Abstract
Pseudomonas aeruginosa biofilms are intrinsically resistant to antimicrobial chemotherapies. At present, very little is known about the physiological changes that occur during the transition from the planktonic to biofilm mode of growth. The resistance of P. aeruginosa biofilms to numerous antimicrobial agents that are substrates subject to active efflux from planktonic cells suggests that efflux pumps may substantially contribute to the innate resistance of biofilms. In this study, we investigated the expression of genes associated with two multidrug resistance (MDR) efflux pumps, MexAB-OprM and MexCD-OprJ, throughout the course of biofilm development. Using fusions to gfp, we were able to analyze spatial and temporal expression of mexA and mexC in the developing biofilm. Remarkably, expression of mexAB-oprM and mexCD-oprJ was not upregulated but rather decreased over time in the developing biofilm. Northern blot analysis confirmed that these pumps were not hyperexpressed in the biofilm. Furthermore, spatial differences in mexAB-oprM and mexCD-oprJ expression were observed, with maximal activity occurring at the biofilm substratum. Using a series of MDR mutants, we assessed the contribution of the MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY efflux pumps to P. aeruginosa biofilm resistance. These analyses led to the surprising discovery that the four characterized efflux pumps do not play a role in the antibiotic-resistant phenotype of P. aeruginosa biofilms.
Pseudomonas aeruginosa is a leading cause of nosocomial infections that are difficult to eradicate because of the organism's inherent resistance to a multitude of antibiotics. Initially, this intrinsic resistance was believed to result primarily from low outer membrane (OM) permeability (27). However, in bacteria grown planktonically, it is now known to be the combined action of multidrug resistance (MDR) pumps and decreased OM permeability that confers this resistance (20). In P. aeruginosa, four efflux pumps have been described, MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY (17, 18, 32, 34). The genes encoding these pumps are arranged as operons, with the first gene encoding a membrane fusion protein that is associated with the cytoplasmic membrane (MexA, MexC, MexE, and MexX). The second gene encodes the transporter (MexB, MexD, MexF, and MexY) thought to export the substrate across the inner membrane. The third gene encodes an OM protein (OprM, OprJ, and OprN) that facilitates passage of the substrate across the OM. Together, the three pump proteins form a channel that traverses the inner membrane and the OM and allows the target to be effluxed directly from the cytoplasm to the extracellular environment. In the case of the mexXY operon, there is no gene encoding the OM component; rather, MexXY appears to share the OprM channel with MexAB (1, 25). However, Westbrock-Wadman et al. (49) suggest that OprM is not the OM channel for AmrAB (MexXY).
Among these four pumps, MexAB-OprM is the only one believed to be constitutively expressed in wild-type P. aeruginosa. As such, MexAB-OprM contributes to the intrinsic resistance of this organism to a number of antimicrobial agents, including tetracycline, chloramphenicol, quinolones, novobiocin, macrolides, trimethoprim, β-lactams and β-lactamase inhibitors (11, 16, 18, 19, 34). Moreover, hyperexpression of mexAB-oprM has been found in MDR clinical isolates that result because of mutations acquired in a repressor gene, mexR (35, 51). In contrast, it is thought that mexCD-oprJ, mexEF-oprN, and mexXY are not expressed during normal laboratory growth. (Note that Westbrock-Wadman et al. [49] suggested that MexXY was expressed in the lab but at much reduced levels.) Expression of mexCD-oprJ only occurs in strains with mutations in nfxB, which encodes a repressor of this system (29, 41). These strains are resistant to chloramphenicol, macrolides, novobiocin, quinolones, tetracycline, and cephems, but exhibit hypersusceptibility to many β-lactams (13, 23). The mexEF-oprN operon is expressed in nfxC-type mutants and is positively regulated by the activator MexT (15, 17). It has been proposed that MexT requires a cognate effector to become active, and in nfxC mutants this effector molecule is expressed at levels sufficient for MexT activation (15). Strains with mutations in nfxC exhibit increased resistance to chloramphenicol, quinolones, trimethoprim, and carbapenems (10, 17). The most recently described P. aeruginosa efflux pump, MexXY, has been found to impart resistance to aminoglycosides, erythromycin, and fluoroquinolones (1, 25, 49).
The intrinsic resistance of P. aeruginosa to numerous antimicrobial agents is even more pronounced when this organism is found growing in a biofilm. Antimicrobial resistance is a trait typical of most biofilm organisms and it has been speculated that biofilms are the causative agent of up to 65% of bacterial infections (36). Biofilms are thought to become recalcitrant to antimicrobial assault through a number of different mechanisms. In some instances, increased resistance may be caused by poor diffusion of antibiotics through the biofilm polysaccharide matrix (45). However, studies have shown that many antibiotics diffuse completely through the biofilm but with a reduced rate of transfer (7, 26, 45). In addition, the physiology of biofilm cells is remarkably heterogeneous and varies according to the location of individual cells within the biofilm (44). Cells located at the biofilm surface presumably have adequate supplies of nutrients and are metabolically active, while deeply embedded cells are likely to be metabolizing more slowly due to potential nutrient and oxygen limitation. Because many antimicrobial agents require actively metabolizing cells to be effective, the presence of slow growing or dormant cells is thought to represent a resistant population (4). Finally, it has been suggested that bacteria growing in a biofilm undergo distinct phenotypic changes associated with surface-attached growth that render them more resistant. At present, very little is known about the genotypic and/or phenotypic changes that occur as cells transition from the planktonic to the biofilm mode of growth. Because MDR pumps play an important role in the resistance of planktonic P. aeruginosa to antimicrobial agents, it seems logical that attachment to surfaces and adopting the biofilm mode of growth might signal cells to increase the expression of efflux pumps (2). In this study, the expression of two P. aeruginosa efflux pumps, MexAB-OprM and MexCD-OprJ, was examined throughout the course of biofilm development. Furthermore, we assessed the contribution of the four P. aeruginosa MDR efflux pumps to biofilm resistance to a number of antibiotics.