Abstract
The affinity of [3H]benzylpenicillin for penicillin-binding protein (PBP) 3A was reduced in 25 clinical isolates of β-lactamase-negative ampicillin (AMP)-resistant (BLNAR) Haemophilus influenzae for which the AMP MIC was ≥1.0 μg/ml. The affinities of PBP 3B and PBP 4 were also reduced in some strains. The sequences of the ftsI gene encoding the transpeptidase domain of PBP 3A and/or PBP 3B and of the dacB gene encoding PBP 4 were determined for these strains and compared to those of AMP-susceptible Rd strains. The BLNAR strains were classified into three groups on the basis of deduced amino acid substitutions in the ftsI gene, which is thought to be involved in septal peptidoglycan synthesis. His-517, near the conserved Lys-Thr-Gly (KTG) motif, was substituted for Arg-517 in group I strains (n = 9), and Lys-526 was substituted for Asn-526 in group II strains (n = 12). In group III strains (n = 4), three residues (Met-377, Ser-385, and Leu-389), positioned near the conserved Ser-Ser-Asn (SSN) motif, were replaced with Ile, Thr, and Phe, respectively, in addition to the replacement with Lys-526. The MICs of cephem antibiotics with relatively high affinities for PBP 3A and PBP 3B were higher than those of AMP and meropenem for group III strains. The MICs of β-lactams for H. influenzae transformants into which the ftsI gene from BLNAR strains was introduced were as high as those for the donors, and PBP 3A and PBP 3B showed decreased affinities for β-lactams. There was no clear relationship between 7-bp deletions in the dacB gene and AMP susceptibility. Even though mutations in another gene(s) may be involved in β-lactam resistance, these data indicate that mutations in the ftsI gene are the most important for development of resistance to β-lactams in BLNAR strains.
Haemophilus influenzae is one of the important pathogens causing respiratory tract infection (RTI), acute otitis media (AOM), pneumonia, and purulent meningitis. Ampicillin (AMP) resistance in this organism is due to two well-known mechanisms. One is resistance mediated by the production of TEM-1 (20) and ROB-1 (13) β-lactamases, and the other is decreasing affinity of AMP for penicillin-binding proteins (PBPs) involved in peptidoglycan synthesis (14–16, 19). Strains with resistance due to the latter mechanism are termed β-lactamase-negative AMP-resistant (BLNAR) H. influenzae.
In surveillance studies conducted in the United States the incidence of β-lactamase-producing AMP-resistant (BLPAR) H. influenzae has gradually increased (6, 7, 10) and accounted for 36.4% of all isolates (5) in 1994 and 1995. In contrast, BLNAR isolates remain extremely uncommon in the United States.
In Japan, according to nation wide surveillance studies conducted in 1997 and 1998, the proportion of clinical isolates supposed to be BLNAR has rapidly increased to 28.8% in parallel with the increased prevalence of penicillin (PEN)-resistant Streptcoccus pneumoniae. The characteristic of resistant H. influenzae isolates is that the AMP MIC is ≥1 μg/ml, whereas the MIC for susceptible strains is 0.25 μg/ml. The percentage of BLPAR strains in Japan is low, with 13.9% of the strains isolated from patients with RTIs and AOM being BLPAR strains, whereas 35% of the strains from patients with meningitis are BLPAR strains.
These differences in the prevalence of resistant strains between the two countries are thought to reflect differences in the use of chemotherapeutic agents for the treatment of community-acquired RTIs.
We set out to identify a PBP involved in the mechanism of resistance of BLNAR strains and to identify the mutation(s) in the gene encoding this PBP. Strains for which AMP MICs were ≥1 μg/ml were randomly selected, and their PBP profiles were analyzed. The affinity of [3H]PEN for PBP 3A was decreased in all strains, and those for PBP 3B and PBP 4 were also decreased in some strains. Subsequently, on the basis of the whole genome sequence of H. influenzae Rd (8), the nucleotide sequences of the ftsI and dacB genes, which encode PBP 3 and PBP 4, respectively, were determined. H. influenzae strains for which AMP MICs were ≥1 μg/ml had several common amino acid substitutions in the ftsI gene. Seven-base-pair deletions in the dacB gene were not related to β-lactam resistance.
In addition, elevations of β-lactam MICs and decreased affinities for PBP 3A and PBP 3B were verified in transformants of strain Rd into which the ftsI gene from BLNAR strains was introduced.
Here we report that the main factors in the β-lactam resistance of BLNAR strains are mutations in the ftsI gene encoding PBP 3A and/or PBP 3B, which is involved in septal peptidoglycan synthesis.
MATERIALS AND METHODS
Clinical isolates.
The strains used in this study are listed in Table 1. Twenty-five H. influenzae strains for which the AMP MIC was ≥1 μg/ml were selected from clinical isolates collected by the Japanese Surveillance Group of Community-Acquired Respiratory Tract Infections Caused by S. pneumoniae and Other Pathogens between January 1998 and December 1998. Table 1 also gives the characteristics of the AMP-susceptible and BLPAR strains used as controls. All strains required β-NAD+ (V factor) and hemin (X factor) for growth when incubated at 37°C in room air. The production of β-lactamase was confirmed by a nitrocefin test (Showa Chemical Inc., Tokyo, Japan) with whole cells. Isolates were serotyped by the capsular swelling technique with antisera purchased from Difco Laboratories (Detroit, Mich.).
