Abstract
Understanding peptide presentation by specific MHC alleles is fundamental for controlling physiological functions of T cells and harnessing them for therapeutic use. However, commonly used in silico predictions and mass spectroscopy have their limitations in precision, sensitivity, and throughput, particularly for MHC class II. Here, we present MEDi, a novel mammalian epitope display that allows an unbiased, affordable, high-resolution mapping of MHC peptide presentation capacity. Our platform provides a detailed picture by testing every antigen-derived peptide and is scalable to all the MHC II alleles. Given the urgent need to understand immune evasion for formulating effective responses to threats such as SARS-CoV-2, we provide a comprehensive analysis of the presentability of all SARS-CoV-2 peptides in the context of several HLA class II alleles. We show that several mutations arising in viral strains expanding globally resulted in reduced peptide presentability by multiple HLA class II alleles, while some increased it, suggesting alteration of MHC II presentation landscapes as a possible immune escape mechanism.
A new method for high-resolution HLA peptide presentation profiling reveals plausible immune escape mechanisms.
INTRODUCTION
Decoding antigen presentation in the context of individual human leukocyte antigen (HLA) alleles is central for understanding immune homeostasis and protection from pathogens and underlies the design of immune medicines. Precise and comprehensive analysis of the short peptides presented by the major histocompatibility complex (MHC) molecules is therefore of major interest. The main approaches used currently, analysis of MHC-eluted peptides by liquid chromatography with tandem mass spectrometry (LC-MS/MS) and in silico prediction algorithms, have contributed to the understanding of peptide presentation. However, they do not provide complete presentability landscapes across many HLAs. LC-MS/MS analysis allows the identification of thousands of naturally presented peptides, but it is technically challenging and requires very large numbers of cells (i.e., 108 to 1010) for good coverage (1, 2). Moreover, presentation of peptides with proven T cell reactivity can be missed (2). The limited sensitivity of LC-MS/MS is especially problematic when working with small tissue samples such as human biopsies. Attempting to circumvent these problems, computational prediction methods have been developed and are relatively reliable in identifying strong [median inhibitory concentration (IC50) < 50 nM] MHC I binders (3). While for MHC II the algorithms are also improving (4), the efficiency in predicting MHC-binding peptides is quite variable and limited. In this respect, the recently improved NetMHCIIpan4 shows better performance than conventional binding prediction algorithms (5) but is accurate only for a limited number of alleles, owing to the lack of suitable peptide datasets for training. To circumvent this, a recently published study improved algorithm performance using yeast display peptide libraries (6). Still, there is a big gap from the several HLAs with high-quality in silico prediction scores and the thousands of unique HLA alleles present in the human population.
Predicting antigen presentation by MHC is further complicated by the fact that it is a dynamic process and can change depending on the physiological state of the cell. It is also regulated by tightly controlled chaperones such as HLA-DM (7), dysregulation of which has been linked to autoimmune disease progression (8, 9), while high expression of HLA-DM correlated with improved survival in patients with cancer (10). Thus, an unbiased method, testing pure peptide presentation capacity of the MHC not obscured by other physiological factors, would help in getting the complete picture of all possible MHC ligands present in a given protein. This reductionist approach would provide a basic set of allele-specific peptides (the presentable peptide space) ready for the generation of peptide libraries for screening of T cell reactivities or the generation of pMHC tetramers. Taking this set as a basis, subsets of peptides could be derived by incorporating protein processing and chaperone functions dependent on cellular state and chaperone expression levels.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the infectious agent responsible for the worldwide coronavirus disease 2019 (COVID-19) pandemic with more than 5 million fatalities (11, 12). Several companies are now providing vaccines inducing humoral and cellular responses against SARS-CoV-2, but for long lasting protection, generation of T cell memory will be required (13) even if preexisting T cell immunity to common cold coronavirus might play a role (14–16). Because protection by antibodies is related to protein function (e.g., blocking receptors that are required for viral cell entry) and/or protein localization (surface expression to allow opsonizing antibodies to bind), it has limited target space, increasing selection pressure for pathogen escape. Protection by T cells, on the other hand, relies entirely on T cell receptor (TCR) recognition of pathogen-derived peptides presented by MHC and is mostly independent of physiological function or localization of the target protein. Consequently, while only particular epitopes of surface proteins allow targeting by neutralizing antibodies, many peptides can serve as T cell targets, providing a much bigger epitope space for therapeutic development. A high-resolution map of all SARS-CoV-2 presentable peptides resolved on different HLA alleles would greatly help these efforts.
In this work, using a novel mammalian epitope display system called MEDi, we tested the capacity of several HLA alleles to present SARS-CoV-2 virus peptides. We validated our findings biologically by studying T cell recognition of the SARS-CoV-2 virus in patients with acute COVID-19 and analyzed the impact of mutations carried by the novel SARS-CoV-2 strains. Our results suggest immune evasion based on shifting peptide presentation away from well-recognized CD4 epitopes. Given the importance of CD4 T cells in controlling B cell and CD8 T cell responses in patients with COVID-19, the results described here may help guide the generation of vaccines or therapeutics designed to elicit efficient cellular immunity.
RESULTS
MEDi, a mammalian epitope display platform based on MCR
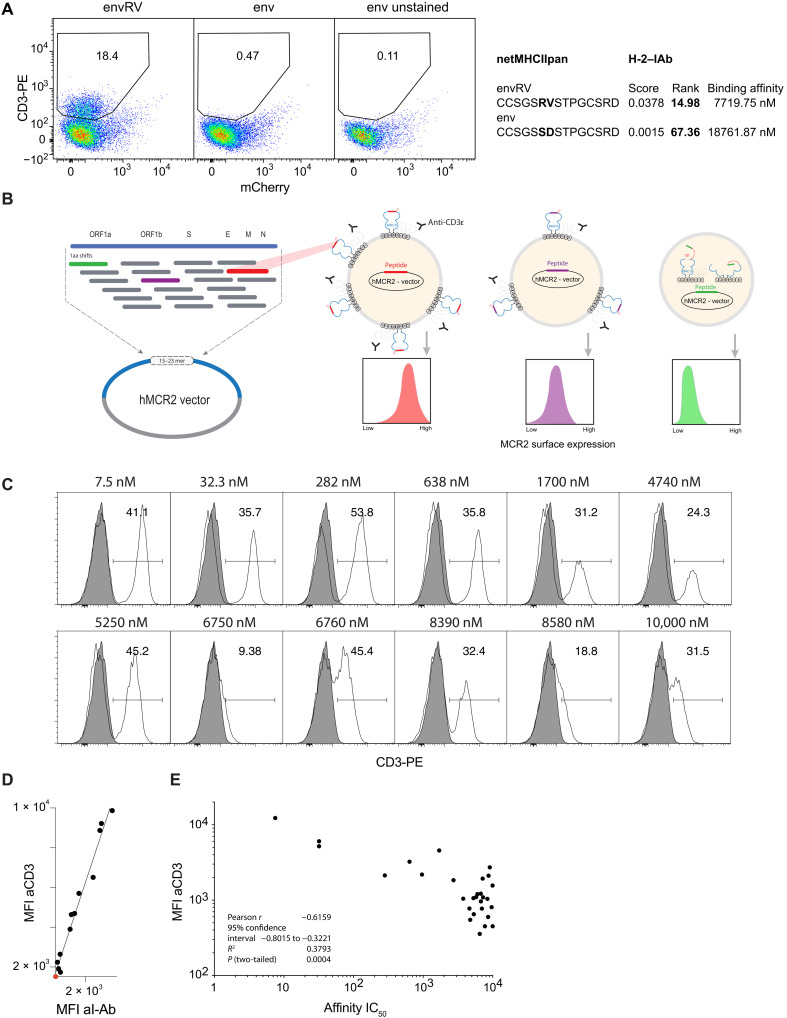
Using the MCR system (fig. S1) in our previous study, we identified an immunogenic, murine leukemia virus envelope protein–derived mutant peptide (MLVenvS126R,D127V, aka envRV) as being efficiently recognized by mouse tumor-infiltrating lymphocytes (17). While MCR2 molecules carrying envRV were expressed well on the surface of the reporter cells, the ones carrying the nonmutated wild-type (WT) peptide (env) could not be detected, consistent with netMHCIIpan affinity predictions (Fig. 1A). Given that MHC molecules without a bound peptide are unstable (18), this observation led to the hypothesis that peptides fitting well into the peptide-binding groove and therefore being efficiently presented by the MHC will also effectively stabilize the MCR2 molecules on the surface of cells. In contrast, peptides not well presented by the MHC destabilize the MCR2 molecules, and therefore, little, if any, cell surface expression will be detected (Fig. 1B). We therefore set out to test our hypothesis and cloned a number of peptides with biochemically tested I-Ab binding affinity ranging from IC50 7.5 to 10,000 nM (Table 1). We transduced them into 16.2X reporter cell line and determined the MCR2 expression by flow cytometry and staining with I-Ab– and CD3-specific antibodies (Fig. 1, C and E). As expected from our previous study, there was a clear linear correlation between both stainings, but CD3 allowed a better separation of the positive and negative populations (Fig. 1D). We therefore used anti-CD3 staining in all further MEDi analysis, with the added advantage of being MHC agnostic and therefore universally usable with all mouse H-2 and human HLA haplotypes. We analyzed MCR2 expression dependence on peptide–I-Ab binding affinity by the mean fluorescence intensity (MFI) of CD3 staining. Consistent with our expectations, the MCR2s carrying peptides with a good I-Ab binding affinity were expressed on the surface at high levels, while MCR2s presenting low-affinity peptides showed lower surface expression (Fig. 1E). Peptides with an affinity below 1 μM (IC50) are considered good MHC binders, and all MCRs carrying these peptides were expressed well on the cell surface. In addition, some peptides with lower MHC binding affinity appeared on the surface, indicating that linking peptides directly to the MHC β chain stabilizes low-affinity peptide-MHC interactions. Being able to test the presentation of these peptides is important, as self-peptides known as targets in autoimmune diseases often bind MHC with low affinity (19). Most peptides with an I-Ab binding below 5 μM IC50 stabilized MCR2 surface expression, while for peptides with lower binding affinity, the MCR expression was variable and generally much lower. Some of the MCR2s carrying peptides with an apparently low affinity (e.g., 8.39 μM) were expressed on the surface at good levels, suggesting that additional factors apart from pure binding affinity (measured in vitro) regulate peptide-MHC interactions. Similarly, the envRV peptide could stabilize MCR2 expression even if its I-Ab binding affinity was predicted by netMHCIIpan to be very low at 7.7 μM, and we needed to add high amounts of envRV peptide for in vitro T cell stimulations by dendritic cells (17).
