Abstract
Quantitative structure–activity relationship models are used in toxicology to predict the effects of organic compounds on aquatic organisms. Common filter feature selection methods use correlation statistics to rank features, but this approach considers only the correlation between a single feature and the response variable and does not take into account feature redundancy. Although the minimal redundancy maximal relevance approach considers the redundancy among features, direct removal of the redundant features may result in loss of prediction accuracy, and cross-validation of training sets to select an optimal subset of features is time-consuming. In this paper, we describe the development of a feature selection method, Chi-MIC-share, which can terminate feature selection automatically and is based on an improved maximal information coefficient and a redundant allocation strategy. We validated Chi-MIC-share using three environmental toxicology datasets and a support vector regression model. The results show that Chi-MIC-share is more accurate than other feature selection methods. We also performed a significance test on the model and analyzed the single-factor effects of the reserved descriptors.
An algorithm based on an improved maximal information coefficient and a redundant allocation strategy, which can terminate feature selection automatically, is presented.
1. Introduction
Quantitative structure–activity relationship (QSAR) models have been applied widely in chemical sciences such as biochemistry, environmental chemistry, food chemistry, and pharmacology.1 Water pollution is a global concern, and developing efficient procedures for assessing the toxicity of organic pollutants to aquatic organisms has become a research priority.2,3 QSAR can model referential activity and toxicity for an unknown compound by computing statistical relationships between biological activities and molecular descriptors (features) for a set of chemical compounds.4
In general, QSAR modeling includes four steps: recording the bioactivity or toxicity of a specific compound, extracting or calculating molecular descriptors, selecting features, and constructing and validating the model. Bioactivities can be obtained by experimental observations, relevant literature, or toxicity databases.5 The quantum chemistry software enable researchers to calculate thousands of theoretical parameters or physico-chemical properties for a chemical molecule,6 like HyperChem, MOPAC, Gaussian, ADF, and Dragon software packages.7 Another package, Parameter Client (PCLIENT), interfaces with various programs to provide calculations for approximately 3000 descriptors,8 we selected PCLIENT for the QSAR modeling in the present study.
The selection of descriptors is the most important step in constructing an efficient QSAR model, as it is essential to remove irrelevant and redundant descriptors.9 Feature selection methods are commonly categorized into three groups: filter, wrapper, and embedded algorithms. The filter algorithm is widely used because of its simplicity and efficiency.10 Univariate filter methods, such as Pearson's correlation coefficient (R), distance correlation coefficient (dCor), and mutual information criterion can eliminate irrelevant features but fail to remove redundancy between features.11 The classical multivariate filter method, minimum redundancy maximal relevance (mRMR),12 considers the maximum correlation between a feature subset and the response variable and simultaneously removes redundancies during the feature selection process. mRMR uses mutual information (I) to characterize the relevance for paired discrete variables, the F statistic for paired discrete versus continuous variables, and Pearson's correlation coefficient R for paired continuous variables. However, the F statistic may not always be appropriate for an unknown population distribution, the R statistic fails to reveal non-linear correlation, and the F and I statistics are not comparable in the mRMR method. What is needed is a measure that can assess the linear and non-linear correlations simultaneously regardless of the distribution of paired variables. Maximal Information Coefficient (MIC) can captures dependence between different types of paired variables, is a major breakthrough in measuring the correlation.13 Its estimation algorithm ApproxMaxMI performs uniform segmentation on one variable and unequal interval discrete optimization on another variable, and make MIC ∈ [0,1] through maximal grid correction. ApproxMaxMI tends to cause excessive segmentation in the direction of optimization for small data, and the MIC value is falsely high. Chen et al. proposed an improved algorithm, Chi-MIC,14 uses the chi-square test to terminate grid optimization and then removes the restriction of maximal grid. Chi-MIC has stronger statistical power and better equitability, so was used in the present study instead of the R statistic.
Another implicit disadvantage of mRMR is that the redundancies within the selected features are not removed properly.15 We used redundancy apportionment in mRMR and formed a new feature selection method, Chi-MIC-share, which can remove many redundancies and terminate feature selection automatically.
Once the refined feature subset is obtained, a statistical model can be applied to evaluate the relationship between these features and molecular bioactivities.16 Support vector regression (SVR) is frequently used in QSAR studies,17–19 and a QSAR model is generally validated by mean square error (MSE) and the coefficient of determination (R2), which we used for internal cross-validation and external independent prediction. The retained descriptors were analyzed for the biological or chemical molecular mechanisms using a significance test and their effects.20
2. Materials and methods
2.1. Datasets
To evaluate the performance of our method, we used three QSAR datasets. Dataset 1 (Table 1) comprises of anesthetic toxicities of 50 phenolic compounds to the free-living ciliate Tetrahymena pyriformis.21 Dataset 2 (Table 2) records anesthetic toxicities of 52 alcohol phenolic compounds to tadpoles,22 and Dataset 3 (Table 3) describes anesthetic toxicities of 85 substituted aromatics to fathead minnows.23 The toxicities in the three datasets are represented by half-maximal growth inhibitory concentration (−log IGC50), concentration (log 1/C), and half-maximal lethal concentration (−log LC50), respectively. Each dataset was divided into a training set and a test set; the compounds in the test set were selected at equal intervals from the dataset in order of toxicity, and the remaining records were used as a training set. Feature selection and modeling were performed only on the training set.
