Abstract
Molecular mimicry of host proteins by pathogens constitutes a strategy to hijack the host pathways. At present, there is no dedicated resource for mimicked domains and motifs in the host–pathogen interactome. In this work, the experimental host–pathogen (HP) and host–host (HH) protein–protein interactions (PPIs) were collated. The domains and motifs of these proteins were annotated using CD Search and ScanProsite, respectively. Host and pathogen proteins with a shared host interactor and similar domain/motif constitute a mimicry pair exhibiting global structural similarity (domain mimicry pair; DMP) or local sequence motif similarity (motif mimicry pair; MMP). Mimicry pairs are likely to be co-expressed and co-localized. 1,97,607 DMPs and 32,67,568 MMPs were identified in 49,265 experimental HP-PPIs and organized in a web-based resource, ImitateDB (http://imitatedb.sblab-nsit.net) that can be easily queried. The results are externally integrated using hyperlinked domain PSSM ID, motif ID, protein ID and PubMed ID. Kinase, UL36, Smc and DEXDc were frequent DMP domains whereas protein kinase C phosphorylation, casein kinase 2 phosphorylation, glycosylation and myristoylation sites were frequent MMP motifs. Novel DMP domains SANT, Tudor, PhoX and MMP motif microbody C-terminal targeting signal, cornichon signature and lipocalin signature were proposed. ImitateDB is a novel resource for identifying mimicry in interacting host and pathogen proteins.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00726-022-03163-3.
Keywords: Host–pathogen protein–protein interaction, Molecular mimicry, Domain, Motif, Infectious diseases
Introduction
In nature, host–pathogen Protein–protein interactions (HP-PPIs) are ubiquitous and essential for initiation and propagation of infectious diseases (Dean Southwood 2019). Pathogenesis involves interactions between the signalling networks of the host and pathogen. Recent studies regarding HP-PPIs focus on the mechanisms employed by pathogens to hijack and exploit the host immune system for their own survival (Mayer et al. 2019; Nicod et al. 2017). Processes for molecular mimicry have evolved to enable the proteins of pathogens to imitate the host proteins, disrupt their interactions, and disturb the signalling pathways (Guven-Maiorov et al. 2016). Thus, the interacting proteins and pathways of the pathogen may be conceived to be in a continuum with those of the host. The mimicry of host antigenic determinants as a survival mechanism was described initially in parasites (Damian 1964). The ability of the pathogen to mimic the host components may be achieved by horizontal gene transfer (Diaz et al. 1997) or convergent evolution (Stebbins and Galan 2000). Molecular mimicry can take four distinct forms as depicted in Table 1 with examples; (a) similarity in both sequence and structure of a full-length protein/ functional domain (Sharp et al. 1997; Dar and Sicheri 2002), (b) structural similarity without an apparent sequence similarity(Standfuss 2015; Burg, et al. 2015), (c) similarity in the sequence of a short linear motif (Wu et al. 2005), (d) similarity of only the binding site architectures or interface mimicry without apparent sequence homology (Huang et al. 2009). The local similarities between epitopes from the infectious agents and antigens present in the host can also lead to autoimmune diseases (Cusick et al. 2012; Venigalla et al. 2020; McClain et al. 2005).
Table 1.
Table depicting the different types of molecular mimicry
| S. no | Types of molecular mimicry | Examples | PDB structure ID |
|---|---|---|---|
| a | Similarity in both sequence and structure of a full-length protein or a functional domain | K3L protein of Vaccinia Virus mimics the S1 domain of human eIF-2-α to bind to human PRK (Sharp et al. 1997; Dar and Sicheri 2002) | K3L S1 domain: PDB ID: 1luz |
| eIF-2-α S1 domain: PDB ID: 1kl9 | |||
| b | Structural similarity without an apparent sequence similarity | Viral proteins (US28 protein of human cytomegalovirus and vMIP-II protein of Kaposi Sarcoma virus) mimics both the structures and interactions of their host counterparts (CXCR4 and CX3CL1, respectively) although they have a very low sequence similarity (Standfuss 2015; Burg, et al. 2015) | US28-CX3CL1 (PDB ID: 4xt1) |
| vMIP-II-CXCR4) PDB ID: 4rws) | |||
| c | Similarity in the sequence of a short linear motif | The LMP1 protein of Epstein–Barr Virus mimics the motif PxQxT of CD40 to interact with TRAF3(Wu et al. 2005) | LMP1-TRAF3(PDB ID: 1zms) |
| TRAF3-CD40(PDB ID: 1fll) | |||
| d | Similarity of only the binding site architectures (interface mimicry) without sequence homology | Map of E. coli, and SopE of S. typhimurium form interfaces with human Cdc42 that mimic the interface between Cdc42 and human intersectin (Huang et al. 2009) | Cdc42-Map (PDB ID: 3gcg), |
| Cdc42-SopE (PDB ID: 1gzs), | |||
| Cdc42-Intersectin (PDB ID: 1ki1) |
Hijacking of host networks by the pathogen essentially works by imitation and competition with endogenous host–host interactions (Franzosa and Xia 2011; Yapici-Eser et al. 2021). It has been observed that a majority of the pathogen proteins bind to the same domain on their respective host target that are otherwise bound by host endogenous proteins (Chen and Xia 2021). In addition to employing mimicking host domains, effectors also employ short linear motifs that bind to host domains with similar specificities as host endogenous proteins, albeit sharing little homology with the latter(Samano-Sanchez and Gibson 2020). However, allosteric modes of binding have also been studied as seen in LegK7 protein of Legionella pneumophila (Lee et al. 2020). Some small molecules have been implicated in inhibition of motif mimicry mediated interactions. Anacardic acid, a small inhibitor has been developed against Anaplasma phagocytophilum effector AmpA that hijacks host cell SUMOylation (Beyer et al. 2015). N-(p-Amylcinnamoyl) anthranilic acid is an inhibitor that has been developed against YxxØ-motif found in multiple viruses that exploit host AP2M1 for intracellular trafficking (Yuan et al. 2020). A CAAX peptidomimetic drug has been developed against CAAX motif which is used by L. pneumophila to exploit host prenylation machinery to facilitate targeting of effector proteins to membrane-bound organelles during intracellular infection (Ivanov et al. 2010). These promising developments suggest that host–pathogen interactions can be targeted using small-molecule inhibitors and peptide mimetics as anti-infective strategies.