TABLE 1.
Serotype, β-lactamase production, PBP affinity, and susceptibility to β-lactam antibiotics of the H. influenzae strains useed in this study
| Assigned group | Strain | Sourcea | Serotype | β-Lactamase | PBP affinity
|
MIC (μg/ml)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBP 3A | PBP 3B | PBP 4 | AMP | CEC | CTX | CRO | CPD | CDR | CDN | CPN | FRM | MEM | |||||
| Control | MT196 | Na | NSb | − | Nc | N | N | 0.125 | 0.5 | 0.008 | 0.004 | 0.016 | 0.25 | 0.008 | 0.008 | 0.25 | 0.031 |
| MT053 | Na | NS | − | N | N | N | 0.25 | 1 | 0.008 | 0.004 | 0.031 | 0.125 | 0.008 | 0.008 | 0.25 | 0.031 | |
| TK422 | Na | NS | − | N | N | N | 0.5 | 2 | 0.031 | 0.008 | 0.063 | 0.5 | 0.016 | 0.016 | 0.5 | 0.063 | |
| TC01 | CSF | b | + | NDd | ND | ND | 8 | 2 | 0.016 | 0.004 | 0.063 | 0.5 | 0.016 | 0.016 | 0.5 | 0.031 | |
| AK01 | CSF | b | + | ND | ND | ND | 8 | 2 | 0.016 | 0.008 | 0.063 | 0.5 | 0.008 | 0.016 | 0.25 | 0.031 | |
| I | ME2014 | Ot | NS | − | De | N | N | 1 | 32 | 0.063 | 0.016 | 0.25 | 2 | 0.031 | 0.063 | 2 | 0.031 |
| ME1547 | Na | NS | − | D | N | N | 1 | 64 | 0.063 | 0.008 | 0.5 | 4 | 0.031 | 0.125 | 1 | 0.063 | |
| ME692 | Na | NS | − | D | N | N | 1 | 32 | 0.063 | 0.016 | 0.5 | 4 | 0.063 | 0.125 | 0.5 | 0.125 | |
| ME1503 | Na | NS | − | D | N | N | 1 | 64 | 0.063 | 0.016 | 1 | 8 | 0.063 | 0.125 | 2 | 0.125 | |
| H17 | Na | NS | − | D | N | N | 1 | 32 | 0.063 | 0.008 | 0.25 | 2 | 0.031 | 0.125 | 1 | 0.125 | |
| N130 | Na | NS | − | D | N | N | 1 | 32 | 0.063 | 0.016 | 0.25 | 2 | 0.031 | 0.125 | 1 | 0.25 | |
| H2 | Na | NS | − | D | N | N | 4 | 64 | 0.25 | 0.031 | 1 | 2 | 0.063 | 0.25 | 1 | 0.025 | |
| ME1495 | Na | NS | + | ND | ND | ND | 64 | 64 | 0.125 | 0.016 | 0.5 | 4 | 0.063 | 0.125 | 1 | 0.031 | |
| H7 | Na | NS | + | ND | ND | ND | 64 | 64 | 0.125 | 0.031 | 4 | 8 | 0.063 | 0.5 | 1 | 0.25 | |
| II | ND01 | CSF | b | − | D | N | N | 1 | 32 | 0.063 | 0.016 | 0.25 | 8 | 0.031 | 0.125 | 2 | 0.25 |
| KK01 | CSF | b | − | D | N | N | 1 | 64 | 0.063 | 0.031 | 0.25 | 4 | 0.031 | 0.125 | 0.5 | 0.125 | |
| ME570 | Na | NS | − | D | D | N | 1 | 64 | 0.063 | 0.016 | 1 | 2 | 0.031 | 0.125 | 1 | 0.25 | |
| ME587 | Na | NS | − | D | D | N | 1 | 64 | 0.063 | 0.008 | 0.25 | 4 | 0.031 | 0.063 | 1 | 0.25 | |
| ME495 | Na | NS | − | D | N | N | 1 | 64 | 0.125 | 0.031 | 1 | 4 | 0.063 | 0.25 | 2 | 0.125 | |
| MT273 | Na | NS | − | D | N | N | 2 | 32 | 0.063 | 0.016 | 0.5 | 2 | 0.031 | 0.125 | 8 | 0.5 | |
| MT160 | Na | NS | − | D | N | N | 2 | 32 | 0.063 | 0.008 | 0.5 | 2 | 0.031 | 0.125 | 2 | 0.5 | |
| ME267 | Ot | NS | − | D | D | N | 2 | 32 | 0.063 | 0.016 | 0.25 | 1 | 0.031 | 0.063 | 2 | 0.25 | |
| ME040 | Ot | NS | − | D | N | D | 2 | 64 | 0.063 | 0.063 | 1 | 4 | 0.063 | 0.25 | 2 | 0.25 | |
| ME1210 | Na | NS | − | D | D | N | 4 | 64 | 0.125 | 0.031 | 1 | 4 | 0.063 | 0.25 | 2 | 0.25 | |
| MT313 | Na | NS | − | D | N | N | 4 | 64 | 0.125 | 0.031 | 1 | 4 | 0.063 | 0.25 | 2 | 0.25 | |
| ME1976 | Na | NS | + | ND | ND | ND | 64 | 32 | 0.063 | 0.016 | 0.25 | 2 | 0.016 | 0.125 | 1 | 0.063 | |
| III | MT066 | Na | NS | − | D | D | N | 2 | 64 | 1 | 0.25 | 4 | 8 | 0.25 | 1 | 2 | 0.25 |
| MT070 | Na | NS | − | D | D | N | 2 | 64 | 1 | 0.25 | 4 | 8 | 0.125 | 1 | 4 | 0.125 | |
| ME1133 | Na | NS | − | D | N | D | 4 | 64 | 1 | 0.25 | 8 | 16 | 0.25 | 2 | 2 | 0.25 | |
| ME870 | Na | NS | − | D | D | N | 4 | 64 | 2 | 0.25 | 4 | 16 | 0.5 | 2 | 4 | 0.5 | |
Na, nasopharynx; Ot, otorrhea; CSF; cerebrospinal fluid.