Toxicities of phenols to Tetrahymena pyriformisa.
| Compound | −log IGC50 (mmol L−1) | Compound | −log IGC50 (mmol L−1) |
|---|---|---|---|
| Phenol | −0.431 | 2-Isopropylphenol | 0.803 |
| *p-Cresol | −0.192 | *3-Chloro-4-fluorophenol | 0.842 |
| m-Cresol | −0.062 | 4-Iodophenol | 0.854 |
| 2,5-Dimethylphenol | 0.009 | 4-tert-Butylphenol | 0.913 |
| 3-Fluorophenol | 0.017 | 2,3,7-Trimethylphenol | 0.93 |
| 3,5-Dimethylphenol | 0.113 | 2,4-Dichlorophenol | 1.036 |
| *2,3-XYLENOL | 0.122 | *2-Phenylphenol | 1.094 |
| 3,4-Dimethylphenol | 0.122 | 3-Iodophenol | 1.118 |
| 2,4-Dimethylphenol | 0.128 | 2,5-Dichlorophenol | 1.128 |
| 2-Ethylphenol | 0.176 | 4-Chloro- 3,5-dimethylphenol | 1.203 |
| 2-Fluorophenol | 0.248 | 2-(tert-Butyl)-4,6-dimethylphenol | 1.245 |
| *2-Chlorophenol | 0.277 | *2,3-Dichlorophenol | 1.271 |
| 3-Ethylphenol | 0.299 | 4-Bromo-6-chloro-2-methylphenol | 1.277 |
| 2,6-Dichlorophenol | 0.396 | 4-Bromo-2,6-dimethylphenol | 1.278 |
| 3,4,5-Trimethylphenol | 0.418 | 2-tert-Butyl-4-methylphenol | 1.297 |
| 4-Fluorophenol | 0.473 | 2,4-Dibromophenol | 1.403 |
| *4-Isopropylphenol | 0.473 | *3,5-Dichlorophenol | 1.562 |
| 2-Bromophenol | 0.504 | 2,4,6-Trichlorophenol | 1.695 |
| 4-Chlorophenol | 0.545 | 4-Bromo-2,6-dichlorophenol | 1.779 |
| 3-Isopropylphenol | 0.609 | 2,6-Di-tert-Butyl-4-methylphenol | 1.788 |
| 2-Chloro- 5-methylphenol | 0.64 | 4-Chloro-2-isopropyl-5-methylphenol | 1.862 |
| *4-Bromophenol | 0.681 | *2,4,6-Tribromophenol | 2.05 |
| 4-Chloro-2-methylphenol | 0.7 | 2,4,5-Trichlorophenol | 2.1 |
| 3-tert-Butylphenol | 0.73 | 2,6-Diphenylphenol | 2.113 |
| 4-Chloro-3-methylphenol | 0.795 | 2,4-Dibromo-6-phenylphenol | 2.207 |
−log IGC50: half-maximal growth inhibitory concentration. *A test set sample.
Toxicities of alcohol phenolic compounds to tadpolesa.
| Compound | log 1/C (mmol L−1) | Compound | log 1/C (mmol L−1) |
|---|---|---|---|
| Methanol | 0.24 | Ethyl isobutanoate | 2.24 |
| Acetonitrile | 0.44 | *Isobutyl acetate | 2.24 |
| *Acetone | 0.54 | Butyl acetate | 2.30 |
| Ethanol | 0.54 | Chloroethane | 2.35 |
| Methyl aminoformate | 0.57 | Ethyl butanoate | 2.37 |
| Isopropyl alcohol | 0.89 | Pentane | 2.55 |
| tert-Butyl alcohol | 0.89 | *Bromoethane | 2.57 |
| *Aldoxime | 0.92 | Chloroethylene | 2.64 |
| Propyl alcohol | 0.96 | 1-Pentene | 2.65 |
| Butanone | 1.04 | Benzene | 2.68 |
| Nitrocarbol | 1.09 | Ethyl pentate | 2.72 |
| Methyl acetate | 1.10 | *Amyl acetate | 2.72 |
| *Ethyl formate | 1.15 | Anisole | 2.82 |
| Neopentyl alcohol | 1.24 | Chloroform | 2.85 |
| Isobutyl alcohol | 1.35 | Iodoethane | 2.96 |
| Ethyl aminoformate | 1.39 | Acetophenone | 3.03 |
| Butyl alcohol | 1.42 | *1,4-Dimethoxybenzene | 3.05 |
| *Ethyl acetate | 1.52 | Phenyl carbamate | 3.19 |
| 3-Pentanone | 1.54 | 1,3-Dimethoxybenzene | 3.35 |
| Diethyl ether | 1.57 | 1-Octanol | 3.40 |
| Isoamyl alcohol | 1.64 | Dimethylbenzene | 3.42 |
| 2-Pentanone | 1.72 | *Butyl valerate | 3.60 |
| *1,3-Dichloro-isopropyl alcohol | 1.92 | Naphthalene | 4.19 |
| Ethyl propionate | 1.96 | 2-Methyl-2-isopropyl phenol | 4.26 |
| Propyl acetate | 1.96 | Azobenzene | 4.74 |
| Acetal | 1.98 | Phenanthrene | 5.43 |
log 1/C: concentration. *A test set sample.
