Abstract
There is a need to identify biomarkers of radiation exposure for use in development of circulating biodosimeters for radiation exposure and for clinical use as markers of radiation injury. Most research approaches for biomarker discovery rely on a single animal model. The current study sought to take advantage of a novel aptamer-based proteomic assay which has been validated for use in many species to characterize changes to the blood proteome after total-body irradiation (TBI) across four different mammalian species including humans. Plasma was collected from C57BL6 mice, Sinclair minipigs, and Rhesus non-human primates (NHPs) receiving a single dose of TBI at a range of 3.3 Gy to 4.22 Gy at 24 h postirradiation. NHP and minipig models were irradiated using a 60Co source at a dose rate of 0.6 Gy/min, the C57BL6 mouse model using an X-ray source at a dose rate of 2.28 Gy/min and clinical samples from a photon source at 10 cGy/min. Plasma was collected from human patients receiving a single dose of 2 Gy TBI collected 6 h postirradiation. Plasma was screened using the aptamer-based SomaLogic SomaScant® proteomic assay technology to evaluate changes in the expression of 1,310 protein analytes. Confirmatory analysis of protein expression of biomarker HIST1H1C, was completed using plasma from C57BL6 mice receiving a 2, 3.5 or 8 Gy TBI collected at days 1, 3, and 7 postirradiation by singleplex ELISA. Summary of key pathways with altered expression after radiation exposure across all four mammalian species was determined using Ingenuity Pathway Analysis (IPA). Detectable values were obtained for all 1,310 proteins in all samples included in the SomaScan assay. A subset panel of protein biomarkers which demonstrated significant (p < 0.05) changes in expression of at least 1.3-fold after radiation exposure were characterized for each species. IPA of significantly altered proteins yielded a variety of top disease and biofunction pathways across species with the organismal injury and abnormalities pathway held in common for all four species. The HIST1H1C protein was shown to be radiation responsive within the human, NHP and murine species within the SomaScan dataset and was shown to demonstrate dose dependent upregulation at 2, 3.5 and 8 Gy at 24 h postirradiation in a separate murine cohort by ELISA. The SomaScan proteomics platform is a useful screening tool to evaluate changes in biomarker expression across multiple mammalian species. In our study, we were able to identify a novel biomarker of radiation exposure, HIST1H1C, and characterize panels of radiation responsive proteins and functional proteomic pathways altered by radiation exposure across murine, minipig, NHP and human species. Our study demonstrates the efficacy of using a multispecies approach for biomarker discovery.
INTRODUCTION
There is a need to identify biomarkers of radiation exposure both for use in development of biodosimetry blood diagnostics for accidental or terrorist radiation exposures and for clinical use as markers of radiation injury. This has resulted in a growing body of literature within the field of biodosimetry, which seeks to characterize novel biomarkers of radiation exposure (1). Blood soluble proteomic biomarkers of radiation exposure are of particular interest as this class of biomarkers are relatively easily assayed and represent active biological processes such as injury response. Key studies using two-dimensional gel electrophoresis coupled with mass spectrometry and liquid chromatography-tandem mass spectrometry have identified novel proteomic biomarkers of radiation exposure in murine and non-human primate (NHP) models (2, 3). These studies complement other characterizations of radiation responsive protein biomarkers using single and multiplexed ELISA, Western and aptamer-based proteomic detection technologies (4–8). Thus, for proteomic biomarker discovery there exist a wide range of complementary platforms, which can be utilized to validate findings across studies. As human validation studies for biodosimetry and radiation injury biomarkers are often unfeasible due to ethical consideration, use of animal models in research studies as surrogates is necessary. Recent studies have demonstrated successful validation of individual protein biomarkers of radiation exposure across multiple species including between mice, minipigs and non-human primates (NHP), and between NHPs and humans (9, 10). As licensing of new medical countermeasures for radiation exposure are guided by the FDA Animal Rule, which allows animal models to be used in place of clinical studies where ethical concerns preclude use of human efficacy studies, use of multiple mammalian species for validation is the current norm (11, 12). Yet there are limitations for extrapolation of comparative human sensitivity for radiation injury, biomarker expression and drug response based on animal model findings and reliance on historical LD50/30 findings for radiation exposure (13). Also, preliminary proof of concept biomarker expression studies are often conducted using a single animal model before verification in a second model of alternate mammalian species, which is time consuming and costly.
In the current study, we sought to characterize changes within the blood proteome due to ionizing radiation exposure in four mammalian species using a novel high throughput, aptamer-based, SomaLogic SomaScan proteomic assay technology, which has been validated for use in a variety of mammalian species (14). Plasma from total-body irradiated (TBI) C57BL6 mice, Sinclair minipigs, Rhesus NHPs and human clinical patients undergoing radiation treatment were pooled. All animal subjects received a single uniform dose of TBI between 3.3 Gy to 4.22 Gy with plasma collected at 24 h postirradiation. Clinical patients received TBI via opposed lateral beams with a midplane dose of 2 Gy, with plasma collection occurring at 6 h postirradiation. These samples were then screened using the SomaScan proteomic assay for changes in expression of 1,310 protein analytes. The overall study focus was to characterize changes in protein expression profiles across species and demonstrate the efficacy and efficiency of a multispecies approach using the latest proteomic technologies to identify new biomarkers of radiation exposure. These study findings highlight key similarities in global proteomic changes resulting from TBI.
MATERIALS AND METHODS
Animal Model NHP
For this study 2 male and 1 female Rhesus monkeys (Macaca mulatta) between 5–8 years of age received a single dose of TBI between 3.85 and 4.22 Gy. Plasma samples were collected at 24 h postirradiation using venipuncture collection. Pre- and postirradiation samples were matched for each individual animal. Animal care was conducted according to the Standard Operating Procedures of CiToxLAB North America under Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) standards.
Animal Model NHP Dosimetry
Animals were exposed to a single dose of gamma-TBI using a 60Co source (Theratron 1000) at a dose rate of approximately 60 cGy/min administered anteroposterior/posteroanterior. The source-to-mid-plane distance was dependent on the size of the animal ranging from 158.5 to 172 cm with a field size of 44.2 × 44.2 cm2. Dosimetry was verified using Landauer nanoDot OSL-based medical dosimeter and a Farmer Ionization Chamber. Mean exposures for the two male NHP animals was 4.22 Gy* and 3.87 Gy, respectively, and for the single female NHP 3.85 Gy (*for this animal, half-dose quantified at 210.9 cGy and TBI calculated at 421.8 cGy).
Animal Model Minipig
For this study 2 male Sinclair minipigs of approximate 3 months of age received a 3.3 Gy single TBI dose. Plasma samples were collected at 24 h postirradiation using venipuncture collection. Pre- and postirradiation samples were matched for each individual animal. Animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) approved facility at the Armed Forces Radiobiology Research Institute (AFRRI). All procedures involving animals were reviewed and approved by the AFRRI Institutional Animal Care and Use Committee (IACUC) and all efforts were made to minimize suffering.
