Abstract
Global climate change is proceeding at an alarming rate with major ecological and genetic consequences for biodiversity, particularly in drylands. The response of species to climate change may differ between intraspecific genetic groups, with major implications for conservation. We used molecular data from 10 nuclear and two chloroplast genomes to identify phylogeographic groups within 746 individuals from 29 populations of Senegalia senegal, a savannah tree species in sub-Saharan Africa. Three phylogroups are identified corresponding to Sudano-Sahelian, Zambezian and Southern African biogeographic regions in West, East and Southern Africa. Genetic diversity was highest in Southern and Zambesian and lowest in the Sudano-Sahelian phylogroups. Using species distribution modeling, we infer highly divergent future distributions of the phylogroups under three climate change scenarios. Climate change will lead to severe reductions of distribution area of the genetically diverse Zambezian (− 41–− 54%) and Southern (− 63–− 82%) phylogroups, but to an increase for the genetically depauperate Sudano-Sahelian (+ 7– + 26%) phylogroups. This study improves our understanding of the impact of climate change on the future distribution of this species. This knowledge is particularly useful for biodiversity management as the conservation of genetic resources needs to be considered in complementary strategies of in-situ conservation and assisted migration.
Subject terms: Biogeography, Genetic variation, Ecological modelling
Introduction
Contemporary patterns of genetic variation have become a crucial aspect of research in biogeography, conservation and evolutionary biology1–3. Major drivers of intraspecific genetic variation include climatic shifts during the Quaternary, ecological gradients, demographic processes and anthropogenic land-use change4–6. Changes in geographic ranges driven by fluctuations in climate have fragmented or reconnected populations of the same species7. This can lead to spatial structuring of genetic diversity in distinct gene pools2 where a phylogeographic discontinuity is interpreted as evidence for range fragmentation during past climatic changes8.
Recent advances in phylogeography and landscape genetics have focused on identifying and understanding genetic structure, including the influence of geographic and environmental heterogeneity, on spatial genetic variation with possible implications for forecasting the distribution of gene pools under climate change3,9,10. There is sufficient evidence that modern climate change is reshuffling the geographic distributions of plant and animal species world-wide11,12. Populations or species react to climate change by either persisting, dispersing to suitable ecological conditions, or going extinct, which influences the amount and distribution of intraspecific genetic diversity13. Despite the increasing evidence for global climate warming, model predictions of how climate will change in the course of the twenty-first century is treated with some level of uncertainty13,14. This is addressed by assuming that emissions will follow one of several scenarios known as the Representative Concentration Pathways (RCPs)15–17. Global climate change projections show a strong warming trend over the twenty-first century and Africa has been identified as one of the regions of the world most vulnerable to climate change18. Particularly in sub-Saharan Africa, temperature is expected to rise by approximately + 2.0 to + 4.5 °C by 210019,20.
The savannah belt of sub-Saharan Africa is affected by significant climatic, ecological and geological barriers21,22, resulting in major partitions into the Sudano-Sahelian (West—Central Africa), Zambezian (East Africa) and Southern African biogeographic regions23 for both plant and animal species21,22. Prominent historical barriers include the Mega Lake Chad, Dahomey Gap, East African Rift valley, Ethiopian highlands, Adamawa Highlands, Namib desert and Kalahari desert21,22,24,25, hindering dispersal and gene flow among regions. Documented evidence shows that tropical savannah and woodland trees potentially played an important role in forest assemblages during the Last Glacial Maxumum (LGM)22. Phylogeographic studies of these tree species have been instrumental in unravelling the role of past environmental changes in the structuring of genetic diversity found in contemporary populations22. Recently, new insights into the geographic distribution and range dynamics of many African savannah tree species have been provided by the analyses of ecological data, while additional insights come from analysing the pattern and distribution of genetic diversity within species8,22,26,27. In some species, intraspecific phylogeographic patterns mirror biogeographic patterns. For example, Prunus africana (Hook.f.) Kalkmanis genetically structures into Sudanian, Zambezian and Southern gene pools26, while genetic discontinuities in the form of parapatric genetic clusters were detected in the Sudanian savannah for Vitellaria paradoxa C.F. Gaertner and Parkia biglobosa (Jacq.) R.Br ex G. Don28,29. However, the absence of clear-cut genetic discontinuities over large distances has been reported for other species, including Adansonia digitata Linn.30, Khaya senegalensis (Desr.) A. Juss.22, Afzelia africana Sm. ex Pers. in the Sudanian, and Afzelia quanzensis Welw. in the Zambezian region8. Although some of these studies investigated the impact of historical perturbations on genetic patterns31, there is a lack of studies into the genetic consequences of climate change for the future distribution of species and their intraspecific groups.
Here, we assess the impact of climate change on intraspecific lineages of a savannah taxon using S. senegal (L.) Britton (Fabaceae, Mimosoideae), commonly known as gum arabic, one of the most important tree species in the tropical woodlands of sub-Saharan Africa both economically and ecologically32. Senegalia senegal has an extensive geographic coverage on the African continent and is common to habitats that experienced past range contraction and expansions due to Pleistocene climatic fluctuation (Fig. 1)33,34, and has ecological attributes such as resilience to adverse environmental conditions35,36. The species is distributed throughout the African arid and semi-arid regions, extending from Senegal along the Sudano-Sahelian zone to the Red Sea and then southwards through the dry savannah and montane areas of the Zambezian region into southern Africa37. A phylogeographic study on S. senegal based on nuclear and plastid genome data has suggested an evolutionary origin in East or Southern Africa and reported a major division separating eastern and southern African populations from those in West and Central Africa, suggesting a recent range expansion starting from East Africa38. Historical distribution and range dynamics of S. senegal indicate variation along climatic and edaphic regimes, separating the eastern and southern ranges from the western and central African ranges27. In addition, population genetic studies of Kenyan and West African populations of S. senegal have shown that anthropogenic perturbations and climatic shifts could impact levels of genetic diversity (GD), population sizes, structure of gene pools, and natural regeneration patterns of the species at regional scale6,39. Investigating the impact of climate change on GD of S. senegal will provide insights into population structure, and can further advance our understanding of the genetic consequences of post-glacial expansion processes and climate warming on the future distribution of genetic variation. We here use a comprehensive data set of nuclear and chloroplast microsatellite data and combine phylogeography, landscape genetics and distribution range modeling to investigate how future climate change will affect the distribution of phylogeographic groups (hereafter referred to as phylogroups) and their gene pools across Africa. Specifically, we address the following questions:
How is phylogeographic structure reflected in levels of genetic variation within populations? Population expansion often leads to the loss of genetic variation due to genetic drift or bottleneck effects. We thus hypothesize that within-population GD declines with distance from the proposed origin of the range expansion towards West Africa40.
