Abstract
Intensive insight into the potential mechanisms of Se-induced Cd tolerance in cucumber seedlings is essential for further improvement of vegetable crop cultivation and breeding to obtain high yields and quality in Cd-contaminated soil. To reveal the ultrastructural and metabolic differences in Se-induced Cd tolerance, we examined the ultrastructures of chloroplasts and root cells and characterised 155 differentially expressed metabolites under Cd and/or Se stress using gas chromatography-mass spectrometry (GC-MS)-based metabolomics. Exogenous Se greatly relieved Cd-caused injuries to the ultrastructures of cucumber leaves and roots; for example, the shapes of chloroplasts treated with Cd + Se improved or even began to return to normal, the nuclei of root cells began to regenerate better and the chromatin was well-distributed compared with plants treated with Cd alone. Metabolite profiling revealed several intermediates of glycolysis and the tricarboxylic acid (TCA) cycle; also, some amino acids were up-accumulated in Cd + Se-treated cucumber seedlings and down-accumulated in Cd-treated cucumber seedlings, such as pyruvic acid, galactose, lactose, glutaric acid and alanine in leaves, glucose-6-phosphate and serine in roots, and lactic acid and glycine in both leaves and roots. These metabolites may play dominant roles in developing Se-mediated Cd tolerance. Moreover, a high level of sugars and polyols, amino acids and organic acids were up-accumulated in Cd-treated plants. Meanwhile, our data suggest that high accumulation of fructose, α-ketoglutaric acid, shikimic acid, fumaric acid and succinic acid in roots is a Cd-specific response, indicating that these metabolites are vital for cucumbers to develop Cd resistance. This study extends the current understanding of the mechanisms of Se in abating Cd contamination in cucumber and demonstrates that metabolomics profiling provides a more comprehensive view of the response of plants to heavy metals.
Intensive insight into the potential mechanisms of Se-induced Cd tolerance in cucumber seedlings is essential for further improvement of vegetable crop cultivation and breeding to obtain high yields and quality in Cd-contaminated soil.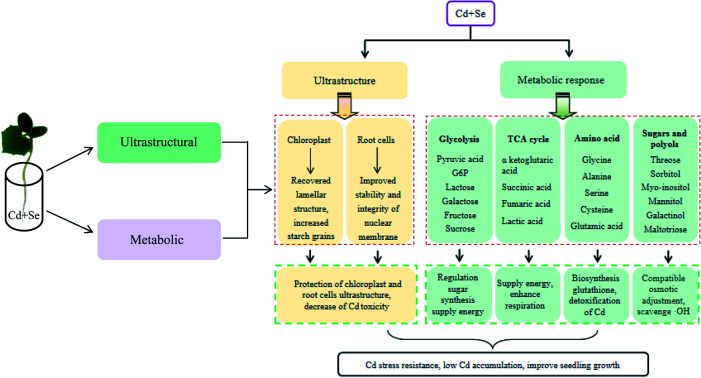
Introduction
Heavy metal pollution caused by anthropogenic activities, such as mining, waste gas and water discharge, sewage irrigation and use of heavy metal products exceeding standards, is receiving increasing attention due to its public hazards to humans and the environment. Cadmium (Cd) is a nonessential element that is toxic to all organisms, including higher plants.1 Cadmium-contaminated soil is a challenge for plants grown in it because Cd is easily absorbed by roots and transported to over-ground parts of plants.2 Subsequently, Cd toxicity can affect plant photosynthesis, respiration and absorption of water and nutrients, thus obviously reducing plant growth and crop yields or even causing death. Accordingly, it is imperative to reveal the phytotoxic mechanisms of Cd and to improve our methods of dealing with polluted soils.3,4
In general, different metals tend to mix in the environment, and the effects of exposure to this mixture can be additive or antagonistic.5–7 Selenium (Se) is an essential element for organisms, while Cd is not; this leads to different accumulations of the two elements in plants. For example, exogenous application of Se could decrease Pb and Cd accumulation in lettuce; moreover, Se could abate Cd accumulation in rice, tobacco and cucumber,8–11 and Se protects against metal-(loid)-induced toxicity and disease manifestations as well as salt stress.12,13 Meanwhile, our previous work also clearly stated that Se is capable of alleviating Cd-inhibited cucumber plant growth, chlorophyll content, and photosynthetic performance; it can also significantly reduce ˙OH, H2O2 and malondialdehyde content as well as Cd accumulation in maize and cucumber. Similarly, Se stimulated the growth of cucumber by enhancing nutrient uptake, balancing carbohydrate contents and altering the expression abundance of some proteins.14–17
Cucumber (Cucumis sativus L.) is a major economic vegetable crop with high consumption that is commonly planted worldwide. Cucumber is also an excellent source of protein, essential vitamins, carbohydrates, crude fibre and other nutrients humans need. With in-depth research on its traits, cucumber is used as a model plant in many studies. However, the research on Se-enhanced growth of cucumber plants mostly focuses on the fields of physiology, biochemistry and proteomics;10,16,17 the underlying stimulatory effects of Se based on the metabolic profiles and ultrastructures of cucumber are not fully understood.
Metabolomics is an important branch of systems biology; it is the quantitative measurement of small molecular compounds and metabolic components in biological specimens. Recently, metabonomics analysis has been widely applied to study the tolerance of plants to abiotic stress.18 Characterising the metabolome of an organism can offer novel insights into its functional performance status related to its phenotype, toxic effects induced by heavy metal toxicity,18 salt stress,19 temperature20 and combined stress of drought and high temperature.21 In the present study, the ultrastructures of chloroplasts and root cells of cucumber seedlings under Cd and/or Se stress were observed, and the metabolic profiles were also analysed using gas chromatography – mass spectrometry (GC-MS) analysis of the leaves and roots of cucumber plants under Cd and/or Se stress to identify the predominant metabolites responsive to Cd and/or Se stress in cucumber. This study will provide insight for understanding the Se-induced ultrastructure and metabolism changes of cucumber plants grown under cadmium exposure.
Materials and methods
Plant materials and chemicals
Cucumber seeds (Jinyan 4) were purchased from Shanxi Agricultural Seed Station. Chemicals were all analytical grade reagents and were obtained from Beijing Chemical Works (Beijing, China), and all other chemicals used in this study were prepared with ultrapure water from a Millipore system unless otherwise specified. Hydrogen peroxide (H2O2, ≥30%), sodium selenite (Na2SeO3, ≥98%), and cadmium chloride (CdCl2, ≥98%) were used for the cucumber hydroponic culture. Na2-EDTA (C10H14N2O8Na2·2H2O, ≥99%), nitric acid (HNO3, ≥70%) and perchloric acid (HClO4, ≥70%) were used for Cd and Se concentration determination, and the extracted acid mixture was prepared according to a volume ratio of HNO3vs. HClO4 of 5 : 1. Methanol (CH3OH, ≥99.5%) extraction liquid (Vmethanol : VH2O = 3 : 1) and 1 mg mL−1 ribitol aqueous solution (C5H12O5, ≥99%) were used for metabolite extraction. Bis(trimethylsilyl)trifluoroacetamide (BSTFA, C8H18F3NOSi2) regent (99%, with 1% chlorotrimethylsilane (TMCS), v/v), and 20 mg mL−1 (in pyridine, C5H5N, ≥99%) methoxyamine hydrochloride (CH6ClNO, ≥98%) were used for derivatization of metabolites. The GC/MS glass vials used for metabolite detection were purchased from Sigma-Aldrich (Beijing, China).
Cucumber hydroponic culture
Seeds of Jinyan 4 cucumber were sterilized in 2% H2O2 for 15 min and washed with deionized water seven times before germination. The experiment protocol of germination, seedling transplant and culture was similar to our previous study.17 This experiment was conducted in the greenhouse of Taiyuan University of Science and Technology, Taiyuan, China.
Treatments and sampling
Based on specific preliminary dose tests, four treatments were formed on the 7th day after transplanting: (1) control, basal nutrient solution (BNS); (2) Se (as Na2SeO3), BNS + 3 μM Se, where Se was added on the 6th day after transplanting; (3) Cd (as CdCl2), BNS + 50 μM Cd; and (4) Cd + Se, BNS + 50 μM Cd + 3 μM Se, where Se was added on the 6th day after transplanting and replaced with Cd + Se on the second day. There were six replicates for each treatment. After seven days of treatment, plants were sampled from each treatment and each replicate, and all plants were washed thoroughly with deionized water; then, the leaves and roots were collected in accordance with Kim and Verpoorte's description22 and stored at −80 °C for metabolite extraction.
Cd and Se concentration determination
After sampling, roots for element determination were soaked in 20 mM Na2-EDTA for 20 min and then rinsed in deionized water. All samples were dried at 70 °C until constant weight. Dried samples were powdered and digested in the acid mixture (HNO3 : HClO4, 5 : 1, v/v).16 The Se and Cd concentrations were determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES) (SPS 1200 VR, Seiko Co., Ltd., Japan).
Ultrastructural examination of leaves and roots
The ultrastructures of the leaves and roots were observed after seven days of treatment; three biological replicates were collected and handled according to Sun et al.23 The specimens were mounted on copper grids and viewed by a transmission electron microscope (JEOL JEM-1230 EX, Japan) at 12 kV with working distances of 15 and 10 mm, respectively.
Metabolite extraction, derivatization and detection
Cucumber leaf and root metabolites were extracted as follows: frozen tissues were taken into 2 mL centrifuge tubes and extracted with 0.4 mL extraction liquid (Vmethanol : VH2O = 3 : 1); then, 20 μL of ribitol (1 mg mL−1 stock in ultrapure water) was added as an internal standard to the sample. The mixture was homogenized in a grinding mill with small steel balls at 45 Hz for 4 min, then ultrasonically processed for 5 min in an ultrasonic machine with ice water; the above procedures were repeated twice. The above mixture was centrifuged at 13 000 rpm for 15 min at 4 °C, and the supernatant (0.35 mL) was transferred into a GC/MS glass vial.
The samples were dried without heating in a vacuum concentrator, followed by adding 40 μL of methoxyamine hydrochloride (20 mg mL−1 in pyridine), and then incubated at 80 °C for 30 min. Finally, 50 μL BSTFA regent (with 1% TMCS, v/v) was added to the sample aliquots, which were then incubated for 2 h at 70 °C.
