Abstract
Pulmonary arterial hypertension (PAH) is a progressive, ultimately fatal cardiopulmonary disease associated with a number of physiologic changes, which is believed to result in imbalances in the intestinal microbiota. To date, comprehensive investigational analysis of the intestinal microbiota in human subjects is still limited. To address this, we performed a pilot study of the intestinal microbiome in 20 PAH and 20 non‐PAH healthy control subjects, recruited from a single center, with each PAH subject recruited simultaneously with a cohabitating non‐PAH control subject. Shotgun metagenomic sequencing was used to analyze the microbiome profiles. There were no differences between PAH and non‐PAH subjects across several measures of microbial abundance and diversity (Alpha Diversity, Beta Diversity, F/B ratio). The relative abundance of Lachnospiraceae bacterium GAM79 was lower in PAH stool samples as compared to non‐PAH control subject' stool. There was no strong or reproducible association between PAH disease severity and global microbial abundance, but several bacterial species (a relative abundance of Anaerostipes rhamnosivorans and a relative deficiency of Amedibacterium intestinale, Ruminococcus bicirculans, and Ruminococcus albus species were associated with disease severity (most proximal right heart catheterization hemodynamics and six‐minute walk test distance) in PAH subjects. Our results support further investigation into the presence, significance, and potential physiologic effects of a PAH‐specific intestinal microbiome.
Keywords: clinical studies, microbial molecular genetics, diet, obesity, lung disease
INTRODUCTION
Pulmonary arterial hypertension (PAH) is a progressive, incurable cardiopulmonary disease that eventually results in right heart failure and death. 1 , 2 , 3 Outcomes in PAH are determined by the degree of cardiac impairment, which is reflected hemodynamically by an impaired cardiac output and corresponding increase in pulmonary vascular resistance (PVR). 4 , 5 , 6 In PAH, a number of systemic physiologic and biochemical changes have been associated with right ventricular failure, including increased circulating inflammatory cytokines, impaired lipid and amino acid metabolism, and increased intestinal edema and permeability. 7 , 8 , 9 , 10 , 11 These metabolic and biochemical changes are believed to be driven in‐part by alterations in composition of the intestinal microbiome, and have been observed in both animal models of PAH and clinical studies of congestive heart failure. 12 , 13 , 14 , 15 , 16 , 17 Abnormalities in the composition of the intestinal microbiome have also recently been described in human PAH, but have yet to be validated or studied in a comprehensive fashion. 18 The aim of our study is to examine the intestinal microbiome composition in a cohort of PAH subjects, to identify PAH‐specific microbial patterns with potential diagnostic and prognostic utility for further development.
METHODS
We performed a prospective cohort study of adult PAH patients between the ages of 18 and 70, screened and enrolled from our center between July 1, 2019 and April 1, 2021. Eligible participants were those who had a body mass index (BMI) < 35 kg/m2 and met the diagnostic criteria for PAH, confirmed by right heart catheterization hemodynamics (mean pulmonary arterial pressure (mPAP) at rest >20 mmHg, pulmonary capillary wedge pressure at rest ≤15 mmHg, and PVR at rest ≥3 Wood units). 1 , 2 Subjects were required to be clinically stable, without changes in their PAH medication regimen for at least 3 months before enrollment. To account for environmental effects on the microbiome, each PAH subject was enrolled simultaneously with a cohabitating, healthy, nonobese volunteer (spouses, siblings, or adult children). Control subjects were eligible if they were nonobese adults between the ages of 18 and 70, did not carry a diagnosis of PAH, did not have known cardiopulmonary disease (defined as known medical conditions affecting the heart or lungs), did not have self‐reported signs or symptoms of cardiovascular disease (chest pain, trouble breathing, coughing, wheezing, passing out, or leg swelling), and shared a living environment (cohabitated) with the primary PAH subject. 19 , 20 , 21
Per the diagnostic criteria for PAH, those with underlying pulmonary obstructive or restrictive disease, based on pulmonary function testing forced vital capacity, forced expiratory volume in 1 s, or total lung capacity ≤60%, were excluded. Those with a diagnosis of chronic thromboembolic pulmonary hypertension were excluded. Given the association between certain etiologies of PAH (such as underlying liver disease, infection with the human immunodeficiency virus, or an underlying inflammatory bowel disease like ulcerative colitis or Crohn's disease) and dysfunction of the intestinal microbiome, subjects with these conditions were also excluded. 22 , 23 Given the known effects of obesity on the composition of the intestinal microbiome, we excluded subjects with class II or greater obesity, as defined by a BMI ≥ 35 kg/m2. 19 , 20 , 21 Both potential subjects with PAH and matched control participants were required to have a BMI below 35 kg/m2, either measured or self‐reported, or they were excluded, and no study participant was believed to have a BMI above this cutoff. Given the known effects of cardiovascular disease on the composition of the intestinal microbiome, those who had clinically significant valvular heart disease, known pericardial disease, known restrictive cardiomyopathy or left ventricular outflow tract obstruction, or evidence of left ventricular dysfunction on trans‐thoracic echocardiography (ejection fraction < 50% or diastolic dysfunction grade II or greater) were excluded. Those with the presence of three or more risk factors for left heart disease (three or more of the following: obesity as defined by a BMI ≥ 30 kg/m2, a history of hypertension, a history of diabetes mellitus, a history of coronary artery disease with either previous myocardial infarction, coronary artery bypass graft, or known coronary artery disease with >50% stenosis in one or more coronary arteries) were also excluded. 24 , 25 , 26 , 27 , 28 Given the association between the intestinal microbiota and underlying pulmonary disease, those with a diagnosis of obesity hypoventilation syndrome (as defined by a BMI ≥ 30 kg/m2 and a resting awake arterial blood gas carbon dioxide measurement >45 mmHg) were excluded. 29 , 30 Given the association between antibiotics, probiotics, and immunosuppressive medications, those taking any antibiotics in the preceding 3 months, probiotics in the preceding week, or on immunosuppressive medications (tacrolimus, methotrexate, prednisone, cyclophosphamide, mycophenolate mofetil, or similar compounds) in the preceding 3 months before enrollment were excluded. 31 , 32 , 33 , 34 , 35 Pregnant women and those with previous intestinal tract resection and/or a diagnosis of short bowel syndrome were excluded. This study conformed to the ethical guidelines of the 1975 Declaration of Helsinki, and as reviewed and approved by our institutional review board (IRB 2019‐0234).
