, Wang L. Overexpression of circ_CELSR1 facilitates paclitaxel resistance of ovarian cancer by regulating miR-149-5p/SIK2 axis. Anticancer Drugs. 2021;32(5):496–507. doi: 10.1097/CAD.0000000000001058. [DOI] [PubMed] [Google Scholar]
- 31.Liu N, Zhang J, Zhang L, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 32.Zhou H, Li J, Lai X, Wang K, Zhou W, Wang J. CircHIPK3 modulates VEGF through MiR-7 to affect ovarian cancer cell proliferation and apoptosis. J BU ON. 2021;26(3):691–697. [PubMed] [Google Scholar]
- 33.Li Y, Lin S, An N. Hsa_circ_0009910: oncogenic circular RNA targets microRNA-145 in ovarian cancer cells. Cell Cycle. 2020;19(15):1857–1868. doi: 10.1080/15384101.2020.1731650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Li X, Yang L, Li M, Zhang Y, Zhang J. CircASH2L promotes ovarian cancer tumorigenesis, angiogenesis, and lymphangiogenesis by regulating the miR-665/VEGFA axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:595585. doi: 10.3389/fcell.2020.595585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Li Y, Zhao W, Liu G, Yang Q. Circ-PGAM1 promotes malignant progression of epithelial ovarian cancer through regulation of the miR-542-3p/CDC5L/PEAK1 pathway. Cancer Med. 2020;9(10):3500–3521. doi: 10.1002/cam4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang LL, Zong ZH, Liu Y, Guan X, Chen S, Zhao Y. CircRhoC promotes tumorigenicity and progression in ovarian cancer by functioning as a miR-302e sponge to positively regulate VEGFA. J Cell Mol Med. 2019;23(12):8472–8481. doi: 10.1111/jcmm.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan X, Zong Z-h, Liu Y, Chen S, Wang L-l, Zhao Y. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–92. doi: 10.1016/j.omtn.2019.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang K, Liu D, Su C. Circ_0007841 accelerates ovarian cancer development through facilitating MEX3C expression by restraining miR-151-3p activity. Aging. 2021;13(8):12058. doi: 10.18632/aging.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Wang J, Li H, Sun Y, Tong X. Downregulation of hsa_circ_0026123 suppresses ovarian cancer cell metastasis and proliferation through the miR-124-3p/EZH2 signaling pathway. Int J Mol Med. 2021;47(2):668–676. doi: 10.3892/ijmm.2020.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Gui R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J Gynecol Oncol. 2020 doi: 10.3802/jgo.2020.31.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Y, Liu X, Zhang S, Chen S, Guan X, Li Q, et al. CircCRIM1 promotes ovarian cancer progression by working as ceRNAs of CRIM1 and targeting miR-383-5p/ZEB2 axis. Reprod Biol Endocrinol. 2021;19(1):1–12. doi: 10.1186/s12958-021-00857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, Luo J, Chen Y, Wu Q. Circ_0061140 knockdown inhibits tumorigenesis and improves PTX sensitivity by regulating miR-136/CBX2 axis in ovarian cancer. J Ovarian Res. 2021;14(1):1–12. doi: 10.1186/s13048-021-00888-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Chi C, Zhou L, Chen Y, Tang X. Circular PVT1 regulates cell proliferation and invasion via miR-149-5p/FOXM1 axis in ovarian cancer. J Cancer. 2021;12(2):611. doi: 10.7150/jca.52234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun X, Luo L, Gao Y. Circular RNA PVT1 enhances cell proliferation but inhibits apoptosis through sponging microRNA-149 in epithelial ovarian cancer. J Obstet Gynaecol Res. 2020;46(4):625–635. doi: 10.1111/jog.14190. [DOI] [PubMed] [Google Scholar]
