Abstract
Background:
Mutations in ITCH, which encodes an E3 ubiquitin-protein ligase, can result in systemic autoimmunity and immunodeficiency. The clinical phenotype and mechanism of disease have not been fully characterized, resulting in a paucity of therapeutic options for this potentially fatal disease.
Objective:
We aimed to: (1) expand the understanding about the phenotype of human ITCH deficiency (2) further characterize the associated immune dysregulation and (3) report the first successful hematopoietic cell transplant (HCT) in a patient with ITCH deficiency.
Methods:
Disease profiling was performed in a patient with multi-system immune dysregulation. Whole exome sequencing (WES) with trio analysis and functional validation of candidate disease variants were performed, including mRNA and protein expression. Analyses to further delineate the immunophenotype included quantitative evaluation of lymphoid and myeloid subsets with flow cytometry and mass cytometry.
Results:
A patient with multi-system immune dysregulation presenting with growth failure, very early onset inflammatory bowel disease (VEO-IBD), arthritis, uveitis, psoriasis and type 1 diabetes mellitus underwent WES, which identified novel compound heterozygous mutations in ITCH. Reduced expression of ITCH mRNA and absent ITCH protein were found. Abnormalities in both lymphoid and myeloid lineages were identified. The patient underwent HCT. He demonstrated excellent immune reconstitution and resolution of many manifestations of his systemic disease.
Conclusions:
Here we report ITCH deficiency with unique clinical features of colonic VEO-IBD, arthritis and uveitis in the setting of immune dysregulation and further characterize the underlying immune dysregulation. We demonstrate that HCT can be an effective, and potentially curative, therapy for ITCH deficiency.
Keywords: hematopoietic cell transplantation, ITCH deficiency, immunodeficiency, autoimmunity, mass cytometry, very-early onset inflammatory bowel disease
INTRODUCTION
ITCH acts as an E3-ubiquitin protein ligase that transfers ubiquitin molecules to protein substrates, targeting them for proteasomal degradation (1, 2). This process serves as a key regulatory component of the immune response by down-regulating inflammatory signaling pathways and, more specifically, T cell-mediated immune responses (3–6). The first link between ITCH mutation and human disease was described in a consanguineous family of 10 affected children with a homozygous truncating frameshift mutation in ITCH. (7). The clinical manifestations of multisystem autoimmunity varied, including enteropathy in two of the ten children. Since then, two additional cases of human ITCH mutation have been described in the literature, including autoimmune manifestations of enteropathy and hepatitis (8, 9). Yet, there remains little mechanistic insight into the intestinal involvement, clinical phenotype and immunophenotype of ITCH deficiency.
While the mechanism and extent of disease in human ITCH deficiency is not well understood, there has been significant work performed in murine models. Itch−/− mice develop autoimmunity with a systemic inflammatory disease of the mucosal surfaces (10). This includes colitis that is mediated through increased IL-17 expression (11) and is associated with gut dysbiosis, with an expansion of Bacteroides species (12). These mice have been noted to have poor antibody responses to viral infection, attributed to a defect in T follicular helper cells (13). Furthermore, Itch−/− mice have increased numbers of activated T cells with CD4+ T cells skewed to a TH2 response (14), as well as elevated levels of autoantibodies and serum immunoglobulins (15). Similar findings were seen in a significant proportion of the previously described human patients, including elevated autoantibodies and clinical manifestations of atopy. These patients were treated with immunosuppression and supportive care, but many had a limited quality of life and 4 of the 12 patients previously reported in the literature were deceased (7–9). Although these insights into the immune dysregulation and clinical progression in human ITCH deficiency have set the foundation for our understanding of this disease, little further mechanistic insight into this deficiency in humans has been generated and no definitive therapy has been established for this debilitating disease. Here, we describe a case of monogenic human ITCH E3 ubiquitin ligase deficiency presenting with unique clinical features of colonic very with this disease. (VEO-IBD), severe arthritis and uveitis as primary components of multi-system immune dysregulation. Following comprehensive immunophenotyping, we describe the first successful treatment of ITCH deficiency with hematopoietic cell transplantation (HCT), which may be a viable therapeutic option for patients with this disease.
METHODS
Genetic Evaluation
Under protocol 2014–010826 approved by the Children’s Hospital of Philadelphia Institutional Review Board, genomic DNA was obtained from the child, his parents and his unaffected brother. The Agilent SureSelect Human All Exon V5 kit, targeting the coding exonic regions of the genome, was used for library preparation. Sequencing was performed using the Illumina HiSeq 2000 with 100bp paired-end reads. Sequence data was assembled and aligned to the reference human genome browser GRCh37/UCSC hg19 using Novoalign (V3.03.01; http://www.novocraft.com). Sequence variants were identified using GATK’s best practices and annotated for allele frequencies using reference databases (1000 Genomes Project, ESP, ExAC and gnomAD) and known disease variants using the Human Genome Mutation Database and NCBI’s ClinVar (16–19). Analysis for the patient included evaluating variants that are de novo, compound heterozygous, homozygous, and X-linked. Identified variants were filtered for rare (minor allele frequency < 1%) or novel missense, nonsense and frameshift mutations
EBV transformation
EBV transformation was performed on 2.9 × 106 patient peripheral blood mononuclear cells (PBMCs) after RBC lysis at the Children’s Hospital of Philadelphia Translational Core Laboratory. Control EBV-transformed cells were provided by Dr. Lieberman at the Wistar Institute. EBV-transformed cells were cultured in DMEM (Gibco) supplemented with 5% FCS (Atlanta) at 37°C and 10% CO2. Healthy donor- and patient-derived EBV-transformed cells were grown for 9 days. Cells were harvested on days 7, 8, and 9.