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 1. To generate pmexA-gfp, the 400-bp SacI-NruI fragment containing the mexA promoter from pPCS952 (40) was cloned into SacI- and SmaI-cut pBluescript KS, creating intermediate plasmid pmexA-BS. The 400-bp SacI-EcoRV fragment from pmexA-BS was then ligated to SacI- and SmaI-cut pJK1, resulting in pmexA-gfp. For pmexA-lacZ, the 400-bp EcoRI-BamHI fragment containing the mexA promoter from pmexA-gfp was cloned into the same sites of pLP170. Plasmid pmexC-gfp was constructed by cloning the mexC promoter from pBSmexC.391 (H. Schweizer, Colorado State University) on a 400-bp KpnI-BamHI fragment into the KpnI-BamHI sites of pJK1. To create pmexC-lacZ, the 400-bp XhoI-BamHI fragment from pBSmexC.391 was ligated into the same sites of pLP170. Plasmid pRSP47 was obtained after cloning a 5-kb BamHI-SstI DNA fragment containing mexC from plasmid pRSP38 (43) into the broad-host-range vector pAK1900 (A. Kropinski, Queen's University). Purification, cloning, electrophoresis, and other manipulations of DNA were performed using standard techniques (12, 38).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Wild type | 14 |
| K767 | Wild-type PAO1 | 22 |
| K1119 | K767 ΔmexAB-oprM; MexAB-OprM deletion strain | 19 |
| OCR1 | K767 nalB; MexAB-OprM-overproducing strain | 22 |
| K1521 | K767 ΔmexCD-oprJ; MexCD-OprJ deletion strain | This study |
| K1536 | K767 nfxB; MexCD-OprJ-overproducing strain | K. Poole, unpublished data |
| K1240 | PAO2375 nfxC; MexEF-OprN-overproducing strain | 10 |
| K1241 | PAO2375 | 10 |
| K1525 | K767 ΔmexXY; MexXY deletion strain | This study |
| Plasmids | ||
| pLP170 | Promoterless lacZ transcriptional fusion vector | 37 |
| pmexA-lacZ | pLP170 containing a mexA-lacZ transcriptional fusion; Ampr | This study |
| pmexC-lacZ | pLP170 containing a mexC-lacZ transcriptional fusion; Ampr | This study |
| pJK1 | Promoterless GFPmut3.1 transcriptional fusion vector, contains an origin of replication for both P. aeruginosa and E. coli; Ampr | B. Iglewski, unpublished data |
| pmexA-gfp | pJK1 containing a mexA-gfp transcriptional fusion; Ampr | This study |
| pmexC-gfp | pJK1 containing a mexC-gfp transcriptional fusion; Ampr | This study |
| poprM | Plasmid poprM contains a 639-bp PCR fragment internal to oprM cloned into the EcoRV site of pBluescript SK; origin of the 660-bp oprM-specific DNA probe | This study |
| pRSP47 | Origin of the 2,000-bp mexCD-specific DNA probe | This study |
Construction of P. aeruginosa mex deletion strains.
For constructing the P. aeruginosa genetic deletions, the deletions were first made in sacB-containing vectors and were subsequently introduced into the chromosome of strain K767 by a previously defined gene replacement approach (39). To construct the mexXY deletion strain K1525, two segments of the mexXY operon were amplified from chromosomal DNA by PCR with Vent DNA polymerase (New England Biolabs) as described previously (43). One primer pair, mexzbl (5′-AAG CTT AAG CTT GCG TTC GCA CTT GAG GTA GAG-3′, with HindIII sites underlined), which anneals to the regulatory gene mexZ, and mexgb1 (5′-ACG CGG ATC CGT TCT CGA CGA TCA CCC ACT C-3′), which anneals to the mexX gene, generated the 5′ portion of the mexXY operon. Another primer pair, mexhfl (5′-ACG CGG ATC CCT GGA TGC TGG TCT ACA CCC T-3′) and mexhbl (5′-A CCG GAA TTC CAC CAG GAA GAA CAG CGG TAC-3′, with the EcoRI site underlined), both of which anneal to the mexY gene, generated the 3′ portion of the mexXY operon. Cloning of these two fragments into HindIII- and EcoRI-restricted gene replacement vector pEX18Tc (H. Schweizer, Colorado State University) generated the mexXY deletion construct, pCSV05, which had 1.7 kb of the mexXY coding sequences removed. Following transformation of Escherichia coli S17-1 (42), pCSV05 was mobilized into K767 via conjugation (33), and transconjugants carrying a copy of pCSV05 were selected on Luria-Bertani (LB) agar containing 100 μg of tetracycline per ml. Subsequent streaking of transconjugants onto LB agar containing 10% (wt/vol) sucrose yielded isolated colonies that had lost pEX18Tc sequences (tetracycline sensitive) and which carried either an unaltered wild-type copy of mexXY or the mexXY deletion. Those carrying a deletion in mexXY were identified by PCR using primers mexzb1 and mexhb1. The mexCD-oprJ deletion strain K1521 was constructed using the mexCD-oprJ deletion construct pRSP05 by a strategy similar to the one described above which was published previously (43). In this instance, however, the selection of transconjugants carrying a copy of pRSP05 was done on LB agar containing 1.5 mg of kanamycin per ml.
β-Galactosidase assays.
Overnight cultures were diluted 1 to 100 in peptone Trypticase soy broth medium (28) and allowed to grow to an optical density of 1.5 to 1.7. Where stated, the medium was supplemented with carbenicillin (200 μg/ml). β-Galactosidase activity was assayed in triplicate as described by Miller (24).
Flow chamber experiments.
The FAB medium used for flowthrough biofilm studies consisted of minimal salts (0.1 mM CaCl2, 0.01 mM Fe-EDTA, 0.15 mM (NH4)SO4, 0.33 mM Na2HPO4, 0.2 mM KH2PO4, 0.5 mM NaCl, and 1 mM MgCl2, added after autoclaving), with 10 mM sodium citrate as the carbon source. Carbenicillin was added at 200 μg/ml for plasmid maintenance. Polycarbonate flowcells (Protofab, Bozeman, Mont.) were employed for biofilm cultivation and microscopic analyses. The flowcells had a channel size of 1.6 by 11.1 by 38 mm, with tapered ends and an overall volume of 1.36 ml. Glass coverslips (40 by 60 mm; no. 1) were used in the flowcells as the biofilm substratum, with silicon tubing utilized throughout the remainder of the system. A peristaltic pump was employed to maintain a flow rate of 0.21 ml/min, yielding a residence time within the flowcells of 6.5 min. A polycarbonate bubble trap was used on each line between the pump and the flowcell to eliminate disruption of the biofilm by air bubbles (6). Three flowcells were run per experiment and were examined on days 4, 6, and 8. At the respective sampling time, a flowcell was stained with a propidium iodide (PI) (2 μM)-Syto85 (5 μM) emulsion (Molecular Probes, Eugene, Oreg.). Syto85 is a cell-permeant nucleic acid stain that stains both live and dead cells, while propidium iodide is a cell-impermeant, intercalating nucleic acid dye generally excluded from viable cells. In our hands, this dual-component emulsion is the most effective for labeling the total cell population. The emulsion (2 ml per flowcell) was slowly injected immediately upstream of the flowcell, allowed to stain cells for 30 min under quiescent conditions, and then flushed with fresh medium for 30 min. After flushing, the flowcell was permanently disconnected from the system prior to microscopic analysis. Each experiment was done in duplicate.
