Abstract
CsgA is a major protein subunit of Escherichia coli biofilms and plays key roles in bacterial adhesion and invasion. CsgA proteins can self-assemble into amyloid nanofibers, characterized by their hierarchical structures across multiple length scales, outstanding strength and their structural robustness under harsh environments. Here, magnetic tweezers were used to study the force spectra of CsgA protein at fibril levels. The two ends of a single nanofiber were directly connected between a magnetic bead and a glass slide using a previously reported tag-free method. We showed that a wormlike chain model could be applied to fit the typical force–extension curves of CsgA nanofibers and to estimate accordingly the mechanical properties. The bending stiffness of nanofibers increased with increasing diameters. The changes in extension of single CsgA fibers were found to be up to 17 fold that of the original length, indicating exceptional tensile properties. Our results provide new insights into the tensile properties of bacterial amyloid nanofibers and highlight the ultrahigh structural stability of the Escherichia coli biofilms.
Magnetic tweezers were used to study the force spectra of CsgA, a major protein subunit of Escherichia coli biofilms, at fibril level.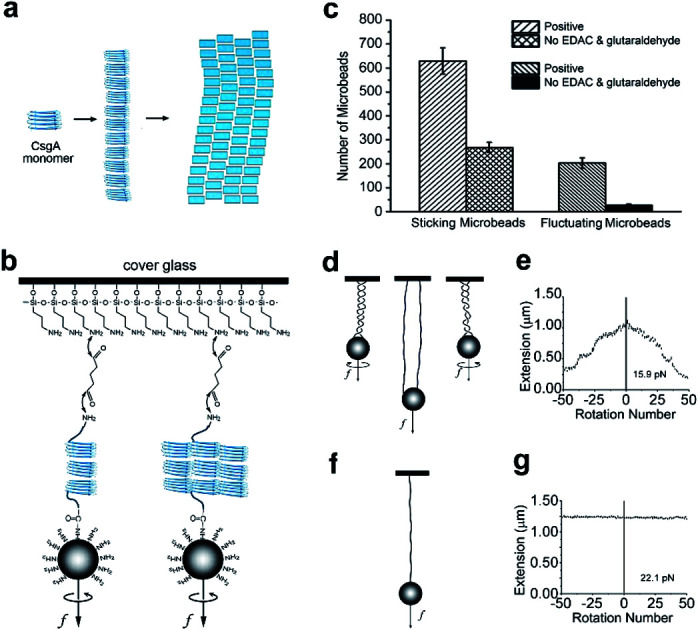
1. Introduction
There are numerous examples of naturally occurring hierarchically self-assembled nanostructures that provide vital physiological functions for living organisms.1–3 Such functional building blocks can serve as inspiration or templates to develop new materials and technologies.4,5 CsgA nanofibers, which are the major protein constituents of Escherichia coli (E. coli) extracellular biofilms, provide adhesion, stiffness and mechanical stability for the biofilms and play critical roles in the binding of host cells for internalization and protection against phage attack.6–8 CsgA protein monomers, which consist of five repeating units with several conservative residues, can fold into a compact β-helix which is capable of self-assembling into CsgA nanofibers (Fig. 1a).9 The cross-β hydrogen-bonded structures within the nanofibers can provide stiffness, strength, and stability like other well-known amyloid nanofibers such as lysozymes and β-lactoglobulin.10–12 By using genetic engineering tools, CsgA fusion protein nanofibers have been designed and explored for many applications, such as underwater adhesives, engineered living materials, and patternable coating materials.13–18
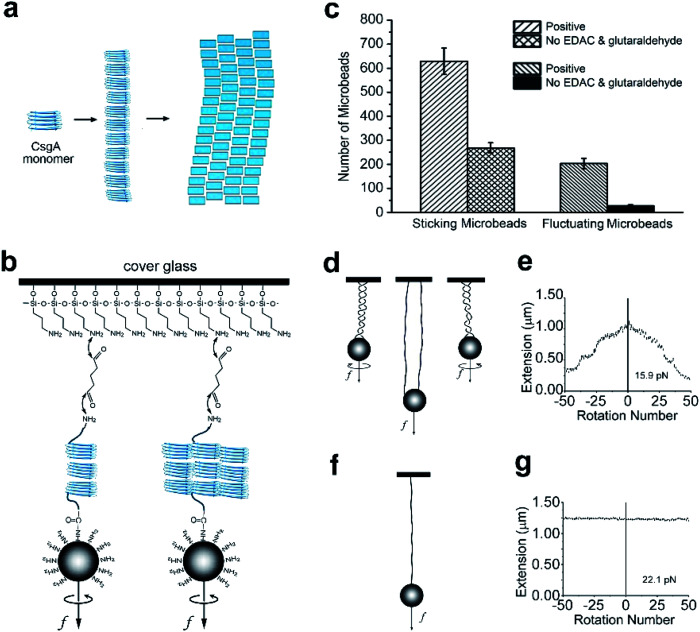
Fig. 1. Self-assembling of CsgA proteins and establishment of single molecule system. (a) A schematic of self-assembling of CsgA proteins. (b) Schematic of the cross-linking of a single CsgA nanofiber. (c) The number of sticking microbeads observed per area in the cross-linking CsgA positive experiment and in the non-linking CsgA control experiment without adding EDAC and glutaraldehyde. (d and e) The relation between extension and rotation number and their schematic diagrams when multiple molecules or nanofibers attached on one microbead and glass. (f and g) The relation between extension and rotation number and their schematic diagram when a single molecule or nanofiber attached to one microbead and glass.