Following consent and enrollment, study participants demographic information (age, sex, race) was collected. For PAH subjects, data were obtained from the medical record, including past medical history, pulmonary function testing results, six‐minute walk testing results, BMI, and right heart catheterization hemodynamics. Due to the significant effects of the Sars‐CoV‐2 pandemic on our institution's clinical research operations, we were unable to obtain BMI values for the non‐PAH control subjects. The REVEAL 2.0 risk score, a multifaceted measure of disease severity in PAH with prognostic utility, was calculated at the time of study enrollment in PAH subjects. 36 Study participants were then provided with stool collection kits, and were required to return kits within 48 h of stool collection. Collected stool samples were stored at −80°C until fecal DNA extraction and sequencing.
For metagenomic sequencing, DNA extraction was performed with the Power Fecal DNA Isolation Kit® (Qiagen). Amplified library generation was performed with Nextgen adapters, and 150 base pairs (bp) DNA paired end reads to a depth of 2.5 Gbp per sample, using an Illumina NovaSeq® sequencing machine (Illumina). Metagenomic taxonomic and functional profiles were generated using the bioBakery3 metagenomic workflow for quality control and functional profiling, and using Kraken2 and Bracken for taxonomic profiling. 37 , 38 , 39 Kneaddata was used to remove reads mapping to the human genome, trim reads where the average of the past four consecutive reads drops below 20, and filter reads <90 bp in length. Functional profiling was performed via HUMAnN3 using default settings. Uniref90 gene families were aggregated to MetaCyc and enzyme commission pathways via HUMAnN3. Taxonomic profiling was performed by assigning sequence reads to the lowest common ancestor using Kraken2. The confidence interval was set to 0.1, and reads mapped to a custom database built from all complete genomes for bacteria, archaea, viruses, fungi, and the GRCh38.p13 human reference genome available in the RefSeq database as of 25 July, 2020. 40 Relative abundances were obtained using Bracken with the threshold for species‐level classification set to t‐10. 41
For microbiome analyses, taxa seen at less than 0.0001% relative abundance were removed before analysis. Differences in Alpha Diversity and the Firmicutes to Bacteroidetes (F/B ratio) between PAH cases and matched controls was assessed using linear mixed effects regression (LMER) as implemented in the LME4 package (Version 1.1.23). Models included case/control status, age, race, and gender as fixed effects, and a random intercept to account for the correlation in the microbiome within matched pairs. p values were obtained using the Satterthwaite approximation to the model degrees of freedom as implemented in the lmerTest package (Version 3.1.2). Alpha diversity estimates were obtained after subsampling without replacement to the lowest observed sequence depth (1,546,755 reads). Beta diversity ordinations were performed on species count matrix using principal component analysis after variance‐stabilizing transformation, as implemented in DESeq. 2 (Version 1.28.1). Differential species abundance was assessed using generalized linear mixed‐effects regression modeling, as implemented in the glmmTMB package (Version 1.0.2). Taxa with a prevalence of <50% were filtered before testing to remove taxa seen in fewer than 10 samples. Counts were modeled as negative binomial distributed responses with a log‐link function, with fixed effects for case/control status, age, race, and gender, and a random intercept denoting matched pairs. Normalization factors were calculated using the geometric mean of pairwise ratios method via the GMPR package (Version 0.1.3), and included as a model offset to account for differences in library sizes between samples. Outlying counts were truncated at the 97th percentile, to improve the performance of count‐based models of microbiome data. 41 Estimates were converted to the log2 scale to facilitate interpretation. Benjamini–Hochberg false discovery rate corrections were applied to the raw p values to control the overall probability of type 1 errors. Volcano plots were generated using the EnhancedVolcano package (Version 1.6.0).
In the PAH cohort, high‐risk subjects were defined as those with a PVR > 5 Wood units. Alpha Diversity, Beta Diversity, and the F/B ratio estimates were generated as described above, and differences between low‐ and high‐risk PAH subjects were tested using linear regression models adjusted for age, race, gender, and BMI. Although digoxin use may affect composition of the intestinal microbiome, only two PAH subjects were on digoxin at the time of study enrollment, and we did not adjust for this covariate in our modeling given the small number of subjects on this medication and to avoid overfitting. 42 Adjusted linear regression models were also constructed to test the relationship between global measures of microbial diversity (Alpha Diversity, Beta Diversity, and F/B ratio) and other metrics of PAH disease severity (mPAP and six‐minute walk distance). Moderated negative binomial regression, as implemented inDESeq. 2, was used to test for differences in the abundance of prevalent microbial species with PAH disease severity (PVR, mPAP, and six‐minute walk distance). Models included terms for age, race, gender, and BMI. Log2‐fold changes were moderated using the apeglm package (Version 1.14.0), and normalization was carried out using the median of ratios method with only positive counts. All analyses were conducted using R Version 4.0.2 (R Foundation for Statistical Computing).
Descriptive data were expressed as median (interquartile range, IQR) for continuous variables, and frequency (percentage, %) for categorical variables. Differences were compared using the nonparametric Mann–Whitney U test or the χ 2 test for continuous or categorical variables respectively. The sample size of this pilot study of 20 subjects per group was chosen based on sample sizes of previously published literature studying the significance of the F/B ratio in various chronic diseases and investigational conditions. 43 , 44 , 45 , 46
Species‐level count matrix and sample metadata have been deposited in Mendeley Data for sharing. 47
RESULTS
A total of 56 eligible subjects (28 pairs) were screened and enrolled, of which 40 (20 pairs) completed all study procedures and were included in the final analysis (Figure 1). A total of 16 consented subjects (8 pairs) failed to complete study procedures and were withdrawn.