How do environmental conditions in the local habitat of S. senegal correlate with genetic differentiation among populations? Landscape characteristics (e.g., barriers to migration), past range changes and habitat fragmentation can influence population genetic structure3,10, and heterogeneity in landscape features has been shown to impact genetic variation in east African populations of Senegalia mellifera24. Thus, the highly contrasting ecological conditions across sub-Saharan Africa are expected to be reflected in patterns of genetic variation. We hypothesize that population genetic differentiation is correlated with geographic and environmental distance resulting in both patterns of isolation by distance (IBD) and isolation by environment (IBE).
What are the genetic consequences of future climate change for S. senegal? Climatic changes may lead to conditions that fall outside the current environmental tolerances of populations and may trigger either a geographic range shift, ecological adaptation or extinction13,40. Assuming niche conservatism (i.e., populations are unable to tolerate the new conditions locally, e.g., through phenotypic plasticity or adaptation), populations will need to track their current environmental tolerances by geographic range shift. Depending on the overall magnitude, direction and rate of climate change, species may suffer potentially severe reduction of suitable habitat and in turn loss of population GD. We predict that the ongoing climate change will lead to a loss of GD due to an overall reduction of range size. We use a population genetic approach, with an improved marker resolution and larger sample size compared to previous attempts38 to refine the phylogeographic structure of African S. senegal. In addition, we model the potential impact of climate change on the distribution of phylogeographic groups of S. senegal using SDMs. This study will increase our understanding of the spatial structuring of genetic variation including how phylogeographic groups may respond under future environmental change scenarios.
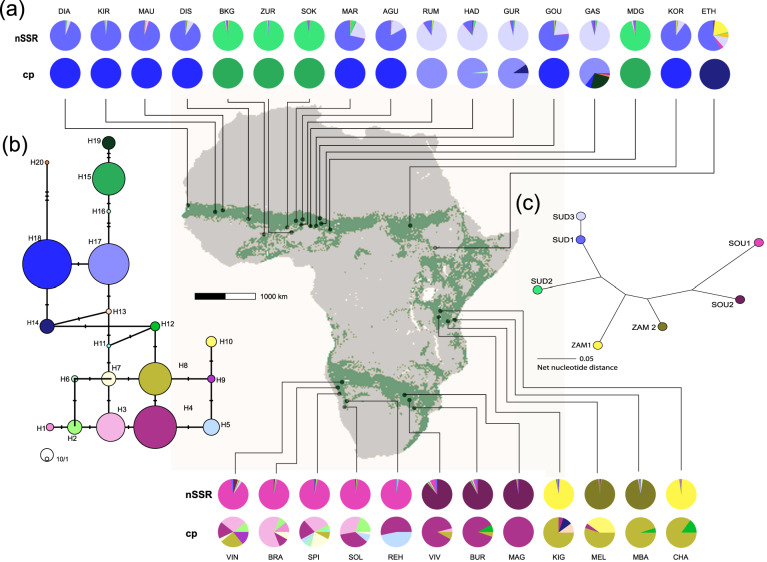
Figure 1.
(a) The Geographic occurrence of populations of Senegalia senegal analyzed, showing the proportional assignment to both, seven gene pools of nSSR and cpDNA haplotypes. Each pie chart represents one population and the colors within the pie chart depict the distinctive haplotypes/gene pools. (b) Median‐joining network of the 20 cpDNA haplotypes (H1-H20). Circle sizes are proportional to haplotype frequencies. Small bars indicate the number of mutational steps in case more than one step occurred. (c) Neighbor-joining tree showing the relationship among seven nSSR gene pools (K = 7) as revealed by STRUCTURE (Fig. 2). The map was downloaded from WORLDCLIM41 and modified manually. The green area in the map background indicates the modeled potential distribution of S. senegal27 generated using ArcGIS Desktop ver. 10.542. See Supplementary Table S1 for population details.
Results
Haplotype distribution and phylogeographic patterns
Two chloroplast (cpSSR) loci yielded 20 haplotypes across the 29 populations from which samples of S. senegal were collected (Fig. 1). Between one and eight haplotypes were found per population. In the Sudano-Sahelian range, except for three populations (HAD, GUR and GAS), all other populations were fixed to a single haplotype. Contrastingly, in both the Zambezian and Southern ranges, almost all populations harboured more than one haplotype.
A strong phylogeographic division between the Zambezian-Southern ranges (haplotypes H1-H11) and the Sudano-Sahelian range (haplotypes H15-H20) is visible in the haplotype network (Fig. 1a). Six mutational steps separate these haplotype groups and they are connected by four intermediate haplotypes (H11-H14) (Fig. 1). The six most common haplotypes indicate different regions they dominate, however, without being restricted to them: in the Zambesian-Southern range, H3 dominated in the Namib, H4 in Kalahari, and H8 in the Zambesian range, whereas for the Sudano-Sahelian group, H17 + H18 dominated in the Sahelian and H15 in the Sudanian region. The genetically intermediate haplotypes either occurred in geographically intermediate populations, like H14, which is dominant in the easternmost Sudano-Sahelian site ETH connecting the Sudano-Sahelian and Zambezian ranges, or intermediate haplotypes were shared between ranges, like H12 which is shared between the Zambezian and Southern range.
As expected for non-recombining plastid markers, population differentiation was very strong (Supplementary Table 2). A non-hierarchical AMOVA for cpSSR showed 73% genetic variation among populations (ФPT = 0.733), whereas hierarchical AMOVAs, using the STRUCTURE clusters as groups (see below) found increasing variance among clusters when more clusters were considered, starting at 43% (ФRT = 0.427) at K = 2, and 48% (ФRT = 0.485) at K = 3, reaching 90% (ФRT = 0.901) at K = 7 (Supplementary Table 2).
