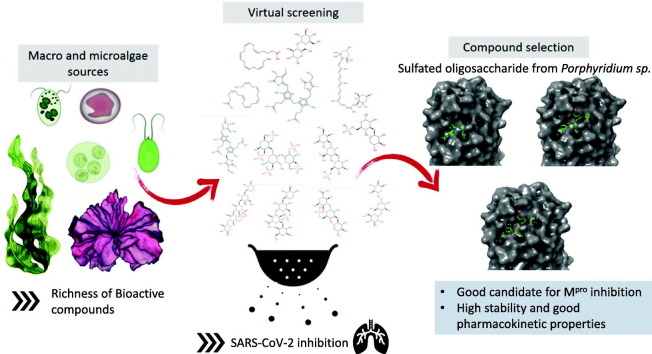
Abstract
The coronavirus pandemic (COVID-19) has created an urgent need to develop effective strategies for prevention and treatment. In this context, therapies against protease Mpro, a conserved viral target, would be essential to contain the spread of the virus and reduce mortality. Using combined techniques of structure modelling, in silico docking and pharmacokinetics prediction, many compounds from algae were tested for their ability to inhibit the SARS-CoV-2 main protease and compared to the recent recognized drug Paxlovid. The screening of 27 algal molecules including 15 oligosaccharides derived from sulfated and non-sulphated polysaccharides, eight pigments and four poly unsaturated fatty acids showed high affinities to interact with the protein active site. Best candidates showing high docking scores in comparison with the reference molecule were sulfated tri-, tetra- and penta-saccharides from Porphyridium sp. exopolysaccharides (SEP). Structural and energetic analyses over 100 ns MD simulation demonstrated high SEP fragments-Mpro complex stability. Pharmacokinetics predictions revealed the prospects of the identified molecules as potential drug candidates.
Keywords: Microalgae, Porphyridium, SARS-CoV-2, Polysaccharide, Docking, Inhibitors
Graphical abstract
1. Introduction
Pharmacotherapy based on natural compounds can be currently considered as a very promising alternative to conventional and chemical therapy (Baklouti et al., 2020; Ben Hlima et al., 2019; Elleuch et al., 2019; Ben Halima et al., 2015; Fendri et al., 2013; Abdelkafi et al., 2005). Known as source of biomacromolecules with promising applications in pharmaceutical formulations, micro and macroalgae are some of the most promising stakeholders in blue biotechnology (Dammak et al., 2018). Their richness in organic compounds accumulated or secreted as primary or secondary metabolites made them been used for long in therapeutic applications and screening for active principles (Ben Amor et al., 2017; Bule et al., 2018; Ameen et al., 2021). They have been found to have antibacterial (Das et al., 2005; Bhadury and Wright, 2004) antifungal, antiprotozoal and antiplasmodial activities (Jaime et al., 2010; Naviner et al., 1999; Kellam and Walker, 1989; Ozdemir et al., 2004; Herrero et al., 2006; Ghasemi et al., 2004) as well as a broad range of antiviral activity. The latter was related mostly to the presence of several active compounds especially sulfated polysaccharides and phycocyanin (Shih et al., 2000; Hasui et al., 1995). Recently, an increasing attention has been paid on polysaccharides as an important class of bioactive natural products and numerous research studies have pointed out the bioactivities of algal polysaccharides against a wide spectrum of viral infections (Raposo et al., 2013; Ahmadi et al., 2015) like Human immunodeficiency virus (HIV), Herpes Simplex Virus (HSV), African swine fever virus (ASFV), and influenza A virus (Flu-A) (Rosales-Mendoza et al., 2020). Nonetheless, the depth of these activities in terms of possible mechanisms against potential drug targets has been poorly studied. More recently, and due to the emergence of the Coronavirus Disease 2019 (COVID-19) caused by the SARS-CoV-2 virus, algal compounds and especially their polysaccharides are exploited at different levels to fight this pandemy (Salih et al., 2021). To challenge highly virulent SARS CoV-2 and its emerging mutations there is a serious demand to ramp up drug discovery pipelines within a short timeframe. Despite the current understanding on the viral structure and cycle, there is no specific treatment for COVID-19 patients. The current vaccination campaign has proven to be less effective against SARS-CoV-2 new variants and the protection effect against COVID-19 infection declined for all vaccine types as observed in United States. Indeed, it was reported that the overall vaccine protection declined from 87.9% to 48.1%. The decline was highest for the Janssen from 86.4% to 13.1%, Pfizer BioNTech was from 86.9% to 43.3%, and Moderna was from 89.2% to 58% (Cohn et al., 2022).
The COVID-19 Main protease (Mpro) is the major protease responsible for functional protein maturation. It can be an attractive primary antiviral target (Li and Kang, 2020). Targeting proteases, such as Mpro, is a common approach for combating viral infections because those proteins are highly conserved between variants as referring to the newly discovered Omicron variant (Ferré et al., 2022). For instance, several in-silico screenings have been conducted to identify Mpro inhibitors both through drug repurposing and drug discovery approaches along with artificial intelligence. In the many drug-repurposing studies, docking simulation-based technologies have been mainly employed and contributed to the identification of several Mpro binders (Jin et al., 2020; Farhat et al., 2022; Ibrahim et al., 2021). In this way, many approved FDA drugs were successfully selected like PF-0730814 and PF-07321332 and entered clinical trials (Owen et al., 2021). Encouraging results of phase of 2/3 were reported (Pfizer, 2021). Paxlovid is a novel oral antiviral drug developed by Pfizer company. It is a combination of PF-07321332 and ritonavir acting specifically against the Mpro of SARS-CoV-2 (Pfizer, 2021). Molecular studies revealed that this small molecule had a strong affinity and stable interaction with the catalytic dyad His41-Cys145 of the protease (Ahmad et al., 2021).
The main goal of our in silico study is to screen a pool of bioactive compounds from some algae which can inhibit Mpro with an efficiency similar to that of Paxlovid. The choice of screened molecules was based on previous studies highlighting their potential antiviral effect and/or bioactivity. Combined virtual docking, molecular dynamics simulation and pharmacokinetics prediction highlighted the most potential candidates to be used as Mpro inhibitor.
2. Materials and methods
2.1. Selection and preparation of algal molecules
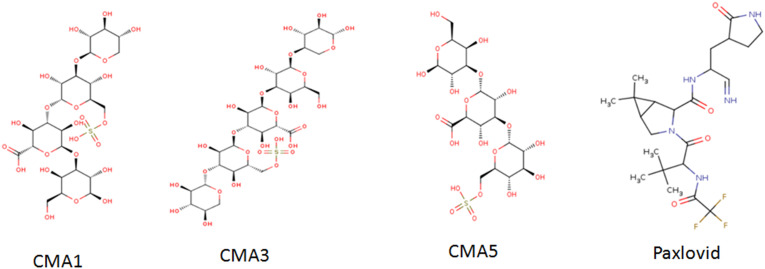
Di, tri, tetra and penta saccharides were constructed according to recent publications on macro and microalgae polysaccharide structures (Table S1). The carbohydrate modelling protocol used by Sapay et al., was employed in our in silico study (Sapay et al., 2013). In brief, the 2D structures of oligosaccharides were built using the Carbohydrate Builder on GLYCAM-Web (www.glycam.org). Oligosaccharides containing deoxy residues were assembled using the tLEaP module from the AMBER16 (Case et al., 2017) package using GLYCAM06 force field parameters (Kirschner et al., 2008) and from literature for sulfate groups (Huige and Altona, 1995). Other molecules than saccharides were obtained from the PubChem repository sample. In total 27 molecules were used for the docking screening, the details of their formula, IUPAC name, molecular mass and pubchem CID are presented in supplementary Table S1. The minimized structures were converted to PDB format followed by conversion to pdbqt one. Three of them were presented as 2D chemical structures in Fig. 1 along with Paxlovid which was used as control for the docking experiments.
