Abstract
The alpha-synuclein mutation E83Q, the first in the NAC domain of the protein, was recently identified in a patient with dementia with Lewy bodies. We investigated the effects of this mutation on the aggregation of aSyn monomers and the structure, morphology, dynamic, and seeding activity of the aSyn fibrils in neurons. We found that it markedly accelerates aSyn fibrillization and results in the formation of fibrils with distinct structural and dynamic properties. In cells, this mutation is associated with higher levels of aSyn, accumulation of pS129, and increased toxicity. In a neuronal seeding model of Lewy body (LB) formation, the E83Q mutation significantly enhances the internalization of fibrils into neurons, induces higher seeding activity, and results in the formation of diverse aSyn pathologies, including the formation of LB-like inclusions that recapitulate the immunohistochemical and morphological features of brainstem LBs observed in brains of patients with Parkinson’s disease.
A NAC domain mutation (E83Q) exacerbates alpha-synuclein aggregation and formation of Lewy body–like inclusions.
INTRODUCTION
The accumulation of fibrillar and aggregated forms of the presynaptic protein alpha-synuclein (aSyn) within Lewy bodies (LBs) and Lewy neurites (LNs) is a characteristic hallmark of many synucleinopathies, including Parkinson’s disease (PD) and dementia with Lewy bodies (DLB). Several missense point mutations in the SNCA gene, which encodes for aSyn, have been linked to familial forms of PD: A53T (1), A30P (2), E46K (3), H50Q (4), A53E (5), A53V (6), and A30G (7). In addition, the G51D mutation has been linked to a form of synucleinopathy with shared characteristics between PD and multiple system atrophy (MSA) (8). Furthermore, SNCA gene multiplications were shown to be sufficient to cause PD and DLB (9, 10). Although aSyn mutation carriers are rare, studies on the biochemical, cellular, aggregation, and toxic properties of these mutants have provided valuable insights into the mechanisms of aSyn aggregation and PD pathology. These studies have also suggested that the various mutations may act via distinct mechanisms.
Recently, Kapasi et al. (11) reported the discovery of a novel SNCA mutation encoding for a glutamic acid–to–glutamine (E83Q) substitution in a patient with DLB and atypical frontotemporal lobar degeneration. Postmortem neuroimaging and histology of the patient’s brain revealed a widespread LB and LN pathology, with severe atrophy of the frontotemporal lobes that correlates with cognitive impairment. As reported in some MSA (12), the brain of the E83Q mutation carrier showed higher LB pathology in the hippocampal neurons than in the substantia nigra where much less pathology was detected. In addition, severe LB pathology was also detected in the cortex and other brain regions, including the thalamus and the basal ganglia. There was no evidence of Tau or TAR DNA-binding protein 43 pathology in the brain of this patient. Notably, the autopsy report of the patient’s father included a diagnosis of Pick’s disease. The presence of the SNCA E83Q mutation in the father was later confirmed (11).
Unlike all previously reported synucleinopathy-related mutations, which invariably occur in the N-terminal region spanning residues 30 to 53, and with most clustering between amino acids 46 and 53 (fig. S1A), the E83Q mutation is within the nonamyloid component (NAC) domain (residues 61 to 95), which plays a critical role in catalyzing aSyn oligomerization and fibril formation (13). Furthermore, the substitution of glutamate by glutamine at residue 83 results in the removal of a negative charge from the highly hydrophobic NAC domain, which contains only three charged residues (two glutamic acid and one lysine). These observations, combined with the fact that this mutation is associated with DLB instead of PD, suggest that it may influence the structure, aggregation, and pathogenicity of aSyn via mechanisms distinct from those of other mutations (11) and, thus, could offer valuable insights into the molecular mechanisms of aSyn pathology and neurodegeneration in synucleinopathies such as DLB.
Here, biochemical and biophysical approaches, including nano–electrospray ionization mass spectrometry (nESI-MS), transmission electron microscopy (TEM), and solid-state nuclear magnetic resonance (ssNMR) spectroscopy, were combined with cellular models to determine the effect of the E83Q mutation on (i) the conformation and membrane binding properties of monomeric aSyn; (ii) the aggregation kinetics of aSyn, as well as the morphology and structural properties of aSyn fibrils in vitro; (iii) the subcellular localization, aggregation, and inclusion formation of aSyn in mammalian cells lines; and (iv) the seeding activity and formation of de novo fibrils and LB-like inclusions in a neuronal seeding model of synucleinopathies (14).
Our in vitro studies demonstrated that the E83Q mutation significantly accelerated aSyn fibrillization. Fibrils generated from E83Q aSyn monomers exhibited distinct structural properties and stability than those formed by the wild-type (WT) protein. Overexpression of E83Q, but not WT aSyn, in mammalian cells [human embryonic kidney (HEK) 293, HeLa or M17] resulted in a gain of toxic functions that are not dependent on aSyn fibrillization but that appear to be associated with increased oligomerization. Last, in a neuronal seeding model of LB formation (14, 15), the E83Q mutation markedly increased the seeding activity of human preformed fibrils (PFFs) and promoted the formation of LB-like inclusions with diverse morphological features, resembling the diversity of aSyn pathology in PD brains (16–18). Unlike mouse PFFs, which induce the formation of diffuse LB-like inclusions, the E83Q PFFs induced the formation of LB-like inclusions with a ring-like organization that recapitulates the immunohistochemical, structural, and morphological features of bona fide brainstem LBs observed in patient’s brains affected by late-stage PD (16, 17, 19–21).
RESULTS
The E83Q mutation disrupts transient, long-range interactions of the aSyn monomer ensembles
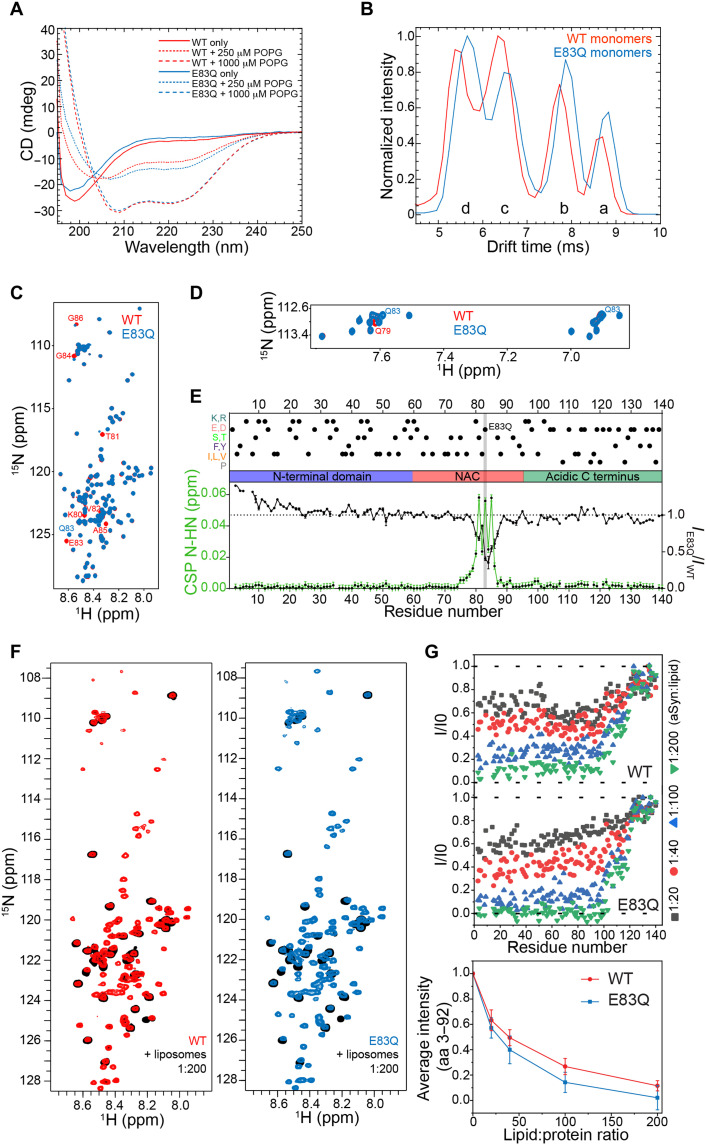
To investigate the effect of the E83Q mutation on the conformation of aSyn monomers, we compared the circular dichroism (CD) spectra of purified WT and E83Q aSyn from Escherichia coli (fig. S2). WT and E83Q aSyn showed identical CD spectra, with a minimum at ~198 nm, consistent with a predominantly disordered conformation for both proteins (Fig. 1A, solid lines).
Fig. 1. E83Q mutation affects the conformational ensembles of free aSyn monomers but not its helical conformation when bound to lipid vesicles.
(A) CD spectra of monomeric WT and E83Q aSyn incubated with or without POPG vesicles. (B) Drift time plot comparison of 8+ charge state in WT (red) and E83Q mutant (blue) aSyn. Both the WT and the E83Q aSyn mutant display four unique conformational populations (labeled “a” to “d” from the most extended to the most compacted, respectively). (C) Backbone amide region of the 1H/15N HMQC of WT (red) and E83Q mutant (blue) aSyn. The most perturbed residues are labeled. ppm, parts per million. (D) Asparagine and glutamine side-chain regions of the 1H/15N HMQC of WT (red) and E83Q mutant (blue) aSyn. The new peaks of Q83 and the perturbed Q79 are labeled. (E) N-HN CSPs (green) and intensity ratios (black) between WT and E83Q aSyn based on the spectrum in (C). Above the distinct domains of aSyn (N-terminal lipid-binding α helix, NAC, and the acidic C terminus), the most representative residues are shown in dots. The mutated residue is highlighted in gray. (F) 1H/15N HSQC of WT (red; left) and E83Q mutant (blue; right) aSyn compared with the respective proteins in the presence of liposomes with a protein:lipid ratio of 1:200 (black). (G) Residue-specific 1H/15N HSQC peak intensity ratios at increasing protein:lipid ratios for WT (top) and E83Q (bottom) aSyn. In the bottom panel, changes in the average intensity of the cross-peak signals of residues 3 to 92 for increasing liposome concentrations are shown for WT (red) and E83Q (blue) aSyn, respectively. Error bars represent SDs. aa, amino acids.
In solution, aSyn exists in an array of dynamic conformations (fig. S1B) (22). To determine whether the E83Q mutation alters the distribution of aSyn monomer conformations, we performed native nESI-MS combined with ion mobility (IM). IM reports on the rotationally averaged shape and size (i.e., compactness) of ionized protein, providing conformational fingerprints per charge state. Data for each mass/charge ratio (m/z) peak was obtained as previously described (23, 24) and visualized in drift time plots. In Fig. 1B, drift time plots of the 8+ charge state of WT and E83Q are overlaid to compare the conformation distributions.
We observed four distinct conformational populations at this charge state for both WT and E83Q monomers. The conformations for the WT monomers were identical to those previously found (23). The E83Q mutant showed a slight shift toward the two more extended conformations (a and b), and within the distributions of compact states (c and d), there was a shift toward the most compact state (d), which was reflected in the disparities of the peak intensities (Fig. 1B). To investigate whether monomer conformations are stabilized or destabilized by the mutation, collisional activation experiments were performed for 7+, 8+, and 11+ charge states. The results are discussed in detail in the Supplementary Materials (fig. S3) and show subtle perturbations in conformational stability, depending on the charge state. Some individual conformational substates are slightly stabilized, while others appear with somewhat reduced stability.
To further delineate the effect of the E83Q mutation on the conformational ensemble of the aSyn monomer in solution, we compared the NMR spectra of 15N-labeled WT and E83Q aSyn proteins. The differences observed in the chemical shifts [chemical shift perturbations (CSPs)] of the 1H/15N HMQC (heteronuclear multiple-quantum coherence) and 1H/13C HSQC (heteronuclear single-quantum coherence) spectra of the mutant were present locally around the mutation (Fig. 1, C to E, and fig. S4, A to C). The CSPs were small even for the mutated residue, as expected for the simple change from an -OH to an -NH2 group upon mutation of a glutamic acid to a glutamine. However, the perturbation was not limited to the immediate vicinity of the site of mutation (Fig. 1D). In the glutamine and asparagine side-chain region (Fig. 1D), we observed, besides the new peak of the new residue Q83, a CSP in one of the protons of residue Q79. This might be attributed to the presence of a polar interaction between Q79 and residue 83 and can also provide a rationale for the more extended CSP for residues N terminal to the site of mutation. Notably, the peak intensity analysis of the same spectra revealed a decrease in intensity in the region of the mutation and an increase in NMR signal intensity for residues 1 to 23 at the N terminus of aSyn (Fig. 1E). Because the 23 N-terminal residues of aSyn are predominantly positively charged, the changes in NMR signal intensities in this region might arise from changes in transient long-range electrostatic interactions of the aSyn ensembles due to the removal of the negative charge at position 83. Thus, both nESI-MS and NMR spectroscopy indicate that the E83Q mutation perturbs the conformational ensemble of aSyn in solution.
The E83Q mutation does not significantly disrupt aSyn interactions with membranes
Since E83Q mutation lies in the distal region of aSyn that binds to membranes (fig. S1B), we investigated whether the E83Q mutation affects aSyn’s binding properties to membranes. It is known that WT aSyn adopts α-helical conformations upon binding to lipid membranes (22, 25). Therefore, we used CD spectroscopy to compare the propensity of WT and E83Q aSyn to bind to liposomes of distinct lipid composition [anionic 1–palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) and DOPE (1,2-fioleoyl-sn-glycero-3-phosphoethanolamine):DOPS (1,2-dioleoyl-sn-glycero-3-phospho-l-serine):DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) (5:3:2)]. As seen in Fig. 1A and fig. S4(D to F), we observed a similar secondary structure transition from unfolded to α-helical conformations for both proteins, irrespective of lipid vesicle composition or ratios of lipids to aSyn.
To probe with residue resolution the binding of WT and E83Q aSyn to membranes, we performed liposome titration experiments against WT and E83Q aSyn monitored by 1H/15N NMR spectroscopy (7). With an increasing liposome concentration, we noticed a gradual decrease in E83Q and WT aSyn NMR signal intensity (Fig. 1F). Residue-specific analysis showed that the most liposome-perturbed region—as evidenced by a strong decrease in NMR signal intensity—comprises for both E83Q and WT aSyn the residues from 3 to 92 (Fig. 1G, top). In contrast, the C-terminal 10 to 20 residues were not broadened even at the highest liposome concentrations (Fig. 1G, top), implying that these residues remained unbound and flexible. A quantitative comparison of the strength of liposome-induced signal broadening further showed that at identical protein:liposome ratios, the NMR signals of E83Q aSyn are more strongly broadened when compared to the WT aSyn (Fig. 1G, bottom). This suggests that the E83Q mutation slightly enhances binding to membranes.
Next, we assessed aSyn membrane-binding properties in cells. We transiently overexpressed E83Q or WT aSyn in HEK293 cells and assessed the aSyn distributions in the cytosolic, membranous, and nuclear subcellular fractions by Western blot (WB) analysis. As shown in fig. S4 (G and H), both E83Q and WT aSyn are predominantly localized in the cytosolic compartment and accumulate minimally in the membrane or nuclear fractions (information related to the antibodies used in this study can be found in fig. S5). Together, our data indicate that although the E83Q mutation is located in the membrane-binding domain, it does not strongly alter its binding to lipid vesicles or intracellular membranes.
The E83Q mutation accelerates the aggregation kinetics of aSyn in vitro
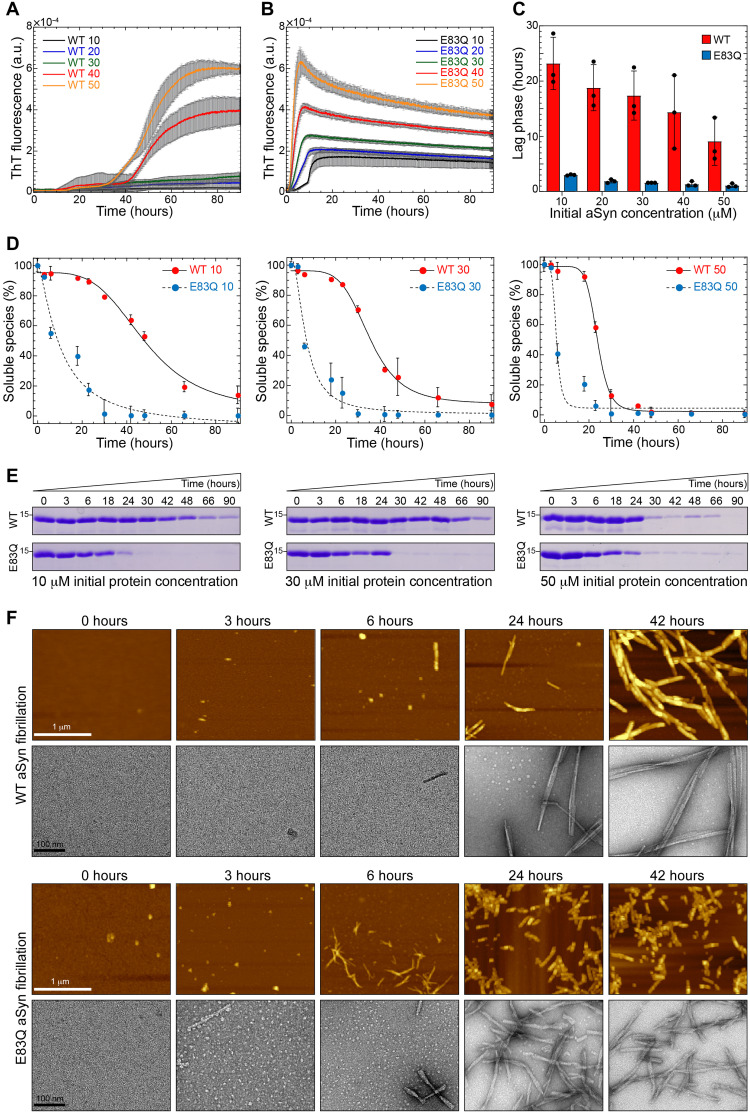
To investigate the effect of the E83Q mutation on aSyn aggregation, we compared the fibrillization kinetics of WT and E83Q aSyn at five initial protein concentrations ranging from 10 to 50 μM. Irrespective of the initial concentration, the E83Q mutant exhibited faster aggregation kinetics relative to the WT protein (Fig. 2, A and B). Analysis of the lag phase from the kinetic curves revealed that the E83Q mutant exhibited ~10-fold faster aggregation kinetics at all concentrations (Fig. 2C). The WT counterpart exhibited a gradual decrease in the lag phase, from ~23 hours at 10 μM to ~11 hours at 50 μM initial aSyn concentration, whereas the E83Q mutant at these concentrations showed lag times of ~2.9 and ~0.9 hours, respectively (Fig. 2C). To further validate the faster aggregation of E83Q relative to WT aSyn, the aggregation of the proteins was monitored in the absence of Thioflavin T (ThT) using the sedimentation assay, namely, by quantifying the remaining soluble aSyn species as a function of time (Fig. 2, D and E). Similar to the data from the ThT aggregation kinetics (Fig. 2, A and B), E83Q showed significantly faster aggregation propensities than WT aSyn at all concentrations (Fig. 2, D and E).
Fig. 2. E83Q aSyn aggregates much faster than WT aSyn.
(A) Aggregation kinetics of WT and (B) E83Q aSyn at different initial concentrations (10, 20, 30, 40, and 50 μM) monitored by ThT fluorescence. a.u., arbitrary units. (C) Bar graph of lag phase extracted from the ThT aggregation kinetics shown by WT (A) and E83Q (B) (mean ± SEM, n = 3). (D) Solubility assay of the WT and E83Q aSyn aggregations at different time points at varying initial concentrations of 10, 30, and 50 μM. (E) SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the same samples from initial concentration. Scale bars, 1 μm (for all the AFM images) and 100 nm (for all the TEM images). (F) AFM and TEM images of time-dependent WT and E83Q aSyn aggregation from 50 μM.
