Abstract
Dysregulated Wnt/β-catenin signaling is implicated in the pathogenesis of many human cancers, including colorectal cancer (CRC), making it an attractive clinical target. With the aim of inhibiting oncogenic Wnt activity, we developed a high-throughput screening AlphaScreen assay to identify selective small-molecule inhibitors of the interaction between β-catenin and its coactivator BCL9. We identified a compound that consistently bound to β-catenin and specifically inhibited in vivo native β-catenin/BCL9 complex formation in CRC cell lines. This compound inhibited Wnt activity, down-regulated expression of the Wnt/β-catenin signature in gene expression studies, disrupted cholesterol homeostasis, and significantly reduced the proliferation of CRC cell lines and tumor growth in a xenograft mouse model of CRC. This study has therefore identified a specific small-molecule inhibitor of oncogenic Wnt signaling, which may have value as a probe for functional studies and has important implications for the development of novel therapies in patients with CRC.
In colorectal cancer, C-1 inhibits oncogenic Wnt activity in vitro and in vivo by blocking β-catenin/BCL9 complex formation.
INTRODUCTION
The Wnt/β-catenin pathway plays critical roles in stem cell renewal, embryonic development, and adult tissue homeostasis (1) due to the importance of Wnt signaling in cellular proliferation and differentiation. One tissue type particularly reliant on Wnt signaling is the intestinal epithelium, which is regenerated by Wnt-induced differentiation of crypt stem cells (2). Aberrant Wnt signaling is involved in the pathogenesis of a wide range of common human cancers including colorectal carcinoma (CRC) among many others (3–10). In many of these cancers, dysregulated Wnt signaling is associated with tumor growth, metastasis, poor prognosis, and lower survival rates (11). In the canonical pathway and in the absence of Wnt ligand, cytoplasmic β-catenin is phosphorylated and targeted for proteasomal degradation by a destruction complex including Axin, adenomatous polyposis coli (APC), protein phosphatase 2A, glycogen synthase kinase 3, and casein kinase 1α (CK1α). Wnt/β-catenin signaling is initiated by the binding of Wnt ligands to the extracellular domain of a Frizzled (Fz) family GTP-binding protein (G protein)–coupled receptor and the low-density lipoprotein receptor–related protein 5 (LRP5), which allows the signal to be transmitted into the cytoplasm and ultimately inhibits the destruction complex (12). Accumulated β-catenin translocates to the nucleus, binds to DNA through T cell factor (TCF)/lymphoid enhancing factor transcription factors, recruits transcriptional coactivators, and initiates gene transduction to achieve a cellular response (13). The two closely related B cell lymphoma 9 (BCL9) and BCL9-like (B9L) genes encode for well-established transcriptional coactivators of β-catenin in the canonical Wnt pathway (14–17). A variety of loss-of-function mutations in APC and Axin, as well as activating mutations in β-catenin itself, enable constitutive Wnt transcription activation and malignant transformation (3, 4, 13). Studies from our laboratory and others have demonstrated that inappropriate activation of Wnt transcriptional activity can also be brought about by low β-catenin turnover (6, 7, 18–20), elevated nuclear BCL9 (21, 22), and decreased levels of miR-30s (an endogenous BCL9 regulator) (23, 24), whereas inhibition of BCL9 expression, or of its binding to β-catenin, is associated with antitumor activity (21, 22).
Because of its important role in a wide range of common cancers, the Wnt/β-catenin pathway has emerged as a desirable therapeutic target (25–27), but development of Wnt-targeted therapies has been hampered by toxicity and off-target effects. β-Catenin interacts with most of its protein partners via the same binding surface (16), so identifying agents that can selectively disrupt its cancer-promoting activities while leaving its homeostatic functions intact is challenging (27). For example, inhibitors of β-catenin/TCF4 complexes have been described but were found to induce severe anemia and generalized wasting (28, 29); this is likely due to the disruption of epithelial tissue integrity caused by the shared binding site of TCF4 and E-cadherin with β-catenin (30). Other inhibitors of Wnt signaling that bind cyclic adenosine monophosphate response element–binding (CREB) protein (31–34), porcupine (35, 36), tankyrase (37), CK1α (38), or Fz receptors (39, 40) are similarly associated with off-target and toxic side effects.
We set out to develop an inhibitor of the β-catenin/BCL9 interaction downstream in the nucleus. One of the fundamental reasons for this approach is that BCL9 activates β-catenin–driven transcription through direct binding of its homology domain 2 (HD2) via a single amphipathic α helix to a distinct site on β-catenin that is not shared with other protein partners (16). Inhibition of β-catenin/BCL9 complexes should therefore not affect interactions of β-catenin with protein partners such as E-cadherin, Axin, and APC, which may substantially reduce toxic side effects. In addition, genetic deletion of BCL9 and B9L in the intestinal epithelium of mice, which effectively eliminates the presence of β-catenin/BCL9/B9L complexes, was not found to cause any phenotypic consequences in the animals, further suggesting that blocking BCL9 may not be harmful to normal intestinal cells (41). This strategy was also appealing because BCL9 is overexpressed in several tumor tissues compared to the cells of origin, potentially revealing a broad therapeutic window, and inhibition of BCL9 expression is associated with antitumor activity (21, 22). In keeping with this, we have previously developed stapled α-helical peptidomimetics of the BCL9-HD2 domain (SAH-BCL9), which demonstrated in vitro and in vivo antitumor activity without detectable side effects, and provided the proof of concept for this strategy (22). SAH-BCL9 peptides did not have pharmacokinetic properties conducive to clinical development due to high binding of serum proteins; therefore, we instead developed a high-throughput AlphaScreen assay to identify small molecules that can disrupt the β-catenin/BCL9 interaction and inhibit oncogenic Wnt signaling. We identified several candidates that inhibited β-catenin/BCL9 complex formation in complementary biophysical assays. Two lead compounds specifically inhibited the expression of Wnt target genes, Wnt activity, and proliferation in CRC cell lines. The top-performing inhibitor in this study was also found to dysregulate cholesterol homeostasis in CRC cells and significantly reduced tumor burden in a Wnt-dependent CRC mouse model. This novel and specific small-molecule inhibitor of the β-catenin/BCL9 interaction may therefore have value as a functional probe to inhibit the Wnt pathway, to investigate the role of Wnt signaling in cholesterol homeostasis, and for the development of novel targeted therapies for patients suffering from Wnt-dependent cancers.
RESULTS
Identification of candidate β-catenin/BCL9 interaction inhibitors
To find small molecules that could disrupt the β-catenin/BCL9 interaction, we developed a high-throughput screening (HTS) assay with AlphaScreen technology (42). The assay platform used full-length recombinant β-catenin bound to a protein A-tagged acceptor bead (AB) via a specific anti–β-catenin antibody and biotinylated BCL9-HD2 peptide bound to a streptavidin-tagged donor bead (DB) (Fig. 1A). When the beads are in proximity because of β-catenin/BCL9-HD2 binding, light is produced by the AB at 615 nm as a result of the DB being excited at 680 nm. Following complex formation, inhibition of the β-catenin/BCL9-HD2 interaction is detected as a reduction in fluorescent emission intensity (Fig. 1B). The assay was miniaturized and adapted for HTS in 1536-well format, with a Z′ factor of 0.84 (Fig. 1C). Two rounds of HTS of a 320,000 small-molecule library were performed independently and confirmed good performance and a Gaussian distribution (Fig. 1, D and E). Compounds with nonselective activity (e.g., metal chelators, oxygen quenchers, and biotin mimetics) were eliminated. After five rounds of dose-response retesting, 240 hits (within 45 scaffold “clusters”) were identified, of which 195 had median inhibitory concentration (IC50) values of <25 μM, and of these, 88 had IC50 values <10 μM.
Fig. 1. Identification of β-catenin/BCL9 inhibitors.
(A) Schematic of the AlphaScreen assay that detects BCL9-HD2/β-catenin complex formation/disruption. (B) Ten-dose response of the AlphaScreen assay with BCL9-HD2 peptide and SAH-BCL9 (stapled BCL9-HD2 peptide). (C) Z′ evaluation of the screen comparing signal upon inhibition with an excess of BCL9-HD2. (D) Frequency plot of the distribution of the activity data from each well normalized to controls and expressed as percentage of inhibition relative to fully inhibited positive controls (no β-catenin in the assay or excess of nonbiotinylated BCL9-HD2 peptide). This frequency plot shows relatively normal distributions for both positive and negative (full binding of β-catenin to BCL9-HD2) controls and centroid of the compound activities, with some tailing to higher activity, consistent with the hit finding ability of this assay. (E) Scatterplot confirming the relative “flatness” of the main centroid of 0% compound activity with slight oscillations likely due to plate-to-plate variability and minor “edge” effects. (F) Ten-dose response AlphaScreen assay of BCL9 peptide (positive control) and five top-performing compounds: E722-2648 (C-1), L814-1428 (C-2), SYN22094413 (C-3), L859-1770 (C-4), and F838-0143 (C-5) titrated against full-length β-catenin (left) and BRD9 (right). (G) ITC of BCL9 peptide, C-1, C-2, C-3, C-4, and C-5. (H) Computational model of a molecularly docked β-catenin/C-1 complex. Ribbon (left), electrostatic surface contoured from −5 to +5 kT/e (middle), and ligand interaction map (right) representations of the lowest energy pose from an extra-precision Glide analysis of C-1 docked into the BCL9 pocket of the 3SL9 structure. All error bars represent means ± SD.
Top hits identified by HTS (table S1) that were commercially available were purchased and examined with a 10-dose response AlphaScreen assay, in which compounds were titrated against full-length β-catenin and against an unrelated protein (BRD9) as a counter-screen to further filter out any unspecific compounds (Fig. 1F). This assay validated 16 compounds that specifically and competitively inhibited β-catenin/BCL9, but not BRD9/ligand complexes (43). The five top-performing compounds include E722-2648 (C-1), L814-1428 (C-2), SYN22094413 (C-3), L859-1770 (C-4), and F838-0143 (C-5) and will be referred to using their shortened names (C-1 to C-5) throughout this manuscript (Fig. 1F). To further confirm specificity and characterize the binding thermodynamics of the top 16 compounds, we used isothermal titration calorimetry (ITC). Dissociation constant (KD) values for the five top-performing compounds ranged between 0.27 and 2.22 μM and were comparable to the BCL9 peptide used as a positive control (2.74 μM) (Fig. 1G and table S2).
Last, extra precision glide docking of C-1 into the BCL9 pocket of the first armadillo repeat of β-catenin (3SL9 structure) showed that the compound filled the central part of the BCL9 cleft (encompassing residues Ser362-Ile369 of BCL9) forming mostly van der Waals interactions with the hydrophobic surface, including residues Ala152-Met174 (Fig. 1H). Together, we identified and validated several compounds that bind to the first armadillo repeat of β-catenin and specifically inhibit the β-catenin/BCL9 interaction in biochemical and biophysical assays.
