Abstract
The global COVID-19 epidemic has spread rapidly around the world and caused the death of more than 5 million people. It is urgent to develop effective strategies to treat COVID-19 patients. Here, we revealed that SARS-CoV-2 infection resulted in the dysregulation of genes associated with NAD+ metabolism, immune response, and cell death in mice, similar to that in COVID-19 patients. We therefore investigated the effect of treatment with NAD+ and its intermediate (NMN) and found that the pneumonia phenotypes, including excessive inflammatory cell infiltration, hemolysis, and embolization in SARS-CoV-2-infected lungs were significantly rescued. Cell death was suppressed substantially by NAD+ and NMN supplementation. More strikingly, NMN supplementation can protect 30% of aged mice infected with the lethal mouse-adapted SARS-CoV-2 from death. Mechanically, we found that NAD+ or NMN supplementation partially rescued the disturbed gene expression and metabolism caused by SARS-CoV-2 infection. Thus, our in vivo mouse study supports trials for treating COVID-19 patients by targeting the NAD+ pathway.
Subject terms: Mechanisms of disease, Cell death
Introduction
Novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global epidemic of coronavirus disease 2019 (COVID-19). Patients with critical COVID-19 develop severe respiratory distress, and many of them died from it1. Although vaccines with high protective potency have been developed, the ability of SARS-CoV-2 to mutate quickly has brought huge uncertainty to the COVID-19 epidemic, which has prioritized the development of drugs for COVID-19 treatment.
Nicotinamide adenine dinucleotide (NAD+) and NADH are essential coenzymes for multiple cellular energy metabolism pathways, such as tricarboxylic acid (TCA) cycle and oxidative phosphorylation2. NAD+ also serves as a substrate for sirtuin deacetylases (SIRTs), cyclic ADP-ribose synthases (including CD38 and CD157), and poly- or mono-(ADP-ribose) polymerases (PARPs), and participate in many important biological processes, including cell survival, aging, inflammation, etc2–4. Besides, NAD+ can be phosphorylated to nicotinamide adenine dinucleotide phosphate (NADP+) by NAD+ kinase (NADK)5. Both NADP+ and its reduced form, NADPH, play critical roles in maintaining redox balance and biosynthesis of fatty acids and nucleic acids6,7. In mammalian cells, NAD+ is synthesized through the de novo pathway and the salvage pathway7.
One of the most striking disturbance in the sera of COVID-19 patients was the increase of metabolites related to de novo production of NAD+8. Whether and how NAD+ metabolism is disrupted in the lungs of COVID-19 patients is still not clear. NAD+ has been proposed as a selective inhibitor for SARS-CoV-2 main protease (Mpro) and multi-functional papain-like protease (PLpro)9,10. Recently, we found that NAD+ supplementation effectively alleviates cell death and inhibits microglia activation in Zika virus (ZIKV)-infected mouse brains11. Therefore, it is intriguing to explore whether NAD+ or its intermediates are protective from SARS-CoV-2 infection.
In this study, we used transcriptomics and metabolomics of lungs to assess the disturbance of gene expression and metabolism after SARS-CoV-2 infection and tested the protective effect of NAD+ and its intermediate, NMN, in lungs in two different mouse models of SARS-CoV-2 infection12,13. Numerous gene expression changes and activation of multiple immune pathways in human samples were imitated in our mouse model. SARS-CoV-2-induced pneumonia could be significantly alleviated by NAD+ and NMN treatment. We used real-time quantitative PCR (RT-qPCR) to validate our transcriptomics results and uncovered the possible mechanisms by which NAD+ and NMN were effective in treating pneumonia caused by SARS-CoV-2 infection. Meanwhile, metabolomics analysis was applied in the lethal mouse model of SARS-CoV-2 infection to reveal the alteration of metabolism after SARS-CoV-2 infection and protective effects of NMN.
Results
Transcriptomics reveals similar immune responses in a mouse model of SARS-CoV-2 infection and COVID-19 patients
To explore potential therapeutic targets for COVID-19, we inspected the differentially expressed genes in a mouse-adapted SARS-CoV-2 (MASCp6) infection mouse model13. Eight- to nine-month-old (aged) female BALB/c mice were intranasally (i.n.) infected with MASCp6 and sacrificed 3 days post infection (dpi). To confirm the infection of MASCp6 in mouse lungs, SARS-CoV-2 proteins and RNA copies were detected in lung tissues. Immunostaining with SARS-CoV-2 spike protein and nucleocapsid protein antibodies indicated that MASCp6 mainly infected airway epithelial cells and some alveolar epithelial cells near the airways (Supplementary Fig. S1). The infection pattern is similar to another mouse-adapted model of SARS-CoV-2, in which receptor-binding domain in the spike protein was remodeled to facilitate its efficient binding to mouse ACE214.
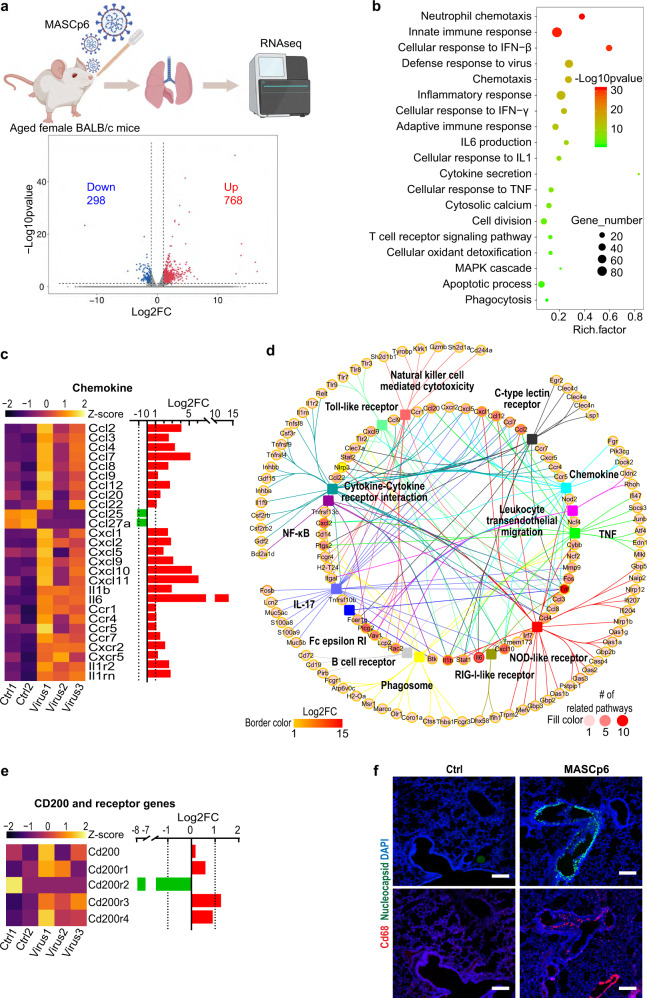
Transcriptomics analysis identified a total of 1066 significantly differentially expressed genes (|log2FoldChange (FC)| > 1, P < 0.05) with 298 downregulated and 768 upregulated genes in the infected lungs (Fig. 1a). Gene ontology (GO) enrichment analysis of significantly upregulated genes revealed the strong activation of immune-related biological processes, including neutrophil chemotaxis, innate immune response, adaptive immune response, cellular response to IFN, TNF, and IL1, as well as apoptotic process and phagocytosis (Fig. 1b). Infection led to robust induction of chemokine and interleukin genes, including Il1b, Il6, Ccl2, Ccl3, Ccl4, Ccl7, Cxcl1, Cxcl2, Cxcl5, Cxcl9, and Cxcl10, and related receptor genes, including Ccr1, Ccr4, Ccr5, Ccr5, Ccr7, Cxcr2, Cxcr5, Il1r2, and Il1rn (Fig. 1c). Many significantly upregulated genes participated in neutrophil chemotaxis, including S100a8 and S100a9 (Fig. 1b; Supplementary Fig. S2a). These results indicated that innate immune response is activated robustly as that in COVID-19 patients15–17.
Fig. 1. Transcriptomics analysis of MASCp6-infected aged mouse lungs.
a Overview of the RNA-seq data. Dot plot shows 1066 significantly differentially expressed (P < 0.05 and |log2FC| > 1) genes with 298 downregulated and 768 upregulated in the infected group. FC, fold change. b GO analysis of significantly upregulated genes. Rich factor represents the ratio of significantly differentially expressed genes to the total genes belonging to the GO term. c Heatmap and bar plot show relative expression levels of significantly differentially expressed genes of chemokine, interleukin, and their receptors. d Network of the genes belonging to significantly-enriched immunity-related KEGG terms of significantly upregulated genes. e Heatmap and bar plot show relative expression levels of CD200 and its receptor genes. f Immunostaining results with SARS-CoV-2 nucleocapsid (green) and macrophage marker (CD68, red) antibodies of lung sections from MASCp6-infected and control aged mice. Scale bars, 200 μm.