NS, not specified.
N, PBP affinity is normal.
ND, not determined.
D, PBP affinity is decreased.
Media and antibiotics.
The medium used for growth and determination of the MIC was Mueller-Hinton agar or broth (Becton Dickinson and Company, Sparks, Md.) supplemented with 2% defibrinated and heat-treated horse blood, 0.5% yeast extract, and 15 μg of β-NAD+ per ml. Chocolate agar II plates purchased from Nippon Becton Dickinson Co. Ltd. (Tokyo, Japan) were used routinely. Plate cultures were incubated at 37°C in an atmosphere of 7% CO2. The β-lactams used in the study were AMP and cefditoren (CDN; Meiji Seika Kaisha, Ltd., Tokyo, Japan), cefotaxime (CTX; Nippon Hoechst Marion Roussel Ltd., Tokyo, Japan), ceftriaxone (CRO; Nippon Roch Ltd., Tokyo, Japan), cefaclor (CEC) and cefcapene (CPN) (Shionogi & Co., Ltd., Osaka, Japan), cefdinir (CDR; Fujisawa Pharmaceutical Co., Ltd., Osaka, Japan), cefpodoxime (CPD; Sankyo Co., Ltd., Tokyo, Japan), faropenem (FRM; Suntory Ltd., Osaka, Japan), and meropenem (MEM; Sumitomo Pharmaceuticals Co., Ltd., Tokyo, Japan).
Antimicrobial susceptibility testing.
The strains were inoculated onto chocolate II agar plates from vials of 10% skim milk stored at −80°C and were incubated overnight. The next day, the strains were suspended in liquid medium (optical density [OD], 0.5 at A560) and diluted 100-fold, and 10 μl of each diluted strain was inoculated onto the susceptibility test plate with a Steers replicator. The MIC was defined as the lowest concentration of antibiotic that inhibited visible growth of the inoculum in comparison with the growth on antibiotic-free plates. Plates were examined after 20 h of incubation in 7% CO2 at 37°C.
PBP profiles and PBP affinities to β-lactams.
Membrane preparations of H. influenzae and PBP binding reactions with [3H]PEN (9.25 MBq; Amersham, Little Chalfont, England) were performed as described previously (1). For bacterial cell cultures, however, cells were Mueller-Hinton in broth containing XV supplements to the logarithmic growth phase (OD, 0.6 at A560). The affinities of several β-lactam antibiotics for PBPs of AMP-susceptible strain H. influenzae TK422 were determined by competition assay. The relative affinity of each compound for PBPs was determined as follows. A 30-μl aliquot of each membrane fraction was incubated with 3 μl of nonradioactive β-lactams diluted to various concentrations for 10 min and then further incubated with 3 μl of [3H]PEN for another 10 min. PBPs were visualized by autoradiography after exposure of the dried gels to Kodak X-OMAT film for 20 days at −80°C.
Transformation.
H. influenzae Rd (ATCC 51907) was grown in XV-supplemented brain heart infusion broth (sBHI) to an OD at 650 nm of 0.5 for about 4 h at 37°C. The cells were chilled on ice for 30 min, harvested by centrifugation at 6,500 rpm in a Hitachi CR20E centrifuge for 10 min, and thoroughly washed five times with SG buffer (10% glycerol, 10% sucrose) at 4°C. The cell pellet was finally suspended in SG buffer to provide a 100-fold increase in the concentration of competent cells, and the cells were stored at −80°C until use.