- 46.Du Z, Wang L, Xia Y. Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway. Cancer Cell Int. 2020;20(1):1–13. doi: 10.1186/s12935-020-01666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou W, Zhang Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract. 2021;217:153275. doi: 10.1016/j.prp.2020.153275. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, Liu W, Yu X, Qi X, Sun C. Circ_0005276 aggravates the development of epithelial ovarian cancer by targeting ADAM9. Eur Rev Med Pharmacol Sci. 2020;24(20):10375–10382. doi: 10.26355/eurrev_202010_23387. [DOI] [PubMed] [Google Scholar]
- 49.Chen S, Wu W, Li Q-h, Xie B-m, Shen F, Du Y-p, et al. Circ-NOLC1 promotes epithelial ovarian cancer tumorigenesis and progression by binding ESRP1 and modulating CDK1 and RhoA expression. Cell Death Discov. 2021;7(1):1–14. doi: 10.1038/s41420-020-00381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Bai H. Effect of circBIRC6 targeting miR-367-3p on cisplatin resistance of ovarian cancer cells. Zhonghua zhong liu za zhi. 2021;43(10):1062–1068. doi: 10.3760/cma.j.cn112152-20200427-00384. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Yao Y, Jin M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging. 2020;12(19):19095. doi: 10.18632/aging.103706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma R, Ye X, Cheng H, Cui H, Chang X. Tumor-derived exosomal circRNA051239 promotes proliferation and migration of epithelial ovarian cancer. Am J Transl Res. 2021;13(3):1125. [PMC free article] [PubMed] [Google Scholar]
- 53.Lu H, Zheng G, Gao X, Chen C, Zhou M, Zhang L. Propofol suppresses cell viability, cell cycle progression and motility and induces cell apoptosis of ovarian cancer cells through suppressing MEK/ERK signaling via targeting circVPS13C/miR-145 axis. J Ovarian Res. 2021;14(1):1–11. doi: 10.1186/s13048-021-00775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karedath T, Ahmed I, Al Ameri W, Al-Dasim FM, Andrews SS, Samuel S, et al. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19(1):1–17. doi: 10.1186/s12885-019-5723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao L, Zhong J, Pang L. Upregulation of circular RNA VPS13C-has-circ-001567 promotes ovarian cancer cell proliferation and invasion. Cancer Biother Radiopharm. 2019;34(2):110–118. doi: 10.1089/cbr.2018.2641. [DOI] [PubMed] [Google Scholar]
- 56.Sun Y, Li X, Chen A, Shi W, Wang L, Yi R, et al. circPIP5K1A serves as a competitive endogenous RNA contributing to ovarian cancer progression via regulation of miR-661/IGFBP5 signaling. J Cell Biochem. 2019;120(12):19406–19414. doi: 10.1002/jcb.29055. [DOI] [PubMed] [Google Scholar]
- 57.Ying H, Zhao R, Yu Q, Zhang K, Deng Q. CircATL2 enhances paclitaxel resistance of ovarian cancer via impacting miR-506-3p/NFIB axis. Drug Dev Res. 2021 doi: 10.1002/ddr.21882. [DOI] [PubMed] [Google Scholar]
- 58.Zhou X, Jiang J, Guo S. Hsa_circ_0004712 downregulation attenuates ovarian cancer malignant development by targeting the miR-331-3p/FZD4 pathway. J Ovarian Res. 2021;14(1):1–13. doi: 10.1186/s13048-021-00859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F, Xu Y, Ye W, Jiang J, Wu C. Circular RNA S-7 promotes ovarian cancer EMT via sponging miR-641 to up-regulate ZEB1 and MDM2. Biosci Rep. 2020;40(7):BSR20200825. doi: 10.1042/BSR20200825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu S-G, Zhou P, Chen J-X, Lei J, Hua L, Dong Y, et al. circ-PTK2 (hsa_circ_0008305) regulates the pathogenic processes of ovarian cancer via miR-639 and FOXC1 regulatory cascade. Cancer Cell Int. 2021;21(1):1–20. doi: 10.1186/s12935-021-01985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, Zhang W, Guo H, Chu D, Zhang R, Guo R. CircLNPEP promotes the progression of ovarian cancer through regulating miR-876-3p/WNT5A axis. Cell Cycle. 