The existing methods of detection of mimicry are simply based on identifying sequence or structure similarity. A previously available database, namely mimicDB (Ludin et al. 2011) provides information about molecular mimicry proteins or epitopes involved in a limited number of human parasites. Another database miPepBase (Garg et al. 2017) lists the experimentally verified mimicry peptides involved in auto-immune disease. A computational pipeline using pBLAST against the human proteome has also been implemented for the prediction of the molecular mimicry candidates in bacterial pathogens (Doxey and McConkey 2013). An interface mimicry-based method, the HMI-PRED server (Guven-Maiorov et al. 2020) carries out structural prediction of given HP-PPIs. However, it is limited due to the requirement of the structure of the microbial protein involved in mimicry.
Similarity between motifs and domains of the host and pathogen proteins does not necessarily indicate their actual interaction. This is further dependent on the proteins having simultaneous expression and being present in the same cellular compartment. The analysis of the PPIs in yeast and human showed that a large majority of the interactions occur between proteins present in the same subcellular compartment (Schwikowski et al. 2000; Gandhi et al. 2006). Studies have also shown that functionally related or interacting proteins from the same pathways share Gene Ontology, and usually constitute a higher co-expression score (Wolfe et al. 2005; Durmus Tekir et al. 2012). Therefore, the resemblance between the experimentally validated host and pathogen interactors of the same host protein may increase the confidence in the identification of molecular mimicry candidates due to colocalization and co-expression of the interacting protein pairs.
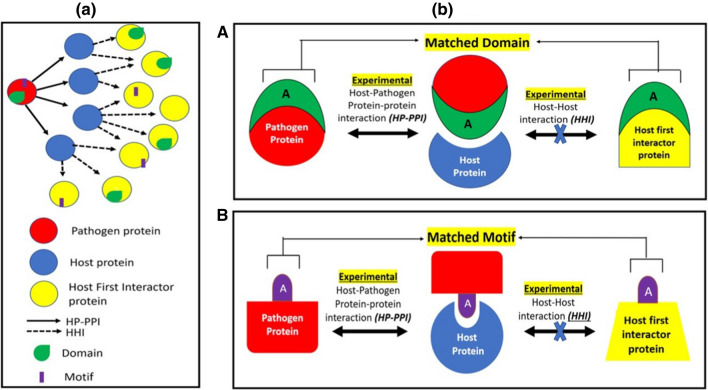
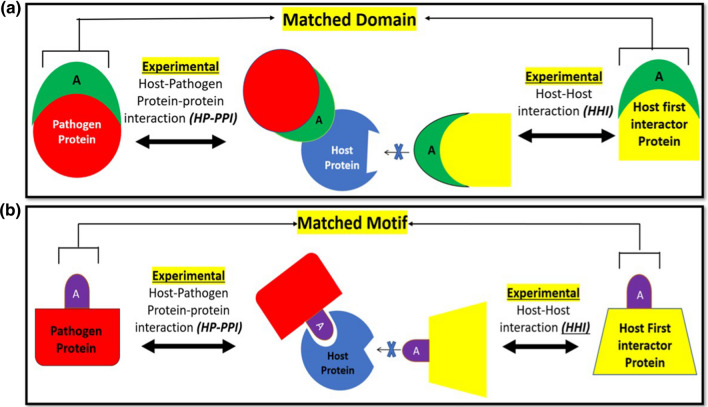
A pair of pathogen and host proteins that interact with the same host protein may share a common domain indicating global structural similarity or a common short linear sequence motif indicating local sequence similarity. This pathogen protein may be involved in molecular mimicry of the host protein. These pathogen and host protein pairs have been termed respectively as domain mimicry pair (DMP) and motif mimicry pair (MMP) as shown schematically in Fig. 1a, b. A competitive mode of binding of both interaction partners to the same protein is shown in Fig. 1b while allosteric mode of binding is depicted in Fig. 2. Delineating the DMPs and MMPs provides information about the host interactions that are likely to be disrupted by pathogen protein mimicry.
Fig. 1.
Panel (a) Domain Mimicry Pair (DMPs) and Motif Mimicry Pairs in Host–pathogen protein–protein interactions and Host–host interaction network: a pathogen protein(red) interacts with many host proteins(blue) that in turn interacts with many other host proteins (yellow). The pathogen protein(red) and host first interactor protein(yellow) with a common interacting host protein (blue) are compared for similar domains (green) and motifs (purple) to determine DMPs and MMPs respectively. Panel (b) A Schematic of individual DMP: a host first interactor protein (yellow) and pathogen protein (red) that interacts with the same host protein (blue) share the same domain ‘A’ (green). B Schematic of individual MMP: a host first interactor protein (yellow) and pathogen protein (red) that interacts with the same host protein (blue) share the same motif ‘A’ (purple)
Fig. 2.
a Schematic of DMP through allosteric binding: a host first interactor protein (yellow) and pathogen protein (red) that interact with the same host protein (blue) share the same domain ‘A’ (green). Here, the pathogen protein and host first interactor protein bind to the same host through the mimicked domain but at different sites i.e., allosterically, thus leading to a change in shape of the host protein binding site. b Schematic of MMP through allosteric binding: a host first interactor protein (yellow) and pathogen protein (red) that interacts with the same host protein (blue) share the same motif ‘A’ (purple). Here, the pathogen protein and host first interactor protein bind to the same host through the mimicked motif but at different sites, i.e., allosterically thus leading to a change in shape of the host protein binding site
In this work, we collated the entire set of experimental HP-PPIs from interaction databases to compute their DMPs and MMPs, which were organized in the form of a publicly available database, ImitateDB available online at http://imitatedb.sblab-nsit.net. The ImitateDB resource can help researchers to search for organism-wise mimicry patterns prominent in the host–pathogen interactome. It houses 1,97,607 DMPs and 32,67,568 MMPs. Several novel potential domain and motif mimics have been identified in our dataset. Specific domains or motifs imitated commonly by many pathogens are likely to be responsible for microbial virulence suitable for drug/vaccine targeting. Thus, ImitateDB constitutes a source of information for molecular imitation in HP-PPIs for researchers in the field of infectious diseases and microbiology.
