Abstract
The application of genome-wide expression profiling to determine how drugs achieve their therapeutic effect has provided the pharmaceutical industry with an exciting new tool for drug mode-of-action studies. We used DNA chip technology to study cellular responses to perturbations of ergosterol biosynthesis caused by the broad-spectrum antifungal agent itraconazole. Simultaneous examination of over 6,600 Candida albicans gene transcript levels, representing the entire genome, upon treatment of cells with 10 μM itraconazole revealed that 296 genes were responsive. For 116 genes transcript levels were decreased at least 2.5-fold, while for 180 transcript levels were similarly increased. A global upregulation of ERG genes in response to azole treatment was observed. ERG11 and ERG5 were found to be upregulated approximately 12-fold. In addition, a significant upregulation was observed for ERG6, ERG1, ERG3, ERG4, ERG10, ERG9, ERG26, ERG25, ERG2, IDII, HMGS, NCP1, and FEN2, all of which are genes known to be involved in ergosterol biosynthesis. The effects of itraconazole on a wide variety of known metabolic processes are discussed. As over 140 proteins with unknown function were responsive to itraconazole, our analysis might provide—in combination with phenotypic data—first hints of their potential function. The present report is the first to describe the application of DNA chip technology to study the response of a major human fungal pathogen to drug treatment.
The N-substituted triazole itraconazole is a broad-spectrum antifungal agent currently available in the form of oral capsules and an oral solution for the treatment and/or prophylaxis of aspergillosis, blastomycosis, disseminated and superficial Candida infections, cryptococcosis, dermatophytosis, histoplasmosis, paracoccidioidomycosis, sporotrichosis, and some forms of coccidioidomycosis (8, 20).
The antifungal activities of itraconazole and related azole derivatives arise from a complex multimechanistic process initiated by the inhibition of two cytochromes P450 involved in the biosynthesis of ergosterol, namely, the P450 that catalyzes the 14α-demethylation of lanosterol or eburicol, encoded by ERG11 (CYP51), and Δ22-desaturase, encoded by ERG5 (CYP61) (12, 31, 32, 33). The sterol-14α-demethylase is the major fungal target for all azole derivatives studied so far. Interaction with CYP51 results in a decreased availability of ergosterol and accumulation of 14-methylsterols and 3-ketosteroids (17, 27, 28, 29, 31, 34, 36).
Ergosterol is an essential component of fungal plasma membranes; it affects membrane permeability and the activities of membrane-bound enzymes. This sterol is a major component of secretory vesicles and has an important role in mitochondrial respiration and oxidative phosphorylation (for reviews, see references 3, 27, and 28). It can thus be expected that changes in ergosterol levels and sterol structure influence the activities of several metabolic pathways.
To examine this further, the effect of itraconazole on the pathogen was studied by analyzing the level of gene expression from essentially every gene in the Candida albicans genome prior to and after treatment of C. albicans with this compound. Transcript profiling with DNA microarrays provides a rapid and systematic method for the high-throughput analysis of gene expression at the level of the whole genome (2). The experiments reported here use the C. albicans Gene Expression Microarray (GEM; Incyte Genomics Inc., Palo Alto, Calif.), which contains over 6,600 DNA fragments. From a drug discovery point of view, the expression patterns generated from the in-parallel analysis of all genes in a microorganism can give clues to the functions of previously uncharacterized genes (target identification), as well as provide information about how drugs achieve their therapeutic effect (mechanism-of-action studies). In addition to providing a much more complete picture of which genes and pathways are involved, gene functions other than those currently known might be discovered (22).
MATERIALS AND METHODS
Strains and media.
C. albicans CAI-4 (ura3::imm434/ura3::imm434) was kindly provided by William Fonzi, Georgetown University (7). SGM medium contains 0.67% (wt/vol) yeast nitrogen base without amino acids (Difco), 0.2% (wt/vol) Ura dropout powder (Bufferad, Inc.), 2% (wt/vol) galactose, and 2% (wt/vol) maltose. Uridine was added at a final concentration of 20 μg/ml to ensure growth of CAI-4. Itraconazole (Janssen Pharmaceutica, Beerse, Belgium) dissolved in dimethyl sulfoxide was added to a final concentration of 10 μM.
Total RNA extraction.
CAI-4 cells were grown overnight at 30°C and 250 rpm to an optical density at 600 nm (OD600) of ∼0.2, the culture was divided in two, and itraconazole was added to one of the cultures. Cells were grown for an additional 24 h and harvested (at an OD600 of ∼1.25 for the nontreated cells and an OD600 of ∼0.8 for the treated cells). Total RNA was prepared with the RNeasy midi kit (enzymatic lysis protocol; Qiagen) according to the manufacturer's instructions. All RNA samples were treated with DNase I (RNase-free; Boehringer) on RNeasy midi columns for 15 min according to the manufacturer's instructions. RNA was precipitated (sodium acetate-ethanol) and the pellet was dissolved in 20 μl of diethyl pyrocarbonate-treated MilliQ water (Millipore). RNA concentrations were determined spectrophotometrically by measuring the absorption at 260 nm in a UV-1601 UV-visible spectrophotometer (Shimadzu).
Microarray preparation.
DNA fragments for microarray fabrication were generated by PCR (http://www.incytegenomics.com/gem/technology/index.shtml). PCR products were purified by gel filtration with Sephacryl-400 (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) equilibrated in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The filtrate was dried and rehydrated in 1/10 volume distilled H2O (dH2O) for arraying of the DNA solutions. The DNA solutions were arrayed by robotics on modified glass slides. After arraying of the DNA solutions, the slides were processed to fix the DNA to the prepared glass surface and washed three times in dH2O at ambient temperature. The slides were then treated with 0.2% I-Block (Tropix, Bedford, Mass.) dissolved in 1× Dulbecco's phosphate-buffered saline (Life Technologies, Gaithersburg, Md.) at 60°C for 30 min. GEMs were rinsed in 0.2% sodium dodecyl sulfate for 2 min, followed by three 1-min washes in dH2O.
Fluorescent labeling of probe for Microarray hybridization.
Only one set of RNAs (treated and non-treated) was used for microarray hybridization experiments. Isolated total RNAs were reverse transcribed with 5′ Cy3- or Cy5-labeled random 9-mers (Operon Technologies, Inc., Alameda, Calif.). The reaction mixtures were incubated for 2 h at 37°C with 1 μg of total RNA, 200 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies, Gaithersburg, Md.), 4 mM dithiothreitol, 1 U RNase inhibitor (Ambion, Austin, Tex.), 0.5 mM deoxynucleoside triphosphates, and 2 μg of labeled 9-mers in a 25-μl volume with enzyme buffer supplied by the manufacturer. The reaction was terminated by incubation at 85°C for 5 min. The paired reactions were combined, and the labeled nucleic acid was purified with a TE-30 column. (Clontech, Palo Alto, Calif.). The eluate was brought to 90 μl with dH2O and precipitated with 2 μl of 1 mg of glycogen per ml–60 μl of 5 M ammonium acetate–300 μl of ethyl alcohol. After centrifugation the supernatant was decanted and the pellet was dissolved in 24 μl of hybridization buffer: 5× SSC, 0.2% sodium dodecyl sulfate, and 1 mM dithiothreitol.