GC-MS analysis was carried out on an Agilent 7890 gas chromatograph system coupled with a Pegasus HT time-of-flight mass spectrometer; the system utilized a DB-5MS capillary column. 1 μL derivatized extract of each sample was injected into the capillary. The temperature-rise was programmed according to the study by He et al.24
Data analysis
The R software platform (http://cran.r-project.org/) and TagFinder software were applied for extraction of raw signals, filtering of the data baselines, deconvolution analysis, peak alignment, peak identification and integration of the peak areas.25 For each identified metabolite, log-transformed response ratios were calculated before the statistical assessment; the expressive abundance of ≥1.5-fold increase or ≤0.5-fold decrease were set as the criteria (e.g. Cd + Se vs. Cd, or Cd vs. control). Differentially expressed metabolites were identified by Simca-P software (version 11.5) by employing the orthogonal projections to latent structures discriminant analysis (OPLS-DA) model; also, some metabolites were searched in available compound libraries, such as KEGG. The principal component analysis (PCA) and the hierarchical clustering analysis (HCA) were performed using the MetaboAnalyst webpage,26 and the metabolic pathway was established according to the KEGG metabolic database.
Results
Cd concentration in cucumber seedlings under Cd and Se application
In the control and Se treatments, the Cd concentration was below the detection limit and consequently is not shown in Table S1.† Exogenous Se (Cd + Se) prominently reduced the concentration of Cd by 23.8% and 36.5% in leaves and roots, respectively, compared with Cd treatment alone. On the other side, Cd exposure remarkably decreased both leaf and root Se concentration compared to Se treatment alone; no significant differences were observed between Cd treatment alone and the control, which were below the detection limit.
Effects of exogenous Se on the ultrastructure of chloroplasts of cucumber seedlings under Cd stress
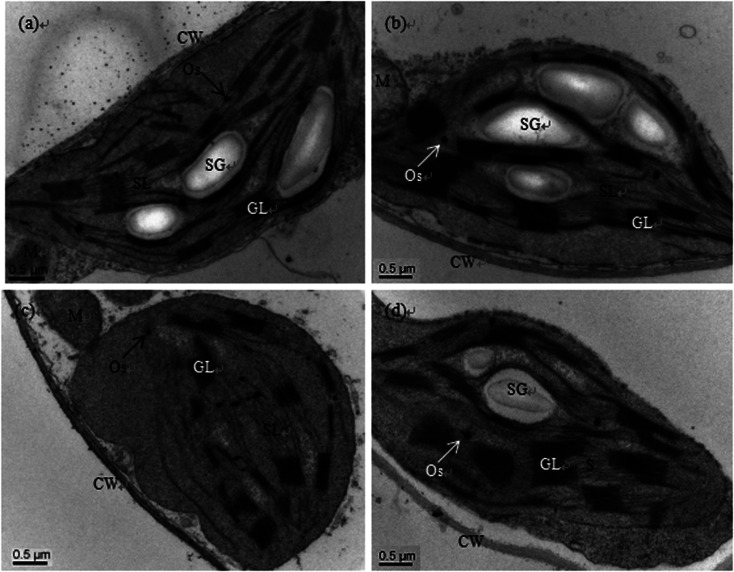
Scanning electron microscopy (SEM) observations showed that there was no clear difference between the control and sole Se treatments; the chloroplast stroma of the cucumber leaf cells was dense, and the lamellae of the grana thylakoids were stacked regularly and formed a continuous whole with the stroma lamellae. Moreover, there were many starch grains in the lamellae (Fig. 1a and b). However, the ultrastructures were deteriorated in the chloroplasts of plants treated with Cd, e.g. the chloroplasts become oval or round, the cytoplasm became sparse, the membrane systems became fuzzy, the stroma lamellae swelled, the thylakoid lamellae became loose, some lamellae even degraded, and the number of starch grains was reduced (Fig. 1c). Although the stromal lamellae and grana were still swollen, similar to Cd treatment alone, addition of exogenous Se (Cd + Se) prevented these Cd-induced deterioration changes to the chloroplasts; the shape of the chloroplasts under Cd + Se treatment became normal, and the structure of the thylakoid membranes was also restored. Additionally, the number of starch grains increased compared with Cd treatment alone (Fig. 1d).
Fig. 1. Transmission electron micrographs of chloroplasts of cucumber seedlings cultured in basic nutrition solution (BNS, CK, (a)), BNS + 3 μM Se (Se, (b)), BNS + 50 μM Cd (Cd, (c)), and BNS + 50 μM Cd + 3 μM Se (Cd + Se, (d)), respectively. Scale bar = 0.2 μm. CW: cell wall, M: mitochondrion, GL: granal lamellae, Os: osmiophilic plastolobuli, SG: starch grain, SL: stromal lamellae. The figure is representative of three different experiments.
Effects of exogenous Se on the root cell ultrastructures of cucumber seedlings under Cd stress
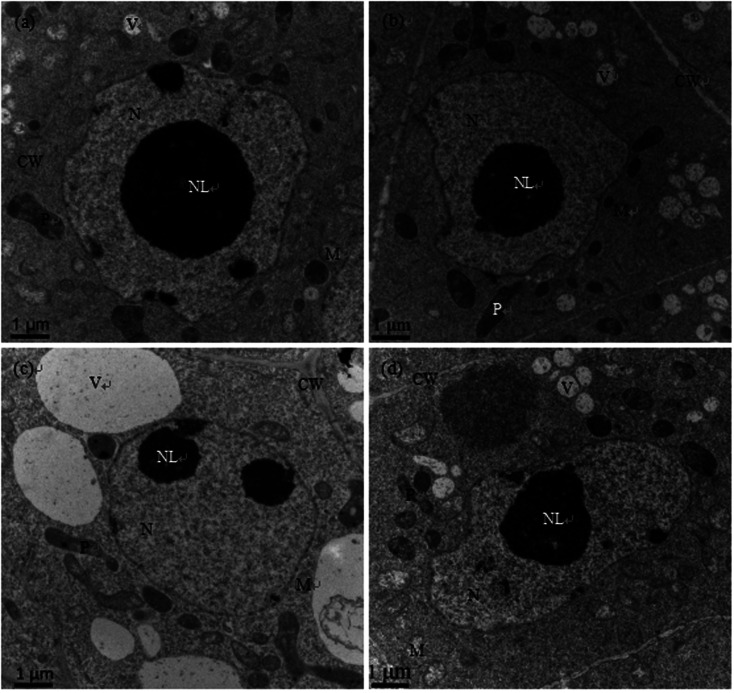
As shown in Fig. 2a and b, the nuclear structures of cucumber root cells under control and Se treatment alone were normal; the nuclei were nearly round, the nuclear membranes were complete and smooth, the nuclear cytoplasm was uniform, and there were abundant organelles and vacuoles in the cytoplasm. However, after Cd treatment, the root meristematic cells exhibited obvious ultrastructural changes; the cell morphology changed, the cell walls thickened, the karyotheca was depressed and deformed, the number of vacuoles significantly decreased while their sizes increased, the cell nuclei even cracked, the mitochondria were swollen and the plastids contained fewer cristae (Fig. 2c). In the presence of Se (Cd + Se), the nuclei and karyotheca were better formed, and chromatin was distributed more uniformly; the nuclear morphology and chromatin returned to normal conditions.
Fig. 2. Transmission electron micrographs of root cells of cucumber seedlings cultured in basic nutrition solution (BNS, CK, (a)), BNS + 3 μM Se (Se, (b)), BNS + 50 μM Cd (Cd, (c)), and BNS + 50 μM Cd + 3 μM Se (Cd + Se, (d)), respectively. Scale bar = 1 μm. CW: cell wall, M: mitochondrion, N: nucleolus, NL: nucleolus, P: plastid, V: vacuole. The figure is representative of three different experiments.
The ultrastructures of root meristem cells exhibited obvious changes after Cd stress, with distorted cells, thickened cell walls, plastids with fewer cristae, swollen mitochondria, increased vacuolar size, and cracked nuclei and karyotheca. In the presence of Se (Cd + Se), the nuclei and karyotheca were better formed, the cytoplasm was denser, and the vacuolar size and the numbers of mitochondria cristae and plastids improved compared with Cd treatment alone (Fig. 2d).
Metabolic changes of cucumber under exogenous Cd and Se exposure
To reveal the molecular mechanisms of Se-mediated underlying Cd tolerance in cucumber seedlings, the effects of Cd (50 μM) and/or Se (3 μM) on the metabolic changes in the leaves and roots were analysed. The total ion chromatograms (TICs) of the leaves and roots of 36 cucumber samples showed that there were apparent chromatographic differences among the different treatments, including Cd and/or Se treatment and the control (Fig. S1†). According to heatmap and PCA analysis (Fig. S2 and S3†), obvious separation among the different treatments was detected. The leaves of the control and treated plants (i.e. Cd, Cd + Se) were distinguished by PC1, representing 22.2% variation; however, in the roots, Cd treatment was separated from the control and Cd + Se treatments by PC1, explaining 22.9% of the variation. Moreover, Cd and Cd + Se in both leaves and roots were separated by PC2; they presented 12.3% and 17.8% variation, respectively (Fig. S3a and b†). At the same time, OPLS-DA clearly showed discrimination and separation among the control, Cd + Se, and Cd treatments both in leaves and roots (Fig. S4†).
Differently expressed metabolites (VIP > 1, P < 0.05) were identified, and the peak values of the control, Cd, and Cd + Se treatments were compared statistically. GC-TOF-MS analysis showed that a large number of metabolites were significantly expressed both in leaves and roots among the control, Cd and Cd + Se treatments. Furthermore, prominent tissue differences of relative abundance were found; 24 and 26 metabolites were up-accumulated or unchanged by Cd + Se vs. Cd treatment alone and down-accumulated by Cd vs. the control in the leaves and roots, respectively (Tables 1 and 2). However, compared with the control, Cd treatment significantly up-accumulated 47 and 68 metabolites in leaves and roots, respectively (Tables 3 and 4).