An open reading frame (ORF) that corresponded to the ftsI gene in several BLNAR strains was amplified by PCR with a sense primer (trans-PBP3S [5′-G1700GACGATTTGGATAACCCATA1720-3′]) and a reverse primer (trans-PBP3R [5′-G3966CTGGATAATTCTGTCTCAGA3946-3′]). PCR was carried out for 30 cycles of incubation, with each cycle consisting of 94°C for 30 s, 52°C for 1 min, and 74°C for 1 min. The PCR product was purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and eluted with 300 μl of distilled water. DNA samples of 5 μl each were added to 50 μl of the ice-chilled competent cells, and the mixture was allowed to stand on ice for 1 min and quantitatively transferred to cuvettes. Samples were electroporated in cuvettees with a 0.1-cm electrode gap by using a GENE PULSER II electroporation apparatus (Bio-Rad Laboratories, Hercules, Calif). The conditions for electropolation were 1.25 kV/cm, 200 Ω, and 25 μF with time constants of 4.7 to 4.8 ms. After that, the cells were immediately mixed with 1 ml of sBHI, incubated at 37°C for 1.5 h, and spread onto selective agar plates containing CEC at 5 and 10 μg/ml or CDR at 2 μg/ml. Colonies grown on selective agar plates were picked at random, and the MICs of the β-lactam antibiotics for the colonies were determined. The PBP affinities of the β-lactams and ftsI gene sequencing were also investigated for several transformants to confirm ftsI gene transfer.
Sequencing.
The 1.0-kb DNA fragment encoding the PBP 3 transpeptidase domain and the 1.3-kb DNA fragment encoding PBP 4 were amplified from the chromosomal DNA of H. influenzae by PCR as reported previously (2). One colony of H. influenzae grown on a chocolate II agar plate was picked and placed in 30 μl of lysis solution, and the mixture was incubated at 60°C for 20 min and then at 94°C for 5 min. Subsequently, 2 μl of the lysate was added to 50 μl of a PCR solution (5 μl of 10 × PCR buffer, 4 μl of a mixture of deoxynucleoside triphosphates at a concentration of 2.5 mM each, 2.5 U of Taq polymerase [Takara Biomedicals, Kyoto, Japan], 0.5 ml of 10 pmol sense and reverse primer per μl). Primers were as follows: the sense primer for ftsI was 5′-G712TTGCACATATCTCCGATGAG732 and the reverse primer was 5′C1762AGCTGCTTCAGCATCTTGC1743; the sense primer for dacB was 5′T13CTTCAATTTCCACCGCACT32, and the reverse primer was 5′-G1550CGACAAACAGTTCAATGAG1531. PCR conditions were as follows: 30 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min. The PCR products were electrophoresed on a 1.2% agarose gel to confirm the presence of the product and were then purified with a Chroma Spin-100 Column (Clontech Laboratories Inc., Palo Alto, Calif.) to prepare a sequencing template.
The sequencing reaction was conducted with a dRhodamine Terminator Cycle Sequencing FS Ready Reaction kit (PE Biosystems, Foster, Calif.). The reaction mixtures were placed in a thermal cycler and denatured at 94°C for 2 min. They were then subjected to 25 PCR cycles each of which consisted of 94°C for 10 s, 50°C for 5 s, and 60°C for 4 min. The nucleotide sequences were determined with an ABI PRISM377 DNA sequencer.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in the present study will appear in the DDBJ/EMBL/GeneBank nucleotide sequence databases under the following accession numbers: for the ftsI gene, AB035737 (strain MT196), AB035738 (strain KK01), AB035739 (strain H2), and AB035740 (strain ME870); for the dacB gene, AB035864 (strain MT196) and AB035865 (strain MT040).
RESULTS
Characterization and β-lactam susceptibility of H. influenzae.
Table 1 shows the source of infection, capsular serotype, β-lactamase production, PBP affinities, and MICs of 10 β-lactam antibiotics for 25 BLNAR strains of H. influenzae used in the present study. Three AMP-susceptible H. influenzae strains and two BLPAR H. influenzae strains were used as controls. Three BLPAR H. influenzae strains for which the CEC MICs were ≥32 μg/ml were and that were thought to have an additional element of non-β-lactamase-mediated resistance were also analyzed.
As described below for the results of DNA sequencing, these strains were classified into three groups according to the mutations identified in the ftsI gene. The MICs of all β-lactam antibiotics tested were higher for the strains classified into groups I and II than for AMP-susceptible strain MT196; the specific increases were as follows: AMP, 8 to 32 times; CEC, 64 to 128 times; CTX, 8 to 32 times; CRO, 2 to 8 times; CPD, 16 to 128 times; CDR, 4 to 32 times; CDN 2 to 8 times; CPN 8 to 64 times; FRM, 2 to 32 times; and MEM, 1 to 16 times. In contrast, four BLNAR strains classified into group III were apparently resistant to cephem antibiotics such as CTX (MIC, 128 to 256 times higher than that for MT194), CPD (MIC, 256 to 512 times higher), and CPN (MIC, 128 to 256 times higher), but not to CRO (MIC, 64 times higher), CDR (MIC, 32 to 64 times higher), and CDN (MIC, 16 to 64 times higher). However, the MICs of AMP (16 to 32 times higher), FRM (8 to 16 times higher), and MEM (4 to 16 times higher) for strains in group III were comparable to those for strains in groups I and II.
Two strains of H. influenzae serotype b isolated from the cerebrospinal fluid of children with meningitis were included in group II.
PBP profiles.