Microarray hybridization and scanning.
Before use, probe solutions were incubated at 65°C for 5 min with mixing. The probe solution was applied to the array, which was covered with a 22-mm2 glass coverslip and placed in a sealed humidified chamber to prevent evaporation. After hybridization at 60°C for 6.5 h the slides were washed in three consecutive washes of decreasing ionic strength.
The microarrays were scanned in both Cy3 and Cy5 channels with GenePix scanners (Axon, Foster City, Calif.) with a 10-μm resolution. The signal was converted into a resolution of 16 bits per pixel, yielding a 65,536-count dynamic range.
Microarray data processing and analysis.
GEMtools software (Incyte Genomics Inc.) was used for image analysis and data visualization. Each element area was determined with a gridding and detection algorithm. The area surrounding each element was used to calculate the local background value, which was subtracted from the total element signal value. Signals from which the background value was subtracted were used to determine differential expression (Cy3/Cy5 ratio) for each element. For these calculations GEM elements were selected according to the following acceptance criteria: (i) each element must have a minimum area of 40% of the average area calculated for all elements on the GEM, and (ii) the element's signal-to-background ratio must be at least 2.5. To be accepted an element must pass both criteria in at least one channel.
A correction factor was applied to account for systematic differences in the probe labels. The ratio of the average Cy3 and Cy5 signals from which background values were subtracted was used to balance the Cy5 signals. In the present study only elements with balanced differential expression (Cy3/balanced Cy5 ratio) higher than 2 were considered in further analyses. Elements for which the homogeneity of the spotted DNA was not confirmed by PCR analysis were excluded. DNA sequences were annotated on the basis of the results of BLAST (n and x) searches against PathoSeq (release, March 2000; Incyte Genomics Inc.) EMBL, SwissProt, and C. albicans genome (release, July 1999; Stanford University) databases. Information on gene functions was also retrieved from the Bioknowledge library (Proteome Inc., Beverly, Mass.).
The correlation coefficient between two different hybridization experiments was determined essentially as described by Eisen et al. (6).
RESULTS AND DISCUSSION
Global gene expression monitoring in C. albicans upon itraconazole treatment: experimental design.
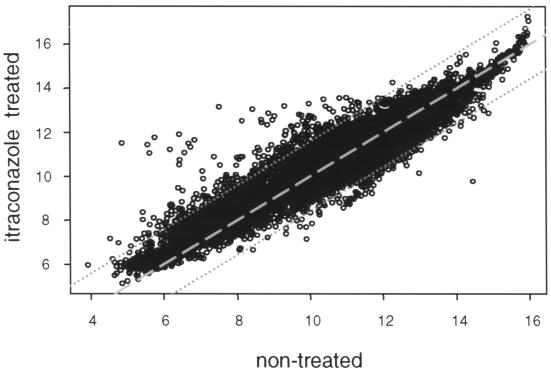
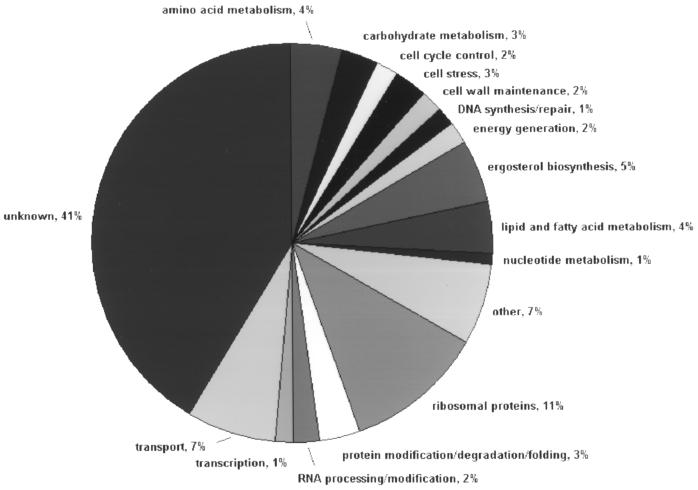
The use of microarray techniques in combination with the C. albicans genome sequence now permits investigation of global changes in gene expression associated with drug treatment. We examined the level of gene expression from essentially every gene in the C. albicans genome simultaneously using the GEM technology offered by Incyte Genomics Inc. Over 6,600 unique open reading frames (ORFs) are represented on a single glass slide. Total RNA was isolated from both nontreated C. albicans CAI-4 cells and cells treated with 10 μM itraconazole as described above. DNA probes for hybridization were prepared by reverse transcription with either Cy3- or Cy5-labeled random nonamers. Experiments were performed in duplicate, and fluorescent signals were detected by scanning. Normalization and ratio determination were performed with GEMTools software. The correlation coefficient between (normalized) fluorescent signals of untreated and treated CAI-4 cells was found to be 0.93 (6) (Fig. 1). A total of 296 genes were identified as responsive, including the majority of genes from the ergosterol biosynthetic pathway: 116 genes showed a more than 2.5-fold decrease in expression upon itraconazole treatment, while 180 showed a more than 2.5-fold increase (Table 1). In-house comparison with other microarray experimental data sets (obtained upon treatment of C. albicans with other compounds or upon comparison of the non-ergosterol-related genes of parental strain CAI-4 and genetic knockouts strains) revealed that the majority of these responses were specific (data not shown). However, the moderate upregulation of many genes involved in protein biosynthesis was found to be a rather unspecific response. To facilitate downstream analysis, differentially expressed genes were divided over known functional classes, as shown in Fig. 2.
FIG. 1.
Correlation graph. Normalized hybridization signals from one representative experiment from Cy5 (untreated wild-type C. albicans cells) and Cy3 (itraconazole-treated C. albicans cells) channels were plotted against each other on a logarithmic scale. The data series have a correlation coefficient of 0.93. Datum points with differential expression less than 2.5-fold fall between the two dotted lines. The dashed line is the perfect diagonal.
TABLE 1.