List of metabolites in leaves of cucumber seedlings whose relative fold changes were significantly down-accumulated (≤0.5) after Cd treatment and up-accumulated (≥1.5) or unchanged (0.5< and <1.5) after Cd + Se-treatment.
| Metabolite name | Similarity | R.T.a | Mass | VIPb | P valuec | Fold changed | Pathway | |
|---|---|---|---|---|---|---|---|---|
| Cd vs. control | Cd + Se vs. Cd | |||||||
| Pyruvic acid | 927 | 7.2052 | 174 | 1.87353 | 0.01560 | 0.35 | 1.98 | Glycolysis |
| Galactose 1 | 939 | 17.7571 | 319 | 2.00540 | 0.00810 | — | 2.40 | Glycolysis |
| Lactose 1 | 801 | 24.6819 | 204 | 1.66282 | 0.03241 | — | 4.99 | Glycolysis |
| Lactic acid | 968 | 7.3206 | 117 | 1.90526 | 0.01573 | — | 0.72 | TCA cycle |
| Oxalic acid | 704 | 8.3484 | 147 | 1.96350 | 0.00734 | — | 1.22 | TCA cycle |
| 3-Hydroxypropionic acid 1 | 757 | 8.5775 | 177 | 2.43025 | 0.00011 | — | 4.59 | TCA cycle |
| 2,3-Dimethylsuccinic acid | 395 | 11.4907 | 221 | 1.82082 | 0.02495 | — | 1.21 | TCA cycle |
| Glutaric acid | 757 | 12.2118 | 55 | 2.52714 | 0.00002 | — | 7.06 | TCA cycle |
| Citramalic acid | 921 | 13.0089 | 247 | 1.82753 | 0.01967 | — | 1.84 | TCA cycle |
| Glycero | 904 | 10.3819 | 117 | 1.89634 | 0.01340 | 0.44 | 1.23 | Sugars and polyols |
| 3-Methylamino-1,2-propanediol 2 | 464 | 11.7196 | 117 | 1.73584 | 0.01805 | — | 1.19 | Sugars and polyols |
| d-Glycerol 1-phosphate | 871 | 16.4002 | 299 | 1.65905 | 0.04984 | — | 1.75 | Sugars and polyols |
| Glucoheptonic acid 3 | 673 | 20.1423 | 319 | 2.14536 | 0.00387 | 0.46 | 2.59 | Sugars and polyols |
| Phytol | 972 | 20.5121 | 143 | 2.17178 | 0.00227 | 0.50 | 1.91 | Sugars and polyols |
| Melibiose 2 | 840 | 26.0556 | 361 | 1.95062 | 0.01075 | — | 4.56 | Sugars and polyols |
| Alanine 1 | 957 | 7.9520 | 116 | 2.01987 | 0.00487 | 0.41 | 1.51 | Amino acid |
| Glycine 2 | 947 | 10.8449 | 174 | 2.41138 | 0.00015 | 0.29 | 4.47 | Amino acid |
| Aminomalonic acid | 676 | 12.9948 | 218 | 2.32434 | 0.00287 | 0.13 | 5.91 | Amino acid |
| Palmitoleic acid | 877 | 19.1547 | 55 | 1.75218 | 0.02081 | — | 1.43 | Lipids |
| Linoleic acid | 923 | 20.8606 | 337 | 1.83842 | 0.01769 | — | 1.90 | Lipids |
| 4-Hydroxybutyrate | 819 | 9.9014 | 233 | 2.47526 | 0.00004 | — | 4.45 | Others |
| Nornicotine | 431 | 13.9813 | 142 | 1.93520 | 0.00797 | — | 2.30 | Others |
| Phthalic acid | 504 | 15.8280 | 304 | 1.77004 | 0.04181 | 0.28 | 2.94 | Others |
| N-Acetyl-beta-d-mannosamine 4 | 771 | 19.8795 | 319 | 1.95543 | 0.00945 | 0.32 | 2.82 | Others |
R.T. represents the retention time.
VIP = variable importance projection, metabolites (VIP > 1) are listed in the table.
P values were calculated according to Student's T-test.
Fold change is represented as the ratio of the peak intensities for the corresponding treatments (n = 6), —: represented, not detected.
List of metabolites in roots of cucumber seedlings whose relative fold changes were significantly down-accumulated (≤0.5) after Cd treatment and up-accumulated (≥1.5) or unchanged (0.5< and <1.5) after Cd + Se treatment.
| Metabolite name | Similarity | R.T.a | Mass | VIPb | P valuec | Fold changed | Pathway | |
|---|---|---|---|---|---|---|---|---|
| Cd vs. control | Cd + Se vs. Cd | |||||||
| α-d-Glucosamine 1-phosphate | 638 | 17.1326 | 226 | 1.63509 | 0.00164 | — | 0.50 | Glycolysis |
| Glucose-6-phosphate 1 | 896 | 21.6314 | 387 | 1.39508 | 0.02454 | — | 0.57 | Glycolysis |
| Glucose-6-phosphate 2 | 931 | 21.8916 | 387 | 1.23427 | 0.04055 | — | 0.60 | Glycolysis |
| 6-Phosphogluconic acid | 670 | 22.5322 | 318 | 1.48791 | 0.00661 | — | 0.55 | Glycolysis |
| Lactic acid | 867 | 7.3366 | 219 | 1.88945 | 0.00009 | 0.36 | 4.09 | TCA cycle |
| Ethanolamine | 883 | 10.3063 | 86 | 1.51962 | 0.00503 | — | 0.76 | Sugars and polyols |
| Ribose | 931 | 15.4504 | 103 | 1.23105 | 0.03573 | — | 0.66 | Sugars and polyols |
| Xylitol | 804 | 15.6796 | 231 | 1.58847 | 0.00229 | — | 0.58 | Sugars and polyols |
| Conduritol β epoxide 2 | 650 | 18.4594 | 220 | 1.52304 | 0.00415 | — | 0.58 | Sugars and polyols |
| N-Acetyl-d-galactosamine 1 | 856 | 19.5938 | 87 | 1.61868 | 0.00126 | — | 0.57 | Sugars and polyols |
| Ribulose-5-phosphate 1 | 758 | 19.9544 | 357 | 1.51388 | 0.00462 | — | 0.50 | Sugars and polyols |
| Lactulose 1 | 832 | 24.6661 | 361 | 1.30364 | 0.03096 | — | 3.80 | Sugars and polyols |
| Lactitol | 707 | 25.3578 | 230 | 1.52403 | 0.00359 | — | 0.50 | Sugars and polyols |
| Oxamic acid | 566 | 10.1376 | 189 | 1.68048 | 0.00105 | 0.25 | 3.04 | Amino acids |
| l-Homoserine 1 | 580 | 12.7268 | 218 | 1.07594 | 0.03957 | 0.09 | 5.21 | Amino acids |
| Glycine 2 | 957 | 10.8446 | 174 | 1.84246 | 0.00034 | 0.59 | 2.17 | Amino acids |
| Serine 1 | 953 | 11.5610 | 218 | 1.86332 | 0.00001 | — | 1.87 | Amino acids |
| 3-Hydroxy-palmitic acid | 498 | 20.7420 | 299 | 1.22242 | 0.03691 | — | 0.63 | Organic acids |
| O-Phosphorylethanolamine | 914 | 16.6380 | 299 | 1.30504 | 0.02247 | — | 0.53 | Organic acids |
| Succinate semialdehyde 1 | 292 | 9.1880 | 132 | 1.84506 | 0.00018 | — | 15.16 | Lipids |
| 2-Deoxyuridine | 526 | 10.4501 | 170 | 1.16267 | 0.03924 | — | 0.52 | Pyridine alkaloid |
| dl-Anabasine 1 | 632 | 12.1288 | 239 | 1.58278 | 0.00166 | — | 0.57 | Pyridine alkaloid |
| Nicotinamide | 467 | 13.4756 | 179 | 1.26266 | 0.03057 | 0.35 | 0.63 | Pyridine alkaloid |
| Maleamate 2 | 449 | 13.8690 | 241 | 1.46397 | 0.00747 | 0.34 | 2.75 | Pyridine alkaloid |
| 2-Hydroxypyridine | 908 | 7.0488 | 152 | 1.76756 | 0.00026 | — | 0.67 | Pyridine alkaloid |
| N-Cyclohexylformamide 2 | 270 | 9.6684 | 227 | 1.61878 | 0.00111 | — | 0.66 | Others |
R.T. represents the retention time.
VIP = variable importance projection, metabolites (VIP > 1) are listed in the table.
P values were calculated according to Student's T-test.
Fold change is represented as the ratio of the peak intensities for the corresponding treatments (n = 6), —: represented, not detected.
List of metabolites whose relative fold changes were significantly up-accumulated (≥1.5) in Cd-treated leaves of cucumber seedlings.