Microscopy.
All flowcells were nondestructively analyzed using a scanning confocal laser microscope (SCLM) equipped with dual photon lasers (Leica Lasertechnik, GmbH, Heidelberg, Germany). Images were collected at 488 and 545 nm to record bacteria emitting green fluorescent protein (GFP) and total cells stained with PI-Syto85, respectively. Scans were taken through the biofilm that had accumulated on the glass surface at varying depths from the substratum to the biofilm surface. Typically, optical sections were taken every 0.5 μm. Five areas were analyzed per flowcell. In each area, a depth profile was constructed based on these compiled scans.
Image analysis.
Each set of confocal images for a given area was further analyzed by image analysis. Percent expression levels were calculated by dividing pixel locations above background in the GFP (green) signal by the same pixel locations above background in the PI-Syto85 (red) signal. These values were generated for each “slice” of the biofilm and then compiled to create a vertical distribution of gene expression. This analysis was done using CellComp software (University of Rochester, Rochester, N.Y.). This software measured the percent similarity between corresponding GFP and PI-Syto85 optical sections. The images were first converted to binary data according to user-specified thresholds. A pixel was marked active if its intensity exceeded the threshold and passive if it did not. Once the images were converted to binary data, correspondence was computed at each scan depth according to the following formula:
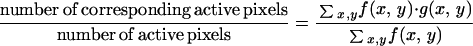 |
where f(x, y) and g(x, y) represent the binary functions of the PI-Syto85 and GFP scans, respectively. The output of CellComp was then fed into a Microsoft Excel Visual Basic macro that graphed correspondence (percent expression) against height.
Normalization.
Percent expression values were normalized due to the variance in promoter strength between the mexA and mexC constructs as well as the difference in signal intensities recorded in the 488- and 545-nm channels. To determine the normalized value for each plasmid construct, 1-ml aliquots from a 24-h culture harboring each construct were washed in phosphate-buffered saline, resuspended in the PI-Syto 85 emulsion for 30 min and then fixed with formalin. Wet mounts were analyzed via confocal microscopy. Ten images were collected for each construct. Images were processed as previously described. Correspondence values were then considered to be the maximal values attainable for each construct. Based on these numbers, percent expression values from the flowcell studies were adjusted accordingly.
Northern blot analysis.
Northern blot analysis was performed as previously described (46). Briefly, RNA was extracted from biofilm and planktonic populations of P. aeruginosa strain PAO1 grown in a minimal biofilm eradication concentration (MBEC) device (MBEC Biofilm Technologies Limited) at 4, 8, 18, and 24 h of growth. Additional biofilm samples were taken following 48, 72, and 96 h of growth to assess transcription in mature biofilms. Fifteen micrograms of RNA from each time point was transferred to a Nytran membrane using a Schleicher and Schuell slot blot apparatus. Transcript accumulation of the mexAB-oprM operon was monitored using a 32P-labeled oprM-specific DNA probe made from a 660-bp HindIII-PstI fragment of poprM. A 32P-labeled 2,000-bp mexCD gene probe generated from KpnI- and SstI-digested pRSP47 was used to measure transcript accumulation of the mexCD-oprJ operon. The blots were exposed to Kodak XAR-5 imaging film and the intensities of the hybridization bands were measured by laser scanning densitometry.
Antibiotic susceptibility tests. (i) Antibiotic preparation.
The following antibiotics were used for antibiotic susceptibility testing: aztreonam (ICN Biomedicals Inc.), chloramphenicol, ciprofloxacin (Bay Leverskusen Pharmaceuticals), erythromycin (Sigma), gentamicin-sulfate (Sigma), piperacillin (Sigma), tetracycline (Sigma), and tobramycin (Sigma). Antibiotics were prepared as stock solutions at 5,120 μg/ml and stored at −80°C. Antibiotics were serial diluted in cation-adjusted Mueller-Hinton broth and ranged in concentration from 1,024 to 2 μg/ml.
(ii) Biofilm susceptibility testing.
MBEC determinations were made using the MBEC device (MBEC Biofilm Technologies Limited) as described previously (5). Briefly, biofilms were grown on the lid of the MBEC device for 6 h in tryptic soy broth to attain a biofilm size of approximately 106 CFU/peg, per the manufacturer's instructions. The lid was then rinsed in 0.9% saline before transfer to antibiotic challenge plates. After antibiotic challenge for 16 to 20 h at 35°C, the biofilm plates were then transferred to 96-well microtiter plates containing 200 μl of cation-adjusted Mueller-Hinton broth/well and sonicated. Biofilm cells were released and allowed to grow overnight at 35°C. Growth in the recovery plates was assessed by turbidity. MBECs are reported as the lowest concentration of antibiotic at which there is no growth in the recovery plate.
(iii) Planktonic susceptibility testing.
MIC assays were performed using the MBEC device as described previously (5). The antibiotic concentration required to prevent growth of the planktonic population was derived by measuring the turbidity at 590 nm after incubation of the cells in antibiotic for 24 h. The MIC was defined as the lowest concentration of antibiotic in which a planktonic population could not be established by shedding of bacteria from the biofilm. Previous work has established that this method of assessing planktonic antibiotic susceptibility patterns produces identical results to NCCLS methods (5).
RESULTS
Expression of mexA-lacZ and mexC-lacZ in planktonically grown P. aeruginosa.
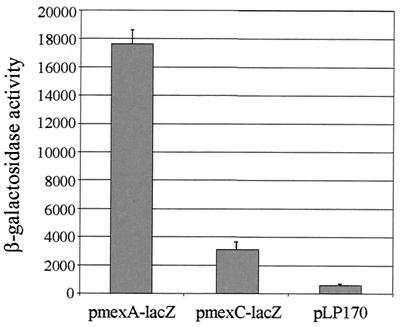
It is generally believed that of the four well-characterized P. aeruginosa efflux pumps, only MexAB-OprM is expressed in wild-type strains; the other pumps remain inactive until mutations have been acquired in repressors of these systems. To examine the basal level of mexAB-oprM and mexCD-oprJ expression in planktonically grown PAO1 cultures, mexA-lacZ and mexC-lacZ fusions were generated and their activities were monitored in strain PAO1. As illustrated in Fig. 1, mexA expression was high in planktonically grown PAO1 (approximately 17,500 Miller units). While mexC was expressed at a much lower level (approximately 3,000 Miller units), it was still significantly higher than the background activity associated with the control vector (approximately 700 Miller units). Thus, under normal laboratory conditions, mexC appears to be expressed at a low level in PAO1.