C-1 and C-2 inhibit β-catenin/BCL9 complex formation and Wnt activity
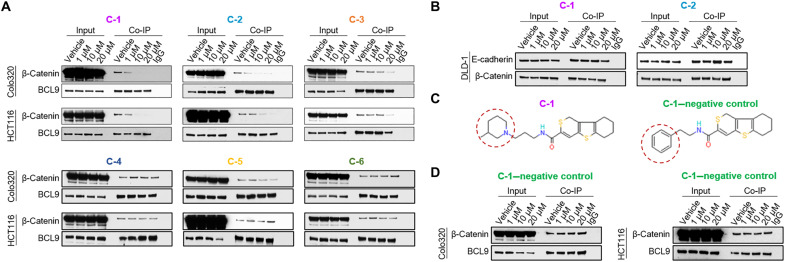
To determine whether these compounds could inhibit β-catenin/BCL9 complexes in cells, we carried out BCL9 coimmunoprecipitation (Co-IP) assays on the well-validated BCL9-dependent CRC cell lines, Colo320 and HCT116 (21, 44). Three of the six compounds that were tested inhibited β-catenin/BCL9 complexes (C-1 to C-3), and three did not (C-4 to C-6) (Fig. 2A). C-1 and C-2 inhibited β-catenin/BCL9 complexes at concentrations as low as 1 μM, while C-3 only appeared to significantly inhibit in vivo β-catenin/BCL9 complexes at a concentration of 20 μM (Fig. 2A). Hence, only C-1 and C-2 were taken forward into further studies. We next investigated whether compounds C-1 and C-2 were specific for BCL9 and did not inhibit β-catenin binding to other essential proteins such as E-cadherin. Accordingly, we performed β-catenin Co-IP assays in DLD-1 cells, which, in contrast to HCT116 and Colo320 cells, express high levels of E-cadherin. Neither compound disrupted the ability of β-catenin to form complexes with E-cadherin in DLD-1 cells (Fig. 2B), supporting their specificity and on-target activity. In addition, as a negative control for our lead compound (C-1), we identified a small molecule called E722-2546, within the same chemical series as C-1, which did not demonstrate activity in the HTS-AlphaScreen (Fig. 2C and table S3). This compound is used as and referred to as the “C-1–negative control” throughout this manuscript. We also confirmed that the C-1–negative control does not demonstrate activity in ITC (fig. S1) and does not inhibit the interaction of β-catenin and BCL9 in Co-IP studies (Fig. 2D).
Fig. 2. In vivo β-catenin/BCL9 complex formation in response to lead compounds.
(A) Immunoblots from BCL9 Co-IP assays of Colo320 and HCT116 cell lines, treated with vehicle, C-1, C-2, C-3, C-4, C-5, or C-6. Input samples (2%) are shown on the left-hand side of each panel. Samples incubated with normal rabbit immunoglobulin G (IgG) antibody are shown on the right-hand side of each panel. (B) Immunoblots from β-catenin Co-IP assays of the DLD-1 cell line, treated with vehicle, and two top-performing compounds (C-1 and C-2). Input samples (2%) are shown on the left-hand side of each panel. Samples incubated with normal rabbit IgG antibody are shown on the right-hand side of each panel. (C) The chemical structures of compound C-1 and the C-1–negative control. The red dashed circles highlight the part of the structure that differs in these two compounds. (D) Immunoblots from BCL9 Co-IP assays of Colo320 and HCT116 cell lines, treated with vehicle or increasing concentrations of the C-1–negative control compound. Input samples (2%) are shown on the left-hand side of each panel. Samples incubated with normal rabbit IgG antibody are shown on the right-hand side of each panel.
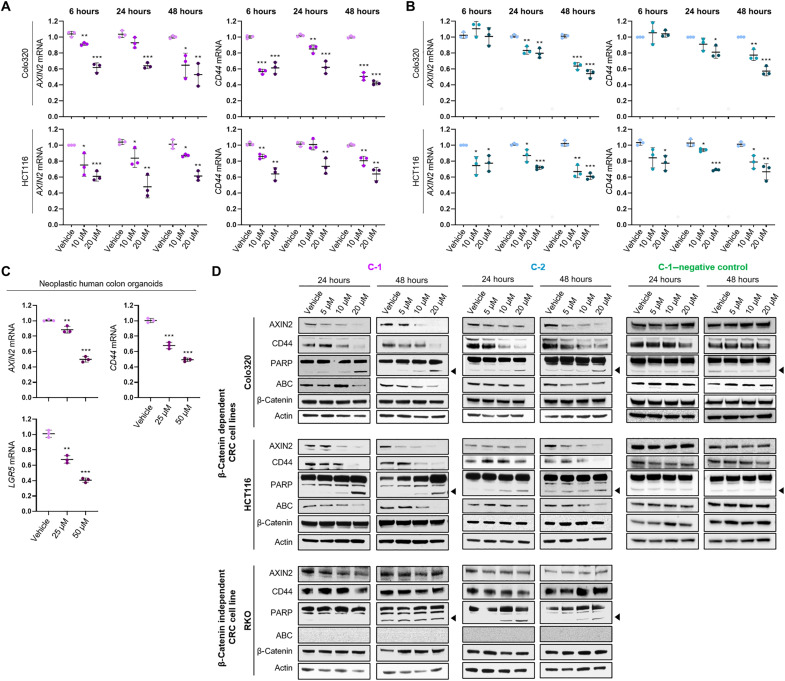
C-1 and C-2 were subsequently tested for their ability to inhibit the expression of bona fide downstream Wnt/β-catenin target genes, AXIN2 and CD44, in the β-catenin/BCL9–dependent CRC cell lines, Colo320 and HCT116, as well as in neoplastic human colonic organoids with a c.4778delA mutation in the APC gene. Both compounds significantly inhibited the expression of AXIN2 and CD44 mRNAs in Colo320 and HCT116 cells in a concentration- and time-dependent manner, as evaluated by reverse transcription quantitative polymerase chain reaction (RT-qPCR) (Fig. 3, A and B). C-1 treatment of neoplastic colonic organoids for 24 hours also significantly inhibited the expression of AXIN2, CD44, and additionally, the bona fide β-catenin/BCL9 target LGR5, in a concentration-dependent manner (Fig 3C). Furthermore, immunoblot analysis demonstrated that C-1 and C-2 treatment significantly decreased the protein expression of AXIN2 and CD44 in the β-catenin–dependent cell lines, Colo320 and HCT116 (Fig. 3D). Treatment with these compounds increased the cleavage of poly(adenosine diphosphate–ribose) polymerase (PARP), suggesting that C-1 and C-2 induced cell death. We also observed decreased abundance of “active” dephosphorylated β-catenin [active β-catenin (ABC)], suggesting that C-1 and C-2 reduce β-catenin stability and activity (Fig. 3D) (45). To further confirm the specificity of these compounds, we also used a β-catenin–independent cell line, RKO, which lacks ABC/TCF-regulated transcription (fig. S2) (46–48). Consistent with this, treatment of RKO cells with either C-1 or C-2 did not affect their expression of AXIN2 or CD44 protein (Fig. 3D). Although C-1 did not induce any changes in PARP cleavage in RKO cells, we observed minor PARP cleavage at 10 and 20 μM C-2, suggesting possible off-target toxicities at high concentrations of this compound. In addition, treatment of CRC cell lines with the C-1–negative control compound did not affect the expression of AXIN2, CD44, and ABC, and did not induce PARP cleavage, further confirming the specificity of our lead compound, C-1 (Fig. 3D).
Fig. 3. The expression of Wnt target genes in response to lead compounds.
(A and B) RT-qPCR of Colo320 and HCT116 cell lines treated with vehicle or increasing concentrations of C-1 (A) and C-2 (B) for 6, 24, and 48 hours for the Wnt target genes AXIN2 (left) and CD44 (right). The data demonstrate the relative fold change normalized to housekeeping genes (B2M and PMM1) and to the vehicle (ΔΔCT). Error bars represent means ± SD of triplicate experiments. (C) RT-qPCR of neoplastic human colon organoids treated with vehicle or increasing concentrations of C-1 for 24 hours for the Wnt target genes, AXIN2, CD44, and LGR5. The data demonstrate the relative fold change normalized to housekeeping genes (GAPDH) and to the vehicle (ΔΔCT). Error bars represent means ± SD of triplicate experiments. (D) Immunoblots of the β-catenin–dependent cell lines, Colo320 and HCT116, and the β-catenin–independent cell line, RKO, treated with vehicle and increasing concentrations of C-1, C-2, or the C-1–negative control compound for 24 and 48 hours. For PARP, the full-length protein (upper bands) and cleaved PARP (lower bands indicated by arrow) are shown. Actin is shown as a loading control. *P > 0.05, **P > 0.01, and ***P > 0.001.
The ability of C-1 and C-2 to inhibit Wnt transcriptional activity was subsequently tested using Colo320 and HCT116 cells transduced with a dual-luciferase Wnt reporter (49). ICG-001, a small-molecule inhibitor of β-catenin/CREB-binding protein complexes, and thus, Wnt activity (34) was used as a positive control. ICG-001, C-1, and C-2 all significantly inhibited Wnt signaling in the reporter cell lines in comparison to the vehicle, whereas the C-1–negative control did not (Fig. 4A).
Fig. 4. Wnt activity and Wnt/β-catenin gene signatures in response to lead compounds.
(A) Wnt reporter assay of Colo320 and HCT116 cells, treated for 16 hours with increasing concentrations of ICG-001 (positive control), the two lead compounds in this study, C-1 and C-2, and the C-1–negative control compound. The data demonstrate the respective firefly/renilla luciferase ratios normalized to that of vehicle-treated cells. Error bars represent means ± SD of triplicate experiments. (B) Gene expression profiling of HCT116 cells treated for 48 hours with vehicle or 20 μM C-1 (in triplicate). (B) Differentially expressed genes between control and C-1–treated HCT116 cells at FDR <0.01. (C) GSEA mountain plots and respective heat maps of the leading edge genes for the β-catenin/TCF target gene signatures in CRCs and colorectal adenomas (50), and for the leading edge genes for the cholesterol homeostasis (MSigDB H) gene signature (51). (D) GSEA mountain plots for the up- and down-regulated gene signatures in HCT116 cells following TCF7L2 KO (52), and the down-regulated gene signature in a BCL9/B9L KO mouse model of CRC (53). *P > 0.05, **P > 0.01, and ***P > 0.001.
To further investigate the ability of C-1 to inhibit Wnt transcriptional activity, we performed gene expression profiling of HCT116 cells treated with vehicle or 20 μM C-1. In principal components analysis, samples clustered on the basis of the treatment condition (fig. S3). We detected 1483 differentially expressed genes at false discovery rate (FDR) < 0.01 (Fig. 4B). Selected genes from the gene expression data were validated with RT-qPCR (fig. S4). Gene set enrichment analysis (GSEA) revealed significant down-regulation of β-catenin/TCF target gene signatures previously described in colorectal adenomas and carcinomas (50) and cholesterol metabolism (Fig. 4C) (51). Consistently, signatures up- and down-regulated in response to TCF7L2 (also known as TCF4) deletion (52) in HCT116 cells were likewise significantly differentially enriched in response to C-1 and those down-regulated in response to BCL9/9L knockout (KO) in a murine model of CRC (Fig. 4D) (53). Furthermore, GSEA of the “Hallmark” database showed significant down-regulation of gene sets related to cell growth and proliferation (e.g., E2F targets, mammalian target of rapamycin signaling, and G2-M checkpoint) (fig. S5A). Notably, cholesterol metabolism and proliferation signatures were also down-regulated in HCT116 cells following TCF7L2 KO (fig. S5B) and ICG-001 treatment (46, 52), suggesting that these are on-target effects of Wnt/β-catenin inhibition. No other signaling signatures were significantly inhibited in response to C-1 treatment. Together, these results highlight the specificity and on-target activity of C-1 in blocking Wnt transcriptional activity without major off-target effects and highlight a role of canonical Wnt activity and C-1 in regulating cholesterol metabolism.