To inspect the induced immune-related genes and pathways by MASCp6 in more detail, we performed KEGG enrichment analysis. Cytokine–cytokine receptor interaction, NOD-like receptor signaling, IL-17 signaling, chemokine signaling, TNF signaling, Toll-like receptor signaling, as well as other immune pathways were detected (Fig. 1d). Among all significantly upregulated genes in these pathways, Tnf, Il1b, Il6, Cxcl2, Cxcl10, and Plcg2 play roles in multiple signaling pathways, suggesting their importance and potential drug targets in SARS-CoV-2 infection (Fig. 1d). In addition, marker genes of neutrophil, macrophage, dendritic cell, B cell, and CD4+ T cell were upregulated in infected mice as that in COVID-19 patients (Supplementary Fig. S2c)18. Moreover, we found two CD200 receptor genes that are responsible for transmitting strong activating signals19,20, Cd200r3 and Cd200r4, were upregulated. This was consistent with the immunofluorescence results showing that substantially more activated macrophages (CD68+) were detected in the infected lung section compared to control (Fig. 1f).
Interestingly, we found that 30.2% (90/298) significantly downregulated genes are long non-coding RNAs (lncRNAs) (Supplementary Fig. S3a). This suggests that lncRNAs may play an important role in SARS-CoV-2-induced immune response, based on the fact that lncRNAs have the capacity to regulate the expression of proinflammatory genes21.
The above results indicated that SARS-CoV-2-infected murine model mimics some of the immune features present in patients15,18,22,23, and it is suitable for testing drugs.
Cell death occurs with the development of immune response in the SARS-CoV-2-infected mice
SARS-CoV-2 infection induces high expression of massive inflammatory genes and even triggers the cytokine storm, which is considered to be the main cause of death in severe COVID-19 patients24. Meanwhile, induction of inflammatory genes in MASCp6-infected mouse lungs might affect cell survival as many of them play roles in the cell death pathways (Supplementary Fig. S2b). In addition, many caspase family members were upregulated after infection (Fig. 2a). The presence of many cleaved caspase 3+ cells confirmed that apoptosis occurred in infected lungs (Fig. 2b).
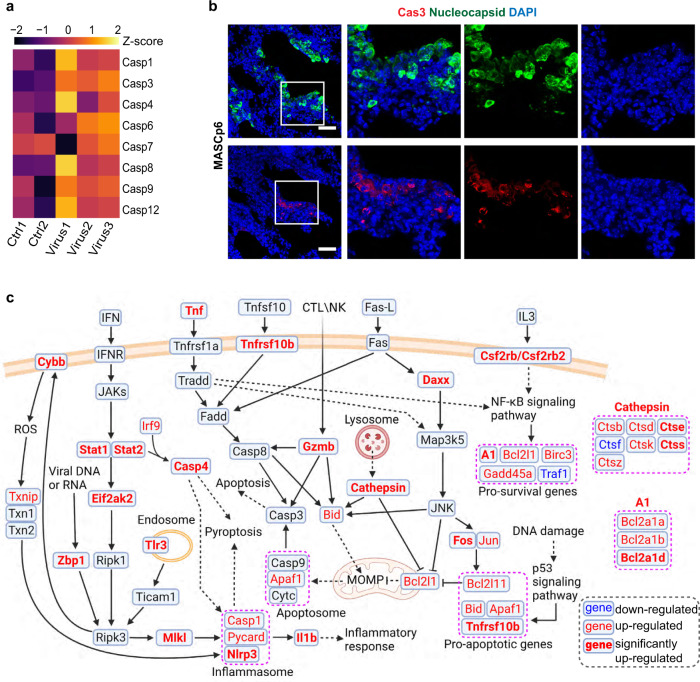
Fig. 2. MASCp6 infection leads to cell death in the lungs.
a Heatmap shows relative expression levels of caspase family genes in the RNA-seq data. b Immunostaining results with SARS-CoV-2 nucleocapsid (green) and activated form of caspase 3 (Cas3, red) antibodies of lung sections from MASCp6-infected and control aged mice. Scale bars, 50 μm. c Schematic diagram (created in BioRender.com) of apoptosis and necroptosis pathways. MOMP, mitochondrial outer membrane permeabilization. Downregulated: P < 0.05 and FC < 1; upregulated: P < 0.05 and FC > 1; significantly upregulated: P < 0.05 and FC > 2.
We summarized the apoptosis and necroptosis pathways combining with our transcriptomics data (Fig. 2c). In the apoptosis pathway, Daxx and Fos were significantly upregulated, which would induce the expression of pro-apoptotic gene, Bcl2l11 (Bim). Other pro-apoptotic genes, Bid, Apaf1, and Tnfrsf10b were induced in the p53 signaling pathway. In addition, the significant upregulation of Gzmb and cathepsin family genes, Ctss and Ctse, suggested the occurrence of mitochondrial outer membrane permeabilization and apoptosome formation25. In the JAK-STAT signaling pathway, Stat1, Stat2, and their target gene, Casp4, and inflammasome member, Nlrp3, were significantly induced. As the sensor component of inflammasome, NLRP3 plays a crucial role in innate immunity and inflammation in response to viral RNA26,27. CASP4 acts as essential effectors of NLRP3 inflammasome-dependent CASP1 activation and IL1B secretion. Both CASP1 and CASP4 can initiate and promote pyroptosis through GSDMD cleavage and activation28. Moreover, a group of pro-survival NF-κB signaling pathway downstream genes, Bcl2a1a, Bcl2a1b, Bcl2a1d, Bcl-XL (Bcl2l1), Birc3, and Gadd45a were also induced, implicating the compensation or rescue response by surviving cells. Thus, cell death or survival depends on the balanced effects of these genes.
Elevated expression of NADPH oxidase genes and PARPs dysregulates NAD system in SARS-CoV-2-infected mice
We noticed that Cybb, the critical component of NADPH oxidase that generates reactive oxygen species (ROS), was significantly induced in MASCp6-infected mouse lungs (Fig. 2c). NADPH oxidase plays an important role in neutrophil extracellular trap formation and phagosome maturation during immune response29. Intriguingly, other NADPH oxidase component genes, Ncf1, Ncf2, Ncf4, and Rac2 were also significantly upregulated, suggesting that more NADPH would be consumed to produce ROS (Fig. 3a).
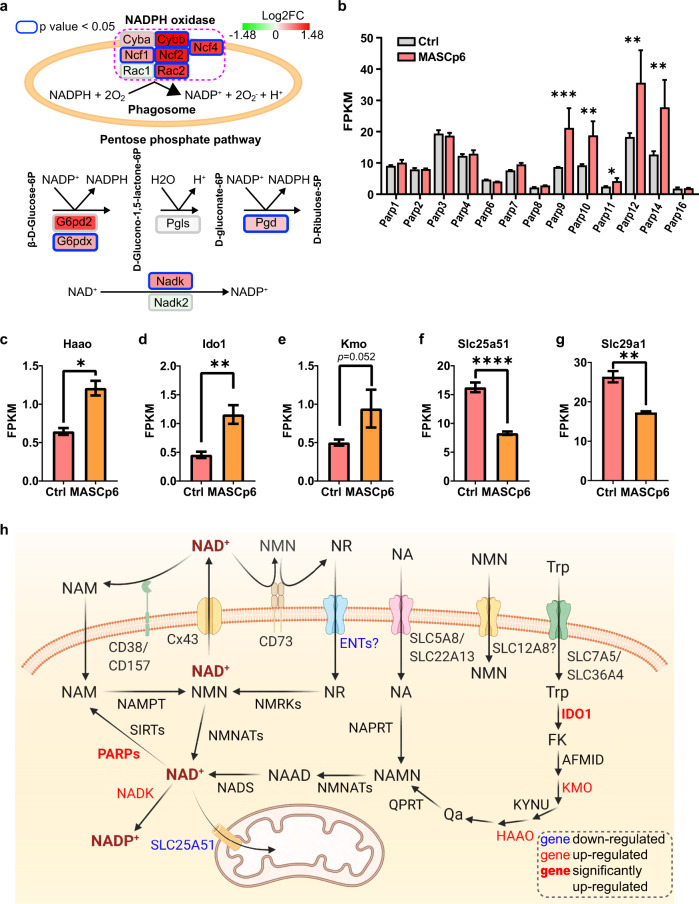
Fig. 3. MASCp6 infection leads dysregulation of NAD(H) and NADP(H) related genes.
a Schematic diagram of NADPH oxidase complex and oxidative phase of pentose phosphate pathway. The fill color of gene represents relative expression level in lungs of MASCp6-infected and control mice. b Bar plot shows relative expression levels of PARPs. c–g Bar plots show relative expression levels of three NAD+ de novo synthesis-related genes, Haao (c), Ido1 (d), and Kmo (e), and two nucleoside transporter genes, Slc25a51 (f) and Slc29a1 (g). h Schematic diagram (created in BioRender.com) of NAD+ metabolism pathway (referred to ref. 3). Downregulated: P < 0.05 and FC < 1; upregulated: P < 0.05 and FC > 1; significantly upregulated: P < 0.05 and FC > 2. NA, nicotinic acid; NAMN, nicotinic acid mononucleotide; NAAD, nicotinic acid adenine dinucleotide; NR, nicotinamide riboside; NAM, nicotinamide; NMN, nicotinamide mononuclotide; Trp, tryptophan; FK, formyl-kynurenine; Qa, quinolinate. All gene expression data are from RNA-seq. All data are shown as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The major source of NADPH in animals is the oxidative phase of pentose phosphate pathway, in which NADPH is produced from NADP+ by glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (PGD)30, while NADP+ is synthesized from NAD+ by NADK5 (Fig. 3a). The induction of G6pd2, G6pdx, Pgd, and Nadk implicated that more NADPH was produced to respond to the increased demand for NADPH in the immune process after MASCp6 infection.
