ABSTRACT
The Mycobacterium tuberculosis genome harbors nine toxin-antitoxin (TA) systems that are members of the mazEF family, unlike other prokaryotes, which have only one or two. Although the overall tertiary folds of MazF toxins are predicted to be similar, it is unclear how they recognize structurally different RNAs and antitoxins with divergent sequence specificity. Here, we have expressed and purified the individual components and complex of the MazEF6 TA system from M. tuberculosis. Size exclusion chromatography–multiangle light scattering (SEC-MALS) was performed to determine the oligomerization status of the toxin, antitoxin, and the complex in different stoichiometric ratios. The relative stabilities of the proteins were determined by nano-differential scanning fluorimetry (nano-DSF). Microscale thermophoresis (MST) and yeast surface display (YSD) were performed to measure the relative affinities between the cognate toxin–antitoxin partners. The interaction between MazEF6 complexes and cognate promoter DNA was also studied using MST. Analysis of paired-end RNA sequencing data revealed that the overexpression of MazF6 resulted in differential expression of 323 transcripts in M. tuberculosis. Network analysis was performed to identify the nodes from the top-response network. The analysis of mRNA protection ratios resulted in identification of putative MazF6 cleavage site in its native host, M. tuberculosis.
IMPORTANCE M. tuberculosis harbors a large number of type II toxin-antitoxin (TA) systems, the exact roles for most of which are unclear. Prior studies have reported that overexpression of several of these type II toxins inhibits bacterial growth and contributes to the formation of drug-tolerant populations in vitro. To obtain insights into M. tuberculosis MazEF6 type II TA system function, we determined stability, oligomeric states, and binding affinities of cognate partners with each other and with their promoter operator DNA. Using RNA-seq data obtained from M. tuberculosis overexpression strains, we have identified putative MazF6 cleavage sites and targets in its native, cellular context.
KEYWORDS: cleavage recognition site, endoribonuclease, gene regulation, maz, rRNA, toxin activity
INTRODUCTION
Toxin-antitoxin (TA) systems are response elements that have evolved in free-living bacteria to combat stress conditions like temperature, antibiotics, and other metabolites, enabling them to survive (1–5). TA systems are typically arranged in a bicistronic manner with elements comprising of an upstream gene encoding a labile antitoxin and a downstream gene encoding stable toxin (1–6). Depending on the nature of the antitoxin and its mechanism of toxin neutralization, TA modules can be categorized into eight distinct types. In different subfamilies, the toxin is either a protein (type I to VII) or a small noncoding RNA (type VIII), which, in the absence of antitoxin, inhibits essential cellular functions such as replication, transcription, translation, etc. (5, 7, 8). The antitoxin is either RNA or protein and inhibits the activity of the toxin by direct binding (type II, III, and VI), preventing expression of toxin by degrading mRNA or sRNA of toxins (type I, V, and VIII), competing with the toxin for its substrate (type IV), or by posttranslational modification by an enzyme antitoxin (type VII) (8–10). The most abundant and well-characterized subfamily is the type II TA systems, where both the toxin and the antitoxin are proteins and antitoxin forms a tight complex with toxin, thereby neutralizing its activity (11).
Previous studies have indicated that TA pairs play critical roles in diverse functions, including plasmid maintenance, phage resistance, phage infectivity, adaptive responses to adverse conditions, antibiotic drug tolerance and persistence, biofilm formation, and microbial pathogenesis (5, 12–20). While it was initially shown that the simultaneous deletion of 10 TA systems in E. coli decreased the number of persisters after exposure to either ciprofloxacin or ampicillin, it was subsequently concluded that the increased drug susceptibility of the TA deletion strain was due to lysogenization by multiple copies of Phi80 phages (21). Notwithstanding this confounding factor, other studies suggest that type II TA systems have a role in bacterial drug tolerance and biofilm formation (15, 16, 22, 23). However, this is a controversial area and more work is required. Additionally, the physiological conditions under which TA modules are activated and their potential roles upon activation need further exploration.
The Mycobacterium tuberculosis genome encodes a large number of type II TA systems, such as mazEF, relBE, higBA, parDE, and vapBC (24–27). Previous studies have reported that the overexpression of the majority of these toxins inhibit growth (24, 28–31) and also contribute to the formation of drug-tolerant populations in vitro (32–34).
The mazEF operon encodes the labile antitoxin MazE and the stable toxin MazF (35–37). MazF toxins are sequence-specific endoribonucleases that cleave free RNA (35–38). The Escherichia coli and Bacillus subtilis MazFs cleave RNAs containing 5′-N↓ACA-3′ and 5′-U↓ACAU-3′ sequences, respectively (7, 38). The M. tuberculosis genome harbors nine TA systems of the MazEF family (24, 27). Previous studies have shown that MazF1, MazF3, MazF4, MazF6, and MazF9 recognize and cleave structurally distinct, unique RNA sequences (39–43). The cleavage specificities of the remaining (MazF2, MazF5, MazF7, and MazF8) have not been studied and therefore are still unknown. The three-dimensional structures of MazEF4 (PDB entry 5XE3), MazEF7 (PDB entry 6A6X), and MazEF9 (6KYT) of M. tuberculosis have been solved (44–46). The MazEF3 and MazEF9 complex adopts a heterohexamer conformation, whereas MazEF7 adopts a heterotetramer conformation. The overall homology among MazF toxins is relatively poor (47). Therefore, structural information on MazEF complexes along with scanning mutagenesis studies (47) and low-resolution electron microscopy (EM) will lead to identification of critical residues that are involved in interactions between toxin and antitoxin.
In the current study, we performed detailed functional and biochemical characterization of the MazEF6 TA system from M. tuberculosis. Using nano-differential scanning fluorimetry (nanoDSF), we have measured the relative stabilities of the MazE6, MazF6, and MazEF6 complex. In addition, size exclusion chromatography coupled to multiangle light scattering (SEC-MALS) was performed to determine the oligomeric status of the free toxin, antitoxin, and the TA complexes, which were in vitro reconstituted using different ratios of toxin and antitoxin. We also performed yeast surface display (YSD) and microscale thermophoresis (MST) to determine the relative binding affinities of the toxin with its cognate full-length antitoxin as well as the C-terminal antitoxin peptide. MST was also employed to determine the relative affinities of antitoxin alone or the toxin-antitoxin complex, assembled in vitro at different ratios, for binding to mazEF6 promoter-operator DNAs. Transcriptome sequencing (RNA-seq) experiments were performed to identify differentially expressed genes upon MazF6 overexpression. A detailed bioinformatic analysis was performed to identify putative MazF6 cleavage sites, cleavage specificities, and substrates in its native, cellular context.
RESULTS
Purification and thermal stability analysis of MazE6, MazF6, and MazEF6 complex.
The MazEF6 complex was purified using nickel nitrilotriacetic acid (Ni-NTA) affinity chromatography following expression in Escherichia coli from the pET-Duet-1 vector, as the MazF6 toxin had an N-terminal 6× His tag (see Fig. S1a in the supplemental material). The proteins MazE6 and MazF6 were individually purified from the pET-15b vector with an N-terminal 6× His tag and C-terminal 3× FLAG tag (Fig. S1b). The buffer conditions were optimized for the purification of the proteins. Removing imidazole and other buffer components was associated with formation of visible aggregates. The aggregation was reduced by addition of 10% glycerol in all the buffers used in the purification process. In the present study, purified proteins MazE6 [(His)6MazE6(FLAG)3] and MazF6 [(His)6MazF6(FLAG)3], MazEF6 complex [MazE6:MazF6(His)6], and the in vitro-assembled MazEF6 complex [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3] of different stoichiometries were subjected to thermal denaturation as described in Materials and Methods.
The unfolding was monitored using intrinsic fluorescence of tryptophan and tyrosine residues with temperature, and the apparent midpoint temperature (Tm) was calculated. Free MazE6 and MazF6 had Tms of 60°C and 50°C, respectively (Fig. 1a). The thermal stability of TA complexes has been reported to be higher than that of the free toxins and antitoxins (48–51). In agreement, we also found a higher thermal stability of the in vivo-assembled MazEF6 complex (Tm = 85°C) compared to the free toxin or antitoxin, indicating stabilization of both the proteins upon complex formation (Fig. 1a). The relative stabilities of the in vitro-assembled MazEF6 complexes were also higher than those of the individual toxin and antitoxin (Fig. 1b). As shown in Fig. 1c, the stability of the complex increased (in the range of 82°C to 91°C), with an increase in the antitoxin concentration. This might be due to the formation of higher-order oligomers (hetero-octamer/hexamers) in the presence of excess MazE6 or MazF6, which is further validated from the SEC-MALS experiments as described below.
FIG 1.
Thermal stabilities of MazEF6 toxin-antitoxin system measured by nanoDSF. (a) Thermal unfolding profiles of 10 μM purified MazE6 [(His)6MazE6(FLAG)3] (green), MazF6 [(His)6MazF6(FLAG)3] (red), and copurified MazEF6 complex [MazE6:MazF6(His)6] (blue). (b) Thermal unfolding profiles at different ratios of in vitro-assembled MazEF6 complexes [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3]. (c) With increase in concentration of the antitoxin, the stability of the complexes increases and the stability saturates at an approximate antitoxin:toxin ratio of 6:1. The error bars in panel c represent the standard errors derived from two independent experiments, each performed in duplicate.
Previous studies have shown that the fluorescence of the tryptophan and tyrosine residues in a protein is strongly dependent on the surrounding environment (52), and changes in the structure of the protein have an effect on both the fluorescence intensity and the emission wavelength.
Analysis of the oligomeric state of MazE6, MazF6, and MazEF6.