Animal Model Minipig Dosimetry
Animals received TBI in a bilateral gamma-radiation field at AFRRI’s Cobalt-60 (60Co) facility at a dose rate of 0.6 Gy/min. Control animals were sham-irradiated and treated in the same manner as the irradiated animals, except the 60Co source was not raised from the shielding water pool.
Animal Model Murine
For this study female, 8–15-week-old C57BL6 mice received a single TBI dose of 3.5 Gy or sham irradiation. Two animals were included in the irradiated group and 5 animals were included in the sham-irradiated group. All mice receiving TBI were confined using a standard pie jig preventing movement. All animal studies were conducted in accordance with the principles and procedures outlined in the NIH Guide for the Care and Use of Animals and procedures were approved by the NIH Lab Animal Safety Program under an approved protocol. Plasma was collected at 24 h postirradiation by cardiac puncture using a heparinized syringe. Mice received 2.5–5.5% Isoflurane anesthesia during cardiac puncture for blood collection.
Animal Model Murine Dosimetry
Murine in vivo models utilized a Pantak X-ray source at a dose rate of 2.28 Gy/min. Dose rate was calibrated based upon the procedures described in American Association of Physicist in Medicine (AAPM) Task Group Report 61 (TG-61) with regard to the following conditions: X-ray tube potential was 300 kV, half-value layer (HVL) is 0.9 mm copper (Cu), homogeneity coefficient (HC) is 0.33, source to surface distance (SSD) was 50 cm with a field size of 20 × 20 cm Dose rate was measured at 2 cm depth in a solid water phantom using a PTW model: N23342 ion chamber and Inovision, model 35040 electrometer.
Human Clinical Samples
All human samples were collected on a previously reported, Institutional Review Boards approved protocol (NCT00096382) for which patients provided written informed consent (15). Plasma samples from 5 melanoma patients receiving a myeloablative regimen including cyclophosphamide 60 mg/kg/d (days −6 and −5), fludarabine 25 mg/m2/d (days −6 through −2), and 2 Gy dose of TBI (day −1) were included. Patients received a TBI dose from a clinical linear accelerator using a 15 MV photon beam with a source-to-surface distance (SSD) of 600 cm and field size of 40 × 40 cm.
Plasma samples were collected at 6 h irradiation and included 1 male and 4 female samples with 2 of the female samples pooled to make the minimum volume. These were compared against 3 samples from healthy adult human controls who had received no exposure.
SomaLogic SomaScan Assay
Approximately 160 ul of plasma per sample was used for the Somalogic SomaScan Assay which uses a novel protein-capture aptamer-based technology (16). For this study the SomaScan HTS Assay 1.3K was used and processed through the Center for Human Immunology at the National Institutes of Health. The assay included measurement of 1,310 protein analytes.
Statistical Analysis
In brief, data were received in the form of Relative Fluorescent Units (RFU) for each of the 1,310 proteins in the SomaScan assay after normalizing for intraplate and interplate variation using the supplied SomaLogic software. These RFU scores for each protein were log2 and z-score transformed. Mean control expression values were compared to each individual radiation treated sample for the murine and clinical samples. Matched sample pre- and postirradiation were used for the NHP and minipig animals. The ratios of respective upregulation or downregulation of each protein was filtered using a (p < 0.05) and 1.3-fold change threshold. Statistical data analysis was performed using R and Ingenuity Pathway Analysis (IPA) (17).
Mouse ELISA
Plasma samples from female, C57BL6 mice between 8–14 weeks of age that received TBI of 2, 3.5 and 8 Gy dose were collected at days 1, 3 and 7 postirradiation, were screened for HIST1H1C expression using a commercially available ELISA kit (#MBS2613293 MyBioSource.com). All samples were run according to manufacturer’s instructions. Statistical analysis of values was done using a Student’s t test and a probability level of p < 0.05 was considered significant. Data is presented as mean ± SEM.
RESULTS
This study sought to identify and correlate changes in the plasma proteome after TBI across multiple mammalian species. To that end, NHP, murine, and minipig research samples from animals receiving a single dose of TBI at a range of 3.3 Gy to 4.22 Gy collected 24 h postirradiation, were matched with the closest available clinical samples of 2 Gy TBI collected 6 h postirradiation. Given the unique nature of the sample set collected for this study, we sought to maximize the probability of gaining useful findings by utilizing a proteomics analysis platform with a highly multiplexed approach. The novel aptamer-based SomaScan proteomic assay technology was used as it has been validated across multiple species including human, murine, canine, feline and a variety of non-human primates (14). This assay also includes analysis of over 1,300 protein analytes using a relatively small volume of sample, and as such, was considered the ideal choice for this study.
Assessment of SomaScan assay data was used for discovery of novel biomarkers of radiation exposure across multiple species. To assess whether significant differences in the baseline proteome existed, control samples for each species were compared using principal component analysis (PCA) and heatmap clustering. As shown in Fig. 1A, the first 2 principal components (PC1 = 31.92% and PC2 = 19.1%) together explain 51% variability among the control samples. Samples showed good clustering by species. The PC1 axes is strongly associated with human and murine species, while PC2 is strongly associated with NHP and human. Samples from minipig clustered close to murine, indicating both these species have relatively similar composition. Similarly, Fig. 1B shows good homology within species but not necessarily between species.
FIG. 1.