NAD+ is essential for multiple metabolic pathways3. We recently found that PARPs, one of main NAD+-responsive signaling family, were significantly upregulated while NAD+ levels were substantially reduced in ZIKV-infected mouse brains11. In MASCp6-infected lungs, Parp9, Parp10, and Parp14, the three PARP members which regulate immune and inflammatory responses3,31, were also significantly induced (Fig. 3b). We therefore explored the expression of other genes involved in NAD+ biosynthetic process and found that some of those required for NAD+ de novo synthesis such as Haao, Ido1, and Kmo, were upregulated (Fig. 3c–e; Supplementary Fig. S4a). This was consistent with the finding in COVID-19 patients8. In addition, the downregulation of mitochondrial NAD+ transporter gene, Slc25a51, and a nucleoside transporter gene, Slc29a1 (Ent1), suggested that both the import of NAD+ from the cytoplasm into the mitochondria and the import of nicotinamide riboside (NR) into cells were hindered (Fig. 3f, g)32–35. Loss of SLC25A51 has been shown to lead to impaired mitochondrial functions34, and a group of mitochondria-encoded genes, including mt-Atp6, mt-Atp8, mt-Co1-3, mt-Cytb, and mt-Nd1-6 were downregulated36, although the expressions of genes associated with the TCA cycle and oxidative phosphorylation were not considerably altered in general (Supplementary Fig. S4b, c). Together with the evidence of significantly upregulated expression of Gzmb and cathepsin family genes, mitochondrial functions can be predicated to be affected in the infected lungs.
The above results suggest that over-consumption of NADPH by upregulation of NADPH oxidase genes would lead to the convert of more NAD+ to NADP+, and NADP+ is transformed into NADPH by G6PD and PGD in the pentose phosphate pathway in the infected lungs. On the other hand, upregulation of PARPs would also lead to reduced NAD+ level. The reduction of NAD+ level would stimulate the compensatory de novo synthesis of NAD+ (Fig. 3h).
NAD+ supplementation alleviates SARS-CoV-2-induced pneumonia
Based on the observations that SARS-CoV-2 infection led to differential expression of NAD+-associated genes in both mouse and human sera (our study and ref. 37), we investigated whether NAD+ supplementation could rescue MASCp6-induced pneumonia. We treated aged (8–9 months old) and young (6–7 weeks old) infected female mice with NAD+ (1 mg/g/day intraperitoneally (i.p.) for 3 days), and sacrificed them on 3 dpi (Fig. 4a).
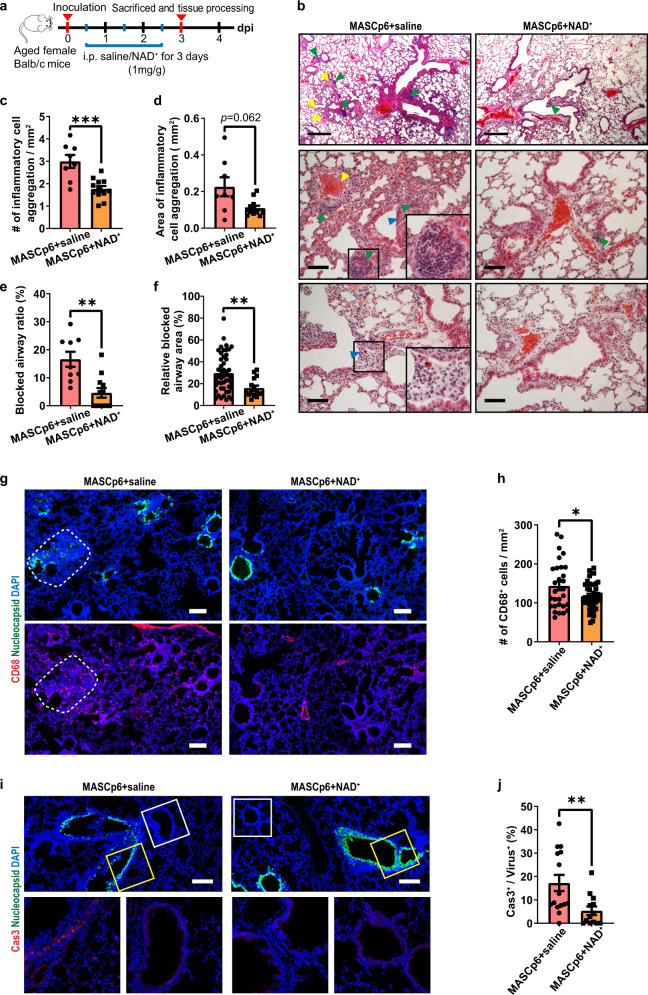
Fig. 4. NAD+ supplementation alleviates pathological phenotypes of MASCp6-induced pneumonia.
a Schematic diagram of experimental design for NAD+ supplementation in MASCp6-infected aged mice. b Hematoxylin and eosin (H&E) staining results show that cell infiltration (green arrowhead), airway blockage (blue arrowhead) and hemolysis (yellow arrowhead) were rescued by NAD+ supplementation in lungs of MASCp6-infected aged mice. Scale bars, 500 μm (top panels) and 50 μm (middle and bottom panels). c–f Quantification of the density (c) and area (d) of inflammatory cell aggregates and the ratio (e) and relative area (f) of blocked airway. t-test (c, d) and Mann–Whitney test (e, f). g Immunostaining results with SARS-CoV-2 nucleocapsid (green) and macrophage marker (CD68, red) antibodies of two adjacent lung sections for each group. White border: excessive aggregation of macrophages. Scale bars, 200 μm. h Quantification of the density of CD68+ cells of saline- and NAD+-administered MASCp6-infected aged mice. t-test. i Immunostaining results with SARS-CoV-2 nucleocapsid (green) and activated form of caspase 3 (Cas3, red) antibodies of two adjacent lung sections for each group. Yellow border: infected airway epithelial cells; White border: airway epithelial cells without virus. Scale bars, 200 μm. j Quantification of the ratio of Cas3+ cells to infected cells of saline- and NAD+-administered MASCp6-infected aged mice. Mann–Whitney test. All quantification data are shown as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n = 3 mice for MASCp6 + saline group, n = 4 mice for MASCp6 + NAD+ group.
For aged mice, pathological features including severe inflammatory cell infiltration, alveolar septal thickening, hemolysis, epithelial damage, airway blockage, and cell death are detected in MASCp6-infected lungs (saline group), similar to that in the hACE2 mouse model (Fig. 4b)38,39. Compared with the saline group, the number and size of inflammatory cell aggregates as well as the ratio and relative area of blocked airway were substantially reduced in the NAD+ treatment group (Fig. 4b–f; Supplementary Fig. S5a). Notably, no hemolysis was detected after treatment with NAD+ (Fig. 4b). For young mice, the pathological features were mild, and eliminated almost entirely by NAD+ treatment (Supplementary Fig. S6b, c). However, comparable levels of viral mRNA as measured by qPCR and fluorescence intensity as measured by immunostaining with the nucleocapsid antibody were detected in saline and NAD+ treatment groups (Supplementary Figs. S5b–d, S6a). This indicates that the treatment relieves the pathological damages, but not viral replication.
Previous studies have shown that SARS-CoV-2 infection induces the infiltration of neutrophils and macrophages in lungs of patients38,40. We found that the number of macrophages was reduced significantly in NAD+-treated aged and young mice (Fig. 4g, h; Supplementary Fig. S6d, e). Meanwhile, NAD+ treatment remarkably suppressed cell death as shown by the reduction of cleaved caspase 3+ cells (68.9%) in aged mice (Fig. 4i, j).