Western Blot
Patient-derived and control EBV-transformed cells (10×105 cells) were lysed in 1% NP-40 lysis buffer for western blot. 10ug cell lysate was run on 4–12% Bis-Tris gel (Invitrogen) and transferred to a PVDF membrane (Millipore). Membrane was probed with mouse anti-Itch (Clone 32/Itch, BD ) and rabbit anti-beta-actin (Abcam), then IR800 and IR680-conjugated secondary anti-mouse and anti-rabbit antibodies. (LI-COR). Membrane was imaged on a LI-COR Odyssey Clx imager.
Real Time PCR
Patient-derived or control EBV-transformed cells were lysed in TRIZOL (Invitrogen) and stored at −80°C. RNA was extracted with chloroform and isopropanol. cDNA was made using the SuperScript™ III First-Strand Synthesis System (Invitrogen). cDNA was amplified, detected, and analyzed using TaqMan Fast Advanced Master Mix with a QuantStudio Real Time PCR System (Applied Biosystems). Primer/probes for Itch (Hs01008308_m1) and Actb (Hs03023943_g1) were purchased from Applied Biosystems.
Flow Cytometry (Comparison of Patient and Healthy Controls)
Ficoll-Paque Plus (GE Healthcare) was used to isolate PBMCs from whole blood. Zombie Aqua was used to distinguish live cells. B cells were identified by physical characteristics and CD19. Subsets were identified by additional antibodies: IgM, IgD, CD27, CD38, CD80 and CD21. T cells were taken from physical gate followed by CD3. Subsets were determined using CD4, CD8, CD45RA, CXCR5, and ICOS. Monocytes were visualized from first the physical gate followed by CD14. CD16 was analyzed from the lymphocyte, monocyte, and neutrophil physical gates. Cells were analyzed on a LSR Fortessa (BD Biosciences) in the Children’s Hospital of Philadelphia Flow Cytometry Core.
Mass Cytometry
Reagents for mass cytometry were purchased or generated using MAXPAR kit (Fluidigm) to custom conjugate antibodies using isotope-loaded polymers. Mass cytometry antibodies in this study are shown in Table E1. Staining was performed as previously published (20). Previously cyropreserved PBMCs from the patient and a healthy control were thawed and incubated with 20μM Lanthanum-139 (Trace Sciences)-loaded maleimido-mono-amide-DOTA (Macrocyclics) in PBS 10 minutes to allow live/dead cell discrimination. Cells were washed in staining buffer and then resuspended in master mix of surface antibodies, incubated for 30 minutes at room temperature and washed twice in staining buffer. Cells were then fixed and permeabilized using the FoxP3 staining buffer kit (eBioscience) and then washed twice with 1X Permeabilization Buffer (eBioscience). Cells were then stained intracellularly for 60 minutes at room temperature. Three more washes were performed in 1X Permeabilization Buffer before being fixed using 1.6% PFA (Electron Microscopy Sciences) solution with Iridium and Osmium overnight at 4 degrees Celsius. Cells were washed twice in PBS and once in dH20 prior to data acquisition on a CyTOF Helios (Fluidigm). For data anaylsis, 50,000 live cells were exported from each participant (ITCH deficient patient and age-matched control) using FlowJo v10 (TreeStar) and data was analyzed using dimensionality reduction algorithm Phenograph within Cytofkit (21). This algorithm defines subpopulations (e.g. clusters) of healthy control and patient cells that share patterns of cell surface and intracellular protein expression in a non-supervised fashion. Live cell cluster identities were then manually annotated using relative expression of key lineage and activation markers.
RESULTS
A two-year-old boy presented with polyarthritis, growth failure and diarrhea, and by 3 years of age, was diagnosed with juvenile idiopathic arthritis. Laboratory studies demonstrated HLA-B27 and ANA positivity, with negative cyclic citrullinated peptide and rheumatoid factor. He later developed psoriasis and significant uveitis with synechiae in the left eye, leading to the diagnosis of juvenile psoriatic arthritis. He had recurrent pneumonias (3), cutaneous abscesses (2), otitis media (>5) and sinusitis (>5). Despite aggressive treatment with anti-inflammatory and immunosuppressive agents (NSAIDS, prednisone, methotrexate and etanercept), his arthritis progressed to include the cervical spine and temporomandibular joints, as well as chronic dislocation of his left patella with left knee contracture, ultimately requiring surgical correction at 13 years of age. The patient had dysmorphic features, including orbital proptosis, flattened mid-facies, small chin, posteriorly-rotated ears, shortened limbs and camptodactyly. He also had severe short stature (height Z-score −5.35) in the context of significant diarrhea and chronic steroid use (Figure 1). Infectious stool studies were negative. Due to the persistent intestinal symptoms, at age 13, he underwent additional evaluation for inflammatory bowel disease (IBD), including fecal calprotectin which was elevated to 689 mcg/g (normal <50 mcg/g) and positive IBD serology with elevated pANCA. Histology from upper endoscopy and colonoscopy revealed esophagitis and gastritis (while on infliximab, methotrexate and prednisone). Despite lack of chronic histopathologic changes, his symptoms, laboratory studies and the fact that these findings were seen while on treatment, prompted the diagnosis of IBD. Over the next few years, while on significant immunosuppressive therapy (detailed below), he developed bloody diarrhea and his fecal calprotectin was elevated to >2000 mcg/g. At age 15, upper endoscopy and colonoscopy demonstrated visual inflammation throughout the colon and histologic evidence of chronic changes, thus making a definitive histologic diagnosis of IBD (Figure 2A).