Fig. 1. Cell surface expression of MCR2 depends on peptide-MHC binding.
(A and C) Flow cytometric analysis of CD3ε expression on 16.2c11 cells transduced with (A) MCR2-envRV or MCR2-env or (C) MCR2 constructs carrying peptides binding I-Ab with different affinity (7.5 nM to 10 μM). In (A), the x axis shows irrelevant, background nuclear factor of activated T cell (NFAT)–mCherry reporter reactivity. Gray histograms, untransduced cells. PE, phycoerythrin. (B) Schematic representation of the peptide library cloning into the human MCR2 vector and of the MEDi principle. ORF1a, open reading frame 1a; aa, amino acid. (D) Comparison of MFI of anti-CD3ε and anti–I-Ab stainings. (E) Correlation of MFI of anti-CD3ε staining (of MCR2+ cells) with the peptide-MHC binding affinity of peptides carried by the MCR2.
Table 1. I-Ab presented peptides cloned in MCR2 vector.
| Peptide | IC 50 | Peptide | IC 50 | |||
| 1 | KSAFQSSVASGFIGF | 7.56 | 16 | VVPDGYKLTGNVLIL | 6520 | |
| 2 | SISKRAYMATTILEM | 31.9 | 17 | YDMFNLLLMKPLGIE | 6750 | |
| 3 | ISGYNFSLSAAVKAG | 32.3 | 18 | LIEDYFEALSLQLSG | 6760 | |
| 4 | IEYAKLYVLSPILAE | 282 | 19 | IEDAKRMIAISAKVA | 6990 | |
| 5 | FSLSAAVKAGASLID | 638 | 20 | LMIAHRVLLSSILES | 7160 | |
| 6 | DQEYHRLIHSLSKTS | 964 | 21 | ILKGVINIWGSGLLQ | 7270 | |
| 7 | SLINSMKTSFSSRLL | 1700 | 22 | VSLIAIIKGIVNLYK | 7740 | |
| 8 | EEALNVALAVVTLLA | 2730 | 23 | IIKYNRRLAKSIICE | 8390 | |
| 9 | TKKSIKEIASSISRL | 3770 | 24 | NKVKSLRILNTRRKL | 8580 | |
| 10 | KVFNTRRNTLLFLDL | 4620 | 25 | RHIVGKPCPKPHRLN | 8700 | |
| 11 | LLNNQFGTMPSLTLA | 4740 | 26 | LKAEAQMSIQLINKA | 9060 | |
| 12 | GLVSQLSVLSSITNI | 5280 | 27 | MGQLISFFGEIPSII | 9640 | |
| 13 | SGDAMARNISSRTLE | 5340 | 28 | ELLDQSDVKEPGVSR | 9970 | |
| 14 | MDLADLFNAQPGLTS | 5780 | 29 | TSAFNKKTFDHTLMS | 10000 | |
| 15 | NSLILLECFVRSSPA | 6030 |
Analysis of SARS-CoV-2 peptide presentability by common HLA alleles
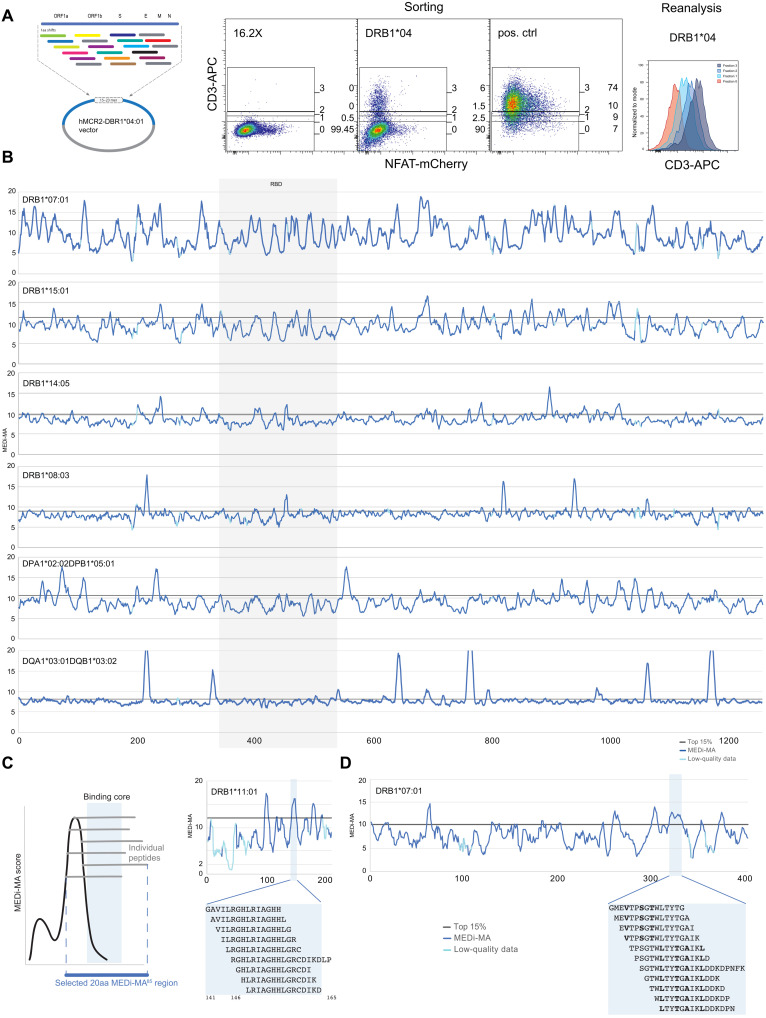
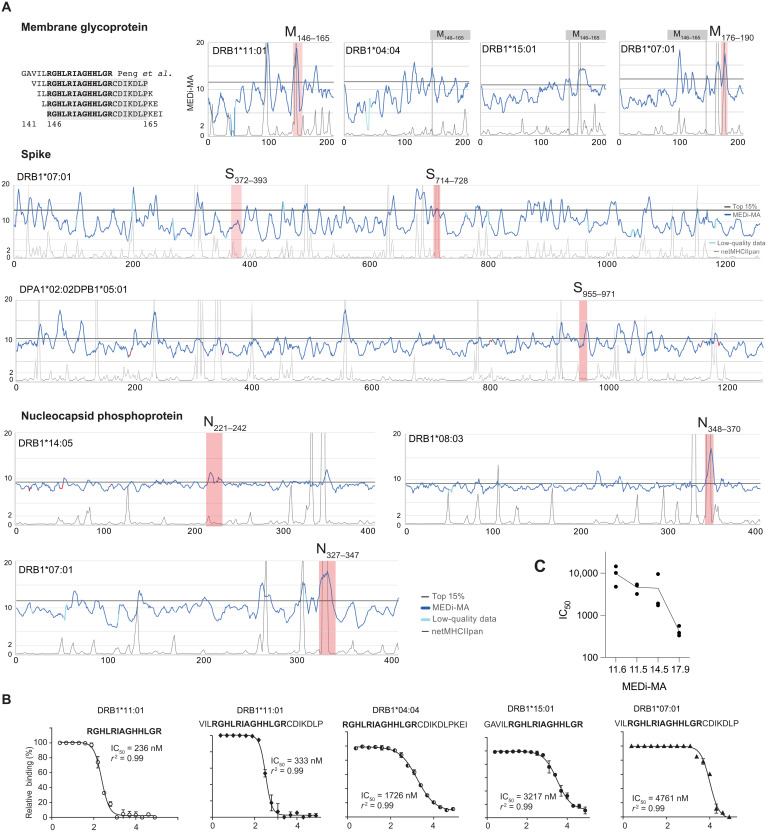
Considering the recent interest in SARS-CoV-2 T cell epitopes effectively presented across the possibly highest number of HLA alleles, we used MEDi to determine the presentability of all peptides encoded in the SARS-CoV-2 genome in the context of some of the most common HLA class II haplotypes. The critical role of CD4 T cell help in supporting B cell and CD8 T cell responses is undisputed and also crucial for COVID-19 protection (20–22). However, a complete picture of the important MHC class II epitopes is missing, as they are more difficult to predict by computer algorithms than MHC class I ligands. To achieve a good resolution, we cloned all possible 15–amino acid peptides derived from the SARS-CoV-2 genome shifted by 1 amino acid (Fig. 2A) into MCR2 vectors containing extracellular domains of several HLAs (Fig. 2). We transduced these libraries into the 16.2X reporter cell line, stained for CD3, and sorted the cells into four fractions (neg, low, mid, and hi) based on the surface expression level of the MCR2 (Fig. 2A). We then determined the peptides carried by the MCR2s in the different fractions by reverse transcription polymerase chain reaction (RT-PCR) and deep sequencing. For each peptide, a MEDi score was calculated and plotted against the position of the starting amino acid of the peptide within the protein (see Materials and Methods). To account for data quality differences related to sorted cell numbers and sequencing depth, we introduced a MEDi quality metric composed of a minimal read count and the coefficient of variation (see Materials and Methods). Figure 2B shows plots of the MEDi score moving average (MEDi-MA; average of five peptides) for the SARS-CoV-2 Spike peptide presentability by a set of six HLA alleles. Peptides derived from particular regions of the protein stabilize surface expression of the MCR better than others, i.e., are being better presented by the MHC. These peptides grouped in regions (“peaks”), indicating that a core MHC-binding epitope was present in a number of peptides starting at several consecutive amino acids (Fig. 2, C and D). This observation is consistent with the fact that, owing to its open peptide-binding groove, MHC class II molecules present peptides of different length (1). Usually, the minimal MHC-binding core is composed of nine amino acids, as shown by the commonly described binding motifs (23), even if residues outside it can also contribute to the MHC binding affinity (24). As expected, MEDi graphs derived from these analyses showed diverse presentation patterns. Each HLA molecule was unique, with regions of specific and promiscuous peptide presentation.
Fig. 2. MEDi analysis of Spike peptide presentability by different HLAs.