Fig. 1.
Chemical structures of the three molecules having the best binding energies and the FDA approved drug Paxlovid.
2.2. Receptor preparation
The three-dimensional crystal structure of SARS-CoV-2 Mpro protease cocrystallized with the inhibitor PF-07321332 (pdb code: 7VH8) was retrieved in pdb format from Protein Data Bank with a resolution of 1.59 Å (Zhao et al., 2021). The ligand was then extracted, and the protein was prepared in Autodock Vina tools 4.2 by removal of water and solvent molecules, addition of polar hydrogens, and partial charge assignment before to be saved as pdbqt format to be included as a receptor in the virtual screening.
2.3. Molecular docking protocol
The docking simulations of algal compounds to the SARS-CoV-2 Mpro receptor protein were performed with Autodock Vina 4.2. Molecular docking scores were set as AutoDock tools of the molecular graphics laboratory software package by keeping the analogue flexible (Seeliger and de Groot, 2010). The grid box center was adjusted X = −17.81, Y = 18.517 and Z = −26.091 with dimensions for SARS-CoX-2 Mpro. Its size was set to 40 × 40 × 50 Angstroms to cover the active site. Paxlovid was redocked in the structure to validate the obtained results and used as control to compare the molecular docking of the algal compounds. Docking results were visualized using LigPlot and Poseview (Wallace et al., 1995; Stierand and Rarey, 2007). Obtained models were examined with PyMol 0.97 and Drug Discovery Studio (v.20.1.0.19295) (Kar and Roy, 2013).
2.4. Molecular dynamics simulation (MD) and free energy landscape analysis
The MD simulations studies were carried out in triplicate on dock complexes for CMA1, CMA3 and control with receptor using the Desmond 2020.1 from Schrödinger, LLC. The triplicate samplings were made using same parameters for each MD run to obtain reproducibility of the results. The OPLS-2005 force field (Bowers et al., 2006; Chow et al., 2008; Shivakumar et al., 2010) and explicit solvent model with the SPC water molecules were used in this system (Jorgensen et al., 1983). NaCl solutions (0.15 M) were added to the system to neutralize the charge and to simulate the physiological environment. Initially, the system was equilibrated using an NVT ensemble for 100 ns to retrain over the protein-ligand complexes. Following the previous step, a short run of equilibration and minimization was carried out using an NPT ensemble for 12 ns. The NPT ensemble was set up using the Nose-Hoover chain coupling scheme (Martyna et al., 1994) with the temperature at 27 °C, the relaxation time of 1.0 ps, and pressure 1 bar maintained in all the simulations. A time step of 2 f. was used. The Martyna-Tuckerman–Klein chain coupling scheme (Martyna et al., 1992) barostat method was used for pressure control with a relaxation time of 2 ps. The particle mesh Ewald method (Toukmaji and Board, 1996) was used for calculating long-range electrostatic interactions, and the radius for the coulomb interactions were fixed at 9 Å. RESPA integrator was used for a time step of 2 f. for each trajectory to calculate the bonded forces. The root means square deviation (RMSD), radius of gyration (Rg), root mean square fluctuation (RMSF) and number of hydrogen (H-bonds) and Solvent accessible surface area (SASA) were calculated to monitor the stability of the MD simulations. The free energy landscape of protein folding bound complexes was measured using Geo_measures v 0.8 (Kagami et al., 2020). Geo_measures include a powerful library of g_sham and form the MD trajectory against RMSD and radius of gyration (Rg) energy profile of folding recorded in a 3D plot using matplotlib python package.
2.5. In-silico Osiris/Molinspiration and ADMET analysis
Osiris and Molinspiration analyses are performed on 2D models and are employed to predict pharmacophore site and biological activity of the tested compounds as well as to determine the drug-likeness score of each ligand (Sander et al., 2009). The acute toxicity in rodent models and chemical classification of the test compounds were predicted by GUSAR (Lagunin et al., 2011). It analyses compounds based on the quantitative neighborhoods of atom descriptors and prediction of activity spectra for substance algorithm and correlates the obtained results with the SYMYX MDL toxicity database. The pharmacokinetic properties of the selected ligands along with Paxlovid were achieved with using the SwissADME, which is an open online tool (http://www.swissadme.ch). The ADME properties define blood–brain barrier (BBB) permeability and human gastrointestinal absorption (GI) as well as substrate or nonsubstrate for permeability to glycoprotein (P-gp) and cytochrome P450 (CYP) (Daina et al., 2017).
2.6. Computational compound toxicity prediction by VEGA Hub software
The selected compounds were subjected to 10 toxicity tests/measurements performed by VEGA software version 1.1.5 using the QSAR (quantitative structure-activity relationship) approach (https://www.vegahub.eu/). The SMILES (Simplified Molecular Input Line Entry Systems) of these compounds were generated (https://cactus.nci.nih.gov/translate/) and inputted into the software for chemo-computational toxicology evaluations.
3. Results and discussion
3.1. Virtual molecular docking
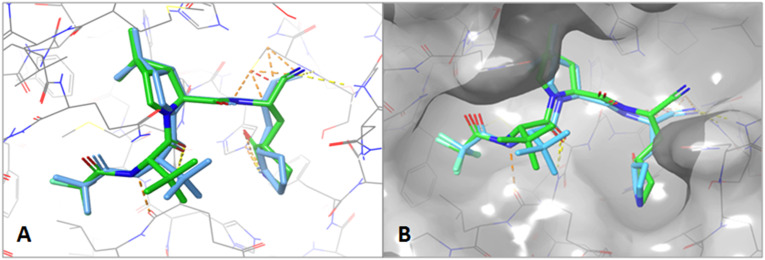
Firstly, the re-docking of the native cocrystal inhibitor (Paxlovid) has been performed for the validation of the whole docking procedure ensuring its reproducibility. The re-docking of this native ligand has shown that it binds to the same site of the main protease (Mpro) of SARS-CoV-2 as the co-crystal ligand binds in the original structure (PDB ID: 7VH8) used for docking (Fig. 2A and B).
Fig. 2.
A close up stick view (A) and surface view (B) of the superimposed Paxlovid position in the active site of Mpro of the solved 3D structure (green) and re-docked structure (blue).