Specific changes in C. albicans gene expression in response to a 24-h treatment with 10 μM itraconazolea
| Gene | Product or function | Responseb |
|---|---|---|
| Ergosterol biosynthesis-linked genes | ||
| Homolog of S.c. ERG2 | C-8 sterol isomerase | +3.9 |
| C.a. ERG9 | Farnesyl-diphosphate famesyltransferase (squalene synthetase) (EC 2.5 1.21) | +4.9 |
| C.a. ERG25 | C-4 methyl sterol oxidase | +4.2 |
| Homolog of S.c. ERG26 | C-3 sterol dehydrogenase (C-4 decarboxylase) | +4.8 |
| C.a. ERG11 | Lanosterol 14 α-demethylase (EC 1.14.14.1) | +12.5 |
| Homolog of S.c. ERG5 | C-22 sterol desaturase (EC 1.14.14.-) | +12 |
| C.a. ERG3 | C5,6-desaturase | +12.4 |
| Homolog of S.c. ERG4 | C-24(28) sterol reductase | +11.6 |
| C.a. ERG6 | Delta-24-sterol C-methyltransferase (EC 2.1.1.41) | +50.3 |
| Homolog of S.c. ERG10 | Acetyl-CoA acetyltransferase (EC 2.3.1.9) | +10.9 |
| Homolog of S.c. IDI1 | Isopentenyl-diphosphate delta-isomerase | +9.2 |
| Homolog of S.c. HMGS | 3-Hydroxy-3-methylglutaryl CoA Synthase (EC 4.1.3.5) | +13.7 |
| C.a. ERG1 | Squalene epoxidase (EC 1.14.99.7) | +13.2 |
| Homolog of C. maltosa NCP1 | NADPH-cytochrome P450 reductase (EC 1.6.2.4) | +6.7 |
| Homolog of S.c. FEN2 | Pantothenate symporter | +5.1 |
| Homolog of S.c. CYB5 | Cytochrome b5 | +4.5 |
| Homolog of S.c. CBR1 | NADH-cytochrome b5 reductase (EC 1.6.2.2) | +2.7 |
| Homolog of S.c. SAM2 | S-Adenosylmethionine synthetase 2 (EC 2.5.1.6) | +5.8 |
| Homolog of S.c. MET6 | Homocysteine methyltransferase (EC 2.1.1.14) | +2.6 |
| Homolog of S.c. COQ7 | Required for derepression of gluconeogenic enzymes and ubiquinone biosynthesis | +3.1 |
| Other genes | ||
| Cell wall maintenance | ||
| C.a. CHT3 | Chitinase | +3.3 |
| Homolog of S.c. MNN4 | Involved in mannosylphosphate transfer from GDP-mannose to N-linked oligosaccharides | +5.1 |
| Homolog of S.c. CWH8 | Involved in generation of the cell wall mannoprotein layer | +3.2 |
| Protein synthesis | ||
| RPS7A, RPL13, RPL30, RPL8B, RPL34B, RPS16, RPL18A, RPL7A, RPL15A, RPS5, RPS10B, RPS19A, SSM1B, RPL26B, RPL14B, RPS29A, RPS11A, ASC1, RPS17B, RPS20, RPL32, RPL9B, RPL27, RPS9B, RPS2, RPLA0, RPL5, RPS27B, RPL11A, RPL35A, RPA4, RPL16A, RPL8A, RPL17B, RPS26A, RPS21, RPS8A, RPS15 | +2.5–+5.2 | |
| Lipid, fatty acid metabolism | ||
| C.a. AUR1 | Phosphatidylinositol ceramide phosphoinositol transferase | +4.6 |
| Homolog of S.c. FAA2 | Long-chain-fatty-acid CoA ligase 2 (EC 6.2.1.3) | +2.5 |
| Homolog of Borago officinalis | Delta-6-desaturase | +7.3 |
| Homolog of S.c. CHO1 | Phosphatidylserine synthase (EC 2.78.8) | +5.8 |
| C.a. PLD1 | Phospholipase D | +2.9 |
| Homolog of S.c. DAK2 | Dihydroxyacetone kinase (EC 2.7.1.29) | −2.8 |
| Homolog of S.c. TES1 | Peroxisoml acyl-CoA thioesterase, | −3.3 |
| Homolog of C. tropicalis FAOT | Long-chain fatty alcohol oxidase | −3.4 |
| Homolog of S.c. PLB1 | Phospholipase B (EC 3.1.1.5) | −3.5 |
| Homolog of S.c. YAT1 | Inner mitochondrial membrane camitine acetylatransferase (EC 2.3.1.7) | −3.6 |
| Homolog of S.c. CAT2 | Peroxisomal and mitochondrial camitine acetyltransferase (EC 2.3.1.7) | −3.8 |
| Homolog of S.c. PDR16 | Involved in lipid biosynthesis and multidrug resistance, belongs to the SEC14 cytosolic factor family | −4.6 |
| Cell stress | ||
| Homolog of S.c. YNL234W | Hemoprotein with similarity to mammalian globins | +3.4 |
| Homolog of S.c. DDR48 | DNA damage-responsive protein, induced by heat shock, DNA damage, or osmotic stress | +2.5 |
| Homolog of S.c. TPS2 | Trehalose 6-phosphate phosphatase (EC 3.1.3.12) | −2.5 |
| Homolog of S.c. HSP104 | Heat shock protein 104 | −3 |
| C.a. TPS1 | Trehalose-6-phosphate synthase (EC 2.4.1.15) | −3.3 |
| Homolog of S.c. TPS3 | Component of trehalose-6-phosphate synthase–phosphatase complex, alternative third subunit | −3.7 |
| Homolog of S.c. RTS1 | Serine-threonine phosphatase 2A (PP2A), B′ regulatory subunit, involved in regulation of stress-related responses and the cell cycle | −4.3 |
| Homolog of S.c. OSM1 | Mitochondrial soluble fumarate reductase involved in osmotic regulation | −4.5 |
| Transport | ||
| Vesicular transport | ||
| C.a. SEC14 | Phosphatidylinositol-hosphatidylcholine transfer protein required for transport of secretory proteins from Golgi complex | +3.3 |
| Homolog of S.c. GYP1 | GTPase-activating protein | +2.7 |
| Homolog of S.c. SEC7 | Component of nonclathrin vesicle coat required for protein trafficking within Golgi complex | −3.3 |
| Small molecule transport | ||
| C.a. CDR4 | Similar to multidrug resistance proteins, member of the ABC superfamily | −2.6 |
| Homolog of S.c. PHO84 | Inorganic phosphate transporter Pho84 | +5.7 |
| Homolog of S.c. YMR162C | Member of the P-type ATPase superfamily of cation-translocating, primary active membrane transporters | +4.1 |
| Homolog of S.c. YOR378W | Belongs to family of the multidrug resistance permeases of MFS | +2.9 |
| C.a. CTR1 | High-affinity copper transporter | −2.5 |
| Homolog of S.c. YHR032W | Member of the multiple antimicrobiotic resistance family of secondary active membrane transporters | −2.6 |
| Homolog of S.c. YPR128C | Peroxisomal protein, member of the mitochondrial carrier family of secondary active membrane transporters | −2.6 |
| Homolog of S.c. SSY1 | Amino acid permease | −2.7 |
| Homolog of S.c. PXA2 | Peroxisomal long-chain fatty acid import protein 1 (peroxisomal ABC transporter 2) | −3.2 |
| Homolog of S.c. PUT4 | Proline permease | −3.7 |
| Homolog of S.c. YOL075C | Member of the ABC superfamily of ATP-driven active membrane transporters | −4.8 |
| Nuclear-cytoplasmic transport | ||
| Homolog of S.c. NUP2 | Nucleoporin | +2.9 |