Previous studies have shown that free toxins and antitoxins of the MazEF family are homodimers (53–55). It has also been reported that toxins and antitoxins form heterohexameric complexes (T2A2T2) (53, 54). Next, we determined the oligomeric status of toxins, antitoxins, and TA complex by SEC-MALS. For individually purified MazE6 [(His)6MazE6(FLAG)3] and MazF6 [(His)6MazF6(FLAG)3], peak 1 is the major peak and corresponds to a homodimer (Fig. 2a and b). Peaks 2 and 3 observed for the purified MazF6 correspond to higher-order aggregates. Two different peaks were observed in the refractive index (RI) trace for the copurified MazEF6 complex [MazE6:MazF6(His)6] (Fig. 2c). The calculated molecular weights of the peaks are shown in Table 1. In the case of the purified MazEF6 complex [MazE6:MazF6 (His)6], peak 1 has the highest mass fraction (90%) corresponding to the mass of a hetero-octamer (T4A4) with molecular mass of 107.1 kDa (Table 1), and the remaining 10% were aggregates (peak 2). The individual monomeric molecular masses of the MazE6 antitoxin and the MazF6 toxin in copurified MazEF6 complex [MazE6:MazF6 (His)6] are 9.0 and 14.0 kDa, respectively. We also measured the oligomeric status of the in vitro-assembled MazEF6 complex from purified toxin and antitoxin [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3] at different ratios, ranging from 1:4 to 4:1 (Fig. 2d to g). We observed a different stoichiometry compared to the copurified complex, which was dependent on the relative molar ratio of MazE6 to MazF6 (Fig. 2 and Table 1). The individual monomeric molecular masses of MazE6 antitoxin [(His)6MazE6(FLAG)3] and MazF6 toxin [(His)6MazF6(FLAG)3] are 14.8 and 17.8 kDa, respectively. At an equal ratio of both toxin to antitoxin as well as at higher antitoxin-to-toxin ratios (MazF6:MazE6, 1:2; 1:4), we observed a hetero-octameric complex, similar to the copurified complex. However, at a higher MazF6-to-MazE6 ratio (MazF6:MazE6, 4:1; 2:1), we observed a hetero-hexameric complex (T2A2T2). In all cases where the complexes were reconstituted in vitro, two additional peaks were observed, where peak 1 corresponds to either free antitoxin or toxin and peak 3 corresponds to higher-order aggregates. These observations explain the high Tm values obtained for the MazEF6 complexes formed at different stoichiometries.
FIG 2.
Oligomeric analysis of purified proteins by SEC-MALS. Traces for refractive index are shown in blue. The molar mass and fits of all traces are plotted as a function of elution time as approximately horizontal red and black lines, respectively. The peaks analyzed for molecular weight determination are numbered in each graph. (a) MazE6 [(His)6MazE6(FLAG)3], (b) MazF6 [(His)6MazF6(FLAG)3], (c) copurified MazEF6 complex [MazE6:MazF6(His)6]. In vitro-assembled MazEF6 complex [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3] at different ratios of MazE6:MazF6, (d) MazE6:MazF6 of 2:1, (e) MazE6:MazF6 of 4:1, (f) MazE6:MazF6 of 1:2, and (g) MazE6:MazF6 of 1:4.
TABLE 1.
The mass fractions and corresponding molecular weights of each peak for the proteins analyzed are representeda
| Protein | Peak | Observed molecular mass (kDa) | Expected molecular mass (kDa) | Mass fraction (%) | Stoichiometry |
|---|---|---|---|---|---|
| MazE6 | 1 | 30 | 29.6 | 100 | A2 (dimer) |
| MazF6 | 1 | 35 | 35.6 | 67 | T2 (dimer) |
| 2 | 100 | 13.9 | Aggregate | ||
| 3 | 592 | 19.1 | Aggregate | ||
| MazEF6 | 1 | 107 | 92 | 90.0 | T4A4 (hetero-octamer) |
| 2 | 1,844 | 10.0 | Aggregate | ||
| MazF6: | 1 | 41 | 32.7 | Free T2/A2 | |
| MazE6 (1:2) | 2 | 122 | 130.4 | 59.1 | T4A4 (hetero-octamer) |
| 3 | 1,488 | 8.2 | Aggregate | ||
| MazF6: | 1 | 40 | 14.7 | Free T2/A2 | |
| MazE6 (1:4) | 2 | 125 | 130.4 | 63.1 | T4A4 (hetero-octamer) |
| 3 | 2,375 | 22.2 | Aggregate | ||
| MazF6:MazE6(2:1) | 1 | 37 | 22.2 | Free T2/A2 | |
| 2 | 97 | 100.8 | 64.1 | T2A2T2 (heterohexamer) | |
| 3 | 3,389 | 13.7 | Aggregate | ||
| MazF6:MazE6 (4:1) | 1 | 33 | 100.8 | 10.8 | Free T2/A2 |
| 2 | 102 | 84.0 | T2A2T2 (heterohexamer) | ||
| 3 | 1,205 | 5.2 | Aggregate |
T, toxin; A, antitoxin. The individual monomeric molecular masses of MazE6 antitoxin [(His)6MazE6(FLAG)3] and MazF6 toxin [(His)6MazF6(FLAG)3] are 14.8 and 17.8 kDa, respectively. The individual monomeric molecular masses of MazE6 antitoxin and MazF6 toxin in copurified MazEF6 complex [MazE6:MazF6 (His)6] are 9.0 and 14.0 kDa, respectively.
Estimation of dissociation constant of MazE6 and MazF6.
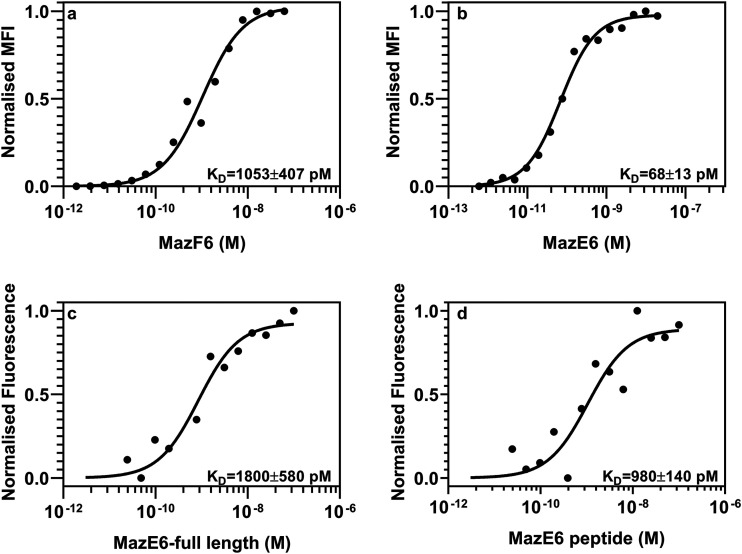
Yeast surface display (YSD) has been used to estimate the dissociation constant between proteins (56). Both MazE6 and MazF6 were individually expressed on the yeast cell surface, and binding was measured with the cognate partner. In the case of yeast surface-displayed MazE6, the KD was around 1,053 pM (Fig. 3a, Table 2). However, when MazF6 was displayed, the KD was 66 pM (Fig. 3b, Table 2). We found a 16-fold difference in the KD depending on the displayed protein. In most type II toxin-antitoxin interactions, the C-terminus of the antitoxin is usually involved in toxin binding. We observed two different populations in the case of binding to surface-displayed MazE6 but not surface-displayed MazF6, indicating the presence of a population with degraded MazE6 in the former (Fig. S2a and b). This might be attributed to the labile nature of the C-terminus of the antitoxins, which is intrinsically disordered (47). We also measured the binding of the toxin MazF6 [(His)6MazF6(FLAG)3] with the C-terminal antitoxin MazE6 peptide using MST. A fixed dimeric concentration of 2 nM of the labeled toxin was titrated with different concentrations of the unlabeled full-length antitoxin [(His)6MazE6(FLAG)3] and antitoxin peptide (15 pM to 0.5 μM). The obtained data indicates that the toxin MazF6 binds with the full-length antitoxin and C-terminal antitoxin peptide MazE6 (residues 48 to 82) with a KD of about 1,800 and 980 pM, respectively (Fig. 3c and d, Table 2). The difference in the apparent affinities measured by YSD and MST is most likely due to the tendency of purified proteins to aggregate as observed from SEC-MALS studies.
FIG 3.
Estimation of dissociation constant between MazE6 and MazF6 using YSD and MST. (a and b) Dissociation constant between MazE6 and MazF6 was measured using YSD coupled to FACS. MazE6 (a) and MazF6 (b) were displayed on the yeast cell surface and titrated with serially diluted purified cognate partner. The binding was measured on a BD Aria III flow cytometer. (c and d) Dissociation constant between full-length MazE6 [(His)6MazE6(FLAG)3] and MazE6 C-terminal peptide with MazF6 [(His)6MazF6(FLAG)3] was measured using MST; 2 nM labeled toxin MazF6 [(His)6MazF6(FLAG)3] was titrated with various concentrations of either full-length antitoxin (c) or the C-terminal antitoxin peptide (residues 48 to 82) (d). Reported standard errors are derived from two independent experiments, each performed in duplicate.
TABLE 2.
Dissociation constants (KD) of MazEF6 determined by YSD and MSTa
| Proteins | KD (pM) |
|---|---|
| MazE6 full lengthb | 1,053 ± 407 |
| MazF6 full lengthb | 68 ± 13 |
| MazE6 full-length antitoxinc | 1,800 ± 580 |
| MazE6 C-terminal peptidec | 980 ± 140 |
Reported standard errors are derived from two independent experiments, each performed in duplicate.
Displayed protein in YSD.
Determined by MST.
Estimation of dissociation constants between antitoxin MazE6 and different ratios of in vitro-assembled MazEF6 complexes with promoter-operator DNA.
We performed MST to measure the binding of the antitoxin MazE6 [(His)6MazE6(FLAG)3] and the in vitro-assembled MazEF6 complexes [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3] of different stoichiometries with the mazEF6 promoter-operator DNA as described in Materials and Methods. A previous study showed that both MazE6 antitoxin and MazEF6 complex can bind to the DNA fragment located 99 bp upstream of the mazEF6 operon (57). In the current study, we probed the binding of MazE6 as well as the complexes with two different lengths of promoter-operator DNA located 100 bp (PO-4) and 50 bp (PO-5) upstream of the mazEF6 operon (Fig. 4 and Fig. S3). In all cases the binding affinities were similar for both PO-4 and PO-5 (Fig. 4 and Fig. S3). The affinity towards the promoter DNAs was lowest for the MazEF6 (1:4) complex and becomes stronger with increase in the antitoxin concentration (Fig. 4 and Fig. S3). The obtained data indicate that with decrease in the MazE6:MazF6 ratios, the affinity towards the PO-4 and PO-5 DNA is reduced. Taken together, this study demonstrates that autoregulation of TA operon is dependent on the relative ratios of MazE6 and MazF6.
FIG 4.
Dissociation constant of promoter DNA fragments as a function of free MazE6 antitoxin and different stoichiometries of toxin:antitoxin complexes. The KD as a function of either free MazE6 antitoxin [(His)6MazE6(FLAG)3] or various ratios of the in vitro-constituted MazEF6 complexes [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3] with promoter DNA, PO-4 (100 bp), and PO-5 (50 bp) was measured using MST. Reported standard errors are derived from two independent experiments, each performed in duplicate.
Transcriptional response to MazF6 overexpression in M. tuberculosis.