Materials and methods
HP-PPI data collection and cleaning
The information regarding the HP-PPIs was collected from different databases namely BioGrid 4.4 (Stark et al. 2006), PHISTO (Durmus Tekir et al. 2013), HPIDb 3.0 (Ammari et al. 2016), MINT (Chatr-aryamontri et al. 2007), IntAct 4.2 (Hermjakob et al. 2004), MPIDB (Goll et al. 2008), UniProt (The UniProt 2017), VirHostNet 3.0 (Navratil et al. 2009), MatrixDB (Launay et al. 2015), I2D 2.9 (Brown and Jurisica 2007), DIP (Xenarios et al. 2000) and InnateDB 5.4 (Lynn et al. 2008). To maintain uniformity in the protein identifiers extracted from different sources, all the identifiers were converted to the corresponding UniProt accession number. Similarly, to maintain uniformity in the syntax/nomenclature of pathogens, all pathogen names were converted into the corresponding NCBI Taxon identifier. The duplicate entries were removed from the data to avoid redundancy. The obsolete entries were either removed or converted into secondary UniProt accession numbers, if available.
Host–host interaction (HHI) data collection
The host proteins directly interacting with the pathogen interacting host proteins (as shown in Fig. 1a) are referred to as the host first interactor proteins. Information about the host first interactor proteins was collected using UniProt. These data were further processed using bioDBnet 2.1 (Mudunuri et al. 2009) to filter out the various non-human interactors of the host proteins and retain only the human interactors of the host proteins.
Domain annotation
The term domain, as used in DMP refers to a tertiary structure (folding unit) which is a distinct functional and/or structural unit of a protein, is independently stable and much larger unit than a motif. Domain annotation was carried out using the NCBI Batch Conserved domain (CD) Search 3.19 (Marchler-Bauer et al. 2011) a sensitive method that searches multiple sequence alignments converted into position-specific scoring matrices (PSSM) using Reverse Position Specific-BLAST. The default parameters of CD search were used with an expect value of 0.01. The domain information was collected in the form of a unique PSSM ID and Domain short name for every unique domain family of all the proteins.
Motif annotation
The term motif, as used in MMP refers to a Prosite pattern. This is a short sequence linear motif corresponding to a short conserved biologically significant region of the sequence. Prosite patterns are represented by sequence signatures, are not independently stable and are much smaller than a domain. The motifs were identified using the Expasy ScanProsite (Castro et al. 2006) API (Application Programming Interface) with the help of a Python script using the get() function to fetch the motifs for a large number of proteins. ScanProsite is considered a standard benchmark for other databases due to the high quality of its motifs (Ferreira and Azevedo 2007). It covers all the important motifs like active sites, binding sites, disulfide bonds and the promiscuous motifs as well like post-translational modification sites that have a role in mimicry mediated host–pathogen interaction and virulence (Kumar et al. 2020). For this study, ScanProsite was run with the “Exclude Profiles” option to eliminate the domains and scan only against the patterns corresponding to short sequence motifs. The motif information for each protein was collected in the form of ScanProsite motif IDs, motif name and pattern.
Identification of shared DMPs and MMPs
The pathogen proteins and host proteins interacting with the same host protein were compared for shared structural domains (global similarity) or short linear sequence motifs (local similarity). This was done using a python script to find the same domain PSSM-IDs and ScanProsite motif IDs between the pathogen protein and host protein interacting with the same host protein.
Identification of COGs (cluster of orthologs) after determining DMPs and MMPs
The pathogen proteins belonging to different strains of the same organism (especially in case of virus) may be orthologs and may lead to redundancy. To determine the number of unique DMPs and MMPs for every pathogen, the orthologous pathogen proteins involved in DMPs and MMPs were clustered into a single COG using eggNOG 5.0 (Huerta-Cepas et al. 2019).
Database and web interface development
After the completion of data processing and formation of domain and motif mimicry pairs, a database was created using MySQL to house the schema and various tables through MySQL Command line client server. Further, an MVC (Model View Controller) web application was created in Node.js using the Express framework. The front end of the web application was developed using HyperText Markup Language (HTML). Embedded JavaScript Templating was used to render HTML with our own set of variables. Cascading Style Sheets (CSS) with JavaScript and jQuery was employed for implementing the various methods and functions.
Data analysis and visualization
Analysis of the database was done using MySQL, Microsoft Excel, R studio and Tableau. MySQL was used to sort the entities and quantitatively measure the frequency of each entity in the entire data. The graphs and highlight tables depicting the analysis were rendered using Microsoft Excel and Tableau. The high-degree pathogen and host proteins were identified using an R script. Pathway and gene ontology annotation of the host proteins was carried out using PANTHER16.0 (Mi et al. 2021), a tool used for functional enrichment of proteins. The Fisher’s exact test with Bonferroni correction for multiple testing (p < 0.05 values) was used to determine statistical overrepresentation during enrichment analysis.
Results and discussion
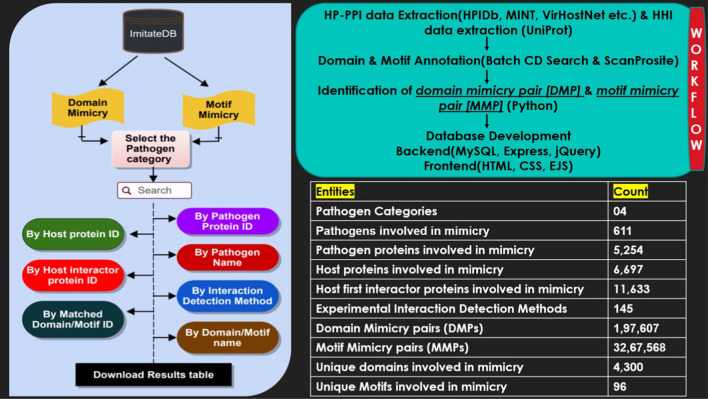
ImitateDB starts with all experimentally determined HP-PPIs. The first interactors of the host proteins were added. These experimentally determined host and pathogen proteins are more likely to be co-expressed and co-localized hence increasing the confidence in the identification of mimicry pairs. The DMPs and MMPs provide information about the matched domains and motifs between the pathogen proteins and first interactor proteins of the respective interacting host proteins. The information in the database is provided for different categories of pathogen like virus, bacteria, and fungi. The pathogens belonging to Protozoa and Amoebozoa are found under the “Others” category. Figure 3 shows the database schema depicting the pipeline followed for all the search options, and the workflow for the development of the database along with the frequency of primary entities.