2021;20(19):2021–2039. doi: 10.1080/15384101.2021.1965723. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Li M, Cai J, Han X, Ren Y. Downregulation of circNRIP1 suppresses the paclitaxel resistance of ovarian cancer via regulating the miR-211-5p/HOXC8 axis. Cancer Manag Res. 2020;12:9159–71. doi: 10.2147/CMAR.S268872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y, Wang J. Circular RNA circTNPO3 regulates paclitaxel resistance of ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol Ther Nucleic Acids. 2020;21:780–791. doi: 10.1016/j.omtn.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):1224–1233. doi: 10.1080/21691401.2019.1593999. [DOI] [PubMed] [Google Scholar]
- 65.Li B, Zhang L. CircSETDB1 knockdown inhibits the malignant progression of serous ovarian cancer through miR-129-3p-dependent regulation of MAP3K3. J Ovarian Res. 2021;14(1):1–15. doi: 10.1186/s13048-021-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W, Wang J, Zhang X, Liu G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:7451. doi: 10.2147/OTT.S220700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo M, Li S, Zhao X, Yuan Y, Zhang B, Guan Y. Knockdown of circular RNA Hsa_circ_0000714 can regulate RAB17 by sponging miR-370-3p to reduce paclitaxel resistance of ovarian cancer through CDK6/RB pathway. Onco Targets Ther. 2020;13:13211. doi: 10.2147/OTT.S285153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H, Guo Y, Zhang Y, Wang D, Zhang G, Hou J, et al. Circ_MUC16 attenuates the effects of Propofol to promote the aggressive behaviors of ovarian cancer by mediating the miR-1182/S100B signaling pathway. BMC Anesthesiol. 2021;21(1):1–14. doi: 10.1186/s12871-021-01517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun D, Liu J, Zhou L. Upregulation of circular RNA circ-FAM53B predicts adverse prognosis and accelerates the progression of ovarian cancer via the miR-646/VAMP2 and miR-647/MDM2 signaling pathways. Oncol Rep. 2019;42(6):2728–2737. doi: 10.3892/or.2019.7366. [DOI] [PubMed] [Google Scholar]
- 70.Chen Y, Ye X, Xia X, Lin X. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26(2):151–161. doi: 10.3233/CBM-190064. [DOI] [PubMed] [Google Scholar]
- 71.Lin X, Chen Y, Ye X, Xia X. Circular RNA ABCB10 promotes cell proliferation and invasion, but inhibits apoptosis via regulating the microRNA-1271-mediated Capn4/Wnt/β-catenin signaling pathway in epithelial ovarian cancer. Mol Med Rep. 2021;23(5):1–9. doi: 10.3892/mmr.2021.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Q-h, Liu Y, Chen S, Zong Z-h, Du Y-p, Sheng X-j, et al. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed Pharmacother. 2019;114:108832. doi: 10.1016/j.biopha.2019.108832. [DOI] [PubMed] [Google Scholar]
- 73.Xie W, Liu L, He C, Zhao M, Ni R, Zhang Z, et al. Circ _ 0002711 knockdown suppresses cell growth and aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian cancer. J Biosci. 2021;46(1):1–11. [PubMed] [Google Scholar]
- 74.Zhang M, Xu Y, Zhang Y, Li B, Lou G. Circular RNA circE2F2 promotes malignant progression of ovarian cancer cells by upregulating the expression of E2F2 protein via binding to HuR protein. Cell Signal. 2021;84:110014. doi: 10.1016/j.cellsig.2021.110014. [DOI] [PubMed] [Google Scholar]