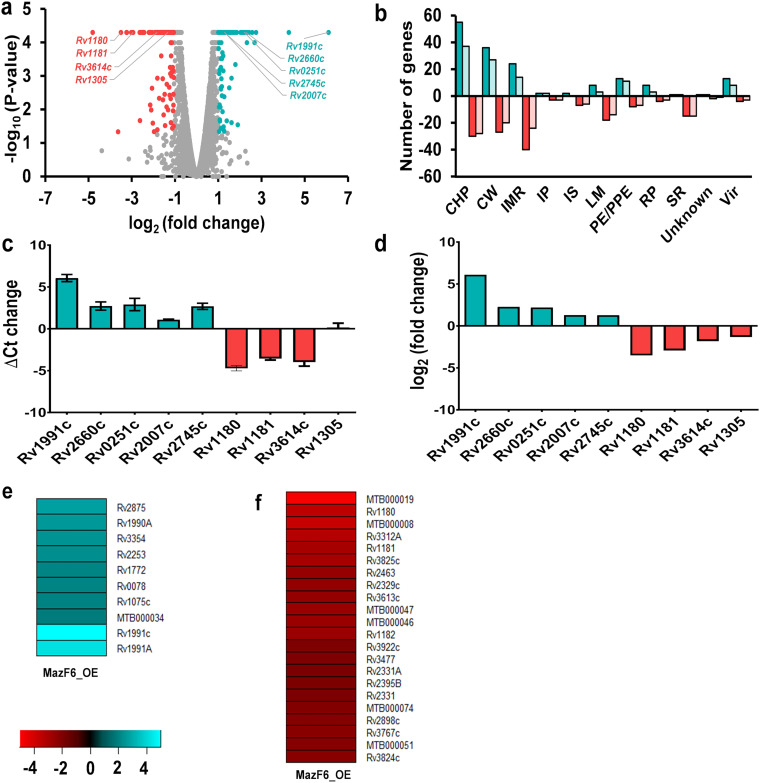
RNA-seq analysis was performed to compare the transcription profiles of control and MazF6 overexpression strains of M. tuberculosis. The overexpression of MazF6 in M. tuberculosis significantly altered the expression of 323 genes (Fig. 5a). These differentially expressed genes (DEGs) were obtained by using a cut-off log2(fold change) of ≥1.0 or ≤−1.0 and P value of <0.05. Among these differentially expressed genes, 163 and 160 transcripts were upregulated and downregulated, respectively (Fig. 5a and Table S2). These DEGs were further annotated according to their functional category. Based on that, approximately 36% and 20% of the genes that are conserved hypotheticals (CHP) are up- and downregulated, respectively (Fig. 5b). For the genes associated with intermediary metabolism and respiration (IMR), 16% and 26% of the genes were upregulated and downregulated, respectively (Fig. 5b). The increased expression of MazF6, Rv2660c, Rv0251c, Rv2007c, and Rv2745c and the decreased expression of Rv1180, Rv1181, and Rv3614c in the overexpression strain were also validated by qPCR using gene-specific primers (Fig. 5c). The expression of Rv1305 slightly varied from the qPCR results. The log2(fold change) of the above-described transcripts from RNA sequencing data are shown in Fig. 5d.
FIG 5.
Effect of overexpression of MazF6 on the transcriptome of M. tuberculosis. (a) The overexpression of MazF6 resulted in differential expression profiles of 323 transcripts. The volcano plot shows the transcripts whose expression were increased (green dots), decreased (red dots), or remained unchanged (gray dots). The x axis and y axis represent log2(fold change) and −log10 P value, respectively. (b) The differentially expressed genes are categorized according to the functional category mentioned in Mycobrowser. Dark green and red bars show the differentially expressed genes due to MazF6 overexpression. The light green and red bars represent the genes specific to MazF6 overexpression compared to other stress conditions, namely, nutrient starvation, enduring hypoxic response (EHR), nonreplicating persisters (NRP), and drug-induced persisters. The y axis represents the number of DEGs for a functional category as mentioned on the x axis. CHP, conserved hypothetical protein; CW, cell wall and cell processes; IMR, intermediary metabolism and respiration; IP, information pathway; IS, insertion sequences and phages; LM, lipid metabolism; PE/PPE, Pro-Glu/Pro-Pro-Glu proteins; RP, regulatory protein; SR, stable RNAs; Vir, virulence, detoxification, and adaptation. (c) The transcript levels of subsets of DEGs were quantified by qPCR using gene-specific primers. The relative expression of these transcripts was obtained after normalization to levels of the sigA housekeeping gene. The data shown in this panel are mean ± standard error change in cycle threshold obtained for each transcript from three independent experiments performed in duplicates. (d) The log2(fold change) of the same transcripts from RNA sequencing data is shown. (e and f) Upregulated and downregulated genes showing ≥4 fold change |log2(fold change)| specific to MazF6 overexpression.
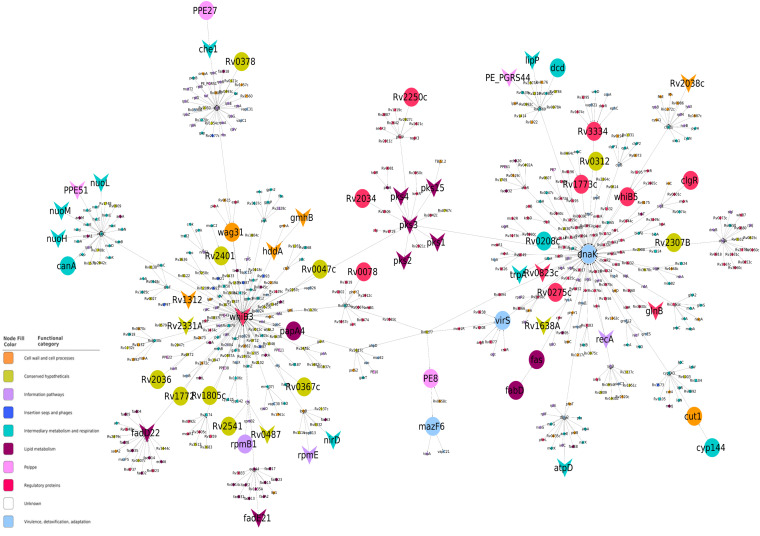
We observed that the transcription profiles obtained in M. tuberculosis upon overexpression of MazF6, while similar to those obtained from bacteria exposed to stress conditions such as nutrient starvation, enduring hypoxic response (EHR), nonreplicating persisters (NRP), and drug-induced persisters (Fig. S4a to d), also exhibited some differences (Fig. 5b). We have only shown the overlap of upregulated genes (Fig. S4a), as the data for genes downregulated under other stress conditions were either not available or nonoverlapping. Among the 231 genes that were specific to MazF6 induction, 107 genes were upregulated and 124 genes were downregulated (Fig. 5b). To further delineate the MazF6-specific effects, a higher threshold of ≥4 fold change. |log2(fold change)| was used, and it was observed that genes belonging to the functional category of stable RNAs, lipid metabolism, and cell wall and cell processes were dominant (Fig. 5e and f). We further performed a network analysis to study the top-ranked variations observed between the control and the MazF6 overexpression strain. The M. tuberculosis protein-protein interaction network consists of 3,686 proteins (nodes) and 34,223 molecular interactions (edges). The subnetwork formed by the top-ranked paths consisted of 582 nodes, of which 62 were DEGs that were upregulated significantly by 2-fold (Fig. 6). The subnetwork was analyzed to identify hub nodes (nodes having a high number of interactions [degree] with other nodes). Among these, transcriptional regulators whiB3, dnaK, and sigB were amongst the highest-degree hubs. The top response network identified 582 genes, which captures the phenotype-related perturbations in M. tuberculosis. The hub nodes with repressed gene expression were whiB3, fadD22, and pks3, whereas the upregulated gene identified was dnaK (Fig. 6). Based on the RNA-seq differential gene expression data, we observed that the expression of heat shock response genes, namely, hsp (Rv0251c) and dnaK, are increased upon overexpression of MazF6. These observations suggest that the presence of free toxin results in metabolic shutdown and induction of stress response pathways of M. tuberculosis (31, 58). The upregulation of the heat shock response genes indicate a proteotoxic stress due to the presence of free toxin in M. tuberculosis. The overexpression of MazF6 toxin also resulted in downregulation of whiB3, a transcriptional regulator that regulates catabolic metabolism and polyketide biosynthesis (59). Further, other proteins belonging to the functional category of regulatory protein, information pathways, or virulence, detoxification, and adaptation, such as sigB, nuoC, dnaA, and phoP, were also identified as hub nodes in the network. These observations suggest that overexpression of MazF6 results in transcriptional reprogramming that enables M. tuberculosis to adapt to different stress conditions and persist in host tissues.
FIG 6.
Top-response network of MazF6 overexpressed strain compared to the wild type. Nodes/genes in the network are colored based upon Mycobrowser functional categories, and the gray arrows represent the directed edges/interactions between the nodes. The nodes are shaped based on the pattern of gene expression where the circle represents the induced/upregulated genes, the arrowhead represents repressed/downregulated genes, and the diamond represents the genes that did not show significant differential regulation.
Mapping of MazF6-dependent cleavage sites.
In order to determine the location of MazF6-dependent cleavages in M. tuberculosis transcripts, RNA cleavage was quantified by comparing counts at each nucleotide in M. tuberculosis expressing MazF6 to the corresponding value in the control sample. The cleavage of an mRNA will result in lower counts at and near the site of cleavage. The regions cleaved by MazF6 were identified by computing a protection ratio (log2 of read counts [+ MazF6:empty vector]) at each position (36) (Fig. S5). It was observed that a majority of transcripts were cleaved upon overexpression of MazF6 in M. tuberculosis (Fig. S5a and c). Individual transcripts were examined, and there were several regions with minima in the protection ratio plot, which suggests cleavage by MazF6 (Fig. S5b and d). Based on the annotation information from the GFF file downloaded from Mycobrowser, 84% of the genes were above the expression threshold (>50 counts). Amongst these, 574 genes had a minimum value of the protection ratio of ≤−1 following MazF6 overexpression, indicating RNA cleavage (Fig. S6a). After enriching for these regions with minimum protection ratio, the average value of the protection ratio minimum was −1.5 and the lowest value was −6.1.
In vivo cleavage specificity of MazF6 in M. tuberculosis.