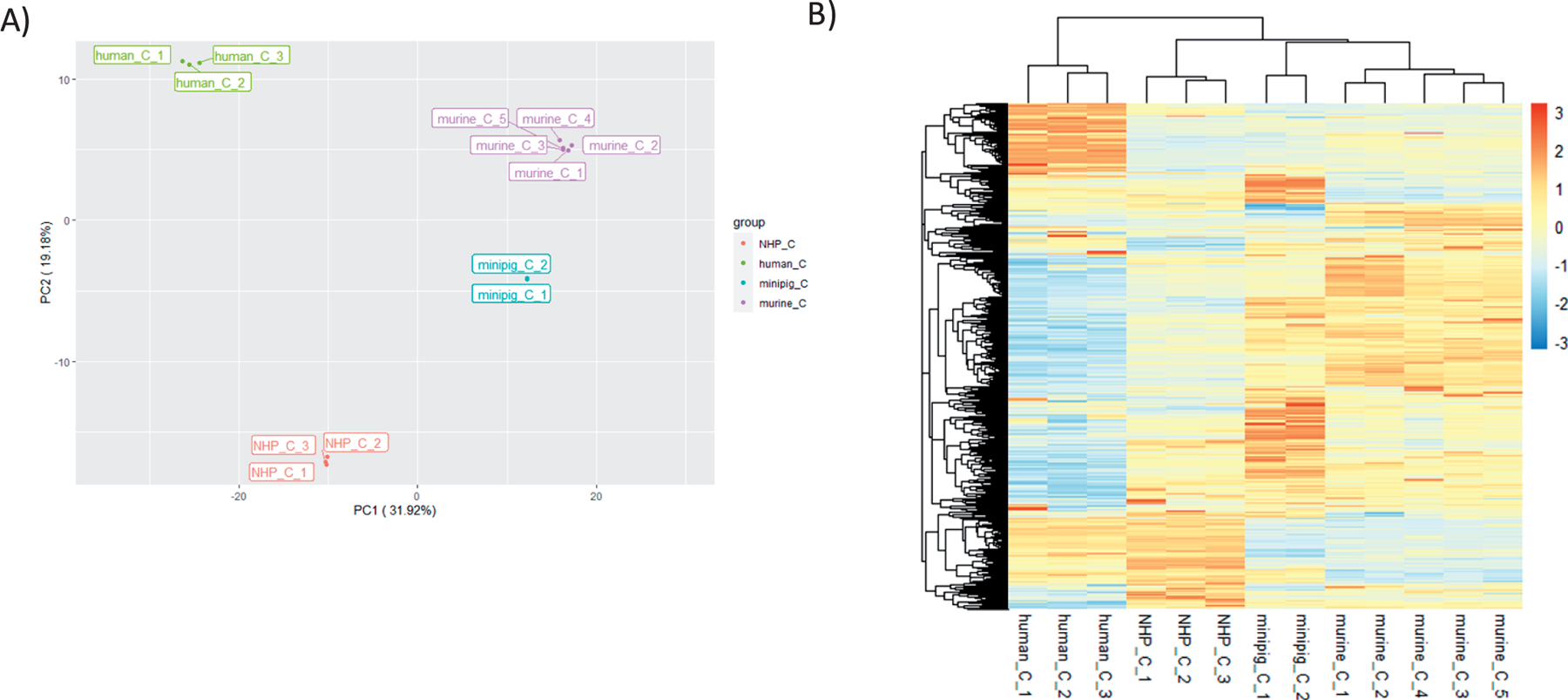
Comparison of proteomic expression values for control samples. Panel A: Principal component analysis (PCA) showing relative clustering of control samples for each species: NHP, human, minipig and murine. Samples from each species are color-coded. Axis percentage labels indicate percent variation on principal component axes 1 and 2. Panel B: Heatmap of Relative Fluorescent Unit (RFU) data for each of the respective controls. Red represents upregulation and blue represents downregulation.
Next, a comparison of protein expression profiles was done using the ratio of treated sample over respective controls. As shown in the PCA plot in Fig. 2A, the first 2 principal components (PC1 = 21.96% and PC2 = 16.9%) together explain only 38.9% of the variability. The treated samples did not cluster as tightly as the controls but still loosely aggregated by species. As shown in Fig. 2A and B, the samples from the treated NHP were tightly clustered as were two of the human samples and the two murine samples. However, the two minipig samples and two of the human samples did not cluster tightly together. Overall, these results indicate radiation treatment has a species independent effect on circulating protein biomarkers.
FIG. 2.
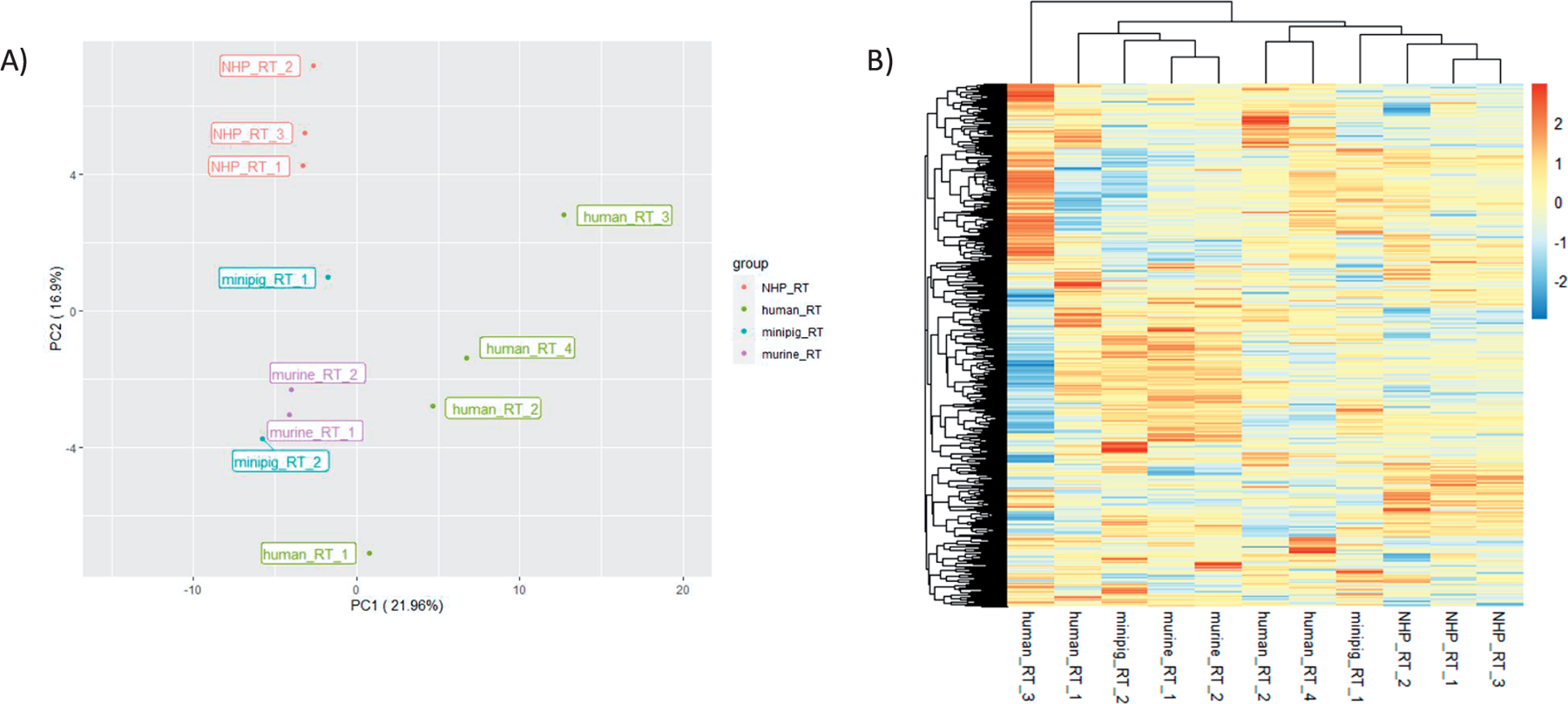
Comparison of proteomic expression values for irradiated samples. Panel A: Principal component analysis (PCA) showing relative clustering of treated samples for each species: NHP, human, minipig and murine. Samples from each species are color-coded. Axis percentage labels indicate percent variation on principal component axes 1 and 2. Panel B: Heatmap of RFU ratio data for treated samples. All values represent ratios of treated samples over their respective controls. Red represents upregulation and blue represents downregulation
To select individual proteins that demonstrated significant changes in expression after irradiation, a ≥ 1.3-fold change and p < 0.05 filter was used for both upregulated and downregulated proteins. Figure 3 lists all individual proteins with at least a 1.3-fold change in expression by species. For NHP, minipig and murine species the listed proteins were consistent across all samples included in the study. The human samples showed a larger number of proteins with changes in expression after TBI compared to the other species and, as such are grouped as proteins showing a ≥ 1.3-fold change in 4/4 samples and in 3/4 samples, respectively. As such, all of samples in the 3/4 grouping would also have the significantly altered proteins from the 4/4 sample list as well.