Genetic population structure
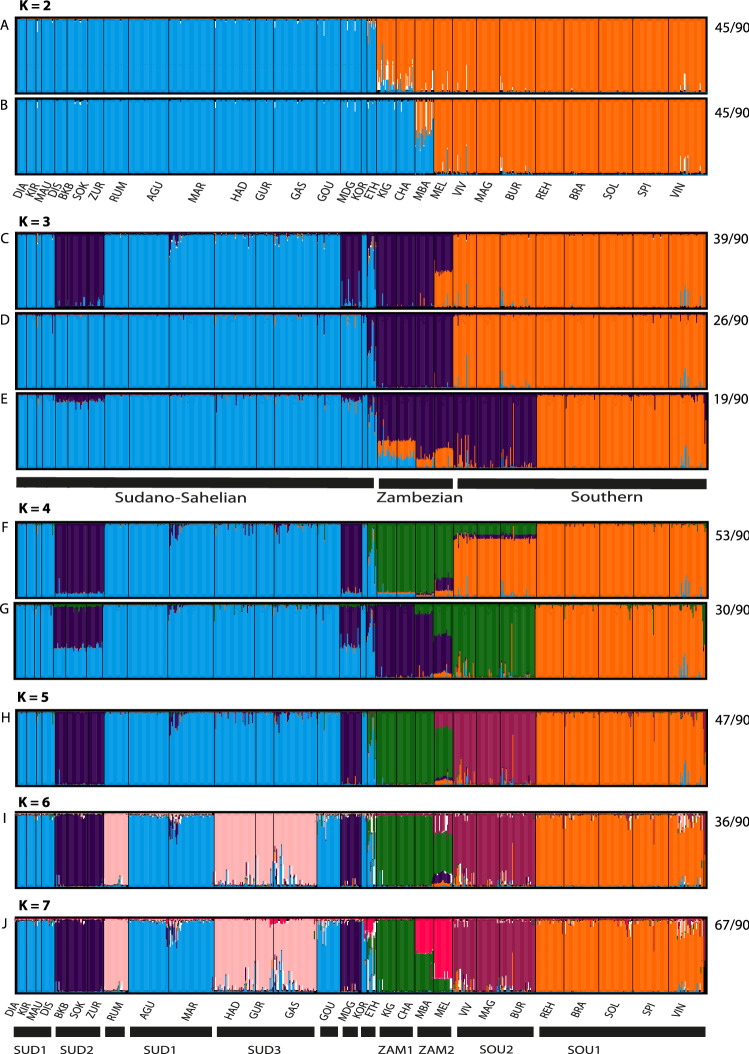
Bayesian analysis of population structure of the combined nuclear and chloroplast data sets yielded a hierarchical pattern across the African range (Fig. 2). The uppermost hierarchical levels were two gene pools as suggested by the method of Evanno et al. (2008) (Supplementary Fig. S1). However, while one pool represented the Sudano-Sahelian region and the other Southern Africa, the Zambezian region grouped equally likely with either of them in two different clustering modes (Fig. 2A,B). The likelihood of data strongly increased for values of K > 2 with (Supplementary Fig. S1) and new emerging clusters generally encompassed whole populations clearly indicating additional biologically relevant and geographically coherent hierarchical genetic structure. At K = 3, three gene pools coincided largely with the Sudano-Sahelian, Zambezian and Southern African biogeographic regions. However, the cluster of the Zambezian range encompassed either only populations from East Africa, or included a few populations from either West- or South-Africa in three different modal solutions (Fig. 2C–E). In all runs at K = 3 the third cluster included all populations of the Zambesian biogeographic range. At K = 4, two contrasting patterns are found in two modes which are resolved at K = 5, where the former Sudano-Sahelian cluster separated into the Sahelian (light blue) and Sudanian (dark blue) biogeographic regions and the Southern African region separated into Namib (SOU1) and Kalahari (SOU2). At K = 6, the Sudano-Sahelian range was further divided in the western-most (SUD1, light blue), the southern (SUD2, dark blue) and the central (SUD3, pink) populations. At K = 7, the Zambesian range split into two gene pools, representing north-western (ZAM1) and south-eastern (ZAM2) populations. At K = 7, the major mode comprised 67/90 (74%) of the STRUCTURE runs (Fig. 2J), which was the highest proportion observed across all levels of K, strongly suggesting that K = 7 can be considered as both comprehensive and consistent.
Figure 2.
Bar plots of individual proportional affiliation to genetic clusters identified by STRUCTURE for the 730 individuals genotyped at ten nSSR and two cpSSR loci for K = 2–7. For K = 2–4 major and minor modes are shown as identified by clumpak43, while for K = 5–7 only the major mode is shown for simplicity. The frequency of modes among 90 runs is given to the right. Vertical bars represent individual samples. See Fig. 1 for geographic location of sites.
With the nuclear data set, non-hierarchical AMOVA revealed 29% variation among populations (FST = 0.287; P ≤ 0.001). Hierarchical AMOVAs revealed increasing variation among clusters with increasing K, ranging from 17% (FRT = 0.171) at K = 2, 20% (FRT = 0.201) at K = 3 up to 25% (FRT = 0.247; Supplementary Table 2) at K = 7.
Genetic variation within populations
Our results show high genetic variation for S. senegal, which is strongly structured across the species’ range in Africa (Fig. 1). Genetic diversity at the population level as indicated by allelic richness (Ar) significantly declined with distance from the assumed East African origin of the range expansion into West Africa (r = − 0.676; r2 = 0.457; p = 0.001, Fig. 3, Table S3). In contrast, genetic variation did not decline between the Zambezian and the Southern subrange (r = − 0.268, p = 0.4). This pattern is consistent with range expansion from East to West with accompanying bottlenecks and genetic drift, while this was not the case for the Eastern and Southern ranges. The sub-range means for nuclear markers (nSSR, Table 1) show that Ar is significantly lower in the Sudano-Sahelian in comparison to the Zambezian and Southern regions, while Apriv is higher in the Zambezian than Sudano-Sahelian and Southern regions, and Fis is higher in the Southern than Sudano-Sahelian and Zambezian regions. Interestingly, sub-range estimates for plastid markers (cpSSR) indicate that the diversity of haplotypes was significantly higher in the Southern range, with a total of 11 haplotypes present compared to eight and six in the Sudano-Sahelian and Zambezian ranges (Table 1, Table S3).
Figure 3.