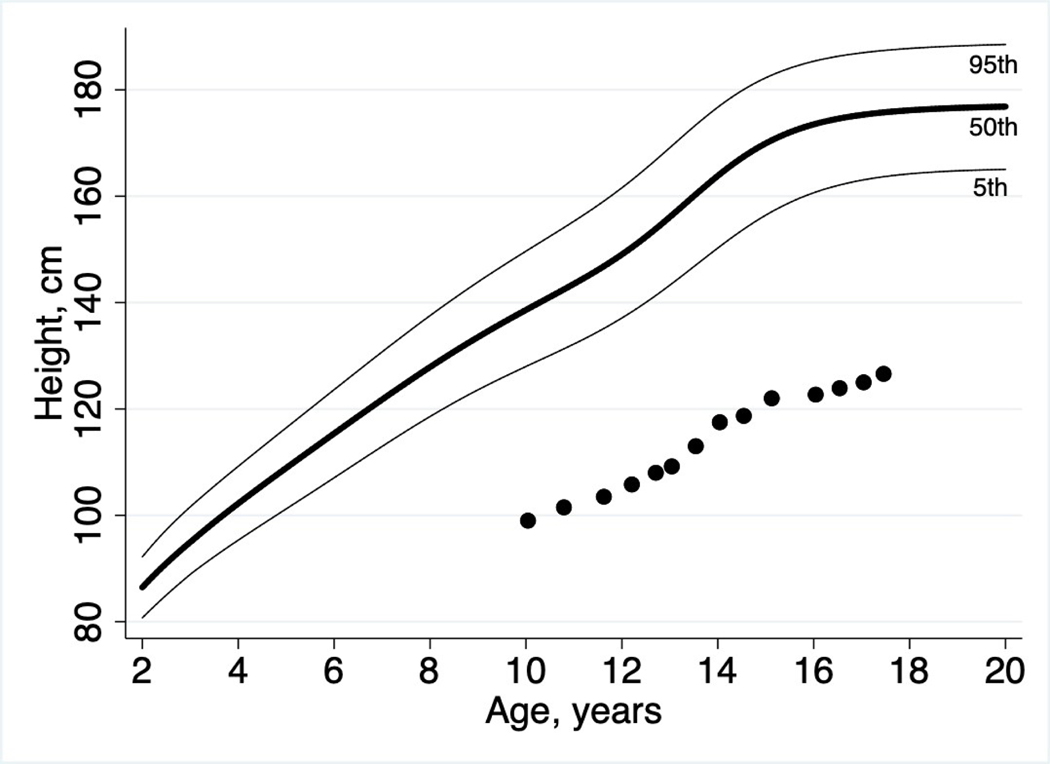
Figure 1: Growth Failure.
Marked growth failure was a key presenting feature in this patient. Height Z-score at the time of presentation to our clinic was −5.35, with proportionately low weight, thus resulting in a normal BMI of 19.06 kg/m2
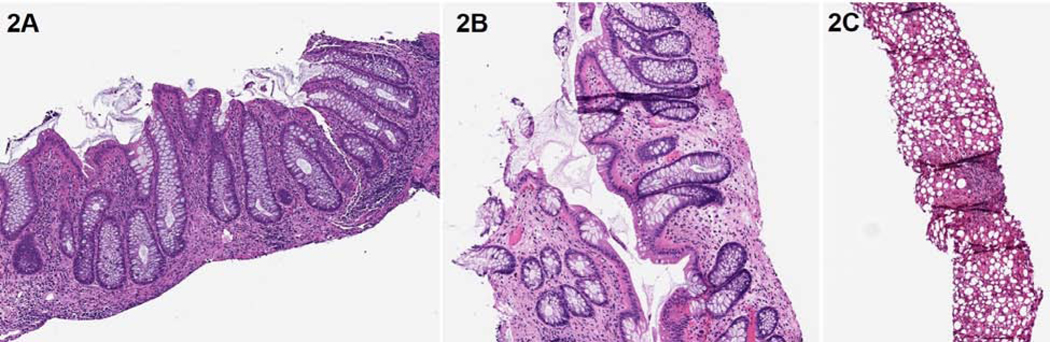
Figure 2: Histologic Findings.
Colonoscopy 3 months prior to HCT demonstrated chronic active pancolitis characterized by crypt architectural distortion and prominent mononuclear infiltrate with focal neutrophilic cryptitis (2A).Colonic biopsy 15 months after HCT demonstrated a net decrease in the inflammatory infiltrate though some crypt architectural distortion remains, consistent with inactive colitis (2B). Liver biopsy 3 years prior to HCT demonstrated steatosis, mild portal inflammation and fibrous expansion (2C)
At age 14, the patient was diagnosed with diabetes with autoantibodies (GAD65+) and hemoglobin A1c of 10.2%, and initiated chronic insulin therapy. Importantly, chronic glucocorticsteroid therapy may have impacted his glucose tolerance. He also had hepatitis characterized by hepatic steatosis and mild portal inflammation despite immunosuppressive therapy with prednisone and golimumab at the time of biopsy (Figure 2C). Liver synthetic function was preserved. The patient had asthma and eczema. Pulmonary function testing at various time points revealed a mixed obstructive and restrictive pattern attributed to a combination of mild interstitial lung disease, asthma, and restrictive lung disease suspected to be related to skeletal dysplasia. He also had obstructive sleep apnea managed with continuous positive airway pressure overnight.