| Metabolite name | Similarity | R.T.a | Mass | VIPb | P valuec | Fold changed | Pathways | |
|---|---|---|---|---|---|---|---|---|
| Cd vs. control | Cd + Se vs. Cd | |||||||
| d-Galacturonic acid 2 | 671 | 18.4902 | 292 | 1.70673 | 0.00117 | 1.61 | — | Glycolysis |
| Galactonic acid | 830 | 18.7519 | 333 | 1.30902 | 0.02551 | 2.60 | — | Glycolysis |
| Galactonic acid | 878 | 18.7944 | 292 | 1.51738 | 0.00999 | 1.51 | — | Glycolysis |
| Oxalacetic acid | 278 | 12.9308 | 68 | 1.37968 | 0.04399 | 299 957 | — | TCA cycle |
| l-Threose 1 | 832 | 12.7645 | 205 | 1.42249 | 0.01424 | 1.64 | — | Sugars and polyols |
| Threitol | 749 | 13.3492 | 217 | 1.80221 | 0.00338 | 189 254 | — | Sugars and polyols |
| d-Arabitol | 775 | 15.9364 | 307 | 1.34576 | 0.03543 | 7.49 | — | Sugars and polyols |
| Mannitol | 942 | 18.1471 | 319 | 1.76594 | 0.00112 | 1.59 | — | Sugars and polyols |
| Sedoheptulose | 555 | 18.1988 | 204 | 1.61517 | 0.01827 | 4.27 | — | Sugars and polyols |
| Sorbitol | 865 | 18.2634 | 217 | 1.44971 | 0.01438 | 1.53 | — | Sugars and polyols |
| Valine | 966 | 9.57236 | 144 | 1.63117 | 0.00341 | 2.78 | — | Amino acids |
| Carbobenzyloxy-l-leucine | 332 | 10.0939 | 171 | 1.62983 | 0.00336 | 5.14 | — | Amino acids |
| Isoleucine | 955 | 10.6617 | 158 | 1.81261 | 0.00055 | 3.63 | — | Amino acids |
| Proline | 896 | 10.7342 | 142 | 1.75822 | 0.00091 | 2.36 | — | Amino acids |
| Serine 1 | 947 | 11.5619 | 204 | 1.38756 | 0.01696 | 2.02 | — | Amino acids |
| Cycloleucine 1 | 347 | 11.655 | 255 | 1.62231 | 0.00388 | 2.68 | — | Amino acids |
| l-Allothreonine 1 | 607 | 11.8983 | 217 | 1.78923 | 0.00065 | 1.74 | — | Amino acids |
| Threonine 1 | 954 | 11.8964 | 219 | 1.88911 | 0.00015 | 2.71 | — | Amino acids |
| O-Acetylserine 1 | 519 | 12.0443 | 174 | 1.67359 | 0.00504 | 4.73 | — | Amino acids |
| Aspartic acid | 947 | 13.6334 | 232 | 1.50107 | 0.01207 | 2.46 | — | Amino acids |
| l-Cysteine | 685 | 14.0558 | 116 | 2.08554 | 0.00004 | 651 149 | — | Amino acids |
| 3-Aminoisobutyric acid 1 | 532 | 12.8712 | 174 | 1.58589 | 0.01316 | 1.60 | — | Amino acids |
| β-Glutamic acid 1 | 395 | 14.6504 | 345 | 1.76216 | 0.00118 | 2.99 | — | Amino acids |
| Glutamic acid | 921 | 14.8229 | 246 | 1.41253 | 0.01601 | 1.94 | — | Amino acids |
| Phenylalanine 1 | 890 | 14.9275 | 218 | 1.53758 | 0.01950 | 12.79 | — | Amino acids |
| Creatine | 573 | 15.0537 | 403 | 1.58464 | 0.01576 | 12.07 | — | Amino acids |
| Asparagine 1 | 898 | 15.4162 | 116 | 1.40902 | 0.04445 | 11 038 471 | — | Amino acids |
| Citrulline 1 | 905 | 17.1051 | 157 | 1.64987 | 0.01145 | 50.24 | — | Amino acids |
| Lysine | 908 | 18.1015 | 156 | 1.83055 | 0.00311 | 8.44 | — | Amino acids |
| Spermidine 2 | 614 | 20.7375 | 200 | 1.38046 | 0.02642 | 4.00 | — | Amino acids |
| 2-Hydroxy-butanoic acid | 252 | 8.34339 | 206 | 1.35095 | 0.04797 | 1.85 | — | Organic acids |
| Malonic acid 1 | 596 | 9.31769 | 147 | 1.58595 | 0.00526 | 1.71 | — | Organic acids |
| Aminooxyacetic acid | 516 | 11.8209 | 247 | 1.49460 | 0.01333 | 1.99 | — | Organic acids |
| Adipic acid | 308 | 13.4852 | 111 | 1.38794 | 0.02267 | 4.90 | — | Organic acids |
| Threonic acid | 924 | 14.0775 | 292 | 1.33816 | 0.02717 | 1.58 | — | Organic acids |
| Gentisic acid | 454 | 16.6609 | 276 | 1.36258 | 0.03474 | 2.52 | — | Organic acids |
| Terephthalic acid | 329 | 16.8005 | 312 | 1.54372 | 0.00730 | 2.85 | — | Organic acids |
| 3,4-Dihydroxyphenylacetic acid | 269 | 17.1885 | 223 | 1.22633 | 0.03820 | 2.75 | — | Organic acids |
| Succinate semialdehyde 1 | 296 | 9.18755 | 132 | 1.44052 | 0.01368 | 1.70 | — | Lipids |
| Citral | 228 | 11.7854 | 164 | 1.66867 | 0.00997 | 10.99 | — | Lipids |
| Hydroxyurea | 463 | 9.86407 | 244 | 1.71827 | 0.00288 | 1.46 | — | Others |
| Catechol | 227 | 11.0355 | 180 | 1.34772 | 0.03911 | 33.49 | — | Others |
| Malonamide 1 | 393 | 13.3798 | 101 | 1.76447 | 0.00064 | 9.48 | — | Others |
| Maleamate 2 | 521 | 13.8687 | 241 | 1.92868 | 0.00100 | 2 436 864 | — | Others |
| Flavin adenine degrad product | 617 | 16.1693 | 174 | 1.72068 | 0.00893 | 5.52 | — | Others |
| Pantothenic acid | 862 | 18.7844 | 201 | 1.94644 | 0.00009 | 2.20 | — | Others |
| Purine riboside | 680 | 22.1714 | 217 | 2.00513 | 0.00000 | 2.18 | — | Others |
R.T. represents the retention time.
VIP = variable importance projection, metabolites (VIP > 1) are listed in the table.
P values were calculated according to Student's T-test.
Fold change is represented as the ratio of the peak intensities for the corresponding treatments (n = 6), —: represented, not detected.
List of metabolites whose relative fold changes were significantly up-accumulated (≥1.5) in Cd-treated roots of cucumber seedlings.
| Metabolite name | Similarity | R.T.a | Mass | VIPb | P valuec | Fold changed | Pathway | |
|---|---|---|---|---|---|---|---|---|
| Cd vs. control | Cd + Se vs. Cd | |||||||
| Pyruvic acid | 955 | 7.2073 | 174 | 1.61302 | 0.00014 | 1.51 | — | Glycolysis |
| d-Glyceric acid | 950 | 11.1479 | 189 | 1.72617 | 0.00009 | 3.58 | 0.44 | Glycolysis |
| Glucose-1-phosphate | 827 | 16.4631 | 217 | 1.59138 | 0.00024 | 1.58 | 0.59 | Glycolysis |
| Shikimic acid | 896 | 16.9190 | 204 | 1.41137 | 0.00268 | 1.79 | 0.52 | Glycolysis |
| Fructose 1 | 807 | 17.5632 | 77 | 1.38707 | 0.00411 | 9.97 | — | Glycolysis |
| Galactose 1 | 923 | 17.7558 | 319 | 1.09648 | 0.04012 | 2.64 | 0.39 | Glycolysis |
| Succinic acid | 932 | 10.9674 | 147 | 1.76705 | 0.00000 | 3.04 | 0.5 | TCA cycle |
| Fumaric acid | 964 | 11.4638 | 245 | 1.68692 | 0.00026 | 4.59 | — | TCA cycle |
| α-Ketoglutaric acid | 844 | 14.3078 | 198 | 1.72267 | 0.00000 | 2.83 | 0.43 | TCA cycle |
| Erythrose 2 | 838 | 12.5637 | 205 | 1.62780 | 0.00070 | 3.13 | 0.62 | Sugars and polyols |
| l-Threose 1 | 871 | 12.7616 | 205 | 1.63895 | 0.00008 | 3.78 | 0.62 | Sugars and polyols |
| Digitoxose 1 | 435 | 14.5575 | 292 | 1.67296 | 0.00002 | 10.56 | 0.47 | Sugars and polyols |
| Lyxose 1 | 782 | 15.1044 | 217 | 1.47580 | 0.00135 | 1.70 | 0.52 | Sugars and polyols |
| Xylose 1 | 937 | 15.2770 | 103 | 1.55379 | 0.00188 | 1.78 | 0.46 | Sugars and polyols |
| Fucose 1 | 797 | 15.9018 | 160 | 1.16512 | 0.02620 | 1.56 | 0.48 | Sugars and polyols |
| Gluconic lactone 1 | 803 | 17.7089 | 220 | 1.64756 | 0.00004 | 2.74 | 0.57 | Sugars and polyols |
| Sorbitol | 869 | 18.2647 | 217 | 1.43477 | 0.00930 | 6.14 | — | Sugars and polyols |
| Isopropyl-beta-d-thiogalactopyranoside | 413 | 19.5198 | 204 | 1.67597 | 0.00002 | 3.29 | 0.17 | Sugars and polyols |
| Myo-inositol | 923 | 19.7238 | 217 | 1.37707 | 0.00483 | 1.54 | 0.43 | Sugars and polyols |
| Glucoheptonic acid 3 | 686 | 20.0160 | 217 | 1.48899 | 0.00081 | 1.51 | 0.50 | Sugars and polyols |
| d-Glucoheptose 1 | 745 | 20.0875 | 319 | 1.65849 | 0.00043 | 3.14 | 0.46 | Sugars and polyols |
| dl-Dihydrosphingosine 1 | 632 | 23.1955 | 321 | 1.42588 | 0.00820 | 2.20 | 0.5 | Sugars and polyols |
| Sucrose | 909 | 24.2913 | 451 | 1.29190 | 0.02344 | 36.11 | — | Sugars and polyols |
| Galactinol 1 | 895 | 26.8080 | 204 | 1.59005 | 0.00014 | 3.24 | 0.30 | Sugars and polyols |
| Maltotriose 1 | 660 | 30.4877 | 361 | 1.64543 | 0.00096 | 47.78 | 0.07 | Sugars and polyols |
| Alanine 1 | 950 | 7.9552 | 116 | 1.63181 | 0.00006 | 2.18 | — | Amino acids |
| N-Methyl-dl-alanine | 638 | 8.9614 | 130 | 1.66118 | 0.00003 | 1.87 | 0.54 | Amino acids |
| 1-Aminocyclopropanecarboxylic acid | 609 | 9.4863 | 202 | 1.66628 | 0.00037 | 2.93 | 0.55 | Amino acids |
| Valine | 967 | 9.5728 | 144 | 1.54758 | 0.00042 | 1.56 | 0.76 | Amino acids |
| 3-Hydroxynorvaline 2 | 368 | 11.5906 | 293 | 1.10472 | 0.03245 | 2.02 | — | Amino acids |
| l-Allothreonine 1 | 635 | 11.9003 | 217 | 1.58211 | 0.00022 | 1.59 | — | Amino acids |
| N-Ethylglycine 1 | 437 | 12.3651 | 218 | 1.32921 | 0.01241 | 2.15 | — | Amino acids |
| β-Alanine 2 | 913 | 12.4680 | 248 | 1.67022 | 0.00003 | 1.69 | 0.42 | Amino acids |
| N-Acetyl-l-leucine 1 | 459 | 13.5145 | 261 | 1.72048 | 0.00000 | 2.70 | 0.45 | Amino acids |
| Aspartic acid | 941 | 13.6366 | 232 | 1.38925 | 0.00415 | 1.71 | — | Amino acids |
| Oxoproline | 829 | 13.6978 | 258 | 1.60554 | 0.00016 | 1.64 | 0.60 | Amino acids |
| 4-Aminobutyric acid 1 | 930 | 13.7692 | 174 | 1.74058 | 0.00004 | 3.12 | — | Amino acids |
| β-Glutamic acid 1 | 290 | 14.6480 | 345 | 1.48911 | 0.00443 | 3.20 | 0.00 | Amino acids |
| Glutamic acid | 917 | 14.8221 | 246 | 1.74821 | 0.00000 | 2.98 | 0.62 | Amino acids |
| Glycocyamine 2 | 225 | 16.0810 | 273 | 1.72689 | 0.00000 | 3.55 | 0.57 | Amino acids |
| Glutamine 1 | 661 | 16.5729 | 156 | 1.27931 | 0.02618 | 14 254 790 | — | Amino acids |
| N-Acetyl-l-phenylalanine 2 | 760 | 17.1019 | 120 | 1.66929 | 0.00067 | 40 255 157 | 0.14 | Amino acids |
| Glycolic acid | 910 | 7.5776 | 177 | 1.22630 | 0.01784 | 1.55 | — | Organic acids |
| Sulfuric acid | 441 | 8.8714 | 281 | 1.44736 | 0.00198 | 1.60 | — | Organic acids |
| Malonic acid 1 | 838 | 9.4241 | 147 | 1.69945 | 0.00033 | 24.56 | 0.43 | Organic acids |
| 4-Hydroxybutyrate | 882 | 9.8991 | 233 | 1.50558 | 0.00080 | 4.13 | — | Organic acids |
| Benzoylformic acid 1 | 634 | 10.4634 | 222 | 1.74493 | 0.00011 | 1 625 262 | 0.11 | Organic acids |
| Tartronic acid | 533 | 11.8036 | 247 | 1.61541 | 0.00170 | 6.17 | 0.37 | Organic acids |
| Glutaric acid | 817 | 12.2055 | 55 | 1.65598 | 0.00085 | 15.41 | 0.24 | Organic acids |
| Threonic acid | 883 | 14.0867 | 292 | 1.79358 | 0.00000 | 7.92 | 0.50 | Organic acids |
| 3-Hydroxy-3-methylglutaric acid | 763 | 14.6068 | 109 | 1.74363 | 0.00000 | 2.27 | 0.57 | Organic acids |
| 2-Ketoadipate 2 | 359 | 15.3140 | 258 | 1.26494 | 0.02976 | 1 230 889 | 0.00 | Organic acids |