To determine whether the E83Q mutation influences early oligomerization events, we monitored changes in the aggregation state of E83Q and WT aSyn as a function of time by atomic force microscopy (AFM) and TEM (Fig. 2F). No aggregate/oligomeric structures were observed immediately after the resuspension of either WT or E83Q monomeric proteins (0 hours). At later time points (from 1.5 to 6 hours), the E83Q showed a slightly enhanced rate of oligomerization and formed only slightly more oligomers at 1.5 hours compared with WT at 3 to 4 hours (fig. S6). At ~24 hours of aggregation, the WT aSyn formed a mixed population of oligomeric and fibrillar structures, whereas the E83Q mutant formed predominantly shorter fibrillar structures (Fig. 2F). After ~42 hours, WT aSyn was observed to form mainly straight and long fibrillar structures, whereas the E83Q mutant formed predominantly fibrillar structures that were significantly shorter (by an average of 180 ± 97 nm) than those of WT aSyn (Fig. 2F). These findings are consistent with the results from the ThT fluorescence-based aggregation kinetics and sedimentation assay, and demonstrate that the E83Q mutation slightly enhances aSyn oligomerization but markedly accelerates aSyn fibrillization and alters the size distribution and dimensions of the final fibrillar structures.
Fibrils of E83Q mutant show distinct morphology, stability, and structural features
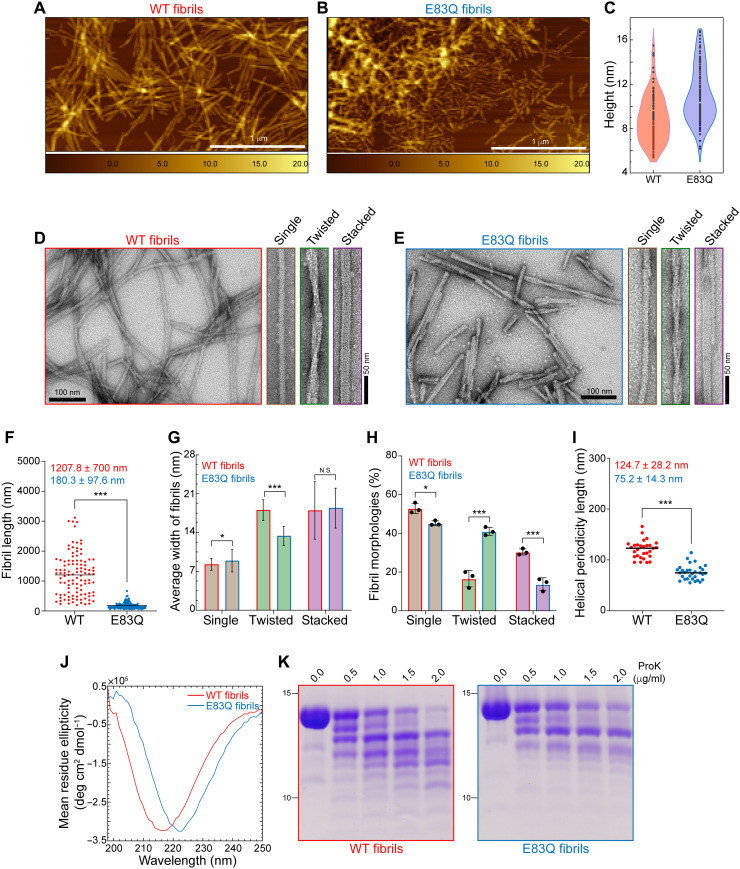
Given that E83 occurs within the core of aSyn fibrils, we speculated that its mutation to glutamine might influence the structural properties or dynamics of aSyn fibrils. To test this hypothesis, AFM and TEM were used to quantify and compare the morphological properties of WT and E83Q aSyn fibrils. As shown in Fig. 3, both WT and E83Q fibrils were polymorphic and had single-filament (twisted) and multifilament (stacked) morphologies. The WT fibrils consistently exceeded 1 μm in length (Fig. 3, A to F, and fig. S7A), whereas E83Q fibrils were much shorter, with an average length of 180 ± 97 nm (Fig. 3, A to F). These observations are highly reproducible using different batches of E83Q aSyn proteins (fig. S7, B to F). Furthermore, WT aSyn showed a broader height distribution, with an average height of 8.9 ± 2.1 nm, whereas E83Q fibrils had an average height of 10.9 ± 2.5 nm. The E83Q and WT aSyn formed fibrils of diverse yet overlapping structures (Fig. 3, D and E). However, an in-depth analysis of the fibril widths of the different polymorphs revealed that the population of twisted fibrils displayed the highest variation in width between WT and E83Q aSyn (WT, 18.0 ± 1.9 nm; E83Q, 13.3 ± 1.8 nm). The other two morphologies exhibited nearly identical widths (single, ~8 nm; stacked, ~17 nm) between WT and E83Q aSyn fibrils (Fig. 3G).
Fig. 3. E83Q aSyn forms fibrils with distinct morphological and structural properties compared with WT fibrils.
(A and B) AFM images of WT (A) and E83Q (B) aSyn fibrils. (C) Violin plot showing the height analysis (WT, 8.9 ± 2.1 nm; E83Q, 10.9 ± 2.5 nm) estimated from (A) and (B). Scale bars, 1 μm. (D and E) TEM images of WT (D) and E83Q (E) aSyn fibrils. TEM montages in (D) and (E) show the single, twisted, and stacked morphologies. Scale bars, 50 or 100 nm. (F) Dot plot showing the length distribution of WT and E83Q fibrils (n = 108 for WT; n = 191 for E83Q fibrils). ***P < 0.0005 (unpaired t test, WT versus E83Q fibrils). (G) Bar graph of TEM-based analysis of average width of different morphologies of WT and E83Q fibrils. WT fibrils [single, 8.2 ± 1.0 nm (n = 87); twisted, 18.0 ± 1.9 nm (n = 30); stacked, 17.9 ± 5.2 nm (n = 47)] and E83Q fibrils [single, 8.8 ± 2.0 nm (n = 79); twisted, 13.3 ± 1.8 nm (n = 68); stacked, 18.4 ± 3.6 nm (n = 29)]. *P < 0.05 and ***P < 0.0005 (unpaired t test, WT versus E83Q fibrils). N.S., nonsignificant. (H) Bar graph displays the distribution of different fibril morphologies between WT and E83Q. The graph represents the mean ± SD of three independent experiments. Two-way analysis of variance (ANOVA) shows a significant statistical interaction between the WT versus E83Q mutation and their different morphological distributions. Multiple comparison analysis with Sidak’s correction. *P < 0.05 and ***P < 0.001. (I) Dot plot showing the helical periodicity length of twisted fibrils of WT and E83Q aSyn (n = 30 for WT; n = 31 for E83Q fibrils). ***P < 0.0005 (unpaired t test, WT versus E83Q fibrils). (J) CD spectra of WT and E83Q aSyn fibrils. (K) SDS-PAGE analysis of proteinase K (ProK) digestion of WT (left) and E83Q (right) aSyn fibrils.
While analyzing the relative frequencies of different morphologies, we observed different distributions of each of the three fibril morphologies between the two proteins (Fig. 3H). In the case of the E83Q mutant, the single-fibril morphologies predominated (~45%), followed by the twisted (~39%) and stacked fibril morphologies (~16%). Although the single-fibril morphologies also predominated (~53%) for the WT protein, the second most common morphology (~29%) was stacked fibrils, followed by twisted fibrils (~18%). Analysis of the helical periodicity of the twisted fibril population showed significant differences between the two proteins. The E83Q fibrils showed an average periodicity length of 75.2 ± 14.3 nm, compared to 124.7 ± 28.2 nm for the WT fibrils (Fig. 3I).
Next, we analyzed the secondary structure of fibrils using CD spectroscopy. The CD spectra of WT and E83Q aSyn fibrils showed the single peak minimum for both fibrils, suggesting the enrichment of β sheet structures. However, a major shift of the CD minimum at 222 nm was observed for E83Q fibrils and at 217 nm for WT fibrils. This suggests the existence of pronounced differences in the arrangement/packing of β sheet structures between aSyn molecules in the fibrils (Fig. 3J). Together, these findings demonstrate that the E83Q mutation significantly alters the distribution and the structural and morphological properties of aSyn fibril conformations.
To further validate our findings, we compared the proteinase K (ProK) digestion profile of WT and E83Q fibrils. To ensure the absence of cross-contamination of oligomeric or monomeric aSyn, we centrifuged the aSyn fibril samples, removed the supernatant, and resuspended the pellet containing the fibrillar structures in phosphate-buffered saline (PBS) (26). After a 30-min incubation of the same concentration of WT and E83Q aSyn fibrils with the increasing concentrations of ProK, the reaction mixtures were visualized by SDS–polyacrylamide gel electrophoresis (SDS-PAGE). The results revealed major differences in the stability and proteolysis pattern between WT and E83Q aSyn fibrils (Fig. 3K). While the protein band at ~15 kDa from WT aSyn degraded almost completely at 2 μg/ml of ProK, the same band from the E83Q mutant displayed a stronger resistance to ProK proteolysis, suggesting greater stability of E83Q aSyn fibrils relative to WT aSyn. Although the band at ~12 kDa was observed in both WT and E83Q samples, additional bands appeared at high concentrations of ProK in the WT, but not in the E83Q, fibril sample. At the highest concentration of ProK (2 μg/ml), we observed 12 bands for WT fibrils and only 8 bands for E83Q fibrils (Fig. 3K and fig. S8, A and B). These differences in the ProK proteolysis profile of E83Q aSyn fibrils are indicative of a distinct fibrillar structure. Together, the data from AFM/TEM imaging and ProK digestion analyses suggest that the fibrils generated from the E83Q-mutated aSyn exhibit distinct morphological and structural properties from their WT counterparts.
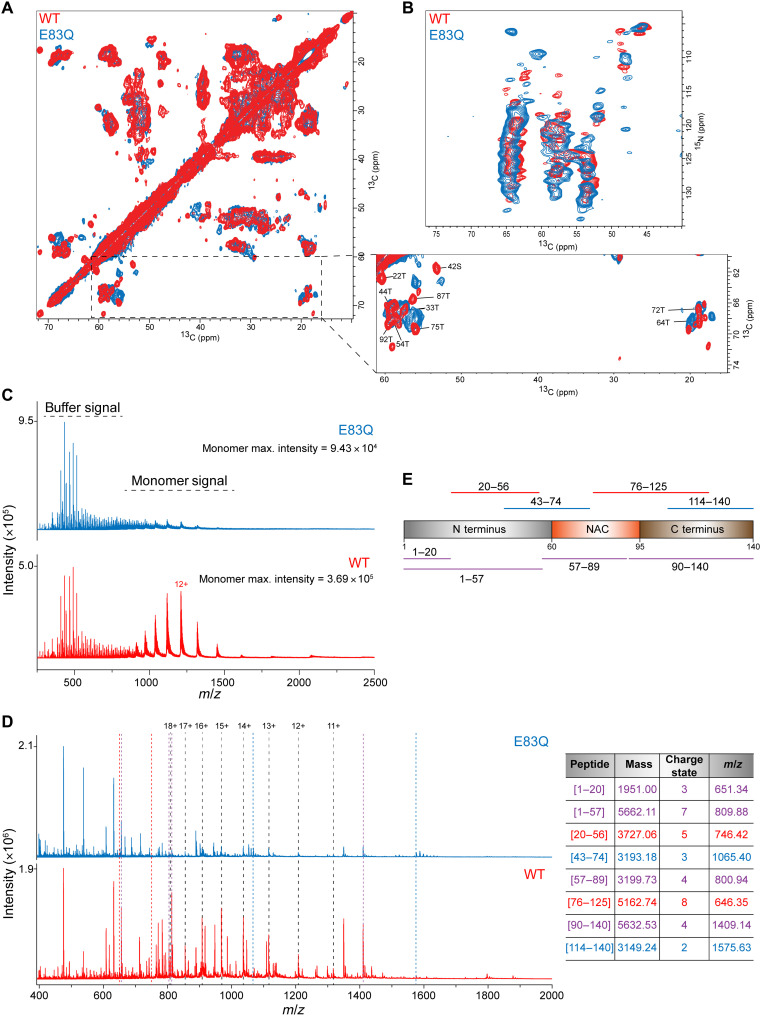
Next, to further probe the differences in the molecular structure of amyloid fibrils formed by E83Q and WT aSyn using ssNMR spectroscopy, we prepared 13C,15N-labeled proteins (fig. S9, A and B) and characterized the aggregated proteins (fig. S9, C and D), which were prepared under identical conditions. First, we established that 13C/15N-labeling of aSyn did not influence the biophysical and aggregation properties of WT or E83Q (fig. S9). Next, we recorded observed high-resolution two-dimensional 13C-13C dipolar-assisted rotational resonance (DARR) and 15N-13Cα (NCA) spectra for both WT and E83Q aSyn (Fig. 4, A and B). DARR and NCA experiments use cross-polarization steps and therefore only detect residues from the rigid cross–β-structure core of amyloid fibrils. The high quality of the DARR and NCA spectra demonstrates that both E83Q and WT aSyn aggregate into structurally well-defined amyloid fibrils. Moreover, a detailed comparison of the cross-peak patterns observed for the two proteins reveals pronounced differences in the position and intensity of many signals. This is particularly apparent in the spectral region of the 13C-13C DARR spectrum, in which Cα-Cβ cross-peaks of threonine and serine residues from the rigid fibrillar core appear (Fig. 4A, zoom). The comparison provides residue-specific support for differences in the molecular structure of the cross–β-structure core of amyloid fibrils formed by E83Q and WT aSyn.
Fig. 4. ssNMR and nESI-MS experiments on WT and E83Q fibrils.
(A and B) Superposition of two-dimensional 13C-13C DARR (A) and NCA spectra (B) of E83Q aSyn (blue) and WT (red) aSyn fibrils. Tentative assignments, which were transferred from previous resonance assignments (BMRB id: 18860), of Cα-Cβ cross-peaks of threonine and serine residues in WT aSyn are indicated in the zoom in of (A). (C) Comparison of intensities of monomers releasing from the fibril during the nESI-MS experiments on fibrils. We also observed the signals of buffer components at a similar intensity between samples. (D) Peptide spectra of WT and E83Q fibrils after 5 min digestion with ProK (0.5 μg/μl). Peaks related to intact monomer charge states are indicated with black dashed lines. The presence of undigested monomer in both cases, even at very low intensity, indicates that secondary cleavage of peptides is kept to a minimum, which is desired to retain structural information from the fibril. Colored dotted lines indicate selected peaks that are present in both conditions (purple), only for WT (red) or only for E83Q (blue). The table lists the selected peaks with their respective mass and linked peptide fragments. Colors are identical to those of the dotted lines in (B). (E) A scheme depicts the full sequence of aSyn and the peptides [from (D)] commonly found for both WT and E83Q (purple), while others were unique to one of the two fibrils (red for WT and blue for E83Q).
Last, to investigate the structural basis underlying the differences in the ProK digestion profile of E83Q and WT aSyn fibrils dynamics, we performed nESI-MS on the digestion products. We first analyzed the fibrils without ProK treatment. We also observed a peak pattern corresponding to monomers, suggesting their release from the fibrils of WT and E83Q aSyn, respectively. It has been shown in other amyloid systems that a dynamic equilibrium exists at the fibril ends involving the dissociation and reassociation of the monomers (27, 28). Here, the stark difference in the concentration of monomers released (about fourfold) indicates a difference in the fibrils’ stability, suggesting that monomers were released from WT aSyn fibril ends more freely than those released from E83Q aSyn fibril ends (Fig. 4C).
We next analyzed the WT and E83Q aSyn fibrils after ProK treatment and detected several peptide peaks (Fig. 4D). The observed peak pattern differed between the WT and E83Q fibrils, with more intense peptide signals seen in the WT fibrils. Following manual deconvolution of the peaks, peptide masses could then be linked to amino acid sequences using the Protein Analysis Work Sheet (PAWS) tool. We observed differences in the peptide regions when mapping the identified peptides onto the aSyn protein sequence, indicating structural discrepancies between WT and E83Q fibrils. We detected peptides commonly found for both WT and E83Q aSyn (representative peptides in purple), while others were unique to one of the two fibrils (representative peptides in red for WT and blue for E83Q; Fig. 4, D and E).
The peptides identified in both WT and E83Q aSyn fibrils were derived from the first half of the N terminus (amino acids 1 to 20) and the entire N terminus (amino acids 1 to 57), in addition to others encompassing nearly the complete NAC domain (amino acids 57 to 89) or the fully intact C terminus (amino acids 90 to 140). In the case of WT aSyn fibrils, peptides were detected that derived from the second half of the N terminus (amino acids 20 to 56), and one peptide was detected that covered a part of the NAC region up to the first half of the C terminus (amino acids 76 to 125). Only the E83Q aSyn fibrils showed a peptide covering the last part of the N-terminal region together with the first half of the NAC domain (amino acids 43 to 74) and a peptide that contained the last part of the C terminus (amino acids 114 to 140). Together, we identified peptides covering the three distinct domains of the aSyn sequence as intact fragments and unique peptides specific for either WT or E83Q aSyn, indicating differences in the accessibility of specific cleavage sites between both structures, resulting from the structural differences between the fibrils. These findings point to significant differences in the dynamics, stability, and structural properties between E83Q and WT aSyn fibrils.
E83Q, but not WT, aSyn is toxic to mammalian cells and increases the formation of puncta structures
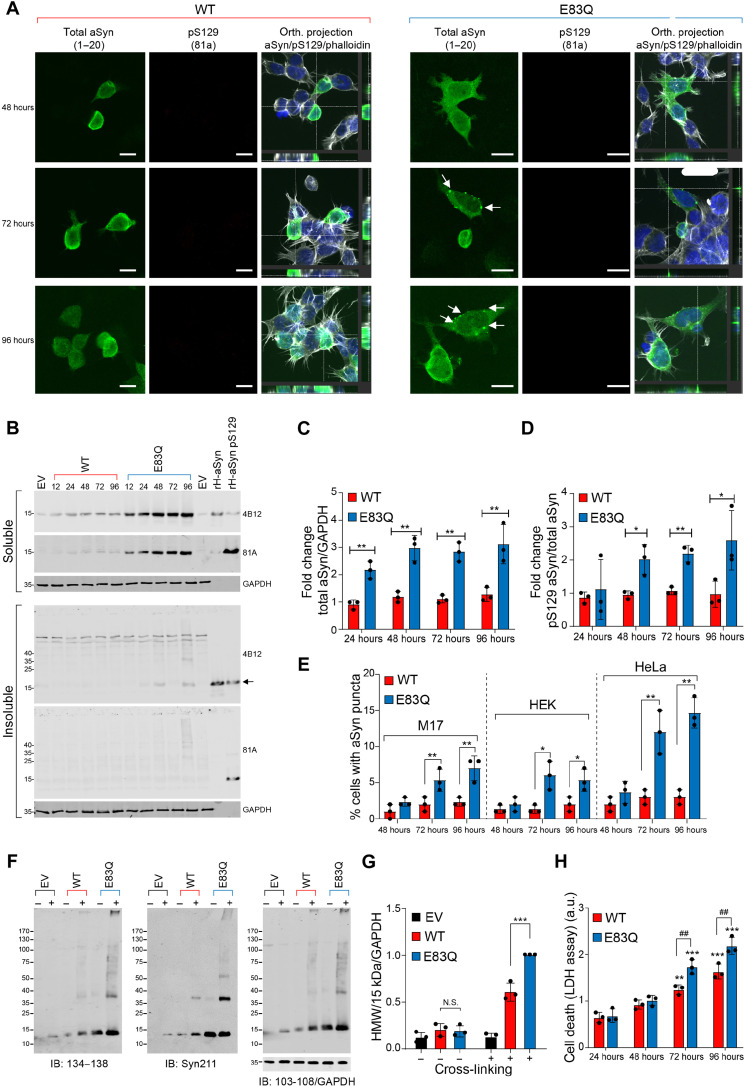
To determine whether the E83Q mutation enhances the aggregation propensity of aSyn and results in the formation of intracellular inclusions, we performed immunocytochemistry (ICC) and confocal imaging in M17, HEK293, and HeLa cells overexpressing WT or E83Q aSyn. The aggregate formation was assessed via ICC between 24 and 96 hours after transfection. Co-staining of the cells using total aSyn antibodies (1 to 20 or 134 to 138) in combination with pS129 antibodies did not reveal the presence of the typical large aSyn aggregates in any of the three mammalian cell lines overexpressing WT aSyn (Fig. 5A and fig. S10, A and B). The overexpression of E83Q aSyn was insufficient to induce spontaneous aggregation of aSyn in either neuronal-like (M17) or non–neuronal-like mammalian cell lines (HEK293 and HeLa; Fig. 5A and fig. S10, A and B). Furthermore, WB analyses showed that aSyn carrying the E83Q mutation was not found significantly enriched in the insoluble cellular fraction (Fig. 5B). Besides, small puncta were consistently observed in cells overexpressing E83Q aSyn, but not in those transfected with WT aSyn. Among the three cell lines tested, these puncta structures were preferentially formed in HeLa cells (Fig. 5B and fig. S10H). However, our ICC demonstrated that these puncta were not positive for pS129, a known marker for aSyn fibrils and pathological aggregates (Fig. 5A and fig. S10, A and B). WB analyses also confirmed the absence of a pS129 signal in the insoluble fractions of the cells overexpressing E83Q aSyn (Fig. 5B and fig. S10, C and D).