The above results indicate that NAD+ supplementation can protect the lung from inflammatory injury, including cell death, caused by SARS-Cov-2 infection in both aged and young mice.
NAD+ supplementation partially salvages the changes in gene expression caused by SARS-CoV-2 infection
To validate the results of RNA-seq and examine whether the changes in gene expression could be rescued by NAD+ supplementation, we performed qPCR to inspect the expression of PARP family genes in the infected lungs (Fig. 5a–e). We found that the reduction of Parp2 expression was rescued almost completely (Fig. 5b). Unexpectedly, the upregulations of Parp9, Parp10, and Parp14 were induced further in the NAD+ treatment group (Fig. 5c–e). This suggests that Parp9, Parp10, and Parp14 are important for host cells to defend against SARS-CoV-2. Host cells had to downregulate the expression of Parp2 to meet the need of NAD+ which was enhanced by elevated expression of Parp9, Parp10, and Parp14.
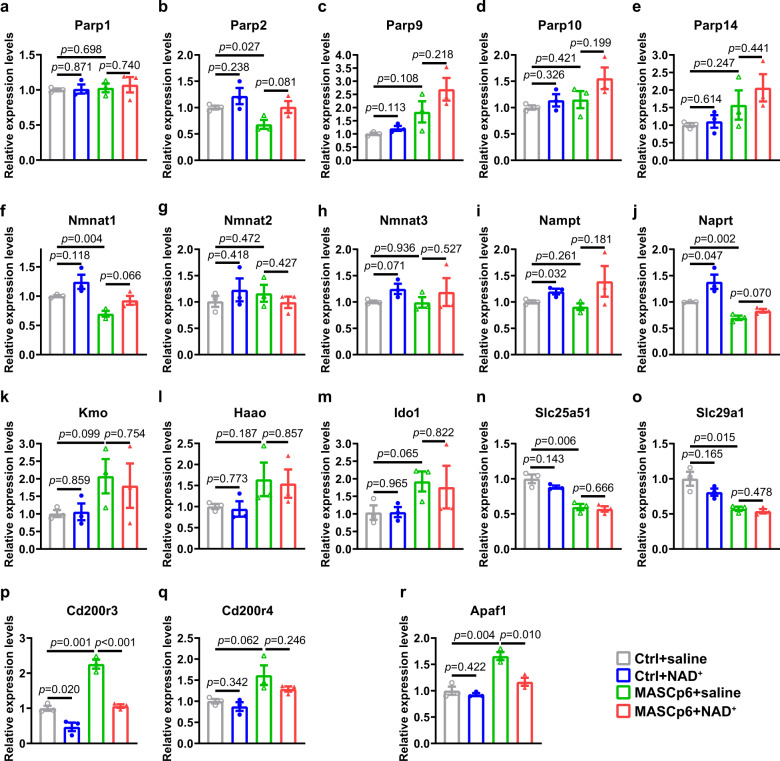
Fig. 5. Dysregulation of genes can be partially rescued by NAD+ supplementation.
a–e qPCR results of relative expression levels of PARP family members as listed. Parp1 (a), Parp2 (b), Parp9 (c), Parp10 (d), and Parp14 (e). f–j qPCR results of relative expression levels of NAD+ salvage pathway-related genes, Nmnat1 (f), Nmnat2 (g), Nmnat3 (h), Nampt (i), and Naprt (j). k–o qPCR results of relative expression levels of three NAD+ de novo synthesis-related genes, Haao (k), Ido1 (l), and Kmo (m), and two nucleoside transporter genes, Slc25a51 (n) and Slc29a1 (o). p, q qPCR results of relative expression levels of CD400 receptor genes, Cd200r3 (p) and Cd200r4 (q). r qPCR results of relative expression levels of apoptosis protease activating factor gene, Apaf1. All quantification data are shown as means ± SEM, t-test. n = 3 mice for each group. Exact P values are indicated.
We also looked at the effect of NAD+ supplementation on the expression of genes associated with NAD+ synthesis (Fig. 5f–m). Interestingly, the downregulation of genes responsible for NAD+ salvage pathway, especially Nmnat1 and Naprt, were partially rescued by NAD+ treatment (Fig. 5f, j), but the genes responsible for NAD+ de novo pathway were not affected (Fig. 5k–m). In addition, NAD+ treatment had no effect on the expression of Slc25a51 and Slc29a1 (Fig. 5n, o).
The upregulation of Cd200r3 was rescued completely while the expression of Cd200r4 was partially suppressed by NAD+ treatment (Fig. 5p, q). These results were matched with reduction of CD68+ cells in the lungs (Supplementary Fig. S5e). This indicates that NAD+ treatment leads to reduced activation of macrophage. Moreover, the upregulation of Apaf1, the apoptosis protease activating factor, was also rescued very significantly by NAD+ treatment (Fig. 5r).
Taken together, our qPCR results confirmed many of our findings from RNA-seq analysis and NAD+ supplementation was beneficial to the maintenance of the NAD system. In addition, severe inflammatory cell infiltration and cell death caused by SARS-CoV-2 infection might be rescued by NAD+ treatment through suppressing the expression of Cd200r3, Cd200r4, and Apaf1, and other related genes untested.
NMN supplementation alleviates SARS-CoV-2-induced pneumonia
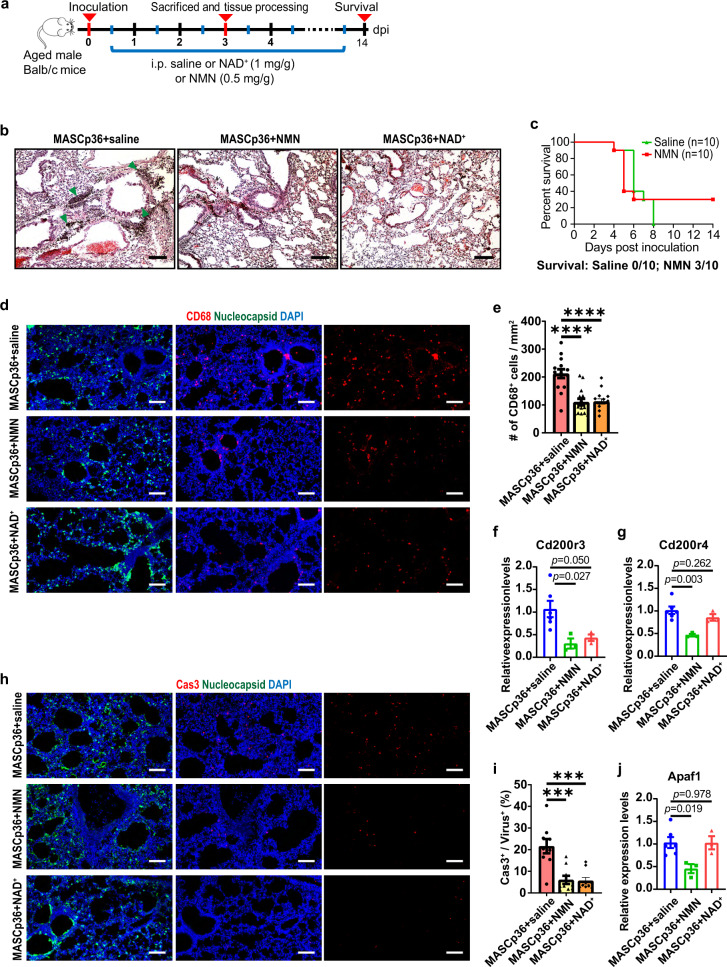
To examine whether NAD+ intermediates have similar therapeutic effects on SARS-CoV-2-induced pneumonia, we adopted a recently developed and more severe infection model in which mice were infected with a mouse-adapted SARS-CoV-2, MASCp3612. Infected mice (8–9 months old) were treated with NMN or NAD+ (Fig. 6a). The dosage of NMN (500 mg/kg/day) was widely used in mouse models41, which is equal to 55 mg/kg/day in humans according to body surface area42. Compared with the severe inflammatory cell infiltration and alveolar septal thickening detected in the saline group, the pathological damage was alleviated obviously in both NMN and NAD+ groups (Fig. 6b). More strikingly, 30% of the mice survived eventually in the NMN group, while 100% of the mice died on 8 dpi in the saline group (Fig. 6c). In addition, both the number of CD68+ cells and the expression of Cd200r3 were reduced significantly in both NMN and NAD+ groups (Fig. 6d–f). Moreover, cell death was substantially suppressed by NMN and NAD+ treatment (72.0% and 73.9%, respectively) (Fig. 6h, i). The suppression of Cd200r4 and Apaf1 expression was significant in the NMN group, but not in NAD+ group of MASCp36-infected lungs (Fig. 6g, j).