| Ciliatine | 290 | 16.2117 | 299 | 1.59016 | 0.00021 | 3.78 | 0.3 | Organic acids |
| 2-Amino-3-(4-hydroxyphenyl)propanoic acid 1 | 301 | 18.0784 | 267 | 1.49834 | 0.00622 | 60.64 | — | Organic acids |
| Gluconic acid 1 | 903 | 18.8386 | 333 | 1.60328 | 0.00144 | 4.39 | 0.42 | Organic acids |
| Citraconic acid 4 | 403 | 11.4894 | 221 | 1.69717 | 0.00001 | 1.63 | 0.75 | Lipids |
| Pelargonic acid | 603 | 11.6675 | 215 | 1.26183 | 0.02841 | 1 059 213 | 0.00 | Lipids |
| 4-Acetamidobutyric acid 2 | 187 | 13.3963 | 159 | 1.23334 | 0.01367 | 8.85 | 0.00 | Lipids |
| Carnitine | 296 | 9.6464 | 70 | 1.46006 | 0.00151 | 2.06 | 0.54 | Others |
| Hydroquinone | 225 | 12.1846 | 174 | 1.17552 | 0.04772 | 1 112 417 | 0.00 | Others |
| Methyl trans-cinnamate | 372 | 12.2422 | 103 | 1.76747 | 0.00000 | 8.05 | 0.39 | Others |
| 4-Aminophenol 1 | 330 | 13.9446 | 254 | 1.39339 | 0.01315 | 4 224 720 | 0.24 | Others |
| Malonamide 2 | 419 | 14.3802 | 329 | 1.52042 | 0.00072 | 3.68 | 0.61 | Others |
| 1,2-Cyclohexanedione 4 | 399 | 15.3965 | 171 | 1.67429 | 0.00002 | 7.06 | 0.46 | Others |
| 5,6-Dimethylbenzimidazole 2 | 269 | 17.1885 | 287 | 1.72488 | 0.00000 | 4.72 | 0.57 | Others |
| N-Acetyl-β-d-mannosamine 4 | 725 | 19.8829 | 103 | 1.50778 | 0.00087 | 3.55 | 0.36 | Others |
| Thioctamide 1 | 270 | 20.5994 | 174 | 1.25682 | 0.02745 | 97 517 | 0.00 | Others |
| Guanosine | 723 | 25.2860 | 245 | 1.14638 | 0.04856 | 3 700 082 | — | Others |
R.T. represents the retention time.
VIP = variable importance projection, metabolites (VIP > 1) are listed in the table.
P values were calculated according to Student's T-test.
Fold change is represented as the ratio of the peak intensities for the corresponding treatments (n = 6), —: represented, not detected.
Metabolic profiles in response to Cd and/or Se in cucumber leaves
The expression quantity of relative expressed metabolites among the control, Cd and Cd + Se treatments was evaluated by the fold change (FC) value. In the leaves of cucumber seedlings, 24 metabolites were up-accumulated or unchanged by Cd + Se vs. Cd treatment but down-accumulated by Cd treatment vs. the control (Table 1). According to the putative biochemical and physiological functions, these metabolites were mainly divided into six categories, namely TCA cycle (25%), sugars and polyols (25%), glycolysis (12.5%), amino acids (12.5%), lipids (8%) and others (17%) (Fig. S5a†). Among these 24 differently expressed metabolites, 18 metabolites showed higher expression after addition of Se (Cd + Se vs. Cd) to Cd-treated cucumber seedlings. The higher expressed metabolites may be related to Se-induced Cd tolerance in cucumber, which includes 3 glycolysis metabolites, pyruvic acid, galactose 1 and lactose 1; 1 TCA cycle-related metabolites, 3-hydroxypropionic acid 1; 4 sugars and polyols, d-(glycerol 1-phosphate), glucoheptonic acid 3, phytol and melibiose 2; 3 amino acids, alanine 1, glycine 2 and aminomalonic acid; 1 lipid, linoleic acid; and 4 others, including 4-hydroxybutyrate, nornicotine, phthalic acid, and N-acetyl-beta-d-mannosamine 4.
Compared with the control, 47 metabolites were up-accumulated under Cd treatment alone (Table 3 and Fig. S5c†), and the numbers of these metabolites in the different categories are as follows: amino acids (20), organic acids (8), sugars and polyols (6), glycolysis (3), lipids (2), TCA cycle-related metabolites (1) and others (7). The Cd-induced expression of some key metabolites was many-fold higher under Cd treatment vs. the control, such as l-cysteine, asparagine 1, oxalacetic acid, threitol, citrulline 1, citral, catechol, and maleamate 2; we assume that these metabolites are related to Cd accumulation.
Metabolic profiles in response to Cd and/or Se in cucumber roots
In roots, 26 metabolites were up-accumulated or unchanged by Cd + Se vs. Cd treatment and down-accumulated by Cd treatment vs. the control (Table 2), of which 31% were related sugars and polyols and 19% were pyridine alkaloids; these metabolites are more likely to play crucial roles in Se-mediated Cd tolerance in cucumber. They also include functional metabolites related to glycolysis (15%), amino acids (15%) and several organic acids (8%), along with TCA cycle-related metabolites (4%), lipids (4%) and others (4%) (Fig. S5b†).
In addition, 68 metabolites were up-accumulated after Cd treatment in roots (Table 4 and Fig. S5d†). Of these, 25% of the metabolites were amino acids, 25% were sugars and polyols, and 19% were organic acids; 7% were related to glycolysis (e.g. pyruvic acid, glucose-1-phosphate, galactose 1), 4% were related to the TCA cycle (α-ketoglutaric acid, succinic acid, fumaric acid), 4% were lipids, and 15% were others, suggesting that Cd treatment induces significant metabolic changes in cucumber seedlings.
Discussion
Currently, Cd contamination remains a global environmental problem due to the strong mobility and toxicity of Cd. Plants grown in Cd-contaminated soil or irrigated by Cd-contaminated water usually contain large amounts of Cd that exceed standards. Furthermore, Cd easily accumulates in edible organs, which consequently causes adverse effects on human health through the food chain.27,28 Cucumber is a vegetable crop that is grown worldwide for safe food production and human health; reliable strategies for reducing cucumber Cd contamination and inducing plant tolerance to Cd are urgently desired. Researchers have found that for large-scale slightly or moderately polluted farmlands, the addition of chemical substances, including allantoin,29 selenium,30 and hydrogen gas,31 can mitigate Cd toxicity and improve the growth of plants. Our previous studies showed that application of Se in hydroponic solution and soil could alleviate Cd toxicity by relieving lipid peroxidation of cucumber leaves induced by Cd stress, abating reactive oxygen species (ROS) accumulation, improving antioxidase and ATPase activities, preventing photosynthetic machinery damage, balancing the concentrations of carbohydrate contents and nutrient elements, and further up-inducing photosynthesis, metabolism or stress-related proteins; this ultimately reduced Cd accumulation in plants and improved Cd tolerance of cucumber seedlings.16,17 The present research further indicates that exogenous Se can mitigate cucumber Cd concentration and translocation.
However, to date, the effects of exogenous Se on the ultrastructural and metabolic patterns of Cd-treated cucumber seedlings have not been studied. Accordingly, to better understand how cucumber adapts to Cd toxicity at the ultrastructural and metabolic levels, in this research, we carried out ultrastructural observation and metabonomics analysis of cucumber seedlings under Cd and/or Se treatment using TEM and GC-MS, respectively. The results revealed the possible differences in the ultrastructural and metabolic profiles of cucumber under Cd and/or Se treatment and the potential specific metabolites that are responsive to Cd and/or Se stress in cucumber seedlings.
Protective effects of exogenous Se on the ultrastructures of chloroplast and root cells damaged by Cd
The chloroplast is not only the location of photosynthesis but is also a sensitive organelle to Cd stress. The TEM results showed that the chloroplasts were deformed and damaged by Cd stress; moreover, in the chloroplasts, the arrangements of the grana and stromal lamellae were disordered, and their number was significantly reduced compared with the control (Fig. 1). These phenomena are similar to those observed in barley.23 Additionally, our previous study reported that Cd stress significantly reduced the photosynthetic rate and chlorophyll content of cucumber.16,17 It can be concluded that Cd-induced changes in chloroplast structure may be the direct cause of damage to the photosynthetic system and the degradation of chlorophyll, which eventually leads to inhibition of photosynthesis. Also, Cd stress significantly reduced the accumulation of starch grains in chloroplasts, which may be caused by the decreased photosynthetic capacity of chloroplasts or excessive energy consumption of cells to resist Cd stress. Keunen et al. also reported that during Cd exposure, Arabidopsis leaves mainly utilised their starch reserves to provide energy for stress defence.2 According to the observation of the ultrastructures of root meristem cells, Cd stress injured the nuclear structures and cracked the karyotheca systems of the cucumber cells; it also reduced the number of vacuoles in the cucumber seedlings (Fig. 2). The addition of exogenous Se significantly alleviated the damage of the chloroplast and root cell structures caused by Cd stress; it recovered the damage of the chloroplast lamellar structure, increased the number of starch grains in the cucumber leaves, and effectively improved the stability and integrity of the nuclear membranes of root meristem cells (Fig. 1d and 2d), indicating that Se can protect the plant growth and ultrastructures of cucumber seedlings from Cd-induced damage. Accordingly, exogenous Se may be beneficial to improve photosynthesis efficiency; as reported in our previous work, exogenous Se (Cd + Se) prominently increased the chlorophyll a, chlorophyll b, and chlorophyll a + b content and improved photosynthetic performance, with higher Pn, Gs, and Tr, in plants treated with Cd + Se compared with those treated solely with Cd.15,16 It can be concluded that the membrane integrity effect of Se addition is a fundamental protective mechanism for Se-induced tolerance of Cd toxicity in cucumber.