(A) Example flow cytometric analysis and sorting of MCR2+ reporter cells, transduced with an MCR2 library and stained for CD3e. On the basis of the surface expression of the MCR2, four fractions (neg, low, mid, and hi) were sorted and reanalyzed. Negative (16.2X) and positive controls are indicated. APC, allophycocyanin. (B) MEDi-MA score graphs for all Spike-derived peptides presented by six different HLAs (dark blue line) are shown. The light blue color indicates data points below the quality threshold (see Materials and Methods). (C and D) Schematics and interpretation of the MEDi graphs. MEDi analysis for the membrane (C) and nucleocapsid (D) proteins with indicated 15–amino acid peptides falling into example peaks above the 85th percentile. Putative anchor residues for DRB1*07:01 are shown in bold. The extended peptides are recognized by some TCRs analyzed in this study.
To distill the best HLA-binding peptides from these data, we selected peptide sequences scoring above the 85th percentile cutoff. As an example, in Table 2, we provide a list of the major presentable peptides derived from the Spike protein, and in the Supplementary Materials, we extend this analysis to other proteins and all peptides derived from the SARS-CoV-2 genome in the context of three HLAs (data file S1). Notably, the spike list contains many peptides overlapping with immunogenic (13, 14) or HLA-eluted (25), therefore naturally presented, peptides described in recent literature.
Table 2. Peptides representative for major presentable regions of Spike.
15aa peptides from the center of peaks having at least 5 consecutive peptides with MEDi-MA scores above the 85th percentile are shown. Sequences overlapping at least 9aa with the minimum sequences of presented peptide clusters identified by Knierman et al. (25) are marked by an asterisk. Peptides overlapping at least 9aa with T cell reactive peptides identified by Peng et al. (13) and Nelde et al. (14) are highlighted in bold.
| Position | DRB10701 | Position | DRB10404 | Position | DRB11501 | Position | DPA10202B10501 |
| 8 | LPLVSSQCVNLTTRT | 7 | LLPLVSSQCVNLTTR | 59 | FSNVTWFHAIHVSGT | 40 | DKVFRSSVLHSTQDL* |
| 27 | AYTNSFTRGVYYPDK* | 62 | VTWFHAIHVSGTNGT | 237 | RFQTLLALHRSYLTP* | 50 | STQDLFLPFFSNVTW* |
| 42 | VFRSSVLHSTQDLFL* | 232 | GINITRFQTLLALHR* | 302 | TLKSFTVEKGIYQTS* | 70 | VSGTNGTKRFDNPVL |
| 60 | SNVTWFHAIHVSGTN | 402 | IRGDEVRQIAPGQTG* | 309 | EKGIYQTSNFRVQPT* | 75 | GTKRFDNPVLPFNDG |
| 111 | DSKTQSLLIVNNATN | 513 | LSFELLHAPATVCGP* | 402 | IRGDEVRQIAPGQTG* | 80 | DNPVLPFNDGVYFAS |
| 202 | KIYSKHTPINLVRDL* | 678 | TNSPRRARSVASQSI | 433 | VIAWNSNNLDSKVGG* | 106 | FGTTLDSKTQSLLIV |
| 230 | PIGINITRFQTLLAL | 683 | RARSVASQSIIAYTM | 470 | TEIYQAGSTPCNGVE* | 111 | DSKTQSLLIVNNATN |
| 239 | QTLLALHRSYLTPGD* | 688 | ASQSIIAYTMSLGAE* | 631 | PTWRVYSTGSNVFQT* | 201 | FKIYSKHTPINLVRD* |
| 309 | EKGIYQTSNFRVQPT* | 854 | KFNGLTVLPPLLTDE* | 677 | QTNSPRRARSVASQS* | 230 | PIGINITRFQTLLAL |
| 314 | QTSNFRVQPTESIVR* | 866 | TDEMIAQYTSALLAG* | 682 | RRARSVASQSIIAYT | 235 | ITRFQTLLALHRSYL* |
| 490 | FPLQSYGFQPTNGVG* | 919 | NQKLIANQFNSAIGK* | 687 | VASQSIIAYTMSLGA* | 495 | YGFQPTNGVGYQPYR |
| 639 | GSNVFQTRAGCLIGA* | 931 | IGKIQDSLSSTASAL* | 692 | IIAYTMSLGAENSVA | 551 | VLTESNKKFLPFQQF* |
| 679 | NSPRRARSVASQSII* | 936 | DSLSSTASALGKLQD* | 752 | LLLQYGSFCTQLNRA | 556 | NKKFLPFQQFGRDIA* |
| 684 | ARSVASQSIIAYTMS | 952 | VNQNAQALNTLVKQL* | 784 | QVKQIYKTPPIKDFG | 680 | SPRRARSVASQSIIA* |
| 689 | SQSIIAYTMSLGAEN* | 957 | QALNTLVKQLSSNFG | 856 | NGLTVLPPLLTDEMI | 704 | SVAYSNNSIAIPTNF |
| 694 | AYTMSLGAENSVAYS | 962 | LVKQLSSNFGAISSV* | 866 | TDEMIAQYTSALLAG* | 782 | FAQVKQIYKTPPIKD* |
| 714 | IPTNFTISVTTEILP | 982 | SRLDKVEAEVQIDRL* | 899 | AMQMAYRFNGIGVTQ* | 812 | PSKRSFIEDLLFNKV* |
| 782 | FAQVKQIYKTPPIKD | 1016 | AEIRASANLAATKMS* | 913 | QNVLYENQKLIANQF | 871 | AQYTSALLAGTITSG* |
| 869 | MIAQYTSALLAGTIT* | 1063 | LHVTYVPAQEKNFTT | 918 | ENQKLIANQFNSAIG* | 918 | ENQKLIANQFNSAIG* |
| 880 | GTITSGWTFGAGAAL | 1170 | SGINASVVNIQKEID* | 961 | TLVKQLSSNFGAISS* | 928 | NSAIGKIQDSLSSTA* |
| 885 | GWTFGAGAALQIPFA | 997 | ITGRLQSLQTYVTQQ | 960 | NTLVKQLSSNFGAIS* | ||
| 896 | IPFAMQMAYRFNGIG* | 1002 | QSLQTYVTQQLIRAA | 1014 | RAAEIRASANLAATK* | ||
| 1008 | VTQQLIRAAEIRASA* | 1007 | YVTQQLIRAAEIRAS* | 1019 | RASANLAATKMSECV | ||
| 1013 | IRAAEIRASANLAAT* | 1012 | LIRAAEIRASANLAA* | 1040 | VDFCGKGYHLMSFPQ | ||
| 1070 | AQEKNFTTAPAICHD | 1017 | EIRASANLAATKMSE* | 1045 | KGYHLMSFPQSAPHG | ||
| 1066 | TYVPAQEKNFTTAPA | ||||||
| 1071 | QEKNFTTAPAICHDG | ||||||
| 1088 | HFPREGVFVSNGTHW | ||||||
| 1134 | NNTVYDPLQPELDSF* |
Validation of MEDi by a competitive peptide-binding assay
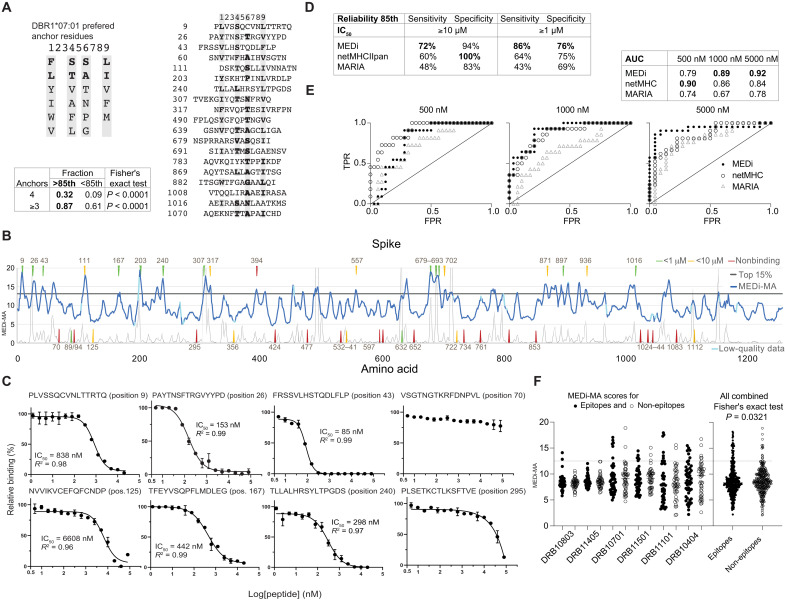
Next, we analyzed peptides from the major peaks in the Spike protein for the presence of a binding motif and found an enrichment of known (23), appropriately spaced anchor residues in most of the selected peptides [Fig. 3A (DRB1*07:01) and fig. S2A (DRB1*15:01)]. The frequency of four or three correct anchor residues was significantly higher in peptides scoring above the 85th percentile cutoff than below, validating our assay. Still, because tethering peptides to the MCR might stabilize some low-affinity interactions not efficiently presented in vivo, we wanted to independently validate and quantify the HLA binding of the peptides defined by MEDi. To this end, we performed measurements of competitive peptide binding by fluorescence polarization (26) for a set of Spike peptides for DRB1*07:01. We selected 43 peptides representing peaks of different heights and “valleys” (Fig. 3B) and considered peptides with IC50 below 10 μM as binders. When IC50 calculation was impossible because of very low peptide binding, it was set arbitrarily to 20 μM. This dataset allowed us to analyze the ability of the MEDi assay to qualify peptides for HLA binding/presentation and compare it to netMHCIIpan (EL rank) and the recent SARS-CoV-2 predictions done by Fast et al. (27) using the novel algorithm called MARIA (28).
Fig. 3. MEDi analysis of Spike peptide presentation by DRB1*07:01 compared to computational predictions and MHC-binding IC50.
