Abstract
The detection to α-synuclein (αS) assemblies as a biomarker of synucleinopathies is an important challenge for further development of an early diagnosis tool. Here, we present proof of concept real-time fast amyloid seeding and translocation (RT-FAST) based on a nanopipette that combines in one unique system a reaction vessel to accelerate the seed amplification and nanopore sensor for single-molecule αS assembly detection. RT-FAST allows the detection of the presence αS seeds WT and A53T variant in a given sample in only 90 min by adding a low quantity (35 μL at 100 nM) of recombinant αS for amplification. It also shows cross-seeding aggregation by adding mixing seeds A53T with WT monomers. Finally, we establish the dependence between the capture rate of aggregates by the nanopore sensor and the initial seed concentration from 200 pM to 2 pM, which promises further development toward a quantitative analysis of the initial seed concentration.
Short abstract
We present a proof of concept of a real-time fast amyloid seeding and translocation method that allows detection of α-synuclein seeds in a sample using protein misfolding amplification inside a nanopipette.
Introduction
The aberrant aggregation of intrinsic-disordered proteins into highly ordered structures rich in β-sheets, called amyloids, is involved in age-related diseases. Among them, α-synuclein (αS), a neuronal presynaptic protein, is involved in the development of Parkinson’s disease (PD).1 Its aggregated forms disrupt the functioning and survival of neurons and represent the main markers of the pathology.2 Indeed, certain aggregate structures (e.g., soluble oligomers) may be present decades before the onset of the first major motor disorders. Today, the accuracy of clinical (or radiological) diagnosis improves with the progression of the disease.3 As a result, these techniques often do not allow diagnosis at the early stages, and they are not able to distinguish PD from other synucleinopathies, such as Lewy body or multiple system atrophy.4 For all these reasons, developing analytical methods for the detection and quantification of αS assemblies in biofluids is an important challenge to propose early diagnosis of synucleinopathies. The most advanced assay, called real-time quaking-induced conversion (RT-QuIC),5,6 allows the detection of αS amyloids directly from a patient’s cerebrospinal fluid (CSF).7 It exploits the self-propagation mechanism of amyloids by seeding the recruitment of the native protein counterpart into the growing assembly. The assay amplifies any seeding-competent αS in the CSF since such proteoforms interact with the recombinant native αS present in the reaction vessel.8 The seeds that are preformed αS aggregates induce misfolding of native αS to form amyloid-type aggregates that bind to the fluorescence reporter thioflavin T (ThT). However, three major drawbacks limit its application in clinical practice and must be tackled before the development of an early diagnostic test. First, the long lag time (several days) is required to reach an aggregate concentration that is readily detected by the bulk readout methodology. Second, RT-QuIC requires the use of a large amount of recombinant αS monomers (100 μL at 10 μM) per reaction condition.7 Last but not the least, it is limited to a binary (positive/negative) readout and thus does not allow the quantification of the initial seed concentration in a sample.
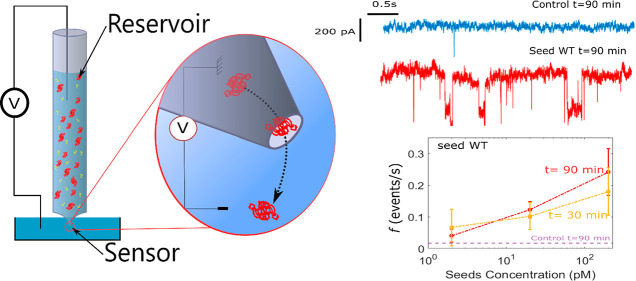
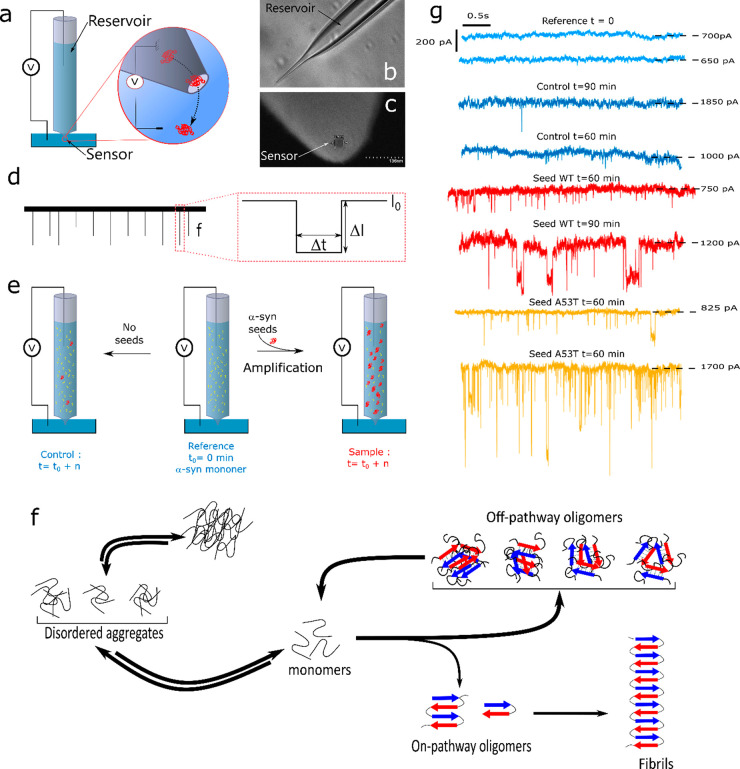
Therefore, an experimental technique is needed for fast, reliable, and inexpensive detection allowing the quantification of αS seeds. To achieve this goal, we have designed a new ultrasensitive in vitro protein misfolding amplification9 assay based on solid-state nanopore technology,10,11 hereafter called real-time fast amyloid seeding and translocation (RT-FAST). The innovative technical aspect of the methodology presented herein is the use of nanopipettes that simultaneously allow an accelerated amyloid seeding reaction and its sensing at the single-molecule scale. Indeed, with the small physical diameter of a glass capillary, nanopipettes provide a reaction vessel with a large surface/volume ratio that favors the amyloid seeding reaction through protein adsorption, conformational change, and desorption process.12−14 On the other hand, nanopipettes have an adjustable pore diameter at the tip side10 allowing for label-free detection of protein aggregates and amyloid fibrils15 by the resistive pulse technique (RPS).16,17 Briefly, the RPS consists of measuring the ionic current perturbation induced by the translocation of an object through the nanopore under a constant voltage. Three parameters are extracted from these current perturbations (Figure 1b): (i) the relative current blockade (ΔI/I0) that depends on the size, the shape, and the conformations of the objects; (ii) the dwell time (Δt) that depends on the net charge, the diffusion coefficient, and the interaction of the object with the nanopore inner wall; and (iii) the capture rate (f) that is relative to the concentration and the diffusion coefficient of the object.