FIG. 1.
Comparison of mexA-lacZ and mexC-lacZ expression in P. aeruginosa strain PAO1 planktonic cultures. Note that while mexC is expressed at a much lower level than mexA, this level is still significantly higher than the background activity from the vector control.
Since carbenicillin is a substrate for the MexAB-OprM pump and this antibiotic was used for plasmid maintenance throughout this study, verification was done to confirm that carbenicillin (200 μg/ml) did not induce hyperexpression of MexAB-OprM. Therefore, we selected our PAO1 (pmexA-lacZ) transformants on medium containing either 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) or carbenicillin. Colonies that appeared blue on X-Gal medium were then monitored for mexA-lacZ activity in the absence of carbenicillin to compare the expression level with that of cells grown in the presence of antibiotic. In all cases, expression levels were similar (data not shown), indicating that the carbenicillin concentration used in these studies did not induce MexAB-OprM hyperexpression.
Analysis of mexAB-oprM expression during P. aeruginosa biofilm formation.
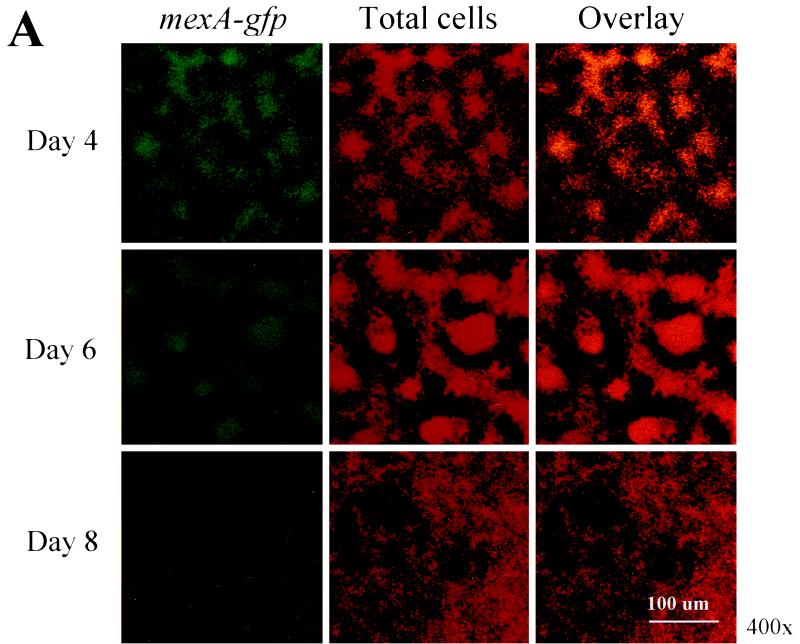
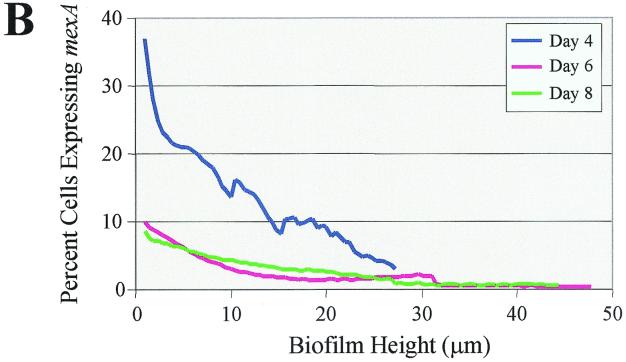
P. aeruginosa biofilms exhibit increased resistance to a number of antibiotics, including tetracycline, chloramphenicol, quinolones, β-lactams, etc., and this resistance profile closely resembles those drugs that are actively effluxed by the MexAB-OprM pump. This led us to wonder whether cells growing in a biofilm overexpress MexAB-OprM, resulting in increased resistance to a number of antimicrobial agents. To monitor mexAB-oprM expression throughout the course of biofilm development, a mexA-gfp fusion was constructed. PAO1 cells harboring mexA-gfp were grown in flowcell chambers and analyzed for mexAB-oprM expression during the course of biofilm development. On days 4, 6, and 8, the biofilms were subjected to nondestructive image analysis using SCLM to determine the percentage of cells expressing mexA-gfp relative to total cells. Analysis of the micrographs shown in Fig. 2A revealed that on day 4, a large number of cells were expressing mexAB-oprM. Nevertheless, as biofilm development proceeded, by days 6 and 8 fewer cells appeared to be expressing mexA-gfp. To more precisely address both spatial and temporal differences in mexAB-oprM expression, we subjected our SCLM images to quantitative analysis using CellComp software. This enabled us to determine the percentage of cells expressing GFP relative to total cells in a particular optical section. For quantitative analysis, the percent expression values from days 4, 6, and 8 were normalized relative to promoter expression in 24-h planktonic cultures. Since mexA-gfp is constitutively expressed, all viable cells in both the planktonic and biofilm populations presumably expressed this fusion. However, the expression level may decrease during the course of biofilm development if the cellular metabolic rate declines. Thus, if 30% of the cells exhibit mexA activity on day 4, it indicates that expression by the remaining 70% is at a level below the 24-h planktonic control. As illustrated in Fig. 2B, >35% of the cells at the substratum (0-μm depth) were expressing mexA on day 4. This number decreased to 10 and 8% on days 6 and 8, respectively, indicating that mexAB-oprM expression decreases over the course of biofilm development.
FIG. 2.
(A) Scanning confocal composite images of biofilms formed by P. aeruginosa strain PAO1 (mexA-gfp) grown for 8 days in a flowthrough chamber. Biofilms were examined on days 4, 6, and 8 to identify cells expressing mexA-gfp (green signal) relative to total cells (red signal). (B) The percentage of cells expressing mexA is plotted as a function of biofilm height. For quantitative analysis, images obtained by SCLM were analyzed using CellComp software.
We were also interested in determining whether mexAB-oprM expression varied according to biofilm height. The results of our analysis revealed that on days 4, 6, and 8, mexA-gfp activity was greatest at substratum and decreased as the height of the biofilm increased (Fig. 2B).
