Abstract
Extended-spectrum β-lactamases (ESBLs), e.g., ESBLs of the TEM or SHV type, compromise the efficacies of expanded-spectrum cephalosporins. An SHV non-ESBL that hydrolyzes only narrow-spectrum cephalosporins can be converted into an SHV ESBL through substitutions at three amino acid positions, 179, 238, or 238–240. In order to improve detection of SHV ESBLs, a novel method, based on real-time PCR monitored with fluorescently labeled hybridization probes and followed by melting curve analysis, was developed. It is able to (i) detect blaSHV genes with high degrees of sensitivity and specificity, (ii) discriminate between blaSHV non-ESBL and blaSHV ESBL, and (iii) categorize the SHV ESBL producers into three phenotypically relevant subgroups. This method, termed the SHV melting curve mutation detection method, represents a powerful tool for epidemiological studies with SHV ESBLs. It even has the potential to be used in the diagnostic microbiology laboratory, because up to 32 clinical isolates can be processed in less than 1 h by starting with just a few bacterial colonies.
The production of extended-spectrum β-lactamases (ESBLs) of the TEM or SHV type by bacterial pathogens is a major threat to the use of the clinically important expanded-spectrum cephalosporins. Since 1983 (19, 20), clinical isolates producing SHV-2 or related ESBLs have increasingly been reported. SHV ESBLs are derived through single amino acid substitutions from a narrow-spectrum cephalosporin-hydrolyzing enzyme, SHV-1. Over 20 SHV ESBLs, designated SHV-2 through SHV-26, have been described in the literature (6, 14, 32) and on an Internet site (http://www.lahey.org/studies/webt.htm). Since the responsible genes are often easily transferable due to their localization on plasmids (34), the situation has recently been called a “plague of plasmids” (12). SHV enzymes have frequently been found in the widespread pathogens Klebsiella, Escherichia, and Salmonella (11), rarely in other members of the family Enterobacteriaceae, and once, recently, in Pseudomonas aeruginosa (26). Phenotypic differences due to various substitutions within the ESBLs were noted early and were found to be responsible for failure of treatment with expanded-spectrum cephalosporins (16, 21).
In order to improve the phenotypic detection of ESBL production, standard susceptibility tests have been refined (3, 13, 15, 17, 36, 37), including two commercially available tests (7, 10). Other investigators developed molecular biology-based methods, such as oligotyping (23) and PCR-restriction fragment length polymorphism analysis (2), for differentiation of different TEM ESBLs, a family of enzymes analogous to the SHV ESBLs. For detection of SHV ESBLs, methods based on single-strand conformation polymorphism analysis (8, 25) and PCR-NheI restriction analysis (30) and two tests based on the ligase chain reaction were elaborated (18, 28).
Despite considerable effort for over a decade, detection of ESBLs still remains a problem due to the notoriously low sensitivities of easy-to-perform susceptibility tests or to the small range of application as well as the laboriousness of molecular biology-based tests (24, 38; D. L. Patterson and V. L. Yu, Editorial response, Clin. Infect. Dis. 29:1419–1422, 1999).
We describe a PCR that uses special fluorescently labeled oligonucleotide hybridization probes on a LightCycler instrument. We demonstrate a rapid, sensitive, and specific method for detection of mutations in all three crucial codons (at positions 179, 238, and 240) of the blaSHV gene in a single reaction. By starting with raw bacterial growth on primary isolation media, this method allows one to conclude in less than 1 h whether a strain harbors an SHV ESBL and, if so, the phenotypic subgroup to which it belongs.
MATERIALS AND METHODS
Bacterial strains. (i) Standard strains.
Five well-characterized (29) derivatives of Escherichia coli DH5α, MPA-1, MPA-2, MPA-5, and MPA-8, carrying multicopy plasmids encoding SHV-1, SHV-2, SHV-5, and SHV-8, respectively, were used as standards to develop and optimize the novel detection method based on analysis with a LightCycler instrument (Roche Diagnostics). SHV β-lactamase-free strain E. coli DH5α was used as a negative control.
(ii) Clinical isolates.
A set of six clinical isolates of Klebsiella pneumoniae or E. coli, described earlier (31) and carrying genes for SHV β-lactamases SHV-1, SHV-2, SHV-2a, SHV-5, SHV-11, and SHV-12, were used to evaluate the novel detection method.
DNA preparation. (i) Preparation of plasmids from standard strains.
Plasmid DNA was prepared with the Qiagen plasmid kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions.