| Homolog of human CNT2 | Na-dependent and purine selective transporter | +2.7 |
| Amino acid metabolism | ||
| Homolog of S.c. UGA1 | 4-Aminobutyrate aminotransferase (EC 2.6.1.19) | −25.6 |
| Homolog of S.c. SAH1 | S-Adenosyl-l-homocysteine hydrolase (EC 3.3.1.1) | +3.9 |
| C.a. GLY1 | Low-specificity threonine aldolase (EC 4.1.2.5) | +2.9 |
| C.a. THR1 | Homoserine kinase (EC 2.7.1.39) | +2.7 |
| Homolog of S.c. PDX3 | Pyridoxine (pyridoxamine) phosphate oxidase (EC 1.4.3.5) | −2.5 |
| Homolog of S.c. HIS2 | Histidinol-phosphatase (EC 3.1.3.15) | −2.6 |
| Homolog of S.c. GCV2 | Subunit of glycine decarboxylase complex (EC 1.4.4.2) functions in the glycine degradation pathway | −3.4 |
| Carbohydrate metabolism | ||
| C.a. AGM1 | N-Acetylglucosamine-phosphate mutase | +2.9 |
| C.a. TKL1 | Transketolase (EC 2.2.1.1) | −2.7 |
| Homolog of S.c. ARA1 | Subunit of NADP+-dependent d-arabinose dehydrogenase | −4 |
| Energy generation | ||
| Homolog of S.c. CBP4 | Ubiquinol-cytochrome c reductase assembly factor | +2.6 |
| C.a. AOX2 | Alternative oxidase (salicythydroxamic acid-sensitive terminal oxidase) | −2.5 |
| Homolog of C. pelliculosa CYB2 | l-Lactate + cytochrome c oxidoreductase (EC 1.1.2.3) | −3.6 |
| Cell polarity and cell structure | ||
| C.a. CDC42 | Cell division control protein 42 homolog | +2.7 |
| Homolog of S.c. LAS1 | Involved in bud formation and cell morphogenesis | +2.6 |
| Homolog of S.c. RAX1 | Involved in determination of budding pattern | +2.9 |
| Cell cycle control | ||
| Homolog of Schizosaccharomyces pombe CDC4 | EF-hand protein involved in cell division | +2.5 |
| Homolog of S.c. CDC48 | Required for homotypic membrane fusion and for ubiquitin-mediated proteolysis | −2.5 |
| Homolog of S.c. BUB3 | Checkpoint protein required for cell cycle arrest in response to loss of microtubule function | −4.3 |
| Signal transduction and mating response | ||
| Homolog of S. pombe RCD1 | Required for induction of conjugation by regulating ste11+ transcription in response to nitrogen starvation | +4.1 |
| Homolog of S.c. MDG1 | Involved in signal transduction and mating response | −2.5 |
| Meiosis (in sexual yeasts) Homolog of S.c. NDT80 | Transcriptional activator of genes required meiotic division (B-type cyclins) and gamete formation (SPS1) | +2.9 |
| Mitosis | ||
| Homolog of S. pombe SPG1 | Inducer of septum formation through the Cdc7p kinase | +3.7 |
| Homolog of S.c. CDC31 | Cell division control protein 31, involved in first phase of spindle pole body duplication | +3.5 |
| Chromatin and chromosome structures | ||
| Homolog of S.c. HAT1 | Histone acetyltransferase (EC 1.48) | −2.6 |
| Homolog of S.c. TBF1 | TTAGGG repeat-binding factor 1 | −4.6 |
| DNA repair and DNA synthesis | ||
| Homolog of S.c. MSH1 | Mitochondrial DNA repair | +4.1 |
| Homolog of S. pombe CRB3 | Damage and replication checkpoint control protein | +2.9 |
| Homolog of S.c. MCM2 | Component of the prereplication complex initiating DNA replication at ARS sequences | +2.7 |
| Homolog of S.c. DPB2 | DNA polymerase epsilon subunit B | −2.7 |
| Homolog of S.c. RAD26 | Involved in transcription-coupled DNA repair | −2.8 |
| Nucleotide metabolism | ||
| C.a. IMH3 | Inosine-5′-monophosphate dehydrogenase (EC 1.1.1.205) | +3.1 |
| C.a. TMP1 | Thymidylate synthase (EC 2.1.1.45) | −3 |
| Homolog of rat MMSDH | Methylmalonate-semialdehyde dehydrogenase (EC 1.2.1.27) | −7.1 |
| Transcription | ||
| Homolog of S.c. RPC10 | Shared subunit of RNA polymerases I, II, and III | −2.5 |
| Homolog of S.c. HIR2 | Histone transcription regulator 2 | −2.5 |
| Homolog of S.c. TYE7 | Transcription factor | −2.7 |
| C.a. CTA1-2 | Possible transcription factor | −2.7 |
| Homolog of S.c. NIP7 | Nucleolar protein required for efficient 60S fibosome subunit biogenesis | +2.6 |
| Homolog of S.c. SEN2 | tRNA-splicing endonuclease subunit (EC 3.1.27.9) | +2.5 |
| Homolog of S. pombe RNH1 | RNase H | +2.5 |
| Homolog of S.c. IFH1 | Involved in regulation of rRNA expression | +2.5 |
| Homolog of S.c. YER146W | Required for maintenance of normal U6 Sn RNA levels | −3 |
| Homolog of S.c. SMM1 | High-copy suppressor of mutations in mitochondrial tRNA (asp) gene | −2.6 |
| Homolog of S.c. RNA15 | Component of pre-mRNA cleavage and polyadenylation factor I | −2.5 |
| Protein modification and protein degradation | ||
| Homolog of S.c. UBC7 | Ubiquitin-conjugating enzyme (EC 6.3.2.19) | +2.7 |
| Homolog of S.c. DPH5 | Diphthine synthase (EC 2.1.1.98) | +2.6 |
| Homolog of S.c. SPC2 | Component of the signal peptidase complex | +2.5 |
| Homolog of S.c. YIM1 | Mitochondrial inner membrane protease | −3.6 |
| Other | ||
| Homolog of S.c. BIO2 | Biotin synthetase (EC 2.8.1.6) | −6.7 |
| Homolog of S.c. BIO3 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase (EC 2.6.1.62) | −3.1 |
| Homolog of S.c. PHO13 | 4-Nitrophenylphosphatase | −3.6 |
| Homolog of S.c. FPR4 | Nucleolar peptidylprolyl cis-trans isomerase | −2.9 |
| Homolog of C. boidinii FDH | Formate dehydrogenase (EC 1.2.1.2) | −3.8 |
| Homolog of S.c. NIT2 | Nitrilase | −3.5 |
| C.a. ALK8 | Cytochrome P450, may be involved in multidrug resistance | −2.9 |
| Homolog of S.c. STR2 | Cystathionine gamma-synthase, part of the C-3 to C-4 transsulfurylation pathway (EC 4.2.99.9) | −2.8 |
| Homolog of S.c. PSP1 | High-copy-number suppressor of temperature-sensitive mutations in Cdc17p DNA polymerase alpha | −2.5 |
| Unknown (n = 143) | ||
| 82 genes of unknown function | +2.5–37.7 | |
| 61 genes of unknown function | −2.5–5.6 | |
Abbreviations: C.a., C. albicans; S.c., S. cerevisiae; CoA, coenzyme A; ARS, autonomously replicating sequence.