FIG. 3.
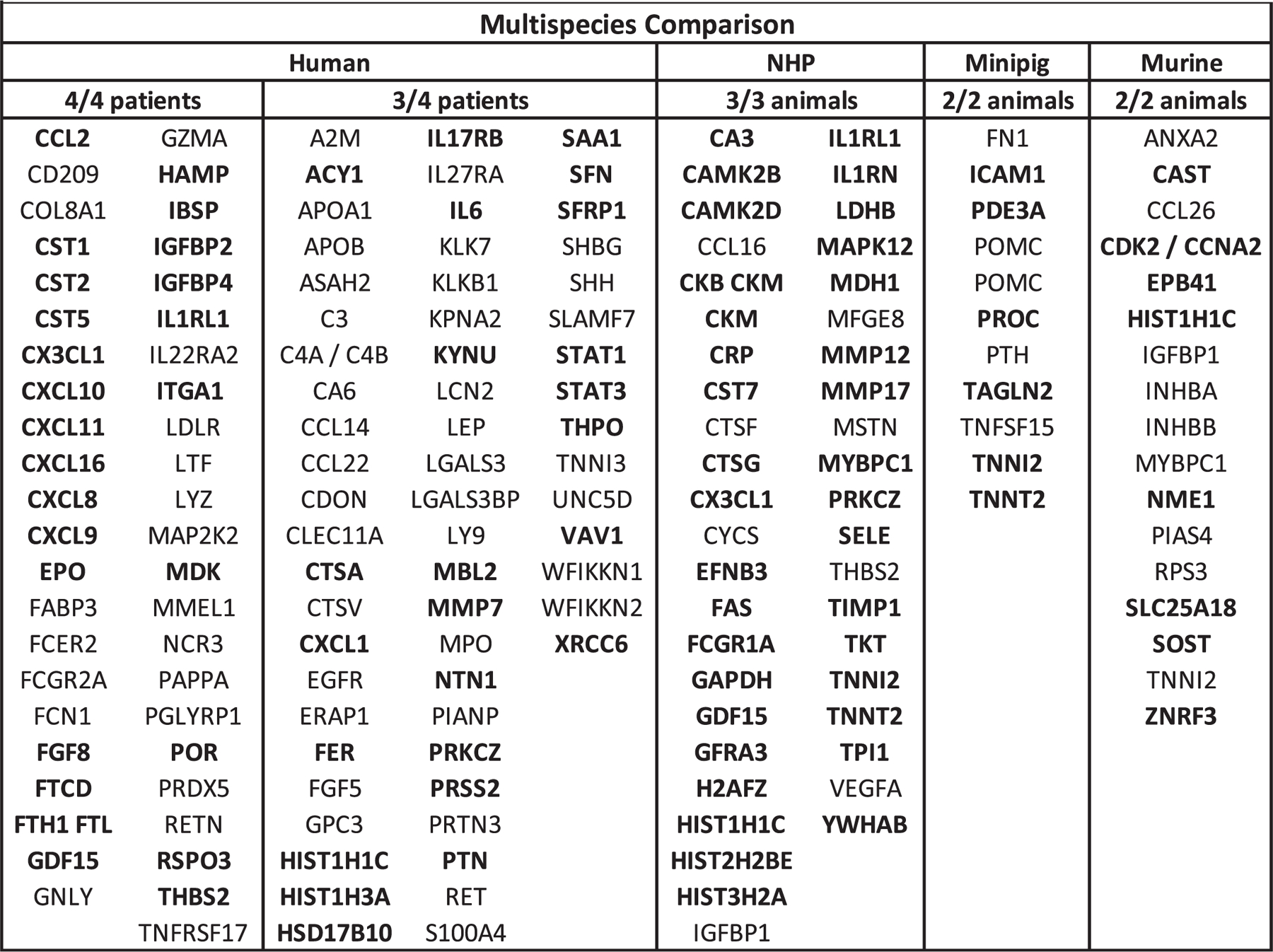
Proteomic expression panels for each species. Shows gene symbols for proteins with a 1.3-fold change (FC) in expression in samples from NHP, minipig and murine species who received a single dose of TBI between 3.3 Gy to 4.22 Gy collected 24 h postirradiation and 2 Gy dose of TBI at 6 h postirradiation for human samples. Bold text proteins are 1.3 FC upregulated over control and unbold text proteins are 1.3 FC downregulated from control.
Radiation-induced expression profile changes compared across species show commonalities when grouped by functional protein family. Specifically, members of the insulin-like growth factor-binding protein (IGFBP) family demonstrated significant changes in expression including IGFBP2 and IGFBP4 in 4/4 human samples and IGFBP1 in all the NHP and murine samples. Members of the cystatin family including cystatin-SN (CST1), cystatin-SA (CST2) and cystatin-D (CST5) were significantly altered in 4/4 human samples and cystatin-F (CST7) in all NHP samples. Similarly, within the cathepsin family, cathepsin A (CTSA) and cathepsin L2 (CTSV) showed altered expression in 3/4 human samples and cathepsin F (CTSF) and cathepsin G (CTSG) in all NHP samples. In the family of matrix metalloproteinases (MMPs), MMP-7 showed altered expression in 3/4 human samples and MMP-12 and MMP-17 in all the NHP samples. Members of the troponin family demonstrated significant changes in expression across all the species examined, with cardiac troponin I (TNNI3) in 3/4 human samples, troponin T (TNNT2) and troponin I type 2 (TNNI2) in all NHP and minipig samples and TNNI2 in all murine samples.
One notable single biomarker, histone H1.2 (HIST1H1C), was shown to be upregulated in 3/4 human samples, and all the NHP and murine samples. As this protein is not an established biomarker of radiation exposure, expression of HIST1H1C was verified by ELISA. Elevation of plasma HIST1H1C over control was shown after 24 h in plasma from C57BL6 mice receiving 3.5 Gy TBI validating the findings of the SomaScan assay. Significant dose dependent elevation of HIST1H1C was also found in the plasma of TBI C57BL6 mice at 2, 3.5 and 8 Gy collected at 24 h postirradiation with (p < 0.05) at 2 Gy and (p < 0.01) at 3.5 and 8 Gy (Fig. 4). This elevation was not seen at later timepoints with no significant elevation of HIST1H1C over control seen from mice receiving 2 and 8 Gy TBI at either day 3 or 7 postirradiation. (Data not shown)
FIG. 4.