(A) Sequence comparison of Spike peptides representative for the major peaks above the 85th percentile containing at least six peptides. Residues matching the HLA-binding consensus are highlighted in gray. Table shows fraction of peptides with four, or at least three, correct anchor residues among peptides above the 85th percentile cutoff or below. (B) MEDi-MA score graphs (dark blue line) for all Spike-derived peptides presented by DRB1*07:01. The horizontal black line indicates the 85th percentile threshold and the light blue color data points on the MEDi-MA graphs below the quality threshold (see Materials and Methods). Arrows indicate peptides chosen for HLA-binding IC50 calculation by the fluorescence polarization assay, color-coded dependent on the result of the binding assay. Thin black line indicates netMHCIIpan prediction scores scaled to fit on the same plot (20/Rank_EL, threshold for weak presentation >2). (C) Results of the competitive peptide-binding fluorescence polarization assay for individual peptides (see also fig. S3). IC50 and R2 values are shown. (D) Reliability of the assay was tested by calculating the sensitivity and specificity. (E) ROC curves of the MEDi-MA, netMHCIIpan, and MARIA scores qualifying peptides as HLA binders. Calculations were done for peptides analyzed in (C) and fig. S3 and positive binding thresholds at IC50 of 500 nM, 1 μM, or 5 μM. TPR, true-positive rate; FPR, false-positive rate. (F) MEDi-MA scores for SARS-CoV-2 epitopes and non-epitopes as defined by Tarke et al. (29) are shown. Gray dotted line indicates a 12.5 MEDi-MA cutoff used.
Eighteen of the 19 peptides (95%) scoring above the 85th percentile cutoff bound to the HLA with IC50 between 85 nM and 10 μM (Fig. 3, B and C, and fig. S3), 12 of them with IC50 below 1 μM. In addition, 5 of the 10 peptides within peaks below the 85th percentile cutoff bound to the HLA (Fig. 3, B and C). From the remaining 14 peptides, which were outside of peaks, 2 (14%) bound to the HLA with low affinity (IC50 6.6 and 1.7 μM; Fig. 3C). This corresponded to a sensitivity of 72% (18 of 25) and specificity of 94% (17 of 18) for the 10 µM IC50 binding threshold and the 85th percentile cutoff. For the 1 μM IC50 binding threshold, a sensitivity of 86% and specificity of 76% were observed (Fig. 3E). netMHCIIpan (cutoff, 85th percentile) had a sensitivity of only 60% (15 of 25) but a specificity of 100% (18 of 18) for the 10 μM IC50 binding threshold and a sensitivity of 64% and specificity of 75% for the 1 μM binding threshold (Fig. 3E). MARIA showed a sensitivity of 48 and 43% with a specificity of 83 and 69%, respectively, for the two thresholds. We also plotted receiver operating characteristic (ROC) curves for different IC50 cutoffs and compared MEDi-MA scores to netMHCIIpan EL rank and MARIA (Fig. 3D). Overall, for DRB1*07:01, the performance of MEDi and netMHCIIpan was comparable, with MEDi performing better for low-affinity peptides [1 and 5 μM IC50 cutoffs: area under the curve (AUC) of 89 to 86% and 92 to 84%, respectively], while netMHCIIpan was better for the 500 nM IC50 cutoff (AUC of 90 to 79%). MARIA showed an AUC of 67 and 78% for low-affinity peptides and 74% for high-affinity peptides (Fig. 3, D and E).
Next, using 30 of these peptides, we performed an unbiased analysis for DRB1*15:01 (fig. S2). Here, because the peptides were chosen according to MEDi data for DRB1*07:01, most peptides corresponded to MEDi scores below the 85th percentile threshold and were not in major peaks (fig. S2, A and B), i.e., they should not be well presented. The majority did not bind the HLA with sufficient affinity (fig. S2C). Nevertheless, 11 of the peptides were in peaks above the threshold, and 9 (82%) bound to HLA. The sensitivity and specificity were 56% (9 of 16) and 86% (12 of 14) for the 10 μM IC50 binding threshold and the 85th percentile cutoff and 71 and 74%, respectively, for the 1 μM binding threshold (fig. S2, C and D). netMHCIIpan (cutoff, 85th percentile) had a poor sensitivity of 37.5% (6 of 16) but a specificity of 100% (14 of 14) for the 10 μM IC50 binding threshold and 57 and 91%, respectively, for the 1 μM binding threshold. MARIA performed similarly to netMHCIIpan. AUC was between 78 and 83% for netMHCIIpan, 72 to 76% for MEDi, and 50 to 73% for MARIA (fig. S2E). Together, also for this well characterized HLA allele, MEDi was much more sensitive than netMHCIIpan and MARIA, however, at a small cost of specificity. These results validate the MEDi platform as a means to select peptides highly presentable by an HLA allele.
Deorphaning TCRs from the bronchoalveolar lavage of acute COVID-19 patients
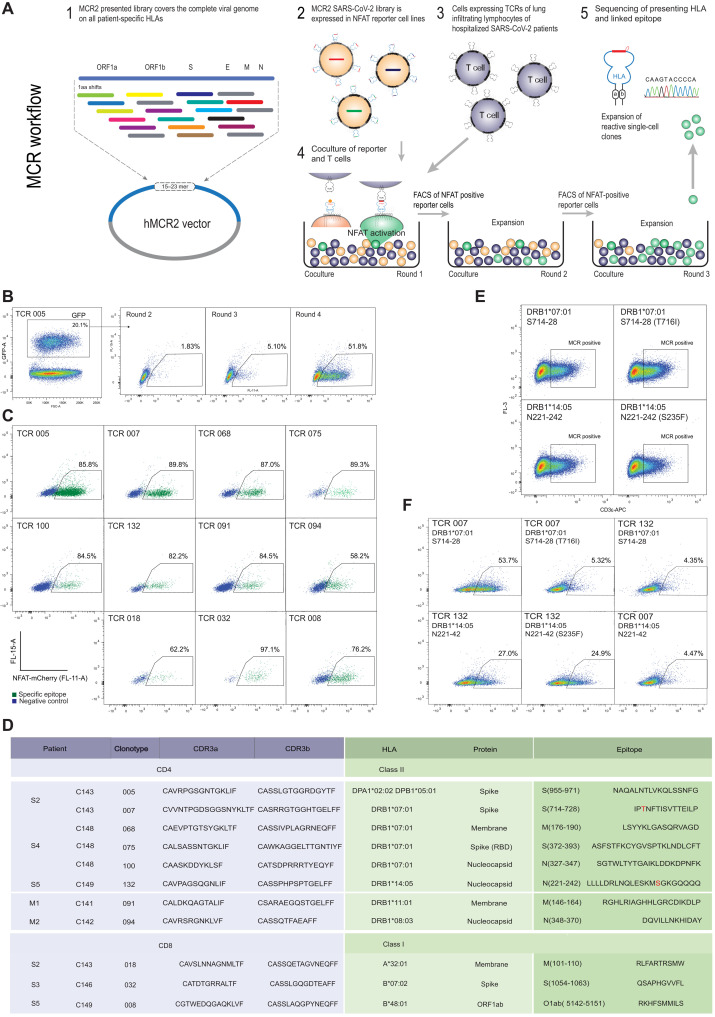
To further test MEDi and the proposed scoring approach for antigen presentability, we extended the analysis to natural T cell targets. We wanted to see what are the MEDi scores of true immunogenic epitopes. First, we compared 49 known T cell epitopes and 49 matched “non-epitopes,” recently described by Tarke et al. (29). Even if the precise HLA restriction of these epitopes was not determined, we still expected to find more “high-scoring” peptides among the real epitopes than non-epitopes. This 98-member pool contained peptides scoring in MEDi above a threshold of 12.5 in five of six HLA alleles analyzed, and they were significantly enriched among real epitopes (Fig. 3F). Although T cell SARS-CoV-2 reactivities, such as the ones described by Tarke et al. have been reported, analyses that comprehensively decode “immune synapses,” including TCR α and β chain sequences, the recognized peptide and the presenting HLA, are sparse. Thus, we used the MCR technology (Fig. 4A) (17) and single-chain trimers (30) linked to the intracellular domain of the TCR zeta chain (SCTz) (31, 32) to deorphan TCRs of enriched clonotypes from the bronchoalveolar lavages (BALs) of patients with COVID-19, recently described by Liao et al. (33). Liao et al. provided high-resolution single-cell data indicating aberrant cellular responses and identified expanded T cell clonotypes, but they decoded neither their antigenic specificity nor the HLA restriction. To address this, we cloned 109 most-enriched TCRs (data file S2), expressed them in a T cell line, and performed an unbiased epitope screening. This included MCR2 libraries containing all possible 23–amino acid SARS-CoV-2–derived peptides (one amino acid shifts through all proteins) and libraries containing all possible 10–amino acid SARS-CoV-2–derived peptides presented in the context of SCTz. This setup allowed for an unprecedented, complete screen of all SARS-CoV-2 peptides in the context of all HLAs from every patient (table S1). Screening these patient-specific MCR2 libraries of approximately 120,000 different peptide-MCR2 combinations and 60,000 peptide-SCTz combinations required at least four rounds of enrichment (Fig. 4B and data file S2) before single-cell clones revealed the specific peptides and the presenting HLA alleles (Fig 4C and data file S2). As expected, not all TCRs showed reactivity against SARS-CoV-2 antigens, but we identified the cognate peptides and the HLA restriction for eight CD4 and three CD8 TCRs (Fig. 4, C and D).
Fig. 4. Deorphaning TCRs from the BAL of patients with COVID-19 by MCR2 screening.
(A) Schematic representation of the TCR deorphaning process. (B) MCR2–SARS-CoV-2+ or SCTz–SARS-CoV-2+ 16.2X reporter cells [green fluorescent protein positive (GFP+)], carrying all possible SARS-CoV-2–derived peptides in the context of all 12 patient-specific HLA alleles (complexity up to 120,000 individual pMHC combinations) were cocultured with 16.2A2 cells transduced with individual TCRs from patients. Responding (NFAT+) reporter cells were sorted, expanded, and cocultured four to five times. FSC-A, forward scatter area. (C) Individual responding reporter clones were isolated and reanalyzed by an additional coculture. (D) Sequences of the deorphaned TCR chains, specific peptides, and HLA restriction. (E) 16.2X reporter cells carrying the MCR2-S714–728 or MCR2-N221–242 were analyzed by flow cytometry for MCR2 expression (by aCD3 staining). (F) 16.2X reporter cells carrying the MCR2-S714–728 or MCR2-S714–728(F716I) (top) and MCR2-N221–242 or MCR2-N221–242(S235F) (bottom) were cocultured with 16.2A2 cells transduced with TCR007 or TCR132, respectively, and NFAT activation was measured on FACS.