Responses are expressed as the fold change relative to the value for the nontreated control strain from one representative experiment. Only those genes whose expression changed 2.5-fold or more are shown. Genes induced by treatment have a positive numerical value, while those that were repressed have a negative value. The responses shown are averages for two or more datum points.
FIG. 2.
Pie chart grouping responsive (more than 2.5-fold change) genes in functional classes (n = 296); 5% of the responsive genes belong to the ergosterol biosynthetic pathway, and 4% are associated with lipid and fatty acid metabolism. Forty-one percent of the responsive genes are of unknown function.
C. albicans CAI-4, the parental strain used in the present study, carries a deletion of the URA3 gene. For this reason, some changes in expression observed in strain CAI-4 upon itraconazole treatment might not be apparent in wild-type (Ura+) Candida cells. Also, as alterations of gene expression in the presence of azoles may be affected by differences in carbon sources, reported changes in expression are relevant for cells grown on galactose and maltose as carbon sources.
Effect on genes in or linked to the ergosterol biosynthetic pathway.
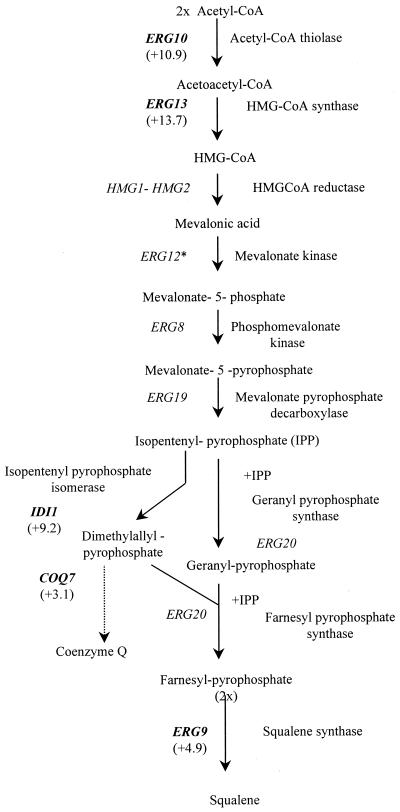
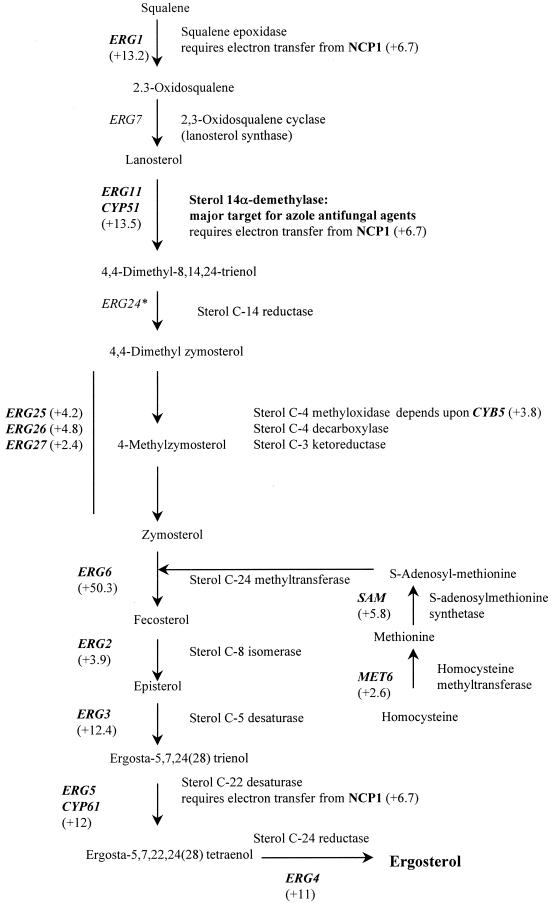
Transcript profiles revealed a global upregulation of ERG genes in response to azole treatment. This is in agreement with previous studies showing that this pathway is the target of azoles and is responsive to modulations in ergosterol levels. A similar observation has recently been reported for Saccharomyces cerevisiae, in which nine ERG genes were found to be responsive upon azole treatment (1). The number and characteristics of the responsive Candida genes. are shown in Table 1. We found intermediate (+3.9- to 5.1-fold) increases in expression for ERG2, ERG9, ERG25 (which depends on CYB5), ERG26, and CYB5 genes and very strong (+9.2 to +50.3 fold) upregulation of the ERG11, ERG5, ERG3, ERG4, and ERG6 genes (all of which act downstream of ERG11) and of the ERG10, ID11, HMGS, and ERG1 genes (all of which act upstream of ERG11) (Fig. 3). In addition, NCP1, which encodes NADP-cytochrome P450 reductase and which is the electron donor for squalene epoxidase (the product of the ERG1 gene), lanosterol 14-demethylase (ERG11), and sterol C-22 desaturase (ERG5), was upregulated 6.7-fold. Previous studies with S. cerevisiae (1, 26) had shown consistent and coordinated increases in the transcript levels of ERG11 and NCP1. In addition, the FEN2 (for fenpropimorph resistance 2) gene was found to be upregulated about fivefold. In S. cerevisiae the product of this gene may be involved in sensing ergosterol levels in the plasma membrane to allow adjustment of ergosterol biosynthesis to match the growth conditions (15). Inhibition of sterol biosynthesis results in increased levels of transcription of some of the genes linked to this pathway, presumably as a result of feedback control (5).
FIG. 3.
Effect of itraconazole (24-h treatment; final concentration, 10 μM) on expression of C. albicans genes involved in the ergosterol biosynthetic pathway; responsive genes are shown in bold; fold changes are shown in parentheses. ∗, not present on DNA chip. CoA, coenzyme A; HMG, 3-hydroxy-3-methylglutaryl.
Previous studies which looked only at the responses of a selection of ERG genes to azole antifungals (9) showed in various Candida spp. upregulation of some early (ERG9, ERG1, ERG7) and late (ERG25, ERG3) genes of the ergosterol pathway, in addition to upregulation of ERG11 itself. As fluconazole-resistant clinical C. albicans isolates which constitutively overexpress ERG11 have been identified (37), ERG gene upregulation has been proposed to play a role in azole resistance (11). Indeed, the ability of C. albicans to tolerate azole treatment (azoles only suppress and do not by themselves eliminate the infection) has partly been attributed to ERG11 upregulation (9).