Their important biological roles and promising applications in material science highlight the significance of probing the mechanical properties of CsgA nanofibers at both bulky and molecular levels. Such research will address fundamental questions regarding the formation and eradication of bacterial biofilms, as well as help to facilitate rational design and utilization of CsgA nanofibers for material research.19 In previous studies, the mechanical unfolding and adhesion mechanisms of a single CsgA protein and similar β-helical proteins had been assessed using both theoretical and experimental approaches.20–22 At single nanofiber level, the relation between adhesion behavior and the dimension of fiber has been demonstrated by simulation study.23 Atomic force microscopy (AFM) was applied to measure the mechanical properties of CsgA nanofiber.24,25 The Young's modulus has been calculated with an assumption that the fiber was homogeneous.24 The axial structure of a linear fiber is quite different from the vertical structure. Therefore the bending stiffness is probably different from the stretching stiffness. Despite these important advances, the persistence length of curli nanofibers (a key parameter for understanding mechanical properties) has not been directly measured by experiments, though it has been analyzed in a coarse-grained model.26 As such, many basic physical and mechanical properties of CsgA nanofibers, particularly at nanofiber level, remains elusive.
The mechanical properties of single proteins have been studied using a number of experimental techniques27 such as optical tweezers,28 magnetic tweezers (MT),29–32 and AFM.33,34 In these approaches, the protein is typically tethered to a microbead or tip via a tag or physisorption, leaving the other end free to interact with another molecule or substrate. The tag, such as antigen–antibody or biotin–streptavidin, requires modification of the protein or a plasmid encoding the protein. The modification and corresponding purification procedures are time-consuming.31,35 In addition, the non-covalent bonds between the tag and proteins may break during stretching, thus affecting experimental results. The physisorption method was used in early experimental measurements,36 but is not appreciated in recent years because it is non-specific. The amino-carboxyl linking is one of the commonly used conjugation methods.37 Applying this tag-free method into the single-molecule force measurement system may improve the efficiency. MT has been widely used to study the mechanical properties of DNA,38 single proteins,29,39 and protein–protein interactions,40,41 and has been demonstrated to possess high resolution, wide force range, and stability.30,42,43
Here, using a robust and tag-free single-molecule strategy, we directly measured the mechanical properties of CsgA nanofibers of different diameters and obtained the force spectra using MT. The number of filaments that made up the nanofibers was estimated based on the persistence lengths. It was found that the mechanical properties of CsgA nanofibers of different diameters varied, and the bending stiffness varied with the diameter. Understanding the mechanical properties of CsgA nanofibers with different dimensions can help to establish the foundation for applying them in both materials science and bio-nanotechnology.