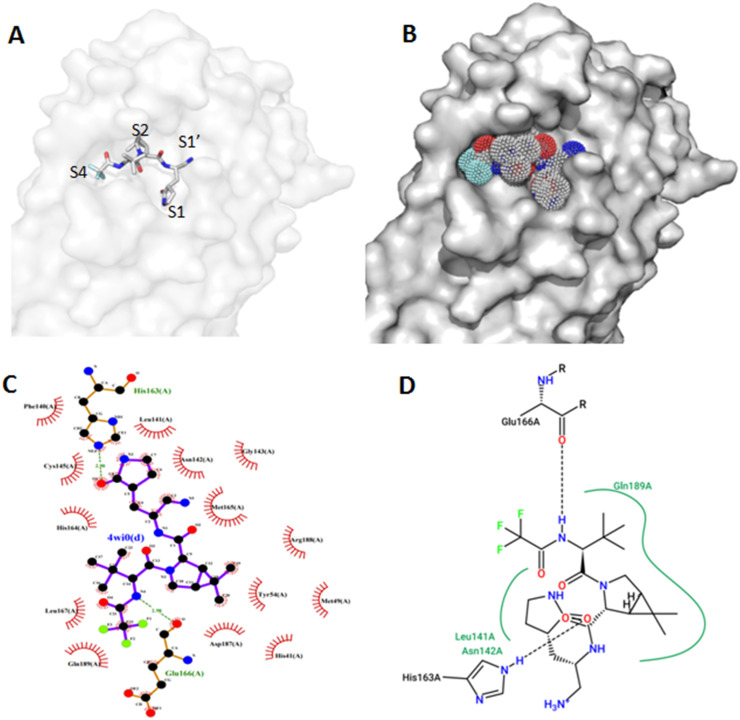
The root mean square deviation (RMSD) value between the co-crystal ligand and re-docked native ligand position is 1.013 Å. Re-docking results depicted that all the major interactions between active site residues and re-docked ligand resembles to the interactions with the co-crystal inhibitor. The binding energy of the re-docked ligand at the active site of Mpro is −8.4 Kcal/mol. As for the published work (Zhao et al., 2021), many residues interact with Paxlovid including His164, Asn142, Gly143, Cys145, His163, Met165, Glu166, Gln189, Arg188, His41, Leu141, Met49, Asp187, Tyr54, Phe140 (Fig. 3C and D). The inhibitor occupies three of four substrate subsites namely S1, S2 and S4 (Fig. 3A and B). In addition, using X-ray crystallography, Zhao et al., 2021 demonstrated that thiol group of catalytic C145 was very close to the inhibitor nitrile carbon and attacks this electrophilic group at P1′ site of the ligand. Hence, the formation of a thioimidate adduct through a standard 1.8-Å C—S covalent bond was possible. Thus, we can consider the molecular docking to be successful.
Fig. 3.
Binding mode of Redocked Paxlovid as stick representation showing S1, S1′, S2 and S4 subsites (A) and as dot surface representation showing fullness of the active site (B). Visualization of docking results interaction of the inhibitor using (C) Ligplotplus and (D) dan Poseview.
After that, a total of 27 major algal molecules have been docked to Mpro (COVID-19 main protease). The choice of these compounds has been notably motivated by the poor knowledge of antiviral potential of sulfated polysaccharides from red microalgae contrary to other ones such as sulfated galactans from red macroalgae. Indeed, there has been a substantial increase in evidence that reveals the antiviral activity of various micro- and macroalgal metabolites like lectins, sulfated polysaccharides, and phycocyanobilins. Recent studies have reported that these compounds demonstrate substantial activity against a wide array of DNA and RNA viruses, including the influenza virus known to be associated with respiratory illnesses (Rosales-Mendoza et al., 2020). So, bioactive molecules from microalgae could serve as a novel therapeutic option to tackle SARS-CoV-2 and alike viruses. SEP from Porphyridium species are known to perform against a wide range of viruses including herpes simplex virus HSV-1 and HSV-2 (Huheihel et al., 2002), varicela zoster virus (VZV), retrovirus (Talyshinsky et al., 2002) murine sarcoma virus (MuSV-124), MuSV/MuLV (murine leukemia virus and hepatitis B virus (HBV)).
Oligosaccharidic units were di- and tri-saccharides from starch, cellulose and glycan as well as di-, tri-, tetra- and penta-saccharides from sulfated exopolysaccharides from Arthrospira sp. and Porphyridium sp. strains. The sulfated exopolysaccharides structures are available in the literature (a total of 15 ligands were constructed). Furthermore, 9 pigments molecules and 4 poly-unsaturated fatty acids were equally tested. The molecular docking indicated that all the compounds used have a substantial binding affinity toward the receptor as compared to the native ligand. The detailed list is depicted in supplementary Table S1. Oligosaccharides presented higher binding energies than other molecule types ranged between −6.4 to −9.9 Kcal/mol and interestingly three sulfated saccharides exhibited higher energies than Paxlovid namely CMA1, CMA3 and CMA5 (Fig. 1). Phycocyanobilin presented a relatively high binding energy (−8.3 Kcal/mol) which could be explored in a future investigation. Similar high binding energy was found in previous in silico work (−8.6 Kcal/mol) highlighting the significant potential of this algae pigment as antiviral agent with good efficiency against SARS-CoV-2 (Pendyala and Patras, 2020). Compounds having binding energy greater than −8.5 Kcal/mol have been taken for further analysis. Two compounds have been taken into consideration as reflected in bold in Table S1 namely sulfated tetra- and penta-sulfated saccharides from Porphyridium sp. The detailed interactions with the different amino acid residues of Mpro have been depicted in Table S2, Fig. 4, Fig. 5 in comparison with the Paxlovid inhibitor (Fig. 3).
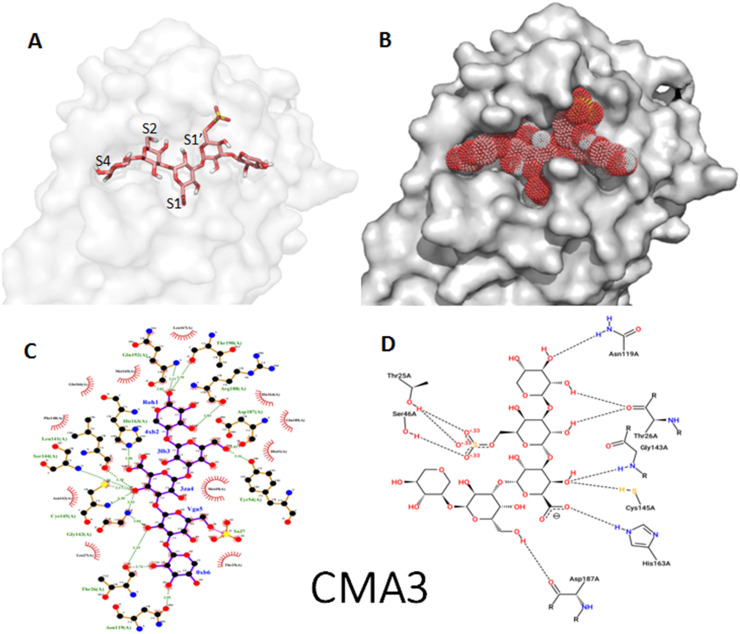
Fig. 4.
Binding mode of the penta-sulphated oligosaccharide (CMA3) as stick representation (A) and as dot surface representation showing fullness of the active site (B). Visualization of docking results interaction of the inhibitor using (C) Ligplotplus and (D) dan Poseview.
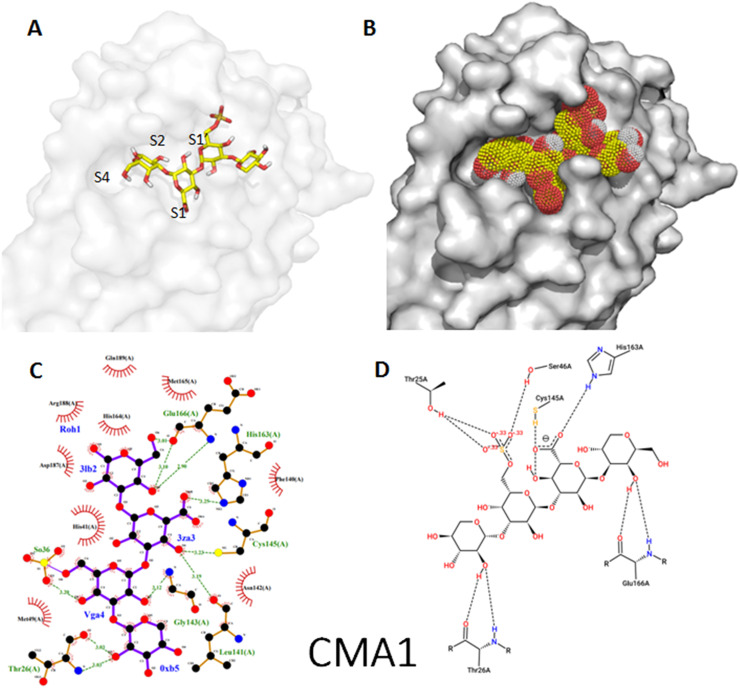
Fig. 5.