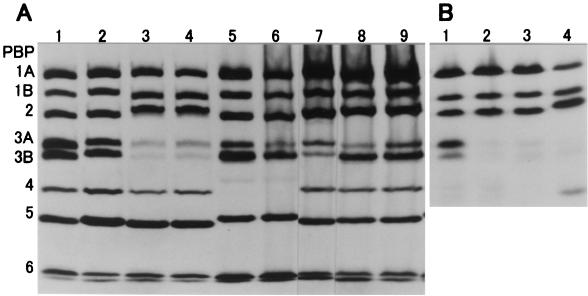
Figure 1A shows the PBP profiles of BLNAR strains and of AMP-susceptible strains MT053 and MT196. PBPs were named according to the proposal of Parr and Bryan (19) from those with the largest molecular weight to those with the smallest molecular weight. In every BLNAR strain for which the AMP MIC was ≥1 μg/ml, the PBP 3A affinity of [3H]PEN was commonly decreased. The affinities to PBP 3B of strains ME066, ME870, and ME587 and PBP 4 of strains MT040 and ME1133 were also decreased.
FIG. 1.
PBP profiles of AMP-susceptible and BLNAR H. influenzae strains (A) and Rd strains transformed with the ftsI gene encoding PBP 3 (B). Membrane fractions were labeled with [3H]PEN for 10 min at 30°C and were then subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and fluorography (I). PBPs were named according to the proposal of Parr and Bryan (19), our data on PBP affinity, and morphologic observations. AMP MICs are as follows: for strain MT053 (lane 1) and MT196 (lane 2), 0.125 μg/ml; for strain MT066 (lane 3), 2 μg/ml; for strain ME870 (lane 4), 4 μg/ml; for strain MT040 (lane 5), 2 μg/ml; for strain ME1133 (lane 6), 4 μg/ml; for strain ME587 (lane 7), 1 μg/ml; for strain KK01 (lane 8), 1 μg/ml; for strain ME495 (lane 9), 1 μg/ml. (B) lane 1, recipient Rd strain of H. influenzae; lane 2, transformed strain Rd3A/ME870-11; lane 3, transformed strain Rd3A/ME870-12; lane 4, BLNAR strain ME870.
Binding of β-lactams to PBPs.
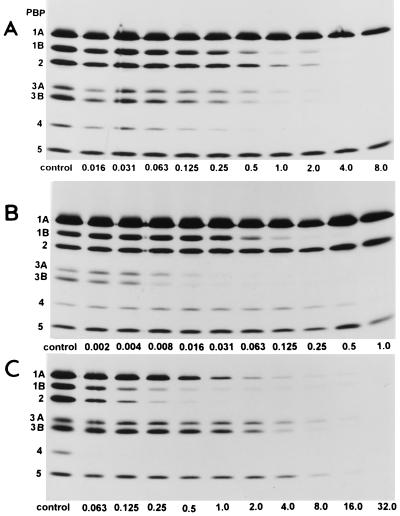
Figure 2 shows fluorograms of PBPs of H. influenzae TK422 pretreated with various concentrations of nonlabeled AMP (Fig. 2A), CTX (Fig. 2B), and FRM (Fig. 2C). PBP affinity patterns varied depending on the chemical structures of the β-lactam antibiotics. The relative orders of the affinities were as follows: for AMP, PBP 4 > PBP 1B = PBP 3A = PBP 3B > PBP 2 > PBP 1A > PBP 5; for CTX, CPD, CDR, and CDN, PBP 3A = PBP 3B > PBP 1B > PBP 4 > PBP 1A = PBP 2 = PBP 5; and for FRM and MEM, PBP 4 > PBP 2 = PBP 1B > PBP 1A > PBP 3A = PBP 3B ≥ PBP 5. In the case of BLNAR strains, the MICs of cephem antibiotics with relatively high affinities for PBP 3A and PBP 3B were more elevated than those of PEN and penem and carbapenem antibiotics.
FIG. 2.
Fluorograms of PBPs of H. influenzae TK422 pretreated with AMP (A), CTX (B), and FRM (C). A 30-μl aliquot of each membrane fraction was incubated with 3 μl of each of the nonradioactive β-lactams diluted to various concentrations for 10 min and then further incubated with 3 μl of [3H]PEN for another 10 min. PBPs were visualized by autoradiography after exposure of the dried gels to X-OMAT film for 20 days at −80°C. The numbers identifying the lanes are concentration (in micrograms per milliliter).
Nucleotide and deduced amino acid sequences of the ftsI gene encoding PBP 3.
On the basis of the whole genome sequence of H. influenzae Rd (8), we assumed that the H. influenzae ftsI gene with 51% homology to the ftsI gene encoding PBP 3 in Escherichia coli encodes PBP 3A or PBP 3B.
The nucleotide sequences of the ftsI gene between 796 and 1,741 bp, which encodes the transpeptidase region of PBP 3, were determined by direct sequencing for 25 BLNAR strains, 2 BLPAR strains, and 3 AMP-susceptible strains. The deduced amino acid sequences (Fig. 3) of AMP-susceptible strain MT196, BLNAR strain H2 (group I), BLNAR strain KK01 (group II), and BLNAR strain ME870 (group III) are aligned only between 931 and 1,710 bp, as was the case for the sequence of the standard AMP-susceptible Rd strains.
FIG. 3.