Fig. 3.
Database schematic: the basic pipeline for search options is represented on the left and the basic workflow as well as the count of entities in the database are shown on the right
Experimental HP-PPI data
Viruses have the highest number of reported HP-PPIs among the different pathogen categories. 5568 pathogen proteins from 629 organisms interacted with 10,078 host proteins with 61,214 HP-PPIs. Of these, 49,249 reported HP-PPIs were of viral origin. In comparison, reported bacterial HP-PPIs were 10,080 while those from other organisms were even fewer. Further, 11,657 host first interactors having 1,03,120 interactions with the host proteins were retrieved as described in the methodology. Domains and motifs of these first interactors were identified and compared with those present in the pathogen proteins. The total as well as the number of unique domains (each corresponding to a unique PSSM-ID) and motifs (each corresponding to unique ScanProsite motif ID) annotated in pathogen proteins and host interactor proteins are shown in (Table 2).
Table 2.
Number of entitiesa for pathogen proteins and host first interactor proteins
| Entities | Pathogen proteins | Host first interactor proteins |
|---|---|---|
| Total count of entities | ||
| Total proteins in HP-PPIs and HHIs | 5568 from 629 pathogens | 11,657 |
| Total domains | 68,838 | 4,78,710 |
| Unique domains | 17,465 | 25,245 |
| Total motifs | 31,594 | 79,944 |
| Unique motifs | 1046 | 1661 |
| Count of entities involved in molecular mimicry | ||
| Total proteins involved in Molecular mimicry | 5254 from 611 pathogens | 11,633 |
| Proteins involved in domain mimicry | 607 from 146 pathogens | 1,558 |
| Proteins involved in motif mimicry | 5254 from 611 pathogens | 11,633 |
| Unique domains mimicked/shared between pathogen and host interactor proteins | 4300 | |
| Unique motifs mimicked/ Shared between pathogen and host interactor proteins | 96 | |
aSince every protein has a unique UniProt accession ID that makes that protein unique and different from other, the count for different proteins is done based on the UniProt accession of each protein. Every pathogen strain (having a unique NCBI taxon identifier) as a different organism and every protein of that organism (having a unique UniProt accession) is considered as a distinct entity. The count of different domains is based on the PSSM-ID of each domain. The count of different motifs is based on the ScanProsite Motif-ID of each motif
MMPs are more numerous in comparison with DMPs
Out of the 5568 pathogen proteins from 629 pathogens, a total of 5254 proteins from 611 pathogens had similar domains or motifs as host interactor proteins (Table 2). The DMPs in the entire database were found to be 1,97,607 whereas the MMPs were found to 32,67,568. The number of DMPs, MMPs, total HP-PPIs, HP-PPIs characterized by mimicked domain and motif for each pathogen category are listed in (Table 3). Viruses showed the highest number of DMPs and MMPs, likely to be due to the preponderance of virus HP-PPIs in the data.
Table 3.
Number of DMPs, MMPs, total HP-PPIs and HP-PPIs characterized by mimicked domains and motifs for each pathogen category
| Pathogen category | Total number of HP-PPIs | Number of HP-PPIs in mimicked domains | Number of HP-PPIs in mimicked motifs | Number of DMPs | Number of MMPs |
|---|---|---|---|---|---|
| Virus | 49,249 | 822 | 39,694 | 1,07,325 | 21,99,131 |
| Bacteria | 10,066 | 221 | 7708 | 5254 | 5,76,195 |
| Fungi | 1869 | 496 | 1833 | 75,729 | 4,88,705 |
| Others | 30 | 12 | 30 | 9299 | 3537 |
| Total | 61,214 | 1551 | 49,265 | 1,97,607 | 32,67,568 |
Interestingly, of the total 61,214 HP-PPIs reported, only 1551 were found to be characterized by domain mimicry whereas 49,265 were found to be characterized by mimicked motifs. The total number of HP-PPIs, the fraction of HP-PPIs characterized by mimicked domains and by motifs were compared across pathogen categories and are shown in Supplementary Figure S1. Motif mimicry dominates in number over domain mimicry across all pathogen categories. Previous studies have also reported the extensive use of motif mimicry by viral proteomes (Davey et al. 2011; Duro et al. 2015; Via et al. 2015; Garamszegi et al. 2013). However, due to the short and ambiguous nature of the motifs, some false positives are also expected. Therefore, the detected MMPs need to be carefully examined and validated in future works.
Domain mimicry
Table 2 shows the number of pathogen proteins and host interactor proteins involved in domain mimicry or being shared between the pathogen and host interactor proteins. As there were multiple instances of every mimicked domain, we looked for unique domains. There were 4300 unique domains shared by the pathogen and host first interactor proteins. The largest number of DMPs were found for the protein Serine Threonine Protein Kinase US3 (UniProt ID: P04413) from human herpesvirus 1 strain 17 (HHV-1). It forms 61,609 DMPs predominantly consisting of STKc_MST3_like domains (PSSM-ID: 270,786). The top 10 pathogens involved in domain mimicry along with the number of DMPs are shown in Supplementary Table S1.
The top 10 most frequent mimicked domains are shown in Supplementary Figure S2 (a). PHA03247 (large tegument protein UL36 domain family) was the most frequent among DMPs. UL36 is an important domain family that is crucial for virus host interaction and host immune evasion (Newcomb and Brown 2010). UL36 is found to be colocalized with host/viral membrane proteins. It aids in the assembly and cell entry of Herpes Simplex Virus (Schipke et al. 2012). The top 10 most frequently occurring mimicked domains in different pathogen categories are shown in Supplementary Table S2. Of these, the DEAD-like helicase domain superfamily was frequent among viral DMPs and has been previously reported as an emerging class of host domains mimicked by viral pathogens (Meier-Stephenson et al. 2018).