Binding mode of the tetra-sulphated oligosaccharide (CMA1) as stick representation (A) and as dot surface representation showing fullness of the active site (B). Visualization of docking results interaction of the inhibitor using (C) Ligplotplus and (D) dan Poseview.
Sulfated penta-saccharide from Porphyridium sp. namely CMA3 has shown the best binding affinity toward Mpro with a binding energy of −9.9 Kcal/mol. The interaction is depicted by generating ligplot and poseview diagrams shown in Fig. 4C and D. Docking study has shown that CMA3 interacts with a total of 23 amino acids; Asn 119, Gln 192, Ser144, His163, Asn142, Cys145, Gly143, His41, Phe140, Thr24, Thr26, Leu27, Thr190, Arg188, Met165, Glu166, Leu167, Asp187, Tyr54, Met49, His164, Leu141 and Gln189. Sixteen H-bonds were found stabilizing the ligand in addition to hydrophobic interactions which enhance CMA3 binding to the active site of SARS-CoV-2 Mpro. As observed for the structure of SARS-CoV-2 Mpro complexed with Paxlovid, the two catalytic residues C145 and H41 are involved in the interactions. The inspection of surface representation shows that, compared to Paxlovid molecule occupying mainly the S4, S2 and S1 subsites, CMA3 inhabits all the four subsites of the substrate-binding pocket (Fig. 4A and B). Thus, it can be predicted that this molecule binds strongly to the entire amino acid residues needed for proper inhibition of the SARS-CoV-2 Main protease.
Sulfated tetrasaccharide from Porphyridium sp. namely CMA1 has shown comparable number of amino acid interactions despite of higher number of hydrogen bonds (10 bonds) compared to Paxlovid (Table S2, Fig. 5C and D). In the same way, and as shown in Fig. 5A and B, CMA1 occupies the four subsites of the catalytic pocket and interact with the catalytic C145 and H41. This second molecule could also bind strongly and inhibit the protease activity. In both cases, sulfate groups established 3 hydrogen contacts with the Thr25 and Ser46. It should be noted that, for some semisynthetic polysaccharides it is documented that higher sulfation means better antiviral activity of molecules (Ghosh et al., 2009).
The higher affinity of CMA3 compared to CMA1, and hence its more potent antiviral activity, may be due to the higher molecular weight of the former providing greater opportunity for multipoint binding to the Mpro of SARS-CoV-2. The glucuronic acid unit occupies the oxyanion hole (S1 subsite) that is stabilized by the backbone NH of G143 and the catalytic C145 through three hydrogen bonds for CMA3 and by the backbone NH of L141 and the catalytic C145 through 2 hydrogen bonds for CMA1. In both cases, the carboxylic group of the glucuronic acid is stabilized by hydrogen bond with the Nε2 atom of H163.
It should be noted here that, only C145 and H41 are engaged directly in the cleavage mechanism of Mpro. An electron transfer between them leads to the nucleophilic attack on the carbonyl carbon atom of the peptide bond, thus leading to a thiohemiketal (THA) intermediate. The remaining of the residues implicated in the substrate interaction would stabilize and favour a step-wise degradation process (Świderek and Moliner, 2020).
Based on these results, the two docked ligands as well as the redocked inhibitor have been selected to find out their system stability, flexibility, and other dynamic properties through 100 ns MD simulation.
3.2. Molecular dynamics simulation (MD) and free energy landscape analysis
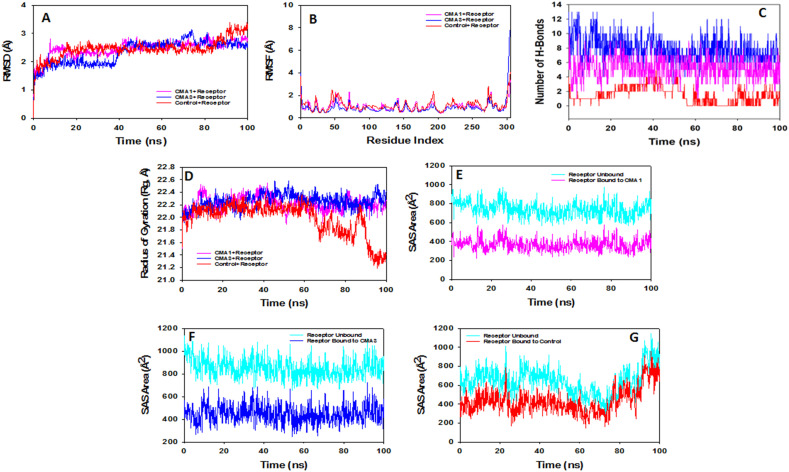
Molecular dynamics and simulation (MD) studies were carried out to investigate the stability and convergence of CMA1 + Mpro and CMA3 + Mpro complexes with comparison to Paxlovid + Mpro one. Simulation of 100 ns displayed stable conformation while comparing the root mean square deviation (RMSD) values. The RMSD of Cα-backbone of the receptor protein (Mpro) bound to CMA1 exhibited a deviation of 0.9 Å (Fig. 6A) while with CMA3 it displayed a deviation 0.7 Å (Fig. 6A). For initial 38 ns there is a decrease in RMSD deviations observed while later than 38 ns the curve is stable till 100 ns for CMA3 bound state (Fig. 6A).
Fig. 6.
Analysis of MD simulation trajectories of 100 ns time scale. (A) RMSD plot displaying the molecular vibration of Cα backbone of Receptor + control (red), Receptor + CMA1 (pink) and Receptor + CMA3 (blue). (B) RMSF plots showing the fluctuations of respective amino acids throughout the simulation time 100 ns for Receptor + control (red), Receptor + CMA1 (pink) and Receptor + CMA3 (blue). (C) Number of hydrogen bonds formed between Receptor + control (red), Receptor + CMA1 (pink) and Receptor + CMA3 (blue) during 100 ns simulation time scale. (D) Radius of gyration plots for the deduction of compactness of protein Receptor + control (red), Receptor + CMA1 (pink) and Receptor + CMA3 (blue). Solvent accessible surface area (SAS Area) displaying the ligand bound and unbound area at the binding pocket (cyan), (E) Receptor + CMA1 (pink), (F) Receptor + CMA3 (blue) and (G) Receptor + control (red).
However, receptor protein with Paxlovid (control + receptor) exhibited much higher deviation 1.5 Å from the 85th ns to the end of the simulation. Large deviation of RMSD is due to unordered structure of the control bound receptor protein while the less deviated RMSD of CMA1 and CMA3 bound receptors signify a good convergence and stable conformations. Therefore, it can be suggested that CMA1 and CMA3 bound to Mpro is quite stable in complex due to the high affinity of the ligand. The plots for root mean square fluctuations (RMSF) displayed small spikes of fluctuation in receptor protein except Paxlovid bound state which leads to un-ordered structure at residues 50, 180 and 225, while the remaining of the residues less fluctuating during the entire 100 ns simulation (Fig. 6B) indicating the stable amino acid conformations during the simulation time. Therefore, from RMSF plots it can be suggested that the structures of receptors were stable during simulation in CMA1 and CMA3 bound conformations. Number of hydrogen bonds between protein and ligand suggests the significant interaction and stability of the complex. The hydrogen bonds showed significant numbers between receptor and CMA1 as well as CMA3 throughout the simulation time of 100 ns (Fig. 6C). A consistent numbers of hydrogen bonds are observed between receptor and CMA1 (average number of 6) and CMA3 (average number of 8) that might facilitate to conform into stable complex (Fig. 6C). In contrast, with control ligand the number of observed hydrogen bonds was significantly less as compared to CMA1 and CMA3 bound receptor. The average numbers are observed to be one which might be the cause of less affinity for the ligand (Fig. 6C). These results are in accordance with the interaction diagrams retrieved after docking experiments.
