Abstract
We investigated the unusual susceptibility to meropenem observed for seven imipenem-resistant clinical isolates of Pseudomonas aeruginosa. These strains were genetically closely related, expressed OprD, as determined by Western blot analyses, and were resistant to imipenem (>5 μg/ml) but susceptible to meropenem (<1 μg/ml). The oprD genes from two isolates were entirely sequenced, and their deduced protein sequences showed 93% identity with that of OprD of strain PAO1. The major alteration consisted of the replacement of a stretch of 12 amino acids, located in putative external loop L7 of OprD, by a divergent sequence of 10 amino acid residues. The oprD gene variants and the wild-type oprD gene were cloned and expressed in a defined oprD mutant. The meropenem MICs for strains carrying the oprD genes from clinical isolates were four times lower than that for the strain carrying the wild-type oprD gene. Imipenem activities, however, were comparable for all strains. Furthermore, meropenem hypersusceptibility was obtained with a hybrid OprD porin that consisted of the PAO1 oprD gene containing loop L7 from a clinical isolate. These results show that the C-terminal portion of OprD, in particular, loop L7, was responsible for the unusual meropenem hypersusceptibility. Competition experiments suggested that the observed OprD modifications in the clinical isolates did not affect antagonism between imipenem and the basic amino acid l-lysine. We further propose that shortening of putative loop L7 of the OprD porin by 2 amino acid residues sufficiently opens the porin channel to allow optimal penetration of meropenem and increase its activity. In contrast, this alteration would not affect susceptibility to a smaller carbapenem molecule, such as imipenem.
Pseudomonas aeruginosa is an opportunistic organism causing difficult-to-treat infections. The high intrinsic resistance of this bacterium to antibiotics results from the complex interaction of several mechanisms, among which the rather impermeable porin pathway plays a key role (40). Substrate-specific transport systems in the outer membrane allow the diffusion of essential nutrients present at low concentrations in the vicinity of the cells and compensate for this low nonspecific permeability. The outer membrane porin OprD facilitates the uptake of basic amino acids (36), small peptides, and carbapenem antibiotics, such as imipenem and meropenem (35). All of these molecules share common binding sites inside the OprD channel (6).
Carbapenems are potent inhibitors of P. aeruginosa because of their high stability to the chromosomally encoded AmpC β-lactamase produced by this species (15). However, resistance of clinical strains to both imipenem and meropenem is increasingly observed as the result of the loss of protein OprD (18, 29), alone or associated with the stable overexpression of the AmpC β-lactamase (16). Overproduction of the MexAB-OprM active efflux system in nalB mutants may also increase the resistance to meropenem but has no effect on the susceptibility of P. aeruginosa to imipenem, which is not a substrate for this pump (13).
In this work, we have studied a group of clonally related clinical isolates of P. aeruginosa showing an unusual susceptibility profile characterized by resistance to imipenem (>5 μg/ml), and susceptibility to meropenem (<1 μg/ml). We provide evidence that alterations in OprD structure account for the dissociation between imipenem and meropenem activities.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
Bacterial strains and plasmids used in this study are listed in Table 1. Primers were synthesized at the University of Geneva oligonucleotide facility. Seven P. aeruginosa isolates were collected from different patients at the University Hospital in Besançon, France, over a period of 3 months. Imipenem MICs for these strains ranged from 5 to 7 μg/ml; meropenem MICs ranged from 0.8 to 1 μg/ml. Imipenem/meropenem MIC ratios ranged from 5 to 8.75.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| S17-1 | pro hsdR (r− m+) chr::RP4-2 (Tc::Mu Km::Tn7) | 32 |
| P. aeruginosa | ||
| PAO1 | Wild type | Laboratory collection |
| PASE1 | PAO1 oprD::ΩTc; Imir Merr | This study |
| PP2, PP63, PP73, PP108, PP118, PP133, PP148 | Imir Mers | Clinical isolates |
| Plasmids | ||
| pαΩ | RSF1010-derived broad-host-range vector; Cmr | 5 |
| pJQ200UC1 | Mobilizable suicide vector for gene replacement; sacB lacZα Gmr | 28 |
| pHP45ΩTc | Vector carrying the Ω interposon for insertional mutagenesis; Tcr Apr | 3 |
| pRK2013 | Tra+ ColE1 replicon Kmr | 4 |
| pUC18 | High-copy-number plasmid; Apr | 38 |
| pUC119 | High-copy-number plasmid; Apr | 37 |
| pSE0 | 1.5-kb PCR fragment carrying the oprD gene of PAO1 in the BamHI site of pUC18 | This study |
| pSE1 | 1.5-kb blunt-ended BamHI fragment of pSE0 in the SmaI site of pJQ200UC1 | This study |
| pSE2 | 2-kb SmaI fragment of pHP45ΩTc carrying the ΩTc interposon in the blunt-ended XhoI site of pSE1 | This study |
| pSE43 | 1.3-kb PCR fragment carrying the oprD gene of PAO1 in the BamHI site of pαΩ | This study |
| pCS2 | 1.3-kb PCR fragment carrying the oprD gene of clinical isolate PP2 in the BamHI site of pαΩ | This study |
| pCS148 | 1.3-kb PCR fragment carrying the oprD gene of clinical strain PP148 in the BamHI site of pαΩ | This study |
| pUCWt | 1.3-kb PCR fragment carrying the oprD gene of PAO1 in the BamHI site of pUC119 | This study |
| pUCPP148 | 1.3-kb PCR fragment carrying the oprD gene of clinical isolate PP148 in the BamHI site of pUC119 | This study |
| pM1Wt | 200-bp ClaI-XbaI fragment in pUCWt containing the 3′ end of the oprD gene replaced by an equivalent fragment from pUCPP148 | This study |
| pM2Wt | pM1Wt mutated for amino acid E399Q by site-specific mutagenesis | This study |
| pαΩM1Wt | 1.3-kb BamHI fragment of pM1Wt in the BamHI site of pαΩ | This study |
| pαΩM2Wt | 1.3-kb BamHI fragment of pM2Wt in the BamHI site of pαΩ | This study |
Imir, imipenem resistant; Merr, meropenem resistant; Mers, meropenem sensitive.