C-1 disrupts cholesterol homeostasis via increased cholesterol esterification and lipid droplet accumulation
We further investigated the effect of C-1 on cholesterol homeostasis since this process was shown to be significantly disrupted in C-1–treated HCT116 cells and in previously published HCT116 TCF7L2 KO and ICG-001–treated datasets. First, cells treated for a series of time points with vehicle, C-1, or the cholesterol transport inhibitor U-18666A (54) were stained with BODIPY to fluorescently label cellular lipids. In vehicle-treated cells, lipids were predominately localized on the plasma membrane at all time points studied (Fig. 5A). This is also the case for cells treated with C-1 or U-18666A for 4 and 8 hours, whereas those treated for 24 and 48 hours demonstrate substantial intracellular lipid accumulation. This effect appears to be more marked in C-1–treated cells than the U-18666A–treated positive control cells, suggesting that C-1 may be a strong modulator of cholesterol trafficking (Fig. 5A). The same effect was seen when HCT116 cells were stained with the cholesterol-specific marker filipin III; cholesterol was predominately localized to the plasma membrane of vehicle-treated cells, whereas C-1 treatment induced accumulation of intracellular cholesterol over time (Fig. 5B). Since cholesterol can be stored in the endoplasmic reticulum (ER) or with triglycerides in lipid droplets (55), we used immunofluorescence to determine in which organelles the cholesterol was accumulating. HCT116 cells treated with vehicle or C-1 for 48 hours were stained for filipin to label cholesterol, in addition to sarcoplasmic/endoplasmic reticulum Ca2+ (SERCA)-ATPase (adenosine triphosphatase) antibody to label the ER, or LipidSpot, stain to label lipid droplets. Filipin did not colocalize with SERCA-ATPase but did colocalize with LipidSpot, indicating that C-1 treatment induces the accumulation of cholesterol within lipid droplets (Fig 5C).
Fig. 5. Cholesterol trafficking and esterification after C-1 treatment.
(A) Representative fluorescent images of lipids (BODIPY) and nuclei (DAPI) in HCT116 cells treated with vehicle, 20 μM C-1, or the cholesterol inhibitor U-18666A for the time points indicated. Scale bars, 50 μm. (B) Representative fluorescent images of filipin III–stained cholesterol in HCT116 cells treated with vehicle and 20 μM C-1 for the time points indicated. Scale bars, 50 μm. (C) Representative fluorescent images of HCT116 cells treated with vehicle or 20 μM C-1 for 48 hours and stained for filipin (cholesterol) and SERCA-ATPase (ER) (left) or LipidSpot (lipid droplets) (right). Scale bars, 50 μm. (D) Measurements of esterified cholesterol in HCT116 and Colo320 cells treated with vehicle and 20 μM C-1 for the time points indicated. Esterified cholesterol is normalized to the cell’s respective viability measurements. Error bars represent means ± SD of triplicate experiments. (E) Viability measurements of HCT116 and Colo320 cells treated with the compounds indicated (20 μM) for 48 hours. Lov, lovastatin; Ava, avasimibe. Error bars represent means ± SD of triplicate experiments. (F) Viability measurements of HCT116 and Colo320 cells treated with the compounds indicated (20 μM) ± cholesterol (chol; 20 μM) for 48 hours. Error bars represent means ± SD of triplicate experiments. (G) Immunoblots of cleaved and uncleaved SREBP2 in HCT116 cells treated with vehicle or 20 μM C-1 for the time points indicated (right) and densitometry analysis of cleaved SREBP2 normalized to respective actin measurements (left). Actin is shown as a loading control. Error bars represent means ± SD of triplicate experiments. *P > 0.05, **P > 0.01, and ***P > 0.001.
The total amount of cholesterol within cells is made up of free, unesterified, biologically active cholesterol and esterified cholesterol (56). We found that the majority of cholesterol in vehicle-treated cells is free and unesterified, whereas C-1–treated cells had significantly increased levels of esterified cholesterol (Fig. 5D); at 24 and 48 hours, respectively, C-1–treated HCT116 cells had 13.1× and 4.7× more esterified cholesterol, while C-1–treated Colo320 cells had 2.7× and 1.6× more esterified cholesterol compared to vehicle-treated cells. Subsequently, to determine whether disruption of cholesterol homeostasis by C-1 may affect cell viability, CRC cell lines were treated with C-1, ICG-001, lovastatin, avasimibe, C-1–negative control, and combinations of these inhibitors. Lovastatin is an inhibitor of hydroxymethylglutaryl–coenzyme A (HMG-CoA) reductase (HMGCR), an enzyme required for cholesterol biosynthesis. The gene encoding this enzyme was found to be significantly down-regulated in C-1–treated HCT116 cells (Fig. 4C). Avasimibe is an inhibitor of acyl-CoA: cholesterol acyltransferase (ACAT), an enzyme that catalyzes the formation of cholesteryl esters. All compounds except for the C-1–negative control significantly reduced cell viability compared to vehicle controls, and combinations of C-1 with either ICG-001, lovastatin, or avasimibe significantly reduced viability compared to treatment with the compounds alone (Fig. 5E). Subsequently, HCT116 and Colo320 cells were treated with vehicle, C-1, lovastatin, or avasimibe, with or without the addition of cholesterol. While addition of cholesterol did not affect the viability of the cells treated with lovastatin or avasimibe, it did partially rescue the viability of C-1–treated cells (Fig. 5F). Together, these results suggest that the effect of C-1 on cell death may be partially mediated through dysregulation of cholesterol synthesis, esterification, and homeostasis.
An increase in cholesterol levels in the ER induces a feedback mechanism limiting the cleavage and therefore activation of the sterol regulatory element–binding protein 2 (SREBP2), a major transcriptional regulator of sterol and fatty acid synthesis and uptake (57). Therefore, we subsequently determined whether this protein is affected by C-1 treatment in HCT116 cells. In line with the transcriptomic profiling (Fig. 4C), C-1–treated cells demonstrated a significant decrease in cleaved SREBP2 at 24 and 48 hours compared to the vehicle-treated cells, while uncleaved SREBP2 levels were not significantly affected (Fig. 5G). This result suggests that a reduction in cleaved SREBP2 may be at least partially responsible for significant down-regulation of the cholesterol homeostasis signature in response to Wnt inhibition. Together, the data suggest that C-1–mediated inhibition of the Wnt/β-catenin/BCL9 transcriptional complex increases intracellular cholesterol and lipid droplet accumulation, significantly increases cholesterol esterification, and disrupts cholesterol homeostasis.
Lead compounds demonstrate antitumorigenic activity in CRC cell lines and xenograft mouse models
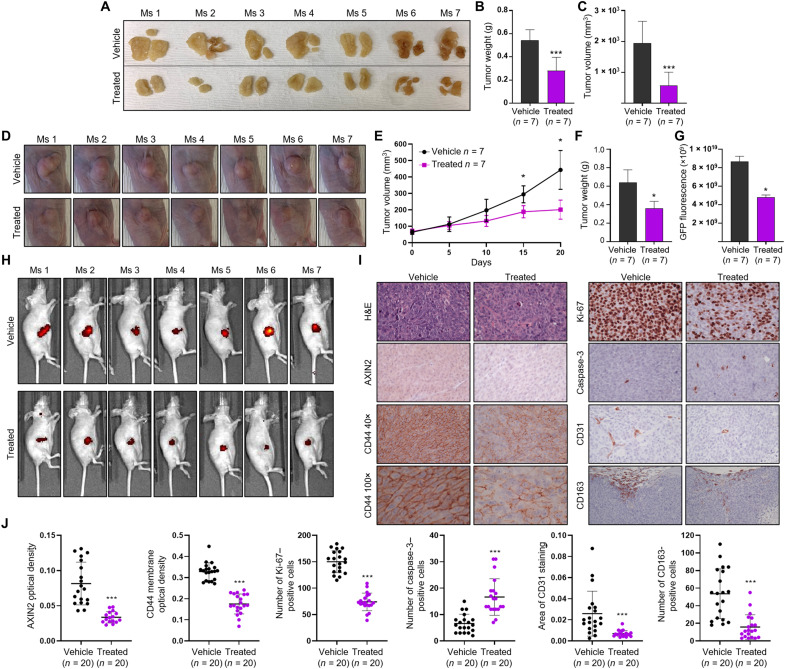
We next determined the functional effects of C-1 and C-2 treatment on CRC cell proliferation. C-1 and C-2 treatment significantly reduced the proliferation of Colo320 and HCT116 cells at all concentrations tested, whereas C-1 and C-2 treatment of RKO cells and C-1–negative control treatment of Colo320 and HCT116 cells did not, further confirming the specificity and on-target activity of our lead compounds (Fig. 6, A and B). We also treated neoplastic human colon organoids with C-1 and found that their proliferation is significantly reduced compared to those treated with the vehicle control (Fig. 6C). Together, C-1 has significant inhibitory activity within colorectal cancer cells and neoplastic human colon organoids. We therefore subsequently sought to investigate whether this compound would be efficacious at reducing tumor growth in a xenograft mouse model of colorectal cancer. First, NOD scid gamma (NSG) mice were transplanted with HCT116 cells in the peritoneal cavity of the animals and, 1 week after engraftment, were treated every other day with vehicle or C-1 (4 mg/kg) via intraperitoneal injections. C-1 treatment significantly reduced tumor growth in the mice compared to the control group, as demonstrated by significant decreases in weight and volume of tumor masses isolated and dissected during autopsy (Fig. 7, A to C). NSG mice have deficient macrophages, so we also carried out a second in vivo study with NCr nude mice to observe the effect of in vivo C-1 treatment on macrophage tumor infiltration. In addition, because NCr mice are hairless, they provide a convenient model in which to measure fluorescently labeled, subcutaneous tumor cells by whole-body imaging. These mice were subcutaneously transplanted with HCT116 cells stably expressing green fluorescent protein (GFP), and after 1 week of engraftment, mice were treated every other day with vehicle or C-1 (3 mg/kg) via intratumoral injections. C-1 treatment significantly reduced tumor growth in these mice compared to those injected with vehicle, as demonstrated by significant decreases in tumor volume measured in vivo (Fig. 7, D and E), whereas the body weight of the animals over the course of the experiment remained stable (fig. S6). We also observed a significant decrease in tumor weight (Fig. 7F) and a significant decrease in GFP fluorescent tumor cells (Fig. 7, G and H) in C-1–treated animals compared to those treated with the vehicle control. Immunohistochemistry (IHC) analysis of tumor tissue sections from these animals demonstrated that C-1–treated animals had significantly lower levels of AXIN2 and CD44, indicating that this compound inhibits oncogenic Wnt signaling in vivo (Fig. 7, I and J). In addition, the pattern of CD44 staining was patchy throughout the membranes of C-1–treated tumor cells, whereas those treated with vehicle demonstrated continuous membranous staining (Fig. 7I). Compared to vehicle-treated mice, tumors from C-1–treated mice demonstrated a significant decrease in proliferation and angiogenesis, measured with Ki-67 staining and CD31 staining of mouse endothelial cells, respectively, while cleaved caspase-3 staining, a marker of apoptosis, was significantly increased (Fig. 7, I and J). Furthermore, C-1 treatment decreased infiltration of protumorigenic M2-like tumor–associated macrophages, as seen with CD163 staining (Fig. 7, I and J). Histologic analysis did not reveal any differences between vehicle- and C-1–treated mice in the architecture of normal liver, kidney, or intestine tissue (fig. S7, A and B). We also did not observe any significant increase in the number of apoptotic bodies or number of cleaved caspase-3–positive cells in the intestine tissue of treated and control mice (fig. S7C). To confirm that the lack of obvious histologic toxicity seen in the xenograft mouse model is not simply a result of C-1 being inactive against murine proteins, we carried out a BCL9 Co-IP assay on the murine Wnt-dependent breast cancer cell line, 4T1, treated with C-1. C-1 inhibited the binding of BCL9 to β-catenin in a dose-dependent manner (fig. S7D), confirming that C-1 is active against mouse β-catenin and BCL9. Although significant further investigation is required to determine whether C-1 demonstrates on-target and/or off-target toxicity, our results suggest that C-1 has antitumor activity in a murine xenograft model of CRC and does not induce general wasting or reveal obvious signs of in vivo toxicity.