Radius of gyration is the measure of compactness of the protein. In this study, receptor Cα-backbone bound to CMA1 displayed stable radius of gyration (Rg) ranging from 22.1 to 22.2 Å (Fig. 6D). Receptor Cα-backbone bound to CMA3 displayed stable as well as lowering of peaks from 22.1 to 22.3 Å (Fig. 6D). On the other hand, the control Paxlovid bound to receptor protein (control + receptor) displayed significant uneven fluctuations and devoid of stability indicating less compact orientation of the protein (Fig. 6D). Followed by Rg analysis, similar patterns were also observed in Solvent Accessible Surface Area (SASA) in both ligand bound and unbound state. It is clearly visible from the Fig. 6E and F that in the unbound state of ligand + receptor protein displayed high surface area accessible to solvent (Fig. 6E and F, cyan) while binding with CMA1 and CMA3, the SASA value lowered as compared to unbound state (Fig. 6E and F, pink and blue). However, a surprisingly plot was observed for control bound to receptor, where, lowering of Rg value observed till 70 ns but later became coincide with the unbound state. That signifies the less ligand affinity and compels out of the binding cavity. The overall study of Rg signifies the ligand CMA1 and CMA3 binding compels the respective proteins to become more compact and less flexible.
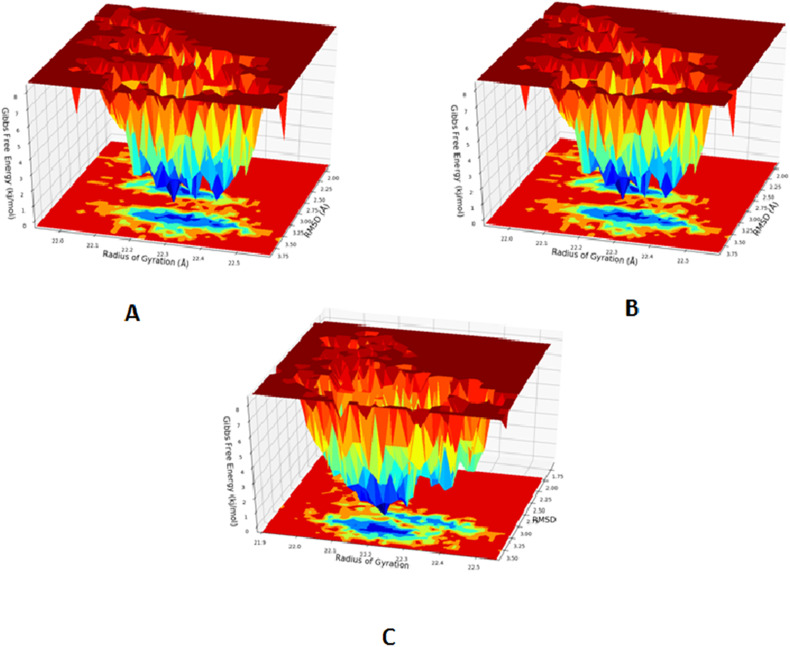
The Free Energy Landscape of (FEL) of achieving global minima of Cα backbone atoms of proteins with respect to RMSD and radius of gyration (Rg) are displayed in Fig. 7 . CMA1 bound to receptor achieved the global minima (lowest free energy state) at 2.7–3.5 Å and Rg of 22.3 Å (Fig. 7A). The FEL envisaged for deterministic behaviour of receptor to lowest energy state owing to its high stability and best conformation at CMA1 bound state. Whereas in case of CMA3 bound with received, the global minimum (lowest free energy state) is achieved at 3.1–3.4 Å and Rg of 22.25 Å (Fig. 7B). On the other hand, global minimum is achieved by paxlovid bound to receptor at high RMSD 3.5 Å and Rg 22.3 Å (Fig. 7C).
Fig. 7.
Free Energy Landscape displaying the achievement of global minima (ΔG, kJ/mol) of (A) receptor in presence of CMA1 with respect to their RMSD (nm) and radius of gyration (Rg, nm); (B). Free Energy Landscape displaying the achievement of global minima (ΔG, kJ/mol) of receptor in presence of CMA3 with respect to their RMSD (nm) and radius of gyration (Rg, nm). (C) Free Energy Landscape displaying the achievement of global minima (ΔG, kJ/mol) of receptor in presence of control with respect to their RMSD (nm) and radius of gyration (Rg, nm).
Therefore, FEL is the indicator of the protein folding to attain minimum energy state, and that aptly achieved due to CMA1 and CMA3 bound state. Further free energy landscape (FEL) of CMA1 and CMA3 bound receptor complexes exhibited a deep basin over areas of increased free energy with the deep blue colour locations represented the local energy minima and actively promoted stable conformations similarly suggested by Singh et al. (2021).
3.3. Pharmacokinetic properties predicted by SwissADME, Osiris and Molinspiration
The toxicity properties of the selected molecules along with Paxlovid were evaluated as potential drug candidates. Firstly, General Unrestricted Structure–Activity Relationships (GUSAR) software was used for quantitative in silico toxicity prediction in rats considering the four types of administration (intraperitoneal, intravenous, oral and subcutaneous). As displayed in Table S3, the LD50 values of CMA1 and CMA3 were higher than Paxlovid for intravenous (IV), oral, and subcutaneous (SC) routes of administration. While the LD50 values through intraperitoneal (IP) route were higher for Paxlovid. According to the OECD chemical classification system, the toxicity profiles of CMA1 and CMA3 are relatively low, they fall for the most in the applicability domain of models and they require high doses to induce toxic responses.
The pharmacophore features and drug-like properties were performed with Molinspiration and Osiris Property Explorer. The cLogP values (which is octanol/water partition coefficient) of CMA1 and CMA3 were found lower than 5.0 (Table 1 ). This finding suggests that these analogues have rational high absorption and permeability.
Table 1.
Drug likeliness properties by SwissADME, Osiris and Molinspiration.
| Drug likeness properties | Paxlovid | CMA1 | CMA3 |
|---|---|---|---|
| Bioavailability and drug-scorea | |||
| Molecular weight g/mol | 498.53 | 730.60 | 864.71 |
| cLogP | 0.98 | −9.49 | −10.73 |
| Solubility | −3.69 | 2.19 | 2.36 |
| TPSA | 131.4 | 396.4 | 455.3 |
| Drug likeness | −36.55 | −4.85 | −4.82 |
| Drug-score | 0.12 | 0.27 | 0.26 |
| Drug likenessb | |||
| GPCR ligand | 0.06 | −0.15 | −1.51 |
| Ion channel modulator | −0.12 | −1.10 | −2.86 |
| Kinase inhibitor | −0.22 | −0.86 | −2.54 |
| Nuclear receptor ligand | −0.21 | −0.85 | −2.62 |
| Protease inhibitor | 0.45 | 0.14 | −0.94 |
| Enzyme inhibitor | 0.03 | −0.05 | −1.67 |
| Pharmacokineticsc | |||
| GI absorption | High | Low | Low |
| BBB permeant | No | No | No |
| P-gp substrate | Yes | Yes | Yes |
| CYP1A2 inhibitor | No | No | No |
| CYP2C19 inhibitor | No | No | No |
| CYP2C9 inhibitor | No | No | No |
| CYP2D6 inhibitor | No | No | No |
| CYP3A4 inhibitor | No | No | No |
| Medicinal chemistryc | |||
| PAINS | 0 | 0 | 0 |
| Brenk | 1 | 0 | 0 |
| Lead-likeness | No | No | No |
| Synthetic accessibility | 3.79 | 6.28 | 6.86 |
GI: gastrointestinal absorption; BBB: blood brain barrier; P-gp: permeability glycoprotein; CYP: cytochrome P450.