Toxicities of aromatics to fathead minnowsa.
| Compound | −log LC50 (mmol L−1) | Compound | −log LC50 (mmol L−1) |
|---|---|---|---|
| Nitrobenzene | 3.02 | *4-Methyl-2,6-dinitroaniline | 4.21 |
| Resorcinol | 3.04 | P-XYLENE | 4.21 |
| 1,4-Dimethoxybenzene | 3.07 | 1,2,4-Trimethylbenzene | 4.21 |
| *3-Methoxyphenol | 3.21 | 3-Methyl-2,4-dinitroaniline | 4.26 |
| p-Toluidine | 3.24 | 4-Chloro-3-methylphenol | 4.27 |
| m-Cresol | 3.29 | *2,4-Dichlorophenol | 4.30 |
| Toluene | 3.30 | 1,3-Dichlorobenzene | 4.30 |
| 2-Methyl-5-nitroaniline | 3.35 | 2,4,6-Trichlorophenol | 4.33 |
| *4-Nitrophenol | 3.36 | 4-Chlorotoluene | 4.33 |
| Benzene | 3.40 | 1,3-Dinitrobenzene | 4.38 |
| 2-Methyl-3-nitroaniline | 3.48 | *1,2-Dichlorobenzene | 4.40 |
| o-Xylene | 3.48 | 2-Phenylphenol | 4.45 |
| Phenol | 3.51 | 4-tert-Butylphenol | 4.46 |
| *2-Methyl-4-nitroaniline | 3.54 | 4-Methyl-3,5-dinitroaniline | 4.46 |
| 2,6-Dimethylphenol | 3.57 | 4-Butylphenol | 4.47 |
| 2-Nitrotoluene | 3.57 | *1-Naphthol | 4.53 |
| p-Cresol | 3.58 | 2,4-Dichlorotoluene | 4.54 |
| 3-Nitrotoluene | 3.63 | 1,4-Dichlorobenzene | 4.62 |
| *4-Amino-2-nitrophenol | 3.65 | 2,4,6-Tribromophenol | 4.70 |
| 4-Hydroxy-3-nitroaniline | 3.65 | 3,4-Dichlorotoluene | 4.74 |
| 4-Fluoronitrobenzene | 3.70 | *1,3,5-Trichlorobenzene | 4.74 |
| 2-Nitroaniline | 3.70 | 4-tert-Amylphenol | 4.82 |
| 2,4-Dinitrotoluene | 3.75 | 2,4,6-Trinitrotoluene | 4.88 |
| *4-Nitrotoluene | 3.76 | 1,2,3-Trichlorobenzene | 4.89 |
| Chlorobenzene | 3.77 | 5-Methyl-2,4-dinitroaniline | 4.92 |
| o-Cresol | 3.77 | *2,4-Dinitro-6-cresol | 4.99 |
| 3-Methyl-2-nitroaniline | 3.77 | 1,2,4-Trichlorobenzene | 5.00 |
| 4-Methyl-3-nitroaniline | 3.77 | 2,3-Dinitrotoluene | 5.01 |
| *4-Methyl-6-nitroaniline | 3.79 | 3,4-Dinitrotoluene | 5.08 |
| 2-Methyl-6-nitroaniline | 3.80 | 2,5-Dinitrotoluene | 5.15 |
| 3-Methyl-6-nitroaniline | 3.80 | *4-Pentylphenol | 5.18 |
| 3-Chlorotoluene | 3.84 | 1,4-Dinitrobenzene | 5.22 |
| 2,4-Dimethylphenol | 3.86 | 4-Phenylazophenol | 5.26 |
| *Bromobenzene | 3.89 | 1,3,5-Trinitrobenzene | 5.29 |
| 3,5-Dinitrotoluene | 3.91 | 2-Methyl-3,6-dinitroaniline | 5.34 |
| 2-Allylphenol | 3.93 | *1,2,3,4-Tetrachlorobenzene | 5.43 |
| 3,4-Dimethylphenol | 3.94 | 1,2-Dinitrobenzene | 5.45 |
| 3-Nitrochlorobenzene | 3.94 | 2,3,4,5-Tetrachlorophenol | 5.72 |
| *2,6-Dinitrotoluene | 3.99 | 1,2,3,5-Tetrachlorobenzene | 5.85 |
| 2-Chlorotoluene | 4.02 | Pentachlorophenol | 6.06 |
| 2,4-Dinitrophenol | 4.04 | *4-Nonylphenol | 6.20 |
| 2-Methyl-3,5-dinitroaniline | 4.14 | 2,3,6-Trinitrotoluene | 6.37 |
| 3-Methyl-2,6-dinitroaniline | 4.18 |
−log LC50: half-maximal lethal concentration. *A test set sample.
2.2. Calculation of molecular descriptors
High-dimensional molecular structure descriptors were obtained from the PCLIENT software package.24 We first drew the structural formula for compounds in the JME editor embedded in PCLIENT, then imported them into the task window and 24 groups of descriptors were calculated. After removing the invalid and duplicate descriptors, 1219, 1323, and 1360 descriptors from the three datasets were used for analysis.
2.3. Chi-MIC-share
Chi-MIC can capture linear and non-linear correlations universally. Similarly to other correlation criteria, it only ranks features by their correlation values with the response variable, and cannot automatically select a subset of features. Although mRMR takes into account both the correlation between features and response variable and the redundancy between features, its method of removing redundancy may cause some loss of prediction accuracy. In addition, mRMR also only sorts features and cannot automatically select a subset of features.25
We used Chi-MIC as an indicator of correlation and redundancy, and replaced redundancy with redundant allocation. Redundant allocation scores of individual features and feature sets are calculated, and feature selection is automatically terminated when the score no longer increases. The process does not depend on the learning machine. The specific process runs as follows:
We have sets of independent features, Ω = {X1, X2, …, Xi, …, Xm}, whose set length (number of elements) is |Ω| = m. If it is assumed that the introduced feature set is S, then the complement of feature set S is ΩS = Ω − S.