(ii) Simplified preparation of DNA from standard strains and clinical isolates.
A loopful of bacteria harvested from an agar plate was suspended in 50 μl of sterile water and heated to 95°C for 10 min. Centrifugation at 16,000 × g was performed for 10 min, just before 1 μl of the DNA-containing supernatant was pipetted into the PCR master mixture.
Mutation detection using FRET.
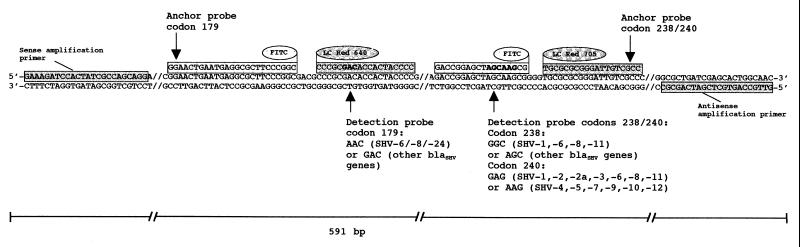
Fluorescence monitoring is based on the concept that a fluorescence signal is generated only if two components, a donor and a acceptor, of a fluorophore system come into contact closely enough to allow fluorescence resonance energy transfer (FRET). This is achieved by attaching the two fluorophores to two oligonucleotide probes designed to hybridize to a target strand leaving a gap no larger than 5 nucleotides wide. Typically, the upstream probe carries the donor fluorophore (fluorescein isothiocyanate [FITC]) at its 3′ end, while the downstream probe is labeled with the acceptor, LightCycler Red 640 or Red 705 (Fig. 1). During the run in the LightCycler instrument, the fluorescein is excited by a light-emitting diode light source and emits light with a wavelength of 640 or 710 nm that excites the acceptor fluorophore. The acceptor fluorophore finally emits light of a greater wavelength, which is measured. This allows monitoring of the amplification process on a per-cycle basis, because the intensity of the FRET signal is proportional to the amount of PCR product generated. Even more important, single mutations can be detected if either the up- or the downstream probe is designed as a shorter “detection probe” spanning the mutation site, while the second one is a longer “anchor probe” (Fig. 1). Known point mutations destabilize binding of the detection probe and, hence, cause a characteristic decrease in the melting temperature (Tm). Being longer, the anchor probe's Tm is always higher than that of the detection probe, thus ensuring FRET.
FIG. 1.
Relative orientations of amplification primers, anchors, and detection probes targeted to three different mutation sites of the β-lactamase gene. The sequences of the detection probes were designed to match that of blaSHV-5 (mutation-prone codons for amino acids 179, 238, and 240 are highlighted in boldface type). The detection probe spanning the mutation site at position 179 was labeled at its 5′ end with LightCycler Red 640 dye and phosphorylated at its 3′ end to block extension. The corresponding anchor carried a fluorescein label (FITC) at its 3′ end. The detection probe covering positions 238 and 240 simultaneously was labeled at its 3′ end with FITC. The corresponding anchor was labeled at its 5′ end with LightCycler Red 705 dye and was phosphorylated at its 3′ end to block extension. The use of two acceptor fluorophores, LightCycler Red 640 and LightCycler Red 705, allowed simultaneous detection of all three important mutations in a single reaction in one tube.
(i) Design of amplification primers.
The forward (24-mer, Tm = 61°C) and the reverse primer (22-mer, Tm = 63°C) were custom synthesized by Microsynth (Balgach, Switzerland). They were used to amplify a 591-bp piece of the blaSHV open reading frame (Fig. 1) spanning nucleotide positions 339 to 929 of the strain with EMBL data bank accession no. X98102.
(ii) Design of fluorogenic probes.
The probe used to detect the mutation at codon 179 was a 19-mer oligonucleotide labeled with LightCycler Red 640 at the 5′ end and phosphorylated at the 3′ end to block extension. Its sequence (Fig. 1) was taken from the defining part of the ESBL blaSHV-5 (Tm = 62°C) and, therefore, is 100% homologous to β-lactamases without mutations at position 179. The corresponding FITC-labeled anchor probe at the 3′ end was a 25-mer (Tm = 67°C) (Fig. 1) that binds to the template strand at a distance of four bases to the bound detection probe.
Analogously, the detection and anchor probes used for detection of the defining mutations at positions 238 and 240 were an 18-mer (tm = 57°C) and a 19-mer (Tm = 62°C), respectively, with their binding sites separated by 3 bp (Fig. 1). Again, the detection probe sequence was designed to fit a double mutant, blaSHV-5, with mutations at positions 238 and 240.