Numerous therapies were attempted in various combinations for his symptoms, with incomplete or poor response, including methotrexate, chronic glucocorticoid therapy, etanercept, infliximab, adalimumab, golimumab, ustekinumab, abatacept and tacrolimus (Figure E1).
The combination of multiple autoimmune disorders (inflammatory arthritis, uveitis, IBD and psoriasis) refractory to standard therapies, as well as dysmorphic features, severe short stature and hearing loss led to high suspicion for an underlying genetic defect. This prompted a multidisciplinary evaluation, including whole exome sequencing (WES) and extensive clinical phenotyping and immunophenotyping.
WES revealed two novel variants in the ITCH gene (NM_001257137) in compound heterozygous status: c.337+2T>C (splice donor) and c.772C>T, p.Arg258* (stop gain) (Figure 3). Neither variant has been reported in the literature or public databases. The c.337+2T>C is located at the canonical splicing donor site, and is predicted to abolish normal splicing. The c.772C>T variant is predicted to result in a premature stop condon (p.Arg258*), and is expected to undergo nonsense-mediated decay. Both variants were predicted to be deleterious by MutationTaster with CADD scores of 25.3 and 31, respectively. The mRNA expression analysis in patient EBV-transformed B cells a significant reduction of over 50% compared to control ITCH mRNA (Figure 4A). In addition, there was no detectable ITCH protein in patient EBV-transformed B cells by western blot analysis (Figure 4B). Both assays confirmed the loss-of-function nature of these two variants and impairment of the ITCH protein function. To date, all reported variants in the ITCH gene have been loss-of-function variants (7–9). Of note, while the patient’s parents were both carriers, neither shared the patient’s clinical features, consistent with the autosomal recessive mode of inheritance of defects in the ITCH gene.
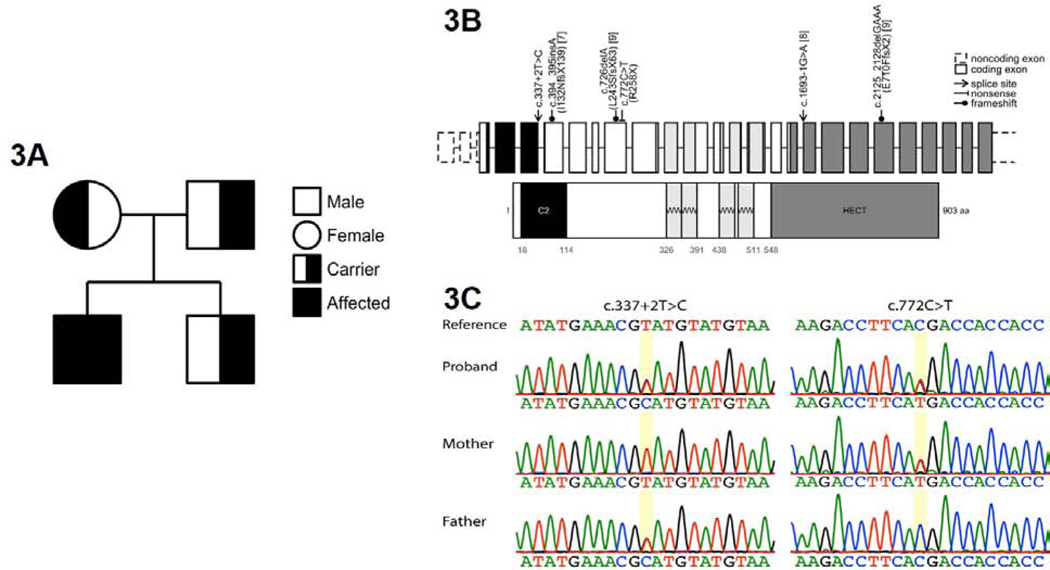
Figure 3: Genetic Evaluation.
The proband demonstrated compound heterozygous mutation in ITCH gene. The proband’s parents and sibling were heterozygous and did not demonstrate the corresponding clinical phenotype (3A). Genetic mutations found in our patient and the previously-published patients are represented here, with corresponding reference numbers (3B). Sanger sequencing was used to confirm the compound heterozygous mutations in this patient (3C).
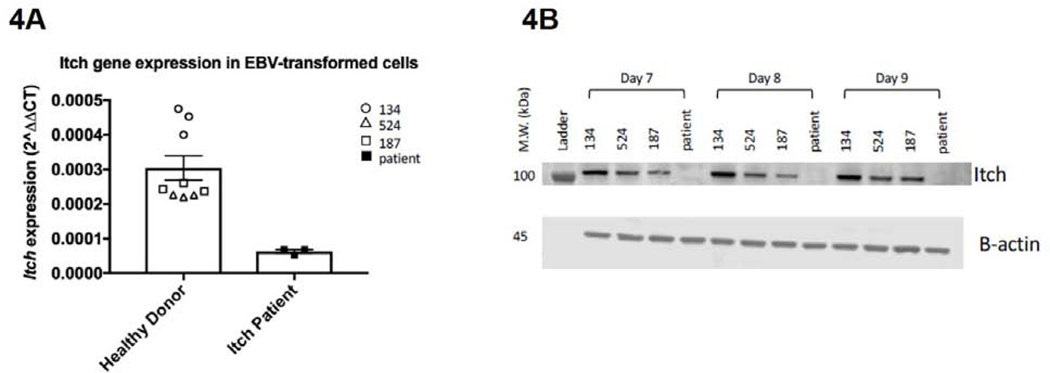
Figure 4: ITCH mRNA and Protein Analysis.