(1) For an introduced feature Xi in S, the score after redundancy allocation is:
 |
1 |
(2) The total score of all features in S after redundancy allocation is:
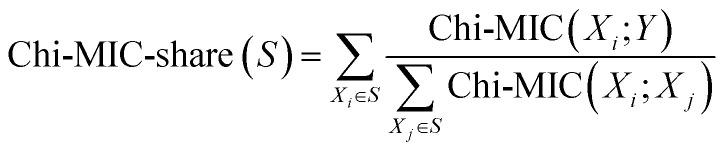 |
2 |
(3) Let the next incoming feature be Xnext, remember D = S + {Xnext}, then |D| = |S| + 1. The standard for introducing the next optimal feature by Chi-MIC-share is:
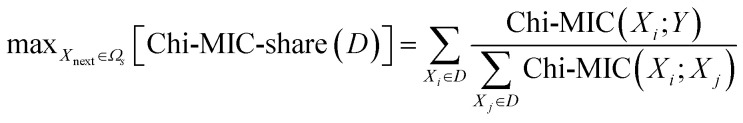 |
3 |
(4) The Chi-MIC-share termination feature criterion is:
| Chi-MIC-share (D) ≤ Chi-MIC-share (S) | 4 |
It should be noted that the correlation score of each feature in D after redundant allocation will be refreshed after the introduction of Xnext. Therefore, with the introduction of features, there is a maximum value for the total correlation score of the feature subset after redundant allocation. Furthermore, Chi-MIC-share does not set the upper limit for feature introduction and can automatically terminate feature introduction without cross-validation, which saves time.
2.4. Model evaluation and interpretation
The independent test evaluation standard uses MSE (eqn (5)) and R2 (eqn (6)). Smaller MSE and larger R2 indicate better prediction ability of the model.
 |
5 |
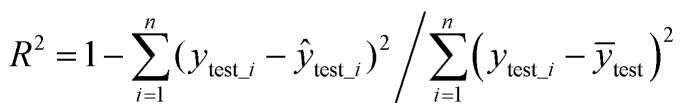 |
6 |
To test whether the regression of the SVR model is significant, we used an F-test. In eqn (7), U is the regression square sum of the model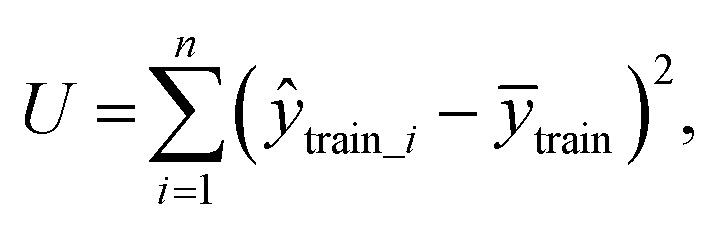 Q is the sum of the residual square of the model
Q is the sum of the residual square of the model 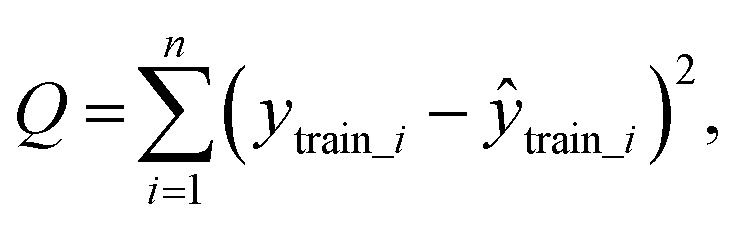 m′ is the number of reserved descriptors, and n is the number of training set samples. If F > Fα(n − m′ − 1), we can assert that the model has significant nonlinear regression at level α (0.01). Furthermore, we used the single-factor effect analysis method to assess the influence trend of the single reserved descriptors on response variable Y.20
m′ is the number of reserved descriptors, and n is the number of training set samples. If F > Fα(n − m′ − 1), we can assert that the model has significant nonlinear regression at level α (0.01). Furthermore, we used the single-factor effect analysis method to assess the influence trend of the single reserved descriptors on response variable Y.20
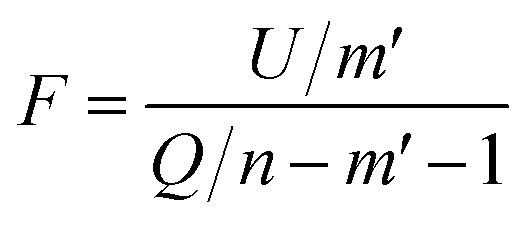 |
7 |
where ytest_i is the experimental value in the test set, 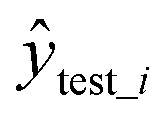 is the predicted value in the test set,
is the predicted value in the test set, 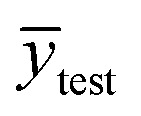 is the mean experimental value of the test set, ytrain_i is the experimental value in the training set,
is the mean experimental value of the test set, ytrain_i is the experimental value in the training set, 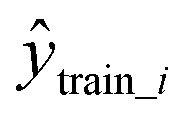 is the predicted value in the training set, and
is the predicted value in the training set, and 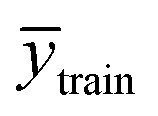 is the mean experimental value of the training set.
is the mean experimental value of the training set.
3. Results and analysis
3.1. Chi-MIC-share feature selection process
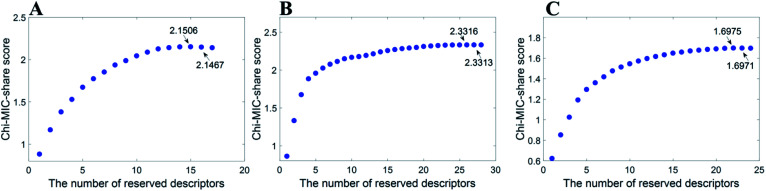
After feature selection by Chi-MIC-share, the reserved descriptors of the three datasets were 15, 27, and 22. Fig. 1 displays the selection processes. The redundancy allocation score no longer increases when features were introduced to a certain number.
Fig. 1. Chi-MIC-share scores for three datasets.