Figure 1.
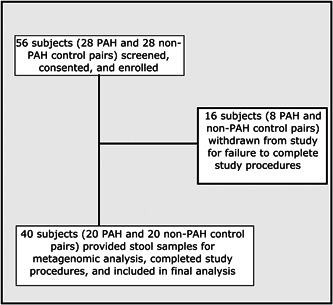
Flow of patients throughout the study.
PAH, pulmonary arterial hypertension
The characteristics of subjects included in the final analyses are shown in Table 1. The majority of subjects were White race (83%). The vast majority of PAH subjects (90%) were females, as compared to the minority of control patients (35% females). The median BMI of study subjects was 29 kg/m2. Although we were unable to measure the BMI of the control subjects, as noted earlier all control subjects had a BMI below 35 kg/m2, and none of the 40 participants in this study had a BMI beyond this threshold. Half of PAH subjects were idiopathic, the remainder had PAH due to connective tissue disease, congenital heart disease, or heritable PAH. Roughly equal proportions of PAH subjects were managed with dual oral therapy, triple oral therapy, or parenteral prostacyclin therapy. Despite having uniformly low REVEAL 2.0 risk scores (median score 6) and robust walk distances (median distance 344 m), PAH subjects had variable hemodynamic measures of disease severity, with a wide range of PVR values from their most proximal right heart catheterization (IQR 3.7–10.1 Wood units). Half of all PAH subjects had a PVR above 5 Wood units on their most recent right heart catheterization. The time between study enrollment and most proximal cardiopulmonary testing was variable across the study cohort (IQR since most proximal walk test of 0–3.5 months, IQR since most proximal right heart catheterization of 0.7–25.7 months).
Table 1.
Baseline characteristics of study cohort
| Clinical variable | PAH (N = 20) | Non‐PAH control (N = 20) |
|---|---|---|
| Median/F (IQR/%) | Median/F (IQR/%) | |
| Age (years) | 53 (42–61) | 52 (37–60) |
| Sex (Female) | 18 (90%) | 7 (35%) |
| BMI (kg/m2) | 29.0 (27.0–33.8) | — |
| Race | ||
| White | 16 (80% | 17 (85%) |
| Black | 3 (15%) | 3 (15%) |
| Other | 1 (5%) | 0 (0%) |
| Etiology of PAH | ||
| Idiopathic | 10 (50%) | — |
| Connective tissue disease associated | 4 (20%) | — |
| Heritable | 4 (20%) | — |
| Congenital heart disease associated | 2 (10%) | — |
| Targeted PAH therapy at the time of enrollment | ||
| Dual oral | 6 (30%) | — |
| Triple oral | 7 (35%) | — |
| Parenteral therapy | 7 (35%) | — |
| NTproBNP (pg/ml) | 118 (108–194) | — |
| Diagnostic mPAP (mmHg) | 47 (35–53) | — |
| Diagnostic PVR (WU) | 9.4 (3.7–9.5) | — |
| Most recent mPAP (mmHg) | 43 (34–45) | — |
| Most recent PCWP (mmHg) | 10 (9–14) | — |
| Most recent CO (L/min) | 5.2 (4.0–5.7) | — |
| Most recent PVR (Wood units) | 5.5 (3.7–10.1) | — |
| High‐risk PVR (>5 Wood units) | 10 (50%) | — |
| Most recent 6‐minute walk test (m) | 344 (256–404) | — |
| Diagnostic FEV1 (%) | 77 (67–86) | — |
| Diagnostic FVC (%) | 83 (73–96) | — |
| Diagnostic FEV1/FVC ratio | 74 (69–83) | — |
| Diagnostic TLC (%) | 94 (80‐102) | — |
| Most recent WHO Functional Class | 2 (1.75–2) | — |
| Most recent REVEAL 2.0 Score | 6 (4–7) | — |
| Time since most recent walk test (months) | 2 (0‐3.5) | — |
| Time since most recent RHC (months) | 7.5 (0.7–25.7) | — |
Note: Data presented as median/frequency (interquartile range/percentage).
Abbreviations: Dual oral: use of oral medication from two of the following three classes: phosphodiesterase‐5 inhibitor/soluble guanylate cyclase activator (sildenafil, tadalafil, riociguat), endothelin receptor antagonist (macitentan, ambrisentan), or oral prostacyclins (treprostinil, selexipag); CO, cardiac output (thermodilution); FEV1, forced expiratory volume in 1 s on pulmonary function testing; FVC, forced vital capacity on pulmonary function testing; mPAP, mean pulmonary arterial pressure; NTproBNP, N‐terminal pro‐type brain natriuretic peptide levels; parenteral therapy, use of either intravenous or subcutaneous treprostinil or intravenous epoprostenol; PCWP, pulmonary capillary wedge pressure; PVR, pulmonary vascular resistance; RHC, right heart catheterization; TLC, total lung capacity on pulmonary function testing; triple oral: use of all three classes of oral medications.
Within the full cohort, there were no significant differences between groups in the number of observed species read counts (Figure 2a), Alpha (Shannon) Diversity (p = 0.3, Figure 2b), no significant clustering by Beta Diversity (Figure 2c), and no significant difference in F/B ratio (p = 0.95, Figure 2d). Only one bacterial species, Lachnospiraceae bacterium GAM79, was significantly different between PAH and control subjects, with a significant deficiency of this bacterial species in the PAH microbiome compared to the non‐PAH control microbiome (log fold change −1.59, p < 0.001) (Figure 3).
Figure 2.