Which mechanism is responsible for the global upregulation of ERG genes in response to azoles remains unclear. One theory postulates that depletion of ergosterol or another sterol formed late in the pathway increases global ERG expression; the other argues that accumulation of an early substrate or toxic sterol by-product induces ERG expression.
Erg1p (squalene epoxidase) activity has been reported to change in response to sterol limitation in S. cerevisiae (19); sterol limitation may thus be at the origin of the upregulation of ERG1 (+13.2-fold) in C. albicans in response to itraconazole treatment. Consistently observed ERG3 and ERG9 upregulation upon treatment of S. cerevisiae with ketoconazole supports the idea of ERG regulation of expression by ergosterol availability (1, 24). However, Dimster-Denk et al. (4) showed decreases in the levels of transcription of the ERG3, ERG4, ERG5, ERG6, and ERG11 genes 21 h after the start of azole treatment in S. cerevisiae. The fact that these observations are in conflict with ours (in C. albicans) and others (in S. cerevisiae [1, 9]) might be explained by the nature of the azole used (miconazole, sulconazole, and fluconazole compared to ketoconazole and itraconazole). Dimster-Denk et al. (4) used promoter fusions as a readout of transcriptional changes and reported that different azole inhibitors have remarkably different effects on gene expression. It was therefore suggested that azoles may have additional bioactivities, that is, bioactivities other than the inhibition of lanosterol demethylase (note that itraconazole was not tested in that study). The large number of genes found to be responsive in our study (296 in total) indeed reflects many changes in transcript levels not directly related to ergosterol biosynthesis in C. albicans.
CYB5, which encodes cytochrome b5-NADH cytochrome b5 reductase in the Cyb5p-Cbrlp complex, was upregulated 4.5-fold in C. albicans upon itraconazole treatment. Expression of CBR1, which encodes NADH cytochrome b5 reductase, was increased 2.7-fold. Studies by Truan et al. (25) showed that the increased sensitivity of an S. cerevisiae ERG11 mutant to azoles could be suppressed by overexpression of cytochrome b5. CYB5 was shown to be involved in several steps of the sterol biosynthesis pathway in yeast (Fig. 3), probably by acting as a cofactor, and was shown to act as an alternative electron carrier able to support ERG11 function (25).
Studies summarized elsewhere (28) point to a 24-alkylated sterol, ergosterol, as being the sterol that best meets the requirements of yeast and fungal membranes. The transmethylation at C-24 is catabolized by the sterol C-24 methyltransferase, the product of ERG6; this enzyme requires S-adenosylmethionine synthesized from methionine by the S-adenosylmethionine synthetase (SAM). We found that both the ERG6 and the SAM2 genes were upregulated (50.3- and 5.8-fold, respectively) upon treatment of C. albicans with itraconazole. In addition, the homocysteine methyltransferase-encoding gene, MET6, was upregulated 2.6-fold (Fig. 3).
Ubiquinone (coenzyme Q) is a lipid that transports electrons in the mitochondrial respiratory chain. The yeast COQ7 gene encodes a protein involved in one or more monoxygenase or hydroxylase steps of ubiquinone biosynthesis (14). The biosynthesis of ubiquinone starts with the conversion of isopentenyl-pyrophosphate into dimethyl allyl-pyrophosphate, both of which are intermediates of the ergosterol biosynthesis pathway (Fig. 3). Our finding that COQ7 is upregulated (+3.1-fold) upon itraconazole treatment of Candida cells is therefore not surprising.
Effect on other genes.
In addition to its role in the yeast plasma membrane, ergosterol is a major component in secretory vesicles and plays a role in mitochondrial respiration and oxygen sensing. Depletion of ergosterol and subsequent accumulation of 14-methylsterols can result in alterations of membrane functions, synthesis and activity of membrane-bound enzymes, and mitochondrial activities, as well as in an uncoordinated behavior of the yeast cell (21, 27, 28). Apart from enzymes involved in ergosterol and lipid biosynthesis, several other enzymes including ATPases, cytochrome c peroxidase, and chitin synthase(s) have been shown to change expression upon azole treatment (for reviews, see references 10 and 27).
Chitin is an important component of the primary septa in yeast. It has been proven that high ergosterol levels inhibit chitin synthases, whereas mutants of C. albicans with low ergosterol contents show increased levels of chitin synthesis (for a review, see reference 27). From these studies it could be inferred that ergosterol biosynthesis inhibitors should affect chitin synthesis. For example, an azole-induced increase in the chitin:total carbohydrate ratio was found in C. albicans grown for 24 h in the presence of itraconazole; the highest ratio was found at an itraconazole concentration of 49.6 nM (27). On the basis of these results, chitin synthases were expected to be upregulated when ergosterol levels drop. However, treatment with itraconazole did not significantly change expression of most known chitin synthases (fold changes were +2.3 for CHSI, −1 for CHS2, +1.7 for CHS3, +1.4 for CHS regulatory factor, +1.4 for SKTS, and 0 [not-change] for CHS5). The increased chitin content may be at the origin of the enhanced expression level (+3.3-fold) of the chitinase-encoding gene, CHT3. A number of other genes involved in cell wall maintenance were found to be significantly and reproducibly upregulated (Table 1, cell wall maintenance).
Screening for high-copy-number suppressors of ketoconazole sensitivity in S. cerevisiae (13) has shown that overexpression of ERG11, cytochrome c oxidase, ribosomal protein L27, or some specific genes of unknown function was sufficient to overcome growth inhibition by ketoconazole. Upon treatment of C. albicans with itraconazole, we found that an overwhelming number of ribosomal proteins, including L27, were moderately upregulated (+2.5- to +5.2-fold change; see Table 1, protein synthesis). Cytochrome c oxidase was only mildly induced (1.3-fold). The previously described effects on cytochrome c oxidase activity were thought to be linked to inhibition of ergosterol biosynthesis (27). It is not surprising that enzymes of fungal mitochondrial membranes are affected: in contrast to animal cells, large amounts of sterols (ergosterol) are found in the inner membranes of fungal mitochondria (3).
Ergosterol plays an important role in lipid and fatty acid biosynthesis. This is reflected in the responsiveness of 12 genes involved in lipid or fatty acid metabolism (Table 1, lipid, fatty acid metabolism). One of the upregulated genes, AUR1, an inositol phosphoryl ceramide synthase, encodes an enzyme involved in fungal sphingolipid biosynthesis. Bammert and Fostel (1) have suggested that there is an interaction between ergosterol and sphingolipid biosynthetic pathways in yeast.