Figure 1.
(a) Sketch showing the two parts of a nanopipette: the reservoir where the αS seeds are amplified and the sensor where αS seeds are detected, (b) optical image of a pipet pulled showing the reservoir and (c) SEM image of the tip aperture, (d) sketch of the current trace and illustration of event parameters (the amplitude ΔI/I0, the dwell time Δt, and the capture rate f), (e) illustration of the RT-FaST experiments, (f) sketch of different pathways of αS aggregation, (g) example of a current trace recorded extracted from different nanopipettes for reference (light blue - nanopipette 1 and 30), the control not seeded (blue - nanopipette 7 and 1), and sample seeded with αS WT (red - nanopipette 11 and 12) and A53T mutant (yellow - nanopipette 15 and 13).
The solid-state nanopores were recently considered investigating protein assemblies.11,18 The quartz nanopipettes were used to detect lysozyme19 and prion20 assemblies based on the enhancement of the current blockade amplitude. In order to improve the detection of the assemblies, a crowded media were considered to discriminate αS fibrils before and after sonication.21 Silicon nitride nanopores (SiNs) were used to detect various proteins. For the αS, four oligomeric species were identified after 3 days of incubation.22 Using a lipid-coated nanopore, Aβ peptide oligomers and fibrils were detected after several days of incubation.23 The common point between these two previous works is that amyloid samples were withdrawn at various time intervals corresponding to different aggregation steps (i.e., monomer, lag phase, elongation phase, and the plateau). These studies also showed the importance of the nanopore coating using phospholipid or Tween 20 to prevent its fouling. The SiN nanopore was found suitable to characterize the morphology of lysozyme, β-lactoglobulin, and BSA24,25 aggregates. Polymer nanopores obtained by a track-etched method on polyethylene terephthalate film were also used to differentiate different protein species (oligomers and protofibrils) during the aggregation process of β-lactoglobulin,26 as well as to discriminate calibrated fibrils of the Aβ-peptide.27 Another study points out the importance of conformational rearrangements and protomer exchanges during the self-assembly of the Aβ peptide.28 The evidence of autofragmentation processes was demonstrated during the aggregation of tau induced by heparin.29 The track-etched nanopore allowed the study of seed-induced reaggregation after protease digestion of β-lactoglobulin.30 In this work, we present the proof of concept of RT-FAST . We demonstrate that nanopipettes are suitable to accelerate the protein misfolding amplification and detect αS assemblies present during the lag phase allowing us to show at a hour time scale the presence of αS assemblies in a sample. Then, we show that the analysis of the assembly kinetic profile can provide quantitative information on the initial seed concentration.
Results and Discussion
Nanopipettes were pulled strictly following the same program to ensure an acceptable reproducibility of the nanopore diameter. Nanopipettes exhibit a tip diameter of about 34 ± 3 nm (Figure SI-1a). In addition, we select only nanopipettes with a similar range of conductance measured by current–voltage dependence at 1 M NaCl (containing PBS 1×, pH = 7.4) (Table SI-1). To prevent the fouling, the inner surface was coated with l-3,4-dihydroxyphénylalanine (l-DOPA).31 The successful coating process is evidenced by a weak modification of the power spectral density (Figure SI-1b). The αS seeds were obtained after 150 min of incubation at 37 °C. At this stage, TEM images show αS species with a typical annular structure but not amyloid fibers (Figure SI-2). This is confirmed by the ThT assay (Figure SI-2c). We selected this type of seed to demonstrate the suitability of RT-FaST for amplification at an early stage of αS aggregation, before the formation of a β-sheet structure (Figure 1f).
The experimental principle of RT-FAST consisted of injecting 35 μL of αS monomer solution (100 nM in NaCl 1 M PBS 1×) in the nanopipette reservoir (internal diameter 700 μm). The control condition contains only αS monomers, the reference is the control at t = 0 min, and the sample is composed of a mixture of αS monomers and WT or A53T seeds (200 pM) (Figure 1e). Every 30 min, a voltage of 500 mV is applied, and the current is recorded during 10 min (Figure 1g).