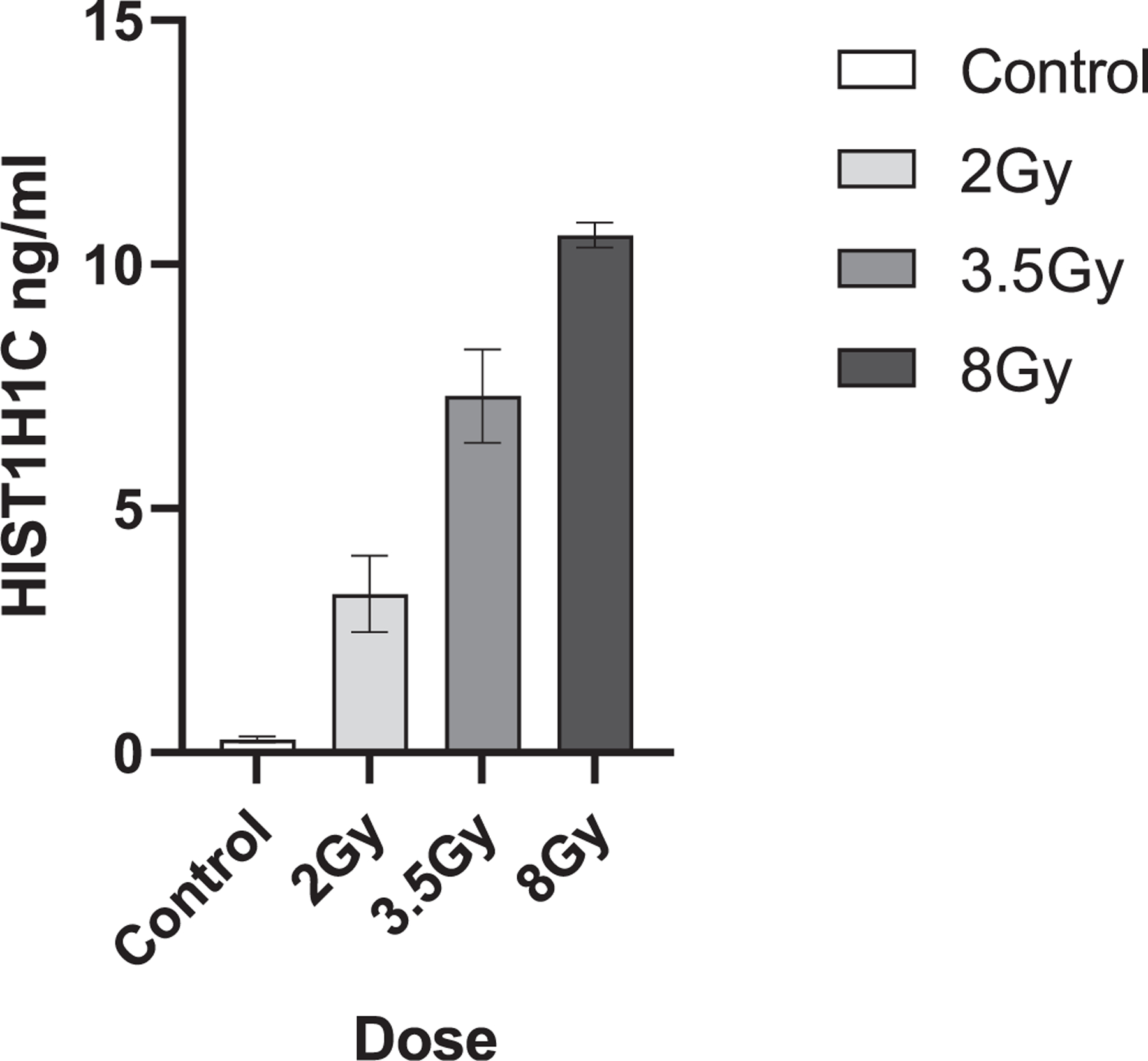
HIST1H1C murine expression. Shows HIST1H1C protein expression (ng/ml) in the plasma of C57BL6 mice at 24 h postirradiation after a 2, 3.5 or 8 Gy dose of TBI. Data represents the mean and SEM of n = 3 animals per timepoint.
Finally, a protein expression pathway analysis was completed for each species using the Ingenuity Pathway Analysis (IPA) platform with filtering for ≥ 1.3-fold change and p < 0.05 using the mean difference of treated over control. As expected, though there was some overlap in significantly altered pathways held in common between species, each species had a unique profile. For top ranked diseases and biofunctions, organismal injury and abnormalities was held in common across all four species. Cancer, cardiovascular and neurological disease were held in common between NHP and murine samples and dermatological disease was held in common across human and NHP samples. Top networks listed for each species were unique, yet all included networks related to cell-to-cell signaling and cellular function (Fig. 5). Expression pathway analysis of molecules highlighted within IPA for organismal injury and abnormalities for both the human and NHP sample cohorts are shown in Fig. 6. In this network built from genes within the NHP cohort, 19 genes were found to overlap within the human cohort. This IPA analysis is demonstrative of the similarities and differences in mammalian response to TBI across species.
FIG. 5.