Fig. 5. Overexpression of E83Q aSyn in immortal mammalian cells does not significantly alter cellular properties.
(A) ICC of M17 cells transfected with either WT or E83Q aSyn plasmids for the indicated time. The aSyn inclusions were not detected in these cells (larger fields of view are depicted in fig. S6H). White arrows, puncta-like structures. Scale bars, 10 μM. (B) WB of soluble and insoluble fractions of M17 cells transfected with either WT or E83Q aSyn for the indicated time. Arrows indicate the low concentration of aSyn in the insoluble fractions from E83Q-expressing cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Graph showing the expression levels of total WT and E83Q aSyn from the M17 soluble fractions. (D) Graph showing the pS129 levels in WT and E83Q aSyn expressing M17 soluble fraction. (E) Graph showing the percentage of cells with WT and E83Q aSyn puncta [as indicated in (A) by white arrows] from all three mammalian cell lines. *P < 0.05, **P < 0.05, and ***P < 0.0005 (ANOVA followed by Tukey post hoc test, WT versus E83Q at each time point). (F) WB of the cross-linking [disuccinimidyl glutarate (DSG)] of HeLa cells overexpressing either an EV, WT, or E83Q aSyn. IB, immunoblot. (G) Graph showing the quantification of high–molecular weight (HMW) aSyn bands detected at and above 25 kDa to the top of the gel from (F). N.S., nonsignificant; ***P < 0.0005 [ANOVA followed by Tukey post hoc test, nontreated cells (WT versus E83Q) and treated cells with DSG (WT+ versus E83Q+)] in both cell lines. “+” cells were treated with DSG; “–” cells were treated with dimethyl sulfoxide (DMSO). (H) Graph displaying the aSyn-mediated toxicity measured by quantifying the concentration of lactate dehydrogenase (LDH) released into the HeLa cell culture media at the indicated times. *P < 0.05 [ANOVA followed by Tukey post hoc test (WT versus E83Q)]. (C to E, G, and H) Data represent means ± SD of a minimum of three independent experiments. ##P < 0.005.
On the other hand, note that aSyn was significantly up-regulated in the soluble cellular fraction of the cell lines overexpressing E83Q (Fig. 5, B to D, and fig. S10, C, D, F, and G). Together, our data show that the E83Q mutation did not promote the formation of pathological-like pS129-positive aggregates in mammalian cell lines; it did, however, increase the number of small, dot-like structures (Fig. 5E), which may represent the accumulation of multimeric/oligomeric species. This finding possibly reflects a greater propensity of E83Q to form oligomers (e.g., dimers and tetramers) intracellularly.
To test this hypothesis and determine the size distribution of the species formed in these puncta structures, we conducted size exclusion chromatography (SEC) on the cellular extracts from HEK and HeLa cells overexpressing either WT or E83Q aSyn. WB analyses of the SEC fractions confirmed that both E83Q and WT aSyn species were mostly detected as monomers (~15 kDa; fig. S11). To investigate the possibility that the E83Q oligomers could be unstable and disassociate on the column, we performed protein cross-linking (29, 30). The SEC analyses of the cellular extracts treated with disuccinimidyl glutarate (DSG) confirmed that in both cell lines overexpressing WT or E83Q, the majority of aSyn species eluted in a similar volume as the unfolded human recombinant aSyn (fig. S12, A and B). We then performed WB analyses to assess the level of oligomerization in cells. In the control cells [empty vector (EV)] treated with DSG or dimethyl sulfoxide (DMSO), the monomeric aSyn was either not observed or only weakly detected, and no oligomeric bands were detected (Fig. 5F and fig. S12C).
In the absence of the cross-linking agent, WT and E83Q aSyn overexpressed in HeLa and HEK cells both were detected as a prominent single band (~15 kDa) corresponding to the molecular weight (MW) of the aSyn monomer. However, DSG treatment of the cells overexpressing WT aSyn revealed the presence of several high–molecular weight (HMW) aSyn species, including a smear above 130 kDa, in addition to the main aSyn monomer band, as previously described (29, 30). The HMW bands and smear above 130 kDa were also detected in the soluble fraction of the cells overexpressing E83Q, with a similar MW as the WT counterparts. As the E83Q mutant is expressed at the protein level, but not at the mRNA level (fig. S10E), at higher steady-state levels than aSyn WT in HeLa cells (Fig. 5B), the level of the HMW signal was normalized to the total aSyn level expressed in cells. This showed significantly higher levels of the HMW aSyn species in cells overexpressing E83Q (Fig. 5, F and G, and fig. S12C). Although the HMW species, such as the dimers and oligomers, seem to be minor species, our data suggest increased oligomerization within cells, especially when overexpressing E83Q aSyn. Last, we assessed cell viability over time in mammalian cells overexpressing WT or E83Q aSyn using the lactate dehydrogenase (LDH) toxicity assay. Our cell death assay showed that overexpression of aSyn WT and E83Q induced a significant increase in toxicity over time, as evidenced by an increase in the loss of plasma membrane permeability from 96 hours for WT overexpressing cells and starting already at 72 hours for E83Q cells (Fig. 5H and fig. S10I). Moreover, in both M17 and HEK cells, E83Q overexpression induced higher cell death than WT aSyn at 96 hours (fig. S10I). These differences could be attributed either to the higher expression levels of E83Q in these cells or to the greater propensity of E83Q aSyn to oligomerize. The HMW bands and smear above 130 kDa were also detected in the soluble fraction of the cells overexpressing E83Q, with a similar MW as the WT counterparts. It remains unclear whether these differences could be attributed solely to the higher expression levels of E83Q in these cells or the greater propensity of E83Q aSyn to oligomerize, or both.
WT and E83Q fibrils preferentially seed the aggregation of monomers with the same sequence
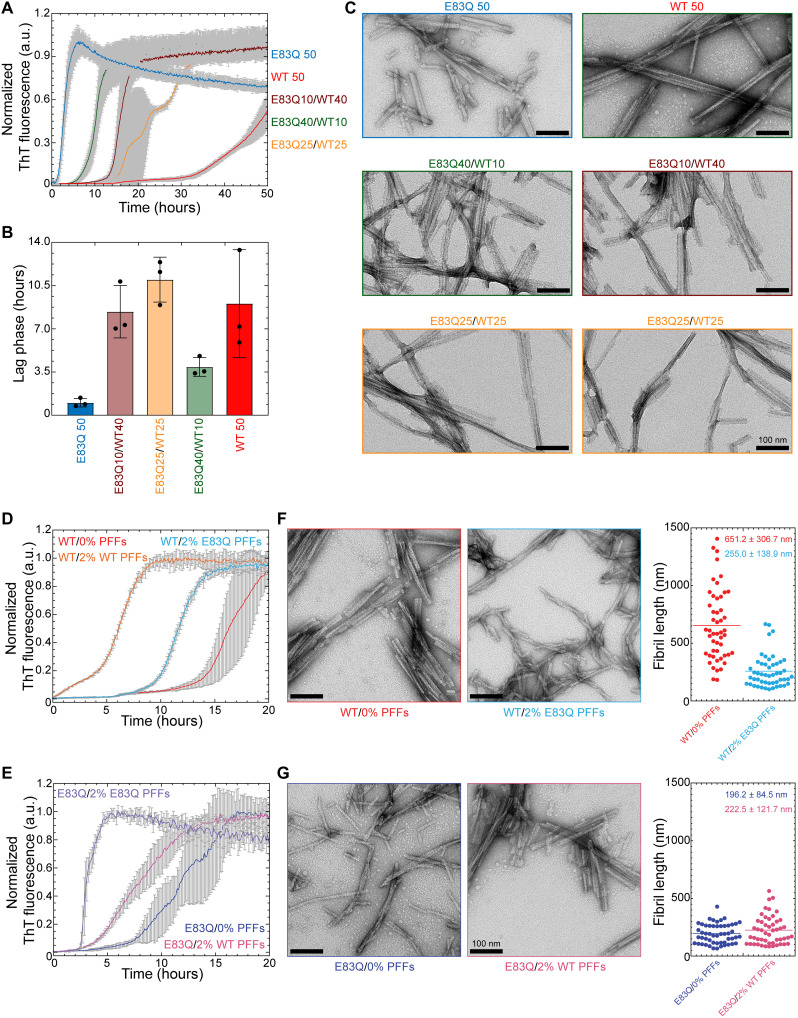
Since the E83Q mutation carrier was heterozygous, we also investigated the aggregation of the E83Q mutant in the presence of WT aSyn at varying molar ratios (4:1, 1:1, and 1:4). As shown in Fig. 6 (A and B), the presence of WT aSyn monomers resulted in a significant and concentration-dependent delay in the aggregation of E83Q monomers, as evidenced by the increase in lag time from ~0.9 hours for only E83Q samples to ~3.9, ~8.4, and ~10.9 hours for samples containing a mixture of 4:1, 1:4, and 1:1 ratio of E83Q:WT aSyn, respectively. EM analysis of the samples at the end of the aggregation process showed fibrils in all the E83Q:WT aSyn mixtures (Fig. 6C). The fibrils formed in these mixtures exhibited a higher propensity to undergo lateral association. The samples containing an equimolar concentration of WT and E83Q monomers showed a multiphasic aggregation profile (Fig. 6A), suggesting a more complex aggregation process involving a complex interplay between different species of both proteins. Despite this, only fibrils were observed at the end of the aggregation process (Fig. 6C).
Fig. 6. WT and E83Q fibrils preferentially seed the aggregation of monomers with the same sequence.
(A) ThT-based aggregation kinetics of cross-species aggregation of human WT (hWT) monomers with hE83Q monomers at varying ratios. (B) Bar diagram showing the lag phase (hours) extracted from the aggregation kinetics [from (A)]. (C) Negatively stained EM images of the fibril samples at the end time point of the ThT kinetics [from (A)]. Scale bars, 100 nm. (D and E) ThT-based aggregation kinetics of seeding of 10 μM concentration of WT monomers (D) or E83Q monomers (E) with 2% of WT PFFs or E83Q PFFs. (F and G) EM analysis of end time point [from (D) and (E)] samples of WT monomers alone and seeded with 2% E83Q PFFs (F) and E83Q monomers alone and seeded with WT PFFs (G) and their fibril length distribution as the dot plot graphs, respectively. Scale bars, 100 nm.
Next, we investigated and compared the ability of WT and E83Q fibrils to seed the aggregation of monomers of both proteins. The primary objective of this experiment was to determine whether E83Q PFFs are able to transmit their morphological and structural properties to the next generation of fibrils and to assess the specificity and efficiency of cross-seeding by WT and E83Q PFFs. As shown in Fig. 6 (D and E) and fig. S13, seeding by PFFs was always more efficient when both the PFF seeds and monomers were of the same sequence, irrespective of the starting concentration of aSyn at 10 μM (Fig. 6, D and E) or 50 μM (fig. S13, A and B). WT aSyn monomers were more efficiently seeded by WT PFFs than E83Q PFFs (Fig. 6D and fig. S13A). Similarly, the E83Q monomers were more efficiently seeded by E83Q PFFs (Fig. 6E and fig. S13B). EM analysis of fibril length in the two-seeded aggregation samples (Fig. 6, F and G, and fig. S13, C and D) showed the WT monomers seeded with E83Q PFFs resulted in fibrils with an average length similar to those observed for the aggregation of E83Q monomers alone. These observations suggest that the E83Q fibrils exhibit distinct conformations, preferentially seed E83Q monomers, and are able to pass some of their morphological and structural properties to WT aSyn monomers. In contrast, when E83Q monomers were seeded with WT PFFs, we observed fibrils predominantly with similar morphology and size distribution as those formed by E83Q monomers. These observations suggest that WT aSyn PFFs can still template E83Q monomer aggregation but do not propagate their structure and morphological properties to E83Q aSyn monomers.
Human E83Q aSyn PFFs show high seeding activity and increased toxicity in primary mouse neurons
Our in vitro biochemical and biophysical experiments established that the E83Q mutation alters the morphology, stability, and structural properties of aSyn fibrils (Figs. 2 to 4). Therefore, we sought to determine to what extent the E83Q mutation could influence the seeding activity of the preformed aSyn fibrils (biophysical characterization of the PFF preparations is shown in fig. S14) in neurons. Given that postmortem neuropathological characterization of the brain of the E83Q mutation carrier showed significantly higher LB pathology in the hippocampal and cortical areas than the substantia nigra (11), we compared the seeding activity of WT and E83Q PFFs in WT hippocampal and cortical primary neuronal cultures. Previous studies from our laboratory (31) and others (32) have shown that aSyn PFF seeding activity correlates with levels of aSyn expression in different neuron types, which explains why aSyn PFFs seed more efficiently in hippocampal neurons compared with cortical neurons, which express lower levels of aSyn. We used a neuronal seeding model, in which the addition of nanomolar concentration of PFFs primary neuronal culture triggers the formation of intracellular aggregates of endogenously expressed aSyn (fig. S15) (14, 15).
As the seeding process requires the internalization of the PFFs into the neurons, we first determined whether the conformational properties of the E83Q PFFs alter the uptake or processing of human aSyn PFFs (e.g., phosphorylation or C-terminal cleavage of PFF) (31). To follow the fate of the PFFs independently of the seeding mechanism, we used aSyn knockout (KO) neurons in which the PFFs are unable to induce seeding because of the absence of endogenous aSyn (fig. S16) (14, 31). ICC combined with confocal imaging confirmed that both human WT (hWT) and human E83Q (hE83Q) PFFs were readily internalized in the neurons via the endolysosomal pathway (lysosomal associated membrane protein 1-positive vesicles) 24 hours after their addition to the primary cultures (fig. S16A), as previously reported in mouse PFF–treated neurons (31, 33).
Next, we quantified the level of PFF internalization by quantifying the amount of internalized HMW (14, 31). The PFF levels were significantly higher in the insoluble fraction of the hE83Q PFF–treated KO neurons than in neurons treated with hWT PFFs, as indicated by the HMW band detected by the pan-synuclein antibody (SYN-1) (fig. S16, B and C). We next investigated whether the E83Q mutation alters the processing of the internalized PFFs. Within 24 hours, both hWT and hE83Q PFFs were C-terminally truncated, as evidenced by the detection of the 12-kDa band. After 3 days, both hWT and hE83Q PFFs were fully cleaved, as shown by the complete loss of full-length aSyn (15 kDa; fig. S16C). The truncated species were cleared over time, but a residual level remained up to 21 days after internalization of the PFFs into the neurons for both hWT and hE83Q PFFs (fig. S16C). Last, the internalized hWT or hE83Q PFFs were never phosphorylated at residue S129 in KO neurons (fig. S16C). Together, our results demonstrate that the conformational properties of E83Q PFFs increase their uptake into neurons without interfering with the processing and clearance once internalized.
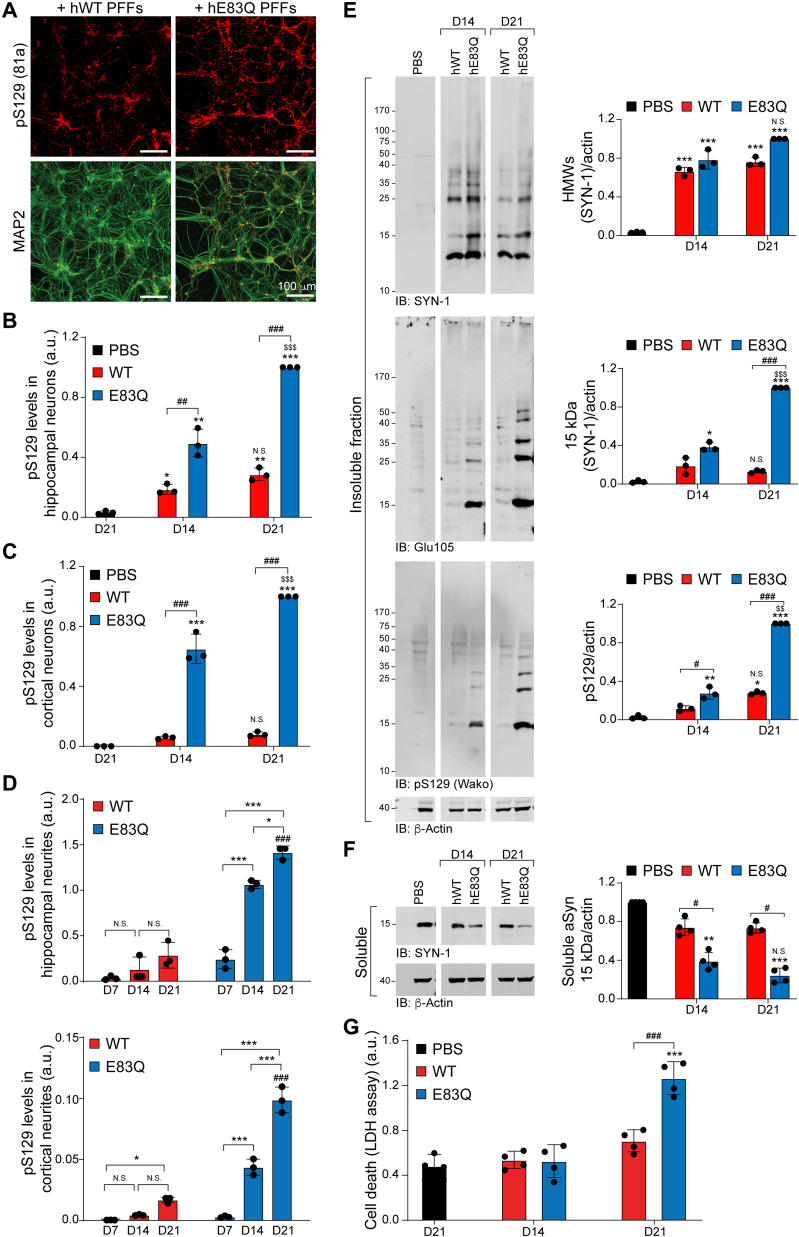
Next, we compared the seeding capacity of the hE83Q and hWT PFFs in WT murine hippocampal neurons. hE83Q or hWT PFFs were added to WT primary culture, and their seeding activity and ability to induce the formation of LB-like inclusions were investigated at day 7 (D7), D14, and D21 of treatment. As previously reported by our group (14) and others (34), the internalized PFF seeds never get phosphorylated at S129 residue at early or late time points, up to 21 days, and the great majority are cleaved at residue 114 within 12 to 24 hours (31). Therefore, they do not interfere with the detection and quantification of newly formed fibrils by pS129 antibodies. ICC confirmed the formation of aggregates immunoreactive to aSyn pS129, both in hWT PFF– or hE83Q PFF–treated neurons at D14 and D21 (Fig. 7A and fig. S17A). Quantification of the pS129 levels by high content imaging [high content analysis (HCA)] demonstrated that more pS129-seeded aggregates were formed in hippocampal neurons treated with hE83Q PFFs than in those treated with hWT PFFs, both at D14 and D21 (Fig. 7, B and D). The level of pS129-seeded aggregates had increased even further from D14 to D21 in hE83Q PFF–treated neurons but not in hWT PFF–treated neurons. Similar to the hippocampal neurons, the addition of hE83Q PFF to the cortical neurons induces the formation of pS129-seeded aggregates that accumulate over time (Fig. 7, C and D, and fig. S17, B and C). The level of pS129-seeded aggregates was also significantly higher in hE83Q PFF–treated cortical neurons than in the hWT PFF–treated cortical neurons at D14 and D21 (Fig. 7, C and D, and fig. S17, D and E). Together, our findings suggest that the E83Q mutation strongly enhances the seeding activity of human aSyn PFFs in both the hippocampal and cortical neurons.