Chemicals and media.
Imipenem (Merck Sharp & Dohme-Chibret, Zürich, Switzerland) and meropenem (Imperial Chemical Industries, plc, Macclesfield, Great Britain) were provided in the form of sterile powders and were dissolved extemporaneously. Strains were routinely grown in Luria-Bertani (LB) medium. Mueller-Hinton (MH) medium or minimal medium (MM) (24) was used for antibiotic susceptibility testing. Solid media were obtained by the addition of agar to a final concentration of 1.5% (wt/vol). Antibiotics were used in selective media at the following concentrations: ampicillin, 100 μg/ml (for Escherichia coli); tetracycline, 10 μg/ml (for E. coli) and 100 μg/ml (for P. aeruginosa); gentamicin, 10 μg/ml (for E. coli) and 20 to 50 μg/ml (for P. aeruginosa); chloramphenicol, 30 μg/ml (for E. coli) and 300 μg/ml (for P. aeruginosa); and kanamycin, 25 μg/ml (for E. coli).
Susceptibility testing.
Susceptibility to antimicrobial agents was determined by the microdilution method with MH broth or MM (12). For analysis of subtle differences in susceptibilities, plates of antibiotic-containing gradients on MH agar were used as described previously (2, 20).
β-Lactamase assay.
Samples (1 ml) of cultures at an optical density at 600 nm of ∼1 were centrifuged and resuspended in 1 ml of permeabilizing buffer (100 mM EDTA, 100 mM phosphate buffer [pH 7.0], 250 μg of lysozyme/ml). β-Lactamase activity was assessed by use of 96-well microtiter plates containing 25 μl of 1 mM nitrocefin per well. A 25-μl sample of prepared bacterial suspension was added to the nitrocefin solution, and the time necessary for a change in color from yellow to red was assessed. This change generally occurs at between 1 and 10 min. After 10 min, the test was considered negative.
RAPD analysis of clinical isolates.
Randomly amplified polymorphic DNA (RAPD) conditions have been described previously (31). PCR amplification products obtained with primers I (GCCCCCAGGGGCACAGT) and II (AGTTCAGCCAC) were electrophoresed in a 2% (wt/vol) agarose gel. Isolates which differed by two or more major bands were considered sufficiently divergent to warrant separate strain designations. Isolates with profiles differing by only one major band or by one or two faint bands were considered subtypes of a common strain.
DNA manipulations.
All DNA techniques followed the protocols outlined by Ausubel et al. (1). Chromosomal DNA was prepared from exponentially grown cells of P. aeruginosa and digested to completion with EcoRI for Southern hybridization (33). After electrophoretic separation, hybridization was performed using as a probe the oprD digoxigenin-11-dUTP-labeled 1.5-kb BamHI DNA fragment extracted from plasmid pSE0. Hybridization was carried out at 42°C in the presence of 50% (vol/vol) formamide. Membranes were developed according to the manufacturer's instructions (Boehringer Mannheim Biochemicals).
DNA sequences were determined from double-stranded templates according to the dideoxy chain termination method (30) using an automated sequencer (Applied Biosystems model 377A). DNA sequences and deduced amino acid sequences were determined and aligned using the PC-GENE software package (Intelligenetics Inc.). Nucleic acid and protein sequence alignments were generated using the CLUSTAL program.
Strain and plasmid construction.
A defined OprD-deficient mutant was constructed as follows. The oprD gene of wild-type strain PAO1 was amplified by PCR using primers D1 (CGCGGATCCATGCGACATGCGTCATGC) and D2 (CGCGGATCCTTACAGGATCGACAGCGG), based on the published sequence (39). The amplified fragment was cloned into the BamHI site of pUC18 and sequenced to confirm that no errors were present in the PCR product. The oprD gene fragment was subsequently extracted from resulting plasmid pSE0 by BamHI digestion, blunt ended with the Klenow fragment, and cloned into the SmaI site of mobilizable suicide vector pJQ200uc1 carrying the sacB gene and a gentamicin resistance marker (28). The resulting construct, pSE1, was used for interposon mutagenesis of the oprD gene by insertion of the SmaI-cleaved ΩTc cassette of pHP45ΩTc (3) into the blunt-ended XhoI restriction site of oprD, which left 0.8 and 0.7 kb of DNA on either side of the ΩTc cassette. The resulting plasmid, pSE2, was subsequently transferred by conjugation into PAO1, and cointegrates were selected on LB agar plates containing gentamicin (50 μg/ml). Tetracycline-resistant and gentamicin-susceptible exconjugants were selected on LB agar plates supplemented with tetracycline (100 μg/ml) and 5% (wt/vol) sucrose. Eight Tcr Gms colonies were analyzed for their genotypes by Southern hybridization with an oprD gene probe. All clones were found to have the chromosomal gene interrupted by a 2-kb fragment containing the ΩTc interposon and were shown to be deficient in OprD expression by immunoblotting of outer membrane preparations with OprD antiserum (2a). One clone, PASE1, was used in all subsequent experiments.