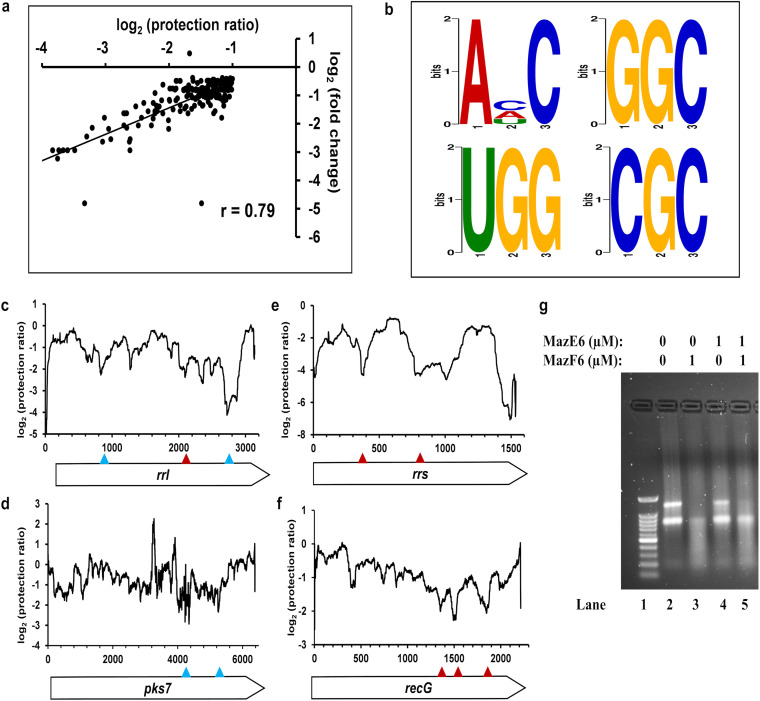
To understand the effect of MazF6 overexpression on the M. tuberculosis genome, we examined the level of differential expression in transcripts with low protection ratios. A significant correlation was observed between the extent of downregulation of the gene due to MazF6 overexpression and the value of its minimum protection ratio (r = 0.79) for cleavage sites with a minimum protection ratio of ≤−1 (Fig. 7a). Amongst the 78 genes with MazF6-dependent cleavage and minimum protection ratio of ≤−1.5, we found that 50% of them have log2(fold change) of ≤−1. The correlation between the minimum protection ratio and log2(fold change) improved further to 0.86 when a cut-off of ≤−1.5 was used for the minimum protection ratio. To determine the sequence specificity of MazF6 toxin, we selected 118 valleys in the M. tuberculosis cleavage profiles that were filtered on the basis of a 1.5-fold increase in the protection ratio within 100 nucleotides upstream and downstream of the minimum protection ratio. This distance from the local minimum was identified based on the distance between the local minimum and its neighboring local maxima (Fig. S6b). These cleavage valleys represent clear, defined regions of MazF6-mediated cleavage. A motif search was performed in the cleavage valleys for putative sites that have at least 25 occurrences and have the additional constraint of being present less than 10 bases upstream or downstream from the putative cleavage site, identified on the basis of local minimum in the protection ratio (Fig. 7b, Table 3). Amongst the various hits obtained for the motifs, an enrichment in the dimer and trimer motifs was found. The results from the dimer motifs suggest an enrichment of CG and UG motifs both upstream and downstream of the cleavage site (Table 3 and Tables S4, S5, S9, and S10). Amongst the trimer motifs AHC (H is A/C/U), GGC and UGG were enriched within 10 nucleotides upstream and downstream of cleavage positions with minimum protection ratios (Table 3). In the trimer motifs, amongst AHC (H is A/C/U), ACC shows the highest propensity for sites upstream of the cleavage site and, thus, was considered among the top four trimer motifs (Table 3). To obtain additional information about the consensus in the surrounding nucleotides for the di- and trimer motifs, the tetra- and pentamer motifs were also searched (Table S4 to S13).
FIG 7.
MazF6 ribonuclease cleavage. (a) Correlation between the log2(protection ratio) (≤−1) and log2(fold change) in transcript level. (b) Sequence logos derived from MEME analysis of cleaved regions identified by RNA-seq in the entire M. tuberculosis transcriptome that are upstream of the cleavage site. (c to f) Sites with ACC and UGG motifs upstream of the cleavage site are indicated with blue and red triangles, respectively. (g) The in vitro ribonuclease activity of the purified MazF6 toxin [(His)6MazF6(FLAG)3] and the in vitro-assembled MazEF6 complex of different stoichiometries [(His)6MazE6(FLAG)3:(His)6MazF6(FLAG)3].
TABLE 3.
List of four highest-frequency di-, tri-, tetra-, and pentamer motifs identified at positions cleaved by MazF6
| Type | Upstream |
Downstream |
||||
|---|---|---|---|---|---|---|
| Top motifs | Propensity of motifs calculated using base frequency | Total no. of motifs upstream of putative cleavage site | Top motifs | Propensity of motifs calculated using base frequency | Total no. of motifs downstream of putative cleavage site | |
| Dimer | CG | 4.11 | 150 | CG | 3.23 | 142 |
| UGa | 5.64 | 90 | UGa | 3.88 | 74 | |
| CCb | 3.40 | 53 | AG | 2.35 | 36 | |
| AA | 5.27 | 42 | GCb | 2.74 | 36 | |
| Trimer | GGC | 8.79 | 37 | GGC | 3.49 | 33 |
| UGGa | 14.21 | 36 | UGGa | 4.16 | 26 | |
| CGC | 7.32 | 33 | ACC | 3.80 | 22 | |
| ACC | 8.53 | 20 | ACG | 3.56 | 16 | |
| Tetramer | GGCG | 24.44 | 19 | GGCG | 31.32 | 14 |
| CCGC | 24.44 | 16 | CCGC | 19.27 | 10 | |
| CUGGa | 47.44 | 13 | CUGG | 30.40 | 9 | |
| CGGG | 24.44 | 11 | GUGGa | 35.08 | 9 | |
Motifs with highest propensity value amongst their respective categories of motifs.
Motifs where actual occurrence in cleaved transcripts is higher than MEME identification.
A major limitation is the noise in the data, due to which we were unable to identify the exact position of the cleavage. A few of the motifs also occur within the identified cleavage sites. An example of a MazF6 cleavage profile is shown for rrl, which encodes 23S rRNA (Fig. 7c). This region of the rrl transcript contained one UGG and two ACC sites, which are upstream of the site with minimum protection ratio and identified as putative cleavage sites of the MazF6 toxin. The occurrence of two UGG motifs upstream of the cleavage site was also found in rrs, the gene encoding 16S rRNA (Fig. 7d). There were several other transcripts that showed cleavage of the mRNA at these putative cleavage sites (Fig. 7c to f). Further, the propensity of the top four di-, tri-, and tetramer motifs was calculated to understand the rate of occurrence of these motifs in the genes with cleavage sites (Table 3). The M. tuberculosis genome is rich in GC content; upon correcting for this, a higher propensity of occurrence amongst the top four motifs was found for motifs containing U and G nucleotides. High propensity values were found for UG, UGG, CUGG, and GUGG for motifs both upstream and downstream from the cleavage sites. This implies that the cleavage due to MazF6 toxin has a degree of sequence specificity, although for any given site, only a small fraction of sites in the genome have low protection ratios.
To probe the activity of the purified toxin MazF6, an in vitro ribonuclease assay was performed with both 16S rRNA and 23S rRNA isolated from M. tuberculosis. We observed that the MazF6 toxin (1 μM) was able to cleave both 16S rRNA and 23S rRNA completely, whereas the antitoxin MazE6 (1 μM) did not have any cleavage activity (Fig. 7g). When the toxin MazF6 was incubated with an equal molar ratio of MazE6 antitoxin (MazF6:MazE6, 1:1), cleavage was reduced with greater inhibition for 16S rRNA than 23S rRNA (Fig. 7g).
DISCUSSION
In M. tuberculosis, around 80 TA modules have been identified so far (60, 61). Most TA systems in M. tuberculosis belong to the vapBC and mazEF families. It has been shown that overexpression of some of these toxins using an inducible vector inhibits the growth of either E. coli, M. bovis BCG, or M. smegmatis (24, 28, 30, 31). Studies have shown that the expression of a subset of toxins is induced in M. tuberculosis upon exposure to different drugs, such as streptomycin, isoniazid, and rifampicin, or stress conditions (24, 30, 31, 62). These observations suggest that these toxins might be important for survival of M. tuberculosis under these conditions.
mazEF was originally discovered on the E. coli chromosome and remains one of the best-characterized TA modules. The E. coli MazEF complex forms a heterohexameric (T2A2T2) structure (53). The C-terminus of each MazE monomer wraps around the interface of the MazF homodimer to neutralize the toxin (53). E. coli MazF specifically cleaves RNA at ACA sequences in single-stranded RNA. The current study involves characterization of the MazEF6 TA system of M. tuberculosis. As expected, thermal unfolding experiments showed that the complexes have higher stability than the individual proteins. The free toxin MazF6 and the antitoxin MazE6 are dimeric in solution. The copurified MazEF6 complexes have a hetero-octameric structure with two dimers of toxin interacting with two dimers of antitoxin (T4A4), unlike MazEF homologues from other organisms (44, 46, 53, 54) but similar to the VapBC TA complexes from M. tuberculosis (63–66). Although the overall sequence similarity among these MazF toxins is relatively poor, the overall structural fold is highly conserved. Structures of MazEF complexes solved from M. tuberculosis, E. coli, or B. subtilis reveal that it forms a heterohexameric complex. In the present study, we observed that altering the ratio of MazF6 to MazE6 resulted in various oligomeric forms of the complexes. We have recently employed Asp and Arg scanning mutagenesis to probe the structure of the M. tuberculosis MazEF6 complex (47). In contrast to the other MazEF6 structures, where the C-terminal region is primarily involved in antitoxin binding, we found that residues 31 to 66 in the middle of the antitoxin contribute significantly to toxin binding (47). This points to an altered complex structure relative to other MazEF complexes. The affinity of interaction between cognate toxin and antitoxin was also estimated for the studied MazEF6 system using YSD and MST. The dissociation constant (KD) for this interaction was found to be in the picomolar range. The physiological relevance of the different oligomers was further tested by their ability to bind to promoter-operator DNA. It was observed that an increase in the toxin:antitoxin ratio lowered the affinity of the complex toward the promoter DNA, indicating the role of stoichiometry of toxin:antitoxin in regulation of the operon.