We found a variety of peptides presented by several HLA alleles. For example, three CD4 T cell clones from severely affected patient C148 recognized peptides from the SARS-CoV-2 proteins Spike (S), membrane glycoprotein (M), and nucleocapsid (N), all presented by DRB1*07:01. TCR091 from patient C141 reacted with the membrane glycoprotein-derived peptide M146–165 presented by DRB1*11:01. In line with a high immunogenicity of this epitope, Peng et al. (13) reported that 32% of patients contained T cells that recognize an overlapping peptide M141–158. Some other peptides that we found were previously shown to be recognized by T cells (29), but we could extend their characterization by precisely determining the HLA restriction and the corresponding TCRs. Still, to independently validate the reactivities of these TCRs, we cocultured our TCR carrying cell lines with peripheral blood mononuclear cells (PBMCs; Sanvitra) carrying correct HLA alleles pulsed with the identified antigenic peptides (fig. S4). This confirmation was particularly important for the previously unknown epitope HLA combinations that we found. All epitopes except one, for which we did not find matching PBMCs, could be confirmed by this assay (fig. S4).
Two of the CD4 T cell–specific peptides identified in our study (S714–728 and N221–242) were mutated in the SARS-CoV-2 B1.1.7 variant first identified in Britain, which rapidly spread because of up to 70% increased transmission rates (34). We transduced reporter cells with MCR2 carrying the WT and mutated S714–728 or N221–242 peptides (Fig. 4E) and found that S714–728(T716I) was not recognized by the TCR007 (Fig. 4F). Recognition of N221–242 peptide was unaffected by the mutation, suggesting that Ser236 was not part of the minimal epitope (Fig. 4F), nor did it affect peptide presentation.
MEDi indicates efficient presentation of immunogenic CD4 T cell epitopes
Next, we looked at the presentability of the CD4 T cell targets identified in our MCR screens. MEDi data indicated good presentability of the TCR091 target peptide region by DRB1*11:01(Fig. 5A and fig. S5). Furthermore, consistent with high reactivity among patients shown by Peng et al. (13), MEDi suggested presentation of this region by other HLA alleles such as DRB1*04:04 and DRB1*15:01 and, to a lower extent, by DRB1*07:01 (Fig. 5A). NetMHCIIpan only predicts DRB1*11:01, but the competitive peptide binding assay confirmed the MEDi results: DRB1*11:01 showed the highest IC50 (236 to 561 nM), followed by DRB1*04:04 (1.7 to 9.5 μM) and DRB1*15:01(3.2 to 5.4 μM), and the lowest was DRB1*07:01 (4.7 to 14 μM) (Fig. 5B and fig. S5). Even if these values do not precisely indicate differences in binding affinity because the competing fluorescent peptides bind the HLAs with different affinities, there was some correlation between the MEDi score and the IC50 (Fig. 5B and fig. S5). This highlights the advantages of MEDi over netMHCIIpan for discovering low-affinity peptide presentation.
Fig. 5. Presentation of immunogenic peptides by MEDi.
(A) MEDi-MA scores (dark blue) are compared to netMHCIIpan prediction scores scaled to fit on the same plot (20/Rank_EL, thin black line, threshold for weak presentation >2). Peptides overlapping by at least 10 amino acids with the CD4 T cell–specific peptides found in this study (Fig. 4D) are indicated as red shades. The 85th percentile threshold is indicated as a black line. M146–165 peptide, recognized by the TCR091 in the context of DRB1*11:01, is indicated in gray for the other alleles. Light blue color indicates data points on the MEDi-MA graphs below the quality threshold. (B) Results of the competitive peptide-binding assay for the indicated peptides and alleles (see also fig. S5). (C) Correlation between the MEDi-MA scores for 15–amino acid peptides and IC50 from (B) and fig. S5.
Next, we analyzed MEDi scores of the other immunogenic peptides found in this study and compared them to netMHCIIpan predictions (Fig. 5). All of the CD4 T cell immunogenic peptides were found in the MEDi peaks, with S955–971 presented by DPA1*02:02/DPB1*05:01 and N221–242 presented by DRB1*14:05 being uniquely identified by MEDi. In addition, seven of the eight peptides passed the 85th percentile cutoff threshold. Only S372–393 showed a peak with lower MEDi scores, suggesting lower-affinity HLA binding. Thus, selecting all immunogenic peptides for screening applications might require an adjustment of the MEDi threshold. Together, these results indicate that MEDi-selected peptides are enriched for immunogenic epitopes and that MEDi has an advantage over in silico predictions for MHC class II alleles, where no high-quality MS results or other training data are available.
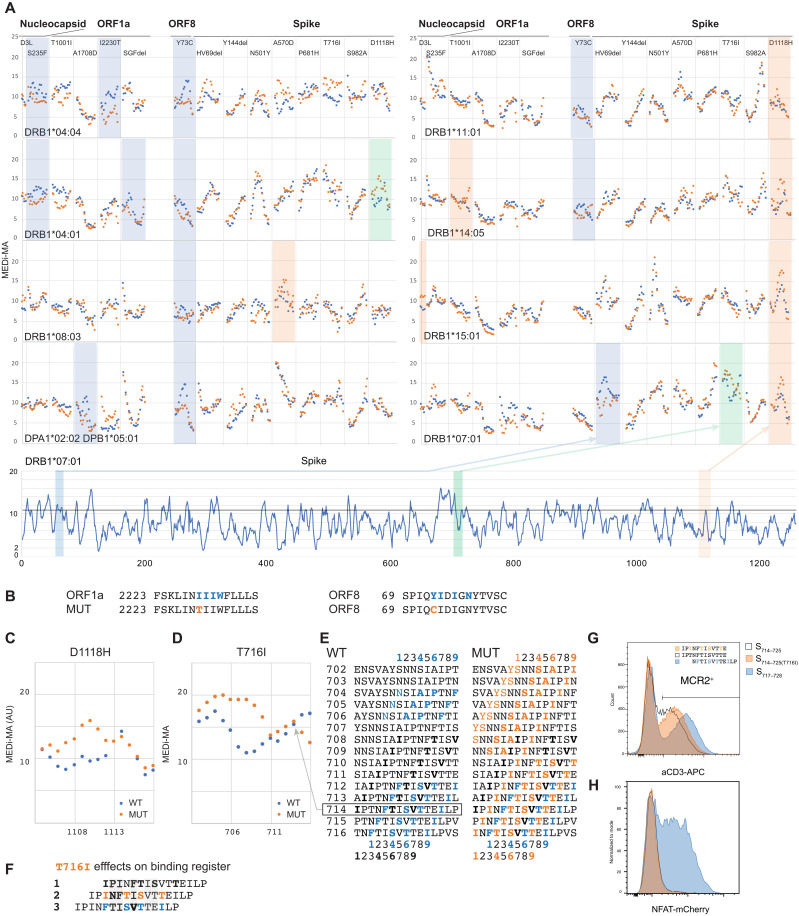
MEDi reveals candidate immune escape mutants
Having established the ability of MEDi to determine presentable peptides, we used it to analyze the effects of 25 mutations present in SARS-CoV-2 variant strains expanding across the globe (Table 3). We generated MCR2 libraries containing mutation-overlapping 15-mer peptides in the context of eight different HLA alleles and performed MEDi analysis. As shown in Fig. 6 and fig. S6, there was a notable HLA-dependent difference in mutant peptide presentability. Open reading frame 8 (ORF8) Y73C and spike R246I mutations abolished peptide presentation by six of eight and five of eight HLA alleles, respectively, suggesting the possibility of immune escape of the virus in patients with these alleles. Several other mutated peptides from nucleocapsid, ORF1a and ORF8, were inefficiently presented by DRB1*04:04 and DRB1*04:01 and DPA1*02:02/DPB1*05:01. For some, the molecular mechanism could be envisioned, e.g., mutations I2230T and Y73C disrupted the N-terminal hydrophobic amino acid stretches constituting a binding motif for DRB1*04:04 (Fig. 6, A and B) (23). In addition, the spike HV69 deletion reduced presentation by DRB1*07:01. The other alleles showed no difference between WT and mutated peptides, with a few exceptions where presentability of mutated peptides was enhanced. In particular, the spike D1118H mutation stabilized binding of several peptides to DRB1*14:05, DRB1*15:01, and DRB1*07:01 and caused a shift in the peptide presentation landscape of DRB1*04:01 (Fig. 6, A and C). In line, the peptide S1111–1130(D1118H) triggered weaker responses in DRB1*04:01-positive patients (35). Similarly, T716I affected the presentation landscape of DRB1*07:01 and abolished T cell reactivity (Fig. 4F). While antibody staining (Fig. 4E) and MEDi-MA scores showed that the 15-mer S714–728(T716I) was presented as well as the WT, they also indicated that mutated peptides starting from Asp702 to Asn710 would be presented substantially better than WT (Fig. 6D). The T716I mutation introduced a new P9 anchor residue at position 716, complementing residues Tyr707/Ser708, Ser711, and Ala713 to form a good DRB1*07:01-binding motif (Figs. 6D and 3A). Furthermore, T716I mutation introduced additional DRB1*07:01-binding motifs, potentially allowing three different presentation registers for peptide S714–728(T716I) (Fig. 6, E and F): first, comprising a weak HLA-binding motif starting at Ile714, with Thr716 directly facing the TCR; second, starting with the mutated Ile716 as a new anchor residue; and, last, where the T716I mutation would be outside the minimal epitope for TCR007. Thus, mutation T716I could abrogate TCR recognition by either of two mechanisms: It could alter peptide presentation on DRB1*07:01, or it could abolish direct TCR007 contacts.
Table 3. Effects of SARS-CoV-2 mutations on presentability of peptides.