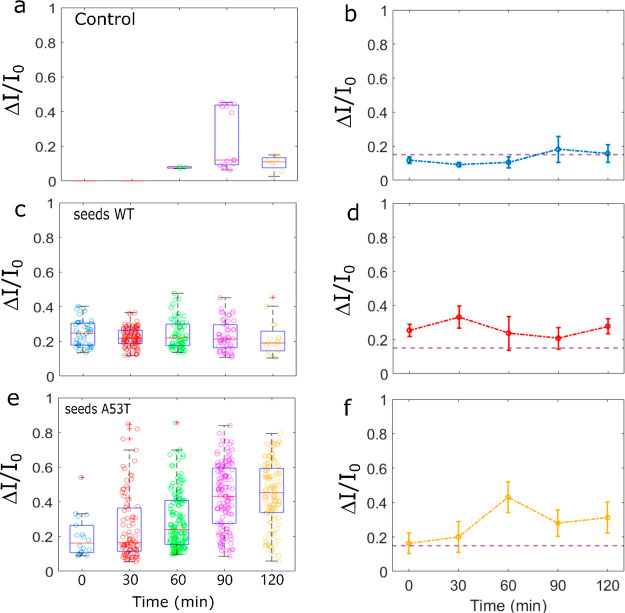
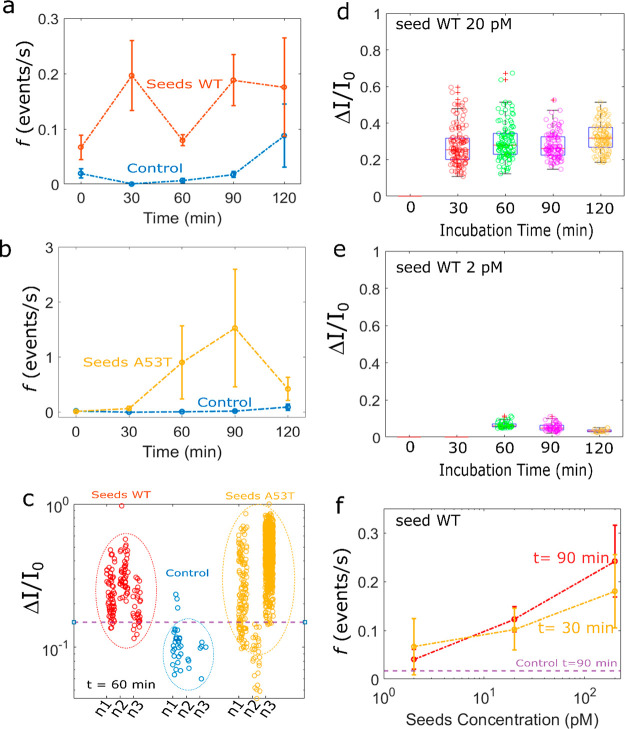
First, we investigated the kinetics of αS aggregation in the nanopipette device without seed addition at 25 °C ± 2 °C. We report in Figures 2a and SI-3 the scatter of ΔI/I0 of the recorded events as a function of time for four different nanopipettes. We noticed that at t = 0 s (the reference), rare events (capture rate f < 0.03 events/s and amplitude ΔI/I0 > 0.2) were detected for 50% of the experiments. These rare events can be attributed to the presence of some αS assemblies at the very early stage since no further purification of the monomeric fraction was carried out after solubilization and filtration. With time, the aggregation process occurs in the nanopipette. The events start to be detectable after 30 or 60 min depending on the experiments. In the absence of seeds, the dependence of the ΔI/I0 distribution on the incubation time is observed suggesting a slight assembly process (Figure 2b). However, we note variability in the different nanopore experiments (Figure SI-3). This is not surprising since the αS aggregation is a stochastic and dynamic process involving numerous pathways. However, we can note that the capture rate is always lower than 0.05 during 90 min. Regardless of the nanopipette and the incubation time, the mean distribution of the ΔI/I0 amplitude is mostly below 0.2. However, the low number of events makes it impossible to provide further information on the aggregate size or morphology. In order to prove that the events come from αS aggregation, we proceed to a set of experiments at T = 30 °C (instead T = 25 °C) (Figure SI-4) in order to accelerate the assembly formation. A rise in the incubation temperature significantly increases the capture rate (f > 0.1 after 30 min). A rise in the incubation temperature significantly increases the capture rate (f > 0.1 after 30 min). In addition, the amplitudes of ΔI/I0 are comparable to those obtained at 25 °C, suggesting that the distribution of the aggregate size is similar. Since the capture rate is mainly due to a diffusion process32f = 2πDCr* (D is the diffusion coefficient, C is the concentration, and r* is the capture radius), the expected ratio is fT=30°C/fT=25°C ≈ 1.13, for a similar capture radius and aggregate size. The experimental results show that the ratio fT=30°C/fT=25°C > 4, suggesting that more aggregates are formed in the nanopipette reservoir due to temperature-accelerated kinetics. For the sample containing 200 pM of αS WT seeds, the current traces recorded after 30 min clearly show larger and more abundant current blockades than for the reference t = 0 min and the control (Figure 1f). This is confirmed by the scatter plot reported for all experiments performed at various times, ranging from 0 to 120 min (Figures 2c and SI-5). We note that events can be readily observed at t = 0 min. A possible explanation could be that these events reflect residual protein assembly induced during the 10 min recording range. The distributions of the amplitude of ΔI/I0 are wide and centered at a value larger than 0.2, regardless of the incubation time (Figure 2d). Compared to the control, the mean values of ΔI/I0 are larger, and the distribution is more spread. This suggests that the detected αS assemblies are larger and heterogeneous. This is supported by the wide range of the dwell time distribution over two orders of magnitude at the millisecond scale. We notice that the millisecond scale for the dwell time is in good agreement with the literature related to the detection of protein aggregates by nanopipette.20,33,34 Regardless of the sample used, the value of ΔI/I0 does not increase with time, as it could be expected for the formation of protofibrils. Previous published data using nanopore were reported with an incubation period of several days, and the aggregates were withdrawn at the different stages of fibril formation (i.e., monomer, lag phase, elongation phase, and plateau).22,23 Here, we show that oligomer species are generated during the lag phase (short time scale dynamics), before the formation of fibrils with a β-sheet structure. At this stage, the oligomer species are highly heterogeneous in terms of size and conformation.35,36 Among them, the majority will not convert to fibrils but dissociate to monomers to further reaggregate.37 Thus, the observation of a wide range of events is consistent with the presence of transient oligomers in association/dissociation equilibrium during the lag phase, as previously reported for the Aβ peptide.28 Interestingly, the capture rate is significantly increased (f > 0.2) as soon as the incubation time is longer than 30 min (Figure 3a). This means that the potentially detectable αS assemblies by our system are more abundant and larger than the ones in the control, thus evidencing, at this stage, the protein misfolding amplification by seed addition.