Osiris.
Molinspiration.
SwissADME.
Solubility is known to be a significant parameter for drug design and pharmacology due to the potential absorption and distribution characteristics. The solubility values of CMA1 and CMA3 were 2.19 and 2.36, respectively compared to −3.69 for Paxlovid (Table 1). Our two compounds were found to have much higher solubility in water than the Paxlovid.
Ion Channel Modulator, Human G-protein coupled receptors (GPCRs) ligands, Nuclear Receptor Ligand, Kinase Inhibitor, Protease Inhibitor and Enzyme inhibitors of the selected ligands were illustrated with the prediction bioactivity scores using online-site Molinspiration (Table 1). Although there are few violations but both molecules can be used as a drug for proper application. As presented in Table 1, metabolic enzymes such as cytochrome P450 (CYP) and the transporter class P-glycoprotein (P-gp) were equally assessed in this study. CMA1 and CMA3 were noticed to be P-gp substrates and were not inhibitors to different tested cytochrome P450 isomers which have a crucial role in drug elimination.
However, skin permeation coefficient (log Kp) indicated that all compounds were impermeable through the skin barrier. Moreover, medicinal chemistry parameters revealed that none of the selected molecules returns any Pan-assay interference compounds (PAINS) alert with also no Brenk alert for both compounds CMA1 and CMA3 (except the Paxlovid which showed one Brenk alert). The synthetic accessibility values of all compounds were in the range of 3.79 for Paxlovid, 6.28 for CMA1 and 6.86 for CMA3 which indicated that they could be easily synthesized for pharmaceutical uses.
3.4. Computational compound toxicity prediction by VEGA Hub
In this section we determined the toxicity of the Paxlovid and the two selected oligosaccharides (CMA1 and CMA3) by VEGA software using the QSAR (quantitative structure-activity relationship) approach. The results were outlined in Table 4S. Toxicity measurements performed by Vega Hub software showed that Paxlovid can be a toxic compound in different assays. It was found to be carcinogen in Carcinogenicity model (CAESAR) and predicted to be an androgen disruptor that interferes with the biosynthesis, metabolism or action of endogenous androgens by the activation of androgen receptors based on Androgen Receptor-mediated effect (IRFMN/COMPARA) model.
Moreover, Paxlovid was predicted to engender Hepatotoxicity based on Hepatotoxicity model (IRFMN). Therefore, this drug is quite unsafe. CMA1 and CMA3 showed no toxicity from any of the studied toxicity model. So, these compounds could be safe and effective antiviral agents as compared to synthetic drugs. It is generally agreed that in silico methods are among the most advisable alternatives for the safety evaluation of chemicals. However, these methods are not capable of entirely substituting in vitro and in vivo testing. It should be noted that Mpro is a conserved protein having no homologs or similar cleavage sites for proteases found in the human proteome, making it an ideal potential drug repurposing and screening target. Thus, drugs targeting Mpro are expected to have less or no side effects and toxicity while still acting as a broad-spectrum antiviral agent. Overall, we demonstrated that oligosaccharide derived from Porphyridium SEP could have a great anti SARS-CoV-2 potential.
The most studied marine algal sulfated polysaccharides are sourced from green macroalgae (ulvans), from red macroalgae (carrageenans and agar), and from brown macroalgae (fucoidans and laminarans). These polysaccharides have been shown to possess antiviral activity against herpes human immunodeficiency virus type1 (HIV-1), chikungunya virus, simplex virus (HSV), cytomegalovirus (CMV), influenza virus, and hepatitis virus. Furthermore, recently their antiviral effect against the COVID-19 pandemic has also been reported (Hans et al., 2021). Various marine sulfated polysaccharides showed inhibitory activity against SARS-CoV-2, including sea cucumber sulfated polysaccharide (SCSP), chondroitin sulfate from sharks, fucoidan and carrageenan which have been shown to prevent SARS-CoV-2 entry into the cell by binding to the S-glycoprotein (Song et al., 2020). Significant antiviral activity was displayed by SCSP with an IC50 of 9.10 μg/mL. An investigation of a pseudotype virus with S glycoprotein confirmed the potential of SCSP to bind to the S glycoprotein and thus to prevent SARS-CoV-2 host cell entry (Geetha Bai and Tuvikene, 2021).
4. Conclusion
The virtual molecular docking study reveals that sulfated penta- and tetra-saccharides from SEP of Porphyridium sp. have a higher binding affinity toward COVID-19 main protease compared to co-crystal ligand inhibitor Paxlovid. Molecular dynamics simulation studies of the top two docked compounds and Paxlovid confirm that they have good stability, flexibility, and binding affinity toward Mpro compared to the inhibitor. From the results, it can be concluded that these compounds can be used for medicinal purposes and are non-toxic. Overall, this in silico study predicts that two oligosaccharides' compounds from Porphyridium SEP, may have the potency to be evolved as an anti-Mpro drug to fight against the novel coronavirus but before that, it must go through under the proper preclinical and clinical trials for further experimental and/or clinical validation.
The following are the supplementary data related to this article.
Details of the macro and microalgal compounds and Paxlovid used for screening and docking analysis with Mpro substrate binding site. Molecules having high binding energies compared to Paxlovid are shown in bold.
Details of interactions between the selected molecules and Paxlovid with residues of the active site as depicted with LigPlot software
Prediction of acute toxicity in silico by GUSAR in rodent models and chemical classification of compounds.
Toxicity assessment of Paxlovid and selected compounds using Vega QSAR model. Notable toxicity measurements are shown in bold character.
CRediT authorship contribution statement
Hajer Ben Hlima: Conceptualization, Investigation, Methodology, Data curation, Formal analysis, Visualization, Writing-original draft preparation, Writing-Reviewing and Editing.
Ameny Farhat: Formal analysis, Visualization.
Sarra Akermi; Data curation, Formal analysis, Visualization.
Bassem Khemakhem: Conceptualization, Investigation, Methodology, Software.
Youssef Ben Halima: Investigation, Methodology, Software.
Philippe Michaud: Resources, Supervision.
Imen Fendri: Conceptualization, Resources, Supervision.
Slim Abdelkafi: Conceptualization, Supervision, Writing-Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work has been carried out under the project (2020−2022) n° PRF-COVID-D5P2 funded by the Tunisian government.