- 75.Cao Y, Xie X, Li M, Gao Y. CircHIPK2 contributes to DDP resistance and malignant behaviors of DDP-resistant ovarian cancer cells both in vitro and in vivo through circHIPK2/miR-338-3p/CHTOP ceRNA pathway. Onco Targets Ther. 2021;14:3151. doi: 10.2147/OTT.S291823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang S, Li Z, Zhu G, Hong L, Hu C, Wang K, et al. RNA-binding protein IGF2BP2 enhances circ_0000745 abundancy and promotes aggressiveness and stemness of ovarian cancer cells via the microRNA-3187-3p/ERBB4/PI3K/AKT axis. J Ovarian Res. 2021;14(1):1–17. doi: 10.1186/s13048-021-00917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan H, Xiang H, Sun B, Feng F, Chen P. Circular RNA-ITCH inhibits the proliferation of ovarian carcinoma by downregulating lncRNA HULC. Reprod Sci. 2020;27(1):375–379. doi: 10.1007/s43032-019-00049-w. [DOI] [PubMed] [Google Scholar]
- 78.Lin C, Xu X, Yang Q, Liang L, Qiao S. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020;20(1):1–13. doi: 10.1186/s12935-020-01420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. 2019;18(1):1–19. doi: 10.1186/s12943-019-1080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun S, Fang H. Curcumin inhibits ovarian cancer progression by regulating circ-PLEKHM3/miR-320a/SMG1 axis. J Ovarian Res. 2021;14(1):1–13. doi: 10.1186/s13048-021-00916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang N, Jin Y, Hu Q, Cheng S, Wang C, Yang Z, et al. Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. J Ovarian Res. 2020;13:1–10. doi: 10.1186/s13048-020-00664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin Y, Wang H. Circ_0078607 inhibits the progression of ovarian cancer via regulating the miR-32-5p/SIK1 network. J Ovarian Res. 2022;15(1):1–13. doi: 10.1186/s13048-021-00931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang N, Yang Z, Jin Y, Cheng S, Yang J, Wang Y. Low expression of circular RNA hsa_circ_0078607 predicts poor prognosis in high-grade serous ovarian cancer. Cancer Manag Res. 2021;13:2877. doi: 10.2147/CMAR.S300738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng Y, Li Z, Yang S, Wang Y, Luan Z. CircEXOC6B suppresses the proliferation and motility and sensitizes ovarian cancer cells to paclitaxel through miR-376c-3p/FOXO3 axis. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2020.3739. [DOI] [PubMed] [Google Scholar]
- 85.Wang Z, Zhang W, Fang J, Xie P, Miao M, Yang H. Circular RNA circEXOC6B inhibits the progression of ovarian cancer by sponging miR-421 and regulating RUS1 expression. Onco Targets Ther. 2020;13:8233. doi: 10.2147/OTT.S243040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gong J, Xu X, Zhang X, Zhou Y. Circular RNA-9119 suppresses in ovarian cancer cell viability via targeting the microRNA-21-5p–PTEN–Akt pathway. Aging. 2020;12(14):14314. doi: 10.18632/aging.103470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen H, Mao M, Jiang J, Zhu D, Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019;12:3869. doi: 10.2147/OTT.S207938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24–33. doi: 10.1016/j.omtn.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Hu Y, Zhu Y, Zhang W, Lang J, Ning L. Utility of plasma circBNC2 as a diagnostic biomarker in epithelial ovarian cancer. Onco Targets Ther. 2019;12:9715. doi: 10.2147/OTT.S211413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li X, Lin S, Mo Z, Jiang J, Tang H, Wu C, et al. CircRNA_100395 inhibits cell proliferation and metastasis in ovarian cancer via regulating miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J Cancer. 2020;11(3):599–609. doi: 10.7150/jca.35041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Li Y, Zhou JH, Shen FR, Shi X, Chen YG. CircATRNL1 activates Smad4 signaling to inhibit angiogenesis and ovarian cancer metastasis via miR-378. Mol Oncol. 2021;15(4):1217. doi: 10.1002/1878-0261.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;112:8–17. doi: 10.1016/j.biocel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 93.Yalan S, Yanfang L, He C, Yujie T. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. 2020 doi: 10.23736/S0031-0808.20.03957-9. [DOI] [PubMed] [Google Scholar]