Figure 2.
Distribution of amplitude of the current blockade recorded for the control (a) and the sample seeded with 200 pM of α-synuclein WT (c) and A53T (e) at as a function of incubation time. The experiments were performed at V = 500 mV in 1 M NaCl, PBS 1× pH 7.4, T = 25 °C using pipet no. 6, no. 11, and no. 15, respectively. Mean of the ΔI/I0 center distribution as a function of the incubation time for the control (b) and the sample seeded with αS WT (d) and A53T (f) obtained from a set of three independent nanopipettes (i.e., nanopipettes Nos. 4, 6, and 7 for the control, nanopipettes Nos. 9, 11, and 12 for samples seeded with αS WT and nanopipettes Nos. 13, 15, and 16 for seeding with αS and A53T). The error bars are the standard deviation.
Figure 3.
Mean of the capture rate as a function of incubation time for the control (in blue) and the sample seeded with αS WT in red (a) and A53T in yellow (b) obtained from a set of three independent nanopipettes (i.e., nanopipettes nos. 4, 6, and 7, for the control, nanopipettes nos. 9, 11, and 12 for the sample seeded with αS WT and nanopipettes nos. 13, 15, and 16 the sample seeded with αS A53T). The error bars are the standard deviation. (c) Distribution of amplitude of the current blockade for the control (blue) and the sample seeded with α-synuclein WT (red) and A53T (yellow) at t = 60 min. Note the violet dash line represents a value of 0.15. obtained from a set of three independent nanopipettes (i.e., nanopipettes nos. 4, 6, and 7 for the control, nanopipettes nos. 9, 11, 12 for the sample seeded with αS WT, and nanopipettes nos. 13, 15, and 16 for the sample seeded with αS A53T). Distribution of amplitude of the current blockade recorded for the sample seeded with (d) 20 pM and (e) 2pM of αS WT as a function of incubation time. The experiments were performed at V = 500 mV in NaCl 1 M, PBS 1× pH 7.4, T = 25 °C using pipet no. 19 and no. 22 respectively. Mean of the capture rate as a function of the initial αS WT seed concentration for incubation times of 30 min (yellow) and 90 min (red). The means were obtained from a set of three independent nanopipettes (i.e., nanopipettes nos. 21, 22, and 23, nanopipettes nos. 17, 18, and 19, and nanopipettes nos. 9, 11, and 12 for initial αS seeds WT concentration 2 pM, 20 pM, and 200 pM, respectively). The error bars are the standard deviation.
The A53T variant of αS is known to promote the aggregation process.38,39 This mutation is associated with early onset Parkinson disease.40 On the other hand, the αS can misfold and mimic the structure of the preformed seeds.41,42 Therefore, we next investigated the impact of an amplification process by A53T seeds and compared the results with those obtained using WT seeds. For this set of experiments, the capture rate (f > 0.1) is larger than the control as soon as the incubation time is longer than 30 min (Figure 3b). The distribution of ΔI/I0 is larger than the control and more spread than the sample containing WT seeds (Figures 2e and SI-6). This suggests that the A53T seeds induce the formation of larger and more heterogeneous aggregates than the WT. The oligomerization of A53T was reported to consume the monomer more rapidly than does the WT.43 On the other hand, the cross-seeding of αS monomers by A53T seed was found to accelerate the elongation process44 as well as to modify the properties of oligomer species.45 Thus, it is plausible that the cross-seeding by A53T produced different populations of oligomeric species compared to the WT during the lag phase. On the basis of only the information given by the mean center of ΔI/I0 distribution from three independent nanopipettes, the presence of seeds in the sample is perfectly detectable. However, the wide variability does not allow distinguishing A53T from WT seeds (Figure 2f).
If we consider now the result as a positive/negative readout of the presence of seeds in the sample, the plot of the ΔI/I0 distribution for the control and the sample after 60 min clearly shows the presence of seeds in the sample with a good reproducibility (Figure 3c). The information from ΔI/I0 distribution is confirmed by the capture rate regardless of the amplitude of the event. This double check is interesting because the seeding phenomenon is characterized by a high degree of variability (Figures SI-5b and 6b) or produces only small aggregates (Figure SI-6d). At this stage, we demonstrated that the RT-FAST provides in less than 90 min a positive/negative response of the presence of seeds in a sample using only 100 nM of monomers. Note that, the attempt of seeding amplification in a 96-well plate by adding 4 μM of monomers does not provide any ThT signal after 144 h (Figure SI-2). Generally speaking, RT-FAST allows decreasing both the amount of added monomer and the reaction time by two orders of magnitude (35 μL, 0.1 μM, and 1 h) compared to the RT-QuIC (100 μL, 10 μM and >100 h). Interestingly, between 60 and 90 min, the capture rate of the sample containing A53T seeds is 10 times larger than the one containing WT seeds (Figure 3a,b). This demonstrates that the RT-FAST is sensitive enough to show the impact of A53T amino acid replacement in the cross-seeding aggregation of the WT form.
We then examined whether the RT-FAST assay could provide quantitative information on the initial seed concentration. To this aim, we performed additional experiments using two different concentrations of WT seeds (2 pM and 20 pM) (Figure 3d,e). For 20 pM of seeds, the events are detected after 30 min (Figure SI-7). For 2 pM, the events are observed only for one-third of the experiments after 30 min, and two-thirds after 60 min, and the ΔI/I0 values are smaller than the one seeded at 200 pM (Figure SI-8). In addition, the center of distribution decreases with the initial seed concentration. This shows that the aggregate size is seed concentration dependent. The plot of the capture rate as a function of the initial WT seed concentration is reported in Figure 3f. At 30 min, the capture rate averaged on several independent experiments cannot be used due to the variability of sample containing 2 pM of WT seeds. At 90 min, all samples can be distinguished from the control (f > 0.01), and the dependence of the capture rate with the initial seed concentration exists. This indicates that the RT-FAST assay is able to provide quantitative information on seed concentration based only on the capture rate.