Fig. 6. NMN supplementation alleviates pathological phenotypes of MASCp36-induced pneumonia.
a Schematic diagram of experimental design for NMN and NAD+ supplementation in MASCp36-infected aged mice. b H&E staining results of lung sections from saline-, NMN-, and NAD+-administered MASCp36-infected aged mice. Green arrowhead: inflammatory cell aggregates. Scale bars, 100 μm. c Survival of saline (n = 10) and NMN (n = 10) administered MASCp36-infected aged mice. d Immunostaining results with SARS-CoV-2 nucleocapsid (green) and macrophage marker (CD68, red) antibodies of two adjacent lung sections for each group. Scale bars, 100 μm. e Quantification of the density of CD68+ cells of lung sections from saline-, NMN-, and NAD+-administered MASCp36-infected aged mice. t-test. n = 3 mice per group. f, g qPCR results of relative expression levels of CD400 receptor genes, Cd200r3 (f) and Cd200r4 (g). t-test. n = 6 mice for saline group and n = 3 mice for NMN and NAD+ groups. Exact P values are indicated. h Immunostaining results with SARS-CoV-2 nucleocapsid (green) and Cas3 (red) antibodies of two adjacent lung sections for each group. Scale bars, 100 μm. i Quantification of the ratio of Cas3+ cells to infected cells of saline-, NMN-, and NAD+-administered MASCp36-infected aged mice. Mann–Whitney test. n = 3 mice per group. j qPCR results of relative expression levels of apoptosis protease activating factor gene, Apaf1. t-test. n = 6 mice for saline group and n = 3 mice for NMN and NAD+ groups. Exact P values are indicated. All quantification data are shown as means ± SEM. ***P < 0.001, ****P < 0.0001.
These results indicate that NMN supplementation can suppress the expression of Cd200r3, Cd200r4, and Apaf1 in the lethal mouse-adapted SARS-CoV-2 mouse model, and alleviate inflammatory injury in the lungs and even death of the infected animals.
NMN supplementation partially rescues the disturbance of metabolism caused by SARS-CoV-2 infection
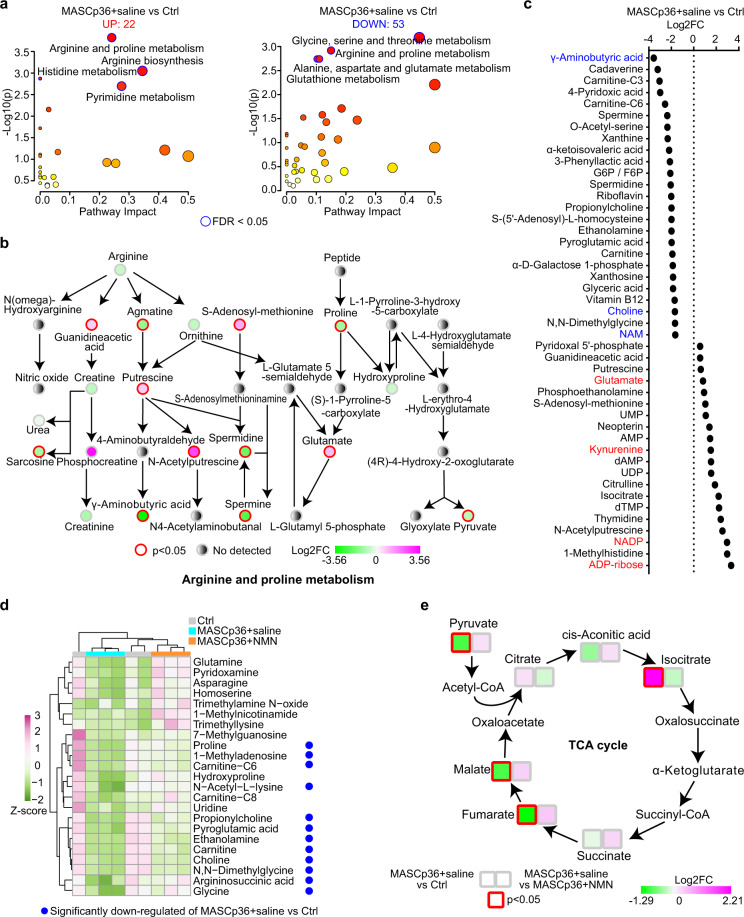
We next investigated in more detail the protective effect of NMN by targeted metabolomics analysis (Supplementary Table S1). We compared the metabolomes between the MASCp36-infected group (MASCp36 + saline) and the control group (uninfected mice of the same sex and age). A total of 75 significantly deregulated metabolites with 22 upregulated (P < 0.05 and fold change > 1.25) and 53 downregulated (P < 0.05 and fold change < 0.8) were detected in infected group (Fig. 7a; Supplementary Fig. S7b). We performed KEGG pathway enrichment analysis and found that the arginine and proline metabolism, glycine, serine and threonine metabolism, alanine, aspartate and glutamate metabolism, arginine biosynthesis, glutathione metabolism, histidine metabolism, and pyrimidine metabolism were significantly affected by MASCp36 infeciton (FDR < 0.05) (Fig. 7a, b; Supplementary Fig. S7c). Unexpectedly, the downregulation of γ-aminobutyric acid (GABA) was most significant one in the lungs after MASCp36 infection (Fig. 7b, c). Intracortical GABAergic dysfunction has been postulated as the cause of seizure in COVID-19 patients43,44, and administration of GABA or GABAa receptor agonists could limit pneumonitis and death in coronavirus-infected mice45. Therefore, GABA is a potential candidate to treat SARS-CoV-2 infection. The significant upregulation of NADP+ was consistent with our transcriptomics analyses, in which Nadk was significantly upregulated (Figs. 3a, 7c), and ADP-ribose was the most significantly upregulated one in the lungs after MASCp36 infection (Fig. 7c). It is possible that the first macrodomain of SARS-CoV-2 nonstructural protein 3 (Nsp3), Mac1, which has de-mono-ADP-ribosylation activity, counters ADP-ribosylation by PARPs46. In addition, the downregulation of choline and upregulation of kynurenine and glutamate in the sera have been reported to be associated with the severity of COVID-19 patients (Fig. 7c)47–50.
Fig. 7. Metabolomics analysis of MASCp36-infected aged mouse lungs.
a KEGG analysis of significantly differential metabolites (P < 0.05; FC < 0.8 or FC > 1.25) between the lungs of saline group of MASCp36-infected mice and control mice. b Schematic diagram depicting the dysregulation of arginine and proline metabolism pathways in response to MASCp36 infection. c Dot plot shows the top 25 most upregulated and top 20 most downregulated metabolites. d Heatmap shows relative abundance of significantly differential metabolites in the NMN group compared with saline group of MASCp36-infected mice. The blue dots mean that the metabolite was significantly downregulated in saline group of MASCp36-infected mice compared with control mice. e Schematic diagram depicting that dysregulation of metabolites belonging to TCA cycle in MASCp36-infected mice was partially rescued by NMN supplementation. Nodes without color-block represent metabolites which were not tested.
We then compared the metabolomes between the saline and the NMN groups of MASCp36-infected mice. A total of 23 significantly differential metabolites were detected in the NMN group, and all of them were upregulated (P < 0.05 and fold change > 1.25), including glutamine, asparagine, proline, choline, and glycine (Fig. 7d). These metabolites participate in the aminoacyl-tRNA biosynthesis, alanine, aspartate and glutamate metabolism, glycine, serine and threonine metabolism and arginine biosynthesis (Supplementary Fig. S7d). Metabolites and proteins in citrate cycle (TCA cycle) were altered in the sera of COVID-19 patients and in vitro SARS-CoV-2 infection model, respectively48,50. We observed that the levels of pyruvate, fumarate and malate were significantly reduced, but isocitrate was significantly increased by MASCp36 infection (Fig. 7e). NMN supplementation could rescue the alterations in TCA cycle metabolites overall (Fig. 7e). One of major dysregulated metabolic pathways of COVID-19 was the tryptophan and kynurenine pathway. Lower level of tryptophan and higher level of kynurenine were found in the sera of severe COVID-19 patients51. Intriguingly, we found that the levels of both tryptophan and kynurenine were increased by NMN supplementation in MASCp36-infected mice (Supplementary Fig. S7e). The elevation of glucose in the plasma was considered as an indicator for the severity of COVID-19 patients47,48. Our results showed the level of glucose had a tendency of increase in the lungs of MASCp36-infected mice, and the level returned to normal in two NMN supplementation mice (Supplementary Fig. S7e). The reduction of GABA was partially rescued, which also helped to limit pneumonitis and seizure (Supplementary Fig. S7e)45. Furthermore, we performed the absolute quantifications of NAD+ and its precursors (Supplementary Table S2). Our results indicated that NAD+, NMN, NR, and NAM were elevated in the lungs of MASCp36-infected mice after NMN supplementation (Supplementary Fig. S7f).