- 94.Gao Y, Zhang C, Liu Y, Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends. 2019;13(2):204–211. doi: 10.5582/bst.2019.01021. [DOI] [PubMed] [Google Scholar]
- 95.Luo L, Gao Y, Sun X. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-alpha. Eur Rev Med Pharmacol Sci. 2018;22(23):8119–8126. doi: 10.26355/eurrev_201812_16503. [DOI] [PubMed] [Google Scholar]
- 96.Hu J, Wang L, Chen J, Gao H, Zhao W, Huang Y, et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505(1):222–228. doi: 10.1016/j.bbrc.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 97.Luo L, Gao Y, Sun X. Circ-ITCH correlates with small tumor size, decreased FIGO stage and prolonged overall survival, and it inhibits cells proliferation while promotes cells apoptosis in epithelial ovarian cancer. Cancer Biomark. 2018;23(4):505–513. doi: 10.3233/CBM-181609. [DOI] [PubMed] [Google Scholar]
- 98.Wang N, Cao Q-X, Tian J, Ren L, Cheng H-L, Yang S-Q. Circular RNA MTO1 inhibits the proliferation and invasion of ovarian cancer cells through the miR-182-5p/KLF15 axis. Cell Transplant. 2020;29:0963689720943613. doi: 10.1177/0963689720943613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu X, Liu D, Wang S, Liu J. Circ_0007444 inhibits the progression of ovarian cancer via mediating the miR-570-3p/PTEN axis. Onco Targets Ther. 2021;14:97. doi: 10.2147/OTT.S266186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zou T, Wang P, Gao Y, Liang W. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22(21):7178–7182. doi: 10.26355/eurrev_201811_16250. [DOI] [PubMed] [Google Scholar]
- 101.Lin W, Ye H, You K, Chen L. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020;20(1):1–11. doi: 10.1186/s12935-019-1071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li L, Yu P, Zhang P, Wu H, Chen Q, Li S, et al. Upregulation of hsa_circ_0007874 suppresses the progression of ovarian cancer by regulating the miR-760/SOCS3 pathway. Cancer Med. 2020;9(7):2491–2499. doi: 10.1002/cam4.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ning L, Lang J, Wu L. Plasma circN4BP2L2 is a promising novel diagnostic biomarker for epithelial ovarian cancer. BMC Cancer. 2022;22(1):1–15. doi: 10.1186/s12885-021-09073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang D, He Y, Wu B, Deng Y, Wang N, Li M, et al. Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in ovarian cancer. J Ovarian Res. 2020;13(1):1–18. doi: 10.1186/s13048-020-0613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Y. Comment on “circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer”, Cancer Lett. 2020 Jan 2; 473 (2020) 139–147. Cancer Lett. 2020;475:1. doi: 10.1016/j.canlet.2019.12.043. [DOI] [PubMed] [Google Scholar]
- 106.Wang YZ. The Limited Functions and Mechanisms of CircASAP1 as a Sponge of Onco-microRNAs. Hepatology. 2021;73(6):2610. doi: 10.1002/hep.31606. [DOI] [PubMed] [Google Scholar]
- 107.Wang Y. Issues of circular RNAs as microRNA sponges. Hepatology. 2020;72(1):365. doi: 10.1002/hep.31303. [DOI] [PubMed] [Google Scholar]
- 108.Wang Y-Z. Comment on “regulation of nuclear factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis?” [Cancer Lett. 509 (2021 May 2) 63–80] Cancer Lett. 2021;2021(516):99–100. doi: 10.1016/j.canlet.2021.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.