3.2. Comparison of feature selection methods
To verify the efficacy of the redundancy allocation concept, we performed feature selection on the three high-dimensional datasets using univariate filter methods R, dCor, Chi-MIC and multivariate filter methods mRMR, Chi-MIC-share. The well-tuned SVR model was used to evaluate the feature subsets refined by feature selection methods. The SVR model used in this paper is derived from the LIBSVM software package (https://www.csie.ntu.edu.tw/∼cjlin/libsvm/index.html). The SVM type is set to epsilon-SVR; the SVR model has three commonly used kernel functions, linear kernel, polynomial kernel, and Radial Basis Function (RBF). Compared with other kernel functions, RBF kernel shows better generalization ability in most cases. Therefore, RBF kernel was used in the experiment. The parameters include penalty parameter c, RBF kernel parameter g, and loss function parameter p. The three parameters were optimized by grid search with 5-fold cross-validation on the training set. The range of each parameter is set according to the software default, and 2 is the base of logarithm, the index range of parameter c is [−1, 6], the index range of parameter g is [−8, 0], and the index range of parameter p is [−1, −8]. Normalize the data to [−1, 1] before training the model to correctly reflect the actual situation of the data.
Since R, dCor, Chi-MIC and mRMR cannot automatically select feature subsets, two heuristic forward selection methods were used to filter the final feature subsets. (a) Introduce one forward feature at a time, and for each feature introduced, 5-fold cross-validation was implemented on the training set by machine learning algorithms such as support vector machine until all features were introduced, and no features were removed in this process. The features with highest accuracy after cross-validation were selected for the optimal feature subset;26 (b) introduce one forward feature at a time, retained those that were useful and eliminated the useless until all features were traversed. In this process, features that cannot improve cross-validation accuracy were eliminated.27 When there are too many features, forward selection methods are time-consuming, so we selected the top 100 features and then performed forward selection methods on the training set.
Table 4 shows that Chi-MIC-share is superior to the reference feature selection methods in all three datasets. There is no obvious difference among the three univariate filter methods because the feature selection process is affected by many factors. First, the univariate screening methods ignore the correlation between features, and at the same time selecting features with strong correlation will lead to deviations in prediction accuracy. Second, heuristic search does not traverse all features, and it is easy to fall into the local optimal. In addition, relying on a learning machine to search for a subset of features may lead to overfitting of training model. The multivariable screening method mRMR does not show advantages over the univariate screening methods, indicating that the redundancy is not removed correctly. Chi-MIC-share considers redundant allocation among features, does not rely on learning machine, and uses a complete search in the feature space. Experimental results show the superiority of this algorithm.
Feature selection and independent prediction accuracy of SVR model.
| Methods | Feature number | Dataset 1 | Feature number | Dataset 2 | Feature number | Dataset 3 | |||
|---|---|---|---|---|---|---|---|---|---|
| MSE | R 2 | MSE | R 2 | MSE | R 2 | ||||
| All | 1219 | 0.1066 | 0.7793 | 1323 | 0.1740 | 0.8389 | 1360 | 0.1709 | 0.7468 |
| R a | 19 | 0.0626 | 0.8686 | 65 | 0.0489 | 0.9658 | 91 | 0.3431 | 0.4541 |
| R b | 20 | 0.0994 | 0.8121 | 18 | 0.0477 | 0.9503 | 37 | 0.3655 | 0.4445 |
| dCora | 49 | 0.0948 | 0.7873 | 88 | 0.0283 | 0.9733 | 100 | 0.2358 | 0.6212 |
| dCorb | 15 | 0.0701 | 0.8368 | 42 | 0.0229 | 0.9767 | 25 | 0.1640 | 0.7518 |
| Chi-MICa | 86 | 0.0985 | 0.7842 | 61 | 0.0561 | 0.9467 | 82 | 0.2488 | 0.5975 |
| Chi-MICb | 27 | 0.1387 | 0.7029 | 34 | 0.0791 | 0.9716 | 15 | 0.4184 | 0.3631 |
| mRMRa | 15 | 0.1339 | 0.7180 | 98 | 0.1088 | 0.8876 | 70 | 0.1686 | 0.7503 |
| mRMRb | 13 | 0.1291 | 0.7188 | 26 | 0.1139 | 0.8578 | 11 | 0.2968 | 0.5607 |
| Chi-MIC-share | 15 | 0.0280 | 0.9590 | 27 | 0.0226 | 0.9750 | 22 | 0.0454 | 0.9367 |
Forward selection method without culling feature.
Forward selection method with culling feature.
3.3. Comparison of the results of this article with references
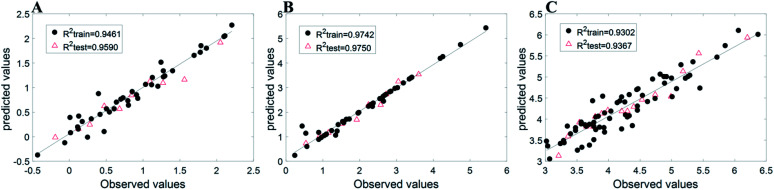
Due to differences in descriptors and evaluation indicators, we simply compared the results with related published reports. For Dataset 1, the MSE of the present work is 0.0280 and R2 is 0.9590; the previously reported values21 are 0.0424 and 0.919, respectively. For Dataset 2, we found that R2 = 0.9750, and a previously reported value22 is 0.8949. For Dataset 3, we found that R2 is 0.9367, and a previously reported value23 is 0.9197. We can see that our independent predictions of SVR based on Chi-MIC-share are better than those reported in the literature. Fig. 2 shows the distribution of observed values and predicted values for the three datasets, confirming the results.
Fig. 2. Observed values and predicted values of three datasets.