All fluorophore-labeled probes were synthesized and purified by reversed-phase high-pressure liquid chromatography by TipMolBiol (Berlin, Germany).
(iii) Sample preparation.
PCR was performed by rapid cycling in a reaction volume of 10 μl with each amplification primer at a concentration of 0.5 μM and each detection and anchor probe at a concentration of 0.2 μM. LightCycler-FastStart DNA Master Hybridization Probe Buffer (Roche Molecular Biochemicals, Mannheim, Germany) was basically used. The final Mg2+ concentration in the reaction mixture was adjusted to 5 mM. To complete the PCR mixtures, 9 μl of the modified master mixture and 1 μl of a DNA preparation were loaded into glass capillary cuvettes (Roche Molecular Biochemicals, Mannheim, Germany). After a short centrifugation (3,000 × g for 10 s), the sealed capillaries were placed into the LightCycler rotor.
(iv) Real-time PCR and melting curve analysis.
After an initial polymerase activation and denaturation step at 95°C for 5 min, the samples underwent 40 amplification cycles, each comprising denaturation (95°C for 20 s), annealing (65°C for 10 s), and extension (72°C for 30 s) in the LightCycler instrument. The temperature transition rates were programmed at 20°C/s; and the fluorimeter gains were set with F1 equal to 1 (at 530 nm, measured in channel 1), F2 equal to 15 (at 640 nm, measured in channel 2), and F3 equal to 45 (at 710 nm, measured in channel 3). Fluorescence was measured at the end of the annealing period of each cycle to monitor the progress of amplification. After completion, a melting curve was recorded by cooling to 35°C at 20°C/s, holding at 35°C for 30 s, and then heating slowly at 0.2°C/s until 85°C. Fluorescence was measured continuously during the slow temperature rise to monitor dissociation of (i) the LightCycler Red 640-labeled detection probe at fluorescence F2 and (ii) the LightCycler Red 705-labeled detection probe at fluorescence F3. Fluorescence signals from both F2 and F3 were plotted automatically in real time versus temperature (T) to produce melting curves for mutations at position 179 (F2 versus T) and at positions 238 and 240 (F3 versus T). Melting curves were then converted, again automatically, into melting peaks by plotting the negative derivative of fluorescence versus T (−dF2/dT versus T and −dF3/dT versus T). The entire process took approximately 40 min.
Control analysis of amplification product.
In order to check the sizes of selected amplification products, the capillaries were opened after the run in the LightCycler instrument and placed upside down in Eppendorf tubes. After a brief centrifugation, 10 μl of each sample was analyzed by agarose gel electrophoresis (0.8% agarose, 1 mg of ethidium bromide per ml; 30 min at 4 V/cm).
RESULTS
Detection of blaSHV mutations in standard strains.
Both fluorescence monitoring of product accumulation and detection of the mutations responsible for the ESBL phenotype in SHV β-lactamases were achieved by DNA amplification and subsequent melting-curve analysis by FRET.
The fluorescence signals rose above the background levels after about 15 cycles. No increase in fluorescence signal was observed with the negative control template of E. coli DH5α during the whole run of 40 cycles (data not shown).
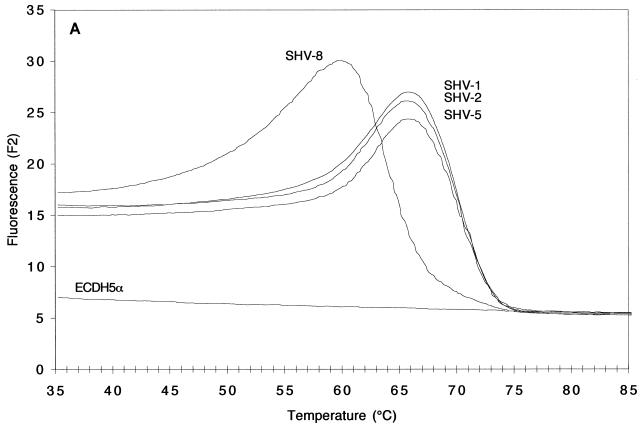
The melting curves obtained with DNA extracted from the standard strains expressing SHV-1, SHV-2, SHV-5, or SHV-8 are depicted in Fig. 2. Figures 2A and 2B illustrate the results obtained with the LightCycler Red 640 dye in channel 2 and targeted to the mutation at codon 179 in SHV-6 and SHV-8. The detection probe, when hybridized to blaSHV-8, started to dissociate at a temperature as low as 60 to 61°C because of the mismatch present (Fig. 2A). In contrast, melting of the same probe from blaSHV-1, blaSHV-2, and blaSHV-5 did not begin before T had risen by 6°C to 66 to 67°C, due to a perfect match. Changes in the fluorescence signals above 75°C were minimal, because all of the probe was dissociated from its target sequence. The peaks obtained after mathematical transformation, at 64°C for blaSHV-8 and 70°C for blaSHV-1, blaSHV-2, blaSHV-5, reflected an equivalent temperature shift of 6°C (Fig. 2B).