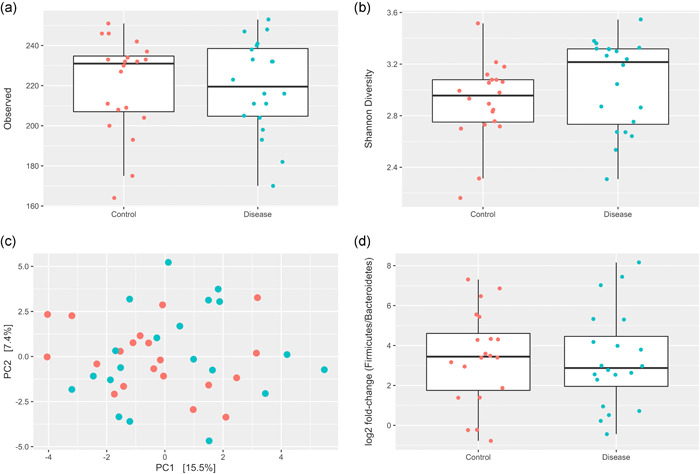
Intestinal microbiota diversity measures between groups. (a) Distribution of observed species read counts. (b) Alpha Diversity (Shannon Index) between PAH and non‐PAH control groups. (c) Beta diversity principal component plot between PAH and non‐PAH control groups. (d) Firmicutes/bacteriodetes ratio between PAH and non‐PAH control groups. PAH, pulmonary arterial hypertension
Figure 3.
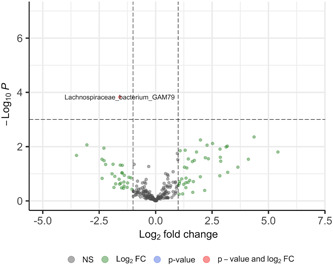
Volcano plot of differential species abundance between PAH and non‐PAH control groups. FC, fold change; PAH, pulmonary arterial hypertension; PVR, pulmonary vascular resistance
When investigating the PAH cohort specifically, there was no significant association between hemodynamic disease severity (mPAP or high‐risk PVR [above 5 Wood units] on most recent right heart catheterization) and any measure of global microbial diversity (Alpha Diversity, Beta Diversity, or F/B ratio) (Table 2). There was a significant clustering by Beta Diversity for six‐minute walk test, but not for mPAP or high‐risk PVR. Among PAH subjects, there were several bacterial species that were associated with multiple different metrics of disease severity (PVR > 5 Wood units and higher mPAP on most recent right heart catheterization, lower six‐minute walk distance on most recent walk test) (Figure 4), specifically an increased abundance of Anaerostipes rhamnosivorans species and a relative deficiency of Amedibacterium intestinale, Ruminococcus bicirculans, and Ruminococcus albus species.
Table 2.
Microbial diversity measures and disease severity in PAH subjects
| Regression model p value | |
|---|---|
| Model for mPAP (mmHg) | |
| Alpha Diversity | 0.846 |
| Beta Diversity | 0.854 |
| F/B ratio | 0.563 |
| Model for PVR > 5 Wood units | |
| Alpha Diversity | 0.591 |
| Beta Diversity | 0.238 |
| F/B ratio | 0.667 |
| Model for six‐minute walk distance (meters) | |
| Alpha Diversity | 0.804 |
| Beta Diversity | 0.031 |
| F/B ratio | 0.668 |
Note: Regression Models are adjusted for age, gender, race, and BMI.
Abbreviations: F/B, Firmicutes/Bacteriodetes ratio; mPAP, mean pulmonary arterial pressure; PVR, pulmonary vascular resistance.
Figure 4.
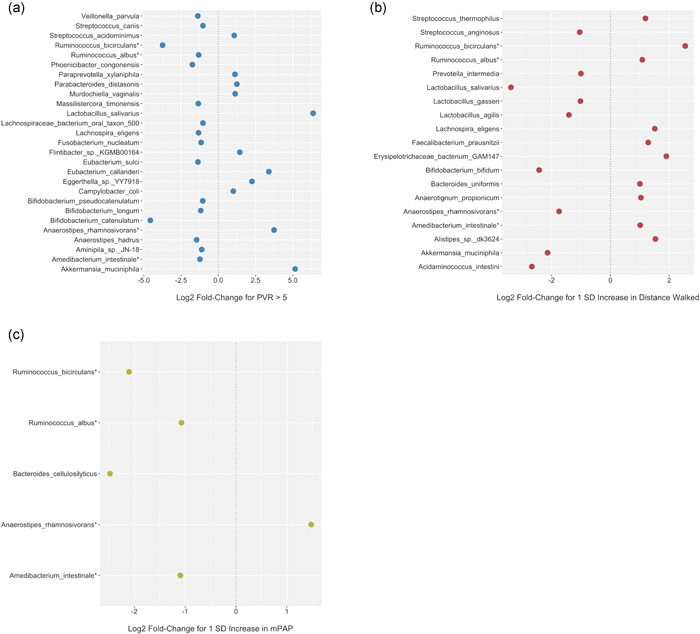
Dot Plot representation of significant differential species abundance in relation to disease severity metrics in PAH group. (a) Significant bacterial species by PVR > 5 Wood units. (b) Significant bacterial species by six‐minute walk distance. (c) Significant bacterial species by mPAP. *Bacterial species (Anaerostipes rhamnosivorans, Amedibacterium intestinale, Ruminococcus bicirculans, and Ruminococcus albus) significant across all three metrics of disease severity. FC, fold change; PAH, pulmonary arterial hypertension; PVR, pulmonary vascular resistance
Although we did not examine the circulating metabolome in our study, we did extract out nonspecific measures of metabolic function (including hemoglobin A1c and circulating lipid profile) and inflammation (C‐reactive protein) from our PAH cohort at the time of study enrollment (Table S1). There was no significant association between any nonspecific measure of metabolic function or inflammation and global measures of microbial diversity.
DISCUSSION
In this prospective cohort study, we identified a significantly lower relative abundance of Lachnospiraceae bacterium GAM79 in PAH subjects as compared to non‐PAH control subjects. We also identified a significant association between PAH disease severity (most proximal PVR, mPAP, and six‐minute walk distance) and an increased abundance of Anaerostipes rhamnosivorans, with a relative lack of Amedibacterium intestinale, Ruminococcus bicirculans, and Ruminococcus albus species, in PAH subject' stool. There were no differences between groups in Alpha Diversity, Beta Diversity, or the F/B ratio, nor any consistent association between these global microbial diversity measures and disease severity in PAH subjects specifically.