Deduced amino acid sequence of part of PBP 3 from the strains listed in Table 1. Only amino acids that differed from the Rd amino acid sequence are shown. Boxes represent the conserved amino acid motifs of Ser-Thr-Val-Lys (STVK), Ser-Ser-Asn (SSN), and Lys-Thr-Gly (KTG).
Strains that were supposed to be BLNAR strains, as shown in Table 2, were classified into three groups on the basis of different amino acid substitutions. His-517 was commonly substituted for Arg-517 in the group I strains, and Lys-526 was similarly substituted for Asn-526 in group II strains. In addition, changes of Asp-350 to Asn-350 and Ser-357 to Asn-357 were found in almost all of these strains.
TABLE 2.
Deduced amino acid substitutions identified in PBP 3s from 25 BLNAR strains and 5 AMP-susceptible strains of H. influenzae
| Assigned group | Strain | Amino acid substitution
|
||||||
|---|---|---|---|---|---|---|---|---|
| Asp-350 | Ser-357 | Met-377 | Ser-385 | Leu-389 | Arg-517 | Asn-526 | ||
| Control | MT196 | |||||||
| MT053 | ||||||||
| TK422 | ||||||||
| TC01 | ||||||||
| AK01 | ||||||||
| I | ME2014 | His | ||||||
| ME1547 | Asn | Thr | His | |||||
| ME692 | Asn | Asn | His | |||||
| ME1503 | Asn | Asn | His | |||||
| H17 | Asn | His | ||||||
| N130 | Asn | Asn | His | |||||
| H2 | Asn | Asn | His | |||||
| ME1495 | Asn | His | ||||||
| H7 | Asn | Asn | IIe | Thr | His | |||
| II | ND01 | Asn | Asn | Lys | ||||
| ME570 | Asn | Lys | ||||||
| ME587 | Asn | Lys | ||||||
| KK01 | Asn | Asn | Lys | |||||
| ME495 | Asn | Asn | Lys | |||||
| MT273 | Asn | Asn | Lys | |||||
| MT160 | Asn | Asn | Lys | |||||
| ME267 | Asn | Asn | Lys | |||||
| MT040 | Asn | Asn | Lys | |||||
| ME1210 | Asn | Asn | Lys | |||||
| MT313 | Asn | Asn | Lys | |||||
| ME1976 | Asn | Lys | ||||||
| III | MT066 | Asn | Asn | IIe | Thr | Phe | Lys | |
| MT070 | Asn | Asn | Ile | Thr | Phe | Lys | ||
| ME1133 | Asn | Asn | Ile | Thr | Phe | Lys | ||
| ME870 | Asn | Asn | Ile | Thr | Phe | Lys | ||
In four strains classified into group III, three amino acid changes, Met-377 to Ile, Ser-385 to Thr, and Leu-389 to Phe, were commonly detected near the Ser-Ser-Asn (SSN) motif, in addition to substitutions of Lys-526 for Asn-526.
Sequencing of the dacB gene encoding PBP 4.
Figure 4 shows the deduced amino acid sequences of the dacB genes encoding PBP 4 in strains TK196, KK01, and MT040 as well strain Rd. Seven base pairs between 943 and 949 bp were deleted from the dacB genes of strains MT040 and ME1133. Because of this deletion, a stop codon was inserted after the C terminus of the 21st amino acid residue from the Ser-Asp-Asn (SDN) motif. PBP 4 was not detected by fluorography in the strains identified to have partial dacB deletions. Among the H. influenzae strains listed in Table 1, common amino acid substitutions in the dacB gene were identified in only two strains.
FIG. 4.
Deduced amino acid sequence of part of PBP 4s from the strains listed in Table 1. Only amino acids that differed from the Rd amino acid sequence are shown. Boxes indicate conserved amino acid sequences of Ser-Thr-Gln-Lys (STQK), Ser-Asp-Asn (SDN), and Lys-Thr-Gly (KTG).
Expression of ftsI gene.
AMP-susceptible strain H. influenzae Rd was transformed with the PCR-amplified ftsI genes from BLNAR strains H 17 (group I), KK01 (group II), and ME870 (group III). Table 3 shows the MICs of eight β-lactam antibiotics for each transformant for which the MICs increased in parallel with those for the donors.
TABLE 3.
Susceptibilities to β-lactam antibiotics of H. influenzae strains transformed with ftsI DNA fragment
| Assigned group | Straina | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | CEC | CTX | CDR | CPD | CDN | CPN | MEM | ||
| Rd (ATCC51097) | 0.25 | 1 | 0.008 | 0.25 | 0.063 | 0.008 | 0.016 | 0.031 | |
| I | Rd3A/H17 | 1 | 16 | 0.063 | 1 | 0.25 | 0.063 | 0.063 | 0.125 |
| II | Rd3A/KK01 | 1 | 16 | 0.063 | 1 | 0.5 | 0.031 | 0.063 | 0.125 |
| III | Rd3A/ME870 | 2 | 16 | 0.5 | 2 | 2 | 0.125 | 1 | 0.25 |
H. influenzae Rd (ATCC 51097) (the recipient) was transformed with PCR-amplified ftsI gene DNA fragments from group I, II, and III strains as the donors. Transformants were selected on plates containing 5 or 10 μg of CEC per ml. As an example of the transformant nomenclature, Rd3A/ME870 corresponds to Rd strain containing the ftsI gene from strain ME870.