Conclusion
In summary, we report an innovative, relevant, and alternative method to the RT-QuIC assay that reduces the time of seeding experiment and provides quantitative information about the initial seed concentration. Using a nanopipette as a device for both seeding and sensing steps, we demonstrate that RT-FAST is suitable to detect the presence αS seeds in a given sample in only 90 min and using a low quantity (35 μL at 100 nM) of recombinant αS for amplification that is interesting to limit the analysis cost. In addition, the assay is sensitive to the initial seed concentration (from 200 pM to 2 pM) that promises further development toward a quantitative analysis of the initial seed concentration. Regarding the variety of synucleinopathies, the RT-FAST is the first step in the development of ultrasensitive in vitro assays to indirectly detect amyloids in patients’ biological fluids as biomarkers based on nanopore technology. Besides that, the RT-FAST is also a promising tool for fundamental investigation of the protein aggregation process including the impact of cross-seeding,46,47 coassembly,48,49 or inhibitors,50,51 leading future advances in the understanding of the lag phase of amyloidogenesis.
Materials and Methods
Materials
l-DOPA (D9628), PBS (P4417), NaCl (71380), and α-synuclein A53T human recombinant (ref S1071) were all purchased from Sigma-Aldrich, and α-synuclein, recombinant human (AS-55555-1000) monomers were purchased from Anaspec.
Nanopipette Pulling and Functionalization
Quartz capillary purchased from Sutter instrument (OD: 1 mm and ID: 0.7 mm) were pulled using a pipet puller P-2000 (Sutter instrument). Pulling parameters used to obtain a tip diameter of 34 nm were HEAT = 700, FIL = 4, VEL = 60, DEL = 150, PUL = 175. We noticed that the pulling parameters were dependent on several factors such as the humidity and pressure of the room as well as the intrinsic features of the P-2000 instruments such as laser alignment.52 The pipettes were filled with pure degassed water following the filling principle described by Sun et al.53 After a complete filling, the nanopipette pipet was coated with the addition of l-DOPA solution (8.5 mg/mL) for 2 h. Then, the nanopipette was carefully washed several times with degassed water to remove the unreacted l-DOPA. The scanning electron microscope (SEM) images were performed on a set of nanopipettes using a Thermo Scientific Quattro ESEM, at high vacuum at 10 kV. The optical microscopy images were obtained using a Leica DM6000 instrument.
Seed Production
Wild type or A53T mutants of recombinant α-synuclein monomers (stock solution 140 μM in PBS 1×) were diluted to 75 μM in 80 μL of PBS in low binding Eppendorf tubes and left to aggregate at 37 °C without shaking. After 150 min of incubation, solutions were diluted in PBS to reach a concentration of 20 nM and stored at −32 °C until the seeding experiments. The seed α-synuclein WT and A53T were characterized by TEM as follows. First, the seed A53T and wild type used for seeding experiments (20 nM, 1 mL) were ultracentrifuged at 75 000 rpm for 30 min at 4 °C. Then, the supernatant was removed, and a 1 mL of seeds (20 nM) was added to pellet. This was repeated three times in order to concentrate the seed sample. Finally, the seed samples were deposited onto Formvar carbon-coated grids, negatively stained with freshly filtered 2% uranyl acetate, and dried. The TEM images were performed using a JEOL 1400 electron microscope at an accelerating voltage of 80 kV.
α-Synuclein Monomers Solubilization
α-Synuclein monomers were solubilized following the protocol described by Pujols et al.54 with slight modifications. Briefly, lyophilized monomers were solubilized as received in PBS 1× at 4 °C and filtered with 0.45 μm PVDF filter. The concentration was determined by measuring the absorbance with a spectrophotometer (JASCO) at 280 nm (extinction coefficient: 5960 M–1 cm–1). Then, the monomer solution was aliquoted to reach a final concentration of 140 μM and stored at −32 °C until their use.
Thioflavine-T (ThT) Fluorescence Assay
A total of 160 μL of α-synuclein monomers (8 μM) was diluted with 80 μL of NaCl 4 M (containing PBS, pH 7.4) and 80 μL of ThT at 26 μM in a low binding Eppendorf tube (final volume = 320 μL). The final concentration of the α-synuclein monomers, NaCl, and ThT are 4 μM, 1 M, and 6 μM, respectively. For the conditions containing seeds (WT and A53T mutant), 3.2 μL of buffer (NaCl 1 M, PBS) was removed and replaced by 3.2 μL of seeds concentrated at 20 nM to reach a final concentration of 0.2 nM. The different solutions were placed in 96-well plates treated with PEG (Corner Brand) in triplicate with a volume of 100 μL in each well. The plate was placed in a Fluoroskan Ascent microplate fluorometer (Thermo Fisher Scientific) to monitor the fibrils formation by following the fluorescence of ThT (λex = 445 nm and λem = 485 nm). The temperature was set to 25 °C without shaking, and one point was measured every 30 min.
Seeding and Resistive Pulse Measurement
Monomers diluted at 200 nM in NaCl 1 M (containing PBS 1×, pH = 7.4) were added directly in the pipet without or with seeds at various concentrations (200 pM, 20 pM, 2 pM equivalent monomer). The working electrode is located inside the pipet, and the ground electrode was placed inside an external reservoir containing only NaCl 1 M PBS 1× solution. A cycle of experiments was composed in two phases. First, a voltage of 500 mV for 10 min was used, and the current was recorded using an EPC10 amplifier (HEKA, Lambrecht, Germany) at 200 kHz and filtered with a Bessel filter at 10 kHz. Second, a break with no voltage applied for 20 min. This cycle was repeated until 2 h of incubation with αS inside the pipet. The current traces at different incubation times were analyzed using a custom-made Labview software (Peak Nano Tool). Briefly, the signal was filtered with a Butterworth filter of 2 kHz (order 1), and then a Savitzky–Golay (order 1) was used to correct the baseline fluctuations. The detection of events was performed using a threshold of 4σ (σ being the standard deviation to the baseline signal). The events were characterized by their relative maximum blockade amplitude (ΔI/I0) as well their dwell time (Δt). Then, statistical analysis was performed using Matlab custom scripts (matlab2021a).