The oprD genes of clinical isolates and of PAO1 were amplified by PCR using primers D23 (GCGGATCCGTAGTTCAAAACCAAAGGAGCAAT) and D2. PCR was carried out with an automated thermal cycler (Perkin-Elmer Cetus GeneAmp PCR system 9600) using either genomic DNA or bacterial suspensions (8) as templates. Reaction mixtures contained 50 pmol of each primer, 250 nmol of each of the four deoxynucleoside triphosphates, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, and 2.5 U of Taq DNA polymerase. Twenty-five cycles were performed for each sample, with a cycle consisting of denaturation at 96°C for 30 s, annealing at the appropriate temperature for 30 s, and elongation at 72°C for 90 s; a final extension was performed for 5 min at 72°C. The resulting fragments were purified on 1% (wt/vol) low-melting-temperature agarose, digested with BamHI, and subsequently cloned into the unique BamHI cloning site of mobilizable plasmid pαΩ (5). E. coli clones carrying plasmids with the insert were detected by colony hybridization with an oprD gene probe. Restriction analysis permitted us to detect the correctly oriented insert. Plasmids were routinely conjugated into P. aeruginosa either from E. coli S17-1 (32) or by triparental mating from DH5α using pRK2013 as a helper plasmid (4). The oprD genes were subsequently expressed in OprD-deficient strain PASE1.
OprD hybrid construction.
The oprD genes of strains PP148 and PAO1 were amplified by PCR and cloned into vector pUC119, yielding plasmids pUCPP148 and pUCWt, respectively. Hybrid gene construction was performed by amplifying the 3′ end of the oprD gene from plasmid pUCPP148 with primers M13R (AGCGGATAACAATTTCACACAGGA) and D25 (CCATCGATGGCACCAAGGTCGACTCC) to introduce a ClaI restriction site into the C-terminal fragment of the oprD gene. Recombinant plasmid pUCWt and the derived PCR product of 200 bp obtained with primers M13R and D25 were subsequently digested with ClaI and XbaI and purified on low-melting-temperature agarose. The purified PCR fragment was then ligated to the plasmid fragment carrying the 5′ downstream sequence of the oprD gene (1.1 kbp) to yield a functional oprD hybrid gene. The restored ClaI site did not alter the deduced amino acid sequence of the oprD gene fragment derived from PP148. Correct construction of the hybrid gene was confirmed by restriction analysis and DNA sequencing.
PCR-mediated site-directed mutagenesis.
Site-directed mutagenesis was performed using as a template pUC119-derived plasmid pM1Wt carrying the oprD hybrid gene construct. Two oligonucleotides, D26 (GCCAATGCCGACCAGGGCGAAGGCGACCAG) and D27 (CTGGTCGCCTTCGCCCTGGTCGGCATTGGC), each complementary to opposite strands of the oprD gene and containing the desired mutation, were extended during temperature cycling by the Pfu DNA polymerase (17). PCR amplification was carried out with 50-μl reaction mixtures containing the recommended buffer, 12 pmol of each primer, 250 μM concentrations of each of the four deoxynucleoside triphosphates, 20 ng of template plasmid DNA, and 2.5 U of Pfu DNA polymerase. Sixteen cycles were performed for each sample, with a cycle consisting of denaturation at 95°C for 30 s, annealing at 45°C for 30 s, and elongation at 68°C for 2 min/kb. After PCR, the product was treated with DpnI to digest the parental DNA template. E. coli DH5α was directly transformed with the nicked plasmid DNA. Clones containing the desired nucleotide substitution were identified by DNA sequencing.
Outer membrane protein and crude lysate preparations.
Outer membranes were prepared either by use of a French pressure cell (SLM Instruments, Inc., Urbana, Ill.) as described previously (22) or with a Sonifier (Branson Instruments, Danbury, Conn.) (21). For crude lysate preparations, cells were grown in LB broth to exponential growth phase (optical density at 600 nm, ∼1.0), and 1 ml of culture containing approximately 3 × 108 cells was centrifuged at 12,000 × g for 2 min. The supernatant was discarded, and the cell pellet was homogenized in 50 μl of 10 mM Tris (pH 7.5)–10% (vol/vol) glycerol.
Gel electrophoresis and immunoblotting.
Outer membrane proteins and crude lysates were analyzed by electrophoresis with a Protean II slab electrophoresis cell (Bio-Rad Laboratories, Richmond, Calif.). Protein fractions were solubilized in sample buffer, heated at 95°C for 5 min, and separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with 10% (wt/vol) acrylamide and 0.1% (wt/vol) piperazine diacrylyl in the running gel at a constant current of 20 mA per gel. The gels were stained with Coomassie blue solution. For immunoblotting, proteins were heated prior to separation by SDS-PAGE at 95°C for 5 min when probed with OprD polyclonal antiserum and at 88°C when OprF monoclonal antiserum MA5-8 (23) was used. Subsequently, proteins were transferred to a nitrocellulose membrane (45-μm-pore size; Bio-Rad) for 2 h by use of a Bio-Rad Mini Trans Blot electrophoretic transfer cell containing 25 mM Tris-HCl (pH 8.3), 192 mM glycine, and 20% (vol/vol) methanol. Immunoblotting was performed (34) with polyclonal OprD (2a) or OprM (41) antiserum or monoclonal OprF antiserum MA5-8 as the primary antibody and subsequently with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G or donkey anti-mouse immunoglobulin G. Detection was performed by the enhanced chemiluminescence method using luminol-H2O2 as a substrate (Amersham International plc, Zurich, Switzerland).
Competition experiments.
Competition experiments with the basic amino acid l-lysine and carbapenems were performed as described previously (6) using MM BM2 containing 20 mM gluconate and supplemented or not supplemented with 50 mM l-lysine (9).
RESULTS
Characterization of clinical strains.
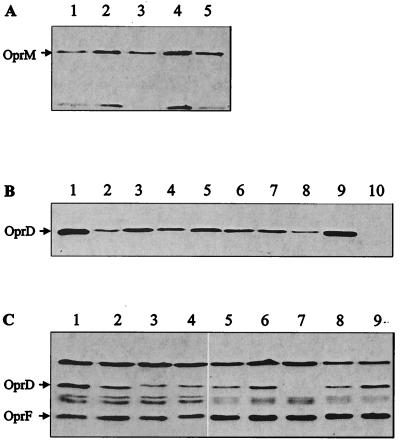
The seven imipenem-resistant meropenem-sensitive strains belonged to serotype O:12 of P. aeruginosa, and RAPD analysis indicated that they shared a similar genotypic background. Six out of the seven strains were resistant to quinolones and chloramphenicol and susceptible to cefpirome (Table 2). PP148 exhibited a different pattern, with susceptibility to quinolones and resistance to cefpirome. All strains except for PP148 were found β-lactamase positive by the nitrocefin test described in Materials and Methods. All seven strains were five- to ninefold more resistant to imipenem than to meropenem (Table 3). Western blot analysis of outer membranes showed comparable OprM expression levels between the clinical strains and PAO1 (Fig. 1A), excluding the absence or decreased expression of the MexAB-OprM pump as an explanation for the meropenem susceptibility. Compared to that in PAO1, OprD expression in all of the clinical strains was reduced (Fig. 1B), a result which could explain their increased resistance to imipenem. nfxC mutants overexpressing the MexEF-OprN efflux pump display decreased OprD expression (14, 19), resulting in increased resistance to carbapenems. However, Western blot analysis with monoclonal OprN antibodies did not reveal MexEF-OprN expression in the strains tested (data not shown).
TABLE 2.
Antibiotic susceptibilities of clinical isolates
| Antibiotic | MIC (μg/ml)a for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PAO1 | PASE1 | PP2 | PP63 | PP73 | PP108 | PP118 | PP133 | PP148 | |
| Ofloxacin | 2 | 2 | >64 | >64 | >64 | >64 | >64 | >64 | 2 |
| Cefpirome | 2 | 2 | 16 | 16 | 16 | 16 | 16 | 16 | 128 |
| Chloramphenicol | 64 | 64 | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 |
| Erythromycin | 256 | 256 | 256 | 256 | 256 | 256 | 256 | 256 | 256 |
MICs were determined by the microdilution method. Strains shown in bold type were selected for further study.
TABLE 3.
Susceptibilities of clinical strains and of strain PASE1 expressing OprD variant proteins
| Antibiotic | MIC (μg/ml)a for:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAO1 | PASE1 | PP2 | PP63 | PP73 | PP108 | PP118 | PP133 | PP148 | PASE1 (pαΩ) | PASE1 (pSE43) | PASE1 (pCS2) | PASE1 (pCS148) | PASE1 (pαΩM1Wt) | PASE1 (pαΩM2Wt) | |
| Imipenem | 2 | 12 | 7 | 5 | 6 | 7 | 7 | 6 | 6 | 12 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Meropenem | 1 | 4 | 1 | 1 | 1 | 1 | 0.8 | 0.8 | 0.8 | 4 | 0.8 | 0.2 | 0.2 | 0.25 | 0.25 |
MICs were determined on antibiotic-containing gradient plates.
FIG. 1.
Western blot analysis of strains used in this study. (A) Outer membrane proteins (OMPs) of clinical strains were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with OprM polyclonal antiserum. Lanes: 1, PAO1; 2, PP2; 3, PP63; 4, PP18; 5, PP148. (B) OMP preparations were probed with OprD polyclonal antiserum. Lanes: 1, PAO1; 2, PP2; 3, PP63; 4, PP73; 5, PP108; 6, PP118; 7, PP133; 8, PP148; 9, PAO1; 10, PASE1. (C) Crude cell extracts were probed with OprD polyclonal antiserum and OprF monoclonal antiserum MA5–8. Lanes: 1, PAO1; 2, PASE1(pSE43); 3, PASE1(pCS2); 4, PASE1(pCS148); 5, PASE1(pαΩM1Wt); 6, PASE1(pαΩM2Wt); 7, PASE1; 8, PASE1(pSE43); 9, PAO1. For each lane in panels A and B, 10 μg of OMPs was loaded; in panel C, 10 μl of crude extract was loaded per lane.
Functional analysis of OprD proteins from clinical strains.
Strains PP2 and PP148 were chosen for detailed OprD analysis because of their different antibiotic susceptibility patterns (Table 2) and slightly different genetic backgrounds, as determined by RAPD analysis. The function of OprD porins from these two strains was analyzed by expressing the respective oprD genes in the OprD-deficient background of strain PASE1. After amplification by PCR, the PP2, PP148, and PAO1 oprD genes, including their own ribosome binding sites, were cloned into the low-copy-number vector pαΩ containing a bacteriophage T4 gene 32-derived expression cassette; this procedure resulted in plasmids pCS2, pCS148, and pSE43, respectively. After transfer into strain PASE1, Western blot analysis showed that plasmid-driven OprD expression was similar in the three constructs (Fig. 1C). In oprD-deficient strain PASE1, imipenem resistance levels conferred by plasmids pCS2, pCS148, and pSE43 were almost identical on imipenem-containing gradient plates. However, on meropenem-containing gradient plates, strain PASE1 harboring plasmid pCS2 or pCS148 was four times more susceptible to meropenem than strain PASE1 harboring plasmid pSE43 (Table 3). This result strongly suggested that OprD alterations were responsible for the unusual meropenem susceptibility phenotype of clinical strains PP2 and PP148.
DNA sequence analysis of oprD genes from clinical strains.
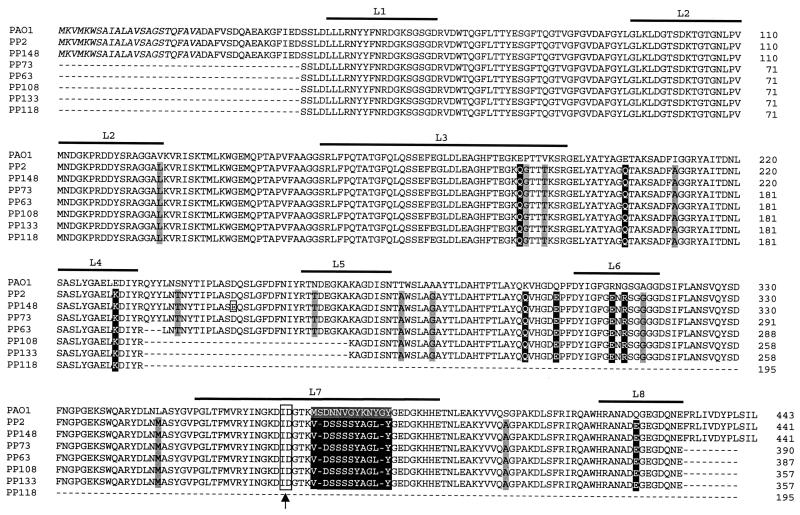
The oprD genes from strains PP2, PP148, and PAO1 were completely sequenced. Partial sequence analysis (Fig. 2) was performed for the oprD genes from the five remaining clinical strains. Nucleic acid sequences of oprD were almost identical in PP2 and PP148: six silent mutations and one neutral mutation were reflected in a unique homologous amino acid substitution, D226E, in the deduced protein sequence. The oprD nucleotide sequences of the clinical strains showed 90.5% identity with that of PAO1, and their deduced amino acid sequences showed 93% identity. The majority of the nucleic acid changes in the oprD genes from the clinical strains were silent mutations and did not affect the amino acid sequence. Twenty single amino acid residues were replaced in the clinical strains; 8 of these modified the charge of the molecule (E162Q, E179Q, E207K, K273Q, Q278E, R287E, G289R, and Q401E). The most striking difference was located in putative loop L7 of the OprD topology model (11). A stretch of 12 amino acid residues (350-MSDNNVGYKNYG-361) present in OprD of PAO1 and designated subsequently as L712 was replaced in OprD from all the clinical strains by a divergent stretch of 10 amino acid residues (350-VDSSSSYAGL-359) designated here as L710 (Fig. 2).
FIG. 2.
Multiple sequence alignment of OprD protein variants from PAO1 and seven clinical strains. Black boxes indicate a change in amino acid charge or replacement of a stretch of residues; grey boxes indicate other amino acid substitutions. The small open box shows the single amino acid change observed in strain PP148. Amino acids in italics represent the N-terminal signal sequence. Locations of putative loops L1 to L8 are shown by bold lines; amino acids in grey boxes in loop L7 indicate the deletion (OprDΔL7) described by Huang et al. (11). The fusion junction in the OprD hybrid proteins is boxed and indicated by an arrow. Numbering includes the signal sequence.
Construction and analysis of an OprD hybrid protein.
To assess the possible role of the divergent L712 and L710 sequences in meropenem susceptibility, we constructed a hybrid protein consisting of the 367 N-terminal amino acid residues from PAO1 OprD and the 74 C-terminal residues of the OprD variant from PP148. The latter fragment contained divergent sequence L710 and two additional altered residues, S378A and Q399E (Fig. 2). DNA sequence analysis was performed to confirm that no errors were introduced by PCR. The resulting hybrid oprD gene was subsequently extracted and cloned into broad-host-range vector pαΩ. The resulting plasmid, pαΩM1Wt, was transferred into strain PASE1. Western blot analysis of outer membrane preparations showed slightly less OprD expression in PASE1(pαΩM1Wt) than in PASE1(pSE43) (Fig. 1C). Carbapenem resistance levels, determined on antibiotic-containing gradient plates, showed that strain PASE1(pαΩM1Wt) retained the meropenem hypersusceptibility phenotype of PASE1(pCS148) without an alteration in imipenem activity (Table 3). The 74 residues containing the C-terminal fragment of the OprD hybrid protein were therefore determined to be responsible for the change in meropenem susceptibility.
Site-directed mutagenesis.
The fused C-terminal fragment of the hybrid protein contained two additional single amino acid substitutions, S378A and Q399E, besides the divergent L710 sequence. In the clinical strains, S380 was replaced by A378, which is less hydrophilic, and Q401, a hydrophilic residue, was replaced by negatively charged E399. To define more precisely the cause of the increased susceptibility to meropenem, in particular, the involvement of the L710 sequence, the glutamate (E) residue at position 399 of the hybrid protein, located on putative external loop L8, was restored to glutamine (Q), present in the wild-type OprD protein, by site-directed mutagenesis. The mutated hybrid gene present on plasmid pαΩM2Wt in strain PASE1 directed OprD expression similar to that of the parental hybrid gene present on plasmid pαΩM1Wt (Fig. 1C). Imipenem and meropenem MICs resulting from the presence of these two plasmids were also comparable (Table 3), suggesting that the shortened L710 loop and not the amino acid change at position 399 of OprD was responsible for the meropenem susceptibility in the clinical strains.
Competition experiments.
In order to confirm that the amino acid substitutions observed in OprD from the clinical strains did not affect common binding sites shared by the basic amino acid l-lysine and carbapenems, competition experiments were carried out. The MICs of imipenem were 0.25 and 1 μg/ml in the absence and in the presence of lysine, respectively, for strain PASE1 carrying plasmid pSE43, pCS148, pαΩM1WT, or pαΩM2WT (Table 4). Meropenem MICs for strain PASE1 carrying plasmid pSE43 were 0.13 and 2 μg/ml in the absence and in the presence of lysine, respectively. A similar 16-fold increase in meropenem MICs in the presence of lysine was observed with OprD variants carried by plasmid pCS148, pαΩM1WT, or pαΩM2WT, suggesting that common binding sites were not affected by the amino acid substitutions. Interestingly, the MIC of imipenem for OprD-deficient strain PASE1 showed a fourfold increase in the presence of lysine, suggesting that common alternate pathways may be used by basic amino acids and imipenem.
TABLE 4.
Effect of basic amino acid l-lysine on imipenem and meropenem susceptibilities of strain PASE1 expressing OprD variant proteins
| Strain | OprD expressiona | MIC (μg/ml)b of the following drug under the indicated conditions:
|
|||
|---|---|---|---|---|---|
| Imipenem
|
Meropenem
|
||||
| BM2 | BM2 + 50 mM l-lysine | BM2 | BM2 + 50 mM l-lysine | ||
| PAO1 | + | 0.25 | 2 | 0.03 | 0.5 |
| PASE1 | − | 2 | 8 | 1 | 2 |
| PASE1(pSE43) | + | 0.25 | 1 | 0.13 | 2 |
| PASE1(pCS148) | + | 0.25 | 1 | 0.03 | 0.5 |
| PASE1(pαΩM1WT) | + | 0.25 | 1 | 0.06 | 1 |
| PASE1(pαΩM2WT) | + | 0.25 | 1 | 0.06 | 1 |
+, expression; −, no expression.
MICs were determined by the microdilution method.
DISCUSSION
The antibacterial activity of meropenem against P. aeruginosa results from different determinants whose effects are combined. The OprD-mediated uptake pathway modulates the rate of penetration of meropenem through the outer membrane. OprD deficiency typically causes a 4- to 16-fold increase in the MICs of carbapenems through an interplay of impermeability and β-lactamase activity (16). Cross-resistance of imipenem-resistant P. aeruginosa clinical isolates to meropenem is generally observed (7, 35). In this study, the analysis of seven imipenem-resistant clinical isolates selected for their unusual meropenem susceptibility gave us the opportunity to perform functional studies on the OprD porin and to demonstrate the involvement of loop L7 in meropenem activity.
Several explanations for the unusual meropenem susceptibility of imipenem-resistant clinical strains were considered and finally ruled out. First, PAO1 derivatives lacking the MexAB-OprM efflux pump are hypersusceptible to meropenem, yet imipenem MICs are not affected (13). However, all seven clinical isolates produced significant levels of OprM in their outer membranes, making this explanation unlikely. Second, Perez et al. (27) described imipenem-resistant, meropenem-susceptible OprD-deficient isolates and suggested a role for porins OprF and OprE in the diffusion of meropenem across the outer membrane. However, analysis of the outer membrane proteins of clinical strains did not reveal overexpression of OprF or OprE. The possibility that a hypothetical but as-yet-unidentified outer membrane channel facilitates meropenem entry into the cells cannot be ruled out completely. The products of at least 15 open reading frames sharing significant sequence similarities with OprD have indeed been identified from the genome of PAO1 (http://www.interchg.ubc.ca/bobh/oprDalignment.html). These OprD homologs might well account for the antagonistic action of lysine toward both imipenem and meropenem in the OprD-deficient strain PASE1. It is therefore conceivable that porins, other than OprD, with less affinity for basic amino acids might be involved in the diffusion process.
Compared to that in strain PAO1, the expression of OprD in all seven clinical isolates was reduced. This result likely explains the resistance of these strains to imipenem. The genetic events leading to the reduced expression of OprD were not explored further; recently, however, overproduction of the MexEF-OprN active efflux system has been shown to be associated with decreased OprD expression in P. aeruginosa (14). Since the expression of this efflux pump was undetectable in our clinical strains, the hypothesis of more efficient penetration of meropenem through an altered OprD structure was put forward. Evidence for this assumption was provided by conferral of the meropenem-hypersusceptible phenotype to the OprD-deficient strain PASE1 with the plasmid-borne oprD genes cloned from the clinical isolates.
OprD is a protein of 420 amino acids, thought to be composed of 16 antiparallel β strands connected by eight short periplasmic turns and eight longer surface loops (11). Numerous single amino acid substitutions were observed for the deduced OprD amino acid sequences of the clinical isolates relative to the PAO1 OprD protein. The N-terminal part of the protein, including putative loops L1, L2, L3, and L5, was less affected by alterations than the C-terminal part. Loops L2 and L3 have been proposed to be implicated in specific binding to imipenem and basic amino acids (10, 25). These regions were indeed conserved in OprD proteins from all clinical isolates. The most striking change observed was the substitution of 12 consecutive residues in loop L7 with a totally divergent stretch of 10 amino acids. The same substitution was found in OprD porins from all seven clonally related clinical isolates. This 10-amino-acid sequence (VDSSSSYAGL) was not present in PAO1 (www.pseudomonas.com). The hybrid construct made of the N-terminal moiety from PAO1 OprD and loops L7 and L8 from the OprD variant of clinical isolate PP148 retained the meropenem-hypersusceptible phenotype. Restoring PAO1 residue Q401 of loop L8 (E399Q in the hybrid construct) had no effect on this phenotype. We therefore concluded that segment L710 of loop L7 was responsible for the meropenem susceptibility profile of our imipenem-resistant clinical isolates.
OprD loop L7, together with loops L5 and L8, presumably contributes to the constriction of the OprD channel opening. The long axes of the elliptical molecules imipenem and meropenem would exceed the exclusion limit of the channel and would have to be aligned with the pore axis for efficient transport. In agreement with our results, Huang et al. (11) reported that a deletion of eight amino acids (349-MSDNNVGY-356) in loop L7 of OprD (Fig. 2) decreased meropenem MICs twofold compared to those for wild-type OprD without affecting imipenem MICs. The same construct increased susceptibilities to cephalosporins, aztreonam, quinolones, and chloramphenicol four- to eightfold (10). However, in our OprD constructs containing the shortened L710 loop, pefloxacin susceptibilities were not affected, probably because nonspecific diffusion across the OprD channel remained unaffected by this alteration.
In our study, the stretch of divergent residues found in loop L710 of OprD from the clinical isolates contained 10 (350-VDSSSSYAGL-359) instead of the 12 (350-MSDNNVGYKNYG-361) amino acids found in loop L712 of strain PAO1 OprD. This feature should result in a shortened loop L7 and a less constricted OprD channel opening. However, the possibility cannot be excluded that the physicochemical properties of the modified loop also play a role in increased meropenem susceptibility, since overall charge and hydrophilicity were altered in loop L710 compared to loop L712. Imipenem (molecular weight, 299) and meropenem (molecular weight, 383.5) differ essentially in the size and the pKa value of their C-2 substituent (13). As measured in proteoliposomes, imipenem diffuses through the OprD channel 10 times faster than meropenem (35). Since the rates of permeation of basic carbapenem compounds are correlated to their molecular weights (26, 35), it is conceivable that permeation of the bulkier meropenem molecule is improved by the shortening of putative loop L7. Alternatively, the absence of the positive charge in loop L710 could affect selectively the interaction with meropenem due to its lower pKa value and the positioning of the positive charge on the molecule relative to these characteristics for imipenem. The results of lysine competition assays also suggested that the alteration in loop L710 did not affect a specific binding site for carbapenems.
The reason for the unusual meropenem susceptibility in imipenem-resistant clinical isolates could be explained by the fact that, in France, meropenem is not listed on the hospital formulary. Furthermore, it can be speculated that the possible channel enlargement provided by modified loop L710 confers a selective advantage on these isolates by favoring the uptake of certain nutrients.
ACKNOWLEDGMENTS
We thank Catherine Cherbulliez for skillful technical assistance, Colette Rossier for performing the sequencing, and Nathalie Lin-Marq for helpful discussions on site-specific mutagenesis. We thank Naomasa Gotoh for performing Western blot analysis with monoclonal antibodies to OprN.