Figure 1B shows the PBP profiles of the recipient Rd strain (lane 1), two transformant RdME870 strains (lanes 2 and 3), and donor and BLNAR strain ME870 (lane 4). The affinities of PBP 3A (molecular weight = 67,000) and PBP 3B (molecular weight = 66,000) decreased simultaneously in these transformants. Recombination of the 2.2-kb DNA fragment containing the ORF of the ftsI gene from strain ME870 with that from the recipient strain was confirmed by DNA sequencing.
DISCUSSION
Meningitis due to BLNAR H. influenzae strains of serotype b was first reported in 1980 (12). Carbenicillin and gentamicin therapy resulted in bacteriologic and clinical cure, while AMP therapy did not. The mechanism of resistance in BLNAR strains was thought to be due to the decreased affinity of AMP for some of the seven kinds of PBPs usually detected in AMP-susceptible H. influenzae (16, 19). Mendelman et al. (15) reported that the affinity of [3H]PEN for PBP 2, PBP 3A, PBP 3B, and/or PBP 4 was decreased in BLNAR strains. Malouin and colleagues (4, 11) have confirmed the relationship between altered PBP 3A and PBP 3B and AMP resistance by transformation with chromosomal DNA from BLNAR strains, but the molecular weight of PBP 3 in the transformant was calculated to be smaller than that in the recipient strain.
Up to the present, there have been no studies on the specific gene mutation(s) that encodes some PBPs and their relationship with β-lactam susceptibility in BLNAR strains.
A new AMP MIC peak that is distinct from that for AMP-susceptible H. influenzae was found at 1 μg/ml for BLNAR strains. The susceptibilities of these strains to other β-lactams, amoxicillin, and cephem antibiotics were also decreased in parallel with that to AMP (K. Ubukata, N. Nakayama, N. Chiba, K. Hasegawa, and Y. Shibasaki, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1440, 1999). We speculated that strains for which the AMP MIC was ≥ 1 μg/ml had acquired a factor(s) for resistance to β-lactam antibiotics.
Our results showed that the affinity for PBP 3A was commonly decreased and mutations were found in the ftsI genes of BLNAR strains. Deduced amino acid substitutions of His-517 for Arg-517 in group I strains and Lys-526 for Asn-526 in group II strains were identified near the conserved amino acids of the KTG motif in the ftsI gene. The MIC of AMP for these strains was no more than 1 to 2 μg/ml, and the MICs of cephem antibiotics were about 2 to 128 times higher for these strains than for the susceptible strains.
In group III strains, three amino acid substitutions, in addition to the substitution of Lys-526, were found near the SSN motif. Met-377 to IIe-377, Ser-385 to Thr-385, and Leu-389 to Phe-389. The susceptibilities of these strains to most cephem antibiotics, such as CTX, CPD, and CPN, were markedly reduced compared to their susceptibilities to AMP, CRO, CDN, FRM, and MEM. Differentiation of the elevation of the MICs of β-lactams in BLNAR strains reflected the high affinities of cephems for PBP 3A and PBP 3B and the relatively low affinities of AMP, FRM, and MEM for these PBPs.
On the basis of the three-dimensional (3D) crystallographic structure of PBP 2X in S. pneumoniae reported previously (3, 18) (PDB entry code, 1PMD), 3D models of PBP 3 in BLNAR and AMP-susceptible strains were constructed. The homology identity between PBP 2X and PBP 3 was 29%. Although the modeling depends on several assumptions, every substitution (Ile-377, Thr-385, Phe-389, His-517, and Lys-526) in the ftsI gene was found at the active-site pocket surrounded by the Ser-Thr-Val-Lys (STVK), SSN, and Lys-Thr-Gly (KTG) motifs. The change of a neutral amino acid (Asn) to a basic amino acid (Lys) may greatly affect the 3D structure of the transpeptidase domain.
As described in the Results section, we thought that the characteristics of the β-lactam MICs for BLNAR strains were supported by the analysis of transformants carrying the ftsI gene from BLNAR strains. However, the PBP 3A and PBP 3B affinities were simultaneously decreased in transformants carrying the 2.2-kb ORF of the ftsI gene. For this reason, two possibilities can be suggested: that PBP 3A and PBP 3B are different forms which are transcribed from the ftsI gene, like PBP 1Bs in E. coli, and that C-terminal processing of PBP 3A occurs, as with PBP 3 in E. coli. The expression and processing of PBP 3A and/or PBP 3B from the ftsI gene should be investigated in the future.
In a nationwide surveillance study conducted by our group in Japan, low-level-resistant BLNAR strains classified into groups I and II usually accounted for about 27% of the H. influenzae isolates from 1998 to 2000. In contrast, the proportions of BLNAR strains classified in to group III have been rapidly increasing, from 2.9% in 1998, to 6.6% in 1999, and to 13.7% in 2000. It is noteworthy that serotype b BLNAR strains are gradually isolated from patients with meningitis (unpublished data). The inappropriate use of oral antibiotics for the treatment of community-acquired RTIs and AOM seems to select for BLNAR strains.