In case of bacteria, viruses and fungi, Rad50 ATPase and SbcC domains were found to be commonly mimicked domains. Both these domains are highly conserved among eukaryotes (humans and fungi), bacteria and viruses, (Cromie and J.C.C., D R Leach 2001; Yoshida et al. 2011) and have been involved in disrupting the host DNA repair pathways (Gagnaire et al. 2017; Lilley et al. 2007). The pathogens with the highest number of DMPs and MMPs in different pathogen categories, i.e., virus, bacteria, fungi, and others are listed in Supplementary data Tables S3, S4, S5 and S6 respectively.
Motif mimicry
Table 2 shows the number of pathogen proteins and host interactor proteins involved in motif mimicry along with the number of unique motifs being mimicked or being shared between the pathogen and host interactor proteins. As there were multiple instances of every mimicked motif, we looked for unique motifs. There were only 96 unique motifs shared by pathogen and host first interactor protein. The largest number of MMPs were found for the Polymerase basic protein 2 from influenza A virus strain A/Wilson-Smith/1933 H1N1. It forms 35,385 MMPs predominantly containing of Protein kinase C or PKC_PHOSPHO_SITE motifs (ScanProsite Motif ID: PS00005). The top 10 pathogens by the count of MMPs are listed in Supplementary Table S7. It was observed that Saccharomyces cerevisiae S288c had the maximum count of DMPs and MMPs though the total number of reported HP-PPIs were very low in comparison with virus or bacteria. The genes that regulate cellular processes in humans have equivalents that control cell division in yeasts, thus facilitating alteration of the host cellular machinery (Cazzanelli et al. 2018).
S. cerevisiae is an opportunistic pathogen as it is found to be associated with cutaneous infections, systemic bloodstream infections and infections of essential organs in immunocompromised or critically ill patients (Perez-Torrado and Querol 2015). Escherichia coli K12 is another opportunistic pathogen in our dataset as it is found to switch over its otherwise dormant pathogenic machinery to exert ill effects on the host under specific conditions such as dysbiosed gut microbiome composition, compromised immune system or a lack of gut microbe competition(Bhat et al. 2019). Thus, HP-PPIs, DMPs and MMPs from these organisms are of interest for the role of mimicry proteins in hijacking of human pathways and development of therapeutics against it.
The total count for the top 10 most frequently occurring motifs in the database is shown in Supplementary Figure S2(b), which indicates the predominance of phosphorylation sites for PKC and casein kinase II (CK2). PKC and CK2 family of serine/threonine kinases plays essential roles in hijacking multiple signalling pathways in humans leading to many viral infections (Keating and Striker 2012). Sites for N-myristoylation, amidation, and N-glycosylation were amongst the most frequently mimicked motifs. N-glycosylation site is a frequently occurring motif used by several pathogen proteins (especially viral glycoproteins) to evade the human immune system (Crispin and Doores 2015; Crispin et al. 2018). The envelope proteins of viruses like HIV-1 are heavily glycosylated and can provide camouflage against the human proteins, leading to alteration of immune recognition (Wagh et al. 2018; Seabright et al. 2019). N-myristoylation motifs, post translational modification sites that have prominent roles in cellular signalling pathways, have been found to be mimicked by viral and bacterial proteins (Davey et al. 2011; Maurer-Stroh and Eisenhaber 2004). A comparative view of the top 10 most frequently mimicked motifs amongst the different pathogen categories is shown in Supplementary Table S8. Additionally, several other commonly mimicked motifs in our data are ABC transporter family signature motif, Q motif, ATP/GTP-binding site motif A (P-loop), arginine-rich motif, ubiquitination site and prenyl group binding site. The number of top 20 mimicked motifs for the top 20 pathogens is shown in Supplementary Table S9.
Mimicry pairs in highly interacting pathogen proteins and host proteins
Several previous studies have shown that essentiality and pathogen fitness are correlated with high number of interactions (Crua Asensio et al. 2017; Ahmed et al. 2018). Therefore, the number of DMPs and MMPs in the top 10 highly interacting pathogen proteins and host proteins were examined (Supplementary Tables S10 and S11, respectively). The top 10 highly interacting pathogen proteins were of viral origin and predominantly formed MMPs. Among host proteins, a few had a very high number of DMPs. It was observed that nuclear factor NF-kappa-B p105 subunit was a part of 491, while Cellular tumor antigen p53 was a part of 180 DMPs.
Chemokine and cytokine, cholecystokinin receptor, epidermal growth factor receptor and platelet-derived growth factor signalling pathways are enriched in host proteins of mimicry pairs
The enriched pathways and processes of the host proteins involved in DMPs and MMPs were annotated. Apart from specific pathways for some autoimmune diseases such as Huntington and Parkinson disease, chemokine and cytokine, cholecystokinin receptor, epidermal growth factor receptor, platelet-derived growth factor signalling pathways, T cell and B-cell activation pathways were enriched among the host proteins constituting the DMPs and MMPs. The enriched pathways along with their corrected p values are listed in supplementary table S12. Similarly, the enriched gene ontologies were determined for the host proteins. The enriched cellular compartments, molecular functions, and biological processes of the host proteins along with their corrected p values are shown in Supplementary tables S13, S14 and S15, respectively.
Selected novel domain and motif mimicry candidates
Several novel candidate mimicry domains like SANT, TCP-1 and Tudor in pathogens were identified from analysis of the ImitateDB data. Some of the novel domain mimics identified in different pathogens along with their functions are shown in Supplementary Table S12. Microbodies C-terminal targeting signal, lipocalin signature and cornichon signature were novel motif mimic candidates. Selected novel mimicry motifs identified in different pathogens along with their functions are shown in Supplementary Table S13.
The ImitateDB web interface
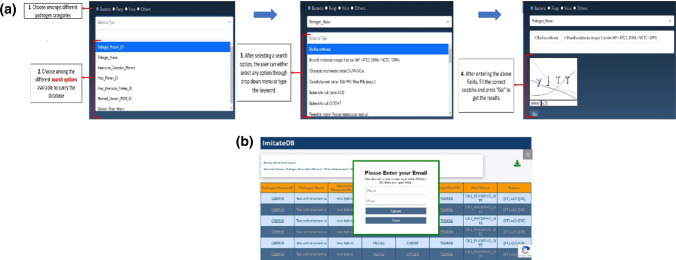
The web interface for the ImitateDB database provides a user-friendly access to the data and allows the user to search for information about DMPs and MMPs using multiple search options. The web interface has a home page that gives an overview of molecular mimicry and procedure for determination of DMPs/MMPs. The interface provides a separate search page to query for domain and motif to search for DMPs and MMPs, respectively. After choosing between domain or motif, the interface provides the next menu to choose among the different categories of pathogens, namely virus, bacteria, fungi and others. In each category, the database can be searched by organism, pathogen protein ID, host protein ID, interaction detection method, host interactor protein ID, matched domain PSSM ID, domain short name, matched motif ID, motif name, or pattern. For easier searching, selection of the category and subcategory leads to the population of a drop-down menu with available options. Additionally, the user can enter a keyword to retrieve the required data. After this selection, the user needs to enter the correct captcha to fetch the results.