EBV-transformed B cell mRNA expression analysis demonstrated less than 50% of healthy control ITCH mRNA in the patient (4A).Western blot analysis on patient EBV-transformed B cells and healthy control cells demonstrated that ITCH protein was not expressed in the patient, but was preserved in the controls. Itch expression was normalized to beta actin. (4B)
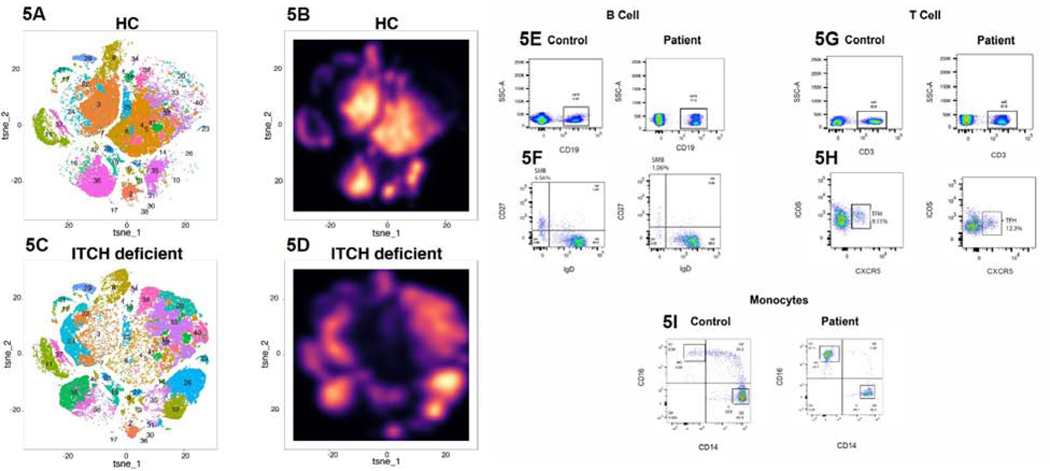
Mass cytometry (CyTOF) using PBMCs was performed to assess immunophenotype on a global scale comparing our patient to healthy controls. In Figure 5, we demonstrate striking alterations in the patient’s PBMC immunophenotype, including both lymphoid and myeloid compartments, as compared to an age-matched healthy control (HC)(Figure 5A−D) (22). Flow cytometry studies were performed in two independent research laboratories to corroborate these findings and assess individual populations more specifically (Figure 5E−I). These studies demonstrated quantitatively normal total B lymphocytes, but abnormal B cell development, as evidenced by lower than expected switched-memory B cells for his age. He had mild T-cell lymphopenia, with elevated T follicular helper cells. The patient also had low total monocytes, with increased non-classical monocytes and decreased classical monocytes. T regulatory cells were evaluated by CyTOF and were overexpressed in the patient as compared to the healthy control (Figure 5A/C, cluster 39). It should be noted that, at the time of these evaluations, the patient was on multiple immunosuppressive therapies, including tacrolimus, abatacept, ustekinumab and prednisone 20 mg daily. Nine months later, after discontinuation of tacrolimus and decrease in prednisone dose to 15 mg daily, clinical flow cytometry demonstrated quantitatively normal T cells. The patient had normal quantitative immunoglobulins, but poor T cell-independent B cell responses to polysaccharide-based pneumococcal vaccine challenge.
Figure 5: Immunophenotyping of ITCH Deficiency.
CyTOF performed on PBMCs from healthy control and ITCH-deficient patient demonstrated significant alterations in immunophenotype on a global scale (5A–5D). In 5A and 5C, clusters are displayed as dot plots,with individual coloration and numbering for each cluster (detailed in Table E2). In 5B and 5D, clusters are displayed as amplitude of cell counts (or “galaxy plots”). Flow cytometry studies demonstrated quantitatively normal B cells (5E), low switched-memory B cells (5F), mildly decreased T cells (5G), elevated T follicular helper cells (5H), low total monocytes, increased non-classical monocytes and decreased classical monocytes (5I).
In the setting of an identified defect in the ITCH gene impacting hemopoietic cells, with evidence of immune dysfunction demonstrated with CyTOF and flow cytometry, as well as prior murine data demonstrating that autoimmunity in Itchy mice is intrinsic to the hematopoietic system, the decision was made to pursue HCT for definitive management of this patient’s multisystem disease (15). An HLA-matched sibling donor was identified as a donor for HCT. Although his sibling donor was a carrier of one of the ITCH gene variants (c.337+2T>C) and had some findings consistent with atopy, including seasonal allergies and eczema, he was otherwise healthy. He underwent a comprehensive evaluation that did not identify evidence of autoimmunity or immune dysregulation, and was therefore deemed a suitable donor source. The patient underwent HCT with donor bone marrow at the age of 16 years. Conditioning included busulfan from day −7 to −4 (daily dosing titrated to goal AUC of 3600–6000 μmole*min/L), fludarabine 40 mg/m2 × 4 from day - 5 to −2 and rabbit thymoglobulin 3 mg/kg x 3 from day −7 to day −5. Graft-versus-host disease (GVHD) prophylaxis included cyclosporine starting on day 1, transitioned to tacrolimus once oral administration could be tolerated, and methotrexate 5 mg/m2/dose on days +1, +3 and +6. Myeloid engraftmen occurred at day +14 and there was no GVHD or other complications. Prednisone was discontinued by 2 months post-HCT.