Fig. 7. hE83Q PFFs have a higher seeding capacity than hWT PFFs in WT hippocampal and cortical primary neurons.
WT hippocampal (A, B, and D to G) or cortical (C and D) neurons were treated with 70 nM hWT or hE83Q aSyn PFFs. Control neurons were treated with PBS. (A to C) The level of seeded aggregates was measured at D14 and D21 in PBS- or PFF-treated hippocampal (A and B) or cortical (C) neurons [microtubule-associated protein 2 (MAP2) positive] by HCA. Seeded aggregates were detected by ICC using pS129 (81a) antibody; neurons were counterstained with the microtubule-associated protein (MAP2) antibody; and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 100 μm. For each independent experiment, a minimum of two wells was acquired per condition, and nine fields of view were imaged per well. (D) HCA analyses measuring the level of pS129 intensity in the neurites of the hippocampal neurons (top) and the cortical neurons (bottom). Segmentation of the neurites and quantification of the LN-like pathology were performed as previously described (14). (E and F) WB of total aSyn, pS129, and actin detected by SYN-1, Glu105 (antibody specific for the detection of mouse aSyn), pS129 Wako, or pS129 MJFR13 antibodies, respectively. In the insoluble fraction, HMW bands corresponding to the newly formed fibrils are detected from 25 kDa up to the top of the gel. (G) Cell death levels were assessed at D14 and D21 in PBS- and PFF-treated hippocampal neurons using LDH release assay. For each independent experiment, triplicate wells were measured per condition. (B to G) The graphs represent means ± SD of a minimum of three independent experiments. ANOVA followed by Tukey post hoc test was performed. *P < 0.05, **P < 0.005, and ***P < 0.0005 (PBS versus PFF). #P < 0.05 and ###P < 0.0005 (hWT versus hE83Q). $$P < 0.005 and $$$P < 0.0005 (D14 versus D21). N.S., nonsignificant (D14 versus D21).
As shown in fig. S17D and reported previously (31, 32), the aSyn PFF seeding level is significantly higher in hippocampal neurons than in cortical neurons. Therefore, we performed the rest of our studies on the biochemical and morphological properties of the newly formed aggregates in primary hippocampal neurons. We further characterized the seeding level in PFF-treated hippocampal neurons by WB analysis (Fig. 7E). Given that SYN-1 detects both the PFFs and the seeded aggregates in the insoluble fraction, the pS129 antibody was used to discriminate the newly formed aSyn aggregates from the exogenously added PFFs, as the latter undergo C-terminal cleavage and are not subjected to phosphorylation at S129 in the neuronal seeding model (14, 31, 35). Consistent with the ICC and HCA measurements, pS129-positive seeded aggregates were barely detected in the insoluble fraction of the hWT PFF–treated neurons. This confirms the low degree of seeding in hWT PFF–treated neurons, which is at the limit of the WB software detection threshold (14, 31, 35).
Conversely, we observed an accumulation of aSyn-seeded aggregates in the insoluble fraction of the hE83Q PFF–treated neurons (Fig. 7E), which were positively stained with SYN-1, which recognizes the 15 kDa and HMW bands at ~23, 37, 40, and 50 kDa, and with a specific antibody against pS129, which recognizes the 15-kDa band and the HMW bands at ~23 and 35 kDa. As measured by HCA, the level of pS129 significantly increased between D14 and D21 in hE83Q PFF–treated neurons. Conversely, pS129-positive seeded aggregates were barely detected at D14 or D21 in hWT PFF–treated neurons (Fig. 7E). The detection of the pS129-positive seeded aggregates in the insoluble fraction was concomitant with the shift of endogenous aSyn from the soluble (Fig. 7F) to the insoluble fraction in the hE83Q PFF–treated neurons. Nevertheless, and despite a trend toward a down-regulation of aSyn in the soluble fraction between D14 and D21, the difference in aSyn signal (SYN-1) was not significant (Fig. 7F).
Last, LDH toxicity assays revealed neuronal cell death at D21 in the hE83Q PFF–treated neurons but not in hWT PFF–treated neurons (Fig. 7G). Together, our findings demonstrate that the E83Q-dependent changes in the structural and dynamic properties of human aSyn PFFs translate into increased uptake, seeding activity, and neurotoxicity.
The E83Q mutation enables the capacity of human aSyn PFFs to induce the formation of LB-like inclusion in neurons
Next, we investigated and compared the shape and morphology of the pS129-positive seeded aggregates formed upon the addition of hWT or hE83Q PFF to the primary culture. It is well established that human fibrils less efficiently seed the aggregation of endogenous mouse aSyn in primary neurons (36, 37). Therefore, the formation and the evolution of aSyn-seeded aggregates in mice primary neurons have been extensively studied using mouse aSyn PFFs (mWT PFFs) (14, 15, 31–34, 36, 38–41).
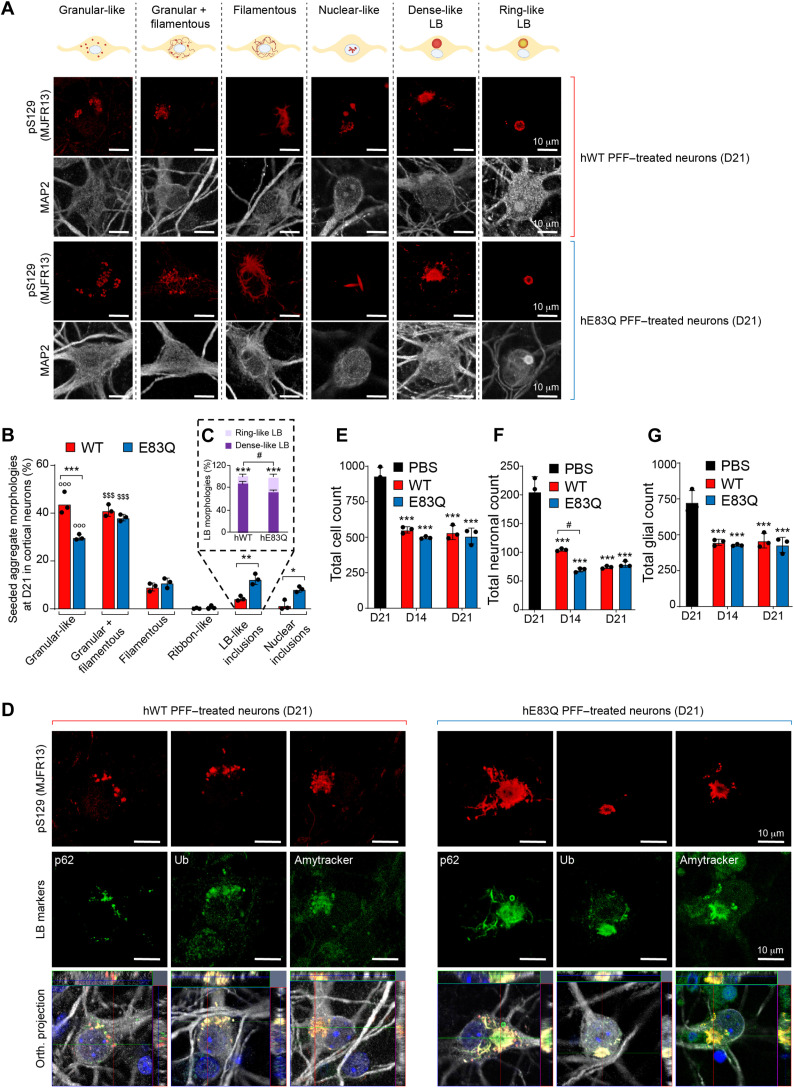
Previously, we and others have shown that the seeded aggregates, up to 14 days of formation, appeared predominantly as long filamentous-like structures that evolve into inclusions at D21, with different structural and morphological properties that we classified as filamentous (~30%), ribbon-like (~45%), or round LB-like inclusions (~22%) (14). In studies using hWT PFFs to induce seeding in mouse primary neurons, the pS129-positive seeded aggregates were mostly detected as small puncta in neurites and as long fibrils in the cell bodies (15, 35, 42). The morphology of hWT PFF–seeded aggregates has never been assessed after D14. Therefore, we compared the morphology of aSyn inclusions formed by the hWT or E83Q seeds to that induced by the addition of mWT PFFs up to D21.
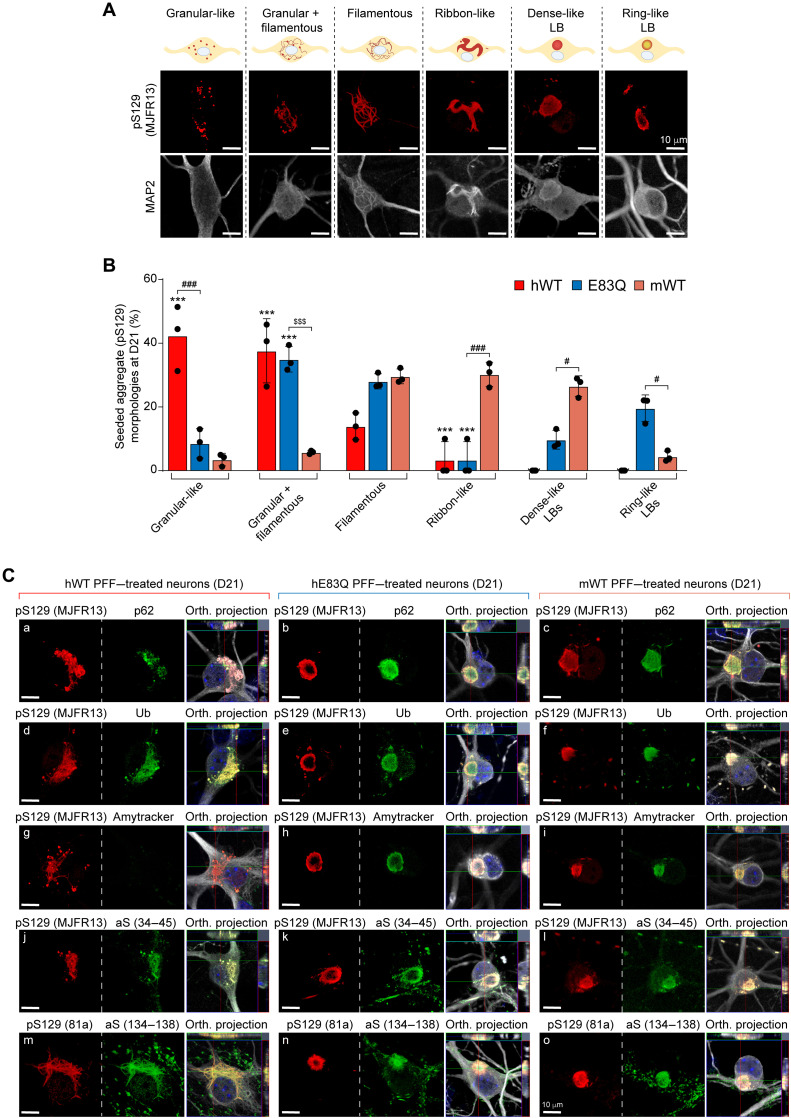
First, on the basis of pS129 staining, confocal imaging revealed that human aSyn PFF induced the formation of seeded aggregates with different types of morphologies than those detected in mWT PFF–treated neurons. The morphological classification of the different types of seeded aggregates formed in human or mouse PFF–treated neurons is depicted in Fig. 8A and fig. S18A, and their relative distribution at D14 and D21, respectively, in fig. S18B (D14) and Fig. 8B (D21).
Fig. 8. Comparative analysis of the morphological properties and diversity of aSyn aggregates and inclusions formed at 21 days after treatment of hippocampal neurons with human E83Q and WT PFFs and mouse WT PFFs.
(A) LB-like pathological diversity was observed in PFF-treated hippocampal neurons. The representative images for the filamentous and the ribbon-like inclusions shown in this panel are also depicted in figs. S13 to S16 with additional markers of the LB-like inclusions. (B) Quantification of the morphologies of the aSyn seeded aggregates observed by ICC in primary hippocampal neurons at D21. A minimum of 100 neurons was counted in each experiment (n = 3). The graphs represent means ± SD of a minimum of three independent experiments. ***P < 0.0005 (ANOVA followed by Tukey post hoc test, mWT PFF–treated neurons versus human PFF (hWT or E83Q)–treated neurons). ###P < 0.0005 (ANOVA followed by Tukey post hoc test, hE83Q PFF–treated neurons versus mWT PFF– or hWT PFF–treated neurons). $$$P < 0.0005 (ANOVA followed by Tukey post hoc test, hWT PFF–treated neurons versus hE83Q PFF–treated neurons). #P < 0.05. (C) Aggregates were detected by ICC using pS129 (MJFR13 or 81a) in combination with p62 (a to c), ubiquitin (Ub) (d to f), Amytracker dye (g to i), total N-terminal aSyn (epitope, 34 to 45) (j to l), or total C-terminal (epitope, 134 to 138) (m to o) aSyn antibodies in hWT PFFs (a, d, g, j, and m), hE83Q PFFs (b, e, h, k, and n), or mWT PFF–treated hippocampal neurons (c, f, i, l, and o). Neurons were counterstained with MAP2 antibody, and the nuclei were counterstained with DAPI. Scale bars, 10 μm.
In mWT PFF–treated neurons at D14, the seeded aggregates appeared predominantly as filamentous-like inclusions [~75%; figs. S18 and S19, A to C (a and b)] detected both in the neurites and neuronal cell bodies (14, 15). In addition to these filamentous aggregates [fig. S19, A (d) and B (d and g)], we observed the formation of aggregates that exclusively appeared either as granular-like aggregates or as a mixture of granular and filamentous aggregates at D14 [figs. S19, A (e, f, h, and i), B (f, h, and i), and C (e, h, and i); S20, A (e, h, and i) and B (f); and S21, A (e, f, h, and i) and B (e to i)] and D21 [Fig. 8B and figs. S19, D (d, e, h and i), E (d, e, h, and i), and F (e, h, and i); S20C (e, h, and i); and S21, C (d, e, h, and i) and D (d, e, h, and i)] for hWT and hE83Q PFF–treated neurons. However, these types of aggregates were not observed in mWT PFF–treated neurons at any time point.
In hWT or E83Q PFF–treated neurons, the ribbon-like structures, which are commonly detected in mouse PFF–treated neurons [~10% of the seeded aggregates at D14 and ~30% at D21 (14)] were not observed at either D14 [figs. S18, A and B; S19, B (c) and C (b); and S20A (c)] or D21 [Fig. 8B and figs. S19, D (b), E (b), and F (b), and S20C (b)]. While the LB-like inclusions were formed at a similar level in hE83Q PFF–treated as in mWT PFF–treated neurons at D14 (~10%; fig. S18, A and B) and D21 (~28%; Fig. 8B), no LB-like inclusions were ever observed in hWT PFF–treated neurons (Fig. 8B and figs. S18 to S21).
Together, our data suggest that the specific conformation adopted by the PFFs generated in vitro from aSyn carrying the E83Q mutation, as shown in our in vitro experiments, appears to interact more favorably with endogenous mouse aSyn monomers, leading to their efficient recruitment to fibrils. These de novo fibrils were able to mature into pathological LB-like inclusions in both cortical and hippocampal primary neurons. These findings show that this single E83Q mutation within the NAC region markedly alters the structure and seeding activity of human aSyn PFFs in manners that facilitate the formation of LB-like inclusions, which was not possible with WT human PFFs.
Seeded aggregates formed upon the addition of hE83Q PFFs recapitulate the biochemical and architectural properties of classical brainstem LBs in hippocampal neurons
To further characterize the newly formed aSyn aggregates, the following LB-like biochemical properties were evaluated: (i) immunoreactivity at D14 and D21 of aSyn-seeded aggregates to ubiquitin (43) and p62 (Fig. 8C and fig. S19, A, B, D, and E) (44); (ii) binding to the fluorescent Amytracker tracer dye, which binds specifically to the β sheet structure of amyloid-like protein aggregates (Fig. 8C and fig. S19, C and F) (45); and (iii) immunoreactivity of aSyn-seeded aggregates to total aSyn (N- and C-terminal antibodies) over time (Fig. 8C and fig. S20). Given that the formation of LB-like inclusions has not yet been investigated in human PFF–treated neurons, we systematically compared the properties and morphological features of the seeded aggregates formed at D14 and D21 in human PFF–treated neurons to those formed in mouse PFF–treated neurons (14).
First, all types of seeded aggregates formed in hWT and hE83Q PFFs or in mWT PFF–treated neurons were positively stained by N- and C-terminal aSyn antibodies (Fig. 8C and fig. S20) or by the ubiquitin antibody (Fig. 8C and fig. S19, B and E). While in the neuronal cell bodies of the mWT PFF–treated neurons, p62 [Fig. 8C and fig. S19, A (a to c) and D (a to c)] and Amytracker dye [Fig. 8C and fig. S19, C (a to c) and F (a to c)] detected all of the seeded aggregates, as previously reported (14), and both markers barely colocalized with the granular-like or the mixture of granular and filamentous aggregates formed after the addition of the human PFFs at D14 [fig. S19, A (e, f, h, and i) and C (e and g to i)] and D21 [Fig. 8C and fig. S19, D (e and g to i) and F (e and g to i)]. The lack of recognition of these structures by p62 and Amytracker was even more pronounced in the hWT PFF–treated neurons [Fig. 8C and fig. S19, A (g to i), C (g to i), D (g to i), and F (g to i)] than in those treated with the E83Q-mutant PFFs. This suggests that some of the pS129-positive species formed by the human PFFs (WT or E83Q) represent nonfibrillar aggregates (i.e., oligomers).
Our morphological analyses revealed a different organization of the LB-like inclusions formed in mWT or hE83Q PFF–treated neurons. While the proportion of the LB-like inclusions detected in both types of neurons was similar at D14 (~10%) and D21 (~28%; Fig. 8B and fig. S18B), a notable difference was seen in the morphology of these inclusions (fig. S18, C to E). In the mWT PFF–treated neurons, most of the LB-like inclusions appeared with a dense core (~80 to 90% at D14 and D21; fig. S18, A and C) positively stained by pS129, p62 (fig. S18C, a), ubiquitin (fig. S18C, c), and the Amytracker dye (fig. S18C, e). In rare cases, LB-like inclusions were organized with a ring-like structure [fig. S18C (b, d, and f)], as previously observed (14). Conversely, in hE83Q PFF–treated neurons, the majority of the LB-like inclusions observed at D14 (~65%; Fig. 8C and fig. S18A) or D21 (~70%; fig. S18, D and E) appeared with a ring-like organization in which the core was no longer detected by pS129 antibodies [fig. S18D (b, d, and f)] or by the Amytracker dye (fig. S18D, f). This may suggest that in these types of LB-like inclusions, aSyn fibrils are preferentially relocated to the periphery rather than the center of the inclusion.
Our findings corroborate those of a recent study that used superresolution microscopy to show the accumulation of pS129-positive aSyn at the periphery of the nigral LB (46). Such organization, with a highly dense and organized shell of phosphorylated aSyn at the periphery of the inclusions, also resembles the brainstem LB inclusions observed in human brain tissues from patients with PD at stages 3 to 5 of disease progression (16, 17, 19–21).
As the diversity of human PD pathology extends beyond the classical halo-like LBs or the diffuse cortical LBs, we also assessed the level of LN-like pathology in the PFF-treated hippocampal and cortical neurons. HCA-based quantification demonstrates that between D14 and D21, the level of the pS129 seeded aggregates significantly increases in both the neurites and the cell bodies of the hE83Q PFF–treated neurons (Fig. 7D and fig. S22). In contrast, in the mPFF-treated hippocampal neurons, the number of neuronal cell bodies containing pS129-positive aggregates reaches its maximum at D14 (fig. S22C), suggesting that in these neurons, the main changes observed between D14 and D21 are associated mainly with their structural reorganization and conversion into LB-like inclusions (Fig. 8 and fig. S18).
Last, the p62 (fig. S18D, b) and ubiquitin (fig. S18D, d) antibodies recognized not only the periphery of the inclusions but also the center. Cytoskeletal proteins such as microtubule-associated protein 2 [MAP2; fig. S18D (b, d, and f)], neurofilaments (NFLs; fig. S21C, f), and the mitochondrial marker Tom20 were also detected both at the periphery and in the center of these inclusions (fig. S21D, f). Our findings align with those of previous studies showing the recruitment and the differential distribution of cytoskeletal proteins and organelles in human LB inclusions from the substantia nigra (17, 47–50), the hippocampal Cornu Ammonis (CA2) region (48, 51), or the stellate ganglion (48).