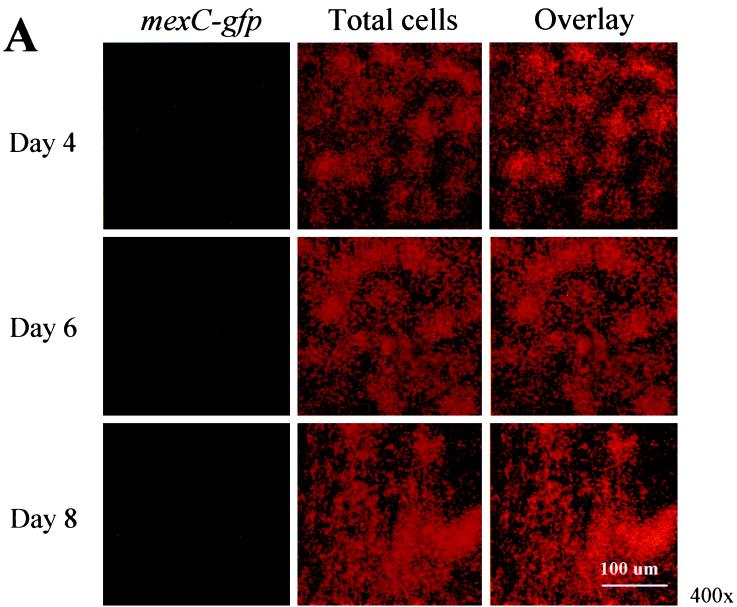
Analysis of mexCD-oprJ expression during P. aeruginosa biofilm formation.
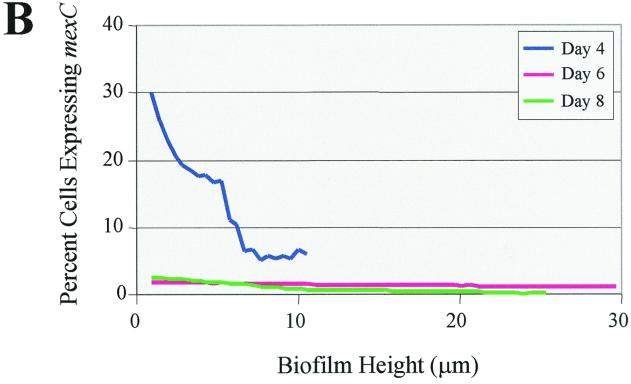
To monitor expression of mexCD-oprJ throughout the course of P. aeruginosa biofilm development, we generated a mexC-gfp fusion. Similar to what was observed for mexA, mexC-gfp activity was maximal on day 4 and markedly decreased on days 6 and 8 (Fig. 3A). When we subjected the SCLM images to quantitative analysis, the decrease in temporal expression was even more striking (Fig. 3B). On day 4, mexC was expressed in approximately 30% of the cells at the substratum, but by days 6 and 8, expression declined to <5% of the total cell population. With respect to spatial expression, mexC-gfp activity was maximal at the substratum and decreased with increasing biofilm height (Fig. 3B). Both the PAO1(mexA-gfp) and PAO1(mexC-gfp) biofilms showed a marked increase in thickness between days 4 and 6 but reached a plateau after day 6 (Fig. 2B and 3B). The biofilms formed by PAO1(pmexA-gfp) were thicker than those of PAO1(pmexC-gfp) in the duplicate experiments. At present, the reason for this difference in thickness is unclear.
FIG. 3.
(A) Scanning confocal composite images of biofilms formed by P. aeruginosa strain PAO1 (mexC-gfp) grown for 8 days in a flowthrough chamber. Biofilms were examined on days 4, 6, and 8 to identify cells expressing mexC-gfp (green signal) relative to total cells (red signal). (B) The percentage of cells expressing mexC is plotted as a function of biofilm height.
mexAB-oprM and mexCD-oprJ transcript accumulation in P. aeruginosa.
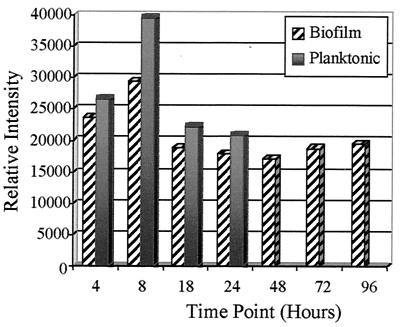
To address further the mexAB-oprM and mexCD-oprJ expression levels in planktonic and biofilm cultures of strain PAO1, Northern blot analysis was performed. For these experiments, biofilm and planktonic cultures were grown in the MBEC device, which produces 96 equivalent biofilm samples (5). Bacteria were grown in the absence of antibiotics in the MBEC device, and at various time points RNA was isolated from both planktonic and adherent populations. As illustrated in Fig. 4, examination of biofilm and planktonic transcript accumulation of mexAB-oprM revealed that this operon is expressed at approximately the same level throughout early batch culture growth, with the exception of a slight increase at 8 h. It is important to note that transcription levels in the MBEC assay biofilm populations are not markedly altered relative to planktonic bacteria.
FIG. 4.
P. aeruginosa mexAB-oprM transcript accumulation. Biofilm and planktonic cultures of strain PAO1 were assayed for the mexAB-oprM message, and relative expression levels at various time points are shown.
In the case of mexCD-oprJ, transcripts corresponding to this operon could not be detected in either biofilm or planktonic cultures of P. aeruginosa strain PAO1. These results further establish that mexCD-oprJ is not hyperexpressed in P. aeruginosa cells during biofilm growth.
Contribution of MexAB-OprM to biofilm antimicrobial susceptibility.
To assess the contribution of MexAB-OprM to biofilm antibiotic resistance, the P. aeruginosa wild type (K767) and strains that either hyperexpressed (OCR1) or lacked (K1119) the MexAB-OprM pump were allowed to form biofilms on the MBEC device for 6 h. Subsequently, the biofilms were subjected to antibiotic challenge for 16 to 20 h. MexAB-OprM played a role in the resistance of biofilms to aztreonam, gentamicin, tetracycline, and tobramycin. This is apparent since biofilms formed by P. aeruginosa OCR1 (mexAB-oprM hyperexpressing) exhibit increased resistance to each of these antibiotics relative to the wild-type strain K767 (Table 2). Similarly, each antibiotic MBEC was decreased for P. aeruginosa K1119 (ΔmexAB-oprM) relative to the K767 wild-type (PAO1). However, even in the absence of MexAB-OprM, K1119 biofilms display extremely high levels of antibiotic resistance relative to planktonic bacteria, indicating that other factors must be contributing to this resistance. Furthermore, it is important to note that the presence or absence of MexAB-OprM has a similar effect on the pattern of antibiotic susceptibility of planktonic cultures (i.e., the values merely mimic those observed in planktonic cells).