Editor: Lotfi Aleya
References
- Abdelkafi S., Chamkha M., Casalot L., Sayadi S., Labat M. Isolation and characterization of a novel Bacillus sp., strain YAS1, capable of transforming tyrosol under hypersaline conditions. FEMS Microbiol. Lett. 2005;252(1):79–84. doi: 10.1016/j.femsle.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Ahmad B., Batool M., Kim M.S., Choi S. Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations. Int. J. Mol. Sci. 2021;22(17):9124. doi: 10.3390/ijms22179124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi A., Zorofchian Moghadamtousi S., Abubakar S., Zandi K. Antiviral potential of algae polysaccharides isolated from marine sources: a review. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/825203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameen F., AlNadhari S., Al-Homaidan A.A. Marine microorganisms as an untapped source of bioactive compounds. Saudi J. Biol. Sci. 2021;28(1):224–231. doi: 10.1016/j.sjbs.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baklouti Z., Delattre C., Pierre G., Gardarin C, Abdelkafi S., Michaud P., Dubessay P. Biochemical characterization of a bifunctional enzyme constructed by the fusion of a glucuronan lyase and a chitinase from Trichoderma sp. Life. 2020;10(10) doi: 10.3390/life10100234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor F., Barkallah M., Eleuch F., Karkouch N., Dammak M., Baréa B., Villeneuve P., Abdelkafi S., Fendri I. Cyanobacteria as sources of marine bioactive compounds: molecular specific detection based on Δ9 desaturase gene. Int. J. Biol. Macromol. 2017;105:1440–1445. doi: 10.1016/j.ijbiomac.2017.07.139. [DOI] [PubMed] [Google Scholar]
- Ben Halima N., Ben Saad R., Khemakhem B., Fendri I., Abdelkafi S. Oat (Avenasativa): oil and nutriment compounds valorization for potential use in industrial applications. J. Oleo Sci. 2015;64(9):915–932. doi: 10.5650/jos.ess15074. [DOI] [PubMed] [Google Scholar]
- Ben Hlima H., Bohli T., Kraiem M., Ouederni A., Mellouli L., Michaud P., Abdelkafi S., Smaoui S. Combined effect of Spirulinaplatensis and Punicagranatum peel extacts: phytochemical content and antiphytophatogenic activity. Appl. Sci. 2019;9(24):5475. doi: 10.3390/app9245475. [DOI] [Google Scholar]
- Bhadury P., Wright P.C. Exploitation of marine algae: biogenic compounds for potential antifouling applications. Planta. 2004;219(4):561–578. doi: 10.1007/s00425-004-1307-5. [DOI] [PubMed] [Google Scholar]
- Bowers K.J., Chow D.E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K. SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE; 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters; p. 43. 2006 Nov 11. [DOI] [Google Scholar]
- Bule M.H., Ahmed I., Maqbool F., Bilal M., Iqbal H.M. Microalgae as a source of high-value bioactive compounds. Front. Biosci. 2018;10(2):197–216. doi: 10.2741/s509. [DOI] [PubMed] [Google Scholar]
- Case D.A., Cerutti D.S., Cheatham T.E., Darden T.A., III, Duke R.E., III, Giese T.J., III, Gohlke H., III, Goetz A.W., III, Greene D., III, Homeyer N., III, Izadi S., III, Kovalenko A., III, Lee T.S., III, LeGrand S., III, Li P., III, Lin C., III, Liu J., III, Luchko T., III, Luo R., III, Mermelstein D., III, Merz K.M., III, Monard G., III, Nguyen H., III, Omelyan I., III, Onufriev A., III, Pan F., III, Qi R., III, Roe D.R., III, Roitberg A., III, Sagui C., III, Simmerling C.L., III, Botello-Smith W.M., III, Swails J., III, Walker R.C., III, Wang J., III, Wolf R.M., III, Wu X., III, Xiao L., III, York D.M., III, Kollman P.A., III . University of California; San Francisco, CA: 2017. AMBER. [Google Scholar]
- Chow E., Rendleman C.A., Bowers K.J., Dror R.O., Hughes D.H., Gullingsrud J., Sacerdoti F.D., Shaw D.E. DE Shaw Research Technical Report DESRES/TR-2008-01. 2008. Desmond performance on a cluster of multicore processors. [Google Scholar]
- Cohn B.A., Cirillo P.M., Murphy C.C., Krigbaum N.Y., Wallace A.W. SARS-CoV-2 vaccine protection and deaths among US veterans during 2021. Science. 2022;375(6578):331–336. doi: 10.1126/science.abm0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(1):1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammak M., Hadrich B., Barkallah M., Hentati F., Ben Hlima H., Pichon C., Denis M., Fendri I., Michaud P., Abdelkafi S. Modelling Tetraselmis sp. growth-kinetics and optimizing bioactive-compound production through environmental conditions. Bioresour. Technol. 2018;249:510–518. doi: 10.1016/j.biortech.2017.10.028. [DOI] [PubMed] [Google Scholar]
- Das B.K., Pradhan J., Pattnaik P., Samantaray B.R., Samal S.K. Production of antibacterials from the freshwater alga Euglenaviridis (Ehren) World J. Microbiol. Biotechnol. 2005;21(1):45–50. doi: 10.1007/s11274-004-1555-3. [DOI] [Google Scholar]
- Elleuch F., Ben Hlima H., Barkallah M., Baril P., Abdelkafi S., Pichon C., Fendri I. Carotenoids overproduction in Dunaliella sp.: transcriptional changes and new insights through lycopene cyclase regulation. Appl. Sci. 2019;9(24):5389. doi: 10.3390/app9245389. [DOI] [Google Scholar]
- Farhat A., Ben Hlima H., Khemakhem B., Ben Halima Y., Michaud P., Abdelkafi S., Fendri I. Apigenin analogues as SARS-CoV-2 main protease inhibitors: in-silico screening approach. Bioengineered. 2022;13(2):3350–3361. doi: 10.1080/21655979.2022.2027181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendri I., Bouaziz M., Labat M., Sayadi S., Abdelkafi S. Olive fermentation brine: biotechnological potentialities and valorization. Environ. Technol. 2013;34:181–193. doi: 10.1080/09593330.2012.689364. [DOI] [PubMed] [Google Scholar]
- Ferré V.M., Peiffer-Smadja N., Visseaux B., Descamps D., Ghosn J., Charpentier C. Omicron SARS-CoV-2 variant: what we know and what we don't. Anaesth. Crit. Care Pain Med. 2022;41(1) doi: 10.1016/j.accpm.2021.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha Bai R., Tuvikene R. Potential antiviral properties of industrially important marine algal polysaccharides and their significance in fighting a future viral pandemic. Viruses. 2021;13(9):1817. doi: 10.3390/v13091817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi Y., Yazdi M.T., Shafiee A., Amini M., Shokravi S., Zarrini G. Parsiguine, a novel antimicrobial substance from Fischerella ambigua. Pharm. Biol. 2004;42(4–5):318–322. doi: 10.1080/13880200490511918. [DOI] [Google Scholar]
- Ghosh T., Chattopadhyay K., Marschall M., Karmakar P., Mandal P., Ray B. Focus on antivirally active sulfated polysaccharides: from structure–activity analysis to clinical evaluation. Glycobiology. 2009;19(1):2–15. doi: 10.1093/glycob/cwn092. [DOI] [PubMed] [Google Scholar]
- Hans N., Malik A., Naik S. Antiviral activity of sulfated polysaccharides from marine algae and its application in combating COVID-19: mini review. Bioresour. Technol. Rep. 2021;13 doi: 10.1016/j.biteb.2020.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasui M., Matsuda M., Okutani K., Shigeta S. In vitro antiviral activities of sulfated polysaccharides from a marine microalga (Cochlodiniumpolykrikoides) against human immunodeficiency virus and other enveloped viruses. Int. J. Biol. Macromol. 1995;17(5):293–297. doi: 10.1016/0141-8130(95)98157-T. [DOI] [PubMed] [Google Scholar]
- Herrero M., Ibáñez E., Cifuentes A., Reglero G., Santoyo S. Dunaliella salina microalga pressurized liquid extracts as potential antimicrobials. J. Food Protec. 2006;69(10):2471–2477. doi: 10.4315/0362-028X-69.10.2471. [DOI] [PubMed] [Google Scholar]