We had previously shown that the overexpression of putative MazF homologs (MazF3, MazF6, and MazF9) is bacteriostatic in M. bovis BCG and M. tuberculosis (31). In another study, it was shown that overexpression of M. tuberculosis MazF6 inhibits the growth of E. coli in a bactericidal manner (31, 62, 67). The discrepancy in results could be attributed to (i) different levels of expression obtained from either isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible or anhydrotetracycline-based expression vectors, (ii) presence of endogenous antitoxin in M. bovis BCG, or (iii) different cellular targets in E. coli and mycobacteria. In the present study, analysis of the transcriptional response to the overexpression of MazF6 in M. tuberculosis resulted in differential gene expression of 323 transcripts that was associated with increased transcript levels of regulatory proteins and other non-cognate toxins. It was also found that toxin overexpression led to the upregulation of transcripts involved in cell wall and cell processes, such as mpt70 and mpt83. mpt70 is soluble secreted antigen, while mpt83 is an exported lipoprotein associated with the bacterial surface (68). Previous studies have shown that although mpt83 and mpt70 are expressed at low levels in vitro, they are highly immunogenic during infection of mice (68). Further, mpt70 was shown be upregulated in response to gamma interferon (IFN-γ) or nutrient/oxygen deprivation of in vitro-infected macrophages (69). Several other lipoprotein genes, such as lpqS, lpqD, and lppP, that have been previously reported to be associated with adaptive responses to different growth conditions and have been linked to M. tuberculosis virulence, also show an increase in their expression levels due to toxin overexpression (70, 71). The genes involved in intermediary metabolism and respiration have also been induced, such as trxB1, cyp144, cysK2, cysA2, and cysA3. Previous studies have shown the expression of major antioxidant genes, including thioredoxins (trxB1, trxB2, and trxC), was increased under acidic conditions (72). The cysK2 gene is overexpressed in various dormancy models under conditions such as oxidative stress and hypoxia (73). The sulfate transporters in M. tuberculosis such as cysA2 and cysA3 also have been found to be upregulated in dormancy (74). Interestingly, there is upregulation of non-cognate antitoxins such as mazE3, vapB17, vapB31, vapB42, and parE2 due to MazF6 overexpression. This suggests that transcriptional responses due to toxin overexpression have similarities with the ones observed in dormancy and other stress conditions, although there are also distinct differences (Fig. 5b).
We hypothesize that overexpression of MazF6 in M. tuberculosis results in shutdown of metabolism due to growth inhibition (31). To adapt to this metabolic shutdown, the expression of various stress-responsive genes is induced, explaining the similarities with bacteria exposed to different stress conditions, such as nutritional stress or low-oxygen stress (66, 75, 76). Additionally, there were a significant number of genes whose RNA levels were specifically altered (≥4 |log2(fold change)|) upon MazF6 toxin induction, which belonged to functional categories of stable RNAs, lipid metabolism, and cell wall and cell processes. This is contrary to a recent study that showed that transcriptional upregulation of TA systems is not correlated with toxin release or activity (77). In agreement, the profiles obtained from the M. tuberculosis VapC21 overexpression strain were also similar to M. tuberculosis exposed to different stress conditions, such as nutritional or low-oxygen stress (34). The changes in expression profiles upon overexpression of MazF6 could be attributed to increased expression of transcriptional regulators such as sigB, sigC, sigD, sigE, virS, and csoR and hypothetical proteins in MazF6 overexpression strains. In concordance with previous studies, we have observed that upregulation of sigB and virS transcriptional regulators is associated with upregulation of genes involved in lipid metabolism (kasA); cell wall and cell processes (wag31, mmpS3, Rv0559c, lpqS, and ctpG); PE/PPE (Rv3514, Rv3508, and Rv2490c); intermediary metabolism and respiration (aroG, argC, Rv2672, and pdc); and regulatory proteins (clgR) and virulence, detoxification, and adaptation (hsp, dnaK, and ahpC) (78–80). A similar trend was observed for csoR, which caused the upregulation of genes such as cysK2, mprA, and argR, which were previously shown to be downregulated under csoR deletion (81).
Amongst the top response network hubs, whiB3 was identified in the MazF6 overexpression strain. whiB3 is a redox-sensitive transcriptional regulator that regulates the expression of several genes encoding polyketide synthases, such as pks2 and pks3 (59, 82). In agreement, pks3 is also identified as a hub node in the top-response network, showing the interaction with whiB3 in the network. Similar to pks3, another gene involved in lipid metabolism, fadD22, was also identified as a hub node. Among the other hub nodes in the top-response network, genes involved in virulence, detoxification, adaptation, and lipid metabolism were also differentially expressed upon overexpression of MazF6 toxin.
We also applied a paired-end RNA sequencing-based method for mapping and quantifying the cleavage of endoribonucleases to map cleavage sites that result from the overexpression of MazF6 by calculation of the mRNA protection ratio (36). Apart from the 16S and 23S rRNA genes, which showed low protection ratios, there were several other genes belonging to polyketide synthases, such as pks1, pks2, and pk3, genes involved in lipid metabolism such as fadD9, which also showed significantly lower protection ratio. In contrast, the genes that were upregulated under stress conditions, such as mpt70, mpt83, trxB1, lpqD, lpqS, cyp144, and cysK2, show a high protection ratio with MazF6 toxin overexpression. Similarly, higher protection ratios were also observed for sig family, virS, and csoR transcriptional regulators.
The analysis revealed several motifs in the M. tuberculosis mRNA and rRNA that may be putative recognition sites for MazF6 cleavage (Table 3; see also Tables S4 to S13 in the supplemental material). Since the M. tuberculosis genome is GC rich, we normalized our data accordingly and the top motifs were identified. The highest propensity values for trimer and tetramer are UGG and CUGG, respectively, located upstream of the putative cleavage site. These sites differ from those identified earlier using primer extension and MORE-seq approaches for MazF6 (40, 42). This might be attributed to the use of different strategies and host strain used in these studies. Using a combination of phage MS2 RNA and CspA, an RNA chaperone, it has been shown that MazF6 cleaves RNA at UU↓CCU or CU↓CCU (40). Further, in a recent RNA sequencing study, MORE-seq revealed that the MazF6 toxin also cleaves RNA at U↓CCUU and identified a cleavage site in 23S rRNA within the helix/loop 70 and in 16S rRNA at the anti-Shine Dalgarno sequence (42). In our study, we have performed experiments in M. tuberculosis to determine the cleavage site. It is possible that we may have identified a few secondary cleavage sites due to the presence of endogenous RNase or increased expression of other toxins, such as VapC18, in our experiments (67).
Cleavages at loci identified earlier to be cleaved by MazF6 toxin (Rv1545 and Rv1685c) as well as those that were resistant to cleavage (Rv0490 and Rv0685) were not consistent with our results (42) (Fig. S7). This is because either no local minima at these loci were identified during the present protection ratio array search or, even if they were identified, the protection ratio was not more negative than −1.5, which was used as a cut-off in our analysis. Therefore, these loci were filtered out from downstream analysis (Table S14). In addition, the above-described study identified U↓CCUU as the cleavage site, in contrast to the present study. We identified multiple positions for cleavage based on the local minimum values. A possible reason for the discrepancy between the two sets of results could be because in the previous study, MazF6 overexpression was carried out in E. coli and a 50-fold enrichment (+MazF6 cells/-MazF6 cells) in the 5′ end of the putative targets was used as a cut-off to identify the cleavage site. In contrast, in the present study, overexpression was carried out in the native context in M. tuberculosis and the MazF6 cleavage sites were identified by computing a protection ratio (log2 of read counts [+ MazF6:empty vector]) at each position (36). We also partially validated our cleavage results by in vitro ribonuclease activity of purified MazF6 toxin with 16S rRNA and 23S rRNA isolated from M. tuberculosis. The major difficulties with accurate identification of cleavage sites from the protection ratio are the noise in the data and possible exonuclease degradation following the initial cleavage; these can be partially overcome by averaging over a large number of putative sites, as we have attempted to do in the present study. Thus, further work is required to confirm which of these motifs are likely to be the correct motifs that occur at the site of cleavage. Both in the present as well as earlier studies, only a small fraction of sites containing the cleavage motif(s) actually appear to be cleaved in vivo. This suggests that other features, such as RNA secondary and tertiary structure, are likely to be important modulators of MazF6-mediated cleavage in vivo.
MATERIALS AND METHODS
Plasmids and host strains.
The mazE6 and mazF6 genes were cloned individually under the control of the T7 promoter in the pET-Duet-1 vector for purification of the MazEF6 complex. For expression of MazF6 and MazE6, genes were individually cloned under the control of the T7 promoter in pET-15b vector. E. coli host strain GB05-dir was used for in vivo recombination of the pET-15b vector backbone with the maz genes. GB05-dir harbors an arabinose-inducible recombination machinery (83). E. coli host strain BL21-(λDE3) pLysE was used for expressing the proteins. The primer sequences used for cloning are reported in Table S1 in the supplemental material.
Protein expression and purification.
The toxin, antitoxin, and complex were purified as described previously (34). Briefly, cultures were grown in TB medium and induced with 1.0 mM IPTG at an optical density at 600 nm (OD600) of 0.6 for 5 h at 37°C for MazE6/MazF6 (His)6 complex expression, for 16 h at 20°C for (His)6MazE6(FLAG)3 expression, and 7 h at 20°C for (His)6MazF6(FLAG)3 expression. Cells were harvested by centrifugation (1,800 × g, 20 min, 4°C). The pellet was resuspended in lysis buffer, pH 8.0 (10 mM HEPES, 100 mM NaCl, 100 mM l-arginine, 10% glycerol containing protease inhibitor tablet [Roche]) and sonicated, followed by centrifugation at 25,000 × g, 30 min, 4°C. The cell lysate was incubated with 2 mL Ni-NTA Sepharose 6 Fast Flow (GE Healthcare) for 4 h at 4°C. The unbound fraction was removed, and the resin was washed with 25 column volumes of wash buffer (10 mM HEPES, 100 mM NaCl, 100 mM l-arginine, 50 mM imidazole, 10% glycerol, pH 8.0). The complex was then eluted with 1 mL of elution buffer (10 mM HEPES, 100 mM NaCl, 100 mM l-arginine, 10% glycerol, and a gradient of imidazole [100 mM to 900 mM], pH 8.0). The eluted fractions were subjected to 15% Tricine SDS-PAGE, and the protein concentration was determined by absorbance measurements. All of the proteins were stored in storage buffer (10 mM HEPES, 100 mM NaCl, 100 mM l-arginine, 500 mM imidazole, 10% glycerol, pH 8.0, with additional cOmplete protease inhibitor cocktail [Roche] added for antitoxins) at −80°C after concentration.
The final concentration of MazEF6 [MazE6:MazF6(His)6] complex was 2 mg/ml, and the corresponding yield was 2 mg/liter. The concentrations of purified MazE6 [(His)6MazE6(FLAG)3] and MazF6 [(His)6MazF6(FLAG)3] were 1 mg/mL and 0.1 mg/mL, and their corresponding yields were 2 mg/liter and 0.3 mg/liter, respectively. The subsequent studies were performed using the tagged version of the proteins as described above.
NanoDSF studies.