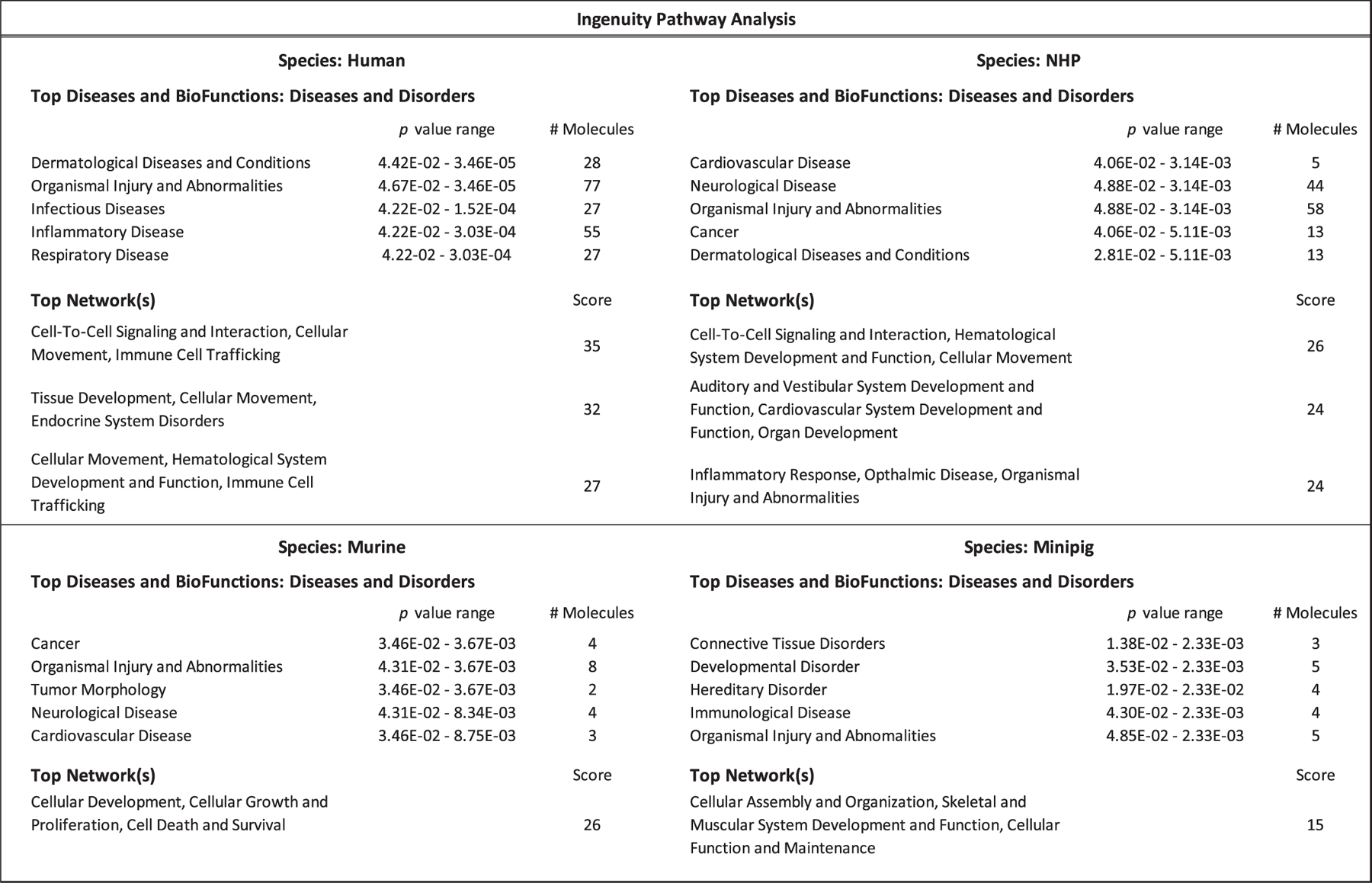
Ingenuity Pathway Analysis. Summary of key pathways with altered expression after irradiation in multiple mammalian species as determined by ingenuity pathway analysis (IPA). NHP, minipig and murine samples at a range of 3.3 Gy to 4.22 Gy TBI, collected 24 h postirradiation and human samples receiving 2 Gy TBI collected 6 h postirradiation were included. IPA filters were inclusive of 1.3 FC proteins with (p < 0.05).
FIG. 6.
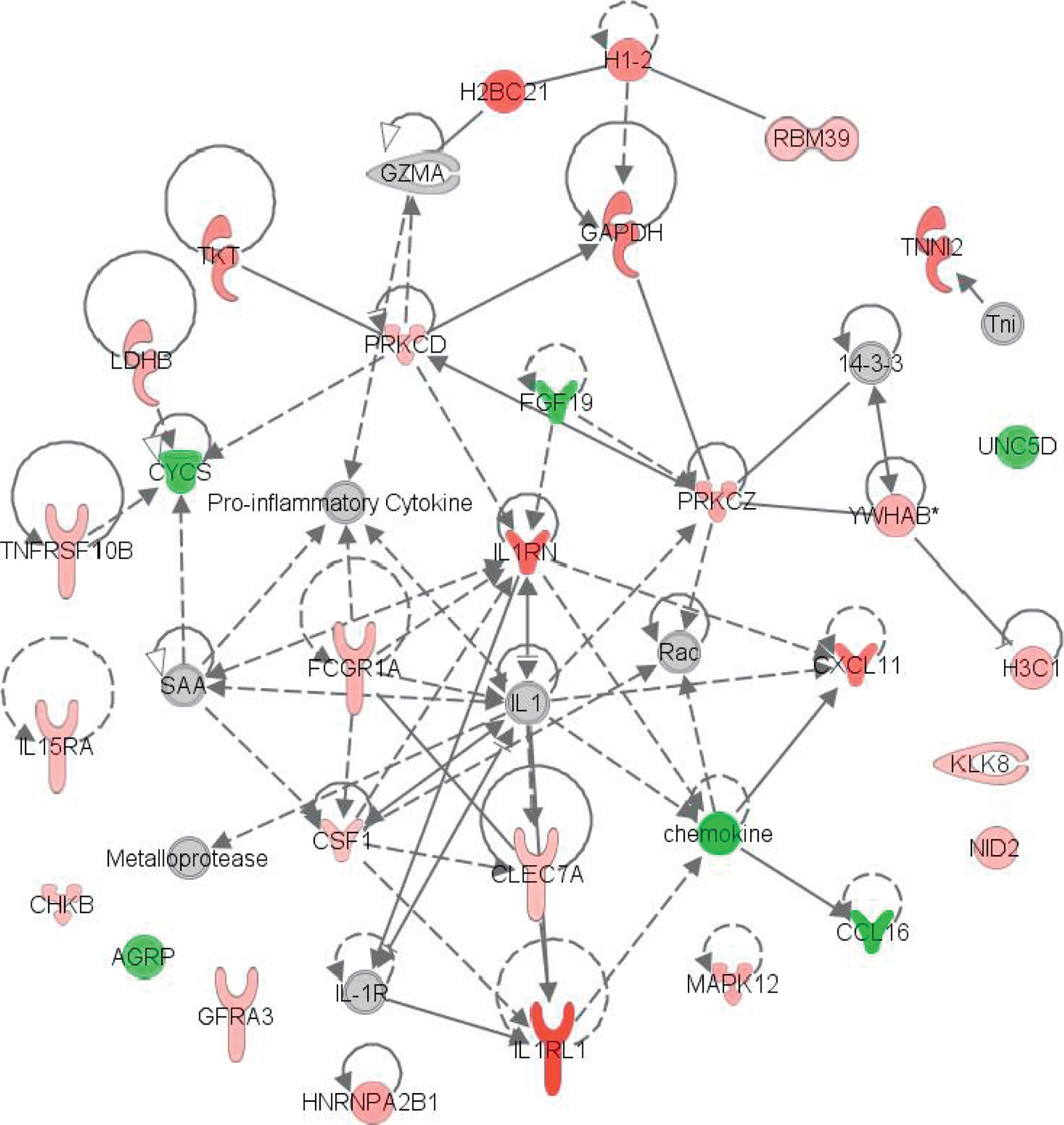
IPA expression pathway analysis. Includes genes held in common across human and NHP samples within the ‘‘Organismal Injury and Abnormalities’’ networks in IPA. Genes present in the sample dataset are red for upregulation and green for downregulation. NHP samples at approximately 4 Gy TBI, collected 24 h postirradiation and human samples receiving 2 Gy TBI collected 6 h postirradiation were included.