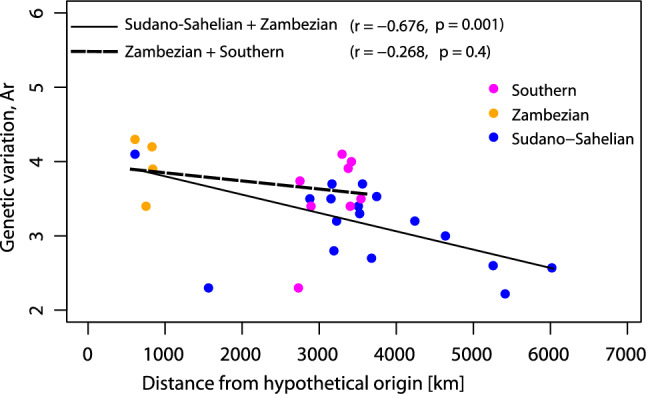
Genetic variation (rarified allelic richness, Ar) of populations of Senegalia senegal as a function of distance from assumed East African origin of the range expansion into the Sudano-Sahelian region and between Eastern and Southern ranges only.
Table 1.
Sub-range genetic diversity estimates at ten nuclear SSR loci and two cpSSR loci for Senegalia senegal in three biogeographic regions. Given are the mean values across populations (with different letters indicating significant differences according to ANOVA and posthoc test) and region estimates in which regions are treated as populations. See Supplemental Table S3 for population level estimates.
| nSSR | cpSSR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | N | Ar | Na | Apriv | Ho | He | Fis | A | P | Ne | Rh | Dv | D2sh |
| Mean values across populations within region | |||||||||||||
| Sudano-Sahelian | 22.4 | 3.14b | 4.1b | 1.24b | 0.62a | 0.56a | − 0.12b | 1.35b | 0.18a | 1.08b | 0.12b | 0.05b | 0.38a |
| Zambezian | 23.0 | 3.95a | 5.93a | 10a | 0.62a | 0.62a | 0.0008ab | 2.75ab | 0.5a | 1.58ab | 0.90ab | 0.34a | 0.64a |
| Southern | 23.6 | 3.54ab | 6.38a | 2.75b | 0.42b | 0.54a | 0.20a | 4.13a | 0.5a | 2.7a | 1.55a | 0.50a | 0.62a |
| Region estimates | |||||||||||||
| Sudano-Sahelian | 374 | 9.11 | 12.4 | 33 | 0.579 | 0.626 | 0.075 | 8 | 6 | 3.135 | 4.488 | 0.683 | 3.163 |
| Zambezian | 80.9 | 13.48 | 13.5 | 54 | 0.588 | 0.763 | 0.23 | 6 | 2 | 1.616 | 5.000 | 0.386 | 0.824 |
| Southern | 267.1 | 9.84 | 13 | 46 | 0.409 | 0.646 | 0.368 | 11 | 8 | 3.536 | 8.146 | 0.720 | 1.094 |
N, number of samples per location; Ar, rarified allelic richness (Mousadik and Petit, 1996); Na, mean number of alleles per locus per population; Ho, observed heterozygosity; He, expected heterozygosity; Apriv, number of private alleles; Fis, inbreeding coefficient; A, number of haplotypes; P, number of private haplotypes; Ne, effective number of haplotypes; Rh, haplotypic richness; Dv, genetic diversity; D2sh, mean genetic distance between individuals.
Geographic and environmental drivers of genetic structure
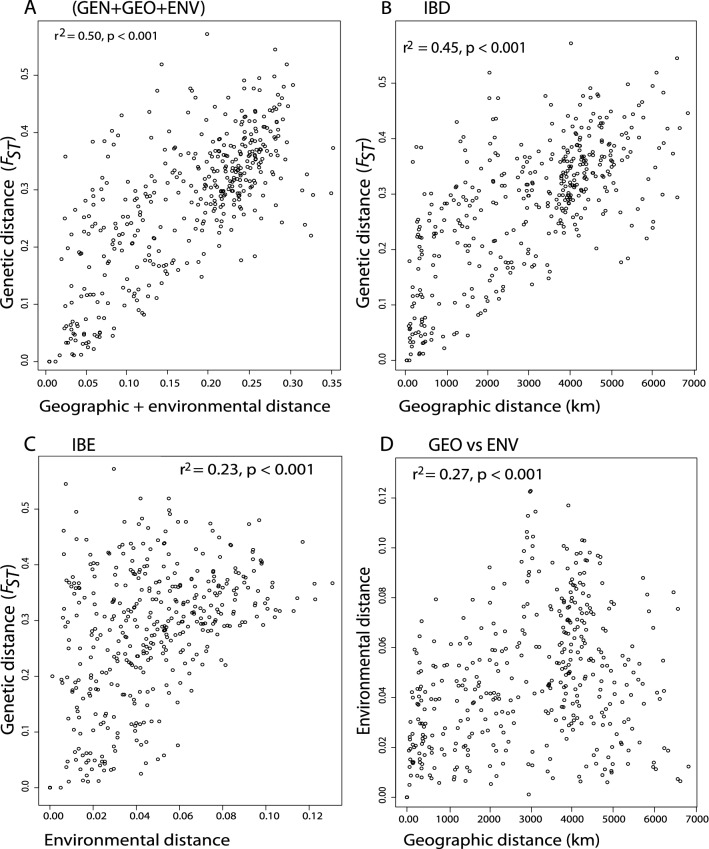
The range-wide MMRR analysis shows that geographic and environmental distances were both associated with genetic distances (Fig. 4; Table 2). Geography and environment jointly explained 50% (r2 = 0.50, p < 0.001) of variation (Fig. 4a) with geography accounting for the largest proportion of variation (45%) in genetic distance (Fig. 4b). The signal for IBE was driven by the precipitation of the wettest month (Bio13), which had a significant association with genetic distances in contrast to the other climatic and soil variables considered in the analysis (Fig. 4c, Table 2). The analysis also revealed a weak but significant correlation between geographic and environmental distances (r2 = 0.267, p < 0.001, Fig. 4d) with Bio13 and monthly variability in potential evapotranspiration (PETseasonality) as the most important.
Figure 4.
Multiple matrix regression with randomization analysis on Senegalia senegal in Africa. Scatterplots show patterns of (A) the combined effects of geographic and environmental distances on genetic distance; (B) Isolation by distance; (C) Isolation by environment; (D) the relationship between geographic and environmental distances.
Table 2.