TABLE 2.
Contribution of the MexAB-OprM efflux operon to antimicrobial susceptibility of P. aeruginosa biofilms
| Antibiotic | K767 (wild type)
|
K1119 (ΔmexAB-oprM)
|
OCR1 (K767 nalB; hyperexpresses mexAB-oprM)
|
|||
|---|---|---|---|---|---|---|
| MBEC (μg/ml) | MIC (μg/ml) | MBEC (μg/ml) | MIC (μg/ml) | MBEC (μg/ml) | MIC (μg/ml) | |
| Aztreonam | 1,024 | 32 | 512 | 64 | >1,024 | 32 |
| Chloramphenicol | 1,024 | 128 | 1,024 | 64 | >1,024 | 1,024 |
| Ciprofloxacin | 16 | <2 | <2 | <2 | 8 | <2 |
| Erythromycin | 1,024 | 32 | 1,024 | 8 | >1,024 | 256 |
| Gentamicin | 16 | 4 | 4 | <2 | 32 | 4 |
| Piperacillin | 1,024 | 32 | 1,024 | 32 | >1,024 | 128 |
| Tetracycline | 512 | 64 | 128 | 64 | >1,024 | 256 |
| Tobramycin | 64 | <2 | 16 | <2 | 512 | <2 |
Contribution of MexCD-OprJ to biofilm antimicrobial susceptibility.
To investigate if mexCD-oprJ expression is induced following transition to the biofilm mode of growth, we compared the biofilm antibiotic resistance profile of P. aeruginosa K767 versus K1521 (K767 ΔmexCD-oprJ). As illustrated in Table 3, there was no difference observed in the MBECs for either of these strains. Moreover, examination of the biofilm resistance of K1536 (K767 nfxB), in which mexCD-oprJ is hyperexpressed, revealed no increase in resistance to the antibiotics tested (Table 3). These findings indicate that even when overexpressed, mexCD-oprJ does not contribute to the antimicrobial resistance of P. aeruginosa biofilms.
TABLE 3.
Contribution of the MexCD-OprJ efflux operon to antimicrobial susceptibility of P. aeruginosa biofilms
| Antibiotic | K767 (wild type)
|
K1521 (K767 ΔmexCD-oprJ)
|
K1536 (K767 nfxB; hyperexpresses mexCD-oprJ)
|
|||
|---|---|---|---|---|---|---|
| MBEC (μg/ml) | MIC (μg/ml) | MBEC (μg/ml) | MIC (μg/ml) | MBEC (μg/ml) | MIC (μg/ml) | |
| Aztreonam | 1,024 | 32 | 1,024 | 16 | 1,024 | 64 |
| Chloramphenicol | 1,024 | 128 | 1,024 | 128 | 1,024 | 256 |
| Ciprofloxacin | 16 | <2 | 16 | 2 | 16 | 2 |
| Erythromycin | 1,024 | 32 | 1,024 | 64 | 1,024 | 256 |
| Gentamicin | 16 | 4 | 64 | 4 | 64 | 4 |
| Piperacillin | 1,024 | 32 | 1,024 | 32 | 1,024 | 32 |
| Tetracycline | 512 | 64 | 512 | 128 | 512 | 128 |
| Tobramycin | 64 | <2 | 32 | 2 | 32 | 2 |
Contribution of MexEF-OprN to biofilm antimicrobial susceptibility.
As shown in Table 4, biofilms of P. aeruginosa strain K1240 (mexEF-oprN hyperexpressing) exhibit little difference in antibiotic resistance compared to those of parental strain K1241, with one exception. Ciprofloxacin resistance was shown to be much greater in strain K1240 biofilms. These findings suggest that while MexEF-OprN increases resistance to ciprofloxacin, it does not contribute to the innate antibiotic resistance of P. aeruginosa biofilms.
TABLE 4.
Contribution of the MexEF-OprN efflux operon to antimicrobial susceptibility of P. aeruginosa biofilms
| Antibiotic | Strain K1241 (parental strain expressing mexEF-oprN)
|
Strain K1240 (PAO2375 nfxC; hyperexpresses mexEF-oprN)
|
||
|---|---|---|---|---|
| MBEC (μg/ml) | MIC (μg/ml) | MBEC (μg/ml) | MIC (μg/ml) | |
| Aztreonam | >1,024 | 64 | 1,024 | 32 |
| Chloramphenicol | >1,024 | 64 | >1,024 | 64 |
| Ciprofloxacin | 2 | <2 | 64 | <2 |
| Erythromycin | >1,024 | 128 | >1,024 | 128 |
| Gentamicin | 8 | 4 | 4 | <2 |
| Piperacillin | >1,024 | 16 | >1,024 | 8 |
| Tetracycline | >1,024 | 32 | 1,024 | 64 |
| Tobramycin | 128 | <2 | 64 | <2 |
Contribution of MexXY to biofilm antimicrobial susceptibility.
The antibiotic-resistant nature of biofilms was shown to be independent of the last well-characterized efflux pump, MexXY. MBECs for K1525, a ΔmexXY derivative of K767, did not significantly differ from those for wild-type K767 (Table 5), indicating that this pump does not contribute to the antibiotic resistance of P. aeruginosa biofilms.
TABLE 5.
Contribution of the MexXY efflux operon to antimicrobial susceptibility of P. aeruginosa biofilms
| Antibiotic | K767 (wild type)
|
K1525 (K767 ΔmexXY)
|
||
|---|---|---|---|---|
| MBEC (μg/ml) | MIC (μg/ml) | MBEC (μg/ml) | MIC (μg/ml) | |
| Aztreonam | 1,024 | 32 | 512 | 16 |
| Chloramphenicol | 1,024 | 128 | >1,024 | 128 |
| Ciprofloxacin | 16 | <2 | 8 | <2 |
| Erythromycin | 1,024 | 32 | >1,024 | 32 |
| Gentamicin | 16 | 4 | 16 | <2 |
| Piperacillin | 1,024 | 32 | >1,024 | 32 |
| Tetracycline | 512 | 64 | 512 | 32 |
| Tobramycin | 64 | <2 | 32 | <2 |
DISCUSSION
The resistance of P. aeruginosa biofilms to numerous antimicrobial agents subject to active efflux from planktonic cells suggests that efflux pumps may substantially contribute to the increased resistance of biofilms. We have discovered that this is not the case, and in fact, using a number of different techniques, we demonstrate that MDR pumps play little role in the innate antibiotic resistance of P. aeruginosa biofilms. Furthermore, expression of MexAB-OprM and MexCD-OprJ is heterogeneous throughout the biofilm population, with maximal expression occurring in cells located at the substratum.