T.K. was supported by a grant from the Swiss National Science Foundation.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 2.Bryson V, Szybalzski W. Microbial selection. Science. 1952;116:45–51. [PubMed] [Google Scholar]
- 2a.Epp, S. F., J.-C. Pechère, and M. Kok. Raising antibodies against OprD, an outer membrane protein of Pseudomonas aeruginosa using translational fusions to MalE. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 3.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 4.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frey J, Mudd E A, Krisch H M. A bacteriophage T4 expression cassette that functions efficiently in a wide range of gram-negative bacteria. Gene. 1988;62:237–247. doi: 10.1016/0378-1119(88)90562-8. [DOI] [PubMed] [Google Scholar]
- 6.Fukuoka T, Masuda N, Takenouchi T, Sekine N, Iijima M, Ohya S. Increase in susceptibility of Pseudomonas aeruginosa to carbapenem antibiotics in low-amino-acid media. Antimicrob Agents Chemother. 1991;35:529–532. doi: 10.1128/aac.35.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fung-Tomc J, Huczko E, Banville J, Menard M, Kolek B, Gradelski E, Kessler R E, Bonner D P. Structure-activity relationships of carbapenems that determine their dependence on porin protein D2 for activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:394–399. doi: 10.1128/aac.39.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh N, Tsujimoto H, Poole K, Yamagishi J, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, Hancock R E. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1993;175:7793–7800. doi: 10.1128/jb.175.24.7793-7800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H, Hancock R E. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1996;178:3085–3090. doi: 10.1128/jb.178.11.3085-3090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Jeanteur D, Pattus F, Hancock R E. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol Microbiol. 1995;16:931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen J H, Ferraro M J, Craig W A, Doern G V, Finegold S M, Fung-Tomc J, Hansen S L, Hindler S, Reller L B, Swenson J M, Tenover F C, Testa R T, Wikler M A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow anaerobically. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 13.Köhler T, Michéa-Hamzehpour M, Epp S F, Pechère J C. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob Agents Chemother. 1999;43:424–427. doi: 10.1128/aac.43.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Kocjancic-Curty L, Pechère J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 15.Livermore D M. Carbapenemases. J Antimicrob Chemother. 1992;29:609–613. doi: 10.1093/jac/29.6.609. [DOI] [PubMed] [Google Scholar]
- 16.Livermore D M. Interplay of impermeability and chromosomal beta-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2046–2048. doi: 10.1128/aac.36.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundberg K S, Shoemaker D D, Adams M W, Short J M, Sorge J A, Mathur E J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991;108:1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 18.Lynch J M, Drusano G L, Mobley H L. Emergence of resistance to imipenem in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:1892–1896. doi: 10.1128/aac.31.12.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michéa-Hamzehpour M, Auckenthaler R, Regamey P, Pechère J C. Resistance occurring after fluoroquinolone therapy of experimental Pseudomonas aeruginosa peritonitis. Antimicrob Agents Chemother. 1987;31:1803–1808. doi: 10.1128/aac.31.11.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michéa-Hamzehpour M, Furet Y X, Pechère J C. Role of protein D2 and lipopolysaccharide in diffusion of quinolones through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:2091–2097. doi: 10.1128/aac.35.10.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michéa-Hamzehpour M M, Pechère J C, Plésiat P, Köhler T. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:2392–2396. doi: 10.1128/aac.39.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutharia L M, Hancock R E. Characterization of two surface-localized antigenic sites on porin protein F of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:381–386. doi: 10.1139/m85-073. [DOI] [PubMed] [Google Scholar]
- 24.Nicas T I, Hancock R E. Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B, and gentamicin. J Bacteriol. 1980;143:872–878. doi: 10.1128/jb.143.2.872-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochs M M, Bains M, Hancock R E. Role of putative loops 2 and 3 in imipenem passage through the specific porin OprD of Pseudomonas aeruginosa. J Bacteriol. 2000;44:1983–1985. doi: 10.1128/aac.44.7.1983-1985.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohba F, Nakamura-Kamijo M, Watanabe N, Katsu K. In vitro and in vivo antibacterial activities of ER-35786, a new antipseudomonal carbapenem. Antimicrob Agents Chemother. 1997;41:298–307. doi: 10.1128/aac.41.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez F J, Gimeno C, Navarro D, Garcia-de-Lomas J. Meropenem permeation through the outer membrane of Pseudomonas aeruginosa can involve pathways other than the OprD porin channel. Chemotherapy. 1996;42:210–214. doi: 10.1159/000239444. [DOI] [PubMed] [Google Scholar]
- 28.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 29.Quinn J P, Dudek E J, DiVincenzo C A, Lucks D A, Lerner S A. Emergence of resistance to imipenem during therapy for Pseudomonas aeruginosa infections. J Infect Dis. 1986;154:289–294. doi: 10.1093/infdis/154.2.289. [DOI] [PubMed] [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segonds C, Bingen E, Couetdic G, Mathy S, Brahimi N, Marty N, Plésiat P, Michel-Briand Y, Chabanon G. Genotypic analysis of Burkholderia cepacia isolates from 13 French cystic fibrosis centers. J Clin Microbiol. 1997;35:2055–2060. doi: 10.1128/jcm.35.8.2055-2060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 33.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:52–57. doi: 10.1128/aac.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 37.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Yoneyama H, Yoshihara E, Nakae T. Nucleotide sequence of the protein D2 gene of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1791–1793. doi: 10.1128/aac.36.8.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura F, Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol. 1982;152:636–642. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziha-Zarifi I, Llanes C, Köhler T, Pechère J C, Plésiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]