Chimerism evaluation by single tandem repeats (STRs) at one month showed 97% donor cells; at 2 months he was 98% overall, with 100% myeloid, 57% T, 97% NK and 99% B. Tacrolimus prophylaxis was discontinued by 4 months post-HCT. Immunologic evaluation at 12 months post-transplant demonstrated good immune reconstitution with normal overall T cell counts, demonstration of normal thymic output and near normal T cell proliferation to phytohemaglutinin (Table 1). T cell receptor beta chain variable region spectratyping performed at 3 and 28 months post-HCT demonstrated a mainly polyclonal repertoire. His B cell counts are higher than expected for age, with switched memory B cell present in low numbers, a common finding in the post-HCT period. It is notable that he never developed hypogammaglobulinemia and his immunoglobulin levels have decreased to within the normal range since HCT. As mentioned above, prior to transplant the patient had a dysregulated humoral response with autoantibodies present but poor response to polysaccharide based challenge. An improved ability for appropriate response to vaccine challenge is suggested by his normal isohemagluttinin titers post-HCT. His NK cell counts were normal. His chimerism analysis by STRs at 1 year demonstrated 97% total, 99% myeloid, 86% T cell and 99% B cell donor engraftment. By 2 years post-HCT, the patient had normal T, B and NK cell counts, with a normal phytohemagglutinin (PHA) proliferative response and protective titers to inactivated vaccine challenge (Table 1).
Table 1:
Clinical Immunology Lab Findings Prior to and Following HCT
| Reference Range | Pre-HCT | 3 months post HCT | 1 year post HCT | 2 years post HCT | |
|---|---|---|---|---|---|
| CD3 (cells/μL) | 700–2100 | 1802 | 311 | 1171 | 1153 |
| CD4 (cells/μL) | 300–1400 | 1208 | 112 | 551 | 673 |
| CD8 (cells/μL) | 200–900 | 504 | 167 | 557 | 384 |
| CD4/CD45RA (cells/μL) | 41–1121 | 712 | 14 | 282 | -- |
| CD4/CD45RO (cells/μL) | 153–582 | 453 | 90 | 247 | -- |
| CD19 (cells/μL) | 100–500 | 499 | 172 | 1740 | 1182 |
| CD19+/CD27+/IgD- (cells/μL) | 9–102 | 13 | 0 | 3 | -- |
| NK (cells/μL) | 90–600 | 279 | 406 | 254 | 179 |
| IgG (mg/dL) | 635–1775 | 1240 | 755 | 744 | 809 |
| IgA (mg/dL) | 70–486 | 152 | 102 | 80 | 104 |
| IgM (mg/dL) | 71–237 | 132 | 54 | 42 | 57 |
| TREC (cells/μL) | ≥3061 | -- | 638 | 8663 | 9135 |
| PHA (% of control) | n/a | -- | 28% of control | 46% of control | >50% of control |
| Isohemagluttini ns | n/a | -- | -- | 1:16 | -- |
| Tetanus Titer (IU/mL) | ≥ 1 | 2.8 | |||
| Chimerism:PB MC | n/a | -- | 99% | 97% | -- |
| Chimerism:T | n/a | -- | 88% | 86% | -- |
| Chimerism:B | n/a | -- | 100% | 99% | -- |
| Chimerism:M | n/a | -- | 100% | 99% | -- |
Table 1: Clinical immunologic parameters pre- and post-HCT. Age-specific reference ranges are provided where appropriate. Post-transplant, the patient demonstrated excellent immune reconstitution, with normal T cell counts by 1 year and normal T cell proliferation to phytohemaglutinin by 2 years, as well as excellent response to tetanus vaccine challenge with protective titers at 2 years post transplant.
Upon clinical evaluation at both 1 and 2 years post-transplant, the patient remains off of all immunosuppressive therapy and there has been no recurrence of arthritis, uveitis, psoriasis or IBD. Repeat upper endoscopy and colonoscopy at 15 months post-transplant demonstrated ongoing mild crypt architectural distortion, but without any evidence of active inflammation (Figure 2B). His mobility and activity tolerance have improved significantly. The patient’s asthma, eczema and allergic rhinitis are all well-controlled on maintenance therapies. Glycemic control was improved post-transplant and insulin therapy was briefly stopped from 11–20 months post-transplant, but long-acting insulin therapy was resumed at 20 months post-transplant. The patient continued with appropriate weight gain and normal BMI and grew 7 cm in height in the 2 years post-transplant, but was unable to reach full adult height potential given advanced pubertal status at the time of transplant.
DISCUSSION
Here, we expand the understanding of the phenotype of human ITCH deficiency to include pan-enteric VEO-IBD, severe arthritis and uveitis in the context of multi-system immune dysregulation and describe the first successful treatment with HCT in human ITCH deficiency. We further delineate the immunophenotype as a dysregulation of both lymphoid and myeloid compartments, with mild T cell lymphopenia and poor B cell maturation, as well as decreased total monocytes with increased non-classical monocytes and decreased classical monocytes. Notably, myeloid defects have not been previously described in human ITCH deficiency.