2. Results and discussion
2.1. Development of a robust and tag-free attachment method for measurement
To simplify the labeling process in force spectroscopy experiments, we applied a tag-free approach for any proteins with natural amino, and carboxyl ends. For the MT experiments, we first linked one end of the CsgA nanofiber to an amino-functionalized magnetic microbead (Dynabeads, M270) through the covalent reaction mediated by EDAC (1-(3-dimethylaminopropyl)-3-ethylcarbodiimide, Thermo Fisher) between the carboxyl group of the protein nanofiber and the amino group of the microbead. Then, the magnetic beads were loaded onto a flow cell covered by glutaraldehyde-coated glass slide to adhere to the CsgA nanofiber through a Schiff base reaction (Fig. 1b). After linking, the buffer in the flow cell was replaced with the MES (2-(N-morpholino)ethanesulfonic acid, Sigma) buffer (pH = 5.5). The fibril filaments that make up the CsgA nanofibers are considered to form hydrogen bonds between each other.9
We observed a large number of fluctuating beads using microscopy (Fig. 1c), indicating most of the CsgA nanofibers were successfully tethered to the microbeads and glass. In the control experiments without adding EDAC and glutaraldehyde, there were much fewer fluctuating microbeads than in the positive experiments, suggesting that much fewer fibers were tethered to magnetic microbeads (Fig. 1c). The number of tethered fluctuating microbeads in the control experiments was only ∼13% that of the positive tests, indicating that the linking method was effective. In addition, we clearly observed a larger number of sticking microbeads (including stationary microbeads and fluctuating microbeads) in positive experiments than in the control experiments. This observation highly suggested that the amino-functionalized beads could bind to glutaraldehyde-coated glass surface, and thus could be used as reference microbeads in MT measurements.
The initial step of single-molecule experiments was to make sure the molecule was in a single molecular state. In our sample, it was surmised that multiple molecules or nanofibers might be attached to one microbead after linking. To select single nanofiber-tethered microbeads, the magnetic beads were rotated. In some experiments, the extension decreased when the rotation number increased positively (in a counter-clockwise direction) or negatively (in clockwise direction), suggesting the formation of coils or braids containing multiple nanofibers attached to one microbead and glass slide (Fig. 1d and e). We noted that the nanofibers tethered to the same microbead might have different contour lengths. Based on previous experiments, when extension did not change upon varying the rotation number, it was an indication that the magnetic bead had only a single tether to the glass slide (Fig. 1f and g).44 We thus selected this kind of nanofibers for further assessment of their mechanical properties (Fig. 1d–g and S1†).
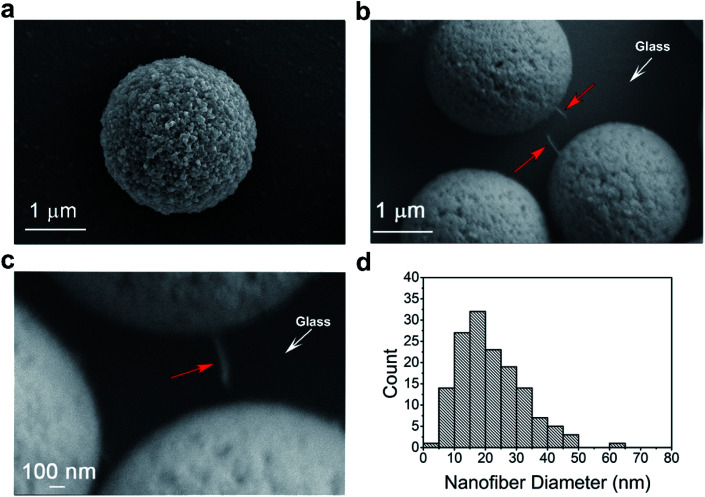
To further confirm the linking effects between nanofibers and microbeads, we performed scanning electron microscopy (SEM) to visualize the CsgA nanofibers and paired microbeads. The magnetic beads were bound to the fibers and tethered to the glass as in the MT experiments. The beads were turned aside by a permanent magnet to reveal the tethered nanofiber. SEM images indeed validated the feasibility of the linking method. For a control sample in which plain microbeads and nanofibers were simply mixed together, no nanofibers were found to be attached to the microbeads (Fig. 2a). However, for the sample using the tag-free linking method, one end of the nanofiber was found to be firmly attached to the microbead, and the other end was adhered to the glass surface, which was the bottom plate of the sample (Fig. 2b and c). We used ImageJ software to measure the diameter of the nanofibers. In Fig. 2b, the length of each pixel on the original photo was measured to be 5.6 nm (the thickness of platinum coating was approximately 2 nm). The diameter was taken as the mean value of data measured at different positions on a fiber. The results showed that the diameters of the CsgA nanofibers varied widely. Among 146 pieces of CsgA nanofibers that were measured, the diameters of the nanofibers ranged from 4 to 64 nm (Fig. 2d) and averaged at 21.7 ± 10.4 nm.
Fig. 2. CsgA nanofibers observed by SEM. (a) The non-crosslinked microbead. (b and c) A single CsgA nanofiber (red arrow) sticking to the bead and the glass surface. (d) Distribution of the diameters of the 146 single fibers.
2.2. Mechanical properties of CsgA nanofibers with different diameters
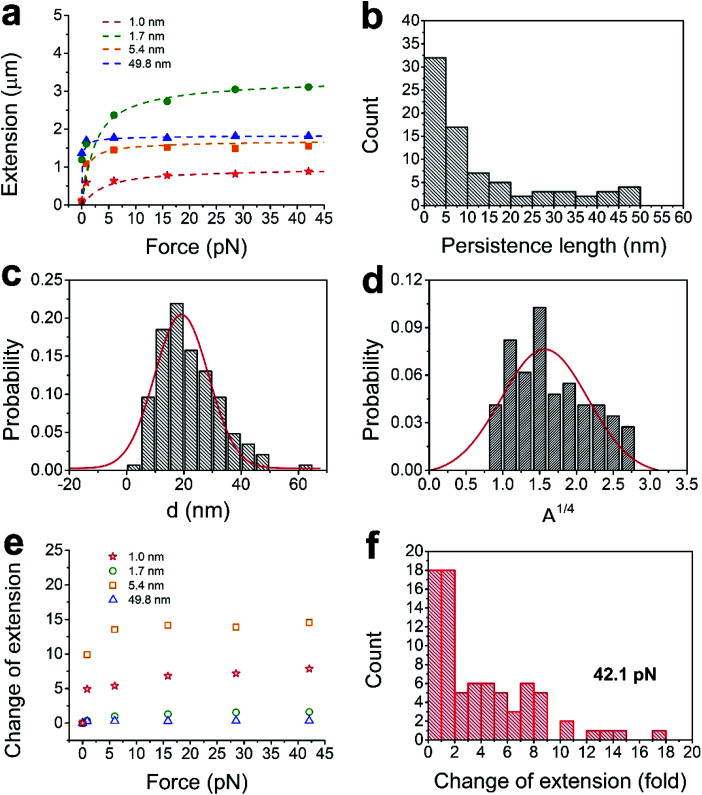
The diameters of the CsgA nanofibers observed by SEM varied widely, and we next explored how much the stiffness of these nanofibers was affected by these variations. Samples for MT were prepared following the same linking procedures as the samples for SEM. The mechanical properties of CsgA nanofibers were characterized using single-molecule MT. Among the 78 single nanofibers studied by MT experiments, the force–extension curves of 73 fibers were fitted by the wormlike chain model with R2 > 0.8 (coefficient of determination). These results indicate that the fibers are flexible and exhibit typical behaviors of a wormlike chain polymer. We selected the force–extension curves of four nanofibers with persistence lengths of 1.0, 1.7, 5.4, and 49.8 nm (Fig. 3a). As determined from the curve fittings, the persistence lengths of the 78 nanofibers were found to range from 0.9 to 49.8 nm (Fig. 3b).
Fig. 3. Relations among force–extension, persistence lengths, and diameters. (a) Four typical force–extension measurements of single CsgA nanofibers and the fitted curves (dotted lines) by the wormlike chain model. (b) Distribution of the persistence lengths. (c and d) The distributions of the diameters and the fourth roots of persistence lengths. (e) Relations between force and change of extension of the 4 CsgA nanofibers corresponded to the symbol in (a). (f) Distribution of the changes in extension for 78 CsgA nanofibers at 42.1 pN.
The persistence lengths of the CsgA nanofibers obtained from the MT experiment were much shorter than that of an amyloid fiber made from insulin.45 The persistence length is the length along the backbone of the chain over which random bends occur. This indicates that CsgA fibers are more flexible or easier to be bent than insulin fibers. Moreover, the insulin fibers were made from a solution that was heated at 60 °C for 24 h and then stored at room temperature for one week, whereas our CsgA nanofibers were made from a CsgA solution in only a few hours under room temperature. Both proteins self-assembled into nanofibers with dimensions that increased with time.13 The dimensions, including diameter, are key to accurate measurement of bending stiffness, and consequently, persistence length.
The persistence lengths measured for the nanofibers spanned a relatively large range. This may be caused by the variances in the number of filaments in the nanofibers, given that the persistence length changes with the number of molecules or filaments46 constituting the polymer. The number of filaments in a nanofiber is indicated by its diameter, which increases during self-assembly even within a short period of time. From the classical relation between the diameter d and the persistence length A:
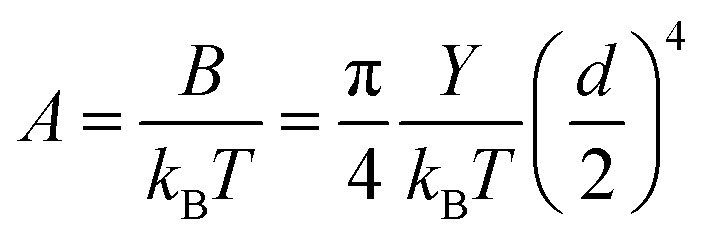 |
1 |
Here, B is the bending stiffness, Y is the Young's modulus, kB is the Boltzmann constant, and T is the absolute temperature.47 For a structurally uniform material, the Young's modulus is a fixed value, regardless of its size. Here, the Young's modulus of the CsgA nanofiber is assumed to be a constant. Then, A is proportional to the fourth power of d. Although A and d were measured from different nanofibers, those nanofibers were produced by the same procedures and were considered to be identical. The distributions of d and A1/4 both roughly follow Gaussian distribution (Fig. 3c and d). This shows that the A value indeed changes with d, and B depends on the fourth power of d. A is proportional to the square of the cross-sectional area of the nanofiber. In our experiments, as A increases from 0.9 to 49.8 nm, which is about the square of 7, the cross-sectional area also increases by 7 folds, i.e., the thickest nanofiber we tested was composed of 7 filaments. In addition, the bending stiffness of CsgA nanofiber increased with the fourth power of the diameter.
Clearly, the distribution of A1/4 and the distribution of d do not perfectly match, which is probably due to off-centered attachment of the magnetic microbead to the nanofiber, or due to the one-filament linkage in a fiber (Fig. 1b). The diameter of magnetic microbeads was 2.8 μm, and the majority of the measured CsgA nanofibers were smaller than this length. Thus, part of the extension change measured at low forces was caused by rotation that occurred in eccentric magnetic microbeads.29 Also, one filament is covalently linked, and other filaments are non-covalently attached and follow the linked filament during the force measurements. This situation may lead to diverse distribution of the force–extension results.
We estimated the Young's modulus of the CsgA nanofiber using the average diameter and average persistence length. The median persistence length was ∼7 nm in the MES buffer. The average diameter of pure CsgA nanofiber in aqueous solution was 1.39 ± 0.48 nm based on our previous report,17 which is more suitable than the SEM measurements here, given that the SEM sample was dry and sputtered with platinum nanoparticles. Therefore, the average Young's modulus 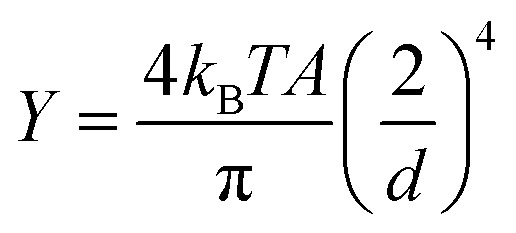 is ∼154.6 MPa, according to the above equation. The lower limit is ∼22.1 MPa from our data, whereas the upper limit value varies based on diameter.
is ∼154.6 MPa, according to the above equation. The lower limit is ∼22.1 MPa from our data, whereas the upper limit value varies based on diameter.
As one of the representatives of amyloid nanofibers, curli nanofibers have similar structures to other amyloid nanofibers.48,49 Thus, our method may also be applied to other amyloid nanofibers. We noted that the Young's moduli of amyloid nanofibers measured by AFM varied widely.16,24,45,50,51 In these studies, the diameters of the fibers were much smaller than the diameter of blunt-ended AFM tips. The fibers usually lay on a flat surface during AFM measurements. It was difficult to align the round tip onto the vertex point of the round fiber, which may have led to inaccuracy. In addition, the force applied by AFM was at the side of the round nanofiber and only acted on the local part of the nanofiber instead of applying uniform stress. The value obtained by our method reflects the bending stiffness of the amyloid nanofiber instead of the Young's modulus. This probably explains the large differences in the values (from MPa to GPa) obtained in previous studies, because of the different diameters of the measured nanofibers.
In order to investigate the tensile properties, we further analyzed the relation between force (f) and change in extension of CsgA nanofibers (Fig. 3e). The change in extension was determined by ΔL/L0, where L0 was the extension of the nanofibers at f = 0 pN, and ΔL was the difference between extensions under f = 0 pN and an applied force. The change in extension increased with the force until it reached a plateau. At 42.1 pN, about 50% of the CsgA nanofibers exhibited a change in extension from 0.1 to 2, and a few were distributed across a range of 10 to 16, which indicated that these fibers were highly flexible (Fig. 3f). The largest change in extension at 42.1 pN was found to be 17.1, which is close to the theoretical strain of 19.5 reported in a previous study for a single CsgA protein using SMD simulations.20 This is consistent with the nature of both single CsgA nanofibers and CsgA proteins, which consist of β-helices, i.e., β-sheets connected by loops in a single tether. In addition, multiple CsgA proteins have connecting overlaps in the fiber, and, therefore, the ratio of 17.1 is slightly shorter than the theoretical strain. In contrast, typical DNA increases only about 4 times in the force range.52,53 Taken together, the results imply that the CsgA nanofibers have excellent tensile properties.
3. Conclusions
In summary, we have demonstrated that the force–extension relation of 90% of the CsgA nanofibers follows the wormlike chain model. We experimentally measured the persistence lengths of CsgA nanofibers and estimated the number of filaments in nanofibers based on the persistence length analysis. The bending stiffness of the nanofiber depends on the fourth power of its diameter. The nanofiber exhibits outstanding tensile strength, with 17-time extension of its original length before breaking. Our study here therefore provides new insights into the tensile properties of curli nanofibers and lays the foundation for exploiting the outstanding mechano-chemical properties of amyloid nanofibers for diverse applications.
Author contributions
C. Z. and B. X. conceived the project. J. L., Y. L., K. Z., K. D., B. X. and C. Z. performed the experiments. B. X., J. L., Y. L. and C. Z. analyzed the data. J. L., Y. L., P. G., C. Z. and B. X. wrote the paper.
Conflicts of interest
There are no conflicts to declare.
Abbreviations
- AFM
Atomic force microscopy
- MT
Magnetic tweezers
- SEM
Scanning electron microscopy
- EDAC
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide
- SMD
Steered molecular dynamics
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (11772133, 11372116) and by the Fundamental Research Funds for the Central Universities (HUST 0118012051). We thank Professors Jie Yan (National University of Singapore), Mian Long, Jianhua Wu, Ying Fang, Ms Quan Long, and Mr Zhen Li for technical assistance and helpful discussions.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra02749a
References
- King C. Y. Tittmann P. Gross H. Gebert R. Aebi M. Wuthrich K. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler S. Estrada L. Lagos R. Baeza M. Castilla J. Soto C. J. Biol. Chem. 2005;280:26880–26885. doi: 10.1074/jbc.M502031200. [DOI] [PubMed] [Google Scholar]
- Fowler D. M. Koulov A. V. Alory-Jost C. Marks M. S. Balch W. E. Kelly J. W. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Knowles T. P. Mezzenga R. Adv. Mater. 2016;28:6546–6561. doi: 10.1002/adma.201505961. [DOI] [PubMed] [Google Scholar]
- Wei G. Su Z. Reynolds N. P. Arosio P. Hamley I. W. Gazit E. Mezzenga R. Chem. Soc. Rev. 2017;46:4661–4708. doi: 10.1039/C6CS00542J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U. Barlev M. Seijffers R. Oelschlager T. A. Hacker J. Ron E. Z. Infect. Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U. Oelschlaeger T. A. Hacker J. Ron E. Z. FEMS Microbiol. Lett. 2002;212:55–58. doi: 10.1111/j.1574-6968.2002.tb11244.x. [DOI] [PubMed] [Google Scholar]
- Johansson C. Nilsson T. Olsen A. Wick M. J. FEMS Immunol. Med. Microbiol. 2001;30:21–29. doi: 10.1111/j.1574-695X.2001.tb01545.x. [DOI] [PubMed] [Google Scholar]
- Sleutel M. Van den Broeck I. Van Gerven N. Feuillie C. Jonckheere W. Valotteau C. Dufrene Y. F. Remaut H. Nat. Chem. Biol. 2017;13:902. doi: 10.1038/nchembio.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isa L. Jung J. M. Mezzenga R. Soft Matter. 2011;7:8127–8134. doi: 10.1039/C1SM05602F. [DOI] [Google Scholar]
- Jordens S. Isa L. Usov I. Mezzenga R. Nat. Commun. 2013;4:1917. doi: 10.1038/ncomms2911. [DOI] [PubMed] [Google Scholar]
- Jung J. M. Gunes D. Z. Mezzenga R. Langmuir. 2010;26:15366–15375. doi: 10.1021/la102721m. [DOI] [PubMed] [Google Scholar]
- Chen A. Y. Deng Z. T. Billings A. N. Seker U. O. S. Lu M. Y. Citorik R. J. Zakeri B. Lu T. K. Nat. Mater. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C. Gurry T. Cheng A. A. Downey J. Deng Z. T. Stultz C. M. Lu T. K. Nat. Nanotechnol. 2014;9:858–866. doi: 10.1038/nnano.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Pu J. An B. Li Y. Shang Y. Ning Z. Liu Y. Ba F. Zhang J. M. Zhong C. Adv. Mater. 2018;30:1705968. doi: 10.1002/adma.201705968. [DOI] [PubMed] [Google Scholar]
- Li Y. Li K. Wang X. An B. Cui M. Pu J. Wei S. Xue S. Ye H. Zhao Y. Liu M. Wang Z. Zhong C. Nano Lett. 2019;19:8399–8408. doi: 10.1021/acs.nanolett.9b02324. [DOI] [PubMed] [Google Scholar]
- Cui M. Qi Q. Gurry T. Zhao T. An B. Pu J. Gui X. Cheng A. A. Zhang S. Xun D. Becce M. Briatico-Vangosa F. Liu C. Lu T. K. Zhong C. Chem. Sci. 2019;10:4004–4014. doi: 10.1039/C9SC00208A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L. Li Y. Chen S. Li K. Huang Y. Zhu Y. Sun Z. Zhang J. He Y. Cui M. Wei S. Qiu F. Zhong C. Liu W. ACS Appl. Mater. Interfaces. 2019;11:32373–32380. doi: 10.1021/acsami.9b08656. [DOI] [PubMed] [Google Scholar]
- Mao X. Li K. Liu M. Wang X. Zhao T. An B. Cui M. Li Y. Pu J. Li J. Wang L. Lu T. K. Fan C. Zhong C. Nat. Commun. 2019;10:1395. doi: 10.1038/s41467-019-09369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBenedictis E. P. Keten S. Soft Matter. 2019;15:1243–1252. doi: 10.1039/C8SM02046A. [DOI] [PubMed] [Google Scholar]
- Knowles T. P. J. Buehler M. J. Nat. Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- Buehler M. J. Keten S. Nano Res. 2008;1:63–71. doi: 10.1007/s12274-008-8006-7. [DOI] [Google Scholar]
- Wang A. Keten S. npj Comput. Mater. 2019;5:29. doi: 10.1038/s41524-019-0154-7. [DOI] [Google Scholar]
- Abdelwahab M. T. Kalyoncu E. Onur T. Baykara M. Z. Seker U. O. S. Langmuir. 2017;33:4337–4345. doi: 10.1021/acs.langmuir.7b00112. [DOI] [PubMed] [Google Scholar]
- Oh Y. J. Hubauer-Brenner M. Gruber H. J. Cui Y. D. Traxler L. Siligan C. Park S. Hinterdorfer P. Sci. Rep. 2016;6:33909. doi: 10.1038/srep33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Wang A. DeBenedictis E. P. Keten S. Nanotechnology. 2017;28:464002. doi: 10.1088/1361-6528/aa8f72. [DOI] [PubMed] [Google Scholar]
- Seol Y. Neuman K. C. Cell. 2013;153:1168. doi: 10.1016/j.cell.2013.04.047. [DOI] [PubMed] [Google Scholar]
- Kim J. Zhang C. Zhang X. Springer T. A. Nature. 2010;466:992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Zeng X. Lu C. Yan J. Nanotechnology. 2017;28:414002. doi: 10.1088/1361-6528/aa837e. [DOI] [PubMed] [Google Scholar]
- Berghuis B. A. Kober M. van Laar T. Dekker N. H. Methods. 2016;105:90–98. doi: 10.1016/j.ymeth.2016.03.025. [DOI] [PubMed] [Google Scholar]