Fig. 6. The effect of lead compounds on the proliferation of CRC cell lines.
The proliferation of the β-catenin–dependent cell lines, Colo320 and HCT116 (A), and the β-catenin–independent cell line, RKO (B), was measured after treatment with vehicle and increasing concentrations of C-1, C-2, or the C-1–negative control for 24, 48, 72, and 96 hours. Error bars represent means ± SD of triplicate experiments. (C) The proliferation of neoplastic human colon organoids treated with vehicle and increasing concentrations of C-1 for 24 and 48 hours (left). Representative images of the organoids treated with the concentrations of C-1 indicated for 24 hours (right). Black arrows indicate apoptotic organoids.
Fig. 7. The effect of C-1 on tumor growth in a xenograft mouse model of CRC.
(A) Images of dissected tumors from mice treated with vehicle (n = 7) or C-1 (n = 7) via intraperitoneal injections. (B and C) Tumor weight and tumor volume of mice treated with vehicle (n = 7) or C-1 (n = 7) via intraperitoneal injections. Error bars represent means ± SD. (D) Images of subcutaneous tumors, taken on day 20 of the study, from mice treated with vehicle (n = 7) or C-1 (n = 7) via intratumoral injections. (E) Body weight (top) and tumor volume (bottom) of mice treated with vehicle (n = 7) or C-1 treated (n = 7) via intratumoral injections. Error bars represent means ± SD. (F and G) Tumor weight and tumor GFP fluorescence measurements recorded from mice treated with vehicle (n = 7) or C-1 (n = 7), via intratumoral injections. Error bars represent means ± SD. (H) GFP fluorescence images of intratumoral vehicle and C-1–treated mice taken on day 20. (I) Representative hematoxylin and eosin (H&E) and IHC stains of AXIN2, CD44, Ki-67, cleaved caspase-3, CD31, and CD163 in tumor tissue from intratumoral vehicle- and C-1–treated mice. Scale bars, 50 μm. (J) Quantification of AXIN2, CD44, Ki-67, CD31, cleaved caspase-3, and CD163 immunostains in the tumors of intratumoral vehicle- and C-1–treated mice. Plots represent individual data points with error bars representing means ± SD. *P > 0.05 and ***P > 0.001.
DISCUSSION
Here, we established an AlphaScreen-based HTS and identified small-molecule inhibitors of the β-catenin/BCL9 interaction, which were further validated for activity and specificity in secondary biophysical assays. Two compounds, C-1 and C-2, specifically inhibited β-catenin/BCL9 complex formation, its downstream transcriptional activity, and proliferation in colorectal cancer cell lines. Furthermore, C-1 manifested antitumorigenic activity in vivo and was associated with decreased host angiogenesis and reduced infiltration of protumorigenic M2-like tumor–associated macrophages.
The Wnt/β-catenin pathway is an attractive target for clinical therapy, but its targeting has proven challenging because of the vital role that β-catenin plays in maintaining adult tissue homeostasis and because of β-catenin’s common interaction surface (27). Small-molecule, peptide, and antibody inhibitors targeting different stages of the Wnt/β-catenin pathway have been described but are almost invariably associated with off-target and toxic side effects (28, 29, 34–39, 58, 59). We set out to develop an inhibitor that targets β-catenin/BCL9 complexes because (i) BCL9 binds to β-catenin via a site that is not shared by other protein partners, (ii) we previously confirmed the feasibility of this approach with BCL9 peptidomimetics, and (iii) knockout of BCL9 and B9L in the intestinal epithelium of mice is well tolerated. In keeping with this, in mice treated with C-1, we have not observed any obvious signs of toxicity such as general wasting or altered intestinal, liver, and kidney cellular morphology and tissue architecture, which have been described for other Wnt/β-catenin inhibitors. Significant additional studies are, however, necessary to meticulously examine on-target and off-target toxicity, as well as an investigation into systemic routes of injection. To this end, we plan to develop and identify chemical derivatives of C-1 that have increased potency and do not affect normal tissues.
Targeting protein-protein interactions (PPIs), such as that of β-catenin and BCL9, is considered technically challenging, not only because the interface at which proteins interact is often relatively large and flat but also because of the very diverse shapes, sizes, and binding affinities of PPIs (60). However, significant advances in screening methodologies over the past decade have now demonstrated that this is possible, and PPI inhibitors have been developed for clinical use, including those against LFA-1, inhibitor of apoptosis proteins, bromodomains, and BCL family proteins (60). Moreover, many research groups are pursuing both peptide (30, 61–63) and nonpeptide approaches (64–66) to target β-catenin/BCL9 complexes. The AlphaScreen-based assay used in this study has many advantages over other screening techniques, including high sensitivity and the absence of radioactive components, and it does not require large fluorescent tags that may sterically disrupt the interaction being studied (42). While many different types of screening assays can be used to identify potent PPI inhibitors, it is crucial that they are validated to eliminate false positives, confirm binding specificity, and ensure on-target activity. We validated our candidate compounds using a BRD9/ligand counter-screen, ITC, β-catenin–dependent and –independent cell lines, neoplastic human colon organoids, another β-catenin–binding partner (E-cadherin), an inactive compound from the same chemical series, and unbiased transcriptomic analysis for off-target activity, which all indicated specificity and on-target activity of C-1. Compounds identified in HTS are not usually ready for direct preclinical utilization, and this is also the case for C-1. This molecule has limited solubility in aqueous solutions, which was challenging for in vivo administration; while C-1 demonstrated promising efficacy via intraperitoneal and intratumoral dosing, we anticipate that improved potency of the C-1 series will be required to enable in vivo efficacy through either oral or intravenous dosing.
In an extension of our previous work on the development of SAH-BCL9 peptides, refined BCL9 peptides have been generated recently with improved and promising properties, but robust in vitro and in vivo validation of these peptides is still required to determine whether they are promising candidates for therapeutic use (30, 62, 63). In terms of small-molecule approaches, a class of β-catenin/BCL9 inhibitors based on a generic scaffold structure that mimics “critical binding elements” of the interaction interface was generated and extensively optimized to inhibit Wnt reporter activity and expression of Wnt target genes at low micromolar concentrations (64–66). However, the authors note that the presence of multiple carboaromatic rings in this optimized compound suggests a potential risk of high binding to serum albumin, which would affect its further development. Moreover, these compounds have not yet been tested in vivo. An HTS approach by De La Roche et al. (67) identified carnosic acid, a natural antioxidant, as an inhibitor of β-catenin/BCL9 complexes; however, this was revealed to have substructural features of a pan assay interference compound (67–69). Although all these studies provide (i) new insight, (ii) molecular proof of feasibility, and (iii) functional necessity for disruption of β-catenin/BCL9 complexes, well-established inhibitors of β-catenin/BCL9 complexes do not yet exist. Our HTS combined with secondary validation assays allowed us to identify several new small molecules, two of which specifically inhibited the β-catenin/BCL9 interaction and downstream Wnt activity in CRC cells and one that displays antitumorigenic activity in a mouse model of CRC. Further efforts in our laboratory will focus on (i) optimizing the potency and pharmacokinetics of C-1, as well as other candidate scaffolds, (ii) evaluating the on-target and off-target toxicity of C-1 and its derivatives using sensitive biochemical assays, and (iii) analyzing the in vitro and in vivo efficacy of our compounds in other Wnt-dependent cancer models, and (iv) given the Wnt pathway functions in regulating tumor microenvironment, we will use the CT26 syngeneic model of CRC to determine the effects of compound treatment on angiogenesis and immune cell infiltration (61).
Future efforts will also include fully determining the mode of action of C-1 on cholesterol homeostasis. Other research groups have found that Wnt signaling regulates cholesterol homeostasis and lipid droplet formation, with tight feedback mechanisms existing between these processes (70), and it is becoming increasingly apparent that dysregulated cholesterol homeostasis is linked with oncogenesis. Stearoyl-CoA desaturase (SCD), for example, which, in our gene expression analysis, was the most significantly down-regulated gene related to cholesterol homeostasis in C-1-treated cells, has been found to be a requirement for cancer cells to proliferate in lipid-depleted environments (71). Cholesterol and sphingolipids are crucial components of plasma membranes and lipid rafts, which play important roles in signal transduction and membrane and receptor trafficking (72), and lipid rafts have been found to initiate oncogenic signal transduction pathways in cancer cells responsible for promoting tumor progression, angiogenesis, epithelial-mesenchymal transition, migration, and cell survival (73). These findings are relevant to the oncogenic Wnt pathway since lipid rafts are also important for the initiation of Wnt signaling in which Wnt ligands bind to cell surface receptors Fz and LRP5. In addition, lipid raft–colocalized CD44, an established Wnt target, is thought to be involved in cancer cell adhesion and migration (73, 74). In keeping with this, we observed patchy membranous CD44 staining in the tumor cells of C-1–treated mice, whereas those of vehicle-treated mice displayed uniform, continuous staining.