↓, loss of presentation; ↑, higher presentation; ⟵, shift in presentation landscape.
| Strain | Mutation | DRB1*04:04 | DRB1*04:01 | DRB1*08:03 |
DPA1*02:02/ DPB1*05:01 |
DRB1*11:01 | DRB1*14:05 | DRB1*15:01 | DRB1*07:01 |
| B.1.1.7 | |||||||||
| D3L | ↑ | ||||||||
| S235F | ↓ | ↓ | |||||||
| T1001I | |||||||||
| A1708D | ↓ | ||||||||
| I2230T | ↓ | ||||||||
| SGF del | ↓ | ||||||||
| Y73C | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | |||
| HV69 del | ↓ | ||||||||
| Y144 del | |||||||||
| A570D | ↑ | ||||||||
| P681H | |||||||||
| T716I | ⟵↑ | ||||||||
| S982A | |||||||||
| D1118H | ⟵↑ | ↑ | ↑ | ↑ | ↑ | ||||
| B.1.1.7/351 | N501Y | ||||||||
| B.1.351 | |||||||||
| L18F | |||||||||
| D80A | |||||||||
| D215G | |||||||||
| 242-244del | ↓ | ||||||||
| R246I | ↓ | ↓ | ↓ | ↓ | ↓ | ||||
| K417T | |||||||||
| B.1.351/526 | E484K | ||||||||
| B.1.526 | A701V | ||||||||
| B.1.526 | D614G | ||||||||
| B.1.298 | Y453F |
Fig. 6. MEDi reveals candidate immune escape mutants.
(A) Micro MCR2 libraries, containing all 15 (15 amino acid long) peptides spanning the individual mutations were cloned for each HLA and transduced into the 16.2X reporter cells. Individual MEDi-MA scores for the WT (blue dots) and mutated (orange dots) peptides are shown. For context, the MEDi scores for the full Spike protein are shown below. Blue, orange, and green shaded squares indicate differences seen in all repeat experiments (n = 2 or 3). (B) Example peptide sequences from ORF8 and ORF1a with indicated starting residues and the MHC-binding motif for DRB1*04:04 are shown. (C) Detailed view of the MEDi-MA scores for the WT and D1118H Spike-mutated peptides in the context of DRB1*04:01. (D) Detailed view of the MEDi-MA scores for the WT and T716I Spike-mutated peptides in the context of DRB1*07:01. AU, arbitrary units. (E) Fifteen-mer peptides spanning the T716I mutation with indicated starting residues and the different DRB1*07:01-binding motifs. (F) S714–728 peptide sequences with indicated different binding registers forced by several DRB1*07:01-binding motifs present in the WT and/or mutated peptide. TCR-facing residues are highlighted in gray. (G) FACS analysis and sorting of reporter cells transduced with DRB1*07:01-MCR2 carrying the 12-mer peptides: S714–725, S714–725(T716I), and S717–728. (H) Reporter cells from (F) were cocultured with 16.2A2 cells transduced with TCR007, and NFAT activation was measured by flow cytometry.
To answer this question, we cloned 12-mer peptides S714–725, S714–725(T716I), and S717–728 into the DRB1*07:01-MCR2 and cocultured MCR2+ reporter cells with TCR007 T cells. As shown in Fig. 6G, all constructs were expressed well, with S717–728 reaching the highest levels indicating best presentation. TCR007 recognized S717–728 but not S714–725 (Fig. 6H). This indicates that T716I abrogated TCR recognition of the S714–728 15-mer indirectly. Change of the presentation register appears as a likely reason, but steric hindrance cannot be excluded at this point.
These results suggest several mechanisms of peptide presentation modulation and highlight the ability of the MEDi platform to decipher molecular details underlying possible viral immune escape strategies. Comprehensive analyses of the arising viral mutants, studying the relation of presentability and immunogenicity, will be important for the development of future therapeutics.
DISCUSSION
Identifying the specificity of pathogen-reactive lymphocytes is crucial for the fields of therapeutics and vaccine development. While protection from viral infections is mostly attributed to B cell and CD8 T cell effector functions, the balance between enabling and restricting them decides the life and death of the host. Thus, understanding the CD4 T cell reactivity, which orchestrates these responses, is critical, and deep knowledge of epitope presentation by HLA class II would greatly help clinical developments. However, owing to the limited sensitivity of MS and varying accuracy of the in silico methods, it is difficult to generate peptide presentation landscapes across multiple HLA alleles. With MEDi, we provide a powerful alternative approach based on functional cell surface expression of the MCR2 molecules. MEDi is HLA agnostic thanks to the association of MCR2 with the CD3 chains. It allows unbiased, fast, and affordable testing of all antigen-derived peptides for their ability to be presented by an HLA on the surface of a mammalian cell in a more physiological context than yeast display systems (6, 36). We show proof-of-concept experiments, indicating that antigenic peptides usually reside within the MEDi high regions (some missed by prediction algorithms), provide a list of presentable SARS-CoV-2 peptides for several different HLA alleles, and describe potential mechanisms of viral immune evasion.
We validated MEDi results biochemically and biologically through presentation analysis of immunogenic epitopes found by “deorphaning” (37) TCRs from more than a hundred T cells enriched in the lungs of patients with acute COVID-19. Highlighting the importance of CD4 T cells, we found that among the enriched TCRs, 8 of 47 (17.0%) CD4-derived ones and 3 of 63 (4.7%) of the CD8-derived ones recognized SARS-CoV-2 peptides.
The appearance of mutated SARS-CoV-2 with higher transmissibility raises important questions about the selective pressure that gave rise to the fitter variants and the role of immune escape in their evolution. While viral escape from antibody-mediated neutralization has been well documented for many diseases (38, 39), including COVID-19 (40–42), much less is known about a potential selective pressure to evade T cell reactivity. Understanding HLA presentation and TCR recognition of mutant and WT epitopes is critical in this regard. One of the recent studies using peptide pools suggested little effect of the novel mutations on peptide immunogenicity (43). Another, using MHC class I tetramer staining, described that only 1 of the 52 screened mutations was located in a CD8 epitope and concluded that most CD8 T cell responses in COVID-19 convalescent individuals target conserved epitopes (44). However, Agerer et al. (45) showed that several mutant peptides exhibited diminished MHC-I binding and were less well recognized by CD8 T cells, suggesting possible immune escape. In line, we found that several mutations present in the emerging SARS-CoV-2 variant strains reduced presentability of the affected peptides by several HLA class II alleles. Furthermore, two of the eight immunogenic peptides found in our study were targeted by the arising mutations. Both mutations were just outside the minimal epitopes, but one affected TCR recognition anyways. We tested two different mechanisms of escape and found that Thr716 was not directly bound by the TCR but that the T716I mutation likely altered peptide presentation by enabling binding in a different register. This evasion strategy could affect all T cells recognizing this peptide, so the T716I mutation might provide a bigger advantage for the virus than appreciated so far. In line, previous studies have shown the influence of so-called peptide-flanking residues on HLA binding and T cell recognition (46). Furthermore, given the optimal peptide length for MHC class II being 18 to 20 amino acids (24), it is very likely that most peptides, comprising the HLA-binding core starting at Phe718, will include Thr/Ile716. Notably, these results indicate a molecular mechanism of escape from TCR recognition, but they do not directly imply clinically relevant immune evasion. For this to happen, a particular set of HLA alleles would have to be present in the patient, where the peptide is mainly presented by one allele, and the epitope would have to be among the major drivers of the T cell response. While such a scenario is feasible, it was not studied here.
The biggest advantage of MEDi is that it is easily scalable to the thousands of alleles present in humans, including DP and DQ for which predictions are not as good, and enables peptide presentability studies with patient-specific HLA alleles for which no good training data are available. Consistently, the immunogenic spike S955–969 peptide presented by DPA1*02:02/DPB1*05:01 and N221–242 presented by DRB1*14:05, both MEDi high, were not well predicted by netMHCIIpan. Our results also indicate that direct attachment of the peptides limits the potentially confounding effects of the allele-intrinsic differential stability of the “empty” conformation. MEDi provides good results for unstable alleles, such as DRB1*15:01, as well as alleles expected to be more stable such as the DRB1*04:04 or DP. However, allele-intrinsic differential stability does affect MEDi score distribution and range to some degree (peak height and valley levels), reflecting the physiological features of the alleles.
Furthermore, with MEDi, we quickly provide presentability information for any immunogenic peptide across multiple HLA alleles, which can also support further training of predictive models similar to Rappazzo et al. (6). This is exemplified by the very immunogenic membrane protein peptide M146–165, recognized by TCR091 in the context of DRB1*11:01, and shown by MEDi to be also presentable by several other HLAs not predicted by netMHCIIpan. Predictions by MARIA were consistent with these MEDi results (27). However, some peptides identified by MEDi may not be efficiently presented in vivo owing to natural peptide processing and the actions of chaperons such as HLA-DM. Another limitation of MEDi is that, except for glycosylation naturally performed by the reporter cell line and deamidation mimicked by the introduction of new amino acids (D and E), posttranslational modifications cannot be easily studied. Furthermore, broad use of MEDi is currently limited to MHC class II, because SCTz fused to different peptides were all expressed on the cell surface, irrespective of correct folding (fig. S7). Unfortunately, monoclonal antibodies specifically distinguishing the native HLA conformation from the misfolded one are not available for many alleles, precluding the broad use of SCTz for MEDi-type applications at present.
The results presented in this study validate the MEDi platform and provide insights into the molecular mechanisms of SARS-CoV-2 peptide presentation and potential escape from T cell recognition. MEDi should help in closing the gaps in peptide presentation landscapes for thousands of HLA alleles and be useful for the development of novel therapeutical approaches beyond prevention of COVID-19 or treatment of patients with SARS-CoV-2.
MATERIALS AND METHODS
Cell lines, molecular cloning, and retroviral transduction
Cell lines, molecular cloning procedures, and retroviral transduction used in this study were described previously (17).
Coculture of TCR-carrying reporter cells with peptide-pulsed PBMCs
TCR-carrying reporter cells were cocultured in triplicates with peptide-pulsed PBMCs (purchased at Sanvitra) at a ratio of 1:20 in standard cell culture conditions for 6 and 9 hours (fig. S4). Peptide concentrations of 10, 5, 2, 1 μg/ml were used. In addition, coculture with PBMCs pulsed with one of the nonspecific peptides (10 μg/ml) was used as a negative control and coculture with MCR2+ reporter cells as a positive control. Cells were washed, stained with anti–CD69-allophycocyanin and anti-human CD4-phycoerythrin. Data were acquired on a BD LSRFortessa and analyzed with FlowJo. TCR+ reporter cells were gated by size and hCD4 expression. CD69 gates were set according to the reporter PBMC cocultures without peptides. Analysis of variance (ANOVA) with Tukey’s multiple comparisons test was done with Prism software.