Expression of MexAB-OprM and MexCD-OprJ in planktonic and biofilm cultures.
Despite the general conviction that MexAB-OprM is the only efflux pump active in wild-type strains of P. aeruginosa, from these studies it appears that MexCD-OprJ is also expressed. In both planktonic and biofilm cultures, transcriptional fusions to lacZ and gfp, respectively, revealed mexCD-oprJ activity. Expression of mexC-lacZ in a non-MDR strain was observed earlier; however, these researchers were unable to detect OprJ using Western blot analysis (32). In this study, we were not able to detect mexCD-oprJ transcripts in either planktonic or biofilm populations. Thus, it appears that mexC activity is detectable in strain PAO1 only after fusion to genes encoding stable proteins (LacZ and GFP). Alternatively, because the mexC-lacZ reporter is on a multicopy plasmid, it is possible that the NfxB repressor is titrated out, allowing for mexCD-oprJ expression.
During the course of mature biofilm development, we discovered that mexAB-oprM and mexCD-oprJ expression does not increase but rather decreases both temporally and spatially. Using mexA- and mexC-gfp fusions, temporal studies showed that of the 3 days examined, expression of these operons was maximal on day 4 and decreased markedly on days 6 and 8. These findings were further substantiated by the fact that no increase in mexAB-oprM transcript accumulation was observed in P. aeruginosa biofilms during a 4-day period (Fig. 4). Moreover, mexAB-oprM transcription levels in the biofilm closely resembled those observed in planktonic bacteria. In previous studies, the MexAB-OprM efflux pump was found to be growth phase regulated, with maximal expression occurring in late log phase (9). Although these researchers did not monitor MexAB-OprM during later stages of growth, it is possible that the progressive decrease in expression we observed in the flowcells is reflective of an overall decline in metabolic activity. Regardless, our results indicate that mexAB-oprM and mexCD-oprJ are not hyperexpressed in P. aeruginosa biofilms.
Differential expression of mexAB-oprM and mexCD-oprJ in biofilms.
It is well established that chemical gradients exist within biofilms (48, 50) and that these gradients have profound effects on the physiological state of the cells within the biofilm. Cells at the periphery have access to nutrients and less problems with accumulation of metabolic wastes compared to more deeply embedded cells. Consequently, peripheral cells are more metabolically active (44). One would expect that the gene expression patterns of cells within the biofilm would reflect this physiological heterogeneity, and in fact, evidence does exist to support the notion of spatially differentiated gene expression (44, 46, 48, 49). Somewhat surprisingly, we discovered that mexAB-oprM and mexCD-oprJ expression was greatest at the biofilm substratum, where cells are presumably metabolizing more slowly. Although the principal function of MDR efflux pumps has yet to be firmly established, it has been proposed that secondary metabolities are the natural substrates for the pumps (31). Recently, the autoinducer signal molecule N-(3-oxododecanoyl)-homoserine lactone was discovered to be one of the first examples of an endogenous natural product subject to efflux by MexAB-OprM (8, 30). At the substratum, cells are in close proximity with one another as well as with the surface to which they are attached, which would limit diffusion. Therefore, increased expression of MexAB-OprM and MexCD-OprJ at this location may be required to ensure sufficient efflux of secondary metabolites, thereby preventing toxic accumulation.
This localized gene activity may partially account for our inability to detect mexCD-oprJ transcripts in cells within the biofilm. For the transcript accumulation analysis, RNA from the entire biofilm population was examined. If mexCD-oprJ is only weakly expressed by a fraction of the population and the message is not very stable, it may not be detectable using this assay. Conversely, we were able to observe mexCD-oprJ expression in P. aeruginosa biofilms using a multicopy vector and stabilized reporter protein in conjunction with confocal microscopy. In these studies, SCLM in conjunction with reporter fusions to stable gfp enabled us to precisely monitor gene expression at distinct locations within the biofilm. These findings underscore the benefit of utilizing several different methods for analyzing gene expression.
Contribution of MDR to biofilm resistance.
A comparison of biofilms formed by cells in which MexAB-OprM was expressed, overexpressed, or completely absent demonstrated that the pump plays a minor role in resistance to aztreonam, gentamicin, tetracycline, and tobramycin (Table 2). The contribution of the MexAB-OprM pump to P. aeruginosa biofilm resistance was investigated earlier using the same deletion (K1119) and hyperexpression (OCR1) strains and three of the antibiotics tested here, namely ciprofloxacin, tetracycline, and tobramycin (3). Previous results revealed no difference in biofilm resistance to ciprofloxacin between the MexAB-OprM deletion and overproducing strains. While we found that the biofilm formed by the MexAB-OprM-overexpressing strain (OCR1) exhibited increased resistance to ciprofloxacin compared to the deletion strain, it was still more sensitive than the wild type. Furthermore, the observed increase in biofilm resistance to tetracycline associated with MexAB-OprM overexpression (Table 2) is consistent with earlier findings found at higher antibiotic concentrations (3). With regard to tobramycin, Brooun et al. (3) saw similar planktonic sensitivities with the MexAB-OprM deletion and hyperexpression strains; however, they did not observe the increased biofilm resistance associated with MexAB-OprM expression seen here (Table 2). It is possible that variations in the experimental parameters used in the two studies, for example, the duration of antibiotic challenge (6 versus 20 h), account for these differences. For the present study, it is important to note that although MexAB-OprM increased biofilm resistance to some antibiotics, similar trends were observed in planktonic cultures. Furthermore, biofilms formed by the MexAB-OprM deletion mutant (K1119) still displayed very high levels of antibiotic resistance compared to planktonic cells.
We also investigated the contribution of the other three characterized efflux pumps to P. aeruginosa biofilm resistance. In almost every instance, deletion and/or hyperexpression of the pump did not markedly alter biofilm antibiotic resistance, with the one exception being the MexEF-OprN hyperexpression strain (K1240), which exhibited increased resistance to ciprofloxacin. Together, these findings indicate that elevated expression of the four MDR pumps does not account for the overall intrinsic resistance of P. aeruginosa biofilms to antibiotics. These findings are similar to those of Maira-Litran and coworkers (21), who observed no difference in the expression of the mar (multiple antibiotic resistance) efflux operon in E. coli grown as a biofilm or planktonically. Nonetheless, there are at least six additional uncharacterized loci with homology to RND pumps that may still play a role in resistance (47).