Exogenous Se utilizes different strategies at the metabolome level for combating Cd stress
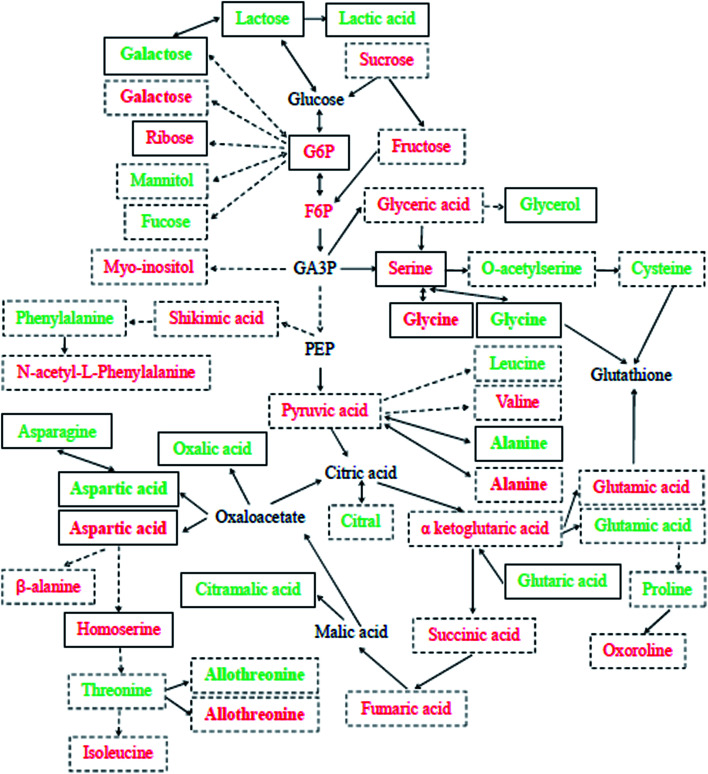
To date, no detailed metabolic evaluation of Se-improved cucumber tolerance to Cd contamination has been fully explored. In the present research, the effects of exogenous Se on the large-scale metabolic profiling of cucumber leaves and roots under Cd stress were examined. PCA analysis and the OPLS-DA model were used to identify all significantly different metabolites (VIP > 1, P < 0.05) among the three treatments (Fig. S3 and S4†); the results showed that there was a clear separation between Cd and the control as well as between Cd + Se and Cd treatment, indicating that Se and/or Cd exposure affected cucumber seedlings to a certain extent. By peak value comparison among the control, Cd and Cd + Se treatments, we found that 50 metabolites were significantly up-accumulated or unchanged by Cd + Se vs. Cd treatment and down-accumulated by Cd vs. the control, and 115 metabolites were significantly up-accumulated by Cd vs. the control. Furthermore, these differently expressed metabolites were identified by GC-TOF-MS; in addition, coupled with metabolic pathway analysis, a comprehensive schematic of Se-mediated tolerance to Cd toxicity in cucumber seedlings was proposed (Fig. 3), which may provide some new insight into the potential mechanisms underlying Se-improved tolerance to Cd contamination.
Fig. 3. Schematic of Cd + Se or Cd stress-responsive primary metabolites in the leaves (green) and roots (red) of cucumber seedlings. Each detected metabolite framed with continuous lines was significantly down-accumulated in Cd-treatment and up-accumulated or unchanged in Cd + Se treatment, and those framed with dotted lines were significantly up-accumulated in Cd-treated cucumber seedlings. Metabolites in bold were checked in both leaves and roots. The relative fold changes in abundance are shown in Tables 1–4.
Se + Cd treatment accumulated relatively higher levels of glycolysis metabolites in leaves, including pyruvic acid, galactose 1 and lactose 1 (Table 1). Pyruvic acid is an important intermediate in the sugar metabolism of all living cells and is also a key intermediate in the mutual transformation of a large number of substances in vivo, involving in several metabolic pathways. Generally, pyruvic acid can be made from glucose by glycolysis and converted back into carbohydrates (such as glucose) by gluconeogenesis or transformed into fatty acids by reaction with acetyl-CoA. Moreover, α-d-glucosamine 1-phosphate, glucose-6-phosphate 1, glucose-6-phosphate 2 and 6-phosphogluconic acid were up-accumulated in roots by Cd + Se treatment and down-accumulated under Cd treatment alone (Table 2); these are intermediates of the conversion of glucose to pyruvic acid through glycolysis. Lactose can be made from glucose and galactose, and lactose can also be degraded into galactose.
TCA cycle-related metabolites were largely responsible for the Se-induced resistance against Cd stress in cucumber. In leaves, lactic acid, oxalic acid, 3-hydroxypropionic acid 1,2,3-dimethylsuccinic acid, glutaric acid and citramalic acid were up-accumulated under Cd + Se vs. Cd treatment and down-accumulated under Cd treatment vs. the control (Table 1). Lactic acid is not only a metabolite produced by anaerobic metabolism but is also an important energy carrier. Glucose will be converted to lactic acid and then enter the tricarboxylic acid cycle (TCA cycle). Thus, lactic acid is probably the most important energy source in the TCA cycle. The data showed that lactic acid was up-accumulated by Cd + Se treatment in roots, 4.09 fold higher than with Cd treatment alone (Table 2). It is enticing to suggest that this high lactic acid expression in cucumber leaves and roots is a specific response to Cd + Se treatment; its higher accumulation in cucumber may facilitate greater Cd tolerance.
Sugars and polyols, organic acids and amino acids are intermediary metabolites that play roles of signals, regulators and antioxidants, amongst other functions.32–34 It was reported that under Cd stress, these intermediary metabolites exert some important functions in plants; for example, the specific accumulation of sugars, amino acids and organic acids can be used to identify important pathways of Cd detoxification in forage grasses.35,36 In the present study, a large number of sugars and polyols were up-accumulated or showed no obvious change after addition of Se (Cd + Se) compared with Cd treatment alone and were down-accumulated under Cd treatment vs. the control. In leaves, the sugar and polyol metabolite with the most significant change in concentration by Cd + Se was melibiose 2, which increased 4.56 fold compared to Cd treatment alone, followed by glucoheptonic acid 3, phytol, d-glycerol 1-phosphate, and 3-methylamino-1,2-propanediol 2. Meanwhile, in the roots, ribulose-5-phosphate 1, lactulose 1, lactitol, etc. were upregulated (Tables 1 and 2). In this sense, the high expression of these sugar-related metabolites may provide energy for developing Se-induced Cd tolerance in cucumber.
Amino acids and lipids are also essential metabolites that are responsible for Se-induced Cd tolerance in the leaves and roots of cucumber seedlings. The addition of Se significantly affected the expression of amino acids derived from pyruvic acid, e.g. the expressive abundance of alanine exhibited an obvious increase, as did glycine and aminomalonic acid, which were characterized by significantly high accumulation in Cd + Se leaves compared to Cd treatment alone (4.47 and 5.91 fold, respectively); moreover, in roots, oxamic acid, l-homoserine 1, glycine 2 and serine 1 were up-accumulated by Cd + Se treatment and down-accumulated under Cd treatment alone (Tables 1 and 2). Hediji reported that the level of alanine was reduced by 25% in young Cd-treated tomato leaves compared to a control;37 these results are similar to those of the present study (Tables 3 and 4). Meanwhile, Wu et al. reported that plants can adjust osmotic stress via accumulating high concentrations of compatible solutes, including glycine, proline and betaine.19 Although glycine is known as a metabolite with compatible solute properties, more importantly, it is also a component of glutathione biosynthesis, which is part of the antioxidant system.20,38 In the present study, the high abundance of alanine and glycine under Cd + Se treatment compared with Cd treatment alone may be important for Se-enhanced cucumber resistance to Cd toxicity. These results indicate that exogenous Se can induce high expression of amino acid and lipid-related metabolites, facilitating cucumber tolerance to Cd toxicity.
Comparative metabonomics reveals key metabolites associated with Cd tolerance
The metabolite changes in response to Cd treatment differed between tissues within the same species, which is similar to salt stress.39Fig. 3 summarizes the effects of Cd and/or Se on the primary metabolism in the leaves and roots of cucumber. An abundance of sugars (fructose, sucrose, myo-inositol and mannitol), several TCA cycle intermediates and pyruvic acid were highly accumulated after Cd treatment in both leaves and roots. Simultaneously, the expression levels of some amino acids and organic acids showed significant increases after Cd treatment compared with the control (Fig. S4c, d,†Tables 3 and 4). Furthermore, the expressive abundance of some metabolites differed between different cucumber tissues after Cd and/or Se stress.