Thermal unfolding of both purified and in vitro-assembled MazEF6 complex, MazE6 antitoxin, and MazF6 toxin were performed using nanoDSF (Prometheus NT.48). The sample was filled into capillaries by capillary force action and then placed into the instrument as described previously (84, 85). All the thermal unfolding experiments were carried out with 10 μM of each protein. Briefly, the samples were excited at 280 nm and emission was detected at 330 nm and 350 nm to derive the F350/F330 ratio for data analysis, with a bandwidth of measurement set at 5 nm. The first derivative of the ratio () is normalized using equation 1:
| (1) |
where y is the first derivative of the observed fluorescence (F350/F330 ratio), ymin is value of the first derivative minimum, and ymax is the value of the first derivative maximum. F350/F330 ratio was used to monitor protein unfolding. At the midpoint (Tm) of the thermal unfolding transition, the fraction unfolded was 0.5 and the first derivative of F350/F330 will be either at a maximum or a minimum depending on whether F350/F330 is increasing or decreasing with temperature. This in turn will depend on the environment of the tryptophan residues in the folded and unfolded states.
SEC-MALS.
MazEF6 complexes and MazE6 and MazF6 proteins were analyzed on a Superdex-200 analytical gel filtration column (GE Healthcare) equilibrated in elution buffer (10 mM HEPES, 100 mM NaCl, 100 mM l-arginine, 500 mM imidazole, pH 8.0) with in-line UV (Shimadzu), MALS (mini DAWN TREOS; Wyatt Technology orp), and refractive index detectors (WATERS 24614) for molecular weight analysis. For each run, 100 μg of each protein was injected. UV, MALS, and RI data were collected and analyzed using ASTRA software (Wyatt Technology).
Binding studies of MazE6 with MazF6 using yeast surface display coupled to flow cytometry.
The affinity of MazE6 and MazF6 was measured using YSD. The proteins (either full-length MazF6 or MazE6) were expressed on the yeast cell surface, and the level of expression was measured using anti-hemagglutinin (HA) antibody and goat anti-chicken antibody conjugated to Alexa fluor 488 against the yeast cell surface displayed protein as explained earlier (86, 87). Binding was measured by incubating the yeast cells expressing either full-length MazE6 or MazF6 with different concentrations of its cognate partner (0.5 pM to 1 μM) having a 3× FLAG tag. The bound protein amount was detected by the anti-FLAG antibody (1:300 dilution) and rabbit anti-mouse conjugated to Alexa fluor 633 secondary antibody (1:1,600 dilution). In all binding studies, the dimeric concentration of proteins was used. The binding was measured on a BD Aria III FACS machine, and binding mean fluorescence intensity (MFI) was fitted against the concentration of added ligand to estimate the dissociation constant, KD, using the equation Y = Bmax × X/(KD + X), where Y is the MFI of binding at each protein concentration, X, in nanomolars (nM), Bmax is the maximum MFI of binding, and KD is the equilibrium dissociation constant, in nM.
Binding studies of toxin to full-length antitoxin and C-terminal antitoxin peptide by MST.
The toxin MazF6 was buffer exchanged in 10 mM HEPES, pH 8.4, and labeled using the Monolith protein labeling kit RED-NHS (NanoTemper Technologies) according to the manufacturer’s instructions. Labeled MazF6 was used as target at a dimeric concentration of 2 nM and titrated with different concentrations of unlabeled full-length and C-terminal (residues 48 to 82) antitoxin peptide MazE6, ranging from 15 pM to 0.5 μM. The measurement was done at LED/excitation power setting 40 to 60% and two MST power settings, medium and high. Data were analyzed using MO Affinity Analysis software (version 2.2.5; NanoTemper Technologies) at different standard MST-off times (88–90).
Binding studies of MazE6 alone or different ratios of in vitro-assembled MazEF6, with its cognate promoter-operator DNA, by MST.
The antitoxin MazE6 was buffer exchanged in 10 mM HEPES, pH 8.4, and labeled using the Monolith protein labeling kit RED-NHS (NanoTemper Technologies) according to the manufacturer’s instructions. Labeled MazE6 was used as a target at a dimeric concentration of 200 nM and titrated with different concentrations of unlabeled promoter-operator DNA from 10 nM to 25 μM. Two lengths of promoter-operator DNA, 100 bp (PO-4) and 50 bp (PO-5) upstream from the start of the mazEF operon, were used for binding studies. The labeled antitoxin at a dimeric concentration of 200 nM was incubated with different concentrations of unlabeled toxin MazF6 (50, 100, 200, 400, and 800 nM) to form different stoichiometries of MazEF6 complexes (MazE6:MazF6, 4:1, 2:1, 1:1, 1:2, and 1:4). Subsequently, the relative binding of the in vitro-assembled complexes with both PO-4 and PO-5 DNA was determined. The measurement was done at LED/excitation power setting 20 to 40% and two MST power settings, medium and high. Data were analyzed using MO Affinity Analysis software (version 2.2.5; NanoTemper Technologies) at different standard MST-off times (88–90).
RNA sequencing experiments.
The gene encoding mazF6 was PCR amplified using gene-specific primers and cloned into an anhydrotetracycline (Atc) inducible vector, pTetR-int (30). The recombinant plasmid pTetR-int-mazF6 was electroporated into M. tuberculosis H37Rv, and transformants were selected on 7H11 plates supplemented with kanamycin. The expression of mazF6 was induced by the addition of Atc at an OD600 of ∼0.2 for 18 h. Total RNA was isolated from M. tuberculosis harboring either pTetR-int or pTetR-int-mazF6 using Qiagen RNAeasy columns as previously described (91). The isolated RNA was submitted to Agrigenome Pvt. Ltd. for RNA-seq analysis as previously described (30, 34). The quality of raw data obtained from the control and overexpression strain was assessed using FastQC v0.11.8 (92). The adaptor sequences were removed from QC-passed reads using Trim Galore v0.6.4, and the cleaned reads were subjected to further analysis (93). The reference M. tuberculosis H37Rv genome was downloaded from the ENSEMBL database. Reads were indexed using Bowtie2, and the trimmed paired-end reads were aligned to the reference genome with TopHat v2.1.1 using default parameter settings (94, 95). The resulting mapped files in SAM format were converted to BAM format using SAMtools v1.9 c. The transcripts were assembled using Cufflinks v2.2.1 and were further merged into a cohesive set using Cuffmerge. The differentially expressed transcripts were identified using Cuffdiff (96). This provided an average expression value for an individual gene in the form of fragment per kilobase of transcript per million mapped reads (FPKM) as the output. The ratio of FKPM values was calculated for each transcript relative to the vector control to obtain the fold change values. The Cuffdiff output of gene counts was subsequently analyzed for differential gene expression using the CummeRbund (R-package) software. The volcano plots for the identified transcripts were plotted using the basic plot function in R software.
qPCR analysis.
For qPCR analysis, total mRNA was isolated from M. tuberculosis harboring either pTetR-int or pTetR-int-mazF6. Approximately 1 μg of the mRNA was subjected to DNase I treatment followed by cDNA synthesis using Superscript III reverse transcriptase. The synthesized cDNA was subsequently used as a template for qPCR using gene-specific primers and SYBR green mix. The obtained data were normalized to the transcript levels obtained for the housekeeping gene sigA as described previously (91).
Network analysis.
A previously reported global protein-protein interaction network for M. tuberculosis (MtbPPIN) was used as a base network to generate a condition-specific network (97). This MtbPPIN is a complex directed graph of the genome-wide interconnections between proteins in an organism. In this graph/network, protein-coding genes are represented as nodes, and interactions between the nodes are represented as edges that are assigned directions based on evidence from the literature or databases such as KEGG or STRING (98, 99). A condition-specific PPI network was generated by integrating gene expression data onto the base MtbPPIN in the form of weights to the nodes and edges of the network. The node weight for gene i (NWi) was the absolute log2(fold change), calculated using equation 2 as described previously (100):
| (2) |
Here, Ti and Ci are the gene expression of gene i in the test condition and control condition, respectively. Edge weights (EWi,j) between the interacting nodes i and j were then calculated using the following (equations 3 and 4):
| (3) |
| (4) |
Here, NWi and NWj are the node weights of the edge-forming interacting nodes, EWmax and EWmin are the maximum and minimum edge weights of the entire network, respectively, and EW′ij is the inversed edge weight. To identify perturbations in a given condition (compared to its control), a sensitive network mining algorithm was used that (i) computes all-to-all shortest paths in the weighted network using Djikstra’s algorithm (implemented in the Zen library), (ii) computes path scores by taking the summation of all edge scores in that path, and (iii) normalizes and ranks them to identify the most perturbed paths and constitutes the top-response network (101). A stringent threshold (top-ranked 0.013 percentile of all paths, to capture top ∼500 genes from the network) was used to identify the highest perturbations. The generated response networks were visualized using Cytoscape 3.7.1 (34, 102).
RNA-seq data analysis for per-base coverage.
The mapped alignment files for the control and MazF6-overexpressed condition were used to calculate the per-base coverage using SAMtools depth (103). The depth was calculated individually for all the replicates under both control and overexpressed conditions. The depth was then normalized within the replicates of control and overexpressed conditions by dividing the counts at each nucleotide position by the total counts of that replicate and multiplying them by a scaling factor (equation 5). In this case, the scaling factor used was M. tuberculosis genome size.
| (5) |
where i is the replicate number.
The experiment for the control and MazF6 overexpressed condition was performed in triplicates, so the normalized count averages across the replicates were performed at each position individually for both conditions (equation 6).
| (6) |
The normalized count averages for both control and overexpressed conditions were further used to calculate the protection ratio. The ratio at each nucleotide position was calculated as the log2 transformed toxin overexpression (OE) missing control ratio (equation 7).
| (7) |
Since most analyses with the protection ratio involved taking the minimum protection ratio in a gene, it is important that the genes have sufficient counts in the control strain for accurate calculation. The control counts at each nucleotide were added for individual transcripts and further normalized with respect to the transcript length (tlength) (equation 8).
| (8) |
where t represents the individual transcript in the control and i represents the replicate number. These normalized counts for each transcript were then averaged across the replicates (equation 9).
| (9) |
To ensure that all cleaved regions were transcribed and of high quality, any transcript that was below the cut-off of 50 counts in the control strain was removed. Based on the gene names, their positions, and gene ontology categories extracted from Mycobrowser, 3,505 transcripts passed the filter of a minimum of 50 counts in the control strain.
Identification of MazF6 cleavage sites and cleavage motifs in M. tuberculosis.
To identify the regions cleaved following MazF6 overexpression, Gaussian smoothing of the protection ratio array was conducted using the smoothed data function in Matlab R2019b. The Gaussian filter method was used in this function with a smoothing factor of 0.2. Further, this smoothed protection ratio data was searched for local minima. Only those positions with local minimum value of protection ratio of ≤−1.5 were considered for subsequent analyses. The method used for the identification of cleavage sites was implemented as described previously, with a few modifications as described below (36). The respective positions for local minima were searched for true minima in the array of the unsmoothed protection ratio. To ensure that the cleavage sites surrounding these local minima were well defined, the upstream and downstream regions within 100 nucleotides were examined. The first position on each side of a local minimum that had a protection ratio >0.5 larger than the protection ratio at the local minimum was identified. The region between these boundaries was defined as the cleaved region. Stretches containing local minima that failed to meet the above-described criteria were removed from the subsequent analyses.