In C. albicans grown for 6 h in the presence of 0.1 μM miconazole, a shift from monounsaturated (oleic acid) to diunsaturated (linoleic acid) fatty acids was observed, suggesting a stimulation of the delta-9-desaturase, a product of the OLE1 gene (27). Compared with the contact time needed to inhibit ergosterol synthesis, a longer time was needed to interfere with fatty acid desaturation. The effects on delta-9-desaturase depend on the concentration and contact time: in C. albicans grown for 16 h in the presence of 10 nM miconazole (27) or 100 nM itraconazole (30), the shift from oleic acid (18:1) to linoleic acid (18:2) was replaced by a shift from oleic to palmitic acid (16:0), suggesting an interference with fatty acid elongation and/or the fatty acid desaturase. We found that the level of OLE1 expression was reduced 1.3-fold. Thus, changes in delta-9-desaturase activity may again result from changes in membrane fluidity as a result of azole-induced changes in the nature of the membrane sterols.
FAA2 was upregulated 2.5-fold. The product of the FAA2 gene, the acyl coenzyme A synthase (fatty acid thiokinase [long-chain] acyl-activating enzyme), acts on fatty acids from C-6 to C-20 and is involved in chain elongation.
It has previously been demonstrated that the level of induction of cell stress (heat shock) genes is determined by the ratio of the saturated fatty acid content to the unsaturated fatty acid content of fungal membranes (18). Therefore, the observed changes in the level of induction of cell stress genes (Table 1, cell stress) might originate from azole-induced alterations of the ratio of the saturated fatty acid content to the unsaturated fatty acid content.
Ergosterol concentrations are highest in the plasma membrane and in secretory vesicles (3). Altered expression of genes whose products are involved in vesicular transport is therefore to be expected (Table 1, transport).
From our analysis it is clear that further studies should be carried out with 4-aminobutyrate–2-oxoglutarate aminotransferase. In S. cerevisiae, this aminotransferase is the product of UGA1. In itraconazole-treated C. albicans the homologous gene is downregulated about 25.6-fold (Table 1, amino acid metabolism), and its precise role in how itraconazole exerts its effect is unknown.
Effect on drug transporter proteins.
Failure to accumulate azole antifungals has been identified as a major cause of resistance posttreatment in several fungal isolates and species that are less sensitive to azole antifungal agents and other ergosterol biosynthesis inhibitors (16, 23, 35). Failure to accumulate antifungal agents can be the consequence of impaired drug influx or enhanced drug efflux. There is now a consensus that efflux pumps are a common cause of decreased intracellular content of ergosterol biosynthesis inhibitors (for a review, see reference 16). Two types of efflux pumps are known: membrane transport proteins belonging either to the major facilitator superfamily (MFS) or to the ATP-binding cassette (ABC) superfamily. Proteins of the MFS are energized by the proton motive force, and the ABC-type transporters utilize ATP as their source of energy. In C. albicans only the pumps encoded by CaMDR1, Flu1, CDR1, and CDR2 were shown to be involved in the mechanisms of azole efflux (16). Our observations do suggest that expression of some small-molecule transporters changes (among them members of the MFS and ABC families; Table 1); but we did not observe upregulation of CaMDR1, CDR1, or CDR2 after 24 h of treatment of C. albicans with itraconazole. One C. albicans gene with homology to S. cerevisiae YJR015w (whose protein has an unknown function) was upregulated fourfold upon itraconazole treatment. This gene does show significant similarity to known multidrug resistance proteins.
Effect on proteins of unknown function.
Over 140 proteins of unknown function were responsive to treatment with itraconazole under the test conditions used. These data might provide the first hints of a possible function of those proteins in azole drug resistance and sensitivity. One ORF (Genbank accession no. AJ304854) encoding a hypothetical 25.3-kDa protein whose gene is in the TIM23-ARE2 intergenic region and which showed similarity to the S. cerevisiae YNR018w ORF was upregulated over 37-fold (the highest level of upregulation after that for ERG6). Two genes not showing homology to any gene in the sequence databases (i.e., putatively C. albicans-specific genes; Genbank accession no. AJ298306 and AJ298308, respectively) were upregulated 24- and 17-fold, respectively; another ORF (Genbank accession no. AJ298307) with 58% identity to C. albicans ERG25 was upregulated fourfold. These genes might thus be strongly linked to either azole or ergosterol function, and study of their putative roles in ergosterol biosynthesis is ongoing. Upregulation of another ORF (Genbank accession no. AJ298309) with 80% identity to C. maltosa CYP52 (N-alkane-inducible cytochrome P450) was found to be increased 2.8-fold. To the best of our knowledge, the CYP52 multigene family has so far been described only for Candida maltosa and Candida tropicalis, not for C. albicans.
Conclusion.
Thousands of genes are being discovered for the first time by sequencing the genomes of various organisms, reminding us that many processes remain to be explored at the molecular level. DNA microarrays provide a natural vehicle for this exploration. The potential of DNA microarrays to elucidate mechanisms of action is supported by our findings showing that the majority of genes in the ergosterol biosynthetic pathway are responsive to itraconazole, a drug known to target this pathway. Inhibition of sterol biosynthesis results in increased levels of transcription of most of the genes in the pathway, presumably as a result of reduced feedback inhibition. In addition, we identified genes of unknown function which are strongly upregulated upon azole treatment of Candida, thus providing first hints of their putative function in this pathogen. As the C. albicans genome sequence is gradually becoming available in both private and public databases, genome-wide transcript profiling will soon be a versatile tool that can be used to provide an understanding of the modes of action of novel antifungal compounds.
However, as not all cellular processes are controlled at the level of gene expression (transcription), protein profiling will certainly prove to be a helpful complementary approach. In addition, one must bear in mind that genes which are expressed at a very low level might be missed by this approach, but as the technology improves, this might be remedied in the near future.
ACKNOWLEDGMENTS
We thank Ing. Ronald de Hoogt (Janssen Research Foundation) for technical support and Jackson Wan (R. W. Johnson Pharmaceutical Research Institute) for helpful advice.
This work was supported by IWT grant 960192 from the “Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie” (Brussels, Belgium).