Note that the proteome of purified LBs or LB-enriched preparations from human brain tissues (52–54) showed that 65 to 80% of the proteins overlap with the proteome of the LB-like inclusions that form in our neuronal model (14, 31). Together, our findings demonstrate that the PFFs generated from the E83Q mutant induced the formation and maturation of LB-like inclusions in hippocampal neurons that appear to recapitulate some of the morphological, biochemical, and architectural features of LBs found in the brain of patients with PD and related synucleinopathies.
The E83Q mutation induced the formation of seeded aggregates with distinct morphology in cortical neurons
Last, we investigated and compared the seeding activity of WT and E83Q PFFs in cortical neurons. As shown in Fig. 9, both WT and E83Q human aSyn PFFs induced the formation of pS129-positive aggregates of different morphologies (Fig. 9, A and B), similar to those observed in the hippocampal neurons (Fig. 8, A and B).
Fig. 9. Morphological properties and diversity of aSyn aggregates and inclusions formed in hWT or E83Q PFF–treated cortical neurons.
(A to C) Quantification of the morphologies of the aSyn seeded aggregates at D21 (see also fig. S17). A minimum of 100 neurons were counted in each experiment. (C) Quantification of the LB-like inclusions (dense core versus ring-like structure) at D21. A minimum of 50 neurons were counted in each experiment. (D) ICC was performed using MAP2 (neurons) and pS129 (MJFR13) with p62, ubiquitin antibodies, or the Amytracker dye. The nuclei were stained with DAPI. The dense core LB-like inclusions were stained by pS129, p62, ubiquitin, and the Amytracker dye, while those with a ring-like organization were stained by p62 and ubiquitin, but not by pS129 or Amytracker. Scale bars, 10 μm. (E to G) Cell death quantification at D14 and D21. ICC was performed using NeuN antibody to stain the neuronal nuclei and DAPI to label the nuclei of all cells in the primary culture (fig. S18). For each experiment, three fields of view (FOVs) per condition were acquired (~400 to 1000 cells per FOV). The total cell population count (E) was based on the number of DAPI-positive nuclei and the count of neurons (F) on the number of NeuN-positive nuclei. Cells were counted as glial (G) if they were DAPI positive and NeuN negative. (B to G) The graphs represent means ± SD of a minimum of three independent experiments. ANOVA followed by Tukey post hoc test was performed. (B) ***P < 0.0005, **P < 0.005, and *P < 0.05 (hWT versus E83Q). °°°P < 0.0005 (granular-like aggregates versus other types of seeded aggregates). $$$P < 0.0005 (granular and filamentous-like aggregates versus other types of seeded aggregates). (C) ***P < 0.0005 (dense core versus ring-like). #P < 0.05 (hWT versus E83Q). (E to G) ***P < 0.0005 (PBS versus hWT or E83Q). #P < 0.05 (hWT versus E83Q).
The newly formed aggregates in hWT or E83Q PFF–treated cortical neurons appeared predominantly as granular-like aggregates (~45% hWT; ~30% E83Q) or as a mixture of granular and filamentous-like aggregates (~41% hWT; ~35% E83Q) that accumulate in the neuronal cell bodies (Fig. 9, A and B, and fig. S23). As for the filamentous-like aggregates (~10% hWT; ~10% E83Q), they were found both in the neurites and the neuronal cell bodies (Fig. 9, A and B, and fig. S23).
While no LB-like inclusions were ever observed in hippocampal neurons treated with the hWT PFF, approximately 5% of newly formed aggregates in cortical neurons were detected as LB-like inclusions (Fig. 9, A and B, and fig. S23). The proportion of LB inclusions was significantly higher in cortical neurons treated with E83Q PFF (~15%). Moreover, at D21, the majority of the LB-like inclusions in hWT PFF–treated (~85%) and E83Q PFF–treated (~70%) neurons exhibited a dense core (Fig. 9C and fig. S23). In addition, a significantly higher number of LB inclusions with a ring-like organization were detected in cortical neurons treated with E83Q (~30%) than with hWT (~15%) PFFs (Fig. 9 and fig. S23).
All types of seeded aggregates formed in hWT and hE83Q PFF–treated cortical neurons were positively stained by the ubiquitin antibody and the Amytracker dye (Fig. 9D and fig. S23). However, p62 antibody detected all of the seeded aggregates but not the granular-like aggregates formed in cortical neurons. This is in line with our findings in the hippocampal neurons [Fig. 8C and fig. S19, A (g to i), D (g to i), and F (g to i)], suggesting that some of the pS129-positive species formed by the human PFFs (WT or E83Q) may represent nonfibrillar aggregated forms of aSyn. Furthermore, the LB-like inclusions formed in the cortical neurons upon hWT PFF treatment were also positive for all the LB markers. This confirms that these inclusions (Fig. 9 and fig. S23) shared the same properties as those formed in the E83Q PFF–treated cortical (Fig. 9 and fig. S23) or hippocampal neurons (Fig. 8 and figs. S18 to S21).
We observed distinct patterns of seeding activity in cortical versus hippocampal neurons. First, the extent of seeding and LB-like inclusion formation after the addition of the E83Q PFF was significantly lower in the cortical neurons (~15%; Fig. 9B) than in the hippocampal neurons (~30%; Fig. 8B). These differences might be due to the reduced aSyn levels and seeding activity of the aSyn PFFs in cortical neurons (fig. S17D) (31, 32). Second, although hE83Q PFFs induced the formation of predominantly ring-like inclusions (~70%; Fig. 8 and fig. S18, C to E) in hippocampal neurons, the dense LB–like inclusions were dominant in cortical neurons (~70%; Fig. 9C). Last, both WT and E83Q PFFs induced the formation of nuclear pS129-positive aSyn aggregates in cortical neurons, with more nuclear aggregates detected in E83Q-PFF cortical neurons (~8%) compared with WT PFF–treated cortical neurons (~2%; Fig. 9, A and B, and fig. S23). These nuclear aggregates were not observed in the PFF-treated hippocampal neurons (Fig. 8 and figs. S18 to S21).
Last, we observed that compared to the control primary cortical cultures treated with PBS, the total number of cells [4′,6-diamidino-2-phenylindole–positive (DAPI+) cells] was significantly reduced by ~40% in those treated with hWT or E83Q PFF (Fig. 9E). At D14, both the neuronal (NeuN+ cells) and the glial (NeuN− cells) populations were markedly affected, with ~40% of cellular loss in the primary cortical cultures treated with hWT PFF (Fig. 9, F and G). Neuronal cell loss was even significantly higher (~60%) in the E83Q PFF–treated primary cortical neurons (Fig. 9F). No further increase in cell death was quantified between D14 and D21 in the glial or neuronal cell population (Fig. 9, E to G). Furthermore, although the cortical neurons bearing LB-like inclusions and especially those with a ring-like organization showed, by confocal imaging, signs of advanced cell death [i.e., cell bodies swelling and cytoplasm vacuolization; fig. S23, A (f), B (f), C (b, c, and f), D (b and c), and E (b to d)]; barely any DNA fragmentation was identified in these neurons by the TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) cell death assay (TUNEL+NeuN+ and pS129+ neurons; fig. S24). This suggests that the neurodegeneration associated with the LB formation and/or maturation in cortical neurons might involve alternative nonapoptotic cell death processes. Together, these findings suggest that the differences in aSyn levels and cellular environment are key determinants of LB formation and maturation.
DISCUSSION
The E83Q mutation markedly accelerates aSyn aggregation and forms fibrils of distinct morphology, stability, and conformation
Given that the NAC domain is essential for the aggregation of aSyn and that the E83Q mutation results in a reduction in charge within an already highly hydrophobic domain, it is expected that this mutant exhibited a ~10-fold faster aggregation than WT aSyn (over the concentration range of 10 to 50 μM; Fig. 2, A and B). Furthermore, previous studies have shown that substitution of E83 by alanine enhances aSyn aggregation (55) and interferes with the ability of dopamine to inhibit aSyn aggregation in vitro (56). A comparison of the lag phase of E46K (~40 hours at 7.5 μM to ~15 hours at 100 μM) (57, 58) and H50Q aggregation kinetics (~20 hours at 50 μM to ~25 hours at 70 μM) (59, 60) with E83Q lag phase (~3 hours at 10 μM to ~0.9 hours at 50 μM) suggests that the E83Q has the highest aggregation propensity among all the known aSyn mutants.
Our comparison of the biophysical properties of the WT and E83Q fibrils shows that the E83Q mutant forms fibrils with different core structures and dynamic properties. This is reflected in our findings, where the E83Q protein formed shorter fibrils (average length, ~180 nm) that exhibit increased stability, slower monomer release, and distinct morphological, structural, and ProK resistance properties compared with WT fibrils. Furthermore, our ssNMR comparative studies provided residue-specific insights suggesting major differences in the molecular structures of fibrils formed by hE83Q and WT aSyn (Fig. 4, A and B). These observations are consistent with recent cryo-EM studies demonstrating that other familial mutants [H50Q (60), A53T (61), and E46K (58)] form fibril structures/folds that differ from the various polymorphs observed for WT aSyn (62, 63). In all the cryo-EM structures of WT aSyn fibrils, the E83 residue is exposed on the fibril surface, except for polymorphs 2a and 2b (fig. S1C). In most fibril structures, the E83 residue is not engaged in interfilament interactions, suggesting that the acidic group of glutamic acid interacts with the solvent. However, in the case of the E83Q mutation, removing the negative charge from E83 due to the glutamine substitution may disrupt the long-range hydrophilic interactions with the solvent along the fibril axis.
It was recently reported that hE46K fibrils had the cross-seeding ability that templated the fibrillation of hWT monomers to form fibrils that inherited the structural and pathological features of the hE46K fibril strain (64). Here, we also showed that E83Q PFFs have the ability to pass their morphological properties to hWT monomers, although they showed preferential seeding of E83Q monomers.
The E83Q mutation promotes the accumulation and oligomerization of aSyn
Despite the fact that we did not see fibril-like inclusions or LB-like pathology in mammalian cell lines, our data showed that overexpression of E83Q aSyn induced the formation of small, dot-like aggregates in a fraction of the transfected cells (Fig. 5, A to E, and fig. S10). Using the cross-linking approach, we established that the formation of these puncta structures correlates with the accumulation of multimeric/oligomeric aSyn species in the soluble fractions (Fig. 5, F and G) and increased toxicity (Fig. 5H and fig. S10). These findings demonstrate that the E83Q mutation promotes the formation of aSyn oligomers. However, the fact that the E83Q mutant is expressed at higher levels than WT aSyn makes it difficult to determine whether the increased oligomerization is simply due to the differences in expression levels or the inherent aggregation properties of the E83 mutant.
As observed with many of the aSyn PD-linked mutations (E46K, H50Q, and A53T) (65–71), the overexpression of E83Q in mammalian cell lines (HEK, HeLa, or M17 neuroblastoma cells) was not sufficient to induce the formation of pS129-positive aggregates or inclusions that share the morphological and biochemical features of bona fide LBs (Fig. 5, A and D, and fig. S10). The lack of a simple correlation between the in vitro aggregation propensities of aSyn PD-linked mutations (e.g., E83Q aSyn; Fig. 2) and the extent of LB pathology formation in the brain or inclusion formation in cells and neurons suggests that additional cellular factors or stressors play important roles in regulating the intrinsic aggregation propensity of aSyn in cells and LB pathology in neurons. These findings also suggest that the different synucleinopathy-related mutations may exert their actions via distinct mechanisms.
The E83Q-mutant aSyn PFFs induce the formation of LB-like inclusions in primary neurons
Several studies have consistently shown that WT human aSyn PFFs exhibit significantly reduced seeding activity than WT mouse aSyn PFFs in primary neurons. Previous studies have also demonstrated the existence of a species barrier that renders human aSyn fibrils less efficient at seeding endogenous aSyn in primary mouse neurons (36, 37) and rodents (36). This explains why the vast majority of neuronal seeding models are based on the use of mouse aSyn PFFs (figs. S25 and S26). Note that the primary structure of mouse and human aSyn sequences differ by seven amino acids. Here, we showed that in both hippocampal and cortical neurons, the E83Q human aSyn PFFs not only exhibit a much higher seeding activity than WT human aSyn PFFs at D14 and D21 (Fig. 7, B and C, and fig. S17, D and E) but also form even higher levels of pS129 pathologies than mouse aSyn PFFs at D21 (figs. S17, D and E, and S26). In our hands, the differences in seeding are not due to differences in the internalization pathway or processing of the fibrils (fig. S16).
Although human aSyn PFFs have been shown to induce the formation of filamentous aggregates in cells, there are no reports in the literature to date demonstrating the induction of LB-like inclusions by human aSyn PFFs in primary neurons. Here, we showed that in primary hippocampal neurons, hE83Q PFFs, but not hWT PFFs, induce the de novo formation of fibrils that can mature into pathological LB-like inclusions as observed in neurons treated with mouse PFFs. These observations suggest that the E83Q mutation induces the formation of fibrils with conformational properties that render them more efficient at recruiting and seeding the aggregation of endogenous mouse aSyn monomers, leading to the accelerated formation of LB-like inclusions in the hippocampal neurons. Conversely, in cortical neurons, the formation and maturation of LB-like inclusions not only were limited to the neurons treated with hE83Q PFFs but also occurred, albeit at a much lower level, in neurons treated with hWT PFFs. Consistent with previous findings, in cortical neurons, both endogenous aSyn and seeding activity levels are much lower than in hippocampal neurons. Therefore, our data suggest that intrinsic cellular properties, other than the endogenous level of aSyn or its misfolded properties, contribute to the cellular vulnerability to LB pathology formation and maturation, as also suggested in human synucleinopathies (72, 73). Together, our findings demonstrate that in the primary neuronal seeding model, human PFFs generated from E83Q aSyn could (i) overcome the reported species barrier (36, 37) and (ii) enhance the capacity of the human PFFs to trigger the de novo formation of pS129-positive fibrils capable of converting into LB-like inclusions.
The E83Q mutation induced the formation of LB inclusions that resemble many of the features of some bona fide LB pathologies
To further assess the effect of the E83Q mutation on the morphological diversity and biochemical composition of the LB-like inclusions, we assessed their immunoreactivity to well-known LB markers. In both cortical and hippocampal neurons, the LB-like inclusions formed upon hE83Q PFF treatment were positively stained for the classical markers used to define bona fide LBs in human pathology, including pS129 aSyn, p62, ubiquitin, and the amyloid tracer dye (Amytracker). In addition, other LB markers (17, 46, 50), such as cytoskeletal proteins (e.g., MAP2 and NFL) and mitochondria (e.g., Tom20), were also found to colocalize with the LB-like inclusions formed in hE83Q PFF–treated neurons. Notably, the distribution of these markers within the LB-like inclusions greatly resembled their distribution in LBs in the brain tissues of patients with PD (44, 46–48, 50, 74, 75).
The addition of E83Q PFFs induced the formation and maturation of seeded aggregates with distinct morphological and organizational features (Figs. 7 to 9 and figs. S18 to S23) in cortical and hippocampal neurons. Morphological classification based on pS129 staining revealed that at D21, the E83Q PFF–seeded neurons were populated either by filamentous- and/or granular-like inclusions (~70% in hippocampal neurons; ~85% in cortical neurons) or by LB-like inclusions (~30% in hippocampal neurons; ~15% in cortical neurons). In both cortical and hippocampal E83Q PFF–treated neurons, we were able to detect LB-like inclusions with the characteristic ring-like appearance of brainstem/nigral LBs (16, 19–21, 44, 46, 76) in addition to the LB-like inclusion with a dense core resembling cortical LBs (16, 19, 74, 77, 78). The relative distribution of the ring- and dense core–like inclusions depended on the type of neurons, with the ring-like inclusions mainly detected in hippocampal neurons (~70%), and the dense core–like inclusions in cortical neurons (~80%). In comparison, the LB-like inclusions observed in mouse PFF–treated neurons presented mostly with a dense core (~80 to 90% at D14 and D21; Fig. 8 and fig. S18, C and D), and only a few presented with a ring-like organization. This suggests that the remodeling of the newly seeded fibrils into LB-like inclusions of distinct morphologies is driven not only by the structure of the PFF seeds but also by neuronal cell type–dependent properties. These findings are consistent with the immunohistochemical-based studies that revealed that the morphology of the LBs varies according to their location in human brain tissues from PD and synucleinopathies (17, 44, 50, 72, 73). Last, at D21, the level of LN-like pathology detected in E83Q PFF–treated neurons was higher than that in the mouse PFF–treated neurons (Fig. 7, A and D, and fig. S22). This demonstrates that the use of PFFs generated from aSyn carrying the E83Q mutation in the neuronal seeding model allows the reproduction of some of the aSyn pathological diversity that may be relevant to the aSyn pathological heterogeneity observed in PD and synucleinopathies (17), including the formation of the halo-like LBs, the diffuse cortical LBs, and the LN-like aggregates in the neuritic extensions.
Recently, the organizational complexity of ring-like brainstem LBs was further characterized via microscopy (46, 76). Using aSyn antibodies specifically against pS129, full-length aSyn, or C-terminal truncated aSyn, these studies revealed a distribution of concentric layers of aSyn species in LBs, with pS129 detected mainly at the periphery. In contrast, the C-terminal truncated aSyn species accumulated in the center core of the inclusions. The NFL cytoskeletal proteins and mitochondria, on the other hand, accumulated at the outer shell (46). To determine to what extent the E83Q inclusions resemble the polymorphism of LB pathology in human brains, we examined the distribution of aSyn species and various known components of LBs using ICC (Fig. 8 and figs. S18 to S21 and S27). The LB-like inclusions formed in the hE83Q or mWT PFF–treated neurons as dense core inclusions showed a homogeneous and even distribution pattern of the pS129 aSyn, p62, ubiquitin, and the Amytracker signals across the inclusion as previously reported for the cortical LB (74, 77, 78). Conversely, in the LB-like inclusions with a ring-like appearance, pS129 aSyn (Fig. 8 and fig. S18 to S21) and the Amytracker signal are preferentially localized at the periphery of these inclusions (fig. S27). In contrast, p62 and ubiquitin were detected at both the periphery and the core of the ring-like inclusions as NFL cytoskeletal proteins and mitochondria (fig. S27). Such organization resembles the brainstem LB inclusions observed in human brain tissues from patients with stage 3 to 5 PD (16, 17, 20, 21, 44, 46, 76). These findings further illustrate the power of using the neuronal seeding model as a platform to investigate the sequence (e.g., PD-linked mutations) and molecular and cellular determinants of LB formation and to screen for modifiers of aSyn pathology formation and toxicity.
In this study, we investigated the effect of the first NAC domain DLB-linked mutation on the structure of aSyn (monomers and fibrils), its aggregation (oligomerization and fibril formation) in vitro and in cells, and fibril seeding activity in primary neurons. Our results show that the E83Q mutation (i) perturbs the conformational ensembles of free aSyn without impairing its ability to bind membranes in vitro and intracellularly, (ii) markedly accelerates the aggregation kinetics of aSyn in vitro, and (iii) alters the dynamics and core structure of aSyn fibrils. The E83Q fibrils are shorter in length, more stable, and exhibit a ProK digestion profile distinct from that of WT aSyn. In the cell-based overexpression models, the E83Q mutation did not affect the cellular distribution of aSyn, but it enhanced aSyn oligomerization and promoted the formation of pS129-negative puncta structures. Last, studies in our neuronal seeding models showed that (i) the E83Q mutation markedly increases the seeding activity of human PFFs and (ii) the E83Q mutation can restore the capacity of PFFs to induce the formation of LB-like inclusions in mouse hippocampal neurons. Most notable is the fact that the E83Q fibrils induced the formation of aSyn aggregate structures with a spectrum of morphologies similar to some of the Lewy pathologies observed in human PD brains, including the classical brainstem LB structures. In both cellular models, overexpression of E83Q or seeding with E83Q PFFs was associated with neurodegeneration and apoptosis. Together, our findings suggest that (E83Q) unlocks the pathogenicity of human aSyn fibrils and recapitulates some of its pathological diversity by promoting its oligomerization, increasing its toxicity, and inducing the formation of more pathogenic fibrillar structures. Therefore, exploring the effects of this mutation in various models of synucleinopathies could yield novel insights into the molecular mechanisms underpinning aSyn-induced neurodegeneration and aSyn pathology formation and spreading within the brain.