In this study, C-1 induced accumulation of intracellular lipid droplets, significantly increased the production of cholesterol esters, and depleted cholesterol and lipids within the plasma membrane. C-1 decreased the viability of CRC cells, but viability was decreased even further when C-1 treatment was combined with the HMGCR inhibitor lovastatin or the ACAT inhibitor avasimibe. Addition of cholesterol partially rescued the viability of C-1–treated cells but not of lovastatin- or avasimibe-treated cells. These results suggest that C-1 may partially mediate antitumor activity via disruption of lipid rafts and/or altered cell membrane fluidity and integrity. These data are supported by a previous finding in which cholesterol-depleting agents decreased lipid raft levels and induced apoptosis in breast and prostate cancer cells, while cholesterol repletion restored cell viability (75). Our findings are also supported by Lee et al. (76), who found that avasimibe suppressed the growth and metastasis of prostate cancer cells, and Agarwal et al. (77), who demonstrated that lovastatin augmented sulindac-induced apoptosis in CRC cell lines and concluded that this statin may amplify chemopreventive effects of other drugs. Most available cholesterol synthesis inhibitors target HMGCR or SREBP (78), both of which were found to be significantly down-regulated by C-1 in gene expression studies and/or immunoblotting. Although the exact mechanism by which Wnt inhibition leads to cholesterol esterification and accumulation is yet to be described, we found that C-1 induces an SREBP2-mediated feedback mechanism that reduces the expression of cholesterol biosynthesis and transport genes, which may further disrupt its homeostasis.
Our data from this study are therefore consistent with a model in which C-1 inhibits not only β-catenin/BCL9 complex formation, oncogenic Wnt signaling, proliferation, and survival of CRC cells but also cholesterol transport and homeostasis, which may add significant therapeutic value to this compound when combined with other small molecules (Fig. 8). In conclusion, we have demonstrated the potential for small molecules, such as C-1, to function as inhibitors of oncogenic Wnt signaling via disruption of β-catenin/BCL9 complex formation. We anticipate that forthcoming efforts may lead to the identification of novel, more efficacious, and bioavailable therapeutic agents for patients with Wnt-dependent cancers.
Fig. 8. Model of C-1 mechanism of action.
Our data from this study are consistent with a model in which C-1 treatment specifically inhibits β-catenin/BCL9 complex formation in CRC cells and reduces cell proliferation and survival, which would otherwise be amplified by oncogenic Wnt signaling in the absence of C-1. C-1 treatment also increases cholesterol esterification and intracellular accumulation of lipid droplets and cholesterol (1), which decreases activation (i.e., cleavage) of SREBP2 (2) and subsequently disrupts the cholesterol homeostasis gene expression signature in the cell (3). These processes are concurrent with the depletion of lipids and cholesterol from the plasma membrane, which may decrease the number of lipid rafts and membrane fluidity/integrity.
MATERIALS AND METHODS
Experimental design
A study screening 320,000 compounds with a high-throughput AlphaScreen assay was conducted to identify small-molecule inhibitors of β-catenin and BCL9 complex formation. Hits were validated using secondary biophysical assays and subsequently tested in vivo using Co-IP studies in colorectal cancer cell lines. Lead compounds were validated for on-target, in vitro activity by qPCR and Western blotting for “bona fide” Wnt–β-catenin targets in addition to Wnt reporter assays. Gene expression profiling was carried out for the top-performing compound in this study to determine its effect on the expression of Wnt–β-catenin target gene signatures. Cholesterol-detecting assays were used to explore how the lead compound dysregulates cholesterol homeostasis. To determine the effect of compound treatment on in vivo tumor growth, mice implanted with colorectal cancer cells subcutaneously or within the peritoneum were randomized into two groups and treated with the lead compound or vehicle. Seven mice were used in each group. The researchers who performed the in vivo treatment injections and assessed the animals were blinded. End points were defined by the standard of the Institutional Animal Care and Use Committee (IACUC) at the Dana-Farber Cancer Institute. IHC was performed on mouse tumor sections to measure proliferation, angiogenesis, apoptosis, infiltration of M2-like macrophages, and expression of Wnt targets. The researchers who performed the IHC were blinded to which tumor sections were from vehicle- and lead compound–treated animals. No data were excluded in any of the experiments.
Protein expression and purification
Constructs of human CTNNB1 (full-length and region 141-305, referred to as “short protein”) in the pET28 vector were overexpressed in Escherichia coli BL21(DE3) in lysogeny broth medium in the presence of kanamycin (50 mg/ml). Cells were grown at 37°C to an optical density of 0.8, cooled to 17°C, induced with 500 μM isopropyl-1-thio-d-galactopyranoside, incubated overnight at 17°C, collected by centrifugation, and stored at −80°C. Cell pellets were microfluidized at 18,000 psi in buffer A [25 mM Na3PO4 (pH 7.4), 500 mM NaCl, 5% glycerol, 10 mM imidazole, and 10 mM β-mercaptoethanol (BME)], and the resulting lysate was centrifuged at 13,000 rpm for 30 min. Nickel–nitrilotriacetic acid beads were mixed with lysate supernatant for 30 min and washed with buffer A. Beads were transferred to a fast protein liquid chromatography–compatible column, and the bound protein was washed with 15% buffer B [10 mM Na3PO4 (pH7.4), 200 mM NaCl, 5% glycerol, 250 mM imidazole, and 10 mM BME] and eluted with 100% buffer B. To cleave the tag from the short construct, His-3C was added to the eluted protein and incubated at 4°C overnight. Samples were concentrated and passed through a Superdex 200 10/300 column in buffer C [20 mM Hepes (pH 7.5), 200 mM NaCl, 5% glycerol, and 1 mM tris(2-carboxyethyl)phosphine (TCEP)]. Relevant fractions were pooled, concentrated, and frozen at −80°C.
AlphaScreen HTS
The AlphaScreen assay (PerkinElmer) (42) was developed to monitor the β-catenin/BCL9 interaction in a microtiter plate format. The assay platform used full-length recombinant β-catenin (1 to 781 amino acids, 88 kDa) bound to a protein A–tagged AB via a specific monoclonal anti–β-catenin antibody (Abcam, #16051), and an N-terminal biotinylated BCL9-HD2 peptide (AnaSpec) (Biotin-LSQEQLEHRERSLQTLRDIQRM) bound to a streptavidin-tagged DB. The beads (#6760617MR) and AlphaScreen TruHits Kits/reagents (#6760627M) were purchased from PerkinElmer. The concentrations of reagents used for the pilot screen were as follows: 10 nM β-catenin, 25 nM biotinylated BCL9 peptide, β-catenin antibody (50 ng/ml), ABs (5 μg/ml), and DBs (5 μg/ml). The assay buffer contained 50 mM MES (pH 6.5), 150 mM NaCl, 0.01% bovine serum albumin, 1 mM dithiothreitol, and 0.1% Tween 20. Replicas of an HTS of a 320,000 small-molecule library, performed independently at the Sanford-Burnham Medical Research Institute (La Jolla, CA), were carried out using compounds at a final concentration of 10 μM [0.5% dimethyl sulfoxide (DMSO)] in 4 μl per well assay in a 1536-well plate format, with an incubation time of 90 min. The 643 primary hits from this screen were subsequently tested in triplicate against the TruHit counter-screen to eliminate compounds with nonselective activity (e.g., metal chelators, oxygen quenchers, and biotin mimetics), which eliminated 21 hits. The 622 “specific” (nonartifactual) hits that passed this counter-screen (<50% inhibitory activity) were then taken into five-point dose-response (40, 20, 10, 5, and 2.5 μM final concentrations) estimations of potency (IC50) in triplicate. Thirty-three compounds from the HTS (table S1) that demonstrated inhibitory activity were purchased and subjected to a 10-point dose-response AlphaScreen assay as described above with β-catenin and BCL9. The compounds were also tested in a counter-screen assay with recombinant BRD9 bromodomain binding to a custom-synthesized BRD9 ligand as previously described (43).
Isothermal titration calorimetry
All calorimetric experiments were carried out in 20 mM Hepes (pH 7.5), 150 mM NaCl, 0.5 mM TCEP, and 3.0% DMSO at 25°C using an Affinity ITC from TA Instruments (New Castle, DE) equipped with autosampler. Short protein or buffer (10 μM; for control) in the calorimetric cell was titrated by injecting 3 μl of 100 μM ligand solution in 200-s intervals with stirring speed at 75 rpm. The resulting isotherms were subtracted against buffer runs and fitted with a single-site model to yield thermodynamic parameters of ΔH and ΔS, stoichiometry, and KD using NanoAnalyze software (TA Instruments).
Computational docking
Canonical smiles were submitted for ligand preparation by LigPrep-2.5 in the Schrödinger Maestro Suite (2016). Receptor grids were calculated encompassing the BCL9-binding pocket in CTNNB1/BCL9 complex (Protein Data Bank, 3SL9), which was preprocessed, optimized, and refined with the OPLS3e force field using the following parameters (van der Waals scaling factor of 1.0 and charge cutoff of 0.25). Unconstrained molecular docking of ligands was performed using XP Precision, flexible ligand sampling, and Epik state penalties. Ten post-docked minimized poses per ligand were calculated using Glide.
Cell culture
The human colorectal cancer cell lines Colo320 and DLD-1 were cultured in RPMI 1640 medium, and the HCT116 and RKO cell lines were cultured in McCoy’s and Dulbecco’s modified Eagle’s medium (DMEM), respectively. The murine 4T1 breast cancer cell line was cultured in RPMI 1640 medium. All cell culture media were supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were cultured at 37°C with 5% CO2 in a humidified incubator. All cell lines used in this study were authenticated and confirmed mycoplasma-free. For all experiments, cells were treated with compounds at the specified concentrations and lengths of time, and cells treated with an equivalent concentration of DMSO were used as vehicle controls.