While the previously-published cases of human ITCH deficiency have been heterogeneous in both presentation and severity of disease, there are several consistent clinical features among all of the previously-described patients and the patient presented in this study (7–9). Universal features include short stature, developmental delay and dysmorphic facial features and nearly all patients, including our patient, have had relative macrocephaly and chronic lung disease (Table 2). Autoimmune inflammatory cell infiltration of multiple organs, including lungs, pancreas, liver and gut has been noted to underlie many of the clinical features of this condition. However, unique to our patient’s phenotype were severe arthritis, uveitis and chronic colonic inflammation, as compared to the published studies that described the GI disease to be limited to lymphocytic inflammation of the small bowel, demonstrating the extent and heterogeneity of the clinical phenotype of human ITCH deficiency. Interestingly, the colitis noted in this patient is consistent with prior murine studies, including a recent study which found that Itch deficiency led to spontaneous colitis and increased susceptibility to colitis-associated colon cancer (CACC) in mice. The authors demonstrated that the Itch deficient mice had higher levels of IL-17 in the inflamed colon mucosa and in the cells that had infiltrated tumor cells. Together, the investigators concluded that the Itch is essential for inhibition of IL-17, which is a key component of colonic inflammation and CACC (11).
Table 2:
Comparison of clinical findings in published ITCH-deficient patients
| Our Patient (n=1) | Lohr et al (n=10) | Kleine-Eggebrecht et al (n=1) | Brittain et al (n = 1) | |
|---|---|---|---|---|
| Hepatomegaly and/or splenomegaly | − | + (9/10) | + | − |
| Short Stature | + | + (10/10) | + | + |
| Developmental delay | + | + (10/10) | + | + |
| Motor Delay | + | + (10/10) | + | + |
| Cognitive Delay | + | + (10/10) | + | − |
| Dysmorphic features | + | + (10/10) | + | + |
| Relative macrocephaly | + | + (9/10) | + | + |
| Chronic lung disease | + | + (9/10) | + | + |
| Hypotonia | + | + (6/10) | + | − |
| Pancytopenia | − | − | + | − |
| Acute liver failure | − | − | + | − |
| Autoimmune disease | + | + (6/10) | + | − |
| Hypothyroidism | − | + (4/10) | + | − |
| Hepatitis | + | + (3/10) | + | − |
| Enteropathy and/or IBD | + (pancolitis) | +(2/10) (enteropathy) | + (enteropathy) | − |
| Diabetes mellitus | + | + (1/10) | + | − |
| Arthritis | + | − | − | − |
| Uveitis | + | − | − | − |
| Psoriasis | + | − | − | − |
Table 2: A comparison of clinical phenotyping of the currently-published cases of human ITCH mutation are summarized here. Of note, short stature, developmental delay (of various types), dysmorphic facial features, relative macrocephaly and chronic lung disease were found in all reported cases. Other features were variable.
While there is thus far a limited understanding about the immunophenotype associated with human ITCH deficiency, there have been many in vitro studies and in vivo murine studies exploring this topic. Classically, the immune dysregulation in ITCH deficiency is thought to be mediated via T cell dysregulation. It is important to note that our patient was on multiple immunosuppressive medications that may have affected his initial immunophenotype, particularly his T cell lymphopenia, and changes in his treatment regimen could have impacted his repeat immunologic studies. However, his effector-skewed T cells were consistent with the shift towards activated-memory CD4 and CD8 T cells as seen in Itch−/− mice (14). Furthermore, a recent study by Liu et al. demonstrated that Itch plays a role in B cell development (23), which appears to be consistent with the decreased switched-memory B cells seen in our patient.
Prior work in Itch-deficient mice demonstrated that adoptive transfer of bone marrow cells from Itch−/− mice into sub-lethally irradiated Rag1−/− mice could recapitulate the immune disorder of the donor mice (15). These data support a disease mechanism that is intrinsic to hematopoietic cells. This understanding, together with our patient’s clinical and immunophenotype, motivated the decision to proceed with HCT in our patient. To the best of our knowledge, HCT has not previously been attempted, and this is the first reported definitive therapeutic intervention with HCT in a patient with ITCH deficiency. This treatment has been highly successful thus far to improve and/or reverse many of his clinical symptoms, and is anticipated to prevent the development of life-threatening complications of his disease. Notably, the patient’s arthritis, uveitis, IBD and psoriasis remain in remission off of immunosuppressive therapy at 2 years post-transplant. He does require ongoing therapy for restrictive lung disease, asthma, allergies, and diabetes. However, these conditions are pathophysiologically distinct from the aforementioned autoimmune conditions and, in the case of restrictive lung disease caused by skeletal dysplasia and likely islet cell destruction caused by autoimmune diabetes, would not be expected to be reversed by HCT.
This study describes severe multi-system immune dysregulation in ITCH deficiency and altered function of nearly all hematopoietic lineages. With such protean effects on hematopoietic cells in human ITCH deficiency, HCT can be life-saving. While ITCH deficiency has been linked with enteropathy, the GI involvement in the prior cases in the literature are not histologically or phenotypically consistent with classic IBD. This current case should further prompt clinicians to consider ITCH mutations in the list of genetic causes of IBD and in cases of unexplained multi-system immune dysregulation with the aforementioned clinical features. Moreover, this may lead to further insight into the potential phenotypes of similar defects in this pathway involving ubiquitin conjugation, proteasomal degradation and apoptosis. This case also underscores the importance of considering HCT as a therapeutic approach in patients with ITCH deficiency and related defects early in the course of disease, to prevent debilitating or fatal complications. With currently-available sequencing technologies, together with a multidisciplinary team approach, it is likely that more single-gene defects will be identified and result in targeted therapeutic strategies that can be life-saving for children with multi-system immune dysregulation.
Supplementary Material
HIGHLIGHTS.
What is already known about this topic? Human ITCH deficiency results in systemic autoimmunity and immunodeficiency. The clinical phenotype has previously been described, but the immunophenotype has been poorly characterized and no definitive therapy has been reported for this devastating condition.