The website also has an instructions manual to help the users to query the database and an interpretation manual to help the users to interpret the results. An expanded view of the search panel in the database is shown in Fig. 4a. The results are displayed in the form of a table that can be downloaded. The results are externally integrated using hyperlinked domain PSSM ID, ScanProsite motif ID, protein ID and PubMed ID. The download feature for bulk files has been restricted due to download constraints. For queries yielding results between 10,000 and 1,00,000 records, the user is provided with the results by email using an in-built mailer (as shown in Fig. 4b), that pops up after clicking the download button. The users are advised to check the spam folder for mails from ImitateDB. For queries yielding results above 1,00,000 records, the user can contact the ImitateDB team to obtain the results.
Fig. 4.
a The ImitateDB web interface: expanded view of the search panel of the web interface showing the steps to query the ImitateDB database. b Receive large result files by email: expanded view of the mailer popped up on the ImitateDB interface
Literature validation of mimicry using selected examples
The following selected examples from ImitateDB could be validated from the literature as instances of domains/motifs that are mimicked by the pathogen to bind to the host and also serve as the site of interaction:
DMP: ImitateDB determined a DMP consisting of protein K3L (UniProt ID: P20639) of Vaccinia Virus which mimics the S1 domain of human eIF-2-α (UniProt ID: P05198) to interact with the human PRK (UniProt ID: P19525). Competitive binding experiments suggested that PKR recognizes and interacts with K3L and eIF2α by a common mode due to homology between the S1 domain of K3L and eIF2α (Sharp et al. 1997; Dar and Sicheri 2002; Beattie and T.J., Paoletti E 1991).
MMP: The ImitateDB MMP consisting of protein BHRF1(UniProt ID: P0C6Z1) of Epstein Barr virus (EBV) that mimics the BH2 motif of human Bcl-xl (UniProt ID: Q07817) to interact with the human BAK1(UniProt ID: Q16611). The BHRF1–Bak complex (PDB ID: 2xpx) and Bcl-xl–Bak complex (PDB ID: 1bxl) shows the competitive mode of binding of BHRF1 and Bcl-xl through the BH2 motif on BH3 peptide of BAK (Kvansakul and Hinds 2013).
Other modes of mimicry-mediated binding
There can be instances where the mimicked domain/motif between the interacting host and pathogen protein is not the actual site of interaction. As an example, ImitateDB contains LegAS4 protein (UniProt ID: Q5ZUS4) of L. pneumophila that competes with human histone methyltransferase/H3K9 (UniProt ID: O43463) to bind to human HP1γ (UniProt ID: Q13185), a possible transcriptional repressor of heterochromatin-like complexes. The SET domain of H3K9 is structurally mimicked. However, the binding at HP1γ is not through the SET domain. SET domain mimicry is used by LegAS4 to target human heterochromatin-1 to activate host rDNA transcription as proved through chromatin immunoprecipitation assays (Li et al. 2013). Therefore, in certain cases, the mimicry in the DMP/MMP may be incidental.
Concluding remarks
ImitateDB integrates experimental PPI data with structural/sequence similarity to bring higher confidence to prediction of mimicry between host and pathogen proteins. While this method increases the confidence in prediction for mimicry pairs, the limitation is that the mimicry candidates will only be identified for those organisms and proteins for which the experimental PPIs have been reported. Therefore, the DMPs and MMPs can also be expanded to include predicted PPIs. Apart from this, host and pathogen proteins interact in similar ways as host-endogenous proteins through interface mimicry which is not visible in either domain or motif form.
The curated information in ImitateDB will help in identifying frequent, unique, and novel mimicry domains/motifs among the mimicking host and pathogen proteins. Additionally, MMPs or DMPs allows us to easily identify and model the host protein motif or domain at which the competition for interaction is taking place. The disruption of these HP-PPIs can be regarded as a strategy for developing novel broad-spectrum therapeutics against multiple infectious diseases.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
ST acknowledges Netaji Subhas University of Technology for grant of Teaching cum Research Fellowship. SB acknowledges Indian Council of Medical Research for project Grant ID 2021-6412.
Abbreviations
- HP-PPI
Host–pathogen protein–protein interaction
- HHI
Host–host interaction
- DMP
Domain mimicry pair
- MMP
Motif mimicry pair
- SANT
Swi3, Ada2, N-Cor, and TFIIIB
- CD search
Conserved domain search
- PSSM
Position-specific scoring matrices
- COG
Cluster of orthologs
- PKC
Protein kinase C
- CK2
Casein kinase II
Author contributions
ST: carried out data cleaning, enrichment and organisation, development of the database, analysis, and manuscript preparation. VB: developed the backend and front end of the web interface. TM: carried out data acquisition and cleaning. SB: was involved in conception, design, analysis, and supervision of the study. The manuscript was reviewed by all the authors.
Data availability
The authors confirm that the data supporting the findings of this study are available at http://imitatedb.sblab-nsit.net. The python script for the identification of DMP/MMP is available at the following link: https://github.com/sblab/ImitateDB.git. At present, Imitate DB contains information from HP-PPIs up to April 2021. We plan to update this database regularly as new releases of the contributing databases become available. Apart from collating all the HP-PPIs from different databases, we are working towards mining the HP-PPIs from literature for computation of additional DMPs and MMPs. We plan to compute DMPs and MMPs for predicted HP-PPIs. Work is also ongoing for providing information on DMPs/MMPs for Sars-CoV-2.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Research involving human participants and/or animals. It is declared that no human participants and/or animals are used in this work.
Informed consent
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sonali Tayal and Venugopal Bhatia share equal first authorship.