MATERIALS AND METHODS
Recombinant overexpression and purification of hWT and hE83Q aSyn
Recombinant overexpression and purification of hWT and hE83Q aSyn were performed as described previously (26). pT7-7 plasmids encoding hWT aSyn were used for transformation in BL21(DE3) E. coli cells on an ampicillin agar plate. A single colony was transferred to 200 ml of Luria broth (LB) medium containing ampicillin (100 μg/ml; small-scale culture; AppliChem, A0839) and incubated overnight at 180 rpm at 37°C. The following day, the preculture was used to inoculate 6 liters of LB medium containing ampicillin (100 μg/ml; large-scale culture) with the starting absorbance at 600 nm of big culture between 0.05 and 0.1. Upon the absorbance at 600 nm approaching 0.4 to 0.6, aSyn protein expression was induced by the addition of 1 mM 1-thio-β-d-galactopyranoside (AppliChem, A1008), and the cells were further incubated at 180 rpm at 37°C for 4 to 5 hours. Cells were harvested by centrifugation at 4000 rpm using JLA-8.1000 rotor (Beckman Coulter, Bear, CA) for 30 min at 5°C. The harvested pellets were stored at −20°C until use. Cell lysis was performed by dissolving the bacterial pellet in 100 ml of buffer A (40 mM tris-HCl, pH 7.5) containing protease inhibitors [1 mM EDTA (Sigma-Aldrich, catalog no. 11873580001) and 1 mM phenylmethylsulfonyl fluoride (PMSF; AppliChem, A0999)], followed by ultrasonication (Vibra-Cell VCX 130, Sonics, Newtown, CT) at the following parameters: 8 min; cycle: 30 s on, 30 s off; amplitude 70%. After lysis, centrifugation at 12,000 rpm at 4°C for 30 min was performed to collect the supernatant. This supernatant was collected in 50-ml Falcon tubes and placed in boiling water (100°C) for ~15 min. This solution was subjected to another round of centrifugation at 12,000 rpm at 4°C for 30 min. The supernatant obtained at this step was filtered through 0.45-μm filters and injected into a sample loop connected to HiPrep Q Fast Flow 16/10 (Sigma-Aldrich, GE28-936-543). The supernatant was injected at 2 ml/min and eluted using buffer B (40 mM tris-HCl, 1 M NaCl, pH 7.5) from gradient 0 to 70% at 3 ml/min. All fractions were analyzed by SDS-PAGE, and the fractions containing pure aSyn were pooled, flash frozen, and stored at −20°C until use. Fractions containing the purified aSyn were loaded on a reverse-phase HPLC (high-performance liquid chromatography) C4 column [PROTO 300 C4 10 μm, Higgins Analytical; buffer A, 0.1% trifluoroacetic acid (TFA) in water; buffer B, 0.1% TFA in acetonitrile], and the protein was eluted using a gradient from 35 to 45% buffer B over 40 min (15 ml/min). The purity of the elution from HPLC was analyzed by ultraperformance liquid chromatography (UPLC) and ESI-MS. Fractions containing highly purified aSyn were pooled, snap frozen, and lyophilized.
Preparation of WT and E83Q aSyn monomers
The lyophilized aSyn of the required quantity was resuspended in PBS buffer and followed by the filtration protocol as described before (26). Briefly, the lyophilized aSyn following resuspension was adjusted to pH ~7.2 to 7.4 and filtered using 100-kDa spin filters. The filtrate containing monomers was estimated for the concentration by measuring the absorbance at 280 nm by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and using an extinction coefficient of 5960 M−1 cm−1 predicted using the sequence (ProtParam, ExPASy). Monomeric aSyn (both WT and E83Q) was prepared as described above and followed any kind of further experiments.
Preparation of WT and E83Q aSyn PFFs seeds for cell studies
WT and E83Q aSyn fibrils were prepared by dissolving 4 mg of lyophilized recombinant aSyn in 600 μl of 1× PBS, and the pH was adjusted to ~7.2 to 7.4. The solution was filtered through 0.2-μm filters (Merck, SLGP033RS), and the filtrate was transferred to black screw cap tubes. This tube was incubated in a shaking incubator at 37°C and 1000 rpm for 5 days. After 5 days, the formation of fibrils was assessed by TEM and Coomassie staining as described previously (26). For the preparation of sonicated seeds, WT and E83Q aSyn fibrils were subjected to sonication for 20 s, at 20% amplitude, 1-s pulse on, and 1-s pulse off (Sonic Vibra-Cell, Blanc-Labo, Switzerland) only in mentioned experiments. The amount of monomers and oligomers released from sonicated fibrils was quantified by filtration following the published protocol (26). The structure of the fibrils was monitored by TEM and the number of released monomers and oligomers after sonication by Coomassie staining analysis.
ThT fluorescence–based aggregation kinetics
For the setup of ThT aggregation kinetics, a 400-μl master mix solution (3× 110 μl) containing five different final concentrations of WT or E83Q aSyn monomers (10, 20, 30, 40, and 50 μM) was prepared in 1× PBS (Gibco), respectively, to which a final ThT concentration of 20 μM was added. For the comparison of aggregation kinetics of 13C,15N WT and 13C,15N E83Q, a 400-μl master mix solution (3× 110 μl) containing three different final concentrations of 13C,15N WT or 13C,15N E83Q aSyn monomers (10, 30, and 50 μM) and 20 μM ThT in 1× PBS (Gibco) was prepared.
For the assessment of aggregation of copresence of the mixture of WT and E83Q monomers, a 400-μl master mix solution (3× 110 μl) encompassing a ratio of WT:E83Q monomer concentration (40:10, 25:25, and 10:40 μM) was prepared in 1× PBS (Gibco), respectively, to which a final ThT concentration of 20 μM was added.
For the cross-seeding aggregation experiments, 2% WT or E83Q PFFs (w/v, relative to the monomer concentration) was added to the 400-μl master mix solution containing the final concentration (50 μM) of WT or E83Q aSyn monomers in 1× PBS, respectively, to which a final ThT concentration of 20 μM was added.
For all the ThT-based aggregation assays, 110 μl of replicates from each master mix solution was added to the three individual wells in a black 96-well optimal bottom plate (Costar), with each well preadded with six SiLibeads ceramic beads (Sigmund Lindner) with a diameter of 1.0 to 1.2 mm. The plate was sealed with Corning microplate tape and transferred to a FLUOstar OPTIMA plate reader (BMG Labtech). The parameters used were as follows: continuous orbital shaking at 600 rpm at 37°C; ThT fluorescence was monitored every 300 s using excitation at 450 ± 10 nm and emission at 480 ± 10 nm. At the end point of the assays, the samples were analyzed by EM.
Solubility assay
As described for the ThT aggregation kinetics experiments, an 800-μl master mix solution (5× 150 μl) containing three different final concentrations of WT or E83Q aSyn (10, 30, and 50 μM) was prepared, respectively. Three individual samples for each concentration were set as replicates. A volume of 150 μl each from the master mix solution was added to the four individual wells, respectively, onto a black 96-well optimal bottom plate (Costar), with each well preadded with six SiLibeads ceramic beads (Sigmund Lindner) with a diameter of 1.0 to 1.2 mm. The plate was sealed with Corning microplate tape and transferred to a FLUOstar OPTIMA plate reader (BMG Labtech). Aggregation was induced on the 96-well plate by using the parameter used for ThT aggregation kinetics such as continuous orbital shaking at 600 rpm at 37°C. As shown in Fig. 2 (D and E), at every time point (0, 3, 6, 18, 24, 30, 42, 48, 66, and 90 hours), nearly 70 μl of sample was taken from one well of the plate after pausing the plate reader. Of the 70 μl drawn, 25 μl was used for the EM/AFM imaging analysis. Forty microliters from the remaining was centrifuged at 100,000g for 30 min at 4°C. After carefully removing the supernatant, one part of it was used for the UPLC analysis and another part was used for the SDS-PAGE analysis.
Oligomer quantification at different aggregation time points
For the oligomer quantification assay at the different time points of the aggregation, the centrifugation-filtration protocol was used (26). Briefly, a reaction mix volume of 30 and 50 μM WT and E83Q aSyn monomers in PBS, respectively, was kept at 37°C under 600-rpm shaking conditions. At different time points (0, 1.5, 3.0, 4.5, 6.0, 18, 24, and 36 hours), a 60-μl aliquot of sample was taken for each condition and ultracentrifuged at 100,000g for 30 min at 4°C to remove the sedimentable pellets. The remaining supernatant was collected and added to 100-kDa spin filters (MRCF0R100) and centrifuged at 15200g for 10 min at 4°C. The filtrate containing monomers was collected and flash frozen immediately for later concentration estimation. For the retrieval of oligomers, 60 μl of PBS was added to the same spin filter, pipetted up-down four to five times, inversed, and spun at 400g for 3 min at 4°C. Five microliters of the retrieved oligomer samples was used for EM grid preparation immediately after collection at every time point. The remaining oligomeric samples were flash frozen immediately. The concentration of monomers and oligomers was estimated using bicinchoninic acid (BCA) assay after thawing back the flash-frozen samples collected at different time points. For the BCA assay, microplate measurements were carried out using BCA protein assay reagents (Pierce, catalog no. 23227). Briefly, 15-μl triplicates of known concentrations (from 10 to 1000 μg/ml) of bovine serum albumin (standard) in PBS and an equal volume of retrieved aSyn monomers and oligomers from the different time points were pipetted into microplate wells, to which 200 μl of BCA working reagent was added and incubated at 37°C for 60 min. Absorbance at 562 nm was measured using a FLUOstar OPTIMA plate reader (BMG Labtech).
ProK digestion of WT and E83Q fibrils
The fibrils used for the ProK digestion were prepared as above and not subjected to sonication. Briefly, 10 μg of fibrils was taken in 20 μl of 1× PBS, added with increasing concentrations (0.0, 0.5, 1.0, 1.5, and 2.0 μg/ml) of ProK and incubated for 37°C for 30 min. Following incubation, the reaction was quenched by directly adding 5 μl of 5× Laemmli buffer to the reaction mixture and boiling the samples for 8 min at 95°C. The digested fibrillar products were separated by SDS-PAGE using precast 12% bis-tris gels (Bio-Rad, Criterion XT) with MES running buffer (Bio-Rad). Bands were visualized using the Coomassie blue staining.
Negative staining: TEM
EM analyses of protein samples were performed as described previously (26). Briefly, 5 μl of protein samples was placed on glow-discharged Formvar and carbon-coated 200 mesh–containing copper EM grids. After about a minute, the samples were carefully blotted using filter paper and air dried for 30 s. These grids were washed three times with water and followed by staining with 0.7% (w/v) uranyl formate solution. TEM images were acquired by Tecnai Spirit BioTWIN electron microscope. Widths of different fibril morphologies (single, twisted, and stacked) and periodicity lengths of twisted fibrils were measured using the image processing package Fiji (RRID:SCR_002285). The relative frequency of each fibril morphology was estimated by examining and counting randomly chosen all visible fibrils within the TEM images. The number of fibrils quantified for WT is 164 for WT and 176 for E83Q.
MS analysis
MS analyses of proteins were performed by LC-MS on the LTQ system (Thermo Scientific, San Jose, CA). Before analysis, proteins were desalted online by reversed-phase chromatography on a Poroshell 300SB C3 column (1.0 × 75 mm, 5 μm; on the LTQ system; Agilent Technologies, Santa Clara, CA). Protein samples (10 μl), roughly at a concentration of 0.5 to 1 μg, were injected on the column at a flow rate of 300 μl/min and were eluted from 5 to 95% of solvent B against solvent A, linear gradient. The solvent composition was solvent A (0.1% formic acid in ultrapure water) and solvent B (0.1% formic acid in acetonitrile). MagTran software (Amgen Inc., Thousand Oaks, CA) was used for charge state deconvolution and MS analysis.
Native nESI-IM-MS experiments
For the monomer experiments, WT and E83Q aSyn were redissolved in 20 mM aqueous ammonium acetate (Sigma-Aldrich, St. Louis, MO, USA) solution at pH 6.8 with a final protein concentration of 20 μM. Native IM-MS experiments were performed on a Synapt G2 HD mass spectrometer (Waters Corporation, Wilmslow, UK) equipped with a direct infusion nESI source. In-house generated gold-coated borosilicate capillaries were used to introduce a few microliters of sample into the instrument. The most important experimental parameters were as follows: spray capillary, 1.5 to 1.7 kV; source temperature, 30°C; sample cone, 25 V; extraction cone, 1 V; trap collision energy, 5 V; transfer collision energy, 0 V; trap bias, 40 V; IM wave velocity, 300 m/s; and IM wave height, 35 V. Gas pressures in the instrument were as follows: source, 2.6 mbar; trap cell, 0.024 mbar; IM cell, 3.0 mbar; and transfer cell, 0.025 mbar. For collision-induced unfolding experiments, identical instrument settings were used, but the trap collision energy (the acceleration voltage into the trap cell) was increased in a stepwise manner in 5-V increments. Mass spectra were analyzed and visualized using MassLynx V4.1 (Waters Corporation, Wilmslow, UK) and Origin 8 (OriginLab Corporation, Northampton, MA, USA). Drift time profiles were generated by selecting the upper half of mass spectral peaks, i.e., the m/z range from half height to half height of each peak, to avoid overlap with adduct peaks. Drift times from a traveling wave IM cell can be converted into collision cross section (CCS) values, the rotationally averaged projection area of the protein, by calibration. Denatured cytochrome C from equine heart, denatured ubiquitin from bovine erythrocytes, and denatured myoglobin from equine skeletal muscle (all purchased from Sigma-Aldrich, St. Louis, MO, USA) were used to calibrate IM measurements of αSyn based on literature CCS values (79) at a final concentration of 10 μM in 50:49:1 H2O/acetonitrile/formic acid. Acetonitrile (≥99.8%) and formic acid (99%) were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
nESI-MS analysis of fibril samples
For the fibril experiments, WT and E83Q fibrils were separated from eventual remaining monomer using centrifugal filters with a cutoff of 100 kDa (MilliporeSigma, Burlington, MA, USA). Samples were incubated with ProK (0.5 μg/ml) for 5 min at 37°C. After incubation, the samples were frozen at −80°C and thawed immediately before the measurement. Control samples were measured directly, while an additional C18 ZipTip (MilliporeSigma, Burlington, MA, USA) purification step was necessary for the digested fibril samples. Measurements were performed with identical experimental parameters as for the monomer samples. Mass spectra were analyzed and visualized as described above, and peptide masses were calculated and assigned to peptide fragments using PAWS (Genomic Solutions Ltd., Cambridgeshire, UK), with a mass tolerance of ±250 parts per million.
Preparation of small unilamellar vesicles
The preparation of small unilamellar vesicles (SUVs) was carried out as previously described (69). Briefly, the negatively charged phospholipid 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1′-racemic glycerol), denoted 16:0/18:1 phosphatidylglycerol and hereafter referred to as POPG, was purchased from Avanti Polar Lipids Inc. (Alabaster, AL). A stock of POPG at 10 mg/ml in chloroform was prepared in a brown glass vial and stored at −20°C. The SUVs were prepared using the extrusion method. Briefly, 100 μl of POPG from the stock was added to 900 μl of chloroform in a new glass vial to have a solution of 1 mg/mL POPG. The solution mixture was dried gently using a nitrogen stream to form a thin film on the wall of the glass vial. Any remaining chloroform was removed by placing the vial under a chemical hood overnight with the glass vial left opened. The phospholipids were then resolubilized in 1× PBS to reach their final concentrations of 1.2 mM and followed by water bath sonication for 30 min. The solution was then extruded through Avestin LiposoFast (Avestin Inc., Ottawa, ON) using a membrane pore size of 100 nm. The size and homogeneity of the resulting vesicles were assessed by dynamic light scattering measurements (Stunner).
For another liposome preparation, 5 mg of DOPE:DOPS:DOPC 5:3:2 (w/w; Avanti Polar Lipids) were resuspended in 0.8 ml of methanol:chloroform 1:1, evaporated under a nitrogen stream, lyophilized overnight, and resuspended in 0.52 ml of Hepes buffer [50 mM Hepes, 100 mM NaCl (pH 7.4), 0.02% NaN3]. The resulting turbid sample was sonicated until transparency (15 min, 10 s on, 20 s off).
Far-ultraviolet CD spectroscopy
Protein samples (10 μM) in 1× PBS loaded onto a 1-mm path length quartz cuvette were analyzed alone or mixed with POPG vesicles using the following protein:lipid molar ratios: 1:0.5, 1:1, 1:10, 1:25, 1:50, and 1:100. CD spectra were recorded at room temperature (RT) using a Chirascan spectropolarimeter (Applied Photophysics) with the following parameters: temperature, 20°C; wavelength range, 198 to 250 nm; data pitch, 0.2 nm; bandwidth, 1 nm; scanning speed, 50 nm/min; and digital integration time, 2 s. The final CD spectra were a binomial approximation on an average of three repeat measurements. For the measurement of aSyn in the presence of another liposome mixture [DOPE:DOPS:DOPC (5:3:2)], individual WT and E83Q aSyn-to-liposome ratios were pipetted (ratio: 1:10, 1:20, 1:40, 1:100, and 1:200) at a protein concentration of 50 μM and the respective liposome concentrations from a 12.5 mM liposome stock in Hepes buffer [50 mM Hepes, 100 mM NaCl (pH 7.4), 0.02% NaN3] to a total volume of 12 μl. For CD measurement, the sample was diluted in 48 μl of deionized water and transferred to a 0.02-cm path length FireflySci cuvette for a final protein concentration of 10 μM. CD data were collected from 190 to 260 nm by using a Chirascan-plus qCD spectrometer (Applied Photophysics, Randalls Rd., Leatherhead, UK) at 20°C, 1.5 time per point (s) in 1-nm steps. The datasets were averaged from three repeats. All spectra were baseline corrected against buffer in deionized water and smoothened (window size, 8).
NMR spectroscopy
Isotopically labeled WT and E83Q aSyn were purified following the same purification protocol as described above in the “Recombinant overexpression and purification of hWT and hE83Q aSyn” section, except the usage of the minimal medium instead of LB medium. Briefly, E. coli BL21(DE3) cells expressing the aSyn E83Q and WT were first grown in LB medium and then transferred to minimal medium containing 13C-labeled glucose and 15N-labeled ammonium chloride for production of 13C,15N-labeled E83Q and WT protein before induction. After protein expression, bacteria were lysed by sonication, and the lysate was subjected to 100°C boiling for ~15 min and further followed by the next steps of purification protocol such as anion exchange chromatography, reversed-phase HPLC, and lyophilization. The purity of the lyophilized 13C,15N-labeled E83Q and WT protein was analyzed by UPLC and ESI-MS.
NMR experiments were measured on a Bruker 800-MHz spectrometer equipped with a 5-mm triple-resonance, pulsed-field z-gradient cryoprobe using two-dimensional 1H,15N SOFAST HMQC2 and 1H,13C HSQC3 pulse sequences for monomer characterization at 15°C. All experiments were performed in Hepes buffer [50 mM Hepes, 100 mM NaCl (pH 7.4), 0.02% NaN3] with 5% (v/v) D2O. The sample concentration for natural abundance 1H,13C-HSQC experiments was 300 μM for both WT and E83Q aSyn. Spectra were processed with TopSpin 3.6.1 (Bruker) and analyzed using Sparky 3.115 (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). The combined 1H/15N CSP was calculated according to (((δH)2+ (δN/10)2)/2)1/2.
Liposome preparation and titration by NMR spectroscopy
Twenty-five milligrams of DOPE:DOPS:DOPC 5:3:2 (w/w; Avanti Polar Lipids) was resuspended in 4 ml of methanol:chloroform 1:1, evaporated under a nitrogen stream, lyophilized overnight, and resuspended in 2.59 ml of Hepes buffer [50 mM Hepes, 100 mM NaCl (pH 7.4), 0.02% NaN3]. The resulting turbid sample was sonicated with a fine-tip sonicator (MS 72) until transparency (five to seven times 30-s pulse on and 30-s pulse off at 60% amplitude).