Acknowledgments
This work was funded by the Agence Nationale de la Recherche (ANR-19-CE42-0006, NanoOligo). The authors thank B. Tinland (CINAM, Marseille) and M. Lepoitevin (ENS, Paris) for the discussion about nanopipette fabrication and filling.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c01404.
The authors declare no competing financial interest.
Supplementary Material
References
- Baba M.; Nakajo S.; Tu P. H.; Tomita T.; Nakaya K.; Lee V. M.; Trojanowski J. Q.; Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 1998, 152 (4), 879–884. [PMC free article] [PubMed] [Google Scholar]
- Mehra S.; Sahay S.; Maji S. K. α-Synuclein misfolding and aggregation: Implications in Parkinson’s disease pathogenesis. Biochimica et biophysica acta. Proteins and proteomics 2019, 1867 (10), 890–908. 10.1016/j.bbapap.2019.03.001. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Fernandez R. Role of DaTSCAN and clinical diagnosis in Parkinson disease. Neurology 2012, 78 (10), 696–701. 10.1212/WNL.0b013e318248e520. [DOI] [PubMed] [Google Scholar]
- Russo M. J.; Orru C. D.; Concha-Marambio L.; Giaisi S.; Groveman B. R.; Farris C. M.; Holguin B.; Hughson A. G.; LaFontant D.-E.; Caspell-Garcia C.; et al. High diagnostic performance of independent alpha-synuclein seed amplification assays for detection of early Parkinson’s disease. Acta neuropathologica communications 2021, 9 (1), 179. 10.1186/s40478-021-01282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.-Y.; Jang H.-S.; Green A. J. E.; Choi Y. P. RT-QuIC-based detection of alpha-synuclein seeding activity in brains of dementia with Lewy Body patients and of a transgenic mouse model of synucleinopathy. Prion 2020, 14 (1), 88–94. 10.1080/19336896.2020.1724608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborio G. P.; Permanne B.; Soto C. Sensitive detection of pathological prion protein by cyclic amplication of protein misfolding. Nature 2001, 411, 810. 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- Fairfoul G.; McGuire L. I.; Pal S.; Ironside J. W.; Neumann J.; Christie S.; Joachim C.; Esiri M.; Evetts S. G.; Rolinski M.; et al. Alpha-synuclein RT-QuIC in the CSF of patients with alpha-synucleinopathies. Ann. Clin Transl Neurol 2016, 3 (10), 812–818. 10.1002/acn3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelise N.; Schmitz M.; Llorens F.; Villar-Pique A.; Cramm M.; Thom T.; Silva Correia S. M.; Cunha J. E. G.; Mobius W.; Outeiro T. F.; Alvarez V. G.; Banchelli M.; D'Andrea C.; Angelis M.; Zafar S.; Rabano A.; Matteini P.; Zerr I. Seeding variability of different alpha synuclein strains in synucleinopathies. Ann. Neurol. 2019, 85 (5), 691–703. 10.1002/ana.25446. [DOI] [PubMed] [Google Scholar]
- Saborio G. P.; Permanne B.; Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. nature 2001, 411 (6839), 810–813. 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- Xue L.; Yamazaki H.; Ren R.; Wanunu M.; Ivanov A. P.; Edel J. B. Solid-state nanopore sensors. Nat. Rev. Mater. 2020, 5 (12), 931–951. 10.1038/s41578-020-0229-6. [DOI] [Google Scholar]
- Houghtaling J.; List J.; Mayer M. Nanopore-Based, Rapid Characterization of Individual Amyloid Particles in Solution: Concepts, Challenges, and Prospects. Small 2018, 14, 1802412. 10.1002/smll.201802412. [DOI] [PubMed] [Google Scholar]
- Assarsson A.; Linse S.; Cabaleiro-Lago C. Effects of polyamino acids and polyelectrolytes on amyloid β fibril formation. Langmuir: the ACS journal of surfaces and colloids 2014, 30 (29), 8812–8818. 10.1021/la501414j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigolato F.; Arosio P. The role of surfaces on amyloid formation. Biophys. Chem. 2021, 270, 106533. 10.1016/j.bpc.2020.106533. [DOI] [PubMed] [Google Scholar]
- Mohammad-Beigi H.; Shojaosadati S. A.; Marvian A. T.; Pedersen J. N.; Klausen L. H.; Christiansen G.; Pedersen J. S.; Dong M.; Morshedi D.; Otzen D. E. Strong interactions with polyethylenimine-coated human serum albumin nanoparticles (PEI-HSA NPs) alter α-synuclein conformation and aggregation kinetics. NANOSCALE 2015, 7 (46), 19627–19640. 10.1039/C5NR05663B. [DOI] [PubMed] [Google Scholar]
- Houghtaling J.; List J.; Mayer M. Nanopore-Based, Rapid Characterization of Individual Amyloid Particles in Solution: Concepts, Challenges, and Prospects. Small 2018, 14 (46), e1802412. 10.1002/smll.201802412. [DOI] [PubMed] [Google Scholar]
- Wen C.; Dematties D.; Zhang S.-L. A Guide to Signal Processing Algorithms for Nanopore Sensors. ACS sensors 2021, 6 (10), 3536–3555. 10.1021/acssensors.1c01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makra I.; Gyurcsányi R. E. Electrochemical sensing with nanopores: A mini review. Electrochem. Commun. 2014, 43, 55–59. 10.1016/j.elecom.2014.03.007. [DOI] [Google Scholar]