References
- Ahmed H, et al. Network biology discovers pathogen contact points in host protein–protein interactomes. Nat Commun. 2018;9(1):2312. doi: 10.1038/s41467-018-04632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammari MG et al (2016) HPIDB 2.0: a curated database for host-pathogen interactions. Database (Oxford) 2016 [DOI] [PMC free article] [PubMed]
- Beattie E, T J, Paoletti E. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology. 1991;183(1):419–22. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- Bennett MK, et al. The syntaxin family of vesicular transport receptors. Cell. 1993;74(5):863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Beyer AR, et al. The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol. 2015;17(4):504–519. doi: 10.1111/cmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat MI, et al. Escherichia coli K12: An evolving opportunistic commensal gut microbe distorts barrier integrity in human intestinal cells. Microb Pathog. 2019;133:103545. doi: 10.1016/j.micpath.2019.103545. [DOI] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8(5):R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg JS, et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015;347(6226):1113–7. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzanelli G, et al. The Yeast Saccharomyces cerevisiae as a model for understanding RAS proteins and their role in human tumorigenesis. Cells. 2018;7(2):4. doi: 10.3390/cells7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, et al. MINT: the molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–4. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Xia Y. Structural profiling of bacterial effectors reveals enrichment of host-interacting domains and motifs. Front Mol Biosci. 2021;8:626600. doi: 10.3389/fmolb.2021.626600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chill JH, et al. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003;11(7):791–802. doi: 10.1016/S0969-2126(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Crispin M, Doores KJ. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr Opin Virol. 2015;11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M, Ward AB, Wilson IA. Structure and Immune recognition of the HIV glycan shield. Annu Rev Biophys. 2018;47:499–523. doi: 10.1146/annurev-biophys-060414-034156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, C JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–74. doi: 10.1016/S1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- Crua Asensio N, et al. Centrality in the host-pathogen interactome is associated with pathogen fitness during infection. Nat Commun. 2017;8:14092. doi: 10.1038/ncomms14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42(1):102–111. doi: 10.1007/s12016-011-8294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damian RT. Molecular mimicry: antigen sharing by parasite and host and its consequences. Am Nat. 1964;98(900):129–149. doi: 10.1086/282313. [DOI] [Google Scholar]
- Dar AC, Sicheri F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell. 2002;10(2):295–305. doi: 10.1016/S1097-2765(02)00590-7. [DOI] [PubMed] [Google Scholar]
- Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem Sci. 2011;36(3):159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- de Castro E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–5. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Southwood SR. Host-pathogen interactions. Encycl Bioinform Comput Biol. 2019;3:103–112. [Google Scholar]
- Diaz A, Ferreira A, Sim RB. Complement evasion by Echinococcus granulosus: sequestration of host factor H in the hydatid cyst wall. J Immunol. 1997;158(8):3779–3786. [PubMed] [Google Scholar]
- Doxey AC, McConkey BJ. Prediction of molecular mimicry candidates in human pathogenic bacteria. Virulence. 2013;4(6):453–466. doi: 10.4161/viru.25180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmus Tekir S, Cakir T, Ulgen KO. Infection strategies of bacterial and viral pathogens through pathogen-human protein–protein interactions. Front Microbiol. 2012;3:46. doi: 10.3389/fmicb.2012.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmus Tekir S, et al. PHISTO: pathogen-host interaction search tool. Bioinformatics. 2013;29(10):1357–1358. doi: 10.1093/bioinformatics/btt137. [DOI] [PubMed] [Google Scholar]
- Duro N, Miskei M, Fuxreiter M. Fuzziness endows viral motif-mimicry. Mol Biosyst. 2015;11(10):2821–2829. doi: 10.1039/C5MB00301F. [DOI] [PubMed] [Google Scholar]
- Ferreira PG, Azevedo PJ. Evaluating deterministic motif significance measures in protein databases. Algorithms Mol Biol. 2007;2:16. doi: 10.1186/1748-7188-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Xia Y. Structural principles within the human-virus protein–protein interaction network. Proc Natl Acad Sci U S A. 2011;108(26):10538–10543. doi: 10.1073/pnas.1101440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire A, et al. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15(2):109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- Gandhi TK, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38(3):285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Garamszegi S, Franzosa EA, Xia Y. Signatures of pleiotropy, economy and convergent evolution in a domain-resolved map of human-virus protein–protein interaction networks. PLoS Pathog. 2013;9(12):e1003778. doi: 10.1371/journal.ppat.1003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, et al. miPepBase: a database of experimentally verified peptides involved in molecular mimicry. Front Microbiol. 2017;8:2053. doi: 10.3389/fmicb.2017.02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll J, et al. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24(15):1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, et al. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108(5):1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino E, Salguero I, Kearsey SE. Cellular regulation of ribonucleotide reductase in eukaryotes. Semin Cell Dev Biol. 2014;30:97–103. doi: 10.1016/j.semcdb.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Guven-Maiorov E, Tsai CJ, Nussinov R. Pathogen mimicry of host protein–protein interfaces modulates immunity. Semin Cell Dev Biol. 2016;58:136–145. doi: 10.1016/j.semcdb.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Guven-Maiorov E, et al. HMI-PRED: a web server for structural prediction of host-microbe interactions based on interface mimicry. J Mol Biol. 2020;432(11):3395–3403. doi: 10.1016/j.jmb.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Hermjakob H, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–5. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, et al. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol. 2009;16(8):853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov SS, et al. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem. 2010;285(45):34686–34698. doi: 10.1074/jbc.M110.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, et al. Sirt1 carboxyl-domain is an ATP-repressible domain that is transferrable to other proteins. Nat Commun. 2017;8:15560. doi: 10.1038/ncomms15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating JA, Striker R. Phosphorylation events during viral infections provide potential therapeutic targets. Rev Med Virol. 2012;22(3):166–181. doi: 10.1002/rmv.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, et al. Role of host-mediated post-translational modifications (PTMs) in RNA virus pathogenesis. Int J Mol Sci. 2020;22(1):323. doi: 10.3390/ijms22010323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvansakul M, Hinds MG. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis. 2013;4:e909. doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay G, et al. MatrixDB, the extracellular matrix interaction database: updated content, a new navigator and expanded functionalities. Nucleic Acids Res. 2015;43:D321–7. doi: 10.1093/nar/gku1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, et al. The Legionella kinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation. Proc Natl Acad Sci U S A. 2020;117(25):14433–14443. doi: 10.1073/pnas.2000497117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, et al. SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO Rep. 2013;14(8):733–740. doi: 10.1038/embor.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15(3):119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ludin P, Nilsson D, Maser P. Genome-wide identification of molecular mimicry candidates in parasites. PLoS ONE. 2011;6(3):e17546. doi: 10.1371/journal.pone.0017546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DJ, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849(7):836–844. doi: 10.1016/j.bbagrm.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber F. Myristoylation of viral and bacterial proteins. Trends Microbiol. 2004;12(4):178–185. doi: 10.1016/j.tim.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Mayer KA, et al. Hijacking the supplies: metabolism as a novel facet of virus-host interaction. Front Immunol. 2019;10:1533. doi: 10.3389/fimmu.2019.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain MT, et al. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med. 2005;11(1):85–89. doi: 10.1038/nm1167. [DOI] [PubMed] [Google Scholar]
- Meier-Stephenson V, et al. DEAD-box helicases: the Yin and Yang roles in viral infections. Biotechnol Genet Eng Rev. 2018;34(1):3–32. doi: 10.1080/02648725.2018.1467146. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49(D1):D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudunuri U, et al. bioDBnet: the biological database network. Bioinformatics. 2009;25(4):555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratil V, et al. VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 2009;37:D661–8. doi: 10.1093/nar/gkn794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol. 2010;84(18):9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod C, Banaei-Esfahani A, Collins BC. Elucidation of host-pathogen protein–protein interactions to uncover mechanisms of host cell rewiring. Curr Opin Microbiol. 2017;39:7–15. doi: 10.1016/j.mib.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Anand A, Kai T. Tudor domain proteins in development. Development. 2012;139(13):2255–2266. doi: 10.1242/dev.073304. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Querol A. Opportunistic strains of saccharomyces cerevisiae: a potential risk sold in food products. Front Microbiol. 2015;6:1522. doi: 10.3389/fmicb.2015.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Santiago P, et al. (2017) Plant and yeast cornichon possess a conserved acidic motif required for correct targeting of plasma membrane cargos. Biochimica et biophysica acta. Mol Cell Res. 1864;10:1809–1818. doi: 10.1016/j.bbamcr.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Samano-Sanchez H, Gibson TJ. Mimicry of short linear motifs by bacterial pathogens: a drugging opportunity. Trends Biochem Sci. 2020;45(6):526–544. doi: 10.1016/j.tibs.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Schipke J, et al. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol. 2012;86(7):3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein–protein interactions in yeast. Nat Biotechnol. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Seabright GE, et al. Protein and glycan mimicry in HIV vaccine design. J Mol Biol. 2019;431(12):2223–2247. doi: 10.1016/j.jmb.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Witzel JE, Jagus R. Homologous regions of the alpha subunit of eukaryotic translational initiation factor 2 (eIF2alpha) and the vaccinia virus K3L gene product interact with the same domain within the dsRNA-activated protein kinase (PKR) Eur J Biochem. 1997;250(1):85–91. doi: 10.1111/j.1432-1033.1997.00085.x. [DOI] [PubMed] [Google Scholar]
- Spiess C, et al. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14(11):598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J. Structural biology. Viral Chemokine Mimicry Sci. 2015;347(6226):1071–1072. doi: 10.1126/science.aaa7998. [DOI] [PubMed] [Google Scholar]
- Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol Cell. 2000;6(6):1449–1460. doi: 10.1016/S1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venigalla SSK, Premakumar S, Janakiraman V. A possible role for autoimmunity through molecular mimicry in alphavirus mediated arthritis. Sci Rep. 2020;10(1):938. doi: 10.1038/s41598-019-55730-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via A, et al. How pathogens use linear motifs to perturb host cell networks. Trends Biochem Sci. 2015;40(1):36–48. doi: 10.1016/j.tibs.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Wagh K, et al. Completeness of HIV-1 envelope glycan shield at transmission determines neutralization breadth. Cell Rep. 2018;25(4):893–908 e7. doi: 10.1016/j.celrep.2018.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver TM, et al. The EZH2 SANT1 domain is a histone reader providing sensitivity to the modification state of the H4 tail. Sci Rep. 2019;9(1):987. doi: 10.1038/s41598-018-37699-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T, Dasgupta S, Nordström K. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol Microbiol. 2000;38(2):392–400. doi: 10.1046/j.1365-2958.2000.02138.x. [DOI] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of "guilt-by-association" within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, et al. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J Biol Chem. 2005;280(39):33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- Xenarios I, et al. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28(1):289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, et al. The Phox homology (PX) domain, a new player in phosphoinositide signalling. Biochem J. 2001;360(Pt 3):513–530. doi: 10.1042/bj3600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici-Eser H, et al. Neuropsychiatric symptoms of COVID-19 Explained by SARS-CoV-2 proteins' mimicry of human protein interactions. Front Hum Neurosci. 2021;15:656313. doi: 10.3389/fnhum.2021.656313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Claverie JM, Ogata H. Mimivirus reveals Mre11/Rad50 fusion proteins with a sporadic distribution in eukaryotes, bacteria, viruses and plasmids. Virol J. 2011;8:427. doi: 10.1186/1743-422X-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura SH, Hirano T. HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments? J Cell Sci. 2016;129(21):3963–3970. doi: 10.1242/jcs.185710. [DOI] [PubMed] [Google Scholar]
- Yuan S, et al. Viruses harness YxxO motif to interact with host AP2M1 for replication: a vulnerable broad-spectrum antiviral target. Sci Adv. 2020;6(35):eaba7910. doi: 10.1126/sciadv.aba7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available at http://imitatedb.sblab-nsit.net. The python script for the identification of DMP/MMP is available at the following link: https://github.com/sblab/ImitateDB.git. At present, Imitate DB contains information from HP-PPIs up to April 2021. We plan to update this database regularly as new releases of the contributing databases become available. Apart from collating all the HP-PPIs from different databases, we are working towards mining the HP-PPIs from literature for computation of additional DMPs and MMPs. We plan to compute DMPs and MMPs for predicted HP-PPIs. Work is also ongoing for providing information on DMPs/MMPs for Sars-CoV-2.