Human colonic organoids
Human colon adenoma samples were obtained from the Department of Surgery, under approval (protocol 13-189) by the Internal Review Board of the Dana-Farber Cancer Institute, Boston, MA, USA, and used to make neoplastic human colon organoids. The organoids used in this study have a c.4778delA mutation, in coding exon 15 of the APC gene, resulting in protein truncation at AA1650 (p.K1593Sfs*57; COSMIC ID: COSV57342520). The colonic tissue after surgery was rinsed with ice-cold phosphate-buffered saline (PBS) in a 90-mm petri dish, washed with ~20 ml of ice-cold PBS in a 50-ml tube by vigorous shaking, and then rinsed again with ice-cold PBS in a 90-mm petri dish. After washing, tissue was transferred to a 35-mm petri dish in a biological tissue culture hood and then minced with fine scissors. One milliliter of collagenase (Invitrogen) solution was added to suspend tissue fragments, and the petri dish was incubated in a cell culture incubator (37°C) with vigorous mixing every 5 to 10 min using a 1000-μl pipette. Once visible, single epithelial units (crypts/pits) were separated from the larger tissue fragments as seen on a phase or dissection microscope. The epithelial units were passed through a 70-μm cell strainer (BD) using a 1000-μl pipette, and the strainer was washed with 9 ml of washing media [penicillin (100 U/ml), streptomycin (0.1 mg/ml), l-glutamine (2 mM), and FBS (10% in DMEM/F-12; Invitrogen) with Hepes]. This filtrate was transferred to a 1.5-ml centrifuge tube, centrifuged at 200g for 5 min, and placed on ice, and the epithelial units were resuspended in Matrigel (15 μl per well; Corning). Fifteen microliters of cell Matrigel suspension was then placed in the center of each well of a 24-well plate using a 20-μl pipette and spread with a pipette tip. To polymerize the Matrigel, plates were incubated upside down to avoid attachment of epithelial units to the plate surface. After 3 to 5 min, plates were returned to the upright orientation, and 500 μl of 50% human L-WRN conditioned medium were added to each well, and the medium was subsequently changed at least every 48 hours. Human L_WRN medium is a 1:1 mix of L WRN conditioned medium and advanced DMEM/F-12 with 20% FBS supplemented with antibiotics Primocin (100 μg/ml; InvivoGen), Normocin (100 μg/ml; InvivoGen); serum-free supplements 1X B27 [Thermo Fisher Scientific (Gibco)] and 1X N2 [Thermo Fisher Scientific (Gibco)]; and chemical supplements 10 mM nicotinamide (Sigma-Aldrich), 500 mM N-acetylcysteine (Sigma-Aldrich), hormone 50 mM [Leu15]-Gastrin (Sigma-Aldrich), growth factor FGF10 (recombinant human) (100 μg/ml; Thermo Fisher Scientific), and 500 nM A-83-01 (Sigma-Aldrich), which is an inhibitor of the transforming growth factor–β receptors ALK4, 5, and 7, and 10 mM rho-associated coiled-coil protein kinase (ROCK) inhibitor Y-27632 (Sigma-Aldrich). For passage, colon organoids were dispersed by trypsin-EDTA and transferred to fresh Matrigel. Passage was performed every 3 to 4 days with a 1:3 to 1:5 split ratio.
Coimmunoprecipitation
The Universal Magnetic Co-IP Kit (Active Motif) was used for the Co-IP studies according to the manufacturer’s protocol. For the BCL9 Co-IP experiments, Colo320, HCT116, and 4T1 cells were treated overnight (16 hours) with either vehicle or 1, 10, or 20 μM compound. Treatment of cells with an equivalent concentration of DMSO was used as a vehicle control. Nuclei were isolated from the treated cells and digested using hypotonic buffer and digestion buffer, respectively. Nuclear protein was collected by centrifugation, and a Bradford-based assay was used to quantify the protein in each sample. A total of 800 μg of each nuclear extract was incubated overnight at 4°C on a rotator, with 3.5 μg of anti-BCL9 (ab37305, Abcam) antibody. For the β-catenin Co-IP experiments, DLD-1 cells were treated in the same way as described above, and cells were lysed using whole-cell lysis buffer. A total of 800 μg of each whole-cell extract was incubated with 3.5 μg of β-catenin (9562L, Cell Signaling Technology) antibody. For all Co-IP assays, incubation of the respective protein samples with normal rabbit immunoglobulin G (IgG) antibody (sc-3888, Santa Cruz Biotechnology) was used as a negative control. Protein G magnetic beads were added to the Co-IP samples and incubated for 1 hour at 4°C on a rotator. The beads were washed four times with washing buffer containing 150 mM NaCl and then resuspended with 2× reducing loading buffer. Input samples (2%) were prepared.
Immunoblotting
For immunoblotting, samples were electrophoresed using NuPAGE, transferred to nitrocellulose membrane, and blocked using 5% nonfat milk. The membrane was subsequently probed overnight at 4°C with antibodies diluted in blocking buffer. Validated antibodies used in this study were as follows: BCL9 (H00000607-M01, Abnova), β-catenin (#610154, BD Transduction Laboratories), E-cadherin (24E10; #3195, Cell Signaling Technology), Axin2 (76G6; #2151, Cell Signaling Technology), CD44 (#5640, Cell Signaling Technology), PARP (#9542, Cell Signaling Technology), ABC (#05-665, Millipore), SREBP2 (#NBP1-54446, Novus Biologicals), and actin–horseradish peroxidase (HRP) (#SC-1615, Santa Cruz Biotechnology). Membranes were incubated with the appropriate species of HRP-conjugated secondary antibodies and imaged using chemiluminescent substrate (34580, Thermo Scientific). Densitometry analysis in this study was performed using ImageJ software, and respective actin loading control measurements were used for normalization.
Proliferation assays
Colo320, HCT116, and RKO cells were seeded into 96-well plates in triplicate (3000 cells per well in 100 μl of respective medium) and treated with vehicle or 5, 10, or 20 μM compound. Human neoplastic colon organoids were seeded into 96-well plates in triplicate and treated with vehicle or 25 or 50 μM compound. CellTiter-Glo 2.0 assay reagent (Promega) was used to measure proliferation according to the manufacturer’s instructions. Luminescence was measured using the Victor X3 multilabel plate reader (PerkinElmer). Proliferation was calculated relative to the respective baseline readings.
Dual-luciferase Wnt reporter assay
To generate stable, dual-luciferase, Wnt reporter cells, lentiviral particles were generated containing a vector with firefly luciferase under the 7XTCF promoter (7TFP; Wnt reporter plasmid), and a vector with Renilla luciferase under the EF1alpha promoter (pLX313-renilla; baseline plasmid). 7TFP was a gift from R. Nusse (Addgene plasmid #24308; http://n2t.net/addgene:24308), and pLX313-renilla luciferase was a gift from W. Hahn and D. Root (Addgene plasmid #118016; http://n2t.net/addgene:118016). Colo320 and HCT116 cells were transduced with lentiviral particles, and the appropriate antibiotics were used for selection. For experiments, 30,000 cells were seeded per well of a white opaque 96-well plate and treated with vehicle or increasing concentrations of the tested compounds for 16 hours. Firefly and Renilla luciferase signals were measured using the Dual-Glo luciferase assay system (#E2920, Promega) according to the manufacturer’s instructions on a Victor X3 multilabel plate reader. Experiments were carried out in triplicate.
RT-qPCR
RT-qPCR was used to examine compound treatment–induced expression changes in the Wnt-target genes AXIN2, CD44, and LGR5 and to validate the gene expression profiling results (IGFBP3, AURKC, PDGFA, CCNE2, SCD, and CYP1A1). Colo320 and HCT116 cell lines were treated with vehicle or 10 or 20 μM compound for 6, 24, and 48 hours. Colon organoids were treated with vehicle or 25 or 50 μM compound for 24 hours. Cells were collected, and total RNA was isolated using TRIzol (Life Technologies). cDNA was synthesized using the iScript Reverse Transcription Supermix kit (Bio-Rad), and subsequent qPCR reactions were run on a CFX96 Real-Time PCR System (Bio-Rad) using PowerUp SYBR Green Master Mix (Applied Biosystems). Expression was normalized to β2 microglobulin (B2M) and phosphomannomutase 1 (PMM1) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (79) using the ΔΔCT method. All primer sequences used were provided by Harvard PrimerBank (80) and are described in table S4.
Gene expression profiling
RNA from triplicate vehicle- or C-1–treated HCT116 samples (20 μM each for 48 hours) was isolated using TRIzol. Libraries were prepared using Roche Kapa stranded mRNA HyperPrep sample preparation kits from 200 ng of purified total RNA according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and RT-qPCR using a Kapa Biosystems library quantification kit according to the manufacturer’s protocols. Uniquely indexed libraries were pooled at an equimolar ratio and sequenced on an Illumina NovaSeq 6000 with paired-end 100–base pair reads by the Dana-Farber Cancer Institute Molecular Biology Core Facilities.
Reads were processed and analyzed (principal components and differential expression analyses) using VIPER (81). β-Catenin/TCF targets gene sets in CRCs and adenomas were based on the intestinal Wnt/TCF4 signature gene set generated by Van der Flier (50). Differential expression analysis of the GSE135328 (52) was used to determine genes down- and up-regulated in HCT116 cells after TCF7L2 KO (top 200 down- or up-regulated genes at FDR < 0.01). BCL9/9L-KO signature components by Moor et al. (53) were used as BCL9/B9L signature. GSEA of these and the Hallmark MSigDB gene sets (51) was performed using recommended settings as previously described (82, 83). Data were visualized as previously described (83).
Fluorescent staining and microscopy
HCT116 cells were seeded into glass-bottomed microwell dishes (MatTek) and treated with vehicle or 20 μM C-1 for 4, 8, 24, and 48 hours. For lipid staining, treated cells were washed with 1× PBS, fixed with 4% paraformaldehyde, and stained using 2 μM BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene) for 15 min. Cells were also incubated with DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride; 1 μg/ml) to stain the nuclei. For cholesterol staining with filipin III, the cholesterol assay kit (Abcam) was used according to the manufacturer’s instructions. HCT116 cells were also stained with filipin in addition to SERCA-ATPase antibody (NB300-581, Novus Biologicals) or LipidSpot (70065-T, Biotium). Fluorescent images were collected using a Nikon inverted live-cell imaging system with a ×40 dry objective and processed with ImageJ. Scale bars represent 50 μm.
Cholesterol/cholesterol Ester-Glo assay
HCT116 and Colo320 cells were treated for 24 and 48 hours with vehicle or 20 μM C-1, and total and free cholesterol were measured with the Cholesterol/Cholesterol Ester-Glo Assay (Promega) according to the manufacturer’s instructions. The luciferase signals were measured on a Victor X3 multilabel plate reader. Experiments were carried out in triplicate. Esterified cholesterol was calculated as total cholesterol (with esterase) − free cholesterol (without esterase). Cholesterol measurements were normalized to the respective viability of the cells, which was measured using the CellTiter-Glo luciferase assay (Promega) according to the manufacturer’s instructions.
Viability assays
Cells were seeded into white opaque 96-well plates and treated for 48 hours with the compounds indicated (20 μM): ICG-001 (Selleck Chemicals), lovastatin (Selleck Chemicals), avasimibe (Millipore Sigma), and water-soluble cholesterol (Millipore Sigma). The CellTiter-Glo assay (Promega) was used to measure the luciferase signals on a Victor X3 multilabel plate reader according to the manufacturer’s instructions. Experiments were carried out in triplicate, and viability was calculated relative to that of vehicle-treated cells.
In vivo mouse xenograft models
For the intraperitoneally treated animals, 2 × 106 HCT116 cells were first injected into the abdomen of 5-week-old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, the Jackson Laboratory). Treatment began 1 week after injection, in which control mice (n = 7) received vehicle, and treated mice (n = 7) received C-1 (4 mg/kg) every other day via intraperitoneal injections. Cohorts were monitored daily. Animals were euthanized after 20 days of treatment, and autopsies were performed. Extracted intraperitoneal tumors were measured with calipers to determine tumor volume (in mm3) using the formula V = 0.5a × b2, where a and b are the long and short dimensions of the tumor, respectively. Tumors were also weighed. All extracted tumors and other tissues were subsequently fixed in 10% formalin.