DISCUSSION
This study sought to characterize changes in the mammalian proteome in response to total-body irradiation across multiple species using a novel high throughput, highly multiplexed proteomic platform. This multispecies approach to characterization of radiation-induced changes to the blood proteome was designed to aid in identification of novel proteomic biomarkers of radiation and to characterize protein expression pathways altered by radiation exposure Though relatively few identical biomarkers showed significant changes in expression after irradiation across species, there was overlap in expression changes of functional protein families held in common across species. In the current study, many of the proteins found to have significantly altered expression after irradiation in the SomaScan assay have previously been shown to be radiation responsive including erythropoietin (EPO) which was upregulated in 4/4 human samples and protein kinase C zeta type (PRKCZ) which was upregulated in 3/4 human samples and all the NHP samples (18, 19). Similarly, many of the functional protein families which demonstrated significant changes in expression across species after irradiation, have also been shown to be radiation responsive including the IGFBP family and cathepsin family, which are involved in DNA double-strand break repair, and the troponin family which are markers for both general and radiation-induced cardiac injury (20–25). Many members of the MMP family have also previously been shown to be radiation responsive (7, 26). However, members of the cathepsin family have not been generally shown to be radiation responsive other than cathepsin C as a marker for renal injury after irradiation (27).
HIST1H1C, which was shown to be upregulated in 3/4 human patient samples and all the NHP and murine samples, has not been established in the literature as a radiation responsive biomarker, though it is known to be involved in DNA damage repair, and one study found radiation-induced HIST1H1C gene expression changes in murine brain tissue (28, 29). This may explain the shortlived elevation of HIST1H1C, which demonstrated dose depended elevation correlative with TBI dose at 24 h postirradiation, but returned to basal values at later timepoints of either day 3 or 7 postirradiation, as most ionizing radiation induced DNA damage are repaired within 48 h postirradiation (30). This biomarker, though its utility is constrained to within 24 h postirradiation, may be a useful addition to the growing number of proteomic biomarkers being developed for use in radiation biodosimetry diagnostics, due to its clear dose dependent elevation profile.
Analysis of key biological pathways altered by radiation exposure represented by the biomarker expression profiles in this study, were also found to be held in common with other proteomic studies. In the current study, the organismal injury and abnormalities pathway in IPA was found to be significantly altered across all 4 animal models and the cancer pathway was significantly altered in the NHP and murine models. In a recently published study looking at proteomic expression profiles in wild Japanese field mice chronically exposed to environmental radioactive contamination within the Fukushima difficult to return zone using the same SomaLogic aptamer-based assay, these pathways were also found to be significantly altered due to radiation exposure (31). In this study pathways related to the inflammatory response were also found to be significantly altered in the human and NHP models. This is validated by numerous other proteomic biomarker studies in murine and NHP models, which have found inflammatory and acute phase response canonical pathways significantly altered after irradiation or identified significantly altered expression levels of specific biomarkers involved in the inflammatory response such as c-reactive protein or serum amyloid A (2, 3, 5, 32, 33).
Limitations of the current study include the challenges of matching sample series across species including health status, received dose and time of collection. Differences in radiation quality and dose rate between the respective TBIs across species also represent a limitation. Though the NHP, murine, and minipig were all healthy animals receiving a single dose of TBI at a range of 3.3 Gy to 4.22 Gy collected at 24 h postirradiation, the samples of TBI human origin were from cancer patients who had received a non-myeloablative chemotherapeutic regimen, 2 Gy TBI, with plasma collected at 6 h postirradiation. A 6-hour timepoint was necessary as patients in the current study were receiving TBI for preparative bone marrow ablation in the form of two fractions of 2 Gy spaced 6 h apart. Due to the scarcity of available clinical samples from irradiated patients, these samples were selected as the best available match for this study. Further, there are dissimilarities in dose equivalency across animal species as evidenced by differences in LD50/30 (13). Due to scarcity of available samples, the current study only validated one biomarker in murine samples. Future studies might characterize additional biomarkers in other minipig, NHP or clinical cohorts.
Though the SomaScan assay was initially designed for homology within human samples, it has been validated for samples from a variety of mammalian species but has not been expressly validated for minipig. Though we obtained values for all 1,317 targets in the SomaScan assay within the minipig samples, there may have been reduced binding complementarity. Also, though the platform has been validated for use across a variety of species, each species may not have identical binding fidelity across each of the 1,317 targets, introducing possible study limitations for comparative expression analysis using an interspecies approach. Despite these potential confounders, the current study was able to characterize changes within the mammalian proteome across multiple species through differentiation of expression changes within functional protein families due to radiation exposure and identify and validate a novel, plasma biomarker of radiation exposure, HIST1H1C. This demonstrates the unique utility of a multispecies approach applied within the same proteomics platform, and as this study used the SomaScan assay version containing ~1,300 targets and the latest SomaScan assay version contains ~7,000 targets, future studies may yield more robust findings.
As the goal of radiation research using animal models is ultimately translation to the clinical setting, inclusion of multiple mammalian species including available human samples, may be an advantageous approach for other similar biomarker expression analyses. As future medical countermeasures for radiation exposure must follow the FDA Animal Rule, characterization of radiation-induced changes within the blood proteome using concurrent mammalian species potentially provides a more efficient methodology for discovery of novel radiation responsive biomarkers for biodosimetry and radiation injury. This pilot study demonstrates the utility and future applications for emerging highly multiplexed proteomics technologies for multispecies studies.
ACKNOWLEDGMENTS
This research was supported in part by funding from the Radiation and Nuclear Countermeasures Program, #Y2-OD-0332–01 NIAID, #AAI12044–001-06000 NIAID, and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, #ZIA SC010373. This work was also supported by the National Institute of Allergy and Infectious Disease, NIH, under HHS Contract HHSN272201500013I awarded to SRI International. The authors would also like to thank the Center for Human Immunology at the Clinical Center at the National Institutes of Health and Brian Sellers for their invaluable scientific and technical assistance with the SomaLogic SomaScan platform.
Footnotes
Sproull M, Nishita D, Chang P, Moroni M, Citrin D, Shankavaram U, Camphausen, K. Comparison of Proteomic Expression Profiles after Radiation Exposure across Four Different Species. Radiat Res. 197, 315–323 (2022).