MEDi procedure and score calculation
Libraries carrying 15–amino acid–long peptides, spanning the entire sequences of the SARS-CoV-2 virus, were cloned as oligonucleotides (Twist Bioscience) into MCR vectors carrying different HLA alleles. 16.2X reporter cell line was transduced with these libraries, and surface expression of the MCR molecules was analyzed by flow cytometry. Four fractions were sorted: Fr.0 (cells expressing no detectable MCRs on the surface), Fr.1 (cells expressing low levels of MCRs), Fr.2 (cells expressing intermediate levels of MCRs), and Fr.3 (cells expressing high levels of MCRs) (see Fig. 3A). For high-quality MEDi scores, it is crucial to sort enough cells per fraction, at least 50 times the library complexity. Peptides carried by the MCRs from sorted cells were amplified from cDNA by RT-PCR using the peptide flanking regions and sequenced on a MiniSeq (Illumina). Sequences from the Illumina output files were trimmed, merged, and translated with the help of the CLC Genomic Workbench program. Counting and further analysis were done with FileMaker Pro18 and Excel (Microsoft).
The individual peptide counts in each fraction were normalized to the total counts in the fraction. For each peptide, a MEDi score was calculated with the following formula: sum_i (Fr_index_i × Fr_count_i)/sum_i(Fr_count_i), with Fr_indexes Fr0 = 1, Fr1 = 2, Fr2 = 4, and Fr3 = 28. MEDi-MA was calculated by averaging MEDi scores for five peptides (−2/−1/0/1/2) and assigned to the middle(0) peptide (see also data file S1, sheet Spike; Fig. 3), except for Fig. 6 and fig. S4, where MEDi-MA was calculated by averaging MEDi scores for three peptides (−1/0/1). The 85th percentile of the MEDi-MA score was calculated for each individual protein.
Statistics
ROC curves were made by plotting the true-positive rate (true positive/number of real positive) versus false-positive rate (false positive/number of real negative). Sensitivity was calculated for two IC50 thresholds (1 and 10 μM) by dividing the number of true positive (peptides having an IC50 above the threshold and scoring above the 85th percentile) by the total number of real positives (peptides having an IC50 above the threshold in the competitive peptide binding assay). Specificity was calculated by dividing the number of true negative (peptides having an IC50 below the threshold and scoring below the 85th percentile) by the total number of real negatives. Fisher’s exact test was calculated with the help of Prism software.
MEDi-MA score quality threshold
MEDi-MA score for a given peptide was considered of good quality if at least 40 reads were collected for a peptide and the MEDi-MA value had a coefficient of variation (CV = Std.Deviation/Average) lower than 0.75.
MCR2 screening
Libraries were generated by cloning all SARS-CoV-2–derived peptides in MCR2 molecules carrying the complete viral genome in 23-mers shifted by one amino acid. For screening, we pooled the libraries at equal ratios, generating a combined patient-specific library of roughly 120,000 different peptide-MCR2 combinations (Table 1).
MCR2 screening was performed as described previously (17). Briefly, MCR2 expressing 16.2X cells have been cocultured with cell clones expressing one specific TCR selected from the work of Liao et al. (33) in a ratio of 1:5 to 1:10. Cells were mixed and cocultured for 8 to 12 hours in a standard tissue culture medium, in the presence of anti-mouse FasL antibodies (13 μg/ml; Bio X Cell) to inhibit induction of cell death during incubation. After harvesting, reporter cells positive for nuclear factor of activated T cell (NFAT) signaling [mutant mCherry: slow fluorescent timer, detection filter 465/30 (FL11-A) from violet laser at 405 nM] have been sorted on a BD FACSAria Fusion Cell Sorter as bulk or as single cells into 96-well plates for further expansion. On average, four to five enrichment rounds per TCR have been performed before single reporter cells have been sorted. Expanded single cells were harvested, and DNA was isolated (KAPA Express Extract), followed by Sanger sequencing of the MCR2 α and β chain including the linked antigen. Whenever overlapping peptides were found in the screen (e.g., Fig. 5A), in Fig. 1D, we listed the common part as the specific peptides recognized by the TCR.
SCTz screening
Single-chain trimers of class I HLAs of all seven patients have been generated by linking the leader sequence, epitope, b2m, and HLA α chain with 3 × G4S linkers and the addition of the intracellular domain from the CD247(zeta chain) molecules. Each α chain was modified/mutated to open the groove of class I by introducing the Y84A mutation in every α chain (47). For SCTz screening, we again used libraries covering the whole SARS-CoV-2 genome with 10-mers shifted by one amino acid, cloned as oligonucleotides into the SCTz vectors.
Fluorescence polarization assay
The MHC II α and β chain extracellular domains were recombinantly expressed with C-terminal Myc and His tag sequences, respectively. For DRB1*15:01, the Myc tag was replaced with a V5 tag. The N terminus of the β chain was fused to class II–associated invariant chain peptide (CLIP) followed by a flexible factor Xa–cleavable linker. Both α and β chains were coexpressed in Chinese hamster ovary (CHO) cells and secreted into the expression medium as a stable CLIP-loaded heterodimer. Heterodimerization of the α and β chains of DRB1*07:01 and DRB1*1501 was forced using a fusion of an engineered human immunoglobulin G1–Fc protein to each chain (48). Following CHO expression, the heterodimer was purified by immobilized metal ion affinity chromatography and size exclusion chromatography. The fluorescence polarization assay was performed as described in (26) with a few modifications. Following factor Xa cleavage, 100 nM HLA was incubated overnight with 25 nM fluorescent probe and various concentrations of the indicated peptide competitor in 100 mM sodium citrate (pH 5.5), 100 mM NaCl, 0.1% octylglucoside, and 1× protease inhibitors (SIGMAFAST) at 30°C. The fluorescent probe for DRB1*04:04, DRB1*07:01, and DRB1*11:01 was PRFV(K/Alexa488)QNTLRLAT. The fluorescent probe for DRB1*15:01 was ENPVVHFF(C/Alexa488Mal)NIVTPR.
Acknowledgments
We thank D. Acker (Repertoire Immune Medicines) for discussions related to MEDi score analysis. We thank A. Coyle and T. Harris (Repertoire Immune Medicines) for discussions and critical reading of the manuscript, A. Schütz and M. Kisielow (ETHZ Flow Cytometry Core Facility) for help with cell sorting, and D. Kollegger and J. Meier for technical assistance.
Author contributions: Conceptualization: F.-J.O. and J.K. Methodology: F.-J.O., M.K., J.S., and J.K. Investigation: F.-J.O., F.R., S.H., C.H.L., N.C., M.L., G.M., Y.H., R.W., O.I., P.P., K.T., and J.K. Resources: M.K. and J.S. Writing (original draft): J.K. Writing (review and editing): F.-J.O., M.K., J.S., and J.K. Supervision: F.-J.O., J.S., and J.K.
Competing interests: F.-J.O., O.I., P.P., K.T., J.S., and J.K. are employees and/or stockholders of Repertoire Immune Medicines. M.K. is an advisor and stockholder of Repertoire Immune Medicines. F.-J.O., F.R., S.H., N.C., M.L., G.M., Y.H., R.W., and J.K. are employees of Repertoire Immune Medicines (Switzerland) AG, formerly Tepthera Ltd. F.-J.O. and J.K. are inventors on a patent application related to this work filed by Repertoire Immune Medicines (WO2022026921) in 30 July 2021, published 3 February 2022.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The reporter cell lines and constructs used in this study can be provided by Repertoire Immune Medicines pending scientific review and a completed material transfer agreement. Requests for the materials should be submitted to J.K.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S2
REFERENCES AND NOTES
- 1.Sofron A., Ritz D., Neri D., Fugmann T., High-resolution analysis of the murine MHC class II immunopeptidome: Immunity to infection. Eur. J. Immunol. 46, 319–328 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kowalewski D. J., Schuster H., Backert L., Berlin C., Kahn S., Kanz L., Salih H. R., Rammensee H.-G., Stevanovic S., Stickel J. S., HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc. Natl. Acad. Sci. U.S.A. 112, E6254–E6256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W., Sher X., Peters B., Systematically benchmarking peptide-MHC binding predictors: From synthetic to naturally processed epitopes. PLOS Comput. Biol. 14, e1006457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen K. K., Andreatta M., Marcatili P., Buus S., Greenbaum J. A., Yan Z., Sette A., Peters B., Nielsen M., Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynisson B., Barra C., Kaabinejadian S., Hildebrand W. H., Peters B., Nielsen M., Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J. Proteome Res. 19, 2304–2315 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Rappazzo C. G., Huisman B. D., Birnbaum M. E., Repertoire-scale determination of class II MHC peptide binding via yeast display improves antigen prediction. Nat. Commun. 11, 4414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloan V. S., Cameron P., Porter G., Gammon M., Amaya M., Mellins E., Zaller D. M., Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature 375, 802–806 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Amria S., Hajiaghamohseni L. M., Harbeson C., Zhao D., Goldstein O., Blum J. S., Haque A., HLA-DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur. J. Immunol. 38, 1961–1970 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z., Reyes-Vargas E., Escobar H., Chang K. Y., Barker A. P., Rockwood A. L., Delgado J. C., He X., Jensen P. E., Peptidomic analysis of type 1 diabetes associated HLA-DQ molecules and the impact of HLA-DM on peptide repertoire editing: Immunodeficiencies and autoimmunity-basic. Eur. J. Immunol. 47, 314–326 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Oldford S. A., Robb J. D., Codner D., Gadag V., Watson P. H., Drover S., Tumor cell expression of HLA-DM associates with a Th1 profile and predicts improved survival in breast carcinoma patients. Int. Immunol. 18, 1591–1602 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Peiris J. S. M., Lai S. T., Poon L. L. M., Guan Y., Yam L. Y. C., Lim W., Nicholls J., Yee W. K. S., Yan W. W., Cheung M. T., Cheng V. C. C., Chan K. H., Tsang D. N. C., Yung R. W. H., Ng T. K., Yuen K. Y., Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matheson N. J., Lehner P. J., How does SARS-CoV-2 cause COVID-19? Science 369, 510–511 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Peng Y., Mentzer A. J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D. I., Paesen G. C., Grimes J. M., Antson A. A., Bayfield O. W., Hawkins D. E. D. P., Ker D.