Multiple matrix regression with randomization (MMRR) model testing for range wide isolation by distance and isolation by environment for Senegalia senegal.
| Variable distance | Coefficient | T-statistic | p-value |
|---|---|---|---|
| Geographic distance | 4.12E-05 | 14.671 | 0.001 |
| Bio13 | 4.52E-04 | 4.617 | 0.006 |
| PETseasonality | 2.23E-08 | 3.723 | 0.067 |
| Soil pH | − 8.20E-04 | − 0.803 | 0.608 |
| Bio8 | − 6.12E-05 | − 0.237 | 0.888 |
| Intercept | 0.111 | 10.624 | 1 |
When the analyses were repeated at the subrange level, significant IBD was detected for the combined Zambezian and Southern ranges (r2 = 0.38) and for the combined Sudano-Sahelian and Zambezian subrange (r2 = 0.29), pointing to an older or more established relationship in Zambezian—Southern than in the Sudano-Sahelian—Zambezian ranges (Supplementary Fig. 2). Accordingly, IBE was detected in Sudano-Sahelian—Zambezian range (r2 = 0.32, p = 0.001) but not significant in the Zambezian—Southern (Supplementary Fig. 2). Environmental distance was partly correlated to geographic distance in the Sudano-Sahelian—Zambezian (r2 = 0.43; p = 0.001). However, our data did not show a significant correlation in the Zambezian—Southern range indicating that both parameters are acting in concert with the other in the Sudanian—Zambezian range but maybe independent in the Zambezian—Southern range (Supplementary Fig. 2).
Habitat suitability modeling
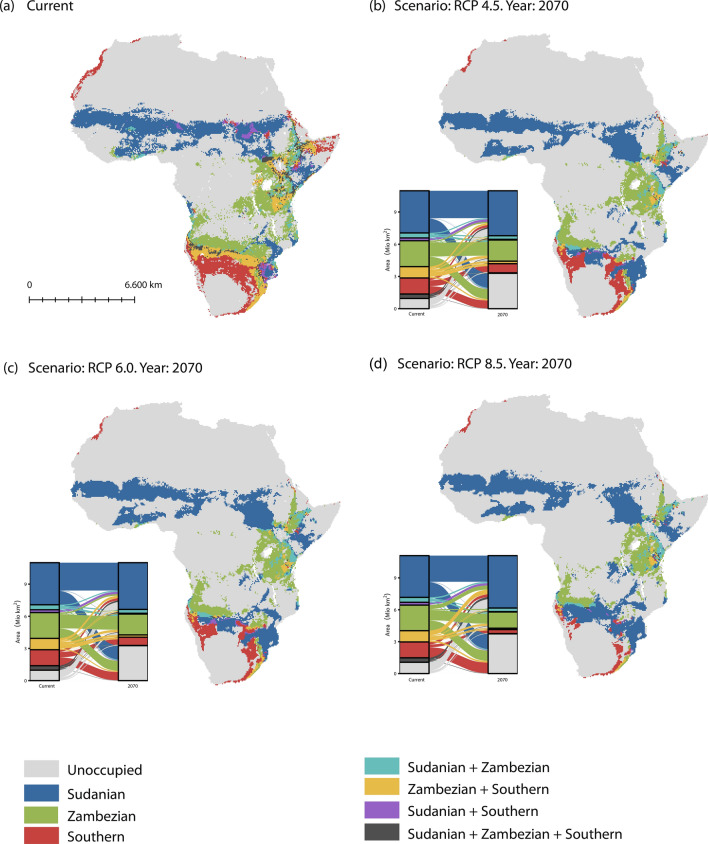
The habitat suitability model results for future projections by 2070 are largely congruent for the CCSM4 and MIROC5 scenarios and only results for CCSM4 are presented (Fig. 5, Table 3). Climate‐based distribution models of S. senegal, built with current conditions, generally indicated the presence of a continuous potential distribution for the species throughout its known distribution range in sub-Saharan Africa (Fig. 5a). The accuracy values obtained for the six ensemble methods indicated a good performance and agreement of the model to the data (Supplementary Table 3). Across the three phylogroups, a decrease of the total suitable area between 22.8% (RCP 6.0) and 26.6% (RCP 8.5) is predicted (Table 3, Fig. 5). However, the moderate reduction of the total range is accompanied by a drastic change of the share and distribution of the three phylogroups. Depending on the scenario, the future total range for the Sudano-Sahelian group may decline by up to 8.4% (RCP 4.5) or increase up to 5.1% (RCP 8.5). However, the area where the Sudano-Sahelian group does not overlap with other groups is predicted to increase for all scenarios by between 6.7% (RCP 4.5) and 25.6% (RCP 8.5). Large parts of this gain of suitable area is located in the Zambezian and Southern range. In contrast, the Zambezian group is predicted to lose about half of its total range (40.7–53.7%), with a loss between 18.3% (RCP 4.5) and 37% (RCP 8.5) predicted for the Zambesian-only areas. Most drastic reductions of suitable area are predicted for the Southern range, declining between 63% (RCP 4.5) and 82% (RCP 8.5) of the total area and between 42.8% (RCP 4.5) and 74.6% (RCP 8.5) considering the Southern-only area. While in general the zones of geographic overlap of the ranges of the different phylogroups are of minor importance, a considerable area of overlap was found between the Zambezian and Southern phylogroup for the potential current distribution, which strongly declined in all future scenarios.
Figure 5.
Potential current and future (2070) distribution of three phylogeographic groups of Senegalia senegal (Sudano-Sahelian, Zambezian and Southern) in Africa according to Biomod2 distribution modelling. (a) Present-day suitability; (b) intermediate emission RCP 4.5 scenario; (c) high emission RCP 6.0 scenario and (d) very high emission RCP 8.5 scenario projected with CCSM4 model. Inset (b–d) are Sankey plots showing how the area occupied by the phylogroups will change for three climate change scenarios. The shape files for the maps were downloaded from WORLDCLIM41. The modeled potential future distribution of S. senegal were generated using ArcGIS Desktop ver. 10.542 .
Table 3.