Synthesis of amino acids, sugars and polyols, and organic acids is known to be beneficial to membrane stability and osmotic adjustment.22,24,40 In this study, the amino acid category was the largest section, followed by sugars and polyols and organic acids. The present results show that threose and sorbitol, which are acknowledged to be compatible osmotic adjustment substances, greatly accumulated after Cd treatment in roots and leaves compared with the control; meanwhile, sucrose, myo-inositol, thiogalactopyranoside, xylose, fucose, galactinol and maltotriose were root-specific compatible solutes, and mannitol, threitol, arabitol and sedoheptulose were leaf-specific solutes (Tables 3 and 4). It was reported that Cd-induced plant growth inhibition is correlated with high accumulation of soluble sugars in tomato and pea37,41 as well as in cucumber,17 and it was suggested that this carbohydrate accumulation is a result of the interference of metals in carbohydrate metabolism. Moreover, galactinol could scavenge ˙OH as efficiently as GSH.33,42 Therefore, the high expression of this sugar in Cd-treated cucumber roots may abate oxidative stress caused by Cd stress. Also, sucrose can be exported to sink organs,2 coinciding with increasing levels of sucrose concomitant to fructose and other sugars after Cd exposure in roots, which may be involved in regulating cucumber Cd toxicity; Cd may have been exported into the vacuoles of roots with the sugars. Fucose is a trehalose dihydrate; trehalose has high chemical stability and a hydrophilic structure,43 and it highly accumulates in response to chilling and freezing.44,45 Xylose has been reported to highly accumulate under cold stress in Arabidopsis;20 it is also a precursor for threitol biosynthesis.46 In the present study, the levels of fucose, xylose, threitol, etc. were up-accumulated after Cd treatment in cucumber seedlings; these metabolites may be contributors to cucumber Cd tolerance.
Some amino acids play important roles in metal tolerance and chelation in plants, and their synthesis pathways are involved in the detoxification of Cd.32,47 For instance, asparagine and lysine are synthesized from aspartate, which is produced by oxaloacetate. Meanwhile, asparagine has also been reported to accumulate metals under toxic metal stress; it can participate in the detoxification of Cd directly or through the biosynthesis of chelating peptides.48,49 The present study indicates that the expressive abundance of aspartic acid and asparagine was high after Cd treatment both in the leaves and roots of cucumber; this indicates that these highly expressed amino acids are substances that accumulate or chelate toxic Cd and enhance the Cd tolerance of cucumber. Moreover, the expressive abundance of valine both in leaves and roots and of isoleucine in leaves significantly increased after Cd treatment, as did that of phenylalanine; these results are similar to previous reports in tomato leaves.37 In addition, our data show that aspartic acid, valine, glutamic acid and allothreonine are the common compatible substances with high expression after Cd treatment both in cucumber leaves and roots, while proline, cysteine, asparagine, serine, phenylalanine, carbobenzyloxy-l-leucine, isoleucine, lysine, spermidine and creatine were leaf-specific. High accumulation of cysteine in Cd-treated leaves is essential for Cd tolerance; it is required for GSH, PCs and methionine synthesis.50 Similarly, it has been reported that proline highly accumulates in Cd-treated tomato leaves51 and that the increased expressive abundance of proline is related to the enhancement of stress tolerance,19 which is crucial for plants to respond to Cd stress.32 Proline is also a direct product of glutamate metabolism (Fig. 3), indicating that Cd stress facilitates proline biosynthesis; this may be due to the positive effects of Cd treatment on the expression of glutamic acid (Tables 3 and 4) and to the preferential use of glutamate in the metabolic route leading to the synthesis of chelators such as phytochelatins or glutathione.52 Consistent with these hypotheses, the present research indicates that glutamic acid increases in both leaves and roots and proline and cysteine increase in leaves simultaneously under Cd stress, suggesting that these amino acids play important roles in Cd tolerance or chelation in cucumber seedlings. These amino acids are also supposed to be compatible solutes which are involved in osmotic regulation, protecting proteins and cell membranes from ROS.53,54 Additionally, the high accumulation of leucine and isoleucine under Cd stress manifests that these branched-chain amino acids play an important role in Cd stress; for instance, they can maintain amino acid homeostasis or promote stress-induced protein synthesis.55,56 Spermidine is another important amino acid for plants grown under stress; it can act as a regulator, signal or antioxidant.57 Simultaneously, spermidine appears to be associated with processes that facilitate Cd accumulation by plants used for Cd phytoextraction.58 The abundance of accumulated spermidine in Cd-treated leaves was high, indicating its importance to Cd tolerance and accumulation in cucumber. The present study also showed that β-alanine, N-acetyl-l-leucine, oxoproline, N-acetyl-l-phenylalanine and some organic acids (glycolic acid, sulfuric acid, malonic acid, threonic acid, and gluconic acid) were highly accumulated in Cd-treated cucumber roots and were root-specific solutes; these amino acids may be involved in mitigation of Cd contamination, as observed in Arabidopsis.59
On the other side, the TCA cycle and glycolysis intermediates were highly accumulated after 7 days of Cd exposure in the leaves and roots of cucumber seedlings. Our data showed that d-galacturonic acid 2 and galactonic acid were highly expressed in leaves after Cd stress (Table 3). As is known, galacturonic acid is the component unit of pectic acid and the major component of pectin in the primary cell wall; it is involved in the metabolism of polysaccharides in the cell wall and affects the structure and function of plant cell walls, with functions in plant growth and development. Meanwhile, Sun et al. reported that the majority of Cd accumulated in the inner epidermis and endodermis of cell walls in barley plants.15 The present results indicate that galacturonic acid and galactonic acid are responsible for developing resistance to Cd stress in cucumber seedlings. The expressive abundance of fructose, galactose, glyceric acid, glucose-1-phosphate, pyruvic acid and shikimic acid in Cd-treated cucumber roots was significantly increased compared with that in the control; these metabolites are the primary intermediates of glycolysis. Meanwhile, succinic acid, fumaric acid and α-ketoglutaric acid in roots and oxalacetic acid in leaves were also expressed more after Cd treatment (Tables 3 and 4); they are the primary intermediates of the TCA cycle and may provide energy for the synthesis of amino acids and proteins.
Conclusions
In conclusion, exogenous Se significantly improved ultrastructural damage induced by Cd in the leaves and roots of cucumber seedlings. For example, chloroplasts under Cd + Se treatment showed more normal morphologies than those treated with Cd alone. The morphology of the root nucleus was better; the chromatin distribution was more uniform, and the cytoplasm was denser. Meanwhile, the size of the cytoplasmic vacuoles and the numbers of starch grains and plasmids almost returned to normal levels. On the other side, cucumber may have different Se-mediated Cd stress tolerance mechanisms, which is highly significant for improving the Cd resistance ability of cucumber and other vegetable crops. In the present study, metabolite profiling revealed that several intermediate products of glycolysis and the TCA cycle and some amino acids were prominently highly accumulated under Cd + Se treatment compared with Cd treatment alone and down-accumulated in Cd-treated cucumber seedlings compared with controls, such as pyruvic acid, galactose, lactose, glutaric acid and alanine in leaves, glucose-6-phosphate and serine in roots, and lactic acid and glycine in both leaves and roots; these may play dominant roles in the development of Se-mediated Cd tolerance. Moreover, high levels of sugars and polyols, amino acids and organic acids were up-accumulated in Cd-treated plants. Meanwhile, our data suggest that the high accumulation of fructose, shikimic acid, succinic acid, α-ketoglutaric acid and fumaric acid in roots is a Cd-specific response, indicating that these metabolites are important to the development of Cd resistance by cucumber. This study may provide a significant metabolic basis for Se-improved cucumber resistance to Cd stress, and the identified metabolites may present a new approach to finding new strategies for Cd detoxification of cucumber.
Abbreviations
- BNS
Basal nutrient solution
- BSTFA
Bis(trimethylsilyl)trifluoroacetamide
- Cd
Cadmium
- F6P
Fructose 6-phosphate
- FC
Fold change
- G6P
Glucose-6-phosphate
- GA3P
Glyceraldehyde 3-phosphate
- GC-TOF-MS
Gas chromatography coupled with time-of-flight mass spectrometry
- GC-MS
Gas chromatography-mass spectrometry
- H2O2
Hydrogen peroxide
- HCA
Hierarchical clustering analysis
- ICP-AES
Inductively coupled plasma atomic emission spectroscopy
- KEGG
Kyoto encyclopedia of genes and genomes
- Na2-EDTA
Disodium ethylenediamine tetraacetic acid
- OPLS-DA
Orthogonal projections to latent structures discriminant analysis
- PCA
Principal component analysis
- PEP
Phosphoenolpyruvic acid
- ROS
Reactive oxygen species
- Se
Selenium
- SEM
Scanning electron microscopy
- TCA
Tricarboxylic acid
- TIC
Total ion chromatogram
- TMCS
Chlorotrimethylsilane
- VIP
Variable importance for the projection