- Meyer N.; Abrao-Nemeir I.; Janot J.-M.; Torrent J.; Lepoitevin M.; Balme S. Solid-state and polymer nanopores for protein sensing: A review. Advances in colloid and interface science 2021, 298, 102561. 10.1016/j.cis.2021.102561. [DOI] [PubMed] [Google Scholar]
- Martyushenko N.; Bell N. A. W.; Lamboll R. D.; Keyser U. F. Nanopore analysis of amyloid fibrils formed by lysozyme aggregation. Analyst 2015, 140 (14), 4882–4886. 10.1039/C5AN00530B. [DOI] [PubMed] [Google Scholar]
- Li W.; Bell N. A. W.; Hernández-Ainsa S.; Thacker V. V.; Thackray A. M.; Bujdoso R.; Keyser U. F. Single protein molecule detection by glass nanopores. ACS Nano 2013, 7 (5), 4129–4134. 10.1021/nn4004567. [DOI] [PubMed] [Google Scholar]
- Chau C. C.; Radford S. E.; Hewitt E. W.; Actis P. Macromolecular Crowding Enhances the Detection of DNA and Proteins by a Solid-State Nanopore. Nano Lett. 2020, 20 (7), 5553–5561. 10.1021/acs.nanolett.0c02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Tong X.; Lu W.; Yu D.; Diao J.; Zhao Q. Label-free detection of early oligomerization of -synuclein and its mutants A30P/E46K through solid-state nanopores. NANOSCALE 2019, 11 (13), 6480–6488. 10.1039/C9NR00023B. [DOI] [PubMed] [Google Scholar]
- Yusko E. C.; Prangkio P.; Sept D.; Rollings R. C.; Li J.; Mayer M. Single-Particle Characterization of A beta Oligomers in Solution. ACS Nano 2012, 6 (7), 5909–5919. 10.1021/nn300542q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamblanco N.; Coglitore D.; Janot J.-M.; Coulon P. E.; Charlot B.; Balme S. Detection of protein aggregate morphology through single antifouling nanopore. Sens. Actuators, B 2018, 260, 736–745. 10.1016/j.snb.2018.01.094. [DOI] [Google Scholar]
- Balme S.; Coulon P. E.; Lepoitevin M.; Charlot B.; Yandrapalli N.; Favard C.; Muriaux D.; Bechelany M.; Janot J.-M. Influence of Adsorption on Proteins and Amyloid Detection by Silicon Nitride Nanopore. Langmuir 2016, 32 (35), 8916–8925. 10.1021/acs.langmuir.6b02048. [DOI] [PubMed] [Google Scholar]
- Giamblanco N.; Coglitore D.; Gubbiotti A.; Ma T.; Balanzat E.; Janot J.-M.; Chinappi M.; Balme S. Amyloid Growth, Inhibition, and Real-Time Enzymatic Degradation Revealed with Single Conical Nanopore. Analytical Chemistry 2018, 90 (21), 12900–12908. 10.1021/acs.analchem.8b03523. [DOI] [PubMed] [Google Scholar]
- Meyer N.; Arroyo N.; Janot J.-M.; Lepoitevin M.; Stevenson A.; Nemeir I. A.; Perrier V.; Bougard D.; Belondrade M.; Cot D.; et al. Detection of Amyloid-β Fibrils Using Track-Etched Nanopores: Effect of Geometry and Crowding. ACS Sensors 2021, 6 (10), 3733–3743. 10.1021/acssensors.1c01523. [DOI] [PubMed] [Google Scholar]
- Meyer N.; Arroyo N.; Baldelli M.; Coquart N.; Janot J. M.; Perrier V.; Chinappi M.; Picaud F.; Torrent J.; Balme S. Conical nanopores highlight the pro-aggregating effects of Pyrimethanil fungicide on Aβ(1–42) peptides and dimeric splitting phenomena. Chemosphere 2022, 291, 132733. 10.1016/j.chemosphere.2021.132733. [DOI] [PubMed] [Google Scholar]
- Giamblanco N.; Fichou Y.; Janot J.-M.; Balanzat E.; Han S.; Balme S. Mechanisms of Heparin-Induced Tau Aggregation Revealed by a Single Nanopore. ACS Sensors 2020, 5, 1158. 10.1021/acssensors.0c00193. [DOI] [PubMed] [Google Scholar]
- Giamblanco N.; Janot J.-M.; Gubbiotti A.; Chinappi M.; Balme S. Characterization of Food Amyloid Protein Digestion by Conical Nanopore. Small Methods 2020, 4, 1900703. 10.1002/smtd.201900703. [DOI] [Google Scholar]
- Karmi A.; Sakala G. P.; Rotem D.; Reches M.; Porath D. Durable, Stable, and Functional Nanopores Decorated by Self-Assembled Dipeptides. ACS Appl. Mater. Interfaces 2020, 12 (12), 14563–14568. 10.1021/acsami.0c00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesa C.; Kowalczyk S. W.; Zinsmeester R.; Grosberg A. Y.; Rabin Y.; Dekker C. Fast translocation of proteins through solid state nanopores. Nano Lett. 2013, 13 (2), 658–663. 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R.-J.; Lu S.-M.; Xu S.-W.; Li Y.-J.; Xu Q.; Ying Y.-L.; Long Y.-T. Single molecule sensing of amyloid-beta aggregation by confined glass nanopores. Chemical Science 2019, 10 (46), 10728–10732. 10.1039/C9SC03260F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubánková M.; Lin X.; Albrecht T.; Edel J. B.; Kuimova M. K. Rapid Fragmentation during Seeded Lysozyme Aggregation Revealed at the Single Molecule Level. Analytical Chemistry 2019, 91 (10), 6880–6886. 10.1021/acs.analchem.9b01221. [DOI] [PubMed] [Google Scholar]