NMR-based liposome titration experiments were performed on a Bruker 600-MHz spectrometer equipped with a 5-mm triple-resonance, pulsed-field z-gradient cryoprobe using two-dimensional 1H,15N HSQC pulse sequences at 15°C (80). All experiments were performed in Hepes buffer [50 mM Hepes, 100 mM NaCl (pH 7.4), 0.02% NaN3] with 100 μM dextran sulfate sodium and 5% (v/v) D2O. The protein concentration for the liposome titration experiments was 40 μM, and we used a liposome stock concentrated at 12.5 mM. Spectra were processed with TopSpin 3.6.1 (Bruker) and analyzed using Sparky 3.115 (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) and CcpNmr Analysis 2.5.
ssNMR spectroscopy
For the analysis of fibrils for ssNMR studies, 13C/15N-labeled E83Q and WT fibrils were prepared by dissolving ~20 mg of lyophilized recombinant aSyn in 4 ml of 1× PBS (Gibco), and the pH was adjusted to ~7.2 to 7.4. The solutions were filtered through 0.2-μm filters (Merck, SLGP033RS), and the filtrates were transferred to a 15-ml Falcon tube. These tubes were incubated in a shaking incubator at 37°C and 1000 rpm for 5 days. After 6 days, the formation of fibrils was assessed by TEM and Coomassie staining. NMR samples contained ~6 mg of U-[13C,15N]–labeled protein packed into a 3.2-mm rotor. ssNMR experiments were recorded on a 950-MHz Bruker Avance III HD standard-bore spectrometer equipped with a 3.2-mm (1H, 13C, 15N) Efree triple-resonance probe (Bruker BioSpin) as previously described (81). 13C-13C dipolar-assisted rotational resonance experiments with a mixing time of 50 ms and hNCA experiments were recorded at 265 K for WT and E83Q aSyn fibrils. The following 90° pulse widths were used: 2 to 3.75 μs for 1H, and 4.5 μs for 13C. 1H decoupling strengths were 80 to 100 kHz. Measurements were performed with a magic angle spinning frequency of 17 kHz. Spectra were processed using TopSpin (Bruker) and analyzed with CcpNmr Analysis (81). 13C-13C correlation DARR and two-dimensional hNCA experiments were processed with a sine bell shift of 2 and 4, respectively.
AFM imaging
AFM was performed on freshly cleaved mica discs that are positively functionalized with 1% (3-aminopropyl)triethoxysilane (APTS) in an aqueous solution for 3 min at RT. For the fibril deposition onto the substrates, 20-μl aliquots each containing a 50 μM protein solution was loaded on the surface of mica discs at predefined time points. Deposition lasted 3 min and was followed by a gentle drying by nitrogen flow. The mica discs were stored in a desiccator for 1 day before imaging to avoid prolonged exposure to atmospheric moisture. AFM imaging was performed at RT by a Park NX10 operating in true noncontact mode that was equipped with a super sharp tip (SSS-NCHR) Park System cantilever. The length and height quantifications were performed manually using XEI software developed by Park Systems Corporation.
Cell culture and transfections
HEK293 HeLa and M17 cells were cultured at 95% air and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; for HEK293 and HeLa) or DMEM-F12 (1:1; for M17) supplemented with 10% fetal bovine serum (Gibco) and penicillin-streptomycin (Thermo Fisher Scientific). According to the manufacturer’s protocol, transfections were performed with Effectene Transfection Reagent (QIAGEN).
WB of HEK293, HeLa, and M17 cells
HEK293, HeLa, or M17 cells were lysed in radioimmunoprecipitation assay buffer [150 mM sodium chloride, 50 mM tris (pH 8.0), 1% NP-40, 0.5% deoxycholate, 0.1% SDS] supplemented with protease inhibitor cocktail, 1 mM PMSF, and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich). Cell lysates were cleared by centrifugation at 4°C for 15 min at 13,000 rpm. The pellet (insoluble fraction) was resuspended in SDS/TBS (tris-buffered saline) supplemented with protease inhibitor cocktail, 1 mM PMSF, and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich) and sonicated using a fine probe (0.5-s pulse at amplitude of 20%, 15 iterations). BCA protein assay was performed to quantify the protein concentration in the soluble and insoluble fractions before the addition of 4× Laemmli buffer. Proteins from the soluble and the insoluble fractions were then separated on a 16.5% or a 18% SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane (Thermo Fisher Scientific) with a semidry system (Bio-Rad), and immunostained.
Subcellular fractionation of mammalian cell lines
Cells from a 10-cm plate were collected by scrapping in 500 μl of fractionation buffer [20 mM Hepes (pH 7.4), 10 mM KCI, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, protease inhibitor cocktail, and phosphatase inhibitor cocktails 2 and 3]. Cell suspensions were incubated for 15 min on ice before being mechanically lysed using a Dounce homogenizer (with type B pestle). Twenty strokes (or until all cells are lysed as verified by microscopy) were performed. Cell extracts were then left on ice for 20 min before being centrifuged at 720g (3000 rpm) for 5 min. The pellet contained nuclei, and the supernatant contained cytoplasm, membrane, and mitochondria fractions. The supernatant (A) was transferred into a fresh tube and kept on ice. The nuclear pellet was washed with 500 μl of fresh fractionation buffer before being mechanically lysed using a Dounce homogenizer (with type B pestle) with a minimum of 20 strokes. Lysates were centrifuged at 720g (3000 rpm) for 10 min. The supernatant (B) was discarded, and the pellet that contained nuclei was resuspended in TBS [pH 7.4, 50 mM tris(hydroxymethyl)aminomethane, and 150 mM NaCl] with 0.1% SDS. Sonication was then performed to shear genomic DNA and homogenize the lysate (3 s on ice at a power setting of 60%). The supernatant (A) that contains the membrane fractions was centrifuged at 40,000 rpm (100,000g) for 1 hour. The supernatant (C) that corresponds to the cytosolic fraction was collected in a fresh tube. The pellet was washed with 400 μl of fractionation buffer and resuspended using a Dounce homogenizer (with type B pestle) with a minimum of 20 strokes. Lysates were centrifuged for 45 min (100,000g), and the pellet was resuspended as membrane fraction in the same buffer used for the nuclei. Protein estimation was carried out using a BCA assay kit, and 30 μg of total protein was loaded onto 16% tricine gels, electrophoresed, and immunostained.
Immunocytochemistry
Immortal cells or primary hippocampal neurons were washed twice with PBS, fixed in 4% paraformaldehyde (PFA) for 20 min at RT, and immunostained as previously described (14). The antibodies used are indicated in the corresponding legend of each figure. The source and dilution of each antibody can be found in fig. S5. The cells plated on coverslips (CS) were then examined with a confocal laser scanning microscope (LSM 700, Carl Zeiss Microscopy) with a 40× objective and analyzed using Zen software (RRID:SCR_013672). The cells plated in black, clear-bottom, 96-well plates were imaged using the IN Cell Analyzer 2200 (with a 10× objective). For each independent experiment, two wells were acquired per tested condition; in each well, nine fields of view were imaged.
SEC and cross-linking assay
After the transfection of the HEK and HeLa cells for 72 hours (i.e., with EV, WT aSyn, and E83Q aSyn), SEC and cross-linking experiments were conducted. Briefly, the medium of both cell cultures was removed, and the cells were washed twice with PBS (pH 7.4). To detach the cells, 2 ml of trypsin was added to each plate, followed by incubation at 37°C for 5 to 7 min. Subsequently, 8 ml of fresh medium was added to quench the reaction, and the cells were transferred into a conical Falcon tube and centrifuged at RT for 2 min at 300g. The supernatant was discarded, the pellet was resuspended in fresh media, and the cells were counted. Cells were spun down once more under the aforementioned conditions; the supernatant was discarded, and the cells were resuspended to a final concentration of approximately 4 × 106 cells per 180 μl of PBS supplemented with phosphatase inhibitor.
To evaluate the major aSyn species after overexpression in mammalian cells (HEK and HeLa), cell extracts were analyzed by SEC. Briefly, 160 μl of cell extracts combined with 1.6 μl of DMSO was injected onto a Superdex 200 10/300 GL column and equilibrated with 1× PBS (pH 7.4). Proteins were eluted at 0.5 ml/min, and 500-μl fractions were collected using the ÄKTA FPLC System (GE Healthcare). The SEC fractions were analyzed by WB, and the SEC chromatograms were replotted with OriginPro software.
The native oligomeric state of E83Q was studied through cross-linking experiments, as follows. DSG was used as the chemical cross-linking reagent and was solubilized in DMSO to an initial concentration of 100 mM. The DSG was then added to the cells for a final concentration of 1 mM; as a control, the same volume of DMSO was added to the cells. After a 30-min incubation at 37°C at 500 rpm, the reaction was quenched by the addition of 1 M tris (pH 7.6), for a final concentration of 50 mM. Following the cross-linking reaction, the HEK and HeLa cells were lysed by a sonication step (amp: Amp 40%; iterations: 2 × 15 s), after which the lysed cells were centrifuged for 30 min at 20,000g at 4°C. The supernatant was collected, and the pellet was snap frozen. The protein concentration was assessed by BCA assay kit before Western blotting of the soluble fraction. All WB analyses related to cross-linking experiments were conducted using NuPAGE 4 to 12% bis-tris gels (Invitrogen). To further assess the size distribution of the native aSyn species upon cross-linking, 160 μl of HEK and HeLa cells resulting from the cross-linking experiments (+1 mM DSG) was analyzed by SEC in similar conditions as those mentioned above.
Primary neuronal culture
Primary hippocampal and cortical neurons were prepared from postnatal D0 pups of WT mice (C57BL/6JRj; Janvier, France) or aSyn KO mice (C57BL/6J; OlaHsd, Envigo) and cultured as previously described (14). All procedures were approved by the Swiss Federal Veterinary Office (authorization numbers VD3392 and VD3694). Briefly, the cerebral hippocampi and the cortexes were isolated stereoscopically in Hanks’ balanced salt solution and digested by papain (20 U/ml; Sigma-Aldrich) for 30 min at 37°C. After inhibiting the papain activity using a trypsin inhibitor (Sigma-Aldrich), the tissues were dissociated by mechanical trituration. The cells were finally resuspended in adhesion media (MEM; 10% horse serum, 30% glucose, l-glutamine, and penicillin-streptomycin; Life Technologies) and plated in 24- or 6-well plates previously treated with 0.1% (w/v) poly-l-lysine in water (Brunschwig, Switzerland) at a density of 300,000 cells/ml (for biochemistry analysis) or 250,000 cells/ml (for ICC/confocal microscopy analysis). After 3 hours, the adhesion medium was removed and replaced with Neurobasal medium (Life Technologies) containing B27 supplement (Life Technologies), l-glutamine, and penicillin-streptomycin (100 U/ml; Life Technologies). Neurons were plated directly in Neurobasal medium in the black, clear-bottom, 96-well plates at a concentration of 200,000 cells/ml.
Primary culture treatment with mouse aSyn fibrils or PBS
PFFs were added to the neuronal cell culture media at different DIV as previously described (14), and all PFF-treated neurons were harvested at days in vitro (DIV) 26 (fig. S15), which ensures that the developmental stage of primary neurons in culture was similar at harvest time. For control neurons, PBS was added at DIV 5.
On the day of the treatment, hWT or hE83Q PFFs or mouse WT PFFs were thawed at RT and diluted to a final concentration of 70 nM using the Neurobasal media collected from wells containing the plated neurons. The leftover Neurobasal media in the wells was then aspirated and replaced by the media containing the diluted PFFs. Treated neurons were cultured for up to 21 days without any further media changes until the end of the treatment (14).
Quantitative HCA
After aSyn PFF treatment, primary hippocampal or cortical neurons plated in black, clear-bottom, 96-well plates (BD) were washed twice with PBS, fixed in 4% PFA for 20 min at RT, and immunostained as described above. Images were acquired using the Nikon 10×/ 0.45 Plan Apo, CFI/60 of the IN Cell Analyzer 2200 (GE Healthcare), a high-throughput imaging system equipped with a high-resolution 16-bit sCMOS (scientific complementary metal-oxide semiconductor) camera (2048 × 2048 pixels), using a binning of 2 × 2. For each independent experiment, duplicated wells were acquired per condition, and nine fields of view were imaged for each well. Images were analyzed using CellProfiler 3.0.0 software (RRID:SCR_007358) to identify and quantify the level of LB-like inclusions (stained with pS129 antibody) formed in MAP2-positive neurons, the number of neuronal cell bodies (costained with MAP2 and DAPI), or the number of neurites (stained with MAP2).
Preparation of soluble and insoluble fractions of primary hippocampal neurons
After the aSyn PFF treatment, primary hippocampal neurons were lysed as previously described (14, 15). Briefly, treated neurons were lysed in 1% Triton X-100/TBS (50 mM tris, 150 mM NaCl, pH 7.5) supplemented with protease inhibitor cocktail, 1 mM PMSF, and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich). After sonication using a fine probe [0.5-s pulse at an amplitude of 20%, 10 iterations (Sonic Vibra-Cell, Blanc Labo)], cell lysates were incubated on ice for 30 min and centrifuged at 100,000g for 30 min at 4°C. The supernatant (soluble fraction) was collected, and the pellet was washed in 1% Triton X-100/TBS, sonicated as described above, and centrifuged for another 30 min at 100,000g. The supernatant was discarded, whereas the pellet (insoluble fraction) was resuspended in 2% SDS/TBS supplemented with protease inhibitor cocktail, 1 mM PMSF, and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich) and sonicated using a fine probe (0.5-s pulse at an amplitude of 20%, 15 iterations).
The BCA protein assay was performed to quantify the protein concentration in the total, soluble, and insoluble fractions before the addition of 4× Laemmli buffer [10% SDS, 50% glycerol, 0.05% bromophenol blue, 0.3 M tris-HCl (pH 6.8), and 20% β-mercaptoethanol]. Proteins from the total, soluble, and insoluble fractions were then separated on 16% tricine gels, transferred onto a nitrocellulose membrane (Thermo Fisher Scientific) with a semidry system (Bio-Rad), and immunostained as previously described (14).
Cell death quantification in mammalian cell lines and primary neurons
Mammalian cell lines (HEK293, HeLa, and M17) overexpressing WT or E83Q aSyn for up to 96 hours or primary hippocampal neurons were plated in 96-well plates and treated with aSyn PFFs (70 nM or up to 500 nM). After 7, 14, and 21 days of treatment, cell death was quantified using complementary cell death assays. Using a CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega, Switzerland), the lactic acid dehydrogenase (LDH) released into culture supernatants was measured according to the manufacturer’s instructions. After a 30-min coupled enzymatic reaction, which results in the conversion of a tetrazolium salt into a red formazan product, the amount of color formed (proportional to the number of damaged cells) was measured using an Infinite M200 Pro plate reader (Tecan) at a wavelength of 490 nm.
In primary cortical neurons, cell death was quantified by the total count of cells and by the TUNEL cell death assay. PFF- or PBS-treated neurons were fixed at the indicated times in 4% PFA for 15 min at RT. Cells were permeabilized in a solution composed of 0.1% Triton X-100 in 0.1% citrate buffer (pH 6.0) and then washed in PBS buffer before incubation with terminal deoxynucleotide transferase (In Situ Cell Death Detection kit; Roche, Switzerland) for 1 hour at 37°C in a solution containing TMR red dUTP. The neurons were stained using an antibody against the NeuN protein, a nuclear neuronal marker. The neurons bearing seeded aggregates were identified using pS129 antibody (MJFR13). The nucleus was counterstained with DAPI at 1:5000 (Sigma-Aldrich, Switzerland). The cells were washed five times in PBS before mounting in polyvinyl alcohol mounting medium with the anti-fading DABCO reagent (Sigma-Aldrich, Switzerland). The cells plated on CS were then examined with a microscope (EVOS, Life Technology) with a 10× objective and analyzed using ImageJ (U.S. National Institutes of Health, Bethesda, MD, USA; RRID:SCR_003070). A minimum of 4000 cells was counted for each condition and for each independent experiment.
Western blotting quantification
The level of total aSyn (15 kDa, 12 kDa, or HMW) or pS129-aSyn was estimated by measuring the WB band intensity using Image Studio Lite (LI-COR, RRID:SCR_013715) and normalized to the relative protein levels of actin; in the case of the cross-linking experiments, the level was normalized to the relative protein levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Statistical analyses
Independent experiments were performed to verify the findings (minimum of three independent experiments), with all attempts at replication successful. No data were excluded from the analysis. The statistical analyses were performed using Student’s t test, one-way analysis of variance (ANOVA) test followed by a Tukey-Kramer or post hoc tests, or two-way ANOVA followed by multiple pairwise comparisons with Sidak’s correction post hoc test using GraphPad Prism 9.1.1 software (RRID:SCR_002798). The data were regarded as statistically significant at P < 0.05.
Acknowledgments
We thank the CIME facility (EPFL) for the use of their EM facility and PTPSP (EPFL) for tremendous support for the use of their CD instrument. We thank F. Kuttler (PTCB, EPFL) for helping optimize the HCA pipeline. We also thank R. Kolla, S. Jagannath, and G. Limorenko for the critical review of the first versions of this manuscript and valuable feedback. We are grateful to S. Nazarov and A. Sadek for help with the generation of fig. S1C. We also thank L. Aeschbach and Y. Jasiqi for help with the generation of plasmids and proteins and the primary neuronal culture related to this work. We would like to express our gratitude to J. Hastings and N. Riguet for help with the statistical analysis.
Funding: M.Z. was supported by the European Research Council (ERC) under the EU Horizon 2020 research and innovation program (grant agreement no. 787679) and by The Michael J. Fox Foundation for Parkinson’s Research (grant ID: MJFF-019033). This work was supported by funding from Ecole Polytechnique Fédérale de Lausanne. S.T.K. and A.-L.M.-M. were also partially supported by funding from Idorsia Pharmaceuticals Ltd.
Author contributions: Conceptualization: H.A.L. Experimental design: H.A.L., M.Z., G.R., A.I.d.O., R.M., F.S., S.T.K., and A.-L.M.-M. Methodology: S.T.K., A.-L.M.-M., R.N.H., P.M., R.M., G.R., A.I.d.O., I.R., M.Z., and S.D. Investigation: S.T.K., A.-L.M.-M., R.N.H., P.M., R.M., G.R., A.I.d.O., M.Z., F.S., and H.A.L. Visualization: S.T.K., A.-L.M.-M., R.N.H., P.M., R.M., S.D., G.R., and A.I.d.O. Supervision: H.A.L., F.S. (nESI-IM-MS), and M.Z. (NMR). Writing (original draft): H.A.L., S.T.K., A.-L.M.-M., P.M., R.N.H., and R.M. Writing (review and editing): H.A.L., S.T.K., A.-L.M.-M., R.N.H., P.M., R.M., S.D., A.I.d.O., M.Z., and F.S.