On the basis of the depth between the local maxima and local minima from the smoothed data, the difference between the local minimum and the neighboring local maximum was calculated. Based on the distribution of these differences, the initial minimum search distance from the local minima was chosen to be 100 nucleotides for motif identification. The sequences matching the above criteria were submitted to MEME (104) for identification of motifs and generation of position weight matrix and sequence logo. Prior to submission to MEME, either the positive- or negative-strand sequence was chosen based on the corresponding gene annotation of orientation in the M. tuberculosis genome. The cleaved regions were analyzed for enrichment in di-, tri-, tetra-, and pentamer sequences. For each MEME search, the number of motifs searched was 20 and the minimum number of sites that should contain the motif was set to 25. The option used to describe the distribution of motif sites was any number of repetitions (anr). Selective differential enrichment function was the objective function in MEME. Only those motifs that occurred fewer than 10 nucleotides upstream or downstream of the putative cleavage site identified on the basis of the criteria described above were finally selected. For searching the top four motifs with maximum number of cleavage sites, the number of conserved bases per position was ≥2 nucleotides for dimer and trimer motifs and ≥3 nucleotides for tetramer and pentamer motifs.
Calculation of propensity of motif sequences in the cleaved transcripts.
The occurrence of a given motif was obtained in the transcripts with cleavage sites identified as described above using custom scripts. Only sequences with 10 nucleotides upstream and downstream of the cleavage site with log2(protection ratio) of ≤−1.5 were considered for motif identification. The total of all occurrences of the most frequent motifs (identified using MEME) was calculated in transcripts with cleavage sites. The fraction of occurrences of a given motif sequence in the set of transcripts with cleavage sites was calculated (Fractioncleaved). Further, the fraction of occurrences of a given motif was calculated in the M. tuberculosis genome (Fractiongenome). This was approximated as a product of fractions of individual bases at each position based on the GC content in M. tuberculosis genome (65.6%). Thus, the propensity of occurrence of a given motif (Propensitymotif) is calculated using equation 10.
| (10) |
In vitro ribonuclease assay with purified toxin MazF6, antitoxin MazE6, and in vitro-assembled MazEF6 complexes.
To further assay the activity of the toxin MazF6, 16S rRNA and 23S rRNA was isolated from M. tuberculosis (OD600 of ∼1.0); 500 ng of gel-purified RNA was then incubated with purified MazF6 toxin and MazE6 antitoxin and in vitro-assembled MazEF6 complex (MazF6:MazE6, 1:1) at final concentrations of 1, 2, and 4 μM.
Footnotes
Supplemental material is available online only.
Contributor Information
Ramandeep Singh, Email: ramandeep@thsti.res.in.
Raghavan Varadarajan, Email: varadar@iisc.ac.in.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Zhang S-P, Wang Q, Quan S-W, Yu X-Q, Wang Y, Guo D-D, Peng L, Feng H-Y, He Y-X. 2020. Type II toxin–antitoxin system in bacteria: activation, function, and mode of action. Biophys Rep 6:68–79. 10.1007/s41048-020-00109-8. [DOI] [Google Scholar]
- 2.Fraikin N, Goormaghtigh F, Van Melderen L. 2020. Type II toxin-antitoxin systems: evolution and revolutions. J Bacteriol 202:e00763-19. 10.1128/JB.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narimisa N, Kalani BS, Amraei F, Mohammadzadeh R, Mirkalantari S, Razavi S, Jazi FM. 2020. Type II toxin/antitoxin system genes expression in persister cells of Klebsiella pneumoniae. Rev Med Microbiol 31:215–220. 10.1097/MRM.0000000000000232. [DOI] [Google Scholar]
- 4.Ramisetty BCM. 2020. Regulation of type II toxin-antitoxin systems: the translation-responsive model. Front Microbiol 11:895. 10.3389/fmicb.2020.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamruzzaman M, Wu AY, Iredell JR. 2021. Biological functions of type II toxin-antitoxin systems in bacteria. Microorganisms 9:1276. 10.3390/microorganisms9061276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA 93:6059–6063. 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, Yamaguchi Y, Inouye M. 2011. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett 585:2526–2532. 10.1016/j.febslet.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JS, Kim W, Suk S, Park H, Bak G, Yoon J, Lee Y. 2018. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol 15:1319–1335. 10.1080/15476286.2018.1532252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lobato-Márquez D, Díaz-Orejas R, García-del Portillo F. 2016. Toxin-antitoxins and bacterial virulence. FEMS Microbiol Rev 40:592–609. 10.1093/femsre/fuw022. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Yao J, Sun YC, Wood TK. 2021. Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol 29:388–393. 10.1016/j.tim.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4:19. 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45:61–79. 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 13.Tripathi A, Dewan PC, Barua B, Varadarajan R. 2012. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc Natl Acad Sci USA 109:12497–12502. 10.1073/pnas.1121217109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen Y, Behiels E, Devreese B. 2014. Toxin-antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog Dis Pathog Dis 70:240–249. 10.1111/2049-632X.12145. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Zhao X, Wang H, Huang X, Duan X, Gu Y, Lambert N, Zhang K, Kou Z, Xie J. 2018. Mycobacterium tuberculosis toxin Rv2872 is an RNase involved in vancomycin stress response and biofilm development. Appl Microbiol Biotechnol 102:7123–7133. 10.1007/s00253-018-9132-0. [DOI] [PubMed] [Google Scholar]
- 16.Ma D, Mandell JB, Donegan NP, Cheung AL, Ma W, Rothenberger S, Shanks RMQ, Richardson AR, Urish KL. 2019. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 10:e01658-19. 10.1128/mBio.01658-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song S, Wood TK. 2020. A primary physiological role of toxin/antitoxin systems is phage inhibition. Front Microbiol 11:1895. 10.3389/fmicb.2020.01895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopatina A, Tal N, Sorek R. 2020. Abortive infection: bacterial suicide as an antiviral immune strategy. Annu Rev Virol 7:371–384. 10.1146/annurev-virology-011620-040628. [DOI] [PubMed] [Google Scholar]
- 19.Picton DM, Luyten YA, Morgan RD, Nelson A, Smith DL, Dryden DTF, Hinton JCD, Blower TR. 2021. The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses. Nucleic Acids Res 49:11257–11273. 10.1093/nar/gkab906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurata T, Saha CK, Buttress JA, Mets T, Brodiazhenko T, Turnbull KJ, Awoyomi OF, Oliveira SRA, Jimmy S, Ernits K, Delannoy M, Persson K, Tenson T, Strahl H, Hauryliuk V, Atkinson GC. 2022. A hyperpromiscuous antitoxin protein domain for the neutralization of diverse toxin domains. Proc Natl Acad Sci USA 119:e2102212119. 10.1073/pnas.2102212119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K. 2017. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964-17. 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401. 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison JJ, Wade WD, Akierman S, Vacchi-Suzzi C, Stremick CA, Turner RJ, Ceri H. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob Agents Chemother 53:2253–2258. 10.1128/AAC.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akarsu H, Bordes P, Mansour M, Bigot DJ, Genevaux P, Falquet L. 2019. TASmania: a bacterial toxin-antitoxin systems database. PLoS Comput Biol 15:e1006946. 10.1371/journal.pcbi.1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tandon H, Sharma A, Wadhwa S, Varadarajan R, Singh R, Srinivasan N, Sandhya S. 2019. Bioinformatic and mutational studies of related toxin–antitoxin pairs in Mycobacterium tuberculosis predict and identify key functional residues. J Biol Chem 294:9048–9063. 10.1074/jbc.RA118.006814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A. 2009. Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol Lett 290:45–53. 10.1111/j.1574-6968.2008.01400.x. [DOI] [PubMed] [Google Scholar]
- 29.Winther K, Tree JJ, Tollervey D, Gerdes K. 2016. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res 44:9860–9871. 10.1093/nar/gkw781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal S, Tiwari P, Deep A, Kidwai S, Gupta S, Thakur KG, Singh R. 2018. System-wide analysis unravels the differential regulation and in vivo essentiality of virulence-associated proteins B and C toxin-antitoxin systems of mycobacterium tuberculosis. J Infect Dis 217:1809–1820. 10.1093/infdis/jiy109. [DOI] [PubMed] [Google Scholar]
- 31.Tiwari P, Arora G, Singh M, Kidwai S, Narayan OP, Singh R. 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat Commun 6:6059. 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 32.Singh R, Barry CE, Boshoff HIM. 2010. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J Bacteriol 192:1279–1291. 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han JS, Lee JJ, Anandan T, Zeng M, Sripathi S, Jahng WJ, Lee SH, Suh JW, Kang CM. 2010. Characterization of a chromosomal toxin-antitoxin, Rv1102c-Rv1103c system in Mycobacterium tuberculosis. Biochem Biophys Res Commun 400:293–298. 10.1016/j.bbrc.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 34.Sharma A, Chattopadhyay G, Chopra P, Bhasin M, Thakur C, Agarwal S, Ahmed S, Chandra N, Varadarajan R, Singh R. 2020. VapC21 toxin contributes to drug-tolerance and interacts with non-cognate VapB32 antitoxin in Mycobacterium tuberculosis. Front Microbiol 11:2037. 10.3389/fmicb.2020.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mets T, Lippus M, Schryer D, Liiv A, Kasari V, Paier A, Maiväli Ü, Remme J, Tenson T, Kaldalu N. 2017. Toxins MazF and MqsR cleave Escherichia coli rRNA precursors at multiple sites. RNA Biol 14:124–135. 10.1080/15476286.2016.1259784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culviner PH, Laub MT. 2018. Global analysis of the E. coli toxin MazF reveals widespread cleavage of mRNA and the inhibition of rRNA maturation and ribosome biogenesis. Mol Cell 70:868–880.e10. 10.1016/j.molcel.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Culviner PH, Nocedal I, Fortune SM, Laub MT. 2021. Global analysis of the specificities and targets of endoribonucleases from Escherichia coli toxin-antitoxin systems. mBio 12:e0201221. 10.1128/mBio.02012-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 12:913–923. 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 39.Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem 281:18638–18643. 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 40.Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol 69:559–569. 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, Woychik NA. 2013. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci USA 110:8501–8506. 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schifano JM, Vvedenskaya IO, Knoblauch JG, Ouyang M, Nickels BE, Woychik NA. 2014. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat Commun 5:3538. 10.1038/ncomms4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schifano JM, Cruz JW, Vvedenskaya IO, Edifor R, Ouyang M, Husson RN, Nickels BE, Woychik NA. 2016. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res 44:1256–1270. 10.1093/nar/gkv1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn DH, Lee KY, Lee SJ, Park SJ, Yoon HJ, Kim SJ, Lee BJ. 2017. Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor. J Biol Chem 292:18832–18847. 10.1074/jbc.M117.807974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen R, Tu J, Tan Y, Cai X, Yang C, Deng X, Su B, Ma S, Liu X, Ma P, Du C, Xie W. 2019. Structural and biochemical characterization of the cognate and heterologous interactions of the MazEF-mt9 TA system. ACS Infect Dis 5:1306–1316. 10.1021/acsinfecdis.9b00001. [DOI] [PubMed] [Google Scholar]