For the intratumoral-treated animals, 2 × 105 HCT116 cells stably transduced with a reporter expressing GFP were first injected subcutaneously into 5-week-old female NCr nude mice (Taconic). Treatment began 1 week after injection, in which control mice (n = 7) received vehicle and treated mice (n = 7) received C-1 (3 mg/kg) every other day via intratumoral injections. Cohorts were monitored daily. Tumor measurements using calipers were also carried out twice a week, and the volume in cubic millimeter was calculated using the formula V = 0.5a × b2. GFP imaging was carried out at day 20 to analyze the tumor burden in the animals, and Living Image software (PerkinElmer) was used to measure the GFP fluorescence from the images. Animals were euthanized after 20 days of treatment, and autopsies were performed. Tumors and other tissues were weighed before being fixed in 10% formalin and embedded in paraffin. Tissue sections were prepared and stained with hematoxylin and eosin and with validated antibodies (all from Cell Signaling Technology unless otherwise stated) for Axin2 (#2151), CD44 (#5640), Ki-67 (#12202), cleaved caspase-3 (#9664), CD31 (#77699), and CD163 (#ab182422, Abcam) using protocols previously described (84). Scale bars represent 50 μm. All animal experiments were approved by and conform to the standard of the IACUC at the Dana-Farber Cancer Institute. Images of IHC-stained tumor and tissue sections were obtained with a Leica DM2000 microscope and ×40 objective unless otherwise stated.
Statistical analysis
For statistical analysis, the two-tailed Student’s t test was used to assess significant differences between compound-treated and control groups. In all figures, error bars represent means ± SD of triplicate experiments unless otherwise stated in the figure legend, and P values are noted by the following: *P > 0.05, **P > 0.01, and ***P > 0.001.
Acknowledgments
We acknowledge and thank members of the DFCI Molecular Biology Core Facilities for assistance with RNA sequencing, members of the DFCI ARF for assistance with in vivo studies, and J. Guerriero for providing the 4T1 cell lines used in this study.
Funding: This work was supported by NIH grants 1R21CA221683 (R.D.C.) and RO1CA196783-01 (R.D.C.) and the Leukemia and Lymphoma Society Screen to Lead Program (R.D.C.).
Author contributions: Conceptualization: R.D.C. Methodology: H.T., T.S., N.S., D.S., J.B., S.D.-P., and R.D.C. Investigation: H.T., T.S., H.-S.S., D.R., R.S.P., P.B., D.A., P.D., K.A., E.G., Z.Y., and N.V. Visualization: H.T., T.S., and R.D.C. Writing (original draft): H.T., T.S., F.G., S.D.-P., H.-S.S., and R.D.C. Writing (review and editing): H.T., T.S., and R.D.C. Supervision: R.D.C. Funding acquisition: R.D.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA sequencing data have been deposited in the NCBI GEO database (GSE182086). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S4
REFERENCES AND NOTES
- 1.Logan C. Y., Nusse R., The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Steinhart Z., Angers S., Wnt signaling in development and tissue homeostasis. Development 145, dev146589 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Clevers H., Nusse R., Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Klaus A., Birchmeier W., Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Sukhdeo K., Mani M., Hideshima T., Takada K., Pena-cruz V., Mendez G., Ito S., Anderson K. C., Carrasco D. R., β-catenin is dynamically stored and cleared in multiple myeloma by the proteasome-aggresome-autophagosome-lysosome pathway. Leukemia 26, 1116–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derksen P. W. B., Tjin E., Meijer H. P., Klok M. D., Mac Gillavry H. D., van Oers M. H. J., Lokhorst H. M., Bloem A. C., Clevers H., Nusse R., van der Neut R., Spaargaren M., Pals S. T., Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc. Natl. Acad. Sci. 101, 6122–6127 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sukhdeo K., Mani M., Zhang Y., Dutta J., Yasui H., Rooney M. D., Carrasco D. E. R., Zheng M., He H., Tai Y., Mitsiades C., Anderson K. C., Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. PNAS. 104, 7516–7521 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S. Y., Xia W., Wang J. C., Kwong K. Y., Spohn B., Wen Y., Pestell R. G., Hung M. C., β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. U.S.A. 97, 4262–4266 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luis T., Ichii M., Brugman M., Kincade P., Staal F., Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia 26, 414–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makena M. R., Gatla H., Verlekar D., Sukhavasi S., Pandey M. K., Pramanik K. C., Wnt/β-catenin signaling: The culprit in pancreatic carcinogenesis and therapeutic resistance. Int. J. Mol. Sci. 20, 4242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhan T., Rindtorff N., Boutros M., Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamos J. L., Weis W. I., The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clevers H., Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kramps T., Peter O., Nellen D., Chatterjee S., Züllig S., Wnt/Wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear β-catenin-TCF complex. Cell 109, 47–60 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Townsley F. M., Cliffe A., Bienz M., Pygopus and legless target Armadillo/β-catenin to the nucleus to enable its transcriptional co-activator function. Nat. Cell Biol. 6, 626–633 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Sampietro J., Dahlberg C. L., Cho U. S., Hinds T. R., Kimelman D., Xu W., Crystal structure of a β-catenin/BCL9/Tcf4 complex. Mol. Cell 24, 293–300 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Sustmann C., Flach H., Ebert H., Eastman Q., Grosschedl R., Cell-type-specific function of bcl9 involves a transcriptional activation domain that synergizes with β-catenin. Mol. Cell. Biol. 28, 3526–3537 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrasco D. R., Tonon G., Huang Y., Zhang Y., Sinha R., Feng B., Stewart J. P., Zhan F., Khatry D., Protopopova M., Protopopov A., Sukhdeo K., Hanamura I., Stephens O., Barlogie B., Anderson K. C., Chin L., Shaughnessy J. D., Brennan C., DePinho R. A., High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 9, 313–325 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Dutta-Simmons J., Zhang Y., Gorgun G., Gatt M., Mani M., Hideshima T., Takada K., Carlson N. E., Carrasco D. E., Tai Y.-T., Raje N., Letai A. G., Anderson K. C., Carrasco D. R., Aurora kinase A is a target of Wnt/beta-catenin involved in multiple myeloma disease progression. Blood 114, 2699–2708 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Sukhdeo K., Mani M., Hideshima T., Takada K., Pena-Cruz V., Mendez G., Ito S., Anderson K. C., Carrasco D. R., Β-catenin is dynamically stored and cleared in multiple myeloma by the proteasome–aggresome–autophagosome–lysosome pathway. Leukemia 26, 1116–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mani M., Carrasco D. E., Yunyu Z., Takada K., Gatt M. E., Dutta-Simmons J., Ikeda H., Diaz-Griffero F., Pena-Cruz V., Bertagnolli M., Myeroff L. L., Markowitz S. D., Anderson K. C., Carrasco D. R., BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 69, 7577–7586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takada K., Zhu D., Bird G. H., Sukhdeo K., Zhao J.-J., Mani M., Lemieux M., Carrasco D. E., Ryan J., Horst D., Fulciniti M., Munshi N. C., Xu W., Kung A. L., Shivdasani R. A., Walensky L. D., Carrasco D. R., Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic wnt signaling. Sci. Transl. Med. 4, 148ra117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J. J., Lin J., Zhu D., Wang X., Brooks D., Chen M., Chu Z. B., Takada K., Ciccarelli B., Admin S., Tao J., Tai Y. T., Treon S., Pinkus G., Kuo W. P., Hideshima T., Bouxsein M., Munshi N., Anderson K., Carrasco R. D., MiR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res. 74, 1801–1813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J. J., Carrasco R. D., Crosstalk between microRNA30a/b/c/d/e-5p and the canonical Wnt pathway: Implications for multiple myeloma therapy. Cancer Res. 74, 5351–5358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker N., Clevers H., Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 5, 997–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Polakis P., Drugging Wnt signalling in cancer. EMBO J. 31, 2737–2746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahn M., Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossmann T. N., Yeh J. T.-H., Bowman B. R., Chu Q., Moellering R. E., Verdine G. L., Inhibition of oncogenic Wnt signaling through direct targeting of β-catenin. Proc. Natl. Acad. Sci. 109, 17942–17947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lepourcelet M., Chen Y. N. P., France D. S., Wang H., Crews P., Petersen F., Bruseo C., Wood A. W., Shivdasani R. A., Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell 5, 91–102 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Kawamoto S. A., Coleska A., Ran X., Yi H., Yang C. Y., Wang S., Design of triazole-stapled BCL9 α-helical peptides to target the β-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction. J. Med. Chem. 55, 1137–1146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura K., Ikoma A., Shibakawa M., Shimoda S., Harada K., Saio M., Imamura J., Osawa Y., Kimura M., Nishikawa K., Okusaka T., Morita S., Inoue K., Kanto T., Todaka K., Nakanishi Y., Kohara M., Mizokami M., Safety, tolerability, and preliminary efficacy of the anti-fibrotic small molecule PRI-724, a CBP/β-catenin inhibitor, in patients with hepatitis c virus-related cirrhosis: A single-center, open-label, dose escalation phase 1 trial. EBioMedicine 23, 79–87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Khoueiry A. B., Ning Y., Yang D., Cole S., Kahn M., Zoghbi M., Berg J., Fujimori M., Inada T., Kouji H., Lenz H.-J., A phase I first-in-human study of PRI-724 in patients (pts) with advanced solid tumors. J. Clin. Oncol. 31, 2501 (2013). [Google Scholar]
- 33.Ko A. H., Chiorean E. G., Kwak E. L., Lenz H.-J., Nadler P. I., Wood D. L., Fujimori M., Inada T., Kouji H., McWilliams R. R., Final results of a phase Ib dose-escalation study of PRI-724, a CBP/beta-catenin modulator, plus gemcitabine (GEM) in patients with advanced pancreatic adenocarcinoma (APC) as second-line therapy after FOLFIRINOX or FOLFOX. J. Clin. Oncol. 34, e15721 (2016). [Google Scholar]
- 34.Emami K. H., Nguyen C., Ma H., Kim D. H., Jeong K. W., Eguchi M., Moon R. T., Teo J.-L., Oh S. W., Kim H. Y., Moon S. H., Ha J. R., Kahn M., A small molecule inhibitor of β-catenin/cyclic AMP response element-binding protein transcription. PNAS. 101, 12682–12687 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funck-Brentano T., Nilsson K. H., Brommage R., Henning P., Lerner U. H., Koskela A., Tuukkanen J., Cohen-Solal M., Movérare-Skrtic S., Ohlsson C., Porcupine inhibitors impair trabecular and cortical bone mass and strength in mice. J. Endocrinol. 238, 13–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madan B., McDonald M. J., Foxa G. E., Diegel C. R., Williams B. O., Virshup D. M., Bone loss from Wnt inhibition mitigated by concurrent alendronate therapy. Bone Res. 6, 17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S. M. A., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., Fazal A., Tomlinson R., Serluca F., Shao W., Cheng H., Shultz M., Rau C., Schirle M., Schlegl J., Ghidelli S., Fawell S., Lu C., Curtis D., Kirschner M. W., Lengauer C., Finan P. M., Tallarico J. A., Bouwmeester T., Porter J. A., Bauer A., Cong F., Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Thorne C. A., Hanson A. J., Schneider U., Tahinci E., Orton D., Cselenyi C. S., Jernigan K. K., Meyers K. C., Hang B. I., Waterson A. G., Kim K., Melancon B., Ghidu V. P., Sulikowski G. A., LaFleur B., Salic A., Lee L. A., III D. M. M., Lee E., Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat. Chem. Biol. 6, 829–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurney A., Axelrod F., Bond C. J., Cain J., Chartier C., Donigan L., Fischer M., Chaudhari A., Ji M., Kapoun A. M., Lam A., Lazetic S., Ma S., Mitra S., Park I. K., Pickell K., Sato A., Satyal S., Stroud M., Tran H., Yen W. C., Lewicki J., Hoey T., Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl. Acad. Sci. U.S.A. 109, 11717–11722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith D. C., Rosen L. S., Chugh R., Goldman J. W., Xu L., Kapoun A., Brachmann R. K., Dupont J., Stagg R. J., Tolcher A. W., Papadopoulos K. P., First-in-human evaluation of the human monoclonal antibody vantictumab (OMP-18R5; anti-Frizzled) targeting the WNT pathway in a phase I study for patients with advanced solid tumors. J. Clin. Oncol. 31, 2540 (2013).23733781 [Google Scholar]