- Huheihel M., Ishanu V., Tal J., Arad S.M. Activity of Porphyridium sp. polysaccharide against herpes simplex viruses in vitro and in vivo. J. Biochem. Biophys. Met. 2002;50(2–3):189–200. doi: 10.1016/S0165-022X(01)00186-5. [DOI] [PubMed] [Google Scholar]
- Huige C.J., Altona C. Force field parameters for sulfates and sulfamates based on ab initio calculations: extensions of AMBER and CHARMm fields. J. Comput. Chem. 1995;16(1):56–79. doi: 10.1002/jcc.540160106. [DOI] [Google Scholar]
- Ibrahim M.A., Abdeljawaad K.A., Abdelrahman A.H., Hegazy M.E.F. Natural-like products as potential SARS-CoV-2 mpro inhibitors: in-silico drug discovery. J. Biomol. Struct. Dyn. 2021;39(15):5722–5734. doi: 10.1080/07391102.2020.1790037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaime L., Rodríguez-Meizoso I., Cifuentes A., Santoyo S., Suarez S., Ibáñez E., Señorans F.J. Pressurized liquids as an alternative process to antioxidant carotenoids' extraction from Haematococcus pluvialis microalgae. LWT-Food Sci. Technol. 2010;43(1):105–112. doi: 10.1016/j.lwt.2009.06.023. [DOI] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- Kagami L.P., das Neves G.M., Timmers L.F.S.M., Caceres R.A., Eifler-Lima V.L. Geo-measures: a PyMOL plugin for protein structure ensembles analysis. Comput. Biol. Chem. 2020;87:107322. doi: 10.1016/j.compbiolchem.2020.107322. [DOI] [PubMed] [Google Scholar]
- Kar S., Roy K. How far can virtual screening take us in drug discovery? Expert Opin. Drug Discov. 2013;8(3):245–261. doi: 10.1517/17460441.2013.761204. [DOI] [PubMed] [Google Scholar]
- Kellam S.J., Walker J.M. Antibacterial activity from marine microalgae in laboratory culture. Br. Phycol. J. 1989;24(2):191–194. doi: 10.1080/00071618900650181. [DOI] [Google Scholar]
- Kirschner K.N., Yongye A.B., Tschampel S.M., González-Outeiriño J., Daniels C.R., Foley B.L., Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydr. J. Comput. Chem. 2008;29(4):622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunin A., Zakharov A., Filimonov D., Poroikov V. QSAR modelling of rat acute toxicity on the basis of PASS prediction. Mol. Informatics. 2011;30(2–3):241–250. doi: 10.1002/minf.201000151. [DOI] [PubMed] [Google Scholar]
- Li Q., Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8(8):1250. doi: 10.3390/microorganisms8081250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyna G.J., Klein M.L., Tuckerman M. Nosé-hoover chains: the canonical ensemble via continuous dynamics. J. Chem. Phys. 1992;97(4):2635–2643. doi: 10.1063/1.463940. [DOI] [Google Scholar]
- Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101(5):4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- Naviner M., Bergé J.P., Durand P., Le Bris H. Antibacterial activity of the marine diatom Skeletonema costatum against aquacultural pathogens. Aquaculture. 1999;174(1–2):15–24. doi: 10.1016/S0044-8486(98)00513-4. [DOI] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei. Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Ozdemir G., Ulku Karabay N., Dalay M.C., Pazarbasi B. Antibacterial activity of volatile component and various extracts of Spirulina platensis. Phytoter. Res. 2004;18(9):754–757. doi: 10.1002/ptr.1541. [DOI] [PubMed] [Google Scholar]
- Pendyala B., Patras A. ChemRxiv. Cambridge Open Engage; Cambridge: 2020. In silico screening of food bioactive compounds to predict potential inhibitors of COVID-19 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp) [Google Scholar]
- Pfizer Pfizer's Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization O Death by 89% in Iterim Analysis of Phase 2/3 EPIC-HR Study. 2021. https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate
- Raposo M.F.D.J., De Morais R.M.S.C., Bernardo de Morais A.M.M. Bioactivity and applications of sulphated polysaccharides from marine microalgae. Mar. Drugs. 2013;11(1):233–252. doi: 10.3390/md11010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Mendoza S., García-Silva I., González-Ortega O., Sandoval-Vargas J.M., Malla A., Vimolmangkang S. The potential of algal biotechnology to produce antiviral compounds and biopharmaceuticals. Molecules. 2020;25(18):4049. doi: 10.3390/molecules25184049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih A.E., Thissera B., Yaseen M., Hassane A.S., El-Seedi H.R., Sayed A.M., Rateb M.E. Marine sulfated polysaccharides as promising antiviral agents: a comprehensive report and modeling study focusing on SARS CoV-2. Mar. Drugs. 2021;19(8):406. doi: 10.3390/md19080406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander T., Freyss J., von Korff M., Reich J.R., Rufener C. OSIRIS, an entirely in-house developed drug discovery informatics system. J. Chem. Inf. Model. 2009;49(2):232–246. doi: 10.1021/ci800305f. [DOI] [PubMed] [Google Scholar]
- Sapay N., Nurisso A., Imberty A. In: Methods in Molecular Biology (Methods and Protocols) Monticelli L, Salonen E, editors. Vol. 924. Humana Press, Totowa, NJ; 2013. Simulation of carbohydrates, from molecular docking to dynamics in water; pp. 469–483. [DOI] [PubMed] [Google Scholar]
- Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24(5):417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S.R., Ho M.S., Lin K.H., Wu S.L., Chen Y.T., Wu C.N., Lin T.Y., Chang L.Y., Tsao K.C., Ning H.C., Chang P.Y., Jung S.M., Hsueh C., Chang K.S. Genetic analysis of enterovirus 71 isolated from fatal and non-fatal cases of hand, foot and mouth disease during an epidemic in Taiwan, 1998. Virus Res. 2000;68(2):127–136. doi: 10.1016/S0168-1702(00)00162-3. [DOI] [PubMed] [Google Scholar]
- Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010;6(5):1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Peng H., Wang Q., Liu Z., Dong X., Wen C., Zhu B. Inhibitory activities of marine sulfated polysaccharides against SARS-CoV-2. Food & Func. 2020;11(9):7415–7420. doi: 10.1039/D0FO02017F. [DOI] [PubMed] [Google Scholar]
- Stierand K., Rarey M. From modeling to medicinal chemistry: automatic generation of two-dimensional complex diagrams. ChemMedChem. 2007;2(6):853–860. doi: 10.1002/cmdc.200700010. [DOI] [PubMed] [Google Scholar]
- Świderek K., Moliner V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 M pro by QM/MM computational methods. Chem. Sci. 2020;11(39):10626–10630. doi: 10.1039/D0SC02823A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talyshinsky M.M., Souprun Y.Y., Huleihel M.M. Anti-viral activity of red microalgal polysaccharides against retroviruses. Cancer Cell Int. 2002;2(1):1–7. doi: 10.1186/1475-2867-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukmaji A.Y., Board J.A., Jr. Ewald summation techniques in perspective: a survey. Comput. Phys. Com. 1996;95(2–3):73–92. doi: 10.1016/0010-4655(96)00016-1. [DOI] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Fang C., Zhang Q., Zhang R., Zhao X., Duan Y., Yang H. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2021;1–5 doi: 10.1007/s13238-021-00883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the macro and microalgal compounds and Paxlovid used for screening and docking analysis with Mpro substrate binding site. Molecules having high binding energies compared to Paxlovid are shown in bold.
Details of interactions between the selected molecules and Paxlovid with residues of the active site as depicted with LigPlot software
Prediction of acute toxicity in silico by GUSAR in rodent models and chemical classification of compounds.
Toxicity assessment of Paxlovid and selected compounds using Vega QSAR model. Notable toxicity measurements are shown in bold character.