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the Key Research and Development Project of Shanxi Province (201903D221066), National Natural Science Foundation of China (31401319), Research Project Supported by Shanxi Scholarship Council of China (2017-083), Laboratory Construction Project of the Shanxi Science and Technology Department of China (2017, 2018, 2019), Program for the Top Young Academic Leaders of Higher Learning Institutions of Shanxi of China (20151006), Shanxi Province Science Foundation for Youths (2015021146), and the Scientific Research Foundation for Doctoral Scholars of Taiyuan University of Science and Technology of China (20182023).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra02866e
Notes and references
- Gzyl J. Gwóźdź E. A. Plant Cell, Tissue Organ Cult. 2005;80:59–67. doi: 10.1007/s11240-004-8808-6. [DOI] [Google Scholar]
- Keunen E. Florez-Sarasa I. Obata T. Jozefczak M. Remans T. Vangronsveld J. Fernie A. R. Cuypers A. Environ. Exp. Bot. 2016;124:64–78. doi: 10.1016/j.envexpbot.2015.11.015. [DOI] [Google Scholar]
- DalCorso G. Farinati S. Maistri S. Furini A. J. Integr. Plant Biol. 2008;50:1268–1280. doi: 10.1111/j.1744-7909.2008.00737.x. [DOI] [PubMed] [Google Scholar]
- Seth C. S. Remans T. Keunen E. Jozefczak M. Gielen H. Opdenakker K. Weyens N. Vangronsveld J. Cuypers A. Plant, Cell Environ. 2012;35:334–346. doi: 10.1111/j.1365-3040.2011.02338.x. [DOI] [PubMed] [Google Scholar]
- Shaw J. R. Dempsey T. D. Chen C. Y. Hamilton J. W. Folt C. L. Environ. Toxicol. Chem. 2006;25:182–189. doi: 10.1897/05-243R.1. [DOI] [PubMed] [Google Scholar]
- Spann N. C Aldridge D. Griffin J. L. Jones O. A. H. Aquat. Toxicol. 2011;105:589–599. doi: 10.1016/j.aquatox.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Muhammad N. Cai S. G. Shah J. M. Zhang G. P. Acta Physiol. Plant. 2016;38(12):277. doi: 10.1007/s11738-016-2296-2. [DOI] [Google Scholar]
- Haghighi M. A Teixeira da Silva J. Commun. Soil Sci. Plant Anal. 2016;47(2):142–155. doi: 10.1080/00103624.2015.1109650. [DOI] [Google Scholar]
- Lin L. Zhou W. H. Dai H. X. Cao F. B. Zhang G. P. Wu F. B. J. Hazard. Mater. 2012;235:343–351. doi: 10.1016/j.jhazmat.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Hawrylak-Nowak B. Dresler S. Wójcik M. Sci. Hortic. 2014;172:10–18. doi: 10.1016/j.scienta.2014.03.040. [DOI] [Google Scholar]
- Liu W. X. Shang S. H. Feng X. Zhang G. P. Wu F. B. Environ. Toxicol. Chem. 2015;34:92–99. doi: 10.1002/etc.2760. [DOI] [PubMed] [Google Scholar]
- Hawrylak-Nowak B. Biol. Trace Elem. Res. 2009;132(1–3):259–269. doi: 10.1007/s12011-009-8402-1. [DOI] [PubMed] [Google Scholar]
- Rahman M. M. Hossain K. F. B. Banik S. Sikder M. T. Akter M. Bondad S. E. C. Rahaman M. S. Hosokawa T. Saito T. Kurasaki M. Ecotoxicol. Environ. Saf. 2019;168:146–163. doi: 10.1016/j.ecoenv.2018.10.054. [DOI] [PubMed] [Google Scholar]
- Sun H. Y. Cao F. B. Wang N. B. Zhang M. Mosaddek A. I. Zhang G. P. Wu F. B. PLoS One. 2013;8(11):e79158. doi: 10.1371/journal.pone.0079158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Y. Chen Z. H. Chen F. Xie L. P. Zhang G. P. Vincze E. Wu F. B. BMC Plant Biol. 2015;15:259. doi: 10.1186/s12870-015-0648-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Y. Dai H. X. Wang X. Y. Wang G. H. Ecotoxicol. Environ. Saf. 2016;133:114–126. doi: 10.1016/j.ecoenv.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Sun H. Y. Wang X. Y. Wang Y. N. Wei Y. Y. Wang G. H. Span. J. Agric. Res. 2016;14(4):e1105. doi: 10.5424/sjar/2016144-10008. [DOI] [Google Scholar]
- Roessner U. Patterson J. H. Forbes M. G. Fincher G. B. Langridge P. Bacic A. Plant Physiol. 2006;142:1087–1101. doi: 10.1104/pp.106.084053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. Z. Cai S. G. Chen M. X. Ye L. Z. Chen Z. H. Zhang H. T. Dai F. Wu F. B. Zhang G. P. PLoS One. 2013;8(1):e55431. doi: 10.1371/journal.pone.0055431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F. Kopka J. Dale W. H. Zhao W. Schiller K. C. Gatzke N. Sung D. Y. Guy C. L. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L. Liang H. J. Shuman J. Shulaev V. Davletova S. Mittler R. Plant Physiol. 2004;134:1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K. Verpoorte R. Phytochem. Anal. 2010;21:4–13. doi: 10.1002/pca.1188. [DOI] [PubMed] [Google Scholar]
- Sun H. Y. Zhang X. H. He X. Y. Ahmed I. M. Cao F. B. Zhang G. P. Wu F. B. Plant Growth Regul. 2014;74(1):93–105. doi: 10.1007/s10725-014-9906-z. [DOI] [Google Scholar]
- He Y. Y. Mao C. X. Chen Z. Y. Wen H. Lu W. Wu H. D. Anim. Nutr. 2016;2(4):351–356. doi: 10.1016/j.aninu.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann A. Strassburg K. Erban A. Kopka J. Bioinformatics. 2008;24:732–737. doi: 10.1093/bioinformatics/btn023. [DOI] [PubMed] [Google Scholar]
- Xia J. Psychogios N. Young N. Wishart D. S. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloui A. Recorbet G. Gollotte A. Robert F. Valot B. Gianinazzi-Pearson V. Aschi-Smiti S. Dumas-Gaudot E. Proteomics. 2009;9:420–433. doi: 10.1002/pmic.200800336. [DOI] [PubMed] [Google Scholar]
- Rizwan M. Ali S. Adrees M. Rizvi H. Zia-ur-Rehman M. Hannan F. Qayyum M. F. Hafeez F. Ok Y. S. Environ. Sci. Pollut. Res. 2016;23:17859–17879. doi: 10.1007/s11356-016-6436-4. [DOI] [PubMed] [Google Scholar]
- Dresler S. Hawrylak-Nowak B. Kovacik J. Pochwatka M. Hanaka A. Strzemski M. Sowa I. Wojciak-Kosior M. Ecotoxicol. Environ. Saf. 2019;170:120–126. doi: 10.1016/j.ecoenv.2018.11.119. [DOI] [PubMed] [Google Scholar]
- Shekari L. Aroiee H. Mirshekari A. Nemati H. J. Plant Nutr. 2019;42(5):529–542. doi: 10.1080/01904167.2018.1554075. [DOI] [Google Scholar]
- Wang B. Bian B. Wang C. Li C. Fang H. Zhang J. Huang D. Huo J. Liao W. PLoS One. 2019;14(2):e0212639. doi: 10.1371/journal.pone.0212639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. S. Dietz K. J. J. Exp. Bot. 2006;57:711–726. doi: 10.1093/jxb/erj073. [DOI] [PubMed] [Google Scholar]
- Sun X. Zhang J. Zhang H. Ni Y. Zhang Q. Chen J. Guan Y. Chemosphere. 2010;78:840–845. doi: 10.1016/j.chemosphere.2009.11.045. [DOI] [PubMed] [Google Scholar]
- Keunen E. Peshev D. Vangronsveld J. Van Den Ende W. Cuypers A. Plant, Cell Environ. 2013;36:1242–1255. doi: 10.1111/pce.12061. [DOI] [PubMed] [Google Scholar]
- Xie Y. Hu L. Du Z. Sun X. Amombo E. Fan J. Fu J. PLoS One. 2014;9(12):e115279. doi: 10.1371/journal.pone.0115279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabêlo F. H. S. Azevedo R. A. Monteiro F. A. Water, Air, Soil Pollut. 2017;228:394. doi: 10.1007/s11270-017-3563-6. [DOI] [Google Scholar]
- Hediji H. Djebali W. Cabasson C. Maucourt M. Baldet P. Bertrand A. Zoghlami L. B. Deborde C. Moing A. Brouquisse R. Chaïbi W. Gallusci P. Ecotoxicol. Environ. Saf. 2010;73(8):1965–1974. doi: 10.1016/j.ecoenv.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Lu S. C. Biochim. Biophys. Acta. 2013;1830:3143–3153. doi: 10.1016/j.bbagen.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q. Li P. Ma S. Indu Rupassara S. Bohnert H. J. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Widodo Patterson J. H. Newbigin E. Tester M. Bacic A. Roessner U. J. Exp. Bot. 2009;60:4089–4103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi R. Munjral N. Gupta A. K. Kaur N. J. Exp. Bot. 2007;61:167–174. doi: 10.1016/j.envexpbot.2007.05.006. [DOI] [Google Scholar]
- Nishizawa A. Yabuta Y. Shigeoka S. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Gómez M. and Lluch C., Trehalose and abiotic stress tolerance, in Abiotic Stress Responses in Plants, ed. P. Ahmad and M. N. V. Prasad, Springer, New York, 2012, pp. 253–265 [Google Scholar]
- Pramanik M. H. R. Imai R. Plant Mol. Biol. 2005;58:751–762. doi: 10.1007/s11103-005-7404-4. [DOI] [PubMed] [Google Scholar]
- Min K. Showman L. Perera A. Arora R. Environ. Exp. Bot. 2018;156:214–227. doi: 10.1016/j.envexpbot.2018.09.011. [DOI] [Google Scholar]
- Walters K. R. Pan Q. Seriannl A. S. Duman J. G. J. Biol. Chem. 2009;284:16822–16831. doi: 10.1074/jbc.M109.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. L. J. Exp. Bot. 2002;53:1–11. doi: 10.1093/jexbot/53.366.1. [DOI] [PubMed] [Google Scholar]
- Lea P. J. Sodek L. Parry M. A. J. Shewry P. R. Halford N. G. Ann. Appl. Biol. 2007;150:1–26. doi: 10.1111/j.1744-7348.2006.00104.x. [DOI] [Google Scholar]
- Costa G. Spitz E. Plant Sci. 1997;128:131–140. doi: 10.1016/S0168-9452(97)00148-9. [DOI] [Google Scholar]
- Cobbett C. Goldsbrough P. Annu. Rev. Plant Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- Chaffei C. Pageau K. Suzuki A. Gouia H. Ghorbel M. H. Masclaux-Daubresse C. Plant Cell Physiol. 2004;45:1681–1693. doi: 10.1093/pcp/pch192. [DOI] [PubMed] [Google Scholar]
- Pavlikova D. Pavlik M. Staszkova L. Motyka V. Szakova J. Tlustos P. Balik J. Ecotoxicol. Environ. Saf. 2008;70:223–230. doi: 10.1016/j.ecoenv.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Bohnert H. J. Nelson D. E. Jensen R. G. Plant Cell. 1995;7:1099–1111. doi: 10.2307/3870060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Ma Y. Huang C. Wan Q. Li N. Bi Y. Planta. 2008;227:611–623. doi: 10.1007/s00425-007-0643-7. [DOI] [PubMed] [Google Scholar]
- Joshi V. Joung J. G. Fei Z. Jander G. Amino Acids. 2010;39:933–947. doi: 10.1007/s00726-010-0505-7. [DOI] [PubMed] [Google Scholar]
- Araújo W. L. Ishizaki K. Nunes-Nesi A. Larson T. R. Tohge T. Krahnert I. Witt S. Obata T. Schauer N. Graham I. A. Leaver C. J. Fernie A. R. Plant Cell. 2010;22:1549–1563. doi: 10.1105/tpc.110.075630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukabe Y. He L. Nada K. Misawa S. Ihara I. Tachibana S. Plant Cell Physiol. 2004;45:712–722. doi: 10.1093/pcp/pch083. [DOI] [PubMed] [Google Scholar]
- Gong G. X. Liu Y. Huang D. Zeng G. Liu S. Tang H. Zhou L. Hu X. Zhou Y. Tan X. Environ. Sci. Pollut. Res. 2016;23:8699–8708. doi: 10.1007/s11356-016-6122-6. [DOI] [PubMed] [Google Scholar]
- Hanson A. D. Rathinasabapathi B. Rivoal J. Burnet M. Dillon M. O. Gage D. A. Proc. Natl. Acad. Sci. U. S. A. 1994;91:306–310. doi: 10.1073/pnas.91.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.