Current suitable area and predicted area (as percentage of current area) for each phylogroups of Senegalia senegal and their co-occurrence for the three climate change scenarios RCP 4.5, RCP 6.0 and RCP 8.5 for the year 2070. Predicted area percentage > 100% and < 100% indicates gain and loss of area, respectively.
| Phylogroup | Current Area (km2) | Percentage (%) of current area for 2070 | ||
|---|---|---|---|---|
| RCP 4.5 | RCP 6.0 | RCP 8.5 | ||
| Total range | 1.17 × 107 | 76.6 | 77.2 | 73.4 |
| Sudano-Sahelian Total | 5.89 × 106 | 91.6 | 95.0 | 105.1 |
| Zambezian Total | 5.0 × 106 | 59.3 | 58.4 | 46.3 |
| Southern Total | 3.76 × 106 | 37.0 | 33.5 | 17.9 |
| Sudano-Sahelian only | 4.56 × 106 | 106.7 | 111.0 | 125.6 |
| Zambezian only | 2.80 × 106 | 81.7 | 80.7 | 63.0 |
| Southern only | 1.73 × 106 | 57.2 | 51.5 | 25.4 |
| Sudano-Sahelian + Zambezian | 5.43 × 105 | 71.8 | 77.0 | 72.4 |
| Sudano-Sahelian + Southern | 3.03 × 105 | 24.8 | 28.6 | 14.0 |
| Zambezian + Southern | 1.22 × 106 | 22.0 | 20.4 | 12.4 |
| Sudano-Sahelian + Zambezian + Southern | 4.87 × 105 | 9.9 | 5.8 | 6.1 |
Discussion
In this study, we used molecular markers to highlight the phylogeography and population-level genetic diversity in the African distribution range of S. senegal. Three major phylogeographic groups were identified in Eastern, Southern and Western Africa. Furthermore, our results provide support for both isolation by distance (IBD) and isolation by environment (IBE) in the genetic structuring of S. senegal. Our SDM projections predict different impacts of climate change on the distribution of the phylogeographic groups under future environmental change scenarios with evolutionary older groups being most drastically affected.
Range-wide heterozygosity levels obtained from this study (He = 0.56) are very similar to levels obtained in previous analyses of microsatellite markers of S. senegal populations in western Africa (He = 0.54–0.56)44 but slightly lower than in the eastern range in Kenya (He = 0.617)6. This may be due to the use of slightly different sets of markers and differences in sample size. Within our study, genetic diversity estimates were highest in the Zambezian and the Southern ranges, the latter showing highest diversity of plastid markers among the subranges. This finding corroborates earlier analyses finding higher diversity in eastern6 and in southern Africa38 compared to Sudano-Sahelian populations in Western Africa. Genetic diversity in Southern Africa may even be underestimated as indicated by the presence of null-alleles in that region. A distance-dependent decline of diversity from Eastern to Western Africa indicates evolutionarily younger populations are affected by, e.g., bottlenecks and genetic drift during range expansion from East to West Africa38. In addition, increasingly unfavorable climatic conditions may have contributed to drift as indicated by the observed IBE. A similar pattern of an East-to-West decline of genetic diversity across Africa was found in P. africana26. However, contrasting patterns with higher diversity in the Sudanian are known for other tree species29,31,45. Thus, the relationships found across sub Saharan Africa corroborate a general pattern of diversity clines that are correlated with climate driven range changes, as e.g. observed in the Mediterranean46.
Population genetic structure and phylogeographic patterns
Although S. senegal currently has a continuous distribution from west (Senegal) to east (Ethiopia) sub-Saharan Africa27, our data show a strong genetic separation between West Africa, i.e. the Sudano-Sahelian biogeographic regions, and East and Southern Africa, i.e. Zambesian and Southern African biogeographic regions. Molecular data were largely congruent between nuclear and plastid genomes at the range wide scale, both showing similar regional genetic structuring and phylogeographic patterns.
Our population level GD data are in line with phylogenetic evidence of a sister group relationship between West- and East-Africa38, and thus corroborate an evolutionary origin of the West African S. senegal in the eastern region. The scenario of colonization from East to West Africa is supported by the relatively small genetic distances (< 4 mutations) from the two dominant West African haplotypes (H17, H18), suggesting that these haplotypes have originated from H8 in the East. Assuming relatively stable environmental conditions during the time frame of range expansion, dispersal from East Africa likely has occurred via Ethiopia-Sudan-northeast Nigeria and westwards to the rest of the Sudano-Sahelian distribution range.
Overall, population differentiation was high at both chloroplast (ФPT = 0.733) and nuclear markers (FST = 0.287) and higher than previous values for regional assessments39,44,47 due to differentiation among phylogroups at the range level, in particular as we included the Namib region in our analysis (Table 1, Supplementary Table 2). However, within regions, we found similar values of differentiation as previously reported for western (0.145)39 and eastern (0.045)47 Africa respectively. The K = 3 partition in STRUCTURE largely reflected the phylogeographic regions of Sudano-Sahelian, Zambezian and Southern ranges. Pairwise FST revealed that the highest differentiations occur between populations from Sudano-Sahelian and Southern ranges, consistent with a range-wide pattern of isolation by distance. Population genetic differentiation can result from evolutionary processes of mutation and genetic drift, demography, anthropogenic disturbances, geographic isolation and limited or restricted gene flow among populations5,6,48. The high GD coupled with endemic gene pools in the Southern range may be due to barriers to gene flow, triggered by the heterogeneous complex mosaic of landscapes38 among the surrounding populations. In addition to demographic history49, spatially heterogeneous landscapes in most cases support greater genetic and species diversity50–52. The presence of mountains (e.g., Spitzkoppe, Erongo region and Brandberg, Naukluft) and inhospitable landscapes (e.g., Khormas highlands that seperates populations Rehoboth and Solitaire) within the sampled range in Namibia, might be acting as a barrier, thus limiting gene flow and facilitating differentiation in local populations. Greater genetic diversity increases the likelihood that appropriate adaptive variation will be available to facilitate adaptation to the new conditions52,53. Our results suggest gene flow limitation between regions resulting in considerable genetic differentiation among phylogeographic lineages. However, populations have remained connected within phylogroups throughout the large, continuous Sudano-Sahelian or Zambezian savannahs.
In deciphering the role of geography and environment as drivers of genetic structure, we found strong relationships between genetic differentiation and both geographic distance (IBD) and environmental distance (IBE), with IBD explaining the majority of genetic differentiation and IBE also contributing significantly. The IBD model assumes that genetic differentiation increases among populations with geographic distance due to limited gene flow and drift54. The strong pattern of geographic isolation observed in our dataset can be explained by either geographic distance, or landscape barriers (e.g., Lake Chad, along the Sudano-Sahelian biogeographic region and the Rift valley in eastern Africa) between populations of S. senegal. IBD patterns are commonly found in large scale analyses of widespread plant species8,55 unless either gene flow or drift are dominating56,57. Isolation by environment (IBE) patterns are caused by environmental heterogeneity and local adaptation related to strong divergent selection58,59. Although IBE patterns for S. senegal were generally weak on a range-wide scale, slightly stronger ecological isolation observed at the subrange level could be explained by either difference in the occupied environmental space, i.e., regional environmental space differentiation27, or selection and local adaptation59–61. Thus, populations that might have dispersed to other suitable ranges within Africa over time adapted to the prevailing local conditions through selection. This result suggests that IBE in S. senegal is primarily driven by differences in temperature during the wettest months in the species local habitat. In addition, different flowering times were observed among population during fieldwork to collect samples. It should be noted that differences in temperature regimes between populations might cause differences in phenology, with reduced overlap of flowering time potentially leading to partial reproductive isolation62. This pattern of reduced overlap in reproductive timing known as isolation by time63 may additionally be a contributing factor driving genetic structure in S. senegal.