Our targeted metabolomics analyses indicate that the disturbed metabolism in the sera of COVID-19 patients was partly reproduced in the lungs of our mouse model, and it could be ameliorated by NMN supplementation.
Discussion
In this study, our transcriptomics and metabolomics analyses demonstrated that some similar immune processes, including cytokine–cytokine receptor interaction, TNF signaling, and IL-6 production, and metabolic pathways were disturbed in SARS-CoV-2-infected mice as those observed in patients and organoid model15,18,23,52. Importantly, NAD+ system was coming under attack in the SARS-CoV-2 infection (as shown by Heer et al.37), and the supplementation of NAD+ and its intermediate, NMN, can effectively protect the lungs from the pathological damage and even death (by NMN). Besides, we found that two CD200 receptor genes, Cd200r3 and Cd200r4, were upregulated in the lungs of SARS-CoV-2-infected mice. The expression level of Cd200r3 coincided with the activation of macrophage and inflammatory injury19,20. CD200R3 and CD200R4 could interact with the activatory adapter protein, DAP12, and transmit strong activating signals19,20. CD200R3 can also function as an activating receptor in mast cells and basophils and plays a role in IgE-independent immune responses, and depletion of basophils with CD200R3 monoclonal antibody ameliorated IgE-mediated systemic and local anaphylaxis53,54. Therefore, Cd200r3 may serve as a potential therapeutic target for the immune storm induced by SARS-CoV-2 infection. Interestingly, we found that NAD+ supplementation could effectively suppress Cd200r3 expression, while NMN supplementation could effectively suppress the expression of both Cd200r4 and Cd200r3. NAD+ intermediate and precursors, including NMN, NR, and NAM, have been shown to have anti-inflammatory effects in different animal models including aging, autoimmune encephalomyelitis, ischemia, Ataxia Telangiectasia and ZIKV infection2,3,55–58. Our results would provide more insights into the underlying mechanism.
NADPH oxidases are responsible for catalyzing the production of ROS which is important for the functions of phagosome during immune response59. As the expressions of NADPH oxidase genes, including Cybb, Ncf2, Ncf4, and Rac2, were significantly upregulated, NADPH would be over-consumed in the lungs of SARS-CoV-2-infected mice. NADPH can provide the reducing equivalents for biosynthetic reactions and the oxidation-reduction, and allow the regeneration of glutathione (GSH)60. Lower levels of NADPH can weaken the red blood cell membrane and trigger hemolysis as the maintenance of red blood cells require large amounts of GSH61. The hemolysis detected in the lungs and the upregulation of genes involved in the production of NADPH and NADP+, including G6pdx, Pgd, and Nadk, suggest the occurrence of NADPH deficiency and compensative effects through the increased expression of genes involved in NADPH synthesis in the infected lungs. Our study supports the notion that oxidative stress generated from ROS occurs in SARS-CoV-2-induced pneumonia62.
Consistent with our study, previous metabolomics studies showed that the level of tryptophan was decreased while the level of kynurenine was increased in the sera from severe COVID-19 patients47,51,63. The kynurenine/tryptophan ratio was higher in severe patients than in mild patients51, suggesting higher IDO activity in severe patients. Indeed, our transcriptomics data showed significantly higher expression level of Ido1 in MASCp36-infected lungs than controls. Although high kynurenine/tryptophan was postulated to correlate with the severity of COVID-19 patients51, kynurenine can inhibit the activities of pathogenic CD4+ T cells64. Our results showed that the levels of both tryptophan and kynurenine were elevated by NMN supplementation, which implicated that the causal relationship between the level of kynurenine and the severity of COVID-19 is uncertain.
Our transcriptomics and qPCR results indicated that a set of PARP family members, including Parp9, Parp10, Parp12, and Parp14, were significantly induced by SARS-CoV-2 infection in mice, similar to those in COVID-19 patients37. PARPs can cleave NAD+ and catalyze poly-ADP-ribosylation or mono-ADP-ribosylation on target proteins, which play important roles in DNA damage repair. However, Nsp3 of SARS-CoV-2 was found to reverse the PARP9-mediated ADP-ribosylation recently65, which may in turn promote PARP activity and increase the demand for NAD+. However, our absolute quantification results showed that the levels of NAD+ precursors, NAM and NA, but not NAD+, NMN, or NR, were significantly downregulated after MASCp36 infection. In addition, the correlation between the decreased NMN levels in patients’ sera and the severity of COVID-19 was reported during the revision of this manuscript50. These results suggest that when the demand for NAD+ increases, NAM and NA were the first two NAD+ precursors to be consumed, followed by NMN and NR. Our absolute quantification results confirmed that the levels of NAD+, NMN, NR, and NAM were increased by NMN supplementation. Moreover, NMN supplementation significantly rescued many metabolites that were significantly reduced after SARS-CoV-2 infection, and helped to maintain the metabolic balance of SARS-CoV-2-infected lungs and protect the mice from death.
In this study, two mouse infection models were adopted. MASCp6-infected mice did not show very significant clinical symptoms that are present in COVID-19 patients, although pathological damages were detected in the lungs13. However, MASCp36 is the lethal mouse-adapted virus, and infected mice exhibit age- and gender-related skewed distribution of mortality akin to severe COVID-1912. Therefore, we used it to validate the therapeutic effects of NAD+ and NMN. Although our preliminary results were in support of clinical trials at the moment, there are fewer data about the dosage and long-term efficacy of NAD+ or NMN as a therapeutic drug in human studies at present. In a patent application, NMN was proposed as an anti-aging drug or healthcare supplement at doses of 1–500 mg/kg/day41. Further trials with different dosages and other clinical indicators remain to be evaluated for optimal therapeutic effect.
Taken together, based on our finding that NAD+ and NMN can alleviate the pathological damage of lungs and even death of SARS-CoV-2-infected mice and other groups’ studies of NAD+-boosting therapy for different diseases in humans2,4, trials for treating COVID-19 patients with NAD+, its intermediates or precursors should be considered.
Materials and methods
Ethics statement
All procedures involving animals were conducted in temperature- and humidity-controlled Biosafety Level 3 laboratory (BSL-3) and approved by the Animal Experiment Committee of Laboratory Animal Center, Beijing Institute of Microbiology and Epidemiology (approval number: IACUC-DWZX-2020-002). All animal experiments were handled in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals.
Mice
All BALB/c mice were purchased from Bejing Vital River Laboratory Animal Technology Co., Ltd. Six- to seven-week-old and 8–9-month-old female mice were used for MASCp6 infection experiments and 8–9-month-old male mice were used for MASCp36 infection experiments.
Virus strains
SARS-CoV-2 strains MASCp6 and MASCp36 were prepared as previously described12,13. All experiments involving infectious SARS-CoV-2 were performed in BSL-3 containment laboratory in Beijing Institute of Microbiology and Epidemiology.
Mouse procedures
For MASCp6-infected mouse model, anesthetized 6–7-week- and 8–9-month-old female BALB/c mice were i.n. treated with a dose of 6 × 103 plaque-forming unit (PFU) of MASCp6 in a total volume of 30 μL in each group. For MASCp36-infected mouse model, anesthetized 8–9-month-old male BALB/c mice were i.n. treated with a dose of 1200 PFU of MASCp36 in a total volume of 30 μL in each group.
The infected mice were i.p. administered with either NAD+ (or NMN) dissolved in 0.9% saline or the equivalent volume of 0.9% saline once a day for 3 consecutive days starting at half a day after inoculation of the virus. The dosages of NAD+ (Sigma, Cat# N7004) and NMN (EffePharm) are 1 mg/g/day and 0.5 mg/g/day, respectively. The mice were euthanized on 3 dpi by isoflurane overdose, and tissue samples were collected for subsequent experiments.
For survival experiment, 8–9-month-old male BALB/c mice were i.n. treated with a dose of 300 PFU for survival experiment of MASCp36 in a total volume of 30 μL in each group. The infected mice were i.p. administered with NMN dissolved in 0.9% saline or the equivalent volume of 0.9% saline once a day until the end of the experiment starting at half a day after inoculation of the virus. The dosage of NMN is 0.5 mg/g/day.
Sample preparation
For histopathological and immunofluorescent staining, the tissues of each mouse were removed and fixed in 4% paraformaldehyde at 4 °C for 2 days. For RNA extraction, the tissues were homogenized and lysed with TRIzol reagent (Life Technologies, 15596018), and RNA was extracted according to the manufacturer’s instructions. For metabolomics, the tissues were weighed and homogenized with 75% ethanol (1:50, tissue weight (mg):volume (μL) of 75% ethanol). The same lobe of the lung from each mouse was collected for the same analysis.
Measurement of viral RNA
Viral RNA (vRNA) was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Cat# 52906) according to the manufacturer’s protocol. vRNA quantification in each sample was performed by qPCR targeting the S gene of SARS-CoV-2 as described previously13.
Transcriptomics
Total RNAs of lungs from MASCp6-infected 8–9-month-old BALB/c mice and control mice were sent to LC-Bio Technology (Hangzhou) for sequencing and analysis. Purified RNA was subjected to the 2× 150 bp paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 (LC-Bio Technology Co., Ltd., Hangzhou, China) following the vendor’s recommended protocol. After preprocessing, the high-quality reads were compared with the mouse reference genome (ftp://ftp.ensembl.org/pub/release-99/fasta/mus_musculus/dna/) by HISAT2 software (https://daehwankimlab.github.io/hisat2/).
The significantly differentially expressed mRNAs were selected with |Log2FC| > 1 and P value < 0.05 by R package DESeq266. GO enrichment analysis and KEGG enrichment analysis of significantly differently expressed genes were performed using the OmicStudio tools at https://www.omicstudio.cn/tool. Gene number and P value of GO and KEGG terms were visualized using the R package ggplot2. The network of the genes among different GO and KEGG terms were visualized using Cytoscape (version 3.7.2). The heatmaps and bar plots of significantly differently expressed genes were drawn with the R package ggplot2 and GraphPad (Prism 7), respectively.
RT-qPCR
1.5 μg of total RNA was used for RT-qPCR using Reverse Transcription Kit (Promega, A3500) according to the manufacturer’s protocol. Then, cDNA was diluted by 1:4 and 1 μL diluted cDNA was used for qPCR with qPCR mix (Bio-Rad, Cat# 1725201). qPCR was performed on Bio-Rad CRX96 Real-Time PCR system.
Histopathological staining and analysis
Four percent paraformaldehyde-fixed lung tissues for histological examination were subjected to alcohol gradient dehydration and embedded in paraffin. Each embedded tissue was sectioned at 5 μm thickness sections. At least three sections of each tissue were used for staining with H&E (Solarbio, Cat# G1120) and examined under a light microscopy. The tissue handling and H&E staining followed standard protocol.
For quantification of inflammatory cell aggregation number and area and alveolar septum relative area, inflammatory cell aggregation was defined by dense cell cluster and dark blue staining with hematoxylin, and the relative area of alveolar septum was calculated as the total area of the substances divided by the area of whole lung section.
Immunofluorescent staining and analysis
Four percent paraformaldehyde-fixed lung tissues were dehydrated in 30% sucrose for 2 days and embedded in optimal cutting temperature compound (O.C.T. compound, SAKURA, 4583, USA). 30 μm sections were sliced with freezing microtome (CM 1950, Leica, Germany). Sections were blocked with blocking buffer (PBS + 10% FBS + 3% BSA + 0.2% Triton X-100) at room temperature for 1 h and incubated with the primary antibodies at 37 °C for 2 h. After washing with PBST (PBS + 0.2% Triton X-100) for 3 times (10 min for each time), the lung sections were subsequently incubated with fluorescent secondary antibodies at room temperature for 1 h, followed by washing with PBST for 3 times (10 min for each time). Primary antibodies include: rabbit anti-cleaved caspase 3 (1:500, Cell Signaling Technology, Cat# 9664), rabbit anti-CD68 (1:1000, Abcam, Cat# ab125212), rabbit anti-SARS-CoV-19 nucleocapsid (1:100, Sino Biological, Cat# 40588-T62), human/mouse anti-SARS-CoV-19 spike (1:100, Sino Biological, Cat# 40150-D001). Secondary antibodies were Alexa Fluor 488-, Alexa Fluor 568-, and Alexa Fluor 647-conjugated goat secondary antibodies against rabbit and human IgG (1:2000; Invitrogen). Nuclei were counterstained with DAPI (Cell Signaling Technology, Cat# 4083s). Fluorescent images were captured using a confocal laser-scanning microscope (Carl Zeiss, LSM700 and LSM980) and analyzed with ZEN3.1 and ImageJ.
For quantification of blocked airway ratio and relative area, blocked airway ratio was calculated as the number of airways having blockage divided by the number of total airways in each section, and the relative area of blockage was calculated as the area of each embolism divided by the area of the corresponding airway. For quantification of the ratio of caspase 3+ cells, the ratio of caspase 3+ cells was calculated as the number of caspase 3+ cells divided by the number of infected cells in the same location of the adjacent section. For quantification of SARS-CoV-2 relative fluorescence intensity and infected area, relative fluorescence intensity was calculated as the total fluorescence intensity of SARS-CoV-2 signal divided by the area of whole lung section. The relative infected area was calculated as the area of minimal convex polygon which can cover all of virus signal divided as the area of whole lung section.
Metabolite extraction and metabolomics
Homogenate of lung tissues was vortexed for 1 min. Then, 100 μL of homogenate was transferred into a tube pre-added with 900 μL of ice-cold 80% methanol, vortexed for 1 min and centrifuged at 14,000 rpm, 4 °C for 15 min. 900 μL of supernatant was collected into a new tube. 500 μL of 80% methanol was added to the pellet, vortexed for 1 min, and centrifuged at 14,000 rpm, 4 °C for 15 min. 500 μL of supernatant was collected and mixed with 900 μL of supernatant collected previously. Supernatant was evaporated to dryness under a speed vacuum concentrator. Dried metabolite pellets were kept at −80 °C until analysis.
Dried metabolite pellets were dissolved in 0.3% formic acid in water. Chromatographic separation was performed on a Nexera UHPLC system (Shimadzu), with a RP-UPLC column (HSS T3, 2.1 mm × 150 mm, 1.8 μm, Waters) and the following gradient: 0–3 min 99% A; 3–15 min 99%–1% A; 15–17 min 1% A; 17–17.1 min 1%–99% A; 17.1–20 min 99% A. Mobile phase A was 0.03% formic acid in water. Mobile phase B was 0.03% formic acid in acetonitrile. The flow rate was 0.25 mL/min, the column was at 35 °C and the autosampler was at 4 °C. Mass data acquisition was performed using an AB QTRAP 6500+ triple quadrupole mass spectrometer (SCIEX, Framingham, MA) in multiple reaction monitoring (MRM) mode for the detection of 260 unique endogenous metabolites as previously described11,50,67,68. Chromatogram review and peak area integration were performed using MultiQuant software v.3.0.2 (SCIEX). Raw peak areas were used as variables for multivariate and univariate statistical data analysis.
Metabolites with significant differences (P < 0.05 and fold change < 0.8 or > 1.25) were used for KEGG enrichment analysis using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). Metabolic pathways with FDR < 0.05 were considered significant.
Supplementary information
Acknowledgements
We thank Dr. John Speakman for his kind comments and editing of the manuscript. This study was supported by grants from the National Natural Science Foundation of China (31921002, 31730108, 32061143026 to Z.X.; 81925025, 81621005 to C.-F.Q.; 82174055 to Y.D.), the Strategic Priority Research Program and Innovation Program of the Chinese Academy of Sciences (XDB32020100, YJKYYQ20200052) to Z.X., the Major Projects of the Ministry of Science and Technology (2021ZD0202300) to Z.X., the Innovation Fund for Medical Sciences (2019-I2M-5-049) from the Chinese Academy of Medical Sciences to C.-F.Q., and Tsinghua University Spring Breeze Fund (2021Z99CFY031) to Z.H. We thank EffePharm (Shanghai) for providing us with NMN and NR gratis.
Author contributions
Z.X., Y.J., and C.-F.Q. conceived the research. Y.J. and Z.X. designed the study and wrote the manuscript. Y.J. and Y.D. designed and performed most of the experiments. Y.J. performed RNA-seq analysis and immunostaining. Y.D. performed live virus and animal assays. H.P. did metabolomics study and analyzed the data. T.M. did H&E staining. H.P. and Z.H. contributed to the ideas. All authors contributed to the writing.
Data availability
The RNA-seq data generated in this study have been deposited in Genome Sequence Archive (BSA) database (https://ngdc.cncb.ac.cn/gsa/) under accession code CRA003464. Metabolomics data are included in Supplementary Tables S1 and S2.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yisheng Jiang, Yongqiang Deng, Huanhuan Pang, Tiantian Ma.
Contributor Information
Yisheng Jiang, Email: ysjiang@genetics.ac.cn.
Zeping Hu, Email: zeping_hu@tsinghua.edu.cn.
Cheng-Feng Qin, Email: qincf@bmi.ac.cn.
Zhiheng Xu, Email: zhxu@genetics.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-022-00409-y.
References
- 1.Pan F, et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katsyuba E, Romani M, Hofer D, Auwerx J. NAD(+) homeostasis in health and disease. Nat. Metab. 2020;2:9–31. doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- 3.Rajman L, Chwalek K, Sinclair DA. Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metab. 2018;27:529–547. doi: 10.1016/j.cmet.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chini CCS, Zeidler JD, Kashyap S, Warner G, Chini EN. Evolving concepts in NAD(+) metabolism. Cell Metab. 2021;33:1076–1087. doi: 10.1016/j.cmet.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGuinness, E. T. & Butler, J. R. NAD+ kinase — a review. Int. J. Biochem.17, 1–11 (1985). [DOI] [PubMed]
- 6.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 7.Xiao W, Wang RS, Handy DE, Loscalzo J. NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxid. Redox Signal. 2018;28:251–272. doi: 10.1089/ars.2017.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migaud M, et al. Metabolomics to predict antiviral drug efficacy in COVID-19. Am. J. Respir. Cell Mol. Biol. 2020;63:396–398. doi: 10.1165/rcmb.2020-0206LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandeel M, et al. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J. Biomol. Struct. Dyn. 2021;39:5129–5136. doi: 10.1080/07391102.2020.1784291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martorana A, Gentile C, Lauria A. In silico insights into the SARS CoV-2 main protease suggest NADH endogenous defences in the control of the pandemic coronavirus Infection. Viruses. 2020;12:805. doi: 10.3390/v12080805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang H, et al. Aberrant NAD(+) metabolism underlies Zika virus-induced microcephaly. Nat. Metab. 2021;3:1109–1124. doi: 10.1038/s42255-021-00437-0. [DOI] [PubMed] [Google Scholar]
- 12.Sun S, et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun. 2021;12:5654. doi: 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu H, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinnon KH, 3rd, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 16.Thompson EA, et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34:108863. doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, et al. S100A8/A9 in inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bost P, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright GJ, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J. Immunol. 2003;171:3034–CD3046. doi: 10.4049/jimmunol.171.6.3034. [DOI] [PubMed] [Google Scholar]
- 20.Voehringer D, Rosen DB, Lanier LL, Locksley RM. CD200 receptor family members represent novel DAP12-associated activating receptors on basophils and mast cells. J. Biol. Chem. 2004;279:54117–54123. doi: 10.1074/jbc.M406997200. [DOI] [PubMed] [Google Scholar]
- 21.Robinson EK, Covarrubias S, Carpenter S. The how and why of lncRNA function: an innate immune perspective. Biochim. Biophys. Acta Gene Regul. Mech. 2020;1863:194419. doi: 10.1016/j.bbagrm.2019.194419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chua RL, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L, et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53:685–696. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben Safta T, et al. Granzyme B-activated p53 interacts with Bcl-2 to promote cytotoxic lymphocyte-mediated apoptosis. J. Immunol. 2015;194:418–428. doi: 10.4049/jimmunol.1401978. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol. 2014;15:1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- 27.da Costa LS, Outlioua A, Anginot A, Akarid K, Arnoult D. RNA viruses promote activation of the NLRP3 inflammasome through cytopathogenic effect-induced potassium efflux. Cell Death Dis. 2019;10:346. doi: 10.1038/s41419-019-1579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji N, et al. Pyroptosis: a new regulating mechanism in cardiovascular disease. J. Inflamm. Res. 2021;14:2647–2666. doi: 10.2147/JIR.S308177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng MY, Miralda I, Armstrong CL, Uriarte SM, Bagaitkar J. The roles of NADPH oxidase in modulating neutrophil effector responses. Mol. Oral. Microbiol. 2019;34:27–38. doi: 10.1111/omi.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, A. M. et al. NAD(+) depletion by type I interferon signaling sensitizes pancreatic cancer cells to NAMPT inhibition. Proc. Natl. Acad. Sci. USA118, e2012469118 (2021). [DOI] [PMC free article] [PubMed]
- 32.Girardi E, et al. Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import. Nat. Commun. 2020;11:6145. doi: 10.1038/s41467-020-19871-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kory N, et al. MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci. Adv. 2020;6:eabe5310. doi: 10.1126/sciadv.abe5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luongo TS, et al. SLC25A51 is a mammalian mitochondrial NAD(+) transporter. Nature. 2020;588:174–179. doi: 10.1038/s41586-020-2741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kropotov A, et al. Equilibrative nucleoside transporters mediate the import of nicotinamide riboside and nicotinic acid riboside into human cells. Int. J. Mol. Sci. 2021;22:1391. doi: 10.3390/ijms22031391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2-induced systemic toxicity. JCI Insight. 2021;6:e145027. doi: 10.1172/jci.insight.145027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heer CD, et al. Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity. J. Biol. Chem. 2020;295:17986–17996. doi: 10.1074/jbc.RA120.015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun SH, et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang RD, et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 41.Nadeeshani H, Li J, Ying T, Zhang B, Lu J. Nicotinamide mononucleotide (NMN) as an anti-aging health product–promises and safety concerns. J. Adv. Res. 2021;37:267–278. doi: 10.1016/j.jare.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei, W., Wu, X. M. & Li, Y. J. Methodology of Pharmacological Experiment. (People’s Medical Publishing House, Beijing, 2010).
- 43.Nikbakht F, Mohammadkhanizadeh A, Mohammadi E. How does the COVID-19 cause seizure and epilepsy in patients? The potential mechanisms. Mult. Scler. Relat. Disord. 2020;46:102535. doi: 10.1016/j.msard.2020.102535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Versace V, et al. Intracortical GABAergic dysfunction in patients with fatigue and dysexecutive syndrome after COVID-19. Clin. Neurophysiol. 2021;132:1138–1143. doi: 10.1016/j.clinph.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian J, Middleton B, Kaufman DL. GABAA-receptor agonists limit pneumonitis and death in murine coronavirus-infected mice. Viruses. 2021;13:966. doi: 10.3390/v13060966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alhammad YMO, et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J. Virol. 2021;95:e01969–20. doi: 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen B, et al. Proteomic and mtabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan S, et al. Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication. Mol. Cell Proteom. 2021;20:100159. doi: 10.1016/j.mcpro.2021.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caterino M, et al. The serum metabolome of moderate and severe COVID-19 patients reflects possible liver alterations involving carbon and nitrogen metabolism. Int. J. Mol. Sci. 2021;22:9548. doi: 10.3390/ijms22179548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao N, et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021;12:1618. doi: 10.1038/s41467-021-21907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giron LB, et al. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front. Immunol. 2021;12:686240. doi: 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamura T, et al. Selective depletion of basophils ameliorates immunoglobulin E-mediated anaphylaxis. Biochem. Biophys. Rep. 2017;9:29–35. doi: 10.1016/j.bbrep.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kojima T, et al. Mast cells and basophils are selectively activated in vitro and in vivo through CD200R3 in an IgE-independent manner. J. Immunol. 2007;179:7093–7100. doi: 10.4049/jimmunol.179.10.7093. [DOI] [PubMed] [Google Scholar]
- 55.Gomes AP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang EF, et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24:566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaneko S, et al. Protecting axonal degeneration by increasing nicotinamide adenine dinucleotide levels in experimental autoimmune encephalomyelitis models. J. Neurosci. 2006;26:9794–9804. doi: 10.1523/JNEUROSCI.2116-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sims CA, et al. Nicotinamide mononucleotide preserves mitochondrial function and increases survival in hemorrhagic shock. JCI Insight. 2018;3:e120182. doi: 10.1172/jci.insight.120182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 60.Buettner GR. Superoxide dismutase in redox biology: the roles of superoxide and hydrogen peroxide. Anticancer Agents Med. Chem. 2011;11:341–346. doi: 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li R, Wang W, Yang Y, Gu C. Exploring the role of glucose6phosphate dehydrogenase in cancer (Review) Oncol. Rep. 2020;44:2325–2336. doi: 10.3892/or.2020.7803. [DOI] [PubMed] [Google Scholar]
- 62.Suhail S, et al. Role of oxidative stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) infection: a review. Protein J. 2020;39:644–656. doi: 10.1007/s10930-020-09935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas T, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5:e140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee, S. M. et al. Inhibition of acute lethal pulmonary inflammation by the IDO-AhR pathway. Proc. Natl. Acad. Sci. USA114, E5881–E5890 (2017). [DOI] [PMC free article] [PubMed]
- 65.Russo LC, et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 2021;297:101041. doi: 10.1016/j.jbc.2021.101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang F, et al. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab. 2018;28:369–382. doi: 10.1016/j.cmet.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li XK, et al. Arginine deficiency is involved in thrombocytopenia and immunosuppression in severe fever with thrombocytopenia syndrome. Sci. Transl. Med. 2018;10:eaat4162. doi: 10.1126/scitranslmed.aat4162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study have been deposited in Genome Sequence Archive (BSA) database (https://ngdc.cncb.ac.cn/gsa/) under accession code CRA003464. Metabolomics data are included in Supplementary Tables S1 and S2.