-S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B. M., Fry J. W., Schwabe N. F., Semple M. G., Baillie J. K., Moore S. C., Openshaw P. J. M., Ansari M. A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-P.; Oxford Immunology Network Covid-19 Response T cell Consortium; ISARIC4C Investigators, Cornall R. J., Conlon C. P., Klenerman P., Screaton G. R., Mongkolsapaya J., McMichael A., Knight J. C., Ogg G., Dong T., Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 21, 1336–1345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelde A., Bilich T., Heitmann J. S., Maringer Y., Salih H. R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., Peter A., Hörber S., Traenkle B., Kaiser P. D., Rothbauer U., Becker M., Junker D., Krause G., Strengert M., Schneiderhan-Marra N., Templin M. F., Joos T. O., Kowalewski D. J., Stos-Zweifel V., Fehr M., Rabsteyn A., Mirakaj V., Karbach J., Jäger E., Graf M., Gruber L.-C., Rachfalski D., Preuß B., Hagelstein I., Märklin M., Bakchoul T., Gouttefangeas C., Kohlbacher O., Klein R., Stevanović S., Rammensee H.-G., Walz J. S., SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 22, 74–85 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sette A., Crotty S., Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184, 861–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisielow J., Obermair F.-J., Kopf M., Deciphering CD4+ T cell specificity using novel MHC–TCR chimeric receptors. Nat. Immunol. 20, 652–662 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Rabinowitz J. D., Vrljic M., Kasson P. M., Liang M. N., Busch R., Boniface J. J., Davis M. M., McConnell H. M., Formation of a highly peptide-receptive state of class II MHC. Immunity 9, 699–709 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Stadinski B. D., Zhang L., Crawford F., Marrack P., Eisenbarth G. S., Kappler J. W., Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc. Natl. Acad. Sci. 107, 10978–10983 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Bert N., Tan A. T., Kunasegaran K., Tham C. Y. L., Hafezi M., Chia A., Chng M. H. Y., Lin M., Tan N., Linster M., Chia W. N., Chen M. I.-C., Wang L.-F., Ooi E. E., Kalimuddin S., Tambyah P. A., Low J. G.-H., Tan Y.-J., Bertoletti A., SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Juno J. A., Tan H.-X., Lee W. S., Reynaldi A., Kelly H. G., Wragg K., Esterbauer R., Kent H. E., Batten C. J., Mordant F. L., Gherardin N. A., Pymm P., Dietrich M. H., Scott N. E., Tham W.-H., Godfrey D. I., Subbarao K., Davenport M. P., Kent S. J., Wheatley A. K., Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 26, 1428–1434 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Sauer K., Harris T., An effective COVID-19 vaccine needs to engage T cells. Trends Immunol. 11, 581807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreatta M., Schafer-Nielsen C., Lund O., Buus S., Nielsen M., NNAlign: A Web-based prediction method allowing non-expert end-user discovery of sequence motifs in quantitative peptide data. PLOS ONE 6, e26781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Brien C., Flower D. R., Feighery C., Peptide length significantly influences in vitro affinity for MHC class II molecules. Immunome Res. 4, 6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knierman M. D., Lannan M. B., Spindler L. J., McMillian C. L., Konrad R. J., Siegel R. W., The human leukocyte antigen class II immunopeptidome of the SARS-CoV-2 spike glycoprotein. Cell Rep. 33, 108454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin L., Stern L. J., Measurement of peptide binding to MHC class II molecules by fluorescence polarization. Curr. Protoc. Immunol. 106, 5.10.1–5.10.12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.E. Fast, R. B. Altman, B. Chen, Potential T-cell and B-cell epitopes of 2019-nCoV. bioRxiv 955484 [Preprint]. 2020. http://biorxiv.org/lookup/doi/10.1101/2020.02.19.955484.
- 28.Chen B., Khodadoust M. S., Olsson N., Wagar L. E., Fast E., Liu C. L., Muftuoglu Y., Sworder B. J., Diehn M., Levy R., Davis M. M., Elias J. E., Altman R. B., Alizadeh A. A., Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 37, 1332–1343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarke A., Sette A., Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2, 100204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen T. H., Lybarger L., Exciting applications of single chain trimers of MHC-I molecules. Cancer Immunol. Immunother. 55, 235–236 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T., He X., Tsang T. C., Harris D. T., SING: A novel strategy for identifying tumor-specific, cytotoxic T lymphocyte-recognized tumor antigens. FASEB J. 18, 600–602 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Joglekar A. V., Leonard M. T., Jeppson J. D., Swift M., Li G., Wong S., Peng S., Zaretsky J. M., Heath J. R., Ribas A., Bethune M. T., Baltimore D., T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat. Methods 16, 191–198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020). [DOI] [PubMed] [Google Scholar]
- 34.A. Rambaut, N. Loman, E. Volz; COVID-19 Genomics Consortium UK, Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations (2020); https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 35.Reynolds C. J., Pade C., Gibbons J. M., Butler D. K., Otter A. D., Menacho K., Fontana M., Smit A., Sackville-West J. E., Cutino-Moguel T., Maini M. K., Chain B., Noursadeghi M.; UK COVIDsortium Immune Correlates Network, Brooks T., Semper A., Manisty C., Treibel T. A., Moon J. C.; UK COVIDsortium Investigators, Valdes A. M., McKnight Á., Altmann D. M., Boyton R., Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science 30, eabh1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W., Boder E. T., High-throughput engineering and analysis of peptide binding to class II MHC. Proc. Natl. Acad. Sci. 107, 13258–13263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerber H.-P., Sibener L. V., Lee L. J., Gee M. H., Identification of antigenic targets. Trends Cancer 6, 299–318 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Kwong P. D., Doyle M. L., Casper D. J., Cicala C., Leavitt S. A., Majeed S., Steenbeke T. D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P. W. I. H., Robinson J., Van Ryk D., Wang L., Burton D. R., Freire E., Wyatt R., Sodroski J., Hendrickson W. A., Arthos J., HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Kleine-Weber H., Elzayat M. T., Wang L., Graham B. S., Müller M. A., Drosten C., Pöhlmann S., Hoffmann M., Gallagher T., Mutations in the spike protein of middle east respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J. Virol. 93, e01381-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J. C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C. D., Caskey M., Robbiani D. F., Rice C. M., Nussenzweig M. C., Hatziioannou T., Bieniasz P. D., Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z., VanBlargan L. A., Bloyet L.-M., Rothlauf P. W., Chen R. E., Stumpf S., Zhao H., Errico J. M., Theel E. S., Liebeskind M. J., Alford B., Buchser W. J., Ellebedy A. H., Fremont D. H., Diamond M. S., Whelan S. P. J., Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomson E. C., Rosen L. E., Shepherd J. G., Spreafico R., da Silva Filipe A., Wojcechowskyj J. A., Davis C., Piccoli L., Pascall D. J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O’Toole A., Pinto D., Colquhoun R. M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V. B., Nix J., Zhang I., Jarrett R. F., Glass W. G., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Croll T. I., Johnson N., Di Iulio J., Wickenhagen A., Ceschi A., Harbison A. M., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Schiuma M., Semple M. G., Openshaw P. J. M., Fadda E., Baillie J. K., Chodera J. D.; ISARIC4C Investigators 20; COVID-19 Genomics UK (COG-UK) Consortium, Rihn S. J., Lycett S. J., Virgin H. W., Telenti A., Corti D., Robertson D. L., Snell G., Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Tarke, J. Sidney, N. Methot, Y. Zhang, J. M. Dan, B. Goodwin, P. Rubiro, A. Sutherland, R. da Silva Antunes, A. Frazier, S. A. Rawlings, D. M. Smith, B. Peters, R. H. Scheuermann, D. Weiskopf, S. Crotty, A. Grifoni, A. Sette, Negligible impact of SARS-CoV-2 variants on CD4 + and CD8 + T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv 433180 [Preprint]. 2021. 10.1101/2021.02.27.433180. [DOI]
- 44.A. D. Redd, A. Nardin, H. Kared, E. M. Bloch, A. Pekosz, O. Laeyendecker, B. Abel, M. Fehlings, T. C. Quinn, A. A. Tobian, CD8+ T cell responses in COVID-19 convalescent individuals target conserved epitopes from multiple prominent SARS-CoV-2 circulating variants. medRxiv (2021); 10.1101/2021.02.11.21251585. [DOI] [PMC free article] [PubMed]
- 45.Agerer B., Koblischke M., Gudipati V., Montaño-Gutierrez L. F., Smyth M., Popa A., Genger J.-W., Endler L., Florian D. M., Mühlgrabner V., Graninger M., Aberle S. W., Husa A.-M., Shaw L. E., Lercher A., Gattinger P., Torralba-Gombau R., Trapin D., Penz T., Barreca D., Fae I., Wenda S., Traugott M., Walder G., Pickl W. F., Thiel V., Allerberger F., Stockinger H., Puchhammer-Stöckl E., Weninger W., Fischer G., Hoepler W., Pawelka E., Zoufaly A., Valenta R., Bock C., Paster W., Geyeregger R., Farlik M., Halbritter F., Huppa J. B., Aberle J. H., Bergthaler A., SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8 + T cell responses. Sci. Immunol. 6, eabg6461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnold P. Y., La Gruta N. L., Miller T., Vignali K. M., Adams P. S., Woodland D. L., Vignali D. A. A., The majority of immunogenic epitopes generate CD4+ T cells that are dependent on MHC class II-bound peptide-flanking residues. J. Immunol. 169, 739–749 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Lybarger L., Yu Y. Y. L., Miley M. J., Fremont D. H., Myers N., Primeau T., Truscott S. M., Connolly J. M., Hansen T. H., Enhanced immune presentation of a single-chain major histocompatibility complex class I molecule engineered to optimize linkage of a C-terminally extended peptide. J. Biol. Chem. 278, 27105–27111 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Gunasekaran K., Pentony M., Shen M., Garrett L., Forte C., Woodward A., Ng S. B., Born T., Retter M., Manchulenko K., Sweet H., Foltz I. N., Wittekind M., Yan W., Enhancing antibody Fc heterodimer formation through electrostatic steering effects. J. Biol. Chem. 285, 19637–19646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Data files S1 to S2