By analyzing changes in the realized environment of S. senegal, we quantified and mapped declining and expanding phylogroups that are projected as a result of the twenty-first century climate change. In principle, the projections across the three future climate scenarios present a complementary pattern of range gain for the Sudano-Sahelian phylogroup and range loss for the Zambezian and Southern phylogroups (Fig. 5). As the rate of loss scales with temperature increase, the ultimate extent of climate change will be critical, especially for the Southern populations of S. senegal. The zones of current geographic overlap between the three phylogroups (e.g., in Sudan-Ethiopia and southern Angola), represent areas where highest levels of genetic variation are to be expected. Anthropogenic climate warming is projected to cause the disappearance of phylogroups in these regions of overlap for the next 50-plus years across all scenarios considered in this study, consequently leading to the loss of GD (Fig. 5). Increase in the mean annual temperature by 1.1 to 6.4 degrees Celsius within the twenty-first century18,64 may cause previously well-adapted genotypes to lose their competitive advantage, by selection favouring other genotypes already present in the population or newly immigrating genetic variants65. In general, whether local population will go extinct will also depend on the presence of phenotypic plasticity and its genetic basis and on small scale opportunities for suitable environmental conditions within the existing range.
The highly genetically diverse Southern populations of S. senegal are predicted to be strongly reduced in extent and go regionally extinct in the future. In contrast, genetically depauperate populations in the Sudano-Sahelian range might not be at high risk of extinction, as they may exist in areas of predicted suitable climate both inside and outside of their currently occupied range with a potential for range expansion (Table 3, Fig. 5). However, our results are valid within the framework of range suitability modeling and do not explicitly consider the dispersal rate for S. senegal. Thus, future suitable area distant from the current range may actually be out of reach by natural means, unless human-assisted migration is considered (see below). Generally, our results show evidence for severe future range loss for the evolutionary older populations (Zambezian and Southern), with a negative impact on unique genetic variation of these phylogroups in comparison to the evolutionary younger and genetically depauperate Sudano-Sahelian phylogroup11. The mid-portion of the currently occupied native range of the Southern phylogroup might be lost to the expanding Sudano-Sahelian phylogroup during the intermediate scenarios and even more severe range retraction during the RCP 8.5 scenario, leaving extant populations as fragments in the Namib region as well as in the Mediterranean scrub of the south-eastern tip of Africa. The extremely localized high-elevation populations that harbour highest levels of genetic diversity within the whole species range occur in the Namib region (Spitzkoppe and Brandberg mountains) and might be at risk. Overall, a range reduction of the Zambezian and Southern phylogroup and potential range extension of the Sudano-Sahelian phylogroup would result in a movement from North to South, in contrast to a South to North movement observed in the northern hemisphere66.
Implications for conservation
There is growing concern over the global rate of environmental change. This situation has raised further concerns about whether organisms, especially plant species can keep track, by migration or evolution, with the predicted changing distribution and spatial arrangement of suitable habitat67,68. Our model highlights that the Zambesian and Southern phylogroups of S. senegal will be at risk due to climate change. Three options may allow them to contend with rapidly changing environments: dispersal, phenotypic plasticity, or adaptation11,69,70. Although S. senegal is known to exhibit potential for long-distance dispersal32,38, it is uncertain whether phylogroups, e.g., populations from the Sudano-Sahelian phylogroup might be able to track a suitable habitat, predicted to occur outside of its currently occupied range in the far Southern range. However, high levels of GD observed for S. senegal in this study might ultimately determine the fate of the species through a rapid adaptive change should, for example, the species be incapable of a plastic response. Greater GD increases the likelihood that appropriate adaptive variation will be available for adaptation to the new conditions. Evolutionary adaptation, however, will need time, and large and reproductively active populations. Therefore, regions with highly diverse extant gene pools predicted to be extirpated or go extinct in the future (e.g. Southern phylogroup), should be high priority areas for both in situ and ex situ conservation. Our model predicted largely non-overlapping distribution areas for the phylogroups based on environmental suitability and thus may guide assisted migration programs. However, while our modeling provides predictions on where the phylgroups would find suitable climate in the future, we propose that common gardens should first be established across different biogeographic regions to prove adaptedness prior to the implementation of assisted migration using the most appropriate seed material71,72.
Additionally, forecasting large-scale species distributions is becoming a crucial component for conservation planning, especially for ecological and commercially relevant species73. If the projected range loss of S. senegal leads to fragmentation, then these changes have severe implications for the species through genetic drift, gene flow and inbreeding depression74,75. This will require enhanced conservation efforts, including proactive and intensive management, to provide greater flexibility for the species to respond successfully and avoid extinction. We hope that our study will provide a clear directive on the genetic consequences of climate change on this economically and ecologically important savannah tree species and that the understanding from the findings will support the development or re-designing of effective conservation strategies in S. senegal, in particular considering intraspecific genetic groups and their individual predicted fate. Furthermore, several other tree species have similarly wide distributions across the drier parts of Africa26,76,77. Therefore, it would be interesting to investigate whether the phylogeographic patterns as well as the potential impact of future climate change observed for S. senegal can serve as a model for other tree species occupying the savannah type environments. Finally, we hope our study will be of interest to biodiversity stakeholders and can be integrated in the conservation programs of the United Nations Environment Programme (UNEP), United Nations Convention to Combat Desertification (UNCCD), Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES), National Biodiversity Strategy and Action Plan (NBSAP) and important plant areas (IPAs).