We did not observe any differences between groups in either Alpha or Beta measures of Diversity, or the F/B ratio, in our study. Comparable measures of global microbial diversity (Alpha and Beta Diversity) between disease and control states has also been seen in a previously published investigation of heart failure patients. 48 This is in contrast to a number of other studies which observed a less diverse intestinal microbiome in PAH and cardiovascular diseases as compared to healthy controls. 16 , 18 , 49 , 50 This inconsistency may be explained by the multitude of potentially confounding factors known affect the composition of the intestinal microbiome including obesity, dietary intake, environmental factors, and even circadian rhythm, which are not consistently accounted for in microbiota clinical research studies and are known to affect global microbial diversity. 19 , 20 , 21 , 51 , 52 In our cohort, we attempted to control for a number of confounding factors by using an extensive exclusionary criteria list and enrolling matched pairs of subjects (who cohabitated in shared living environments), and this rigorous study design may explain why we did not observe any differences in global diversity measures between groups. It is also possible our limited sample size was under‐powered to detect a difference where one existed. Given the complexity of the microbiome, we suggest future microbiota studies in pulmonary vascular disease should be designed to minimize the influence of confounders and address environmental factors.
We identified a significant deficiency in Lachnospiraceae bacterium GAM79 in PAH subjects relative to non‐PAH control subjects. A deficiency in Lachnospiraceae species has also been observed in another analysis of the PAH gut microbiome profile, as well as in analyses of the gut microbial signature in heart failure patients, and this relative deficiency was theorized in these studies to possibly affect the production of butyrate, propionate, and other short‐chain fatty acids. 18 , 49 Lachnospiraceae species are known to produce short‐chain fatty acids such as butyrate and propionate, which are believed to exert a protective effect against cardiovascular diseases, and may theoretically offer protection from the development or progression of pulmonary vascular disease. 53 , 54 , 55 , 56 , 58 , 59 We were unable to comprehensively examine the circulating metabolome of study subjects to confirm this relationship, and further investigation of the unique gut‐derived metabolomic signature in PAH and the significance ofLachnospiracea species is needed to explore this possible mechanism.
In our study there was no association between disease severity (most proximal PVR, mPAP, and six‐minute walk distance) and global measures of microbial abundance (Alpha Diversity, F/B ratio). There was a significant association between Beta Diversity and distance walked, but not hemodynamics (mPAP, high‐risk PVR). Given our limited sample size, the lack of other diversity measures reflecting PAH disease severity, and the comparable global diversity between PAH and non‐PAH control patients in our study, the significance of this isolated finding is unclear and should be interpreted cautiously.
In our PAH cohort, more severe disease was associated with a relative abundance of Anaerostipes rhamnosivorans and decrease in Amedibacterium intestinale, Ruminococcus bicirculans, and Ruminococcus albus species. Many of these species (Anaerostipes rhamnosivorans, Ruminococcus bicirculans, and Ruminococcus albus) are believed to play a role in the metabolic processes of fermentation and short‐chain fatty acid synthesis, which may protect against the progression of cardiopulmonary disease. 57 , 60 , 61 , 62 , 63 A deficiency of circulating short‐chain fatty acids, potentially regulated by bacterial fermentation of dietary fiber, has been recognized as a feature of cardiovascular disease that may stimulate vascular inflammation and remodeling. 16 , 48 , 49 , 58 , 64 Additionally, in a recently published study of the microbiome in PAH, multiple fermentation pathways were enriched in healthy control patients relative to PAH patients based on functional analysis of their gut microbial composition. 18 The intestinal microbiome is known to affect both local and systemic inflammatory responses, and a potential link between intestinal microbial composition and inflammation has been described in other pulmonary diseases such as asthma. 65 , 66 Inflammation is also believed to stimulate the pathogenesis of PAH, and a number of the species we observed as associated with more severe PAH disease in our cohort (Anaerostipes, Ruminococcus) are associated with both short‐chain fatty acid production (butyrate) and augmentation of local and systemic pro‐inflammatory cytokine levels (Interleukin 12, interferon gamma, etc.). 42 , 67 It is possible that specific intestinal gut microbes, such as the ones identified in our study as associated with more severe PAH disease, exert their effects on the pulmonary vasculature through pro‐inflammatory intermediaries such as circulating cytokines and short‐chain fatty acids. Our results support further investigation into the interplay between the intestinal microbiome composition, circulating metabolome, and PAH disease development and progression.
There were several strengths to our study, including the use of a paired enrollment scheme to address the impact of environmental factors on the intestinal microbiome, the application of a comprehensive list of exclusionary criteria to avoid known confounders (coronary artery disease, obesity, use of antibiotics or immunosuppressive medications, comorbid conditions such as liver cirrhosis or inflammatory bowel disease, etc.), the multivariable adjustment of regression models for age, gender, race, and BMI, and the grouped structure of our data set. None of the participants in our study suffered from class II or greater obesity, defined by a BMI above 35 kg/m2. There were also a number of limitations that merit discussion. This was a small single center pilot study with a limited sample size, the significance of our results should be interpreted cautiously, and will require confirmation and validation in additional larger cohorts. As typical of PAH there was a significant gender discrepancy between groups, which was adjusted for in the final analysis but still could have affected our results. We were unfortunately not able to adjust for the exact BMI in the control cohort as we had originally intended, due to the impact of the Sars‐CoV‐2 pandemic in clinical research, and although we excluded significantly obese patients from this study, we cannot exclude that an association between BMI and intestinal microbial composition confounded our results. We did not perform simultaneous cardiopulmonary testing at the time of sample collection, and there was significant variability in time elapsed between certain tests (such as right heart catheterization) and enrollment in the study (median time elapsed 7.5 months). Although we attempted to account for this variability by evaluating microbial composition across multiple metrics of PAH disease severity (both right heart catheterization hemodynamics and walk test distance), the most recent cardiopulmonary testing values may not have been reflective of the true values in some subjects and may have affected our results. We did not examine the circulating metabolome in our cohort, and thus are unable to definitively associate circulating metabolites (such as fermentation products and short‐chain fatty acids) with specific characteristics of the intestinal microbiome in PAH subjects.