- Yu M. Yuan X. Lu C. Le S. Kawamura R. Efremov A. K. Zhao Z. Kozlov M. M. Sheetz M. Bershadsky A. Yan J. Nat. Commun. 2017;8:1650. doi: 10.1038/s41467-017-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Yu M. Hovan L. Zhao Z. Ervasti J. Yan J. ACS Nano. 2018;2:12140–12148. doi: 10.1021/acsnano.8b05721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. L. Dougan L. Rep. Prog. Phys. 2016;79:076601. doi: 10.1088/0034-4885/79/7/076601. [DOI] [PubMed] [Google Scholar]
- Knowles T. P. Fitzpatrick A. W. Meehan S. Mott H. R. Vendruscolo M. Dobson C. M. Welland M. E. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- Chen H. Yuan G. Winardhi R. S. Yao M. Popa I. Fernandez J. M. Yan J. J. Am. Chem. Soc. 2015;137:3540–3546. doi: 10.1021/ja5119368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B. Wu S. Liu S. Rodriguez-Aliaga P. Yu J. Cui S. Nanoscale. 2015;7:2970–2977. doi: 10.1039/C4NR07140A. [DOI] [PubMed] [Google Scholar]
- Xing Y. Chaudry Q. Shen C. Kong K. Zhau H. Chung L. Petros J. A. O'Regan R. M. Yezhelyev M. V. Simons J. W. Wang M. D. Nie S. Nat. Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- Lu C. Le S. Chen J. Byrd A. K. Rhodes D. Raney K. D. Yan J. Nucleic Acids Res. 2019;47:7494–7501. doi: 10.1093/nar/gkz541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G. Le S. Yao M. Qian H. Zhou X. Yan J. Chen H. Angew. Chem., Int. Ed. 2017;56:5490–5493. doi: 10.1002/anie.201700411. [DOI] [PubMed] [Google Scholar]
- Yao M. Qiu W. Liu R. Efremov A. K. Cong P. Seddiki R. Payre M. Lim C. T. Ladoux B. Mège R.-M. Yan J. Nat. Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- Yan J. Yao M. Goult B. Sheetz M. Cell. Mol. Bioeng. 2015;8:151–159. doi: 10.1007/s12195-014-0364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vlaminck I. Dekker C. Annu. Rev. Biophys. 2012;41:453–472. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- Neuman K. C. Nagy A. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B. McLean M. M. Lei X. Marko J. F. Johnson R. C. Sci. Rep. 2016;6:23697. doi: 10.1038/srep23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. F. Knowles T. P. J. Dobson C. M. MacPhee C. E. Welland M. E. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko J. F. Phys. Rev. E. 1997;55:1758. doi: 10.1103/PhysRevE.55.1758. [DOI] [Google Scholar]
- Diaz A. M. Zhang Z. Y. Lee B. Luna F. M. H. Li Sip U. Y. Lu X. Y. Heidings J. Tetard L. Zhai L. Kang H. R. ACS Omega. 2018;3:18304–18310. doi: 10.1021/acsomega.8b02822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D. Hawthorne W. J. Lo J. Dear A. Jain N. Meisl G. Andreasen M. Fletcher C. Koch M. Darvill N. Scull N. Escalera-Maurer A. Sefer L. Wenman R. Lambert S. Jean J. Xu Y. Q. Turner B. Kazarian S. G. Chapman M. R. Bubeck D. de Simone A. Knowles T. P. J. Matthews S. J. Sci. Rep. 2016;6:24656. doi: 10.1038/srep24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenAkker C. C. Engel M. F. Velikov K. P. Bonn M. Koenderink G. H. J. Am. Chem. Soc. 2011;133:18030–18033. doi: 10.1021/ja206513r. [DOI] [PubMed] [Google Scholar]
- Guo S. Akhremitchev B. B. Biomacromolecules. 2006;7:1630–1636. doi: 10.1021/bm0600724. [DOI] [PubMed] [Google Scholar]
- Adamcik J. Berquand A. Mezzenga R. Appl. Phys. Lett. 2011;98:193701–193710. doi: 10.1063/1.3589369. [DOI] [Google Scholar]
- Fu H. Chen H. Zhang X. Qu Y. Marko J. F. Yan J. Nucleic Acids Res. 2011;39:3473–3481. doi: 10.1093/nar/gkq1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B. Zhang H. Johnson R. C. Marko J. F. Nucleic Acids Res. 2011;39:5568–5577. doi: 10.1093/nar/gkr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.