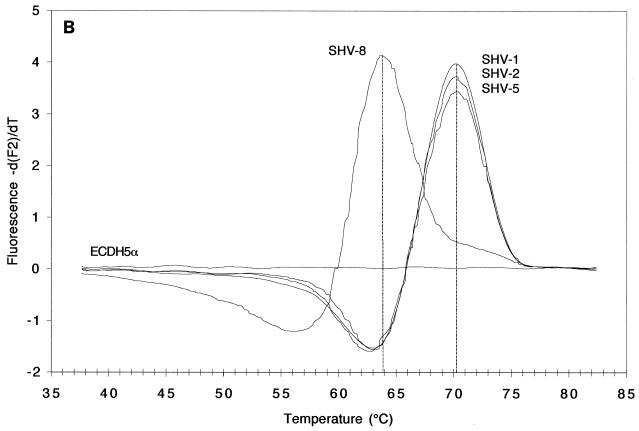
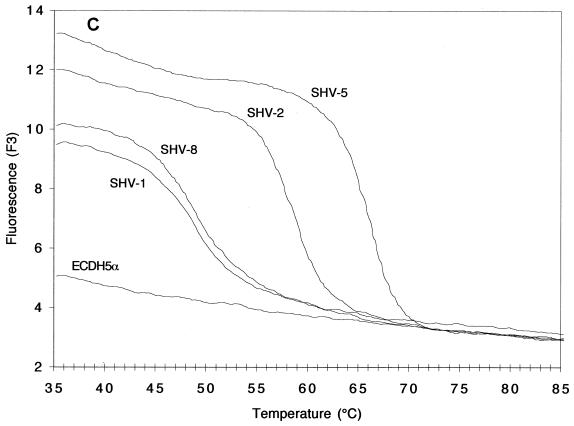
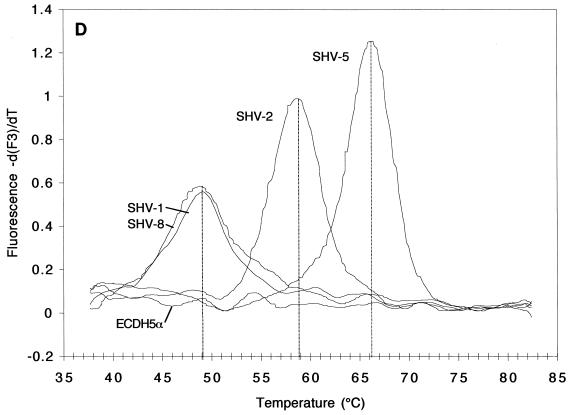
FIG. 2.
Melting curves (A and C; fluorescence F2 or F3 versus T) and melting peaks (B and D; plotted as the negative derivative of fluorescence F2 or F3 versus T) of blaSHV genes in standard strains. Fluorescence F2 (A and B) generated by the fluorophore of the mutation at codon 179 and recorded by channel 2, revealed melting peaks at higher T's for strains harboring blaSHV-1, blaSHV-2, and blaSHV-5 than for those harboring blaSHV-8, the representative with a mutation at codon 179. Fluorescence F3 (C and D), generated by the fluorophore of the mutation at codons 238 and 240 and recorded by channel 3, revealed melting peaks at three different T's: below 50°C for strains harboring blaSHV-1 and blaSHV-8 (two mismatches), ca. 59°C for strains harboring blaSHV-2 (one mismatch), and ca. 66°C for the strain expressing blaSHV-5 (no mismatches).