What does this article add to our knowledge? This article expands understanding about the clinical phenotype (including colitis, arthritis and uveitis) and immunophenotype (both lymphoid and myeloid alterations) of human ITCH deficiency. We report the first successful hematopoietic cell transplantation for management of this condition.
How does this study impact current management guidelines? Clinicians caring for patients with ITCH deficiency may consider hematopoietic cell transplantation for management of multisystem immune dysregulation.
ACKNOWLEDGMENTS
The authors would like to thank the following individuals: Dr. Babette Zemel at the Bionutrition Core Laboratory at Children’s Hospital of Philadelphia for analysis and graphical representation of growth; Dr. Jenny Knight and the Department of Pathology at Prisma Health System for preparing, reviewing and sharing histology slides from the patient’s endoscopies; Dr. Lieberman at the Wistar Institute for providing EBV-transformed control cell lines; and Dr. Nancy B. Spinner and her team in the Division of Genomic Diagnostics at Children’s Hospital of Philadelphia for assistance with genetic sequencing and interpretation.
Sources of Funding:
National Institutes of Health Grant Numbers 1K23DK100461-01A1 (Judith R. Kelsen), 5R01DK111843-02 (Marcella Devoto), 5K12HD043245 (Sarah E. Henrickson), K08AI135091 (Sarah E. Henrickson) and K23DK119585 (Maire Conrad) Ann and Richard Frankel Endowed Chair in Gastroenterology Research, Children’s Hospital of Philadelphia (Judith R. Kelsen)
ABBREVIATIONS:
- HCT
hematopoietic cell transplantation
- IBD
inflammatory bowel disease
- VEO-IBD
very early-onset inflammatory bowel diease
- CyTOF
mass cytometry by time-of-flight
- PBMC
peripheral blood mononuclear cell
- PHA
phytohemagglutinin
- STR
single tandem repeat
Footnotes
Frank R. Wallace Endowed Chair in Infectious Diseases, Children’s Hospital of Philadelphia (Kathleen E. Sullivan)
Clinical Immunology Society (Sarah E. Henrickson)
Burroughs Wellcome Foundation (Sarah E. Henrickson)
Parker Institute for Cancer Immunotherapy (E. John Wherry)
Conflicts of Interest: The authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rathinam VA, Fitzgerald KA. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell. 2016;165(4):792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottin C, Fel A, Butel N, Domont F, Remond AL, Savey L, et al. Anakinra in the Treatment of Patients with Refractory Scleritis: A Pilot Study. Ocul Immunol Inflamm. 2018;26(6):915–20. [DOI] [PubMed] [Google Scholar]
- 3.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat Immunol. 2004;5(9):883–90. [DOI] [PubMed] [Google Scholar]
- 4.Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. 2005;6(5):472–80. [DOI] [PubMed] [Google Scholar]
- 5.Jeon MS, Atfield A, Venuprasad K, Krawczyk C, Sarao R, Elly C, et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21(2):167–77. [DOI] [PubMed] [Google Scholar]
- 6.Matesic LE, Copeland NG, Jenkins NA. Itchy mice: the identification of a new pathway for the development of autoimmunity. Curr Top Microbiol Immunol. 2008;321:185–200. [DOI] [PubMed] [Google Scholar]
- 7.Lohr NJ, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet. 2010;86(3):447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleine-Eggebrecht N, Staufner C, Kathemann S, Elgizouli M, Kopajtich R, Prokisch H, et al. Mutation in ITCH Gene Can Cause Syndromic Multisystem Autoimmune Disease With Acute Liver Failure. Pediatrics. 2019. [DOI] [PubMed] [Google Scholar]
- 9.Brittain HK, Feary J, Rosenthal M, Spoudeas H, Deciphering Developmental Disorders S, Wilson LC. Biallelic human ITCH variants causing a multisystem disease with dysmorphic features: A second report. Am J Med Genet A. 2019;179(7):1346–50. [DOI] [PubMed] [Google Scholar]
- 10.Perry WL, Hustad CM, Swing DA, O’Sullivan TN, Jenkins NA, Copeland NG The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18(2):143–6. [DOI] [PubMed] [Google Scholar]
- 11.Kathania M, Khare P, Zeng M, Cantarel B, Zhang H, Ueno H, et al. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-gammat ubiquitination. Nat Immunol. 2016;17(8):997–1004. [DOI] [PubMed] [Google Scholar]
- 12.Kathania M, Tsakem EL, Theiss AL, Venuprasad K. Gut Microbiota Contributes to Spontaneous Colitis in E3 Ligase Itch-Deficient Mice. J Immunol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao N, Eto D, Elly C, Peng G, Crotty S, Liu YC. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol. 2014;15(7):657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol. 2002;3(3):281–7. [DOI] [PubMed] [Google Scholar]
- 15.Parravicini V, Field AC, Tomlinson PD, Basson MA, Zamoyska R. Itch−/− alphabeta and gammadelta T cells independently contribute to autoimmunity in Itchy mice. Blood. 2008;111(8):4273–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA: (URL: http://evs.gs.washington.edu/EVS/) [July 2017]. [Google Scholar]
- 18.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski KJ. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. [Google Scholar]
- 20.O’Boyle KC, Ohtani T, Manne S, Bengsch B, Henrickson SE, Wherry EJ, et al. Exploration of T-Cell Diversity Using Mass Cytometry. Methods Mol Biol. 2020;2111:1–20. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS Comput Biol. 2016;12(9):e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162(1):184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Zhang Y, Wei Y, Wang Z, Zhu G, Fang Y, et al. The E3 ubiquitin ligase Itch is required for Bcell development. Sci Rep. 2019;9(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.