3.4. Model significance test and single-factor effect analysis
Taking Dataset 1 as an example, the 15 reserved descriptors selected by Chi-MIC-share established the SVR model, F = 21.93 > F0.01(15, 40) = 2.1076, indicating that the model is significant. Table 5 shows the reserved descriptors and the corresponding group names and explanations, and Fig. 3 shows the single-factor effects of the 15 reserved descriptors in Dataset 1.
Fifteen reserved descriptors in Dataset 1.
| Group name | Descriptor name | Explanation |
|---|---|---|
| Molecular properties | BLTF96 | Verhaar model of algae base-line toxicity from MLOGP (mmol l−1) |
| 3D-MoRSE descriptors | Mor30p | 3D-MoRSE-signal 30/weighted by atomic polarizabilities |
| Mor16m | 3D-MoRSE-signal 16/weighted by atomic masses | |
| Mor28m | 3D-MoRSE-signal 28/weighted by atomic masses | |
| Mor18m | 3D-MoRSE-signal 18/weighted by atomic masses | |
| Mor21m | 3D-MoRSE-signal 21/weighted by atomic masses | |
| Geometrical descriptors | SPAN | span R |
| L/Bw | Length-to-breadth ratio by WHIM | |
| WHIM descriptors | Am | A total size index/weighted by atomic masses |
| Atom-centered fragments | H-047 | H attached to C1(sp3)/C0(sp2) |
| C-024 | R–CH–R | |
| 2D autocorrelations | ATS5p | Broto–Moreau autocorrelation of a topological structure-lag 5/weighted by atomic polarizabilities |
| GATS3e | Geary autocorrelation-lag 3/weighted by atomic Sanderson electronegativities | |
| GETAWAY descriptors | R5p+ | R maximal autocorrelation of lag 5/weighted by atomic polarizabilities |
| HATS5u | Leverage-weighted autocorrelation of lag 5/unweighted |
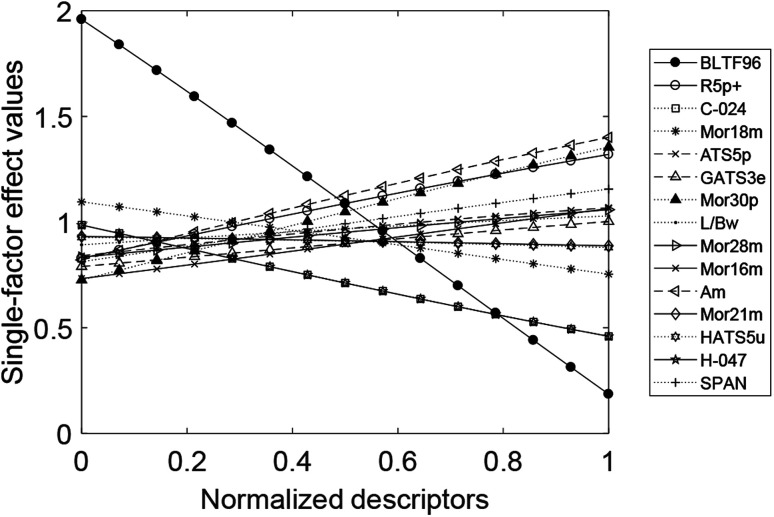
Fig. 3. Single-factor effects of the 15 reserved descriptors in Dataset 1.
For the 15 reserved descriptors, the molecular global descriptors are molecular properties, three-dimensional molecule representation of structure based on electron diffraction (3D-MoRSE) descriptors, geometrical descriptors, and weighted holistic invariant molecular (WHIM) descriptors. The molecular local descriptor is atom-centered fragments. Molecular combination descriptors are two-dimensional autocorrelations and geometry, topology, and atom weights assembly (GETAWAY) descriptors. BLTF96 is the n-octanol/water partition coefficient, which is a parameter in measuring the lipophilicity of organic compounds in water. Experiments have shown that the n-octanol/water partition coefficient is strongly correlated with various toxicological properties of compounds.28 Mor30p, Mor16m, Mor28m, Mor18m, and Mor21m29–34 are atomic polarity parameters and atomic mass parameters; SPAN and L/Bw descriptors35 reflect geometrical features such as molecular surface area, volume, and stereoscopic parameters. H-047 and C-024 36 highlight the importance of hydrogen and carbon atoms in influencing the negative log half-maximal inhibition growth concentration, as they participate in intermolecular interactions through hydrogen bonds in the solid state. ATS5p and GATS3e37,38 are vector descriptors that are based on the two-dimensional structure of a molecule and the properties of atomic pairs. R5p+ and HATS5u39,40 characterize the distribution of atomic properties on a topological structure, which is a combination of geometry, topology, and atomic components. The effects of these parameters on chemical compounds have been reported in the literature.
WHIM descriptors are new three-dimensional molecular property indices, contain information about the molecular structure of a chemical compound in terms of size, shape, symmetry, and atom distribution. Am is the total molecular volume parameter of WHIM descriptors, and our findings demonstrate that its effects cannot be ignored.
Fig. 3 displays the single-factor effect. The factors that are positively correlated with the effects of phenolic compounds on T. pyriformis are Mor30p, Am, R5p+, SPAN, ATS5p, Mor28m, Mor16m, L/Bw, and GATS3e. The factors that are negatively correlated with the effects are BLTF96, H-047, C-024, Mor18m, Mor21m, and HATS5u.
4. Conclusions
When the features of sample contain massive irrelevant or redundant information, the performance of the mathematical model is seriously affected. By constructing more reasonable and reliable correlation or significance metrics, feature selection methods can single out the most suitable feature subset from high-dimensional features, reduce the complexity of the model, avoid over-fitting, and enhance the interpretability of the model. MIC can be used to measure the correlation compactness between variables, has universality and equivalence, and relevant studies have shown its effectiveness.