Remarkable changes in fluorescence F3 of the LightCycler Red 705 dye, targeted to the mutations at codons 238 and 240, were noted, depending on the blaSHV involved (Fig. 2C and D). After mathematical transformation, three characteristic and well-separated Tm peaks became apparent. As expected, the highest Tm of 66°C was produced by the blaSHV-5 template because its sequence matched that of the detection probe 100% (Fig. 2D). With blaSHV-2, the detection probe detached at a temperature that was 7°C lower (59°C) because of a lack of a mutation at codon 240, creating one mismatch. Finally, blaSHV-1 and blaSHV-8, with two mismatches compared to the sequence of the detection probe, dissociated at 49°C, a temperature that was another 10°C lower.
In order to reduce the time and labor necessary for plasmid preparation, we minimized the method for total DNA preparation. The resulting simplified protocol (see Materials and Methods) yielded data of the same quality as those shown in Fig. 2 (data not shown).
Detection of blaSHV genes in clinical isolates.
Clinical isolates were grown and processed by the simplified protocol and then analyzed as described above for the standard strains. Fluorescence F2 emitted by the hybridization probes targeted to the mutation at codon 179 was mathematically transformed and is shown in Fig. 3A. As expected, all clinical isolates tested showed identical melting peaks, because none of them harbored a mutation at codon 179.
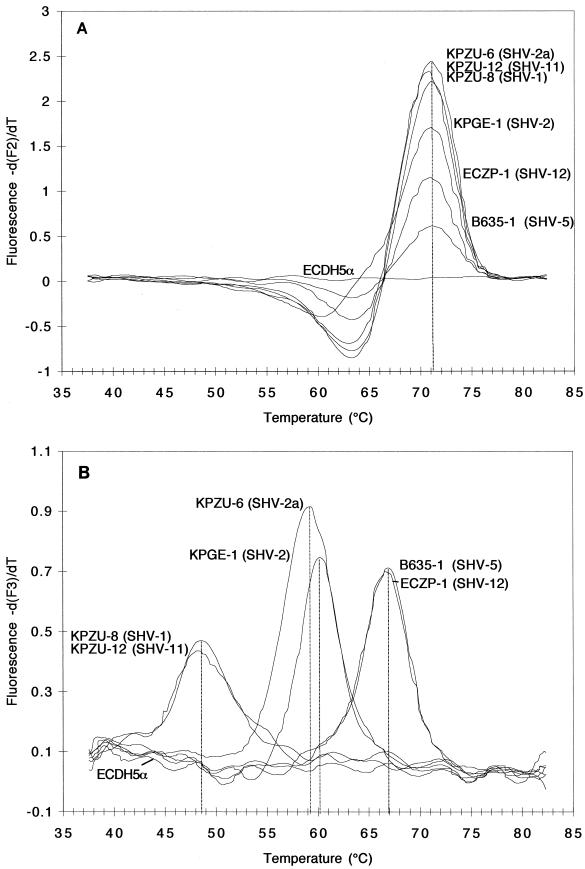
FIG. 3.
Melting peaks (A and B; plotted as the negative derivative of fluorescence F2 or F3 versus T) of blaSHV genes in clinical isolates. Fluorescence F2 (A), generated by the fluorophore of the mutation at codon 179 and recorded by channel 2, revealed identical melting peaks for all clinical isolates with respect to codon 179, indicating that none of them harbored a mutation at this site. Fluorescence F3 (B), generated by the fluorophore of the mutation at codons 238 and 240 and recorded by channel 3, revealed melting peaks at three different temperatures: below 50°C for strains harboring blaSHV-1 and blaSHV-11, ca. 60°C for strains harboring blaSHV-2 and blaSHV-2a, and ca. 67°C for strains expressing blaSHV-5 and blaSHV-12.
Clear temperature shifts were observed, however, in fluorescence F3, targeted to the mutations at positions 238 and 240 (Fig. 3B). Isolates harboring blaSHV-5 and blaSHV-12, two members of the subgroup of SHV ESBLs carrying mutations at both positions 238 and 240, showed the highest Tm peak (67°C) because of 100% identity to the detection probe. Producers of SHV-2 and SHV-2a, which belong to the subgroup of SHV ESBLs carrying one important substitution at position 238, showed an approximately 7°C lower Tm peak at 60°C because of one mismatch relative to the sequence of the detection probe at codon 240. Finally, the producers of SHV non-ESBLs, SHV-1 and SHV-11, which harbored two mismatches compared to the SHV-5-specific sequence of the detection probe, yielded Tm peaks at approximately 49°C.