Methods
Sampling strategy
The African range of S. senegal encompasses tropical woodland, open savannah and semi-desert steppe, encompassing biogeographically the Sudano-Sahelian region that extends from extreme western Africa to the Zambezian region in eastern Africa and then southwards up-to the Kalahari and Namib regions21. We did not consider the small Asian part of the species range. Fresh leaf samples were collected from 746 individuals of S. senegal across 29 locations covering most of its geographic range in Africa (Fig. 1, Supplementary Table 1). We aimed at 20 samples per site, but included populations with > 5 samples, resulting in an average of 25 samples, but correcting for biased sample size by rarefaction (see below). A distance of 10 m was maintained between sampled individuals within populations. Field-collected material was dried in silica gel before DNA extraction. At least one individual per population was deposited as a voucher specimen at the herbaria of the National Centre for Genetic Resources and Biotechnology (NACGRAB), Ibadan, Nigeria, at Leipzig University herbarium (LZ), Germany, and the National Botanical Research Institute, Windhoek, Namibia (Supplementary Table 7).
Genetic variation and population structure
We used ten diploid nuclear simple sequence repeat (nSSR), i.e. biparentally inherited, markers developed for S. senegal, and two universal haploid chloroplast (cpSSR), i.e. maternally inherited, microsatellite markers as described previously39. The final data set consisted of 730 samples for nSSR and 746 for cpSSR. Genotypes of nSSR markers were tested for null alleles using the program FreeNA78 finding null allele frequencies > 0.2 in 4.8% of locus x population combinations, preferentially in Southern African populations. We did not correct for null alleles, as the number and identity of null alleles is unknown. However, the results obtained with null allele corrected data are qualitatively the same (Supplementary Fig. 4). For the nSSR data set, intrapopulation genetic diversity was estimated as the total number of alleles per locus (Na), the average number of alleles per locus over loci (Aavr), the unbiased estimate of expected (He) and observed (Ho) heterozygosity using Arlequin v.3.5.1.379. The number of private alleles (Apriv) was estimated using GDA v.1.080. FSTAT v.1.281 was used to calculate the inbreeding coefficient (Fis) and rarified allelic richness (Ar), thus correcting for different sample sizes by rarefaction82. The package genepop v.1.283 was used to perform exact tests of Hardy–Weinberg equilibrium (HWE).
Population genetic structure was evaluated using a Bayesian clustering approach implemented in STRUCTURE v.2.3.484. For each K ranging from one to 29 (the number of sampling sites), we performed 90 replicate runs with 100,000 steps after a burn-in period of 50,000 steps considering the model of correlated allele frequencies and admixture without prior population information85. The “Evanno approach”85 was used to identify the value of K for the uppermost hierarchical level using Structure Harvester86, but we also scrutinized whether results of other K allowed for a clear biologically interpretation, i.e. whether emerging clusters include several individuals who are strongly assigned to that cluster, as suggested in the STURCTURE manual87. Moreover, as different non-symmetric modes of model outcomes are possible in large and complex data sets87, we used CLUMPAK43 to sort out modes, align clusters across runs and calculate the consensus. We used STRUCTURE both for an analysis with only nSSR data (Fig. 1) and a combined data set of nSSR and cpSSR data (Fig. 2), thus combining all available evidence.
For the cpSSR data, we used HAPLOTYPE v.1.0588 to estimate the mean number of alleles per locus (NacpSSR), number of haplotypes detected per population (A), effective number of haplotypes (Ne), genetic diversity (D2sh), haplotype richness, correcting for sample size (Rh), the number of private haplotypes (P), gene diversity within and over all populations. A parsimony network illustrating genetic relationships among haplotypes was inferred using PopArt89, assuming single-step mutations between alleles. For both nSSR and cpSSR, we quantified genetic differentiation with analyses of molecular variance (AMOVA) using sampling sites and clusters suggested by STRUCTURE as hierarchical levels using GenAlex v.6.90.
We tested for a decline of genetic variation (Ar) with distance from the East African origin of the range expansion into West Africa, with a linear model. A hypothetical location at 0.57°N; 36.38°E between two eastern populations was chosen as the point of origin. We compared population level diversity estimates among the three phylogroups with ANOVA, followed by a TukeyHSD test.
Geographic and environmental drivers of genetic structure
To elucidate the roles of geographic and environmental factors for genetic differentiation, we tested for isolation by distance (IBD) and isolation by environment (IBE). We generated matrices for genetic distance as pairwise FST (Supplementary Table 5) using GenAlex and geographic distance. Environmental distances were generated as Euclidian distances of four bioclimatic and soil variables that have been shown to best predict the current distribution of S. senegal in Africa (Lyam et al. 2020; Supplementary Table 6): mean temperature of the wettest quarter (Bio8), precipitation of the wettest month (Bio13)41, monthly variability in potential evapotranspiration (PETseasonality)91 and soil pH92. To quantify IBD and IBE, we performed Multiple Matrix Regression with Randomization (MMRR) using the R function ‘MMRR’93. This analysis was also done for two subranges (Sudano-Sahelian + Zambezian and Zambezian + Southern) to assess whether the same parameters are relevant in different parts of the range.
Future projection of current climate distribution models
To assess the potential loss of intraspecific genetic diversity due to climate change, we obtained the current potential distribution for the phylogroups of S. senegal generated from a total of 1132 unique occurrence records and four environmental variables27. The occurrence records were assigned to three phylogroups at K = 3 (Supplementary Fig. 3). We used the result of the Bayesian Cluster analysis at K = 3 because the three genetic groups corresponding to the biogeographic zones of Sudano-Sahelian, Zambezian and Southern Africa were strongly genetically differentiated and spatially coherent. The current potential distribution of the three phylogroups was independently projected at a resolution of 30 s (0.93 × 0.93 km = 0.86 km2 at the equator) while the future projections were assessed at a resolution of 10 min (18.6 × 18.6 km = 346 km2 at the equator) to three IPCC climate scenarios. We use the most recently updated scenarios based on different socioeconomic assumptions, also known as the “Shared Socioeconomic Pathways” (SSPs). The SSP2 4.5 (RCP 4.5) and SSP4 6.0 (RCP 6.0) are intermediate scenarios, while the SSP5 8.5 (RCP 8.5) is the high emission scenario. All projections were estimated by ensemble SDMs implemented in Biomod2. The model accuracy was evaluated with kappa, TSS and AUC for each phylogenetic group (Supplementary Table 4). We generated continuous probabilistic maps for the current and future potential distributions of three phylogroups of S. senegal in Africa. To obtain final range changes, we downscaled the future projections by resampling all rasters to the resolution of the current projection and quantified absolute and relative range changes of the phylogroups.
Research permit
The collection of plant material used in this study complied with relevant institutional, national, and international guidelines and legislation, in particular the Nagoya protocol. The following research permits were obtained for sampling for this study. