Abstract
A natural rsbU mutant of Staphylococcus aureus, unable to activate the alternative transcription factor ςB via the RsbU pathway and therefore forming unpigmented colonies, produced first-step teicoplanin-resistant mutants upon selection for growth in the presence of teicoplanin, of which the majority were of an intense orange color. By using an asp23 promoter-luciferase fusion as an indicator, the pigmented mutants were shown to express increased ςB activity. Increased ςB activity was associated with point mutations in rsbW, releasing ςB from sequestration by the anti-sigma factor RsbW, or to promoter mutations increasing the ςB/RsbW ratio. Genetic manipulations involving the sigB operon suggested that the mutations within the operon were associated with the increase in teicoplanin resistance. The upregulation of ςB suggests that a ςB-controlled gene(s) is directly or indirectly involved in the development of teicoplanin resistance in S. aureus. Carotenoids do not contribute to teicoplanin resistance, since inactivation of the dehydrosqualene synthase gene crtM abolished pigment formation without affecting teicoplanin resistance. The relevant ςB-controlled target genes involved in teicoplanin resistance remain to be identified.
Teicoplanin and vancomycin are the drugs of choice against multidrug-resistant methicillin-resistant Staphylococcus aureus. Their antibacterial activity is based on the ability to bind the terminal d-alanyl-d-alanine present in the lipid-II-linked peptidoglycan precursor and in peptidoglycan intermediates, thereby inhibiting transglycosylation and transpeptidation of the cell wall. Though both drugs interact with the same target, teicoplanin anchors to the membrane while vancomycin forms dimers to increase its activity (1). Even though teicoplanin is more active than vancomycin against staphylococci (8), resistance to teicoplanin is more easily acquired than resistance to vancomycin. Teicoplanin resistance is believed to precede vancomycin resistance (reviewed in reference 13). In clinical isolates of S. aureus, teicoplanin resistance was found to emerge during extended teicoplanin treatment (15), suggesting an in vivo selection for resistant mutants. In contrast to the van gene-mediated glycopeptide resistance in enterococci, resistance in S. aureus is not due to acquisition of foreign elements but formed endogenously. Analogously, teicoplanin-resistant mutants can be obtained in vitro by step selection for growth on increasing concentrations of the drug. Such in vitro-selected teicoplanin-resistant mutants may have characteristics similar to those of clinical teicoplanin-resistant isolates, allowing their use to study the genes involved in the resistance mechanism. Except for the work of Shlaes et al. (23), who identified a site in the SmaI-I fragment of the S. aureus chromosome responsible for increase in a 35-kDa protein and PBP 2 production in teicoplanin-resistant S. aureus, few genetic studies of teicoplanin resistance have been done.
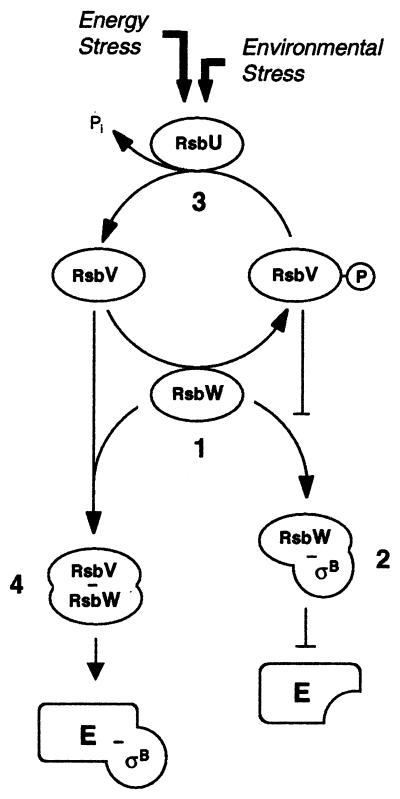
In the process of infection and disease, S. aureus has to adapt to variable external surroundings. One of the triggers responding to environmental stimuli is alternate transcription factors, such as ςB. The S. aureus sigB operon comprises the genes rsbU, rsbV, rsbW, and sigB (17, 26), which modulate ςB activity in a sequential fashion (Fig. 1). RsbW acts as an anti-sigma factor by sequestering ςB through protein-protein interactions, and RsbU controls, via RsbV phosphorylation, the availability of free RsbW to interact with ςB. The widely used pigmentless laboratory strain NCTC8325 and its descendants are natural rsbU mutants (17). They are unable to activate the RsbU-initiated cascade leading to ςB activity, resulting in low ςB activity (10). This may have consequences for the mode of stress response. One of the properties of S. aureus influenced by the sigB operon is pigment formation. The yellow-to-orange color of S. aureus colonies stems from triterpennoid carotenoids. Pigment production, although chromosomally encoded, is an unstable characteristic. It is usually found in strains freshly isolated from natural sources or those which are multiply resistant and tends to be lost in stored organisms. Pigmented variants are more resistant to desiccation than nonpigmented ones (11). Pigment formation in S. aureus is a multistep procedure, involving regulatory genes and several biosynthetic genes (20), of which crtMN catalyze early steps in carotenoid biosynthesis (25). Both the sigB operon and the uncharacterized pig mutation in NCTC8325 derivatives map in the chromosomal SmaI-I fragment (14).
FIG. 1.
Proposed model for the regulation of ςB in S. aureus (adapted from references 22 and 24). Based on the known functions of the RsbUVW homologues from B. subtilis (reviewed in reference 12), it is assumed that the anti-ςB protein RsbW from S. aureus can form mutually exclusive complexes with either ςB or its antagonist, RsbV (step 1). RsbV is normally inactive (RsbV-P) due to phosphorylation by RsbW and is thus unable to complex with RsbW, leaving the latter free to interact with ςB (step 2). When bound to RsbW, ςB is unable to aggregate with the RNA polymerase core enzyme (E) to form an active holoenzyme (E-ςB). Upon stress, the RsbV-P-specific phosphatase activity of RsbU, a positive activator of ςB, becomes activated and thus reactivates RsbV (step 3). Unphosphorylated RsbV interacts and complexes highly specifically with RsbW (step 4), thereby releasing ςB. RsbW, if complexed with RsbV, is unable to bind to ςB, leaving the latter free to form an active ςB-holoenzyme (E-ςB). Even though the exact mode of activation of RsbU in S. aureus remains unclear, there is evidence that its activation differs substantially from that of the RsbU homologue in B. subtilis.
In this study, we identified the sigB operon as one of the preferred mutation sites associated with first-step teicoplanin resistance in a pigmentless S. aureus strain.
MATERIALS AND METHODS
General methods.
All DNA manipulations, basic molecular methods, and handling of Escherichia coli were performed in accordance with standard protocols (21). Genetic manipulation of S. aureus was done as described earlier (17). The general transducing phage 80α was used for transductions. Sequence data were obtained from the website of The Institute for Genomic Research (http://www.tigr.org).
Strains and growth conditions.
The strains used in this study are listed in Table 1. Growth was on Luria-Bertani (LB) agar (Difco) plates at 35°C unless otherwise specified. MIC determinations for antibiotics were performed with the Etest from AB-Biodisk (Solna, Sweden) in accordance with the NCCLS guidelines on Mueller-Hinton agar (Difco) plates with an inoculum of 0.5 McFarland standard. Additionally, MICs of teicoplanin were determined on brain heart infusion (BHI) (Difco) plates as recommended by AB-Biodisk (Etest Technical Guide 11: Etest Application Sheets) with an inoculum of 2 McFarland standard and incubation at 35°C for 48 h. Resistance levels were compared on rectangular plates containing an antibiotic gradient by swabbing a 0.5-McFarland-standard suspension of an overnight culture along the gradient. The antibiotic used for selection of transductants was either erythromycin (20 μg ml−1) or tetracycline (5 μg ml−1). Population analysis profiles were done by plating 0.1 ml of appropriate dilutions of overnight cultures on LB agar plates containing increasing concentrations of teicoplanin and incubating them for 48 h.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH10B | F− φ80dlacZΔM15 recA1 | Gibco |
| BL21 (DE3) | F−ompT gal [dcm] [lon] hsdSB (rB− mB−); with DE3 | Novagen |
| S. aureus | ||
| RN4220 | NCTC8325-4 r m+ (restriction minus; modification plus); rsbU | 16 |
| MSSA1112 | Clinical isolate; bla rsbU+ | 7 |
| Newman | ATCC25904; clinical isolate; high level of clumping factor; rsbU+ | 6 |
| GP266 | RN4220; rsbU+sigB1(Am) Tcr | This study |
| GP268 | BB255; rsbU+V+W+sigB+ Tcr | 10 |
| MB33 | BB255; rsbU asp23+asp23p::pECasp23p-luc+ Emr | 10 |
| MB49 | GP268; asp23+asp23p::pECasp23p-luc+ Emr Tcr | 10 |
| MB51 | RN4220; crtM::pBTcrtM Tcr | This study |
| MB118 | MB33; rsbU rsbW1 Emr | This study |
| MB119 | MB33; rsbU rsbW2(Am) Emr | This study |
| MB127 | MB33; rsbU rsbW3 Emr | This study |
| MB128 | MB33; rsbU rsbW4 Emr | This study |
| MB130 | MB33; rsbU rsbW5(Am) Emr | This study |
| MB132 | MB33; rsbU rsbW6 Emr | This study |
| MB137 | MB33; rsbU sigB2 Emr | This study |
| MB138 | MB33; rsbU rsbW7 Emr | This study |
| MB140 | MB33; rsbU chrX1 Emr | This study |
| MB148 | MB33; rsbU rsbW8 Emr | This study |
| MB158 | MB137; rsbU+sigB1(Am) Emr Tcr | This study |
| MB159 | MB137; rsbU crtM::pBTcrtM Emr Tcr | This study |
| MB161 | MB140; rsbU+V+W+sigB+ Tcr Emr | This study |
| MB162 | MB140; rsbU+sigB1(Am) Emr Tcr | This study |
| MB163 | MB140; rsbU crtM::pBTcrtM Emr Tcr | This study |
| MB213 | MB137; rsbU+V+W+sigB+ Emr Tcr | This study |
| MB215 | MB119; rsbU+sigB1(Am) Emr Tcr | This study |
| Plasmids | ||
| pBC SK(+) | Cmr; E. coli cloning vector | Stratagene |
| pBT | Tcr; 1.6-kb PCR fragment of tet(L) from pHY300PLK into Alw261-digested pBC SK(+) | 10 |
| pEC1 | Apr Emr; 1.45-kb Clal erm(B) fragment of Tn551 in pUC18 | 4 |
| pET-24b(+) | Kmr; E. coli expression vector | Novagen |
| pSP-luc+ | Apr; firefly luciferase casette vector | Promega |
| pPG11 | Apr Tcr; 6.6-kb PstI-EcoRI sigB fragment from strain BB255, including a 252-bp MluI-BstXI fragment of rsbU from strain COL replacing the corresponding region of the sigB operon from strain BB255 and carrying a 1.6-kb PCR fragment of tet(L) from pHY300PLK integrated into a blunted Bsp1191 site downstream of sigB in pUC19 | 10 |
| pETrsbW | Apr; 486-bp PCR fragment of rsbW from strain BB255 in pET24b(+) | This study |
| pSPasp23p | Apr; 1.1-kb fragment of the asp23 promoter from strain COL into pSP-luc+ | 10 |
| pECasp23p-luo+ | Emr; 2.7-kb KpnI-EcoRI asp23p-luc+ fragment of pSP asp23p in pEC1, S. aureus integration vector that inserts into the asp23 promoter (asp23p) | 10 |
| pBTasp23p-luc+ | Tcr; 2.7-kb KpnI-EcoRI asp23p-luc+ fragment of pSPasp23p in pBT, S. aureus integration vector that inserts into the asp23 promoter (asp23p) | 10 |
| pBTcrtM | Tcr; 510-bp PCR fragment of crtM in pBT, S. aureus integration vector that inserts into crtM | This study |
Abbreviations are as follows: Am, amber mutation; Apr, ampicillin resistant; chrX1, unmapped mutation; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant.
Selection of first-step teicoplanin-resistant mutants.
First-step teicoplanin-resistant mutants were selected by plating 0.1 ml of serial 10-fold dilutions of an overnight culture in Luria broth on LB agar plates containing increasing concentrations of the antibiotic. After a 48-h incubation, single colonies appearing at the highest concentration were purified on sheep blood agar without a selective agent and kept for further use.
Generation of GP266.
Electroporation of plasmid pPG11 (Table 1), carrying a functional sigB operon and a tet(L) gene cassette downstream of the sigB operon (10), into RN4220 ΔrsbUVWsigB and screening for double-crossover transformants sensitive to erythromycin and resistant to tetracycline yielded, among others, the unpigmented strain GP266. Strain GP266 carries an amber mutation due to a 2-bp deletion in the 5′ part of sigB (corresponding to positions 2932 to 2933 [accession number Y07645]), which must have occurred accidentially upon selection. Double-crossover integration of the sigB operon, including the tet(L) cassette, was confirmed by Southern blot analysis. The inability of GP266 to produce ςB was confirmed by Western blot analysis using anti-ςB antibodies as probes (data not shown). The tet(L)-tagged sigB1(Am) mutation of GP266 was used to transduce sigB1(Am) into different genetic backgrounds.
Construction of plasmid pBTcrtM used for insertional inactivation.
An internal 510-bp crtM fragment containing a HindIII restriction site was generated by PCR using the upstream primer 5′-CAATATAGGAGGACTAGTATGAC-3′ and the downstream primer (5′-GGAATTCCAACGATTCACCAAGTCTTCTTGCG-3′), including an EcoRI linker, with the italic nucleotides corresponding to positions 211 to 233 and 696 to 720 of the sequence accession no. X73889, respectively. The PCR product was digested with HindIII and EcoRI and cloned into the suicide plasmid pBT (13). The plasmid obtained was transformed by electroporation into RN4220 and subsequently transduced into the strains MB137 and MB140.
Construction of an E. coli vector for overexpression of His-tagged RsbW and generation of anti-RsbW antibodies.
A DNA fragment encoding 486 bp of the rsbW gene was amplified by PCR using the NdeI linker containing the primer 5′-GGAGATATACATATGCAATCTAAAGAAGATTTTATCG-3′ and the XhoI linker containing the primer 5′-GGTGGTGCTCGCTGATTTCGACTCTTTCGC-3′, with the italic nucleotides corresponding to positions 2210 to 2234 and 2677 to 2696 of the sequence accession no. Y07645, respectively. The PCR product was cloned into pET24b to obtain pETrsbW. The junction regions including the PCR product were sequenced to ensure proper ligation and fidelity of the PCR. E. coli strain BL21 (DE3) was transformed with the plasmid obtained. Overexpression and purification of the His-tagged protein was performed using Ni-nitrilotriacetic acid columns (Qiagen, Basel, Switzerland) according to the recommendations of the manufacturer. The purified protein was separated using sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis, and bands containing the protein were cut out of the gels. N-terminal sequencing confirmed the identity of the desired protein. The gel slices containing the protein were injected into rabbits to raise anti-RsbW polyclonal antibodies (BioScience, Göttingen, Germany). The resulting antisera were purified against the immobilized antigen.
Luciferase assay.
Bacterial cells from overnight cultures containing the appropriate antibiotic were diluted in fresh drug-free Luria broth to an an optical density at 600 nm (OD600) of 0.01 and grown at 37°C and 200 rpm. The cells were harvested at different time points by centrifugation at 11,000 × g for 1 min at room temperature, and the cell pellets were resuspended in 0.1 M sodium phosphate buffer (pH 7.0) to an OD600 of 10. Luciferase activity was then determined by rapidly mixing the resuspended cells (10 μl) with an equal volume of luciferase assay reagent (Promega, Madison, Wis.). Luminescence was measured on a Tumer Designs TD-20/20 luminometer (Promega) for 10 s with a delay of 2 s.
RESULTS
Selection of spontaneous first-step mutants with decreased teicoplanin susceptibilities.
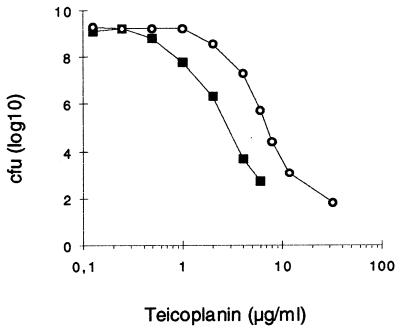
The susceptible strain MB33 (10), derived from NCTC8325, showed heterogeneous susceptibility on plates containing increasing concentrations of teicoplanin and was able to form a few colonies in the presence of up to 8 μg ml−1 of teicoplanin (Fig. 2). While the parental strain was white, more than half of the colonies growing at concentrations over 4 μg ml−1 of teicoplanin produced an intense orange pigmentation. Propagation of those colonies on nonselective plates showed that they retained pigment formation and increased teicoplanin resistance (Fig. 2, strain MB128). Pigment formation in NCTC8325 derivatives, which are natural rsbU mutants, is known to be restored by overexpression of ςB (18). Strain MB33, which harbors a chromosomally integrated reporter system that allows monitoring of the activity of ςB (10), was therefore used in these experiments. First-step teicoplanin-resistant mutants arose with a frequency of 1.3 × 10−7 when MB33 was plated on 7 μg of teicoplanin ml−1. White mutants appeared after 24 h; orange mutants appeared only after 48 h. The ratio of white to orange mutants was approximately 1:3. The teicoplanin MICs for those mutants ranged between 6 and 12 μg ml−1 on Mueller-Hinton agar and between 24 and 32 μg ml−1 on BHI agar (Table 2). The pigmented mutants were generally slightly more resistant than the white ones, as shown on the teicoplanin gradient plate in Fig. 3. Increase in resistance was specific for glycopeptides and more pronounced for teicoplanin than for vancomycin (Table 2). The MICs of other, unrelated antibiotics, such as methicillin, oxacillin, cefoxitin, imipenem, ciprofloxacin, kanamycin, erythromycin, rifampin, and tetracycline, seemed not to be affected (data not shown).
FIG. 2.
Population analysis profiles. Colonies formed from overnight cultures of the parent, MB33 (squares), and its teicoplanin first-step mutant MB128 (circles) were plated on LB agar plates containing increased concentrations of teicoplanin.
TABLE 2.
Phenotypic properties of different S. aureus strains
| Strain | Relevant genotypea | MIC
(μg/ml)b
|
ςB activityc | Color | |||
|---|---|---|---|---|---|---|---|
| Teicoplanin
|
Vancomycin
|
||||||
| MH | BHI | MH | BHI | ||||
| MB33 | BB255; asp23p::pECasp23p-luc+ | 2 | 6 | 2 | 4 | 34.0 ± 3.00 | White |
| MB49 | MB33; rsbU+V+W+sigB+ | 4 | 8–12 | 3 | 4–6 | 301.3 ± 23.8 | Orange |
| MB118 | MB33; rsbW1 | 8 | 24 | 3 | 6 | 632.5 ± 13.8 | Orange |
| MB119 | MB33; rsbW2(Am) | 12 | 38 | 4–6 | 8 | 609.0 ± 43.5 | Orange |
| MB127 | MB33; rsbW3 | 12 | 32 | 4–6 | 8 | 760.3 ± 52.8 | Orange |
| MB128 | MB33; rsbW4 | 6 | 24 | 3 | 6 | 933.8 ± 99.5 | Orange |
| MB130 | MB33; rsbW5(Am) | 8 | 24 | 4 | 8 | 869.5 ± 74.5 | Orange |
| MB132 | MB33; rsbW6 | 8 | 32 | 4 | 8 | 804.0 ± 70.5 | Orange |
| MB137 | MB33; sigB2 | 12 | 32 | 4 | 8 | 671.0 ± 35.5 | Orange |
| MB138 | MB33; rsbW7 | 12 | 32 | 3–4 | 8 | 1,084.0 ± 46.5 | Orange |
| MB140 | MB33; chrX1 | 8 | 16 | 3 | 6 | 31.2 ± 2.80 | White |
| MB148 | MB33; rsbW8 | 12 | 32 | 4 | 8 | 883.8 ± 70.3 | Orange |
| MB158 | MB137; sigB1(Am) | 2 | 4 | 2 | 4 | 0.59 ± 0.08 | White |
| MB159 | MB137; crtM::pBTcrtM | 12 | 32 | 4 | 8 | 704.8 ± 43.8 | White |
| MB161 | MB140; rsbU+V+W+sigB+ | 18 | 32 | 3 | 8 | 315.3 ± 24.3 | Orange |
| MB162 | MB140; sigB1(Am) | 8 | 16 | 3 | 6 | 0.66 ± 0.07 | White |
| MB163 | MB140; crtM::pBTcrtM | 8 | 16 | 3 | 6 | 41.6 ± 3.80 | White |
| MB213 | MB119; rsbU+V+W+sigB+ | 4 | 8–12 | 3 | 4–6 | 313.0 ± 12.8 | Orange |
| MB215 | MB119; sigB1(Am) | 2 | 4 | 2 | 4 | 0.57 ± 0.09 | White |
| Newman | rsbU+V+W+sigB+ | 2 | 6 | 2 | 4 | 354.5 ± 31.5 | Yellow |
| MSSA1112 | rsbU+V+W+sigB+ | 1.5 | 4 | 1.5 | 6 | 272.4 ± 27.3 | Yellow |
Detailed relevant genotypes and phenotypes are listed in Table 1; exact mutation sites are listed in Table 3. chrX1, unmapped mutation.
MICs were obtained using Etest as outlined in Materials and Methods. MH, Mueller-Hinton agar.
ςB transcriptional activity (relative light units) was determined from cells grown to an OD600 of 1.5 by measuring the luciferase activity of Luc+, the product of the luc+ reporter gene fused to the ςB-dependent promoters of asp23 (asp23p). The values shown are the results of four independent assays.
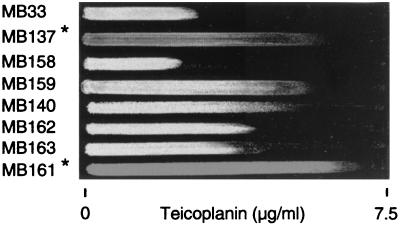
FIG. 3.
Teicoplanin gradient plate. Suspensions (0.5 McFarland standard) of overnight cultures were swabbed on an LB agar plate along an antibiotic gradient as indicated. Growth was monitored after 24 h of incubation. The asterisks indicate pigmented strains. For a detailed description of the strains, refer to Table 1.
Genetic analyses of first-step mutants.
Sequencing of the rsbVWsigB gene region of orange-pigmented mutants revealed nine independent point mutations (Table 3). Two mutants, MB128 and MB148, carried mutations in the putative ribosome-binding site of rsbW, while four mutants, MB118, MB127, MB138, and MB139, had single-nucleotide exchanges in the rsbW gene, resulting in amino acid exchanges. One mutation, rsbW2(Am) of mutant MB119, resulted in a stop codon, while another mutation, rsbW5(Am) of mutant MB130, led to a frame shift within the rsbW open reading frame, introducing a premature stop codon. A single orange mutant, MB137, had an intact rsbW gene but a point mutation in sigB, leading to an amino acid exchange that did not, however, affect the ability of ςB to interact with the RNA polymerase core enzyme. White first-step mutants had intact rsbVW and sigB genes.
TABLE 3.
Mutation sites in orange-pigmented first-step mutants
| Strain | Allele | Mutationa | Remarks |
|---|---|---|---|
| MB118 | rsbW1 | A2350T | Amino acid exchange K44Mb in RsbW |
| MB119 | rsbW2(Am) | C2525G | Amino acid exchange Y102stopb in RsbW |
| MB127 | rsbW3 | C2375G | Amino acid exchange T52Rb in RsbW |
| MB128 | rsbW4 | G2210A | Mutation in putative ribosome-binding site |
| MB130 | rsbW5(Am) | 2527T | Frame shift mutation by T insertion in RsbW |
| MB132 | rsbW6 | G2222T | Amino acid exchange M1Ib in RsbW |
| MB137 | sigB2 | G3115A | Amino acid exchange E148Kc in ςB |
| MB138 | rsbW7 | G2287A | Amino acid exchange R23Hb in RsbW |
| MB148 | rsbW8 | G2209T | Mutation in putative ribosome-binding site |
ςB activity.
The asp23 gene, encoding a protein of yet-unknown function that was shown to be highly expressed upon alkaline stress and heat shock (9, 19), is preceded by three tandem ςB consensus promoters that are known to be under the sole control of ςB (10). The asp23 promoters were found to be suitable for monitoring ςB activity by reporter gene fusion experiments using the firefly luciferase gene luc+ as a reporter gene (10). Experiments with derivatives of wild-type strains, such as Newman and MSSA1112, carrying this reporter system integrated in their genomes, as well as strain MB49, showed that ςB activity increased during the exponential growth phase, reaching a maximal activity during the late exponential growth phase (OD600, 2 to 4), which was followed by a significant decrease thereafter (10). In contrast, in rsbU strains, such as MB33, ςB activity remained low throughout the growth cycle (10) (Fig. 4). All pigmented teicoplanin-resistant mutants derived from MB33, represented in Fig. 4 by strains MB130 and MB137, showed ςB to be highly induced during the early exponential growth phase, reaching a maximal activity at an OD600 of 1, followed by a strong decrease thereafter. In all orange teicoplanin-resistant mutants, the maximal activities of ςB were drastically higher than in the rsbU-negative parent, MB33, irrespective of the mutation site (Table 2). ςB activities were still over twofold higher and were induced in an earlier growth phase than in strains carrying the rsbU wild-type allele (Table 2 and Fig. 4). In contrast, the ςB activities of white first-step mutants, represented by MB140, were as low as those of their parental strains.
FIG. 4.
ςB activities of different S. aureus strains during growth. The expression profiles of asp23p::luc+ during growth of different S. aureus strains, grown in LB medium at 37°C, are shown. Bacterial growth was measured as the OD600 (solid symbols). ςB transcriptional activity was determined by measuring the luciferase activity of Luc+ (open symbols), the product of the luc+ reporter gene fused to the ςB-dependent promoters of asp23 (asp23p). Squares, parental strain MB33 (rsbU); triangles, MB49 (MB33 rsbU+V+W+ sigB+); circles, MB130 [MB33 rsbW5(Am)]; diamonds, MB137 (MB33 sigB2).
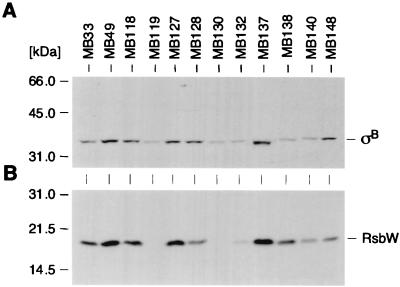
Western blot analyses of RsbW and ςB.
The content of RsbW and ςB was determined by analysis of Western blots from cytoplasmic extracts of cells harvested at an OD600 of 1.5 (Fig. 5). The differences either in the content or in the ratio of ςB and RsbW were remarkable. The parent, MB33, as well as the white teicoplanin-resistant mutant MB140 produced RsbW and ςB in significantly smaller amounts than their rsbU+ relative MB49. Strains such as MB138, harboring a point mutation in RsbW, produced both proteins in a ratio comparable to that of their parent. Strain MB128, carrying a mutation in the proposed ribosome-binding site of rsbW, also produced both proteins but showed a significantly higher ςB/RsbW ratio than the parent. Only very little RsbW was found in the protein fraction of MB132, which had an isoleucine instead of the start methionine, and no RsbW at all could be seen in the protein fractions of MB119 and MB130, harboring mutations in rsbW that lead to premature stop codons. Those mutants produced significantly smaller but still detectable amounts of ςB. Finally, the ςB in the sigB mutant MB137 was abundant and migrated slightly faster than wild-type ςB but was still active, as seen in Table 2.
FIG. 5.
Western blot analyses of RsbW and ςB. Cytoplasmic protein fractions (10 μg/lane) of different S. aureus strains, obtained from cells grown to an OD600 of 1.5, were separated using sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and blotted onto nitrocellulose. The blotted proteins were stained with amido black prior to hybridization to ensure equal loading and were subjected to Western blot analyses using either antigen-purified anti-ςB antibodies (A) or anti-RsbW antibodies (B). The broad-range molecular-weight marker (Gibco-BRL) was used as a size marker. Relevant protein signals are indicated. For a detailed description of the strains, refer to Table 1.
Genetic manipulations showing the influence of the sigB operon on teicoplanin resistance.
Transfer of the rsbU wild-type allele into the white parent, MB33, resulted in the orange-pigmented strain MB49 with an intact sigB operon (10). This strain possessed a slightly increased teicoplanin resistance, though not as high as in the first-step mutants. Teicoplanin MICs obtained for other rsbU wild-type strains, such as Newman and MSSA1112, were in the range of MB33 (Table 2).
Replacement of the active sigB2 allele in the orange first-step mutant MB137 by the defective sigB1(Am) allele, as well as transduction of sigB1(Am) into the orange mutant MB119, yielded the white teicoplanin-susceptible strains MB158 and MB215, respectively, showing virtually no ςB activity (Table 2). In contrast, inactivation of sigB in the white teicoplanin-resistant mutant MB140 resulted in the white mutant MB162, for which the teicoplanin MIC was the same as that of its parent, as expected (Table 2 and Fig. 3).
Introduction of an intact sigB operon into MB140 enhanced the resistance and increased ςB activity 10-fold, as seen in the resulting orange mutant, MB161 (Table 2 and Fig. 3). However, when the wild-type sigB operon was introduced into the resistant orange mutant MB119, which overproduced ςB, the resulting strain, MB213, with a wild-type sigB operon, possessed a resistance level and ςB activity corresponding to those of MB49 (Table 2).
Inactivation of carotenoid biosynthesis genes.
To analyze the impact of carotenoids on teicoplanin resistance and to distinguish it from the effects of the sigB operon, crtM, encoding an early step in S. aureus carotenoid biosynthesis, was inactivated by insertional inactivation in the pigmented first-step mutant MB137, as well as in the white mutant MB140, resulting in strains MB159 and MB163, respectively. The inactivation of crtM resulted in loss of pigment formation in MB137. Resistance to teicoplanin was not affected by crtM in either of the strains (Fig. 3), showing that the increase in ςB activity was the primary responsible effector in teicoplanin resistance.
DISCUSSION
In staphylococci, teicoplanin resistance is assumed to be acquired endogenously in a stepwise manner through mutation and selection after exposure to the glycopeptide (15). Here, we identified the sigB operon as the preferred mutation site leading to first-step teicoplanin resistance in an rsbU mutant. Mutations within the sigB operon were associated with excessively high ςB activities and resulted in teicoplanin MICs two- to sixfold-higher than that for the parental strain (Table 2), classifying all first-step mutants as teicoplanin intermediate resistant according to the interpretive standards of the NCCLS.
Increased resistance was associated in most cases with a single-base-pair mutation (Table 3). All mutants, regardless of the individual mutation site or kind of mutation within the sigB operon, showed essentially the same approximately 20-fold increase in ςB activity in addition to the increase in the MIC. The increase in resistance was significantly higher for teicoplanin than for vancomycin, suggesting that the relevant ςB-controlled gene product(s) interfered more efficiently with teicoplanin than with vancomycin. The ςB activities and MICs clearly exceeded those of the rsbU wild-type strains (Table 2), implying that excessive ςB activity enhances the teicoplanin resistance level of S. aureus. However, the slightly higher teicoplanin MICs observed for MB49 did not reflect the usual teicoplanin susceptibilities found for other rsbU wild-type strains, such as Newman or MSSA1112, which ranged between 1.5 and 2 μg/ml on Mueller-Hinton agar (Table 2). This may suggest that the original rsbU deletion gave rise to second-site mutations or that transduction brought in other genes that may affect susceptibility to teicoplanin. Analogously, in strain BB938, a teicoplanin-resistant transformant, the sigB operon was also identified as the causative agent for increased resistance, but a second cotransducible locus involved in teicoplanin resistance could not be ruled out completely (3).
Conceivable ways to increase ςB activity in S. aureus are mutations resulting in the uncoupling of ςB from its antagonistic protein, RsbW. Accordingly, we identified most of the mutations within the rsbW structural gene (Table 3), which either (i) suppressed the translation of RsbW, (ii) significantly altered the length of the open reading frame of RsbW, or (iii) resulted in the loss of function of RsbW. Inactivation of sigB in orange-pigmented first-step mutants resulted in unpigmented strains that, along with loss of ςB activity, lost the increased resistance against teicoplanin. Transfer of an intact sigB operon into such an orange mutant yielded an orange-pigmented strain possessing a teicoplanin resistance level similar to that found for the rsbU+ derivative MB49. These data strongly suggest that except for the mutation leading to increased ςB activity no further unlinked mutation contributed to teicoplanin resistance in those strains. While increased ςB activity constituted the majority of mutants, other pathways leading to first-step teicoplanin resistance are possible, as shown by the white mutant MB140. Deletion of ςB in MB140 did not affect the teicoplanin resistance level (Table 2 and Fig. 3), indicating that the mutation conferring teicoplanin resistance in MB140 was not linked to the sigB operon. The introduction of an rsbU wild-type allele into MB140 had an additive effect on teicoplanin resistance (Table 2 and Fig. 3), implying that a functional sigB operon contributes positively to the teicoplanin resistance levels of S. aureus NCTC8325 derivatives.
In S. aureus, carotenoid biosynthesis is dependent on the activity of ςB, so that high ςB activity results in strong pigmentation (10). All mutations in the sigB operon leading to increased ςB activity produced in perfect correlation a strong orange pigmentation. By inactivation of crtM, a gene encoding dehydrosqualene synthase, an early step in the carotenoid biosynthesis of S. aureus, we could rule out the possibility that the increased pigment content itself enhances teicoplanin resistance, since the unpigmented crtM mutant MB159 was found to be as resistant as its orange-pigmented donor (Fig. 3). Among the multiple ςB-activated genes, those responsible for modulation of teicoplanin resistance still need to be identified. The sigB operon may well have been the site in SmaI-I identified by Shlaes et al. (23) and postulated to control teicoplanin resistance.
Glycopeptide resistance of S. aureus has been associated in some but not all clinical isolates with thickened cell walls and increased proportions of glutamine-nonamidated muropeptides (5), PBP 2 overproduction, and enhanced production of a 35-kDa membrane protein of still-unknown function (13, 23). In none of the sigB operon mutants investigated could we identify an increase either in PBP 2 or the 35-kDa protein (data not shown).
Besides their role in teicoplanin resistance, such mutations within the sigB operon are of additional interest, as they provide us with new insights and information concerning the regulation and functional sites of ςB and its regulators in S. aureus. Single-base-pair mutations that result in a single-amino-acid exchange and that are sufficient to abolish the ability of RsbW to regulate ςB activity suggest that such amino acid residues are important for RsbW function. The finding that mutants carrying an amber mutation in rsbW produced only small amounts of ςB, even though transcription of rsbUVWsigB was found to be highly abundant and no alterations in the sigB gene itself were uncovered in such strains (data not shown), strongly suggest translational coupling of RsbW and ςB in S. aureus. It is noteworthy that a similar situation has been proposed for the RsbW and ςB homologues of Bacillus subtilis (2). In the absence of its antagonist, RsbW, even small amounts of ςB are likely to be sufficient to produce high ςB activity. The strong decrease in ςB activity observed in orange-pigmented first-step mutants (Fig. 4), beginning from the mid-exponential growth phase, suggests that a further, as-yet-unidentified negative regulator of ςB is present in S. aureus. The unexpected finding that ςB activity also decreases in MB137 (Fig. 4), harboring the mutation in sigB, suggests that the proposed negative regulator interacts with ςB in a way that is different from that of RsbW, as its ability to interact with ςB is not inhibited by the sigB2 mutation.
Taken together, this system provides us with an elegant means for functional studies by generating mutations increasing ςB activity. These mutations are favored because of the rsbU background, and hence low ςB activity, in the original strain used. It remains an open question if and to what extent such mutations would also occur in S. aureus strains harboring a functional sigB operon.
ACKNOWLEDGMENTS
This study was supported by the Swiss National Science Foundation grants 31-52237.97 to B. Berger-Bächi and 31.46762.96 to I. Kullik.
We thank U. Hardegger and E. Huf for excellent technical support. Preliminary sequence data were obtained from The Institute of Genomic Research through the website at http://www.tigr.org. Sequencing of S. aureus COL was accomplished with support from the National Institute of Allergy and Infectious Diseases and the Merck Genome Research Institute.
REFERENCES
- 1.Beauregard D A, Williams D H, Gwynn M N, Knowles D J C. Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob Agents Chemother. 1995;39:781–785. doi: 10.1128/AAC.39.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff M, Roos M, Putnik J, Wada A, Glanzmann P, Giachino P, Vaudaux P, Berger-Bächi B. Involvement of multiple genetic loci in Staphylococcus aureusteicoplanin resistance. FEMS Microbiol Lett. 2001;194:77–82. doi: 10.1111/j.1574-6968.2001.tb09449.x. [DOI] [PubMed] [Google Scholar]
- 4.Brückner R. Gene replacement in Staphylococcus camosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 5.Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureusMu50. Antimicrob Agents Chemother. 2000;44:2276–2285. doi: 10.1128/aac.44.9.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duthie E S, Lorenz L L. Staphylococcus coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 7.Entenza J M, Vouillamoz J, Glauser M P, Moreillon P. Levofloxacin versus ciprofloxacin, flucloxacillin, or vancomycin for treatment of experimental endocarditis due to methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1662–1667. doi: 10.1128/aac.41.8.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felmingham D, Brown D F J, Soussy C J. European glycopeptide susceptibility survey of gram-positive bacteria for 1995. Diagn Microbiol Infect Dis. 1998;31:563–571. doi: 10.1016/s0732-8893(98)00053-4. [DOI] [PubMed] [Google Scholar]
- 9.Gertz S, Engelmann S, Schmid R, Ohlsen K, Hacker J, Hecker M. Regulation of sigma(B)-dependent transcription of sigB and asp23 in two different Staphylococcus aureusstrains. Mol Gen Genet. 1999;261:558–566. doi: 10.1007/s004380051001. [DOI] [PubMed] [Google Scholar]
- 10.Giachino P, Engelman S, Bischoff M. ςB activity depends on RsbU in Staphylococcus aureus. J Bacteriol. 2001;183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grinsted J, Lacey R W. Ecological and genetic implications of pigmentation in Staphylococcus aureus. J Gen Microbiol. 1973;75:259–267. doi: 10.1099/00221287-75-2-259. [DOI] [PubMed] [Google Scholar]
- 12.Hecker M, Volker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtiliscells by the expression of the sigmaB regulon, Mol. Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K. Vancomycin resistance in staphylococci. Drug Res Updates. 1998;1:135–150. doi: 10.1016/s1368-7646(98)80029-0. [DOI] [PubMed] [Google Scholar]
- 14.Iandolo J J. Genetic and physical map of the chromosome of Staphylococcus aureus 8325. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 317–325. [Google Scholar]
- 15.Kaatz G W, Seo S M, Dorman N J, Lerner S A. Emergence of teicoplanin resistance during therapy of Staphylococcus aureusendocarditis. J Infect Dis. 1990;162:103–108. doi: 10.1093/infdis/162.1.103. [DOI] [PubMed] [Google Scholar]
- 16.Kreiswirth B N, Löfdahl S, Bentley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:680–685. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 17.Kullik I, Glachino P. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigBoperon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 18.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureusreveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda M, Ohta T, Hayashi H. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- 20.Marshall J H, Wilmoth G J. Proposed pathway of triterpenoid carotenoid biosynthesis in Staphylococcus aureus: evidence from a study of mutants. J Bacteriol. 1981;147:914–919. doi: 10.1128/jb.147.3.914-919.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Scott J M, Smirnova N, Haldenwang W G. A Bacillus-specific factor is needed to trigger the stress-activated phosphatase/kinase cascade of ςBinduction. Biochem Biophys Res Commun. 1999;257:106–110. doi: 10.1006/bbrc.1999.0418. [DOI] [PubMed] [Google Scholar]
- 23.Shlaes D M, Shlaes J H, Vincent S, Etter L, Fey P D, Goering R V. Teicoplanin-resistant Staphylococcus aureusexpresses a novel membrane protein and increases expression of penicillin-binding protein-2 complex. Antimicrob Agents Chemother. 1993;37:2432–2437. doi: 10.1128/aac.37.11.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 25.Wieland B, Feil C, Gloria-Maecker E, Thumm G, Lechner M, Bravo J-M, Poralla K, Götz F. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4-diaponeurosporene of Staphylococcus aureus. J Bacteriol. 1994;176:7719–7726. doi: 10.1128/jb.176.24.7719-7726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu S, Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureusRNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]