- 41.Deka J., Wiedemann N., Anderle P., Murphy-Seiler F., Bultinck J., Eyckerman S., Stehle J. C., André S., Vilain N., Zilian O., Robine S., Delorenzi M., Basler K., Aguet M., Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res. 70, 6619–6628 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Eglen R. M., Reisine T., Roby P., Rouleau N., Illy C., Bossé R., Bielefeld M., The use of alphascreen technology in HTS: Current status. Curr. Chem. Genomics. 1, 2–10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remillard D., Buckley D., Paulk J., Brien G., Sonnett M., Seo H.-S., Dastierdi S., Wuhr M., Dhe-Paganon S., Armstrong S., Bradner J., Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew. Chem. Int. Ed. Engl. 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang M., Kang Y., Sewastianik T., Wang J., Tanton H., Alder K., Dennis P., Xin Y., Wang Z., Liu R., Zhang M., Huang Y., Loda M., Srivastava A., Chen R., Liu M., Carrasco R. D., BCL9 provides multi-cellular communication properties in colorectal cancer by interacting with paraspeckle proteins. Nat. Commun. 11, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Noort M., Meeldijk J., Van Der Zee R., Destree O., Clevers H., Wnt signaling controls the phosphorylation status of β-catenin. J. Biol. Chem. 277, 17901–17905 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Gaddis M., Gerrard D., Frietze S., Farnham P. J., Altering cancer transcriptomes using epigenomic inhibitors. Epigenetics and Chromatin. 8, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka N., Mashima T., Mizutani A., Sato A., Aoyama A., Gong B., Yoshida H., Muramatsu Y., Nakata K., Matsuura M., Katayama R., Nagayama S., Fujita N., Sugimoto Y., Seimiya H., APC mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer. Mol. Cancer Ther. 16, 752–762 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Parker T. W., Neufeld K. L., APC controls Wnt-induced β-catenin destruction complex recruitment in human colonocytes. Sci. Rep. 10, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuerer C., Nusse R., Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLOS One. 5, e9370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Flier L. G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M. E., Suijkerbuijk S., Van de Wetering M., Marra G., Clevers H., The intestinal Wnt/TCF signature. Gastroenterology. 132, 628–632 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Liberzon A., Birger C., Ghandi M., Jill P., Tamayo P., Jolla L., Jolla L., The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wenzel J., Rose K., Haghighi E. B., Lamprecht C., Rauen G., Freihen V., Kesselring R., Boerries M., Hecht A., Loss of the nuclear Wnt pathway effector TCF7L2 promotes migration and invasion of human colorectal cancer cells. Oncogene 39, 3893–3909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moor A. E., Anderle P., Cantù C., Rodriguez P., Wiedemann N., Baruthio F., Deka J., André S., Valenta T., Moor M. B., Gyorffy B., Barras D., Delorenzi M., Basler K., Aguet M., BCL9/9L-β-catenin signaling is associated with poor outcome in colorectal cancer. EBioMedicine 2, 1932–1943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu F., Liang Q., Abi-Mosleh L., Das A., de Brabander J. K., Goldstein J. L., Brown M. S., Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife 4, e12177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pol A., Gross S. P., Parton R. G., Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soccio R. E., Breslow J. L., Intracellular cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 24, 1150–1160 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Madison B. B., Srebp2: A master regulator of sterol and fatty acid synthesis. J. Lipid Res. 57, 333–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C.-W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., Lum L., Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J., Pan S., Hsieh M. H., Ng N., Sun F., Wang T., Kasibhatla S., Schuller A. G., Li A. G., Cheng D., Li J., Tompkins C., Pferdekamper A. M., Steffy A., Cheng J., Kowal C., Phung V., Guo G., Wang Y., Graham M. P., Flynn S., Brenner J. C., Li C., Villarroel M. C., Schultz P. G., Wu X., McNamara P., Sellers W. R., Petruzzelli L., Boral A. L., Seidel H. M., McLaughlin M. E., Che J., Carey T. E., Vanasse G., Harris J. L., Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. U.S.A. 110, 20224–20229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arkin M. R., Tang Y., Wells J. A., Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chem. Biol. 21, 1102–1114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng M., Jin J. Q., Xia L., Xiao T., Mei S., Wang X., Huang X., Chen J., Liu M., Chen C., Rafi S., Zhu A. X., Feng Y. X., Zhu D., Pharmacological inhibition of β-catenin/BCL9 interaction overcomes resistance to immune checkpoint blockades by modulating Treg cells. Sci. Adv. 5, eaau5240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasper M. A., Glanz M., Oder A., Schmieder P., Von Kries J. P., Hackenberger C. P. R., Vinylphosphonites for staudinger-induced chemoselective peptide cyclization and functionalization. Chem. Sci. 10, 6322–6329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sang P., Zhang M., Shi Y., Li C., Abdulkadir S., Li Q., Ji H., Cai J., Inhibition of β-catenin/B cell lymphoma 9 protein−protein interaction using α-helix–mimicking sulfono-γ-AApeptide inhibitors. Proc. Natl. Acad. Sci. U.S.A. 166, 10757–10762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoggard L. R., Zhang Y., Zhang M., Panic V., Wisniewski J. A., Ji H., Rational design of selective small-molecule inhibitors for β-catenin/B-cell lymphoma 9 protein-protein interactions. J. Am. Chem. Soc. 137, 12249–12260 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Wisniewski J. A., Yin J., Teuscher K. B., Zhang M., Ji H., Structure-based design of 1,4-dibenzoylpiperazines as β-catenin/B-cell lymphoma 9 protein-protein interaction inhibitors. ACS Med. Chem. Lett. 7, 508–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M., Wang Z., Zhang Y., Guo W., Ji H., Moffitt H. L., States U., States U., Structure-based optimization of small-molecule inhibitors for the β-catenin/B-cell lymphoma 9 protein–protein interaction. J. Med. Chem. 61, 2989–3007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De La Roche M., Rutherford T. J., Gupta D., Veprintsev D. B., Saxty B., Freund S. M., Bienz M., An intrinsically labile α-helix abutting the BCL9-binding site of β-catenin is required for its inhibition by carnosic acid. Nat. Commun. 3, 610–680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baell J. B., Holloway G. A., New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Baell J., Walters M. A., Chemistry: Chemical con artists foil drug discovery. Nature 513, 481–483 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Scott C. C., Vossio S., Vacca F., Snijder B., Larios J., Schaad O., Guex N., Kuznetsov D., Martin O., Chambon M., Turcatti G., Pelkmans L., Gruenberg J., Wnt directs the endosomal flux of LDL -derived cholesterol and lipid droplet homeostasis. EMBO Rep. 16, 741–752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lien E. C., Westermark A. M., Zhang Y., Yuan C., Li Z., Lau A. N., Sapp K. M., Wolpin B. M., Vander Heiden M. G., Low glycaemic diets alter lipid metabolism to influence tumour growth. Nature 599, 302–307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munro S., Lipid rafts. Cell 115, 377–388 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Greenlee J. D., Subramanian T., Liu K., King M. R., Rafting down the metastatic cascade: The role of lipid rafts in cancer metastasis, cell death, and clinical outcomes. Cancer Res. 81, 815–817 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Azbazdar Y., Karabicici M., Erdal E., Ozhan G., Regulation of Wnt signaling pathways at the plasma membrane and their misregulation in cancer. Front. Cell Dev. Biol. 9, 1–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y. C., Park M. J., Ye S. K., Kim C. W., Kim Y. N., Elevated levels of cholesterol-rich lipid rafts in cancer cells are correlated with apoptosis sensitivity induced by cholesterol-depleting agents. Am. J. Pathol. 168, 1107–1118 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee H. J., Li J., Vickman R. E., Li J., Liu R., Durkes A. C., Elzey B. D., Yue S., Liu X., Ratliff T. L., Cheng J. X., Cholesterol esterification inhibition suppresses prostate cancer metastasis by impairing the Wnt/β-catenin pathway. Mol. Cancer Res. 16, 974–985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Agarwal B., Rao C. V., Bhendwal S., Ramey W. R., Shirin H., Reddy B. S., Holt P. R., Lovastatin augments sulindac-induced apoptosis in colon cancer cells and potentiates chemopreventive effects of sulindac. Gastroenterology 117, 838–847 (1999). [DOI] [PubMed] [Google Scholar]
- 78.Vona R., Iessi E., Matarrese P., Role of cholesterol and lipid rafts in cancer signaling: A promising therapeutic opportunity? Front. Cell Dev. Biol. 9, 622908 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dowling C. M., Walsh D., Coffey J. C., Kiely P. A., The importance of selecting the appropriate reference genes for quantitative real time PCR as illustrated using colon cancer cells and tissue. F1000Research 5, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spandidos A., Wang X., Wang H., Seed B., PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cornwell M. I., Vangala M., Taing L., Herbert Z., Köster J., Li B., Sun H., Li T., Zhang J., Qiu X., Pun M., Jeselsohn R., Brown M., Liu X. S., Long H. W., VIPER: Visualization pipeline for RNA-seq, a snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics. 19, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sewastianik T., Straubhaar J. R., Zhao J. J., Samur M. K., Adler K., Tanton H. E., Shanmugam V., Nadeem O., Dennis P. S., Pillai V., Wang J., Jiang M., Lin J., Huang Y., Brooks D., Bouxsein M., Dorfman D. M., Pinkus G. S., Robbiani D. F., Ghobrial I. M., Budnik B., Jarolim P., Munshi N. C., Anderson K. C., Carrasco R. D., miR-15a/16-1 deletion in activated B cells promotes plasma cell and mature B-cell neoplasms. Blood 137, 1905–1919 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sewastianik T., Guerrera M. L., Adler K., Dennis P. S., Wright K., Shanmugam V., Huang Y., Tanton H., Jiang M., Kofides A., Demos M. G., Dalgarno A., Patel N. A., Nag A., Pinkus G. S., Yang G., Hunter Z. R., Jarolim P., Munshi N. C., Treon S. P., Carrasco R. D., Human MYD88L265P is insufficient by itself to drive neoplastic transformation in mature mouse B cells. Blood Adv. 3, 3360–3374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S4