Statistical analysis of 10 determinations of the Tm of detection probe LightCycler Red 705 with blaSHV-1 and blaSHV-8 yielded a mean Tm of 48.9 ± 0.3°C. Taking into account the temperature shifts of about 6°C per mismatch, the high degree of reproducibility and the reliability of mutation detection are obvious.
Additionally, monitoring by gel electrophoresis (data not shown) revealed that the amplification products had the expected length of 591 bp and were absent for the negative controls.
Therefore, with the limited number of strains processed in the present study, the novel method reached 100% sensitivity and 100% specificity for detection and discrimination of blaSHV ESBL and blaSHV non-ESBL genes in standard strains and clinical isolates.
DISCUSSION
Although ESBL production likely leads to failure of treatment with expanded-spectrum cephalosporins, MICs for ESBL producers may be increased insignificantly compared to those for fully susceptible variants (16, 21). Consequently, detection of ESBLs in clinical isolates is difficult. Indeed, the investigators in a recent study estimated that up to 33% of ESBLs in Europe may go undetected (22).
A number of easy-to-carry-out tests, mostly based on synergy between clavulanic acid and an expanded-spectrum cephalosporin, were recommended between 1988 and 1996 (10, 15, 17, 36), but their sensitivity and specificity were less than optimal. This prompted the National Committee for Clinical Laboratory Standards to establish a working group to address the problem (1). Although the activities thus evoked led to improved recommendations (27), the principal problems of synergy testing, (i) limited sensitivity and (ii) the requirement for an overnight incubation, remained.
Detection of ESBLs at the genetic level represents an alternative, independent of the degree of gene expression by the strain involved, as recently reviewed (9). One such method, called oligotyping, is based on colony hybridization with specially designed oligonucleotides that discriminate between single-mutation variants (23) of TEM ESBLs. Another one uses single-strand conformational polymorphism analysis-PCR to detect particular blaSHV genes. It is based on the observation that the migration of small single-stranded DNA molecules in nondenaturing gels is affected by conformational changes caused by point mutations (25, 35). Both methods are precise and can detect specific enzymes. A further method, the PCR-NheI test for SHV ESBLs (30), uses conventional PCR with subsequent restriction enzyme digestion.
The disadvantages of the molecular biology-based methods, such as labor expense, costliness, and the lack of general applicability, have outweighed their advantages of the and have so far prevented their broad acceptance.
Most of these disadvantages are overcome by the novel method presented here, termed SHV melting curve mutation detection (MCMD). The method works perfectly with small or single-copy DNA templates obtained from crude extracts of bacterial colonies on plates. Moreover, SHV MCMD is performed in a closed system, with no postamplification manipulations such as restriction digestion or electrophoresis necessary. Consequently, the results are available in less than 1 h after the harvesting of suspected bacterial colonies, and possible end-product contamination and sample tracking errors are eliminated. The proposed SHV MCMD assay has been shown to be reliable, sensitive, and specific. All mutations published so far could easily and clearly be identified by generation of characteristic and well-separated melting peaks. Genes of β-lactamases encoded on uncharacterized low-copy-number plasmids were analyzed equally well as those encoded on high-copy-number plasmids in laboratory mutants. Thus, accurate detection of blaSHV genes is possible regardless of the vastly variable amounts of template DNA available from clinical isolates. The test differentiates between SHV non-ESBLs (SHV-1 and SHV-11) that hydrolyze only narrow-spectrum cephalosporins and SHV ESBLs that are able to inactivate expanded-spectrum cephalosporins. Moreover, although no individual enzymes are determined, the method distinguishes between representatives of all three phenotypically relevant SHV ESBL subgroups. These subgroups are (i) SHV-6 and SHV-8, weak ESBLs that cause only weak ceftazidime resistance; (ii) SHV-2, SHV-2a, and SHV-3, which cause significant resistance to cefotaxime and ceftriaxone and moderate resistance to ceftazidime; and (iii) SHV-4, SHV-5, SHV-9, SHV-10, and SHV-12, which is the subgroup that is the most effective against all expanded-spectrum cephalosporins (33). However, the technology requires a LightCycler instrument, which is not normally part of the standard equipment found in research and diagnostic laboratories at this time.