We relied upon self‐reported cardiopulmonary disease status among the control population, and underlying non‐cardiopulmonary medical conditions (such as chronic kidney disease or diabetes mellitus) within the control population that were not assessed could have affected the significance of our results. We also did not obtain a detailed dietary history from our participants, and differences in fiber and macronutrient consumption between groups, which can affect both intestinal gut microbial composition as well as metabolites such as short‐chain fatty acid levels, may have confounded our results. 68 , 69 The majority of PAH subjects in our study were low‐risk and stable (as evidenced by their low REVEAL 2.0 risk scores and high six‐minute walk distance testing), and our results may not be generalizable to sicker PAH patients. We did not comprehensively study the circulating metabolome of PAH subjects in this study, and are unable to comment on the metabolomic effects of the PAH‐specific microbial compositions we observed.
In summary, this pilot study showed no differences between groups in global measures of intestinal microbial abundance, but a relative deficiency of Lachnospiraceae bacterium GAM79 in PAH as compared to non‐PAH control samples, and a relative increased abundance of Anaerostipes rhamnosivorans and lack of Amedibacterium intestinale, Ruminococcus bicirculans, and Ruminococcus albus species in PAH subjects with more severe disease (higher PVR and mPAP, lower six‐minute walk distance). The physiological effects of these microbial differences may be reflected by differences in the circulating metabolome and short‐chain fatty acid biosynthesis. Our results support further investigation into the complex relationship between the intestinal microbiome and circulating metabolite levels in PAH. Given the complexity of intestinal microbial regulation, these future studies should be designed to minimize the effect of confounding from patient level and environmental factors.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ETHICS STATEMENT
Approved by University of Cincinnati Institutional Review Board (2019‐0234).
AUTHOR CONTRIBUTIONS
Arun Jose was responsible for funding acquisition, conceptualization, data curation, analysis, writing the original draft, and review/editing the manuscript. Walaa E. Hussein and Nicholas J. Ollberding were responsible for data curation and analysis, methodology, and review/editing the manuscript. Senu Apewokin, Jean M. Elwing, and David B. Haslam were responsible for conceptualization, and review/editing of the manuscript.
Supporting information
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
The project described was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health, under Award Numbers 2UL1TR001425 (Arun Jose and Nicholas J. Ollberding) and 2KL2TR001426 (Arun Jose). This study was also supported by the National Cancer Institute of the National Institutes of Health, under Award Number 1K08CA237735‐01A1 (Senu Apewokin), and the Parker B. Francis Fellowship Program (Arun Jose). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Francis Family Foundation.
Jose A, Apewokin S, Hussein WE, Ollberding NJ, Elwing JM, Haslam DB. A unique gut microbiota signature in pulmonary arterial hypertension: A pilot study. Pulm Circ. 2022;12:e12051. 10.1002/pul2.12051
REFERENCES
- 1. Galiè N, Humbert M, Vachiery JL, Gibbs S, Lang I, Torbicki A, Simonneau G, Peacock A, Vonk Noordegraaf A, Beghetti M, Ghofrani A, Gomez Sanchez MA, Hansmann G, Klepetko W, Lancellotti P, Matucci M, McDonagh T, Pierard LA, Trindade PT, Zompatori M, Hoeper M, ESC Scientific Document Group . 2015 ESC/ERS guidelines for the diagnosis and treatment of pulmonary hypertension. Eur Heart J. 2016;37(1):67‐119. [DOI] [PubMed] [Google Scholar]
- 2. Galiè N, McLaughlin VV, Rubin LJ, Simonneau G. An overview of the 6th world symposium on pulmonary hypertension. Eur Respir J. 2018;53, 10.1183/13993003.02148-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montani D, Gunther S, Dorfmuller P, Perros F, Girerd B, Garcia G, Jaïs X, Savale L, Artaud‐Macari E, Price LC, Humbert M, Simonneau G, Sitbon O,. Pulmonary arterial hypertension. Orphannet J Rare Dis. 2013;8(97):1750‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vonk‐Noordegraaf A, Haddad F, Chin KM, Forfia PR, Kawut SM, Lumens J, Naeije R, Newman J, Oudiz RJ, Provencher S, Torbicki A, Voelkel NF, Hassoun PM Right heart adaptation to pulmonary arterial hypertension. J Am Coll Cardiol. 2013;62(25):D22‐33. [DOI] [PubMed] [Google Scholar]
- 5. Andersen S, Nielsen‐Kudsk JE, Vonk Noordegraaf A, de Man FS. Right ventricular fibrosis: a pathophysiologic factor in pulmonary hypertension. Circulation. 2019;139:269‐85. [DOI] [PubMed] [Google Scholar]
- 6. Thenappan T, Ormiston ML, Ryan JJ, Archer SL. Pulmonary arterial hypertension: pathogenesis and clinical management. BMJ. 2018;360:j5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zelniker T, Uhlmann L, Spaich S, Friedrich J, Preusch MR, Meyer FJ, Katus HA, Giannitsis E. Novel biomarkers for risk stratification in pulmonary arterial hypertension. ERJ Open Res. 2015;1(2):00008‐2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al‐Naamani N, Palevsky HI, Lederer DJ, Horn EM, Mathai SC, Roberts KE, Tracy RP, Hassoun PM, Girgis RE, Shimbo D., Post W.S., Kawut S.M., ASA‐STAT Study G. Prognostic significance of biomarkers in pulmonary arterial hypertension. Ann Am Thorac Soc. 2016;13(1):25‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nickel NP, Yuan K, Dorfmuller P, Provencher S., Lai YC, Bonnet S, Austin ED, Koch CD, Morris A, Perros F, Montani D, Zamanian RT, de Jesus Perez VA. Beyond the lungs: systemic manifestations of pulmonary arterial hypertension. Am J Respir Crit Care Med. 2020;201(2):148‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bujak R, Mateo J, Blanco I, Izquierdo‐García JL, Dudzik D, Markuszewski MJ, Peinado VI, Laclaustra M, Barberá JA, Barbas C, Ruiz‐Cabello J. New biochemical insights into the mechanisms of pulmonary arterial hypertension in humans. PLoS One. 2016;11(8):e0160505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yanagisawa R, Kataoka M, Inami T, Momose Y, Kawakami T, Takei M, Kimura M, Isobe S, Yamakado M, Fukuda K, Yoshino H, Sano M, Satoh T. Usefulness of circulating amino acid profile and Fischer ratio to predict severity of pulmonary hypertension. Am J Cardiol. 2015;115(6):831‐836. [DOI] [PubMed] [Google Scholar]