REFERENCES
- 1.Sproull M, Camphausen K. State-of-the-art advances in radiation biodosimetry for mass casualty events involving radiation exposure. Radiat Res 2016; 186(5):423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang W, Yu J, Liu T, Defnet AE, Zalesak S, Farese AM, et al. Proteomics of non-human primate plasma after partial-body radiation with minimal bone marrow sparing. Health Phys 2020; 119(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rithidech KN, Honikel L, Rieger R, Xie W, Fischer T, Simon SR. Protein-expression profiles in mouse blood-plasma following acute whole-body exposure to (137)Cs gamma rays. Int J Radiat Biol 2009; 85(5):432–47. [DOI] [PubMed] [Google Scholar]
- 4.Marchetti F, Coleman MA, Jones IM, Wyrobek AJ. Candidate protein biodosimeters of human exposure to ionizing radiation. Int J Radiat Biol 2006; 82(9):605–39. [DOI] [PubMed] [Google Scholar]
- 5.Ossetrova NISDJ, Blakely WF. C-reactive protein and serum amyloid A as early-phase and prognostic indicators of acute radiation exposure in nonhuman primate total-body irradiation model. Radiat Meas 2011; 46(9):1019–24. [Google Scholar]
- 6.Blakely WF, Ossetrova NI, Whitnall MH, Sandgren DJ, Krivokrysenko VI, Shakhov A, et al. Multiple parameter radiation injury assessment using a nonhuman primate radiation model-biodosimetry applications. Health Phys 2010; 98(2):153–9. [DOI] [PubMed] [Google Scholar]
- 7.Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Multivariate Analysis of radiation responsive proteins to predict radiation exposure in total-body irradiation and partial-body irradiation models. Radiat Res 2017; 187:251–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sproull M, Shankavaram U, Camphausen K. Novel murine biomarkers of radiation exposure using an aptamer-based proteomic technology. Front Pharmacol 2021; 12:633131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha CT, Li X-H, Fu D, Moroni M, Fisher C, Arnott R, et al. Circulating interleukin-18 as a biomarker of total-body radiation exposure in mice, minipigs, and nonhuman primates (NHP). PLoS One 2014; 9(10):e109249–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balog RP, Bacher R, Chang P, Greenstein M, Jammalamadaka S, Javitz H, et al. Development of a biodosimeter for radiation triage using novel blood protein biomarker panels in humans and non-human primates. Int J Radiat Biol 2020; 96(1):22–34. [DOI] [PubMed] [Google Scholar]
- 11.Singh VK, Romaine PLP, Seed TM. Medical Countermeasures for radiation exposure and related injuries: characterization of medicines, fda-approval status and inclusion into the strategic national stockpile. Health Phys 2015; 108(6):607–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrea LD, David RC, William ED, John LE, Judith AH, Oxana S, et al. Challenges and benefits of repurposing products for use during a radiation public health emergency: lessons learned from biological threats and other disease treatments. Radiat Res 2018; 190(6):659–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiCarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, et al. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep 2011; 5(0 1):S32–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SomaLogic, SomaScan. SomaScan Proteomic Assay: Technical White Paper 2015; (DCN 15–310, SSM-002, Rev. 3):[1–14 pp.]. Available from: https://www.somalogic.com/wp-content/uploads/2016/08/SSM-002-Rev-3-SomaScan-Technical-White-Paper.pdf.
- 15.Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U, et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol 2008; 26(32):5233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, et al. Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids 2014; 3(10):e201–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Team RC. R: A language and environment for statistical computing Vienna, Austria: RFoundation for Statistical Computing; 2015. [Google Scholar]
- 18.Ossetrova NI, Stanton P, Krasnopolsky K, Ismail M, Doreswamy A, Hieber KP. Comparison of biodosimetry biomarkers for radiation dose and injury assessment after mixed-field (neutron and gamma) and pure gamma radiation in the mouse total-body irradiation model. Health Phys 2018; 115(6):743–59. [DOI] [PubMed] [Google Scholar]
- 19.Cataldi A, Centurione L, Di Pietro R, Rapino M, Bosco D, Grifone G, et al. Protein kinase C zeta nuclear translocation mediates the occurrence of radioresistance in friend erythroleukemia cells. J Cell Biochem 2003; 88(1):144–51. [DOI] [PubMed] [Google Scholar]
- 20.Wei W, Bai H, Feng X, Hua J, Long K, He J, et al. Serum Proteins as new biomarkers for whole-body exposure to high- and low-let ionizing radiation. Dose Response 2020; 18(1):1559325820914172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chua MW, Lin MZ, Martin JL, Baxter RC. Involvement of the insulin-like growth factor binding proteins in the cancer cell response to DNA damage. J Cell Comm Sign 2015; 9(2):167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skyttä T, Tuohinen S, Boman E, Virtanen V, Raatikainen P, Kellokumpu-Lehtinen PL. Troponin T-release associates with cardiac radiation doses during adjuvant left-sided breast cancer radiotherapy. Radiat Oncol 2015; 10:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzler B, Hammerer-Lercher A, Jehle J, Dietrich H, Pachinger O, Xu Q, et al. Plasma cardiac troponin T closely correlates with infarct size in a mouse model of acute myocardial infarction. Clin Chim Acta 2002; 325(1–2):87–90. [DOI] [PubMed] [Google Scholar]
- 24.Seo HR, Bae S, Lee YS. Radiation-induced cathepsin S is involved in radioresistance. Int J Cancer 2009; 124(8):1794–801. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Wang X, Xu S, Li X, Ma X. Cathepsin B contributes to radioresistance by enhancing homologous recombination in glioblastoma. Biomed Pharmacother 2018; 107:390–6. [DOI] [PubMed] [Google Scholar]
- 26.Olivares-Urbano MA, Griñán-Lisón C, Zurita M, del Moral R, Ríos-Arrabal S, Artacho-Cordón F, et al. Matrix metalloproteases and TIMPs as prognostic biomarkers in breast cancer patients treated with radiotherapy: A pilot study 2020; 24(1):139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasouli HA, Moghadam MM, Tabatabaiefar M, Taslimi F, Sheybani KM, Alidoosti A, et al. Comparing cystatin C changes as a measure of renal function before and after radiotherapy in patients with stomach cancer. Acta Med Iran 2012; 50(1):43–6. [PubMed] [Google Scholar]
- 28.McNamee JP, Bellier PV, Konkle AT, Thomas R, Wasoontarajaroen S, Lemay E, et al. Analysis of gene expression in mouse brain regions after exposure to 1.9 GHz radiofrequency fields. Int J Radiat Biol 2016; 92(6):338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Li Y, Tang M, Peng B, Lu X, Yang Q, et al. Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair. Cell Res 2018; 28(7):756–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacPhail SH, Banáth JP, Yu TY, Chu EH, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int J Radiat Biol 2003; 79(5):351–8. [DOI] [PubMed] [Google Scholar]
- 31.Sproull M, Hayes J, Ishiniwa H, Nanba K, Shankavaram U, Camphausen K, et al. Proteomic biomarker analysis of serum from Japanese field mice (apodemus speciosus) collected within the Fukushima difficult-to-return zone. Health Phys 2021; 121(6):564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Serum Amyloid A as a Biomarker for radiation exposure. Radiat Res 2015; 184(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, Yu J, Liu T, Tudor G, Defnet AE, Zalesak S, et al. Proteomic evaluation of the natural history of the acute radiation syndrome of the gastrointestinal tract in a non-human primate model of partial-body irradiation with minimal bone marrow sparing includes dysregulation of the retinoid pathway. Health Phys 2020; 119(5):604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]