In this paper, we used the redundancy allocation algorithm with Chi-MIC to automatically filter trusted features, then verified the superiority of redundancy allocation over de-redundancy using experimental datasets. Dynamic calculation of share score is an important part of the chi-MIC-share algorithm. First, it does not rely on the learning machine, but only uses the constructed statistics to filter features. Second, based on the overall features, it comprehensively weighs the change of each feature score in the original set after a new feature is introduced, and the impact of such changes on the entire new set. This dynamic adjustment will prevent the subset score from increasing all the time. When a vertex is reached, the feature selection process is also terminated. This may provide a new idea for quantitative research and has certain reference value.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 61701177 and No. 31701164), the Natural Science Foundation of Hunan Province, China (No. 2018JJ3225 and No. 2018JJ3238) and Scientific Research Foundation of Education Office of Hunan Province, China (No. 17A096).
Notes and references
- Zhou W. Fan Y. Cai X. et al., High-accuracy QSAR Models of Narcosis Toxicities of Phenols Based on Various Data Partition, Descriptor Selection and Modelling Methods. RSC Adv. 2016;6(108):106847–106855. doi: 10.1039/C6RA21076G. [DOI] [Google Scholar]
- Gupta S. Basant N. Singh K. P. Predicting aquatic toxicities of benzene derivatives in multiple test species using local, global and interspecies QSTR modeling approaches. RSC Adv. 2015;5(87):71153–71163. doi: 10.1039/C5RA12825K. [DOI] [Google Scholar]
- He J. Tang Z. Zhao Y. et al., The Combined QSAR-ICE Models: Practical Application in Ecological Risk Assessment and Water Quality Criteria. Environ. Sci. Technol. 2017;51(16):8877–8878. doi: 10.1021/acs.est.7b02736. [DOI] [PubMed] [Google Scholar]
- Toropov A. A. Toropova A. P. Pizzo F. et al., CORAL: model for no observed adverse effect level (NOAEL) Mol. Diversity. 2015;19(3):563–575. doi: 10.1007/s11030-015-9587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. Green D. V. S. Luscombe C. N. et al., QSAR workbench: automating QSAR modeling to drive compound design. J. Comput.-Aided Mol. Des. 2013;27(4):321–336. doi: 10.1007/s10822-013-9648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damme S. V., Quantum chemistry in QSAR: quantum chemical descriptors: use, benefits and drawbacks, Ghent University. Faculty of Sciences, 2009 [Google Scholar]
- Liu H. Chen Q. Zhang S. et al., Relationship of mineralization of amino naphthalene sulfonic acids by Fenton oxidation and frontier molecular orbital energies. Chem. Eng. J. 2014;247:275–282. doi: 10.1016/j.cej.2014.03.019. [DOI] [Google Scholar]
- Tetko I. V. Gasteiger J. Todeschini R. Mauri A. Livingstone D. Ertl P. Palyulin V. A. Radchenko E. V. Zefirov N. S. Makarenko A. S. Tanchuk V. Y. Prokopenko V. V. Virtual computational chemistry laboratory - design and description. J. Comput.-Aided Mol. Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- Algamal Z. Y. Lee M. H. A new adaptive L1-norm for optimal descriptor selection of high-dimensional QSAR classification model for anti-hepatitis C virus activity of thiourea derivatives. SAR QSAR Environ. Res. 2017;28(1):75–90. doi: 10.1080/1062936X.2017.1278618. [DOI] [PubMed] [Google Scholar]
- Tang B. Kay S. He H. et al., Toward Optimal Feature Selection in Naive Bayes for Text Categorization. IEEE Trans. Knowl. Data Eng. 2016;28(9):2508–2521. [Google Scholar]
- Guyon I. Elisseeff A. AndrAndr Andr Knowledge and Data EnginSelection. J. Mach. Learn. Res. 2003;3(6):1157–1182. [Google Scholar]
- Peng H. Long F. Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- Reshef D. N. Reshef Y. A. Finucane H. K. et al., Detecting novel associations in large data sets. Science. 2011;334(6062):1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C. Ying Z. Feng L. Zheming Y. A New Algorithm to Optimize Maximal Information Coefficient. PLoS One. 2016;11(6):e0157567. doi: 10.1371/journal.pone.0157567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inf. 2010;29(6–7):476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Dai Z. Wang L. Chen Y. et al., A pipeline for improved QSAR analysis of peptides: physiochemical property parameter selection via BMSF, near-negighbor sample selection via semivariogram, and weighted SVR regression and prediction. Amino Acids. 2014;46(4):1105–1119. doi: 10.1007/s00726-014-1667-5. [DOI] [PubMed] [Google Scholar]
- Wang L. Xing P. Wang C. Zhou X. Dai Z. Bai L. Maximal information coefficient and support vector regression based nonlinear feature selection and QSAR modeling on toxicity of alcohol compounds to tadpoles of rana temporaria. J. Braz. Chem. Soc. 2019;30(2):279–285. [Google Scholar]
- Zhou W. Wu S. Dai Z. et al., Nonlinear QSAR models with high-dimensional descriptor selection and SVR improve toxicity prediction and evaluation of phenols on Photobacterium phosphoreum. Chemom. Intell. Lab. Syst. 2015;145:30–38. doi: 10.1016/j.chemolab.2015.04.010. [DOI] [Google Scholar]