- Dear A. J.; Michaels T. C. T.; Meisl G.; Klenerman D.; Wu S.; Perrett S.; Linse S.; Dobson C. M.; Knowles T. P. J. Kinetic diversity of amyloid oligomers. Proc. Natl. Acad. Sci. U.S.A. 2020, 117 (22), 12087–12094. 10.1073/pnas.1922267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear A. J.; Meisl G.; Šarić A.; Michaels T. C. T.; Kjaergaard M.; Linse S.; Knowles T. P. J. Identification of on- and off-pathway oligomers in amyloid fibril formation. Chemical Science 2020, 11 (24), 6236–6247. 10.1039/C9SC06501F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels T. C. T.; Šarić A.; Curk S.; Bernfur K.; Arosio P.; Meisl G.; Dear A. J.; Cohen S. I. A.; Dobson C. M.; Vendruscolo M.; et al. Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide. Nature Chem. 2020, 12 (5), 445–451. 10.1038/s41557-020-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira G. A. P.; Silva J. L. Alpha-synuclein stepwise aggregation reveals features of an early onset mutation in Parkinson’s disease. Commun. Biol. 2019, 2 (1), 374. 10.1038/s42003-019-0598-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagmeier P.; Meisl G.; Vendruscolo M.; Knowles T. P. J.; Dobson C. M.; Buell A. K.; Galvagnion C. Mutations associated with familial Parkinson’s disease alter the initiation and amplification steps of α-synuclein aggregation. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (37), 10328–10333. 10.1073/pnas.1604645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos M. H.; Lavedan C.; Leroy E.; Ide S. E.; Dehejia A.; Dutra A.; Pike B.; Root H.; Rubenstein J.; Boyer R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science (New York, N.Y.) 1997, 276 (5321), 2045–2047. 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Watanabe-Nakayama T.; Nawa M.; Konno H.; Kodera N.; Ando T.; Teplow D. B.; Ono K. Self- and Cross-Seeding on α-Synuclein Fibril Growth Kinetics and Structure Observed by High-Speed Atomic Force Microscopy. ACS Nano 2020, 14 (8), 9979–9989. 10.1021/acsnano.0c03074. [DOI] [PubMed] [Google Scholar]
- Long H.; Zheng W.; Liu Y.; Sun Y.; Zhao K.; Liu Z.; Xia W.; Lv S.; Liu Z.; Li D.; et al. Wild-type α-synuclein inherits the structure and exacerbated neuropathology of E46K mutant fibril strain by cross-seeding. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (20), e2012435118. 10.1073/pnas.2012435118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway K. A.; Lee S. J.; Rochet J. C.; Ding T. T.; Williamson R. E.; Lansbury P. T. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (2), 571–576. 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo J.; Escalona-Noguero C.; Sot B. Role of α-Synuclein Regions in Nucleation and Elongation of Amyloid Fiber Assembly. ACS Chemical Neuroscience 2020, 11 (6), 872–879. 10.1021/acschemneuro.9b00527. [DOI] [PubMed] [Google Scholar]
- Wood S. J.; Wypych J.; Steavenson S.; Louis J. C.; Citron M.; Biere A. L. alpha-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson’s disease. J. Biol. Chem. 1999, 274 (28), 19509–19512. 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- Vaneyck J.; Segers-Nolten I.; Broersen K.; Claessens M. M. A. E. Cross-seeding of alpha-synuclein aggregation by amyloid fibrils of food proteins. J. Biol. Chem. 2021, 296, 100358. 10.1016/j.jbc.2021.100358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B.; Zhang Y.; Zhang M.; Liu Y.; Zhang D.; Gong X.; Feng Z.; Tang J.; Chang Y.; Zheng J. Fundamentals of cross-seeding of amyloid proteins: an introduction. Journal of Materials Chemistry B 2019, 7 (46), 7267–7282. 10.1039/C9TB01871A. [DOI] [PubMed] [Google Scholar]
- Liu P.; Zhang S.; Chen M.-s.; Liu Q.; Wang C.; Wang C.; Li Y.-M.; Besenbacher F.; Dong M. Co-assembly of human islet amyloid polypeptide (hIAPP)/insulin. Chemical Communications (Cambridge, England) 2012, 48 (2), 191–193. 10.1039/C1CC14285B. [DOI] [PubMed] [Google Scholar]
- Chau E.; Kim J. α-synuclein-assisted oligomerization of β-amyloid (1–42). Archives of Biochemistry and Biophysics 2022, 717, 109120. 10.1016/j.abb.2022.109120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L.; Liu L.; Xi W.; Han Q.; Li Q.; Yu Y.; Huang Q.; Qu F.; Xu M.; Li Y.; et al. Synergistic Inhibitory Effect of Peptide-Organic Coassemblies on Amyloid Aggregation. ACS Nano 2016, 10 (4), 4143–4153. 10.1021/acsnano.5b07396. [DOI] [PubMed] [Google Scholar]
- Torpey J. H.; Meade R. M.; Mistry R.; Mason J. M.; Madine J. Insights Into Peptide Inhibition of Alpha-Synuclein Aggregation. Frontiers in Neuroscience 2020, 14, 561462. 10.3389/fnins.2020.561462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.; Pataillot-Meakin T.; Shibakawa A.; Ren R.; Bevan C. L.; Ladame S.; Ivanov A. P.; Edel J. B. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 2021, 12 (1), 3515. 10.1038/s41467-021-23497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Shigyou K.; Ando T.; Watanabe S. Thermally Driven Approach To Fill Sub-10-nm Pipettes with Batch Production. Analytical Chemistry 2019, 91 (21), 14080–14084. 10.1021/acs.analchem.9b03848. [DOI] [PubMed] [Google Scholar]
- Pujols J.; Peña-Díaz S.; Conde-Giménez M.; Pinheiro F.; Navarro S.; Sancho J.; Ventura S. High-Throughput Screening Methodology to Identify Alpha-Synuclein Aggregation Inhibitors. International Journal of Molecular Sciences 2017, 18, 478. 10.3390/ijms18030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.