REFERENCES
- 1.Asahi Y, Ubukata K. Association of a Thr-371 substitution in a conserved amino acid motif of penicillin-binding protein 1A with penicillin resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2267–2273. doi: 10.1128/aac.42.9.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahi Y, Takeuchi Y, Ubukata K. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob Agents Chemother. 1999;43:1252–1255. doi: 10.1128/aac.43.5.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlier P, Buisson G, Dideberg O, Wierenga J, Keck W, Laible G, Hakenbeck R. Crystallization of a genetically engineered water-soluble primary penicillin target enzyme. The high molecular mass PBP2X of Streptococcus pneumoniae. J Mol Biol. 1993;232:1007–1009. doi: 10.1006/jmbi.1993.1452. [DOI] [PubMed] [Google Scholar]
- 4.Clairoux N, Picard M, Brochu A, Rousseau N, Gourde P, Beauchamp D, Parr T R, Jr, Bergeron M G, Malouin F. Molecular basis of the non-β-lactamase-mediated resistance to β-lactam antibiotics in strains of Heamophilus influenzae isolated in Canada. Antimicrob Agents Chemother. 1992;36:1504–1513. doi: 10.1128/aac.36.7.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doern G V, Brueggemann A B, Pierce G, Horry H P, Jr, Rauch A. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of β-lactamase-positive strains resistant to amoxicillin-clavulanate: results of a national multicenter surveillance study. Antimicrob Agents Chemother. 1997;41:292–297. doi: 10.1128/aac.41.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doern G V, Jorgensen J H, Thornsberry C, Preston D A the Haemophilus influenzae surveillance group. Prevalence of antimicrobial resistance among clinical isolates of Haemophilus influenzae: a collaborative study. Diagn Microbiol Infect Dis. 1986;4:95–107. doi: 10.1016/0732-8893(86)90143-4. [DOI] [PubMed] [Google Scholar]
- 7.Doern G V, Jorgensen J H, Thornsberry C, Preston D A, Tubert T, Redding J S, Maher L A. National collaborative study of the prevalence of antimicrobial resistance among clinical isolates of Haemophilus influenzae. Antimicrob Agents Chemother. 1988;32:180–185. doi: 10.1128/aac.32.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, McKenney K, Sutton G G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 9.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol. 1991;173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen J H, Doern G V, Maher L A, Howell A W, Redding J S. Antimicrobial resistance among respiratory isolates of Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae in the United States. Antimicrob Agents Chemother. 1990;34:2075–2080. doi: 10.1128/aac.34.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malouin F, Schryvers A B, Bryan L E. Cloning and expression of genes responsible for altered penicillin-binding proteins 3a and 3b in Haemophilus influenzae. Antimicrob Agents Chemother. 1987;31:286–291. doi: 10.1128/aac.31.2.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markowitz S M. Isolation of an ampicillin-resistant, non-β-lactamase-producing strain of Haemophilus influenzae. Antimicrob Agents Chemother. 1980;17:80–83. doi: 10.1128/aac.17.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medeiros A A, Levesque R, Jacoby G A. An animal source for the ROB-1 β-lactamase of Heamophilus influenzae type B. Antimicrob Agents Chemother. 1986;29:212–215. doi: 10.1128/aac.29.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendelman P M, Chaffin D O, Kalaitzoglou G. Penicillin-binding proteins and ampicillin resistance in Haemophilus influenzae. J Antimicrob Chemother. 1990;25:525–534. doi: 10.1093/jac/25.4.525. [DOI] [PubMed] [Google Scholar]
- 15.Mendelman P M, Chaffin D O, Musser J M, DeGroot R, Serfass D A, Selander R K. Genetic and phenotypic diversity among ampicillin-resistant, non-β-lactamase-producing, nontypeable Haemophilus influenzae isolates. Infect Immun. 1987;55:2585–2589. doi: 10.1128/iai.55.11.2585-2589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendelman P M, Chaffin D O, Stull T L, Rubens C E, Mack K D, Smith A L. Characterization of non-β-lactamase-mediated ampicillin resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1984;26:235–244. doi: 10.1128/aac.26.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa J, Matsuhashi M. Molecular divergence of a major peptidoglycan synthetase with transglycosylase-transpeptidase activities in Eschericha coli—penicillin-binding protein 1Bs. Biochem Biophys Res Commun. 1982;105:1546–1553. doi: 10.1016/0006-291x(82)90964-0. [DOI] [PubMed] [Google Scholar]
- 18.Pares S, Mouz N, Petillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 19.Parr T R, Jr, Bryan L E. Mechanism of resistance of an ampicillin-resistant, β-lactamse-negative clinical isolate of Haemophilus influenzae type b to β-lactam antibiotics. Antimicrob Agents Chemother. 1984;25:747–753. doi: 10.1128/aac.25.6.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega R, Sadoff H L, Patterson M J. Mechanism of ampicillin resistance in Haemophilus influenzae type B. Antimicrob Agents Chemother. 1976;9:164–168. doi: 10.1128/aac.9.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zijderveld C A L, Aarsman M E G, Nanninga N. Differences between inner membrane and peptidoglycan-associated PBP 1B dimmers of Escherichia coli. J Bacteriol. 1995;177:1860–1863. doi: 10.1128/jb.177.7.1860-1863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]