As illustrated by the antibiotic sensitivity profiles in Tables 2 to 5, there is clearly an enhanced resistance associated with the biofilm mode of growth, yet the mechanisms underlying this phenomenon remain an enigma. Limited diffusion of antibiotics would not likely be a contributing factor, since the biofilms formed on the MBEC device (5) were grown for a short period of time and reached only intermediate thickness. Furthermore, nutrient deprivation leading to a decline in metabolic activity would not play a role because the biofilms were only formed for a 6-h period. Thus, it appears that the innate antibiotic resistance we observed can be attributed to physiological changes in cells associated with the biofilm mode of growth unrelated to MDR efflux.
In summary, the mechanisms underlying biofilm resistance are clearly multifactorial. Early on, phenotypic changes that accompany adherence and biofilm initiation render cells more resistant. As the biofilm matures, cells become encased in a thick polysaccharide matrix, and factors such as antimicrobial penetration, nutrient limitation, and growth rate may become important. Because biofilms tend to be very heterogeneous, MDR pump expression may be influenced by factors such as growth rate and metabolite accumulation, depending on the region of the biofilm. At present, the nature of the genotypic and/or phenotypic changes that cells experience during the course of biofilm development is completely unknown. Our findings reveal that expression of the four well-characterized MDR pumps is not increased in P. aeruginosa biofilms, suggesting that other factors such as decreased membrane permeability and/or alteration of antimicrobial targets must be responsible for the innate resistance to antibiotics by this population of bacteria.
ACKNOWLEDGMENTS
This work is supported in part by a National Institutes of Health (NIH) research grant, AI133713 (to B.H.I.), a Canadian Cystic Fibrosis Foundation (CCFF) research grant (to K.P.), and postdoctoral fellowships from CCFF (to T.D.K. and R.S.). Further support was provided to D.G.S. by the CCFF and to H.C. by NSERC. M.D.P. was supported by AHFMR and NSERC studentships.
We are grateful to H. Schweizer for providing pmexC.391 and HPS952.
REFERENCES
- 1.Aires J R, Kohler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison D G, Gilbert P. Modification by surface association of antimicrobial susceptibility of bacterial populations. J Ind Microbiol. 1995;15:311–317. doi: 10.1007/BF01569985. [DOI] [PubMed] [Google Scholar]
- 3.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640–646. doi: 10.1128/aac.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M R, Allison D G, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- 5.Ceri H, Olson M E, Stremick C, Read R R, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen B B, Sternberg C, Andersen J B, Palmer R J, Jr, Nielsen A T, Givskov M, Molin S. Molecular tools for study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- 7.Darouiche R O, Dhir A, Miller A J, Landon G C, Raad I I, Musher D M. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J Infect Dis. 1994;170:720–723. doi: 10.1093/infdis/170.3.720. [DOI] [PubMed] [Google Scholar]
- 8.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans K, Poole K. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol Lett. 1999;173:35–39. doi: 10.1111/j.1574-6968.1999.tb13481.x. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1990;34:1757–1761. doi: 10.1128/aac.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotoh N, Tsujimoto H, Poole K, Yamagishi J, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickson W G, Misra T K. Nucleic acid analysis. In: Gerhardt P, Murray R G E, Krieg N R, editors. Methods for general and molecular biology. Washington, D.C.: American Society for Microbiology; 1994. pp. 436–460. [Google Scholar]
- 13.Hirai K, Suzue S, Irikura T, Iyobe S, Mitsuhashi S. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:582–586. doi: 10.1128/aac.31.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler T, Epp S F, Curty L K, Pechere J C. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 1999;181:6300–6305. doi: 10.1128/jb.181.20.6300-6305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Curty L K, Pechere J C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 18.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X Z, Zhang L, Srikumar R, Poole K. Beta-lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 21.Maira-Litran T, Allison D G, Gilbert P. An evaluation of the potential of the multiple antibiotic resistance operon (mar) and the multidrug efflux pump acrAB to moderate resistance towards ciprofloxacin in Escherichia coli biofilms. J Antimicrob Chemother. 2000;45:789–795. doi: 10.1093/jac/45.6.789. [DOI] [PubMed] [Google Scholar]
- 22.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 25.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols W W, Evans M J, Slack M P, Walmsley H L. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J Gen Microbiol. 1989;135:1291–1303. doi: 10.1099/00221287-135-5-1291. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohman D E, Cryz S J, Iglewski B H. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki T, Hirai K. Cloning and nucleotide sequence of the Pseudomonas aeruginosa nfxB gene, conferring resistance to new quinolones. FEMS Microbiol Lett. 1992;76:197–202. doi: 10.1016/0378-1097(92)90386-3. [DOI] [PubMed] [Google Scholar]
- 30.Pearson J P, Van Delden C, Iglewski B H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole K. Bacterial multidrug resistance—emphasis on efflux mechanisms and Pseudomonas aeruginosa. J Antimicrob Chemother. 1994;34:453–456. doi: 10.1093/jac/34.4.453. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 33.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 34.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837. doi: 10.1126/science.283.5409.1837. , 1839. [DOI] [PubMed] [Google Scholar]
- 37.Preston M J, Seed P C, Toder D S, Iglewski B H, Ohman D E, Gustin J K, Goldberg J B, Pier G B. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect Immun. 1997;65:3086–3090. doi: 10.1128/iai.65.8.3086-3090.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer H P. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother. 1998;42:394–398. doi: 10.1128/aac.42.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiba T, Ishiguro K, Takemoto N, Koibuchi H, Sugimoto K. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J Bacteriol. 1995;177:5872–5877. doi: 10.1128/jb.177.20.5872-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Puhler A. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 43.Srikumar R, Li X Z, Poole K. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sternberg C, Christensen B B, Johansen T, Toftgaard Nielsen A, Andersen J B, Givskov M, Molin S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storey D G, Ujack E E, Mitchell I, Rabin H R. Positive correlation of algD transcription to lasB and lasA transcription by populations of Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Infect Immun. 1997;65:4061–4067. doi: 10.1128/iai.65.10.4061-4067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 48.Vroom J M, De Grauw K J, Gerritsen H C, Bradshaw D J, Marsh P D, Watson G K, Birmingham J J, Allison C. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl Environ Microbiol. 1999;65:3502–3511. doi: 10.1128/aem.65.8.3502-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westbrock-Wadman S, Sherman D R, Hickey M J, Coulter S N, Zhu Y Q, Warrener P, Nguyen L Y, Shawar R M, Folger K R, Stover C K. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu K D, Stewart P S, Xia F, Huang C T, McFeters G A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziha-Zarifi I, Llanes C, Kohler T, Pechere J C, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]