REFERENCES
- 1.Bammert G F, Fostel J M. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob Agents Chemother. 2000;44:1255–1263. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–7. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 3.Daum G, Lees N D, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Dimster-Denk D, Rine J, Philips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- 5.Dixon G, Scanlon D, Cooper S, Broad P. A reporter gene assay for fungal sterol biosynthesis inhibitors. J Steroid Biochem Mol Biol. 1997;62:2–3. doi: 10.1016/s0960-0760(97)00032-0. , 165–167. [DOI] [PubMed] [Google Scholar]
- 6.Eisen M B, Spellman P T, Brown P O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harousseau J L, Dekker A W, Stamatoullas-Bastard A, Fassas A, Linkesch W, Gouveia J, De Bock R, Rovira M, Seifert W F, Joosen H, Peeters M, De Beule K. Itraconazole oral solution for primary prophylaxis of fungal infections in patients with hematological malignancy and profound neutropenia: a randomized, double-blind, double-placebo, multicenter trial comparing itraconazole and amphotericin B. Antimicrob Agents Chemother. 2000;44:1887–1893. doi: 10.1128/aac.44.7.1887-1893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry K W, Nickels J T, Edlind T D. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob Agents Chemother. 2000;44:2693–2700. doi: 10.1128/aac.44.10.2693-2700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly S L, Quail M A, Rowe J, Kelly D E. Sterol 14α-demethylase: target of the azole antifungal agents. In: Fernandes P B, editor. New approaches for antifungal drugs. Boston, Mass: Birkhauser; 1992. pp. 155–187. [Google Scholar]
- 11.Kontoyiannis D P, Sagar N, Hirschi K D. Overexpression of Ergl 1p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1999;43:2798–2800. doi: 10.1128/aac.43.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb D C, Maspahy S, Kelly D E, Manning N J, Geber A, Bennett J E, Kelly S L. Purification, reconstitution, and inhibition of cytochrome P-450 sterol, Δ22-desaturase from the pathogenic fungus Candida glabrata. Antimicrob Agents Chemother. 1999;43:1725–1728. doi: 10.1128/aac.43.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Launhardt H, Hinnen A, Munder T. Drug-induced phenotypes provide a tool for the functional analysis of yeast genes. Yeast. 1998;14:935–942. doi: 10.1002/(SICI)1097-0061(199807)14:10<935::AID-YEA289>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Marbois B N, Clarke C F. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 15.Mariceau C, Joets J, Pousset D, Guilloton M, Karst F. FEN2: a gene implicated in the catabolite repression-mediated regulation of ergosterol biosynthesis in yeast. Yeast. 1996;12:531–539. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C531::AID-YEA934%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 16.Marichal P. Mechanisms of resistance to azole antifungal compounds. Curr Opin Anti-infective Investig Drugs. 1999;1:318–333. [Google Scholar]
- 17.Marichal P, Gorrens J, Laurijssens L, Vermuyten K, Van Hove C, Le Jeune L, Verhasselt P, Sanglard D, Borgers M, Ramaekers F V S, Odds F, Vanden Bossche H. Accumulation of 3-ketosteroids induced by itraconazole in azole resistant clinical Candida albicans isolates. Antimicrob Agents Chemother. 1999;43:2663–2670. doi: 10.1128/aac.43.11.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews R C, Maresca B, Burnie J P, Cardona A, Carrutu L, Conti S, Deepe G S, Florez A M, Franceschelli S, Garcia E, Gargano L S, Kobayashi G S, McEwen J G, Ortiz B L, Oviedo A M, Polonelli L, Ponton J, Restrepo A. Stress proteins in fungal diseases. Med Mycol. 1998;36(Suppl. 1):45–51. [PubMed] [Google Scholar]
- 19.M'baya B, Fegueur M, Servouse M, Karst F. Regulation of squalene synthase and squalene epoxidase activities in Saccharomyces cerevisiae. Lipids. 1989;24:1020–1023. doi: 10.1007/BF02544072. [DOI] [PubMed] [Google Scholar]
- 20.Odds F C. Antifungal therapy. In: Kibbler C C, Mackenzie D W R, Odds F C, editors. Principles and practice of clinical mycology. Chichester, United Kingdom: John Wiley & Sons; 1996. pp. 35–48. [Google Scholar]
- 21.Parks L W. Metabolism of sterols in yeast. Crit Rev Microbiol. 1978;6:301–341. doi: 10.3109/10408417809090625. [DOI] [PubMed] [Google Scholar]
- 22.Rosamond J, Allsop A. Harnessing the power of the genome in the search for new antibiotics. Science. 2000;287:1973–1976. doi: 10.1126/science.287.5460.1973. [DOI] [PubMed] [Google Scholar]
- 23.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrobiol Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith S J, Crowley J H, Parks L W. Transcriptional regulation by ergosterol in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5427–5432. doi: 10.1128/mcb.16.10.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truan G, Epinat J-C, Rougeulle C, Cullin C, Pompon D. Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypresensitivity in a NADPH-P450 reductase-deficient strain. Gene. 1994;149:123–127. doi: 10.1016/0378-1119(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 26.Turi T G, Loper J C. Multiple regulatory elements control expression of the gene encoding S. cerevisiae cytochrome P450, Ianosterol 14 alpha-demethylase (ERG11) J Biol Chem. 1992;267:2046–2056. [PubMed] [Google Scholar]
- 27.Vanden Bossche H. Biochemical targets for antifungal azole derivatives hypothesis on the mode of action. Curr Top Med Mycol. 1985;1:313–351. doi: 10.1007/978-1-4613-9547-8_12. [DOI] [PubMed] [Google Scholar]
- 28.Vanden Bossche H. Importance and role of sterols in fungal membranes. In: Kuhn P J, Trinci A P J, Jung M J, Goosey M W, Copping L G, editors. Biochemistry of cell walls and membranes in fungi. Berlin, Germany: Springer-Verlag; 1990. pp. 135–157. [Google Scholar]
- 29.Vanden Bossche H. Chemotherapy of human fungal infections. In: Lyr H, editor. Modern selective fungicides. Jena, Germany: Gustav Fischer Verlag; 1995. pp. 431–484. [Google Scholar]
- 30.Vanden Bossche H, Heeres J, Backx L J J, Marichal P, Willemsens G. Discovery, chemistry, mode of action, and selectivity of itraconazole. In: Rippon J W, Fromtling R A, editors. Cutaneius antifungal agents. New York, N.Y: Marcel Dekker, Inc; 1993. pp. 263–283. [Google Scholar]
- 31.Vanden Bossche H, Koymans L. Cytochromes P450 in fungi. Mycoses. 1998;41(Suppl. 1):32–38. doi: 10.1111/j.1439-0507.1998.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 32.Vanden Bossche H, Willemsens G, Marichal P. Anti-Candida drugs: the biochemical basis for their activity. Crit Rev Microbiol. 1987;15:57–72. doi: 10.3109/10408418709104448. [DOI] [PubMed] [Google Scholar]
- 33.Vanden Bossche H, Willemsens G, Cools W, Marichal P, Lauwers W. Hypothesis on the molecular basis of the antifungal activation of N-substituted imidazoles and triazoles. Biochem Soc Trans. 1983;11:665–667. doi: 10.1042/bst0110665. [DOI] [PubMed] [Google Scholar]
- 34.Vanden Bossche H, Marichal P, Le Jeune L, Coene M C, Gorrens J, Cools W. Effects of itraconazole on cytochrome P-450-dependent sterol 14-demethylation and reduction of 3-ketosteroids in Cryptococcus neoformans. Antimicrob Agents Chemother. 1993;37:2101–2105. doi: 10.1128/aac.37.10.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanden Bossche H, Dromer F, Improvisi I, Lozano Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 36.Wheat J, Marichal P, Vanden Bossche H, Le Monte A, Connolly P. Hypothesis on the mechanism of resistance to fluconazole in Histoplasma capsulatum. Antimicrob Agents Chemother. 1997;41:410–414. doi: 10.1128/aac.41.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases on azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;42:1488–1494. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]