- 12. Ranchoux B, Bigorgne A, Hautefort A, Girerd B, Sitbon O, Montani D, Humbert M, Tcherakian C, Perros F. Gut‐Lung connection in pulmonary arterial hypertension. Am J Respir Cell Mol Biol. 2017;56(3):402‐405. [DOI] [PubMed] [Google Scholar]
- 13. Asosingh K, Erzurum S.Mechanisms of right heart disease in pulmonary hypertensionPulm Circ. 2018;8(1):2045893217753121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ryan JJ, Archer SL. The right ventricle in pulmonary arterial hypertension: disorders of metabolism, angiogenesis and adrenergic signaling in right ventricular failure. Circ Res. 2014;115(1):176‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kindlay S, Michel T, Leopold JA. The future of vascular biology and medicine. Circulation. 2016;133(25):2603‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui X, Ye L, Li J, Jin L, Wang W, Li S, Bao M, Wu S, Li L, Geng B, Zhou X, Zhang J, Cai J. Metagenomic and metabolomics analyses unveil dysbiosis of gut microbiota in chronic heart failure. Sci Rep. 2018;8(1):635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Callejo M, Mondejar‐Parreño G, Barreira B, Izquierdo‐Garcia JL, Morales‐Cano D, Esquivel‐Ruiz S, Moreno L, Cogolludo Á, Duarte J, Perez‐Vizcaino F. Pulmonary arterial hypertension affects the rat gut microbiome. Sci Rep. 2018;8(1):9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim S, Rigatto K, Gazzana MB, Knorst MM, Richards EM, Pepine CJ, Raizada MK. Altered gut microbiome profile in patients with pulmonary arterial hypertension. Hypertension. 2020;75(4):1063‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O'Toole PW, Cotter PD. The gut microbiota and its relationship to diet and obesity. Gut Microbes. 2012;3(3):186‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haro C, Rangel‐Zúñiga OA, Alcalá‐Díaz JF, Gómez‐Delgado F, Pérez‐Martínez P, Delgado‐Lista J, Quintana‐Navarro GM, Landa BB, Navas‐Cortés JA, Tena‐Sempere M, Clemente JC, López‐Miranda J, Pérez‐Jiménez F, Camargo A. Intestinal microbiota is influenced by gender and body mass. PLoS One. 2016;11(5):e015409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beam A, Clinger E, Hao L. Effect of diet and dietary components on the composition of the gut microbiota. Nutrients. 2021;13(8):2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60(5):940‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vujkovic‐Cvijin I, Sortino O, Verheij E, Sklar J, Wit FW, Kootstra NA, Sellers B, Brenchley JM, Ananworanich J, Loeff M, Belkaid Y, Reiss P, Sereti I. HIV‐associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat Commun. 2020;11:2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang WHW, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120(7):1183‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen JY, Geng QS, Zhang ZW, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barrington WT, Lusis AJ. Association between the gut microbiome and atherosclerosis. Nat Rev Cardiol. 2018;14(12):699‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lynch S, Oluf P. The human intestinal microbiome in health and disease. New Engl J Med. 2016;2375(24):369‐79. [DOI] [PubMed] [Google Scholar]
- 28. Yamashita T. Intestinal immunity and gut microbiota in atherogenesis. J Atheroscler Thromb. 2017;24(2):110‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xue J, Zhou D, Poulsen O, Imamura T, Hsiao YH, Smith TH, Malhotra A, Dorrestein P., Knight R, Haddad GG. Intermittent hypoxia and hypercapnia accelerate atherosclerosis, partially via trimethylamine‐oxide. Am J Respir Cell Mol Biol. 2017;57(5):581‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Durgan DJ, Ganesh BP, Cope JL, Ajami NJ, Phillips SC, Petrosino JF, Hollister EB, Bryan RM Jr. Role of the gut microbiome in obstructive sleep apnea‐induced hypertension. Hypertension. 2016;67(2):469‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77(6):2367‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raymond F, Déraspe M, Boissinot M, Bergeron M.G., Corbeil J. Partial recovery of microbiomes after antibiotic treatment. Gut Microbes. 2016;7(5):428‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Panda S, El Khader I, Casellas F, López Vivancos J, García Cors M, Santiago A, Cuenca S, Guarner F, Manichanh C. Short‐term effect of antibiotics on human gut microbiota. PLoS One. 2014;9(4):e95476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grazul H, Kanda LL, Gondek D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes. 2016;7(2):101‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787‐8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benza RL, Gomberg‐Maitland M, Elliott CG, Farber HW, Foreman AJ, Frost AE, McGoon MD, Pasta DJ, Selej M, Burger CD, Frantz RP. Predicting survival in patients with pulmonary arterial hypertension: the REVEAL risk score calculator 2.0 and comparison with ESC‐ERS‐based risk assessment strategies. Chest. 2019;156(2):323‐37. [DOI] [PubMed] [Google Scholar]
- 37. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beghini F, McIver LJ, Blanco‐Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles‐Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. Integrating taxonomic, functional, and strain‐level profiling of diverse microbial communities with bioBakery3. ELife. 2021;10:e65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McIver LJ, Abu‐Ali G, Franzosa EA, Schwager R, Morgan XC, Waldron L, Segata N, Huttenhower C. Biobakery: A meta‐omic analysis environment. Bioinformatics. 2018;34(7):1235‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith‐White B, Ako‐Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox ., Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud‐Nissen F, Tolstoy I, Tully RE, Vatsan A., Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acid Res. 2016;44:D733‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. 10.7717/peerj-cs.104 [DOI] [Google Scholar]