In addition, SHV MCMD has the potential for extension as the family of SHV β-lactamases evolves. Any further mutation of phenotypic relevance can be implemented into the scheme of the method simply by designing an appropriate detection-anchor probe and processing the sample DNA in an additional tube. A valuable improvement of this kind will be a probe for a second version of the codon for amino acid Lys240. Lys240 is encoded by AAG in the original blaSHV-5 gene but is encoded by AAA in blaSHV-7 (4) and in two SHV-5 homologues found in K. pneumoniae isolates KPLA-4 and KPGE-2 (31). Since TEM ESBLs are also derived through acquisition of single point mutations, the MCMD method could also be applied for discrimination between TEM non-ESBLs and TEM ESBLs (a study is being planned).
In conclusion, ease, speed, and reliability render the SHV MCMD method a powerful tool for important (5) epidemiological studies concerning SHV ESBLs and make it a serious candidate for implementation into routine diagnostics.
ACKNOWLEDGMENT
This work was supported by the Swiss National Foundation (grant 3200-52532.97).
REFERENCES
- 1.Anonymous. Problems in testing for β-lactamases still unsettled. ASM News. 1995;61:391–392. [Google Scholar]
- 2.Arlet G, Brami G, Decre D, Flippo A, Gaillot O, Lagrange P H, Philippon A. Molecular characterisation by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol Lett. 1995;134:203–208. doi: 10.1111/j.1574-6968.1995.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradford P A, Sanders C C. Development of test panel of β-lactamases expressed in a common Escherichia coli host background for evaluation of new β-lactam antibiotics. Antimicrob Agents Chemother. 1995;39:308–313. doi: 10.1128/aac.39.2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford P A, Urban C, Jaiswal A, Mariano N, Rasmussen B A, Projan S J, Rahal J J, Bush K. SHV-7, a novel cefotaxime-hydrolyzing β-lactamase, identified in Escherichia coli isolates from hospitalized nursing home patients. Antimicrob Agents Chemother. 1995;39:899–905. doi: 10.1128/aac.39.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K. Is it important to identify extended-spectrum β-lactamase-producing isolates? Eur J Clin Microbiol Infect Dis. 1996;15:361–364. doi: 10.1007/BF01690090. [DOI] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter M W, Oakton K J, Warner M, Livermore D M. Detection of extended-spectrum β-lactamases in klebsiellae with the Oxoid combination disk method. J Clin Microbiol. 2000;38:4228–4232. doi: 10.1128/jcm.38.11.4228-4232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanawong A, M'Zali F H, Heritage J, Lulitanond A, Hawkey P M. Characterisation of extended-spectrum β-lactamases of the SHV family using a combination of PCR-single strand conformational polymorphism (PCR-SSCP) and PCR-restriction fragment length polymorphism (PCR-RFLP) FEMS Microbiol Lett. 2000;184:85–89. doi: 10.1111/j.1574-6968.2000.tb08995.x. [DOI] [PubMed] [Google Scholar]
- 9.Cockerill F R. Genetic methods for assessing antimicrobial resistance. Antimicrob Agents Chemother. 1999;43:199–212. doi: 10.1128/aac.43.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormican M G, Marshall S A, Jones R N. Detection of extended-spectrum β-lactamase (ESBL)-producing strains by the Etest ESBL screen. J Clin Microbiol. 1996;34:1880–1884. doi: 10.1128/jcm.34.8.1880-1884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois S K, Marriott M S, Amyes S G B. TEM-and SHV-derived extended-spectrum β-lactamases: relationship between selection, structure and function. J Antimicrob Chemother. 1995;35:7–22. doi: 10.1093/jac/35.1.7. [DOI] [PubMed] [Google Scholar]
- 12.Fierer J, Guiney D. Extended-spectrum β-lactamases—a plague of plasmids. JAMA. 1999;281:563–564. doi: 10.1001/jama.281.6.563. [DOI] [PubMed] [Google Scholar]
- 13.Ho P L, Tsang D, Que T L, Ho M, Yuen K Y. Comparison of screening methods for detection of extended-spectrum β-lactamases and their prevalence among Escherichia coli and Klebsiella species in Hong Kong. APMIS. 2000;108:237–240. doi: 10.1034/j.1600-0463.2000.d01-50.x. [DOI] [PubMed] [Google Scholar]
- 14.Jacoby G A, Medeiros A A. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 16.Karas J A, Pillay D G, Muckart D, Sturm A W. Treatment failure due to extended spectrum β-lactamase. J Antimicrob Chemother. 1996;37:203–204. doi: 10.1093/jac/37.1.203. [DOI] [PubMed] [Google Scholar]