Competing interests: H.A.L. is the founder and chief scientific officer of ND BioSciences, Épalinges, Switzerland, a company that develops diagnostics and treatments for neurodegenerative diseases (NDs) based on platforms that reproduce the complexity and diversity of proteins implicated in NDs and their pathologies. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S27
REFERENCES AND NOTES
- 1.Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L., Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O., AlaSOPro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat. Genet. 18, 106–108 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B., Llorens V., Gomez Tortosa E., del Ser T., Muñoz D. G., de Yebenes J. G., The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Appel-Cresswell S., Vilarino-Guell C., Encarnacion M., Sherman H., Yu I., Shah B., Weir D., Thompson C., Szu-Tu C., Trinh J., Aasly J. O., Rajput A., Rajput A. H., Jon Stoessl A., Farrer M. J., Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov. Disord. 28, 811–813 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Pasanen P., Myllykangas L., Siitonen M., Raunio A., Kaakkola S., Lyytinen J., Tienari P. J., Pöyhönen M., Paetau A., Novel α-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol. Aging 35, 2180.e1-5 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Yoshino H., Hirano M., Stoessl A. J., Imamichi Y., Ikeda A., Li Y., Funayama M., Yamada I., Nakamura Y., Sossi V., Farrer M. J., Nishioka K., Hattori N., Homozygous alpha-synuclein p.A53V in familial Parkinson’s disease. Neurobiol. Aging 57, 248.e7–248.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Liu H., Koros C., Strohäker T., Schulte C., Bozi M., Varvaresos S., Ibáñez de Opakua A., Simitsi A. M., Bougea A., Voumvourakis K., Maniati M., Papageorgiou S. G., Hauser A. K., Becker S., Zweckstetter M., Stefanis L., Gasser T., A novel SNCA A30G mutation causes familial Parkinson’s disease. Mov. Disord. 36, 1624–1633 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Kiely A. P., Asi Y. T., Kara E., Limousin P., Ling H., Lewis P., Proukakis C., Quinn N., Lees A. J., Hardy J., Revesz T., Houlden H., Holton J. L., α-synucleinopathy associated with G51D SNCA mutation: A link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol. 125, 753–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K., Alpha-synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Konno T., Ross O. A., Puschmann A., Dickson D. W., Wszolek Z. K., Autosomal dominant Parkinson’s disease caused by SNCA duplications. Parkinsonism Relat. Disord. 22 (Suppl. 1), S1–S6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapasi A., Brosch J. R., Nudelman K. N., Agrawal S., Foroud T. M., Schneider J. A., A novel SNCA E83Q mutation in a case of dementia with Lewy bodies and atypical frontotemporal lobar degeneration. Neuropathology 40, 620–626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ando T., Riku Y., Akagi A., Miyahara H., Hirano M., Ikeda T., Yabata H., Koizumi R., Oba C., Morozumi S., Yasui K., Goto A., Katayama T., Sakakibara S., Aiba I., Sakai M., Konagaya M., Mori K., Ito Y., Yuasa H., Nomura M., Porto K. J. L., Mitsui J., Tsuji S., Mimuro M., Hashizume Y., Katsuno M., Iwasaki Y., Yoshida M., Multiple system atrophy variant with severe hippocampal pathology. Brain Pathol. e13002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giasson B. I., Murray I. V., Trojanowski J. Q., Lee V. M., A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J. Biol. Chem. 276, 2380–2386 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Mahul-Mellier A. L., Burtscher J., Maharjan N., Weerens L., Croisier M., Kuttler F., Leleu M., Knott G. W., Lashuel H. A., The process of Lewy body formation, rather than simply α-synuclein fibrillization, is one of the major drivers of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 117, 4971–4982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volpicelli-Daley L. A., Luk K. C., Patel T. P., Tanik S. A., Riddle D. M., Stieber A., Meaney D. F., Trojanowski J. Q., Lee V. M., Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba M., Nakajo S., Tu P. H., Tomita T., Nakaya K., Lee V. M., Trojanowski J. Q., Iwatsubo T., Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 152, 879–884 (1998). [PMC free article] [PubMed] [Google Scholar]
- 17.Fares M. B., Jagannath S., Lashuel H. A., Reverse engineering Lewy bodies: How far have we come and how far can we go? Nat. Rev. Neurosci. 22, 111–131 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Kosaka K., Lewy bodies in cerebral cortex, Report of three cases. Acta Neuropathol. 42, 127–134 (1978). [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M. S., Shen J., Takio K., Iwatsubo T., α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M., α-synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y., Halliday G., Can we clinically diagnose dementia with Lewy bodies yet? Transl. Neurodegener. 2, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eliezer D., Kutluay E., Bussell R. Jr., Browne G., Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 307, 1061–1073 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Konijnenberg A., Ranica S., Narkiewicz J., Legname G., Grandori R., Sobott F., Natalello A., Opposite structural effects of epigallocatechin-3-gallate and dopamine binding to α-synuclein. Anal. Chem. 88, 8468–8475 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Stephens A. D., Zacharopoulou M., Moons R., Fusco G., Seetaloo N., Chiki A., Woodhams P. J., Mela I., Lashuel H. A., Phillips J. J., De Simone A., Sobott F., Schierle G. S. K., Extent of N-terminus exposure of monomeric alpha-synuclein determines its aggregation propensity. Nat. Commun. 11, 2820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Runfola M., De Simone A., Vendruscolo M., Dobson C. M., Fusco G., The N-terminal acetylation of α-synuclein changes the affinity for lipid membranes but not the structural properties of the bound state. Sci. Rep. 10, 204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S. T., Donzelli S., Chiki A., Syed M. M. K., Lashuel H. A., A simple, versatile and robust centrifugation-based filtration protocol for the isolation and quantification of α-synuclein monomers, oligomers and fibrils: Towards improving experimental reproducibility in α-synuclein research. J. Neurochem. 153, 103–119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carulla N., Caddy G. L., Hall D. R., Zurdo J., Gairí M., Feliz M., Giralt E., Robinson C. V., Dobson C. M., Molecular recycling within amyloid fibrils. Nature 436, 554–558 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Sánchez L., Madurga S., Pukala T., Vilaseca M., López-Iglesias C., Robinson C. V., Giralt E., Carulla N., Aβ40 and Aβ42 amyloid fibrils exhibit distinct molecular recycling properties. J. Am. Chem. Soc. 133, 6505–6508 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Dettmer U., Newman A. J., Luth E. S., Bartels T., Selkoe D., In vivo cross-linking reveals principally oligomeric forms of α-synuclein and β-synuclein in neurons and non-neural cells. J. Biol. Chem. 288, 6371–6385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fauvet B., Mbefo M. K., Fares M. B., Desobry C., Michael S., Ardah M. T., Tsika E., Coune P., Prudent M., Lion N., Eliezer D., Moore D. J., Schneider B., Aebischer P., El-Agnaf O. M., Masliah E., Lashuel H. A., α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 287, 15345–15364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A.-L. Mahul-Mellier, M. F. Altay, J. Burtscher, N. Maharjan, N. Ait-Bouziad, A. Chiki, S. Vingill, R. Wade-Martins, J. Holton, C. Strand, C. Haikal, J.-Y. Li, R. Hamelin, M. Croisier, G. Knott, G. Mairet-Coello, L. Weerens, A. Michel, P. Downey, M. Citron, H. A. Lashuel, The making of a Lewy body: The role of α-synuclein post-fibrillization modifications in regulating the formation and the maturation of pathological inclusions. bioRxiv 500058 [Preprint]. 2018.
- 32.Courte J., Bousset L., Boxberg Y. V., Villard C., Melki R., Peyrin J. M., The expression level of alpha-synuclein in different neuronal populations is the primary determinant of its prion-like seeding. Sci. Rep. 10, 4895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karpowicz R. J. Jr., Haney C. M., Mihaila T. S., Sandler R. M., Petersson E. J., Lee V. M., Selective imaging of internalized proteopathic α-synuclein seeds in primary neurons reveals mechanistic insight into transmission of synucleinopathies. J. Biol. Chem. 292, 13482–13497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson M. X., Chung C. H., Riddle D. M., Zhang B., Gathagan R. J., Seeholzer S. H., Trojanowski J. Q., Lee V. M. Y., Unbiased proteomics of early Lewy body formation model implicates active microtubule affinity-regulating kinases (MARKs) in synucleinopathies. J. Neurosci. 37, 5870–5884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Q., Takano H., Riddle D. M., Trojanowski J. Q., Coulter D. A., Lee V. M., α-Synuclein (αSyn) preformed fibrils induce endogenous αSyn aggregation, compromise synaptic activity and enhance synapse loss in cultured excitatory hippocampal neurons. J. Neurosci. 39, 5080–5094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luk K. C., Covell D. J., Kehm V. M., Zhang B., Song I. Y., Byrne M. D., Pitkin R. M., Decker S. C., Trojanowski J. Q., Lee V. M.-Y., Molecular and biological compatibility with host alpha-synuclein influences fibril pathogenicity. Cell Rep. 16, 3373–3387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fares M. B., Maco B., Oueslati A., Rockenstein E., Ninkina N., Buchman V. L., Masliah E., Lashuel H. A., Induction of de novo α-synuclein fibrillization in a neuronal model for Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 113, E912–E921 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Froula J. M., Henderson B. W., Gonzalez J. C., Vaden J. H., McLean J. W., Wu Y., Banumurthy G., Overstreet-Wadiche L., Herskowitz J. H., Volpicelli-Daley L. A., Alpha-synuclein fibril-induced paradoxical structural and functional defects in hippocampal neurons. Acta Neuropathol. Commun. 6, 35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson M. X., Sedor S., McGeary I., Cornblath E. J., Peng C., Riddle D. M., Li H. L., Zhang B., Brown H. J., Olufemi M. F., Bassett D. S., Trojanowski J. Q., Lee V. M. Y., Glucocerebrosidase activity modulates neuronal susceptibility to pathological α-synuclein insult. Neuron 105, 822–836.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Becker K., Levine N., Zhang M., Lieberman A. P., Moore D. J., Ma J., Pathogenic alpha-synuclein aggregates preferentially bind to mitochondria and affect cellular respiration. Acta Neuropathol. Commun. 7, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grassi D., Howard S., Zhou M., Diaz-Perez N., Urban N. T., Guerrero-Given D., Kamasawa N., Volpicelli-Daley L. A., LoGrasso P., Lasmezas C. I., Identification of a highly neurotoxic α-synuclein species inducing mitochondrial damage and mitophagy in Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 115, E2634–E2643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran H. T., Chung C. H., Iba M., Zhang B., Trojanowski J. Q., Luk K. C., Lee V. M., α-Synuclein immunotherapy blocks uptake and templated propagation of misfolded α-synuclein and neurodegeneration. Cell Rep. 7, 2054–2065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasegawa M., Fujiwara H., Nonaka T., Wakabayashi K., Takahashi H., Lee V. M., Trojanowski J. Q., Mann D., Iwatsubo T., Phosphorylated alpha-synuclein is ubiquitinated in alpha-synucleinopathy lesions. J. Biol. Chem. 277, 49071–49076 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Kuusisto E., Parkkinen L., Alafuzoff I., Morphogenesis of Lewy bodies: Dissimilar incorporation of alpha-synuclein, ubiquitin, and p62. J. Neuropathol. Exp. Neurol. 62, 1241–1253 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Pretorius E., Page M. J., Hendricks L., Nkosi N. B., Benson S. R., Kell D. B., Both lipopolysaccharide and lipoteichoic acids potently induce anomalous fibrin amyloid formation: Assessment with novel Amytracker stains. J. R. Soc. Interface 15, 20170941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moors T. E., Maat C. A., Niedieker D., Mona D., Petersen D., Timmermans-Huisman E., Kole J., El-Mashtoly S. F., Spycher L., Zago W., Barbour R., Mundigl O., Kaluza K., Huber S., Hug M. N., Kremer T., Ritter M., Dziadek S., Geurts J. J. G., Gerwert K., Britschgi M., van de Berg W. D. J., The subcellular arrangement of alpha-synuclein proteoforms in the Parkinson’s disease brain as revealed by multicolor STED microscopy. Acta Neuropathol. 142, 423–448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashida K., Oyanagi S., Mizutani Y., Yokochi M., An early cytoplasmic change before Lewy body maturation: An ultrastructural study of the substantia nigra from an autopsy case of juvenile parkinsonism. Acta Neuropathol. 85, 445–448 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Forno L. S., Neuropathology of Parkinsonʼs disease. J. Neuropathol. Exp. Neurol. 55, 259–272 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Uryu K., Richter-Landsberg C., Welch W., Sun E., Goldbaum O., Norris E. H., Pham C. T., Yazawa I., Hilburger K., Micsenyi M., Giasson B. I., Bonini N. M., Lee V. M.-Y., Trojanowski J. Q., Convergence of heat shock protein 90 with ubiquitin in filamentous alpha-synuclein inclusions of alpha-synucleinopathies. Am. J. Pathol. 168, 947–961 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wakabayashi K., Tanji K., Odagiri S., Miki Y., Mori F., Takahashi H., The Lewy body in Parkinson’s disease and related neurodegenerative disorders. Mol. Neurobiol. 47, 495–508 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Dickson D. W., Ruan D., Crystal H., Mark M. H., Davies P., Kress Y., Yen S. H., Hippocampal degeneration differentiates diffuse Lewy body disease (DLBD) from Alzheimer’s disease: Light and electron microscopic immunocytochemistry of CA2-3 neurites specific to DLBD. Neurology 41, 1402–1409 (1991). [DOI] [PubMed] [Google Scholar]
- 52.Datta A., Chai Y. L., Tan J. M., Lee J. H., Francis P. T., Chen C. P., Sze S. K., Lai M. K. P., An iTRAQ-based proteomic analysis reveals dysregulation of neocortical synaptopodin in Lewy body dementias. Mol. Brain 10, 36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia Q., Liao L., Cheng D., Duong D. M., Gearing M., Lah J. J., Levey A. I., Peng J., Proteomic identification of novel proteins associated with Lewy bodies. Front. Biosci. 13, 3850–3856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.B. A. Killinger, L. Marshall, D. Chatterjee, Y. Chu, J. H. Kordower, etection and purification of Lewy pathology from formalin fixed primary human tissue using biotinylation by antigen recognition. bioRxiv 2020.11.11.378752 [Preprint]. 2020.
- 55.Waxman E. A., Emmer K. L., Giasson B. I., Residue Glu83 plays a major role in negatively regulating alpha-synuclein amyloid formation. Biochem. Biophys. Res. Commun. 391, 1415–1420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herrera F. E., Chesi A., Paleologou K. E., Schmid A., Munoz A., Vendruscolo M., Gustincich S., Lashuel H. A., Carloni P., Inhibition of alpha-synuclein fibrillization by dopamine is mediated by interactions with five C-terminal residues and with E83 in the NAC region. PLOS ONE 3, e3394 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh D., Mondal M., Mohite G. M., Singh P. K., Ranjan P., Anoop A., Ghosh S., Jha N. N., Kumar A., Maji S. K., The Parkinson’s disease-associated H50Q mutation accelerates α-synuclein aggregation in vitro. Biochemistry 52, 6925–6927 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Boyer D. R., Li B., Sun C., Fan W., Zhou K., Hughes M. P., Sawaya M. R., Jiang L., Eisenberg D. S., The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl. Acad. Sci. U.S.A. 117, 3592–3602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villar-Piqué A., Lopes da Fonseca T., Sant’Anna R., Szegö É. M., Fonseca-Ornelas L., Pinho R., Carija A., Gerhardt E., Masaracchia C., Abad Gonzalez E., Rossetti G., Carloni P., Fernández C. O., Foguel D., Milosevic I., Zweckstetter M., Ventura S., Outeiro T. F., Environmental and genetic factors support the dissociation between α-synuclein aggregation and toxicity. Proc. Natl. Acad. Sci. U.S.A. 113, E6506–E6515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyer D. R., Li B., Sun C., Fan W., Sawaya M. R., Jiang L., Eisenberg D. S., Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 26, 1044–1052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y., Hou S., Zhao K., Long H., Liu Z., Gao J., Zhang Y., Su X. D., Li D., Liu C., Cryo-EM structure of full-length α-synuclein amyloid fibril with Parkinson’s disease familial A53T mutation. Cell Res. 30, 360–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerrero-Ferreira R., Taylor N. M., Arteni A.-A., Kumari P., Mona D., Ringler P., Britschgi M., Lauer M. E., Makky A., Verasdonck J., Riek R., Melki R., Meier B. H., Böckmann A., Bousset L., Stahlberg H., Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8, e48907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B., Ge P., Murray K. A., Sheth P., Zhang M., Nair G., Sawaya M. R., Shin W. S., Boyer D. R., Ye S., Eisenberg D. S., Zhou Z. H., Jiang L., Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long H., Zheng W., Liu Y., Sun Y., Zhao K., Liu Z., Xia W., Lv S., Liu Z., Li D., He K. W., Liu C., Wild-type α-synuclein inherits the structure and exacerbated neuropathology of E46K mutant fibril strain by cross-seeding. Proc. Natl. Acad. Sci. U.S.A. 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guan Y., Zhao X., Liu F., Yan S., Wang Y., Du C., Cui X., Li R., Zhang C. X., Pathogenic mutations differentially regulate cell-to-cell transmission of α-synuclein. Front. Cell. Neurosci. 14, 159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohite G. M., Dwivedi S., Das S., Kumar R., Paluri S., Mehra S., Ruhela N., Narendra A. S., Jha N., Maji S. K., Parkinson’s disease associated α-synuclein familial mutants promote dopaminergic neuronal death in Drosophila melanogaster. ACS Chem. Nerosci. 9, 2628–2638 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Flagmeier P., Meisl G., Vendruscolo M., Knowles T. P., Dobson C. M., Buell A. K., Galvagnion C., Mutations associated with familial Parkinson’s disease alter the initiation and amplification steps of α-synuclein aggregation. Proc. Natl. Acad. Sci. U.S.A. 113, 10328–10333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fares M. B., Ait-Bouziad N., Dikiy I., Mbefo M. K., Jovičić A., Kiely A., Holton J. L., Lee S. J., Gitler A. D., Eliezer D., Lashuel H. A., The novel Parkinson’s disease linked mutation G51D attenuates in vitro aggregation and membrane binding of α-synuclein, and enhances its secretion and nuclear localization in cells. Hum. Mol. Genet. 23, 4491–4509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khalaf O., Fauvet B., Oueslati A., Dikiy I., Mahul-Mellier A. L., Ruggeri F. S., Mbefo M. K., Vercruysse F., Dietler G., Lee S. J., Eliezer D., Lashuel H. A., The H50Q mutation enhances α-synuclein aggregation, secretion, and toxicity. J. Biol. Chem. 289, 21856–21876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mbefo M. K., Fares M. B., Paleologou K., Oueslati A., Yin G., Tenreiro S., Pinto M., Outeiro T., Zweckstetter M., Masliah E., Lashuel H. A., Parkinson disease mutant E46K enhances α-synuclein phosphorylation in mammalian cell lines, in yeast, and in vivo. J. Biol. Chem. 290, 9412–9427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luk K. C., Song C., O’Brien P., Stieber A., Branch J. R., Brunden K. R., Trojanowski J. Q., Lee V. M.-Y., Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 106, 20051–20056 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alegre-Abarrategui J., Brimblecombe K. R., Roberts R. F., Velentza-Almpani E., Tilley B. S., Bengoa-Vergniory N., Proukakis C., Selective vulnerability in α-synucleinopathies. Acta Neuropathol. 138, 681–704 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Y. Y. Fathy, F. J. de Jong, A.-M. van Dam, A. J. M. Rozemuller, W. D. J. van de Berg, Insular cortex sub-region-dependent distribution pattern of α-synuclein immunoreactivity in Parkinson’s disease and dementia with Lewy bodies. bioRxiv 156984 [Preprint]. 2017.
- 74.Gai W. P., Yuan H. X., Li X. Q., Power J. T., Blumbergs P. C., Jensen P. H., In situ and in vitro study of colocalization and segregation of alpha-synuclein, ubiquitin, and lipids in Lewy bodies. Exp. Neurol. 166, 324–333 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Takahashi H., Iwanaga K., Egawa S., Ikuta F., Ultrastructural relationship between Lewy bodies and pale bodies studied in locus ceruleus neurons of a non-Parkinsonian patient. Neuropathology 14, 73–80 (1994). [Google Scholar]
- 76.Prasad K., Beach T. G., Hedreen J., Richfield E. K., Critical role of truncated α-synuclein and aggregates in Parkinson’s disease and incidental Lewy body disease. Brain Pathol. 22, 811–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Irizarry M. C., Growdon W., Gomez-Isla T., Newell K., George J. M., Clayton D. F., Hyman B. T., Nigral and cortical Lewy bodies and dystrophic nigral neurites in Parkinsonʼs disease and cortical Lewy body disease contain α-synuclein immunoreactivity. J. Neuropathol. Exp.Neurol. 57, 334–337 (1998). [DOI] [PubMed] [Google Scholar]
- 78.Harding A. J., Halliday G. M., Cortical Lewy body pathology in the diagnosis of dementia. Acta Neuropathol. 102, 355–363 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Bush M. F., Hall Z., Giles K., Hoyes J., Robinson C. V., Ruotolo B. T., Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Bodenhausen G., Ruben D. J., Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 (1980). [Google Scholar]
- 81.Stevens T. J., Fogh R. H., Boucher W., Higman V. A., Eisenmenger F., Bardiaux B., van Rossum B. J., Oschkinat H., Laue E. D., A software framework for analysing solid-state MAS NMR data. J. Biomol. NMR 51, 437–447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S27