- 42. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson‐Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11(1):76‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, Ni Z, Nguyen TH, Andersen GL. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83(2):308‐15. [DOI] [PubMed] [Google Scholar]
- 44. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katsimichas T, Ohtani T, Motooka D, Tsukamoto Y, Kioka H, Nakamoto K, Konishi S, Chimura M, Sengoku K, Miyawaki H, Sakaguchi T, Okumura R, Theofilis K, Iida T, Takeda K, Nakamura S, Sakata Y. Non‐ischemic heart failure with reduced ejection fraction is associated with altered intestinal microbiota. Circ J. 2018;82(6):1640‐50. [DOI] [PubMed] [Google Scholar]
- 46. Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jose A. Species level count matrix and sample data. Mendeley Data, V1, 2021. 10.17632/rwsvgmpr7y.1
- 48. Kamo T, Akazawa H, Suda W, Saga‐Kamo A, Shimizu Y, Yagi H, Liu Q, Nomura S, Naito AT, Takeda N, Harada M, Toko H, Kumagai H, Ikeda Y, Takimoto E, Suzuki JI, Honda K, Morita H, Hattori M, Komuro I. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLoS One. 2017;12(3):e0174099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kummen M, Mayerhofer CCK, Vestad B, Broch K, Awoyemi A, Storm‐Larsen C, Ueland T, Yndestad A, Hov JR, Trøseid M. Gut microbiota signature in heart failure defined from profiling of 2 independent cohorts. J Am Card Cardiol. 2018;71(10):1184‐86. [DOI] [PubMed] [Google Scholar]
- 50. Luedde M, Winkler T, Heinsen FA, Rühlemann MC, Spehlmann ME, Bajrovic A, Lieb W, Franke A, Ott SJ, Frey N. Heart failure is associated with depletion of core intestinal microbiota. ESC Heart Fail. 2017;4(3):282‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Frazier K, Chang EB. Intersection of the gut microbiome and circadian rhythms in metabolism. Trends Endocrinol Metab. 2020;31(1):25‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill‐Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5(52):s40168‐017‐0267‐5. s40168017‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8(6):1323‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang J, Song L, Wang Y, Liu C, Zhang L, Zhu S, Liu S, Duan L. Beneficial effect of butyrate‐producing Lachnospiraceae on stress‐induced visceral hypersensitivity in rats. J Gastroenterol Hepatol. 2019;34(8):1368‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El‐Osta A, Mackay CR, Kaye DM. High‐fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017;135(10):964‐77. [DOI] [PubMed] [Google Scholar]
- 56. Bartolomaeus H, Balogh A, Yakoub M, Homann S, Markó L, Höges S, Tsvetkov D, Krannich A, Wundersitz S, Avery EG, Haase N, Kräker K, Hering L, Maase M, Kusche‐Vihrog K, Grandoch M, Fielitz J, Kempa S, Gollasch M, Zhumadilov Z, Kozhakhmetov S, Kushugulova A, Eckardt KU, Dechend R, Rump LC, Forslund SK, Müller DN, Stegbauer J, Wilck N. The short‐chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. 2019;139(11):1407‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE, Louis P, Flint HJ, de Vos WM Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaye DM, Shihata WA, Jama HA, Tsyganov K, Ziemann M, Kiriazis H, Horlock D, Vijay A, Giam B, Vinh A, Johnson C, Fiedler A, Donner D, Snelson M, Coughlan MT, Phillips S, Du XJ, El‐Osta A, Drummond G, Lambert GW, Spector TD, Valdes AM, Mackay CR, Marques FZ. Deficiency of prebiotic fibre and insufficient signaling through gut metabolite sensing receptors leads to cardiovascular disease. Circulation. 2020;141(17):1393‐1403. [DOI] [PubMed] [Google Scholar]
- 59. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut Lachnospiraceae. Microorganisms. 2020;8(4):573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bui T, Schols HA, Jonathan M, Stams A, de Vos WM, Plugge CM. Mutual metabolic interactions in co‐cultures of the intestinal Anaerostipes rhamnosivorans with an acetogen, methogen, or pectin‐degrader affecting butyrate production. Front Microbiol. 2019;10:2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Park W. Gut microbiomes and their metabolites shape human and animal health. J Microbiol. 2018;56(3):151‐153. [DOI] [PubMed] [Google Scholar]
- 62. Bui TPN, Mannerås‐Holm L, Puschmann R, Wu H, Troise AD, Nijsse B, Boeren S, Bäckhed F, Fiedler D, de Vos WM Conversion of dietary inositol into propionate and acetate by commensal Anaerostipes associates with host health. Nat Commun. 2021;12(1):4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Christopherson MR, Dawson JA, Stevenson DM, Cunningham AC, Bramhacharya S, Weimer PJ, Kendziorski C, Suen G. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiologic and transcriptomic analysis. BMC Genomics. 2014;15:1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang WH, Li DY, Hazen SL. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol. 2019;16(3):137‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom‐Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159‐66. [DOI] [PubMed] [Google Scholar]
- 66. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, Roberts LK, Wong CH, Shim R, Robert R, Chevalier N, Tan JK, Mariño E, Moore RJ, Wong L, McConville MJ, Tull DL, Wood LG, Murphy VE, Mattes J, Gibson PG, Mackay CR. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- 67. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate‐producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275‐83. [DOI] [PubMed] [Google Scholar]
- 68. Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El‐Osta A, Mackay CR, Kaye DM. High‐fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017;135(10):964‐77. [DOI] [PubMed] [Google Scholar]
- 69. Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235‐43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.