- 17.Katsanis G P, Spargo J, Ferraro M J, Sutton L, Jacoby G A. Detection of Klebsiella pneumoniae and Escherichia coli strains producing extended-spectrum β-lactamases. J Clin Microbiol. 1994;32:691–696. doi: 10.1128/jcm.32.3.691-696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Lee H J. Rapid discriminatory detection of genes coding for SHV β-lactamases by ligase chain reaction. Antimicrob Agents Chemother. 2000;44:1860–1864. doi: 10.1128/aac.44.7.1860-1864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kliebe C, Nies B A, Meyer J F, Tolxdorff N R, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 21.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livermore D M, Yuan M. Antibiotic resistance and production of extended-spectrum β-lactamases amongst Klebsiella spp. from intensive care units in Europe. J Antimicrob Chemother. 1996;38:409–424. doi: 10.1093/jac/38.3.409. [DOI] [PubMed] [Google Scholar]
- 23.Mabilat C, Courvalin P. Development of “oligotyping” for characterization and molecular epidemiology of TEM β-lactamases in members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1990;34:2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKenzie F M, Gould I M. Extended spectrum β-lactamases. J Infect. 1998;36:255–258. doi: 10.1016/s0163-4453(98)94027-0. [DOI] [PubMed] [Google Scholar]
- 25.Mzali F H, Heritage J, Gascoyne-Binzi D M, Snelling A M, Hawkey P M. PCR single strand conformational polymorphism can be used to detect the gene encoding SHV-7 extended-spectrum β-lactamase and to identify different SHV genes within the same strain. J Antimicrob Chemother. 1998;41:123–125. doi: 10.1093/jac/41.1.123. [DOI] [PubMed] [Google Scholar]
- 26.Naas T, Philippon L, Poirel L, Ronco E, Nordmann P. An SHV-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1281–1284. doi: 10.1128/aac.43.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; 10th informational supplement. M100–S10. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 28.Niederhauser C, Kaempf L, Heinzer I. Use of the ligase detection reaction-polymerase chain reaction to identify point mutations in extended-spectrum β-lactamases. Eur J Clin Microbiol Infect Dis. 2000;19:477–480. doi: 10.1007/s100960000285. [DOI] [PubMed] [Google Scholar]
- 29.Nüesch-Inderbinen M T, Hächler H, Kayser F H. New system based on site-directed mutagenesis for highly accurate comparison of resistance levels conferred by SHV β-lactamases. Antimicrob Agents Chemother. 1995;39:1726–1730. doi: 10.1128/aac.39.8.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nüesch-Inderbinen M T, Hächler H, Kayser F H. Detection of genes coding for extended-spectrum SHV β-lactamases in clinical isolates by a molecular genetic method, and comparison with the Etest. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 31.Nüesch-Inderbinen M T, Kayser F K, Hächler H. Survey and molecular genetics of SHV-β-lactamases in Enterobacteriaceae in Switzerland; two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philippon A, Labia R, Jacoby G. Extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randegger C C, Keller A, Irla M, Wada A, Hächler H. Contribution of natural amino acid substitutions in SHV extended-spectrum β-lactamases to resistance against various β-lactams. Antimicrob Agents Chemother. 2000;44:2759–2763. doi: 10.1128/aac.44.10.2759-2763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirot D. Extended-spectrum plasmid-mediated β-lactamases. J Antimicrob Chemother. 1995;36(Suppl. A):19–34. doi: 10.1093/jac/36.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 35.Speldooren V, Heym B, Labia R, Nicolas-Chanoine M H. Discriminatory detection of inhibitor-resistant β-lactamases in Escherichia coli by single-strand conformation polymorphism-PCR. Antimicrob Agents Chemother. 1998;42:879–884. doi: 10.1128/aac.42.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson K S, Sanders C C. Detection of extended-spectrum β-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother. 1992;36:1877–1882. doi: 10.1128/aac.36.9.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson K S, Sanders C C. A simple and reliable method to screen isolates of Escherichia coli and Klebsiella pneumoniae for the production of TEM- and SHV-derived extended-spectrum β-lactamases. Clin Microbiol Infect. 1997;3:549–554. doi: 10.1111/j.1469-0691.1997.tb00306.x. [DOI] [PubMed] [Google Scholar]
- 38.Tzouvelekis L S, Vatopoulos A C, Katsanis G, Tzelepi E. Rare case of failure by an automated system to detect extended-spectrum β-lactamase in a cephalosporin-resistant Klebsiella pneumoniae isolate. J Clin Microbiol. 1999;37:2388. doi: 10.1128/jcm.37.7.2388-2388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]