- Wang L. Dai Z. Zhang H. et al., Quantitative sequence-activity model analysis of oligopeptides coupling an improved high-dimension feature selection method with support vector regression. Chem. Biol. Drug Des. 2014;83(4):379–391. doi: 10.1111/cbdd.12242. [DOI] [PubMed] [Google Scholar]
- Wang L. F. Tan X. S. Bai L. Y. et al., Establishing an Interpretability System for Support Vector Regression and Its Application in QSAR of Organophosphorus Insecticide. Asian J. Chem. 2012;24(4):1575–1578. [Google Scholar]
- Deng X. Chen Y. Tan S. et al., QSAR study on toxicities of alcohol and phenol compounds. Acta Sci. Circumstantiae. 2016;36(12):4490–4499. [Google Scholar]
- Li B. The relationship between anesthesia active for tadpole and structural parameters of organic compounds. Comput. Appl. Chem. 2004;21(2):232–234. [Google Scholar]
- Li Q. Yang Y. Zhang S. DFT study on the structure of aromatic derivatives and its toxicity to fathead minnows. J. Shaanxi Norm. Univ., Nat. Sci. Ed. 2016;044(006):43–47. [Google Scholar]
- Tetko I. V. Gasteiger J. Todeschini R. et al., Virtual Computational Chemistry Laboratory-Design and Description. J. Comput.-Aided Mol. Des. 2005;19(6):453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- Yuan Z. Yang J. Chen Y. A novel feature selection method based on maximum information coefficient and redundancy sharing. Comput. Eng. 2019:1–8. [Google Scholar]
- Ding C. Peng H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinf. Comput. Biol. 2005;3(02):185–205. doi: 10.1142/S0219720005001004. [DOI] [PubMed] [Google Scholar]
- Zhang H. Li L. Luo C. et al., Informative gene selection and direct classification of tumor based on chi-square test of pairwise gene interactions. BioMed Res. Int. 2014:1–9. doi: 10.1155/2014/589290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi I. Hirono S. Liu Q. et al., Simple Method of Calculating Octanol/Water Partition Coefficient. Chem. Pharmaceut. Bull. 1992;40(1):127–130. doi: 10.1248/cpb.40.127. [DOI] [Google Scholar]
- Arab C. M. Modelling of cytotoxicity data (CC50) of anti-HIV 1-[5-chlorophenyl) sulfonyl]-1H-pyrrole derivatives using calculated molecular descriptors and Levenberg-Marquardt artificial neural network. Chem. Biol. Drug Des. 2010;73(4):456–465. doi: 10.1111/j.1747-0285.2009.00790.x. [DOI] [PubMed] [Google Scholar]
- Le T. C. Yan B. Winkler D. A. Robust Prediction of Personalized Cell Recognition from a Cancer Population by a Dual Targeting Nanoparticle Library. Adv. Funct. Mater. 2015;25(44):6927–6935. doi: 10.1002/adfm.201502811. [DOI] [Google Scholar]
- Deshpande S. Goodarzi M. Katti S. B. et al., Topological Features in Profiling the Antimalarial Activity Landscape of Anilinoquinolines: A Multipronged QSAR Study. J. Chem. 2012;2013(1):1–14. [Google Scholar]
- Sun M. Chen J. Cai J. et al., Simultaneously Optimized Support Vector Regression Combined With Genetic Algorithm for QSAR Analysis of KDR/VEGFR-2 Inhibitors. Chem. Biol. Drug Des. 2010;75(5):494–505. doi: 10.1111/j.1747-0285.2010.00958.x. [DOI] [PubMed] [Google Scholar]
- Ghasemi G. Arshadi S. Nemati Rashtehroodi A. Nirouei M. Shariati S. Rastgoo Z. QSAR Investigation on Quinolizidinyl Derivatives in Alzheimer’s Disease. J. Comput. Med. 2013;2013:8. [Google Scholar]
- Duchowicz P. R. Fernandez M. Caballero J. et al., QSAR for non-nucleoside inhibitors of HIV-1 reverse transcriptase. Bioorg. Med. Chem. 2006;14(17):5876–5889. doi: 10.1016/j.bmc.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Massarelli I. Macchia M. Minutolo F. et al., QSAR models for predicting enzymatic hydrolysis of new chemical entities in 'soft-drug' design. Bioorg. Med. Chem. 2009;17(10):3543–3556. doi: 10.1016/j.bmc.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Diez R. P. Duchowicz P. R. Castañeta H. et al., A theoretical study of a family of new quinoxaline derivatives. J. Mol. Graph. Model. 2007;25(4):487–494. doi: 10.1016/j.jmgm.2006.03.004. [DOI] [PubMed] [Google Scholar]
- GonzJournal oSunzJo P. L. Fall Y. et al., Quantitative structure-activity relationship studies of vitamin D receptor affinity for analogues of 1alpha,25-dihydroxyvitamin D3. 1: WHIM descriptors. Bioorg. Med. Chem. Lett. 2005;15(23):5165–5169. doi: 10.1016/j.bmcl.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Yousefinejad S. Honarasa F. Nekoeinia M. et al., Investigation and Modeling of the Solubility of Anthracene in Organic Phases. J. Solution Chem. 2017;46(2):352–373. doi: 10.1007/s10953-017-0568-0. [DOI] [Google Scholar]
- Cheng Z. Zhang Y. Zhou C. QSAR Models for Phosphoramidate Prodrugs of 2′-Methylcytidine as Inhibitors of Hepatitis C Virus Based on PSO Boosting. Chem. Biol. Drug Des. 2011;78(6):948–959. doi: 10.1111/j.1747-0285.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- Zheng F. Zheng G. Deaciuc A. G. et al., Computational neural network analysis of the affinity of lobeline and tetrabenazine analogs for the vesicular monoamine transporter. Bioorg. Med. Chem. 2007;15(8):2975–2992. doi: 10.1016/j.bmc.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]