- 46.Chen R, Zhou J, Sun R, Du C, Xie W. 2020. Conserved conformational changes in the regulation of Mycobacterium tuberculosis MazEF-mt1. ACS Infect Dis 6:1783–1795. 10.1021/acsinfecdis.0c00048. [DOI] [PubMed] [Google Scholar]
- 47.Chandra S, Chattopadhyay G, Varadarajan R. 2021. Rapid identification of secondary structure and binding site residues in an intrinsically disordered protein segment. Front Genet 12:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cherny I, Rockah L, Gazit E. 2005. The YoeB toxin is a folded protein that forms a physical complex with the unfolded YefM antitoxin. Implications for a structural-based differential stability of toxin-antitoxin systems. J Biol Chem 280:30063–30072. 10.1074/jbc.M506220200. [DOI] [PubMed] [Google Scholar]
- 49.Cherny I, Overgaard M, Borch J, Bram Y, Gerdes K, Gazit E. 2007. Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry 46:12152–12163. 10.1021/bi701037e. [DOI] [PubMed] [Google Scholar]
- 50.Nieto C, Cherny I, Khoo SK, de Lacoba MG, Chan WT, Yeo CC, Gazit E, Espinosa M. 2007. The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J Bacteriol 189:1266–1278. 10.1128/JB.01130-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahoo A, Khare S, Devanarayanan S, Jain PC, Varadarajan R. 2015. Residue proximity information and protein model discrimination using saturation-suppressor mutagenesis. Elife 4:e09532. 10.7554/eLife.09532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teale FW, Weber G. 1957. Ultraviolet fluorescence of the aromatic amino acids. Biochem J 65:476–482. 10.1042/bj0650476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamada K, Hanaoka F, Burley SK. 2003. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell 11:875–884. 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 54.Simanshu DK, Yamaguchi Y, Park J-H, Inouye M, Patel DJ. 2013. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol Cell 52:447–458. 10.1016/j.molcel.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zorzini V, Buts L, Sleutel M, Garcia-Pino A, Talavera A, Haesaerts S, De Greve H, Cheung A, van Nuland NAJ, Loris R. 2014. Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics. Nucleic Acids Res 42:6709–6725. 10.1093/nar/gku266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams RM, Mora T, Walczak AM, Kinney JB. 2016. Measuring the sequence-affinity landscape of antibodies with massively parallel titration curves. Elife 5:e23156. 10.7554/eLife.23156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao L, Zhang J. 2008. Biochemical characterization of a chromosomal toxin-antitoxin system in Mycobacterium tuberculosis. FEBS Lett 582:710–714. 10.1016/j.febslet.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 58.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology (Reading) 148:3129–3138. 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 59.Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Steyn AJC. 2007. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci USA 104:11562–11567. 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z. 2011. TADB: a web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res 39:D606–D611. 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sala A, Bordes P, Genevaux P. 2014. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6:1002–1020. 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta A, Venkataraman B, Vasudevan M, Gopinath Bankar K. 2017. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci Rep 7:5868. 10.1038/s41598-017-06003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miallau L, Faller M, Janet C, Arbing M, Guo F, Cascio D, Eisenberg D. 2009. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems VapBC-5 from Mycobacterium tuberculosis. J Biol Chem 284:276–283. 10.1074/jbc.M805061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das U, Pogenberg V, Subhramanyam UKT, Wilmanns M, Gourinath S, Srinivasan A. 2014. Crystal structure of the VapBc-15 complex from mycobacterium tuberculosis reveals a two-metal ion dependent pin-domain ribonuclease and a variable mode of toxin-antitoxin assembly. J Struct Biol 188:249–258. 10.1016/j.jsb.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Lee IG, Lee SJ, Chae S, Lee KY, Kim JH, Lee BJ. 2015. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res 43:7624–7637. 10.1093/nar/gkv689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deep A, Tiwari P, Agarwal S, Kaundal S, Kidwai S, Singh R, Thakur KG. 2018. Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin–antitoxin system: targeting a tRNase to tackle mycobacterial adaptation. Nucleic Acids Res 46:11639–11655. 10.1093/nar/gky924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cintrón M, Zeng JM, Barth VC, Cruz JW, Husson RN, Woychik NA. 2019. Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci Rep 9:5949. 10.1038/s41598-019-41548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hewinson RG, Michell SL, Russell WP, McAdam RA, Jacobs WR. 1996. Molecular characterization of MPT83: a seroreactive antigen of Mycobacterium tuberculosis with homology to MPT70. Scand J Immunol 43:490–499. 10.1046/j.1365-3083.1996.d01-78.x. [DOI] [PubMed] [Google Scholar]
- 69.Clemmensen HS, Dube JY, McIntosh F, Rosenkrands I, Jungersen G, Aagaard C, Andersen P, Behr MA, Mortensen R. 2021. In vivo antigen expression regulates CD4 T cell differentiation and vaccine efficacy against mycobacterium tuberculosis infection. mBio 12:e00226-21. 10.1128/mBio.00226-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rezwan M, Grau T, Tschumi A, Sander P. 2007. Lipoprotein synthesis in mycobacteria. Microbiology (Reading) 153:652–658. 10.1099/mic.0.2006/000216-0. [DOI] [PubMed] [Google Scholar]
- 71.England K, Crew R, Slayden RA. 2011. Mycobacterium tuberculosis septum site determining protein, Ssd encoded by rv3660c, promotes filamentation and elicits an alternative metabolic and dormancy stress response. BMC Microbiol 11:79. 10.1186/1471-2180-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mehta M, Rajmani RS, Singh A. 2016. Mycobacterium tuberculosis WhiB3 responds to vacuolar pH-induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. J Biol Chem 291:2888–2903. 10.1074/jbc.M115.684597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park H-D, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol 48:833–843. 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R, Pushparajan AR, Mundayoor S, Jaleel A, Kumar RA. 2015. Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol Cell Proteomics 14:2160–2176. 10.1074/mcp.M115.051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 76.Rustad TR, Harrell MI, Liao R, Sherman DR. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502-8. 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.LeRoux M, Culviner PH, Liu YJ, Littlehale ML, Laub MT. 2020. Stress can induce transcription of toxin-antitoxin systems without activating toxin. Mol Cell 79:280–292.e8. 10.1016/j.molcel.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee J-H, Karakousis PC, Bishai WR. 2008. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J Bacteriol 190:699–707. 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fontán PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. 2009. The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191:5628–5633. 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh S, Goswami N, Tyagi AK, Khare G. 2019. Unraveling the role of the transcriptional regulator VirS in low pH-induced responses of Mycobacterium tuberculosis and identification of VirS inhibitors. J Biol Chem 294:10055–10075. 10.1074/jbc.RA118.005312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marcus SA, Sidiropoulos SW, Steinberg H, Talaat AM. 2016. CsoR is essential for maintaining copper homeostasis in Mycobacterium tuberculosis. PLoS One 11:e0151816. 10.1371/journal.pone.0151816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJC. 2009. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog 5:e1000545. 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Müller R, Stewart AF, Zhang Y. 2012. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30:440–446. 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 84.Chattopadhyay G, Varadarajan R. 2019. Facile measurement of protein stability and folding kinetics using a nano differential scanning fluorimeter. Protein Sci 28:1127–1134. 10.1002/pro.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chattopadhyay G, Bhowmick J, Manjunath K, Ahmed S, Goyal P, Varadarajan R. 2021. Mechanistic insights into global suppressors of protein folding defects. bioRxiv 10.1101/2021.11.18.469098. [DOI] [PMC free article] [PubMed]
- 86.Najar TA, Khare S, Pandey R, Gupta SK, Varadarajan R. 2017. Mapping protein binding sites and conformational epitopes using cysteine labeling and yeast surface display. Structure 25:395–406. 10.1016/j.str.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 87.Ahmed S, Bhasin M, Manjunath K, Varadarajan R. 2022. Prediction of residue-specific contributions to binding and thermal stability using yeast surface display. Front Mol Biosci 8:800819. 10.3389/fmolb.2021.800819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. 2010. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1:1038. 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 89.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. 2011. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9:342–353. 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seidel SAI, Wienken CJ, Geissler S, Jerabek-Willemsen M, Duhr S, Reiter A, Trauner D, Braun D, Baaske P. 2012. Label-free microscale thermophoresis discriminates sites and affinity of protein-ligand binding. Angew Chem Int Ed Engl 51:10656–10659. 10.1002/anie.201204268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh R, Singh M, Arora G, Kumar S, Tiwari P, Kidwai S. 2013. Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J Bacteriol 195:2839–2851. 10.1128/JB.00038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andrews S. 2010. FastQC. A quality control tool for high throughput sequence data. Babraham Institute, Cambridgeshire, United Kingdom. [Google Scholar]
- 93.Krueger F. 2012. Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. Babraham Institute, Cambridgeshire, United Kingdom. [Google Scholar]
- 94.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mishra S, Shukla P, Bhaskar A, Anand K, Baloni P, Jha RK, Mohan A, Rajmani RS, Nagaraja V, Chandra N, Singh A. 2017. Efficacy of β-lactam/β-lactamase inhibitor combination is linked to WhiB4-mediated changes in redox physiology of Mycobacterium tuberculosis. Elife 6:1–30. 10.7554/eLife.25624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. 2003. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31:258–261. 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bhosle A, Datey A, Chandrasekharan G, Singh D, Chakravortty D, Chandra N. 2020. A strategic target rescues trimethoprim sensitivity in Escherichia coli. iScience 23:100986. 10.1016/j.isci.2020.100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sambarey A, Prashanthi K, Chandra N. 2013. Mining large-scale response networks reveals “topmost activities” in Mycobacterium tuberculosis infection. Sci Rep 3:2302–2314. 10.1038/srep02302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental file 1. Download jb.00058-22-s0001.pdf, PDF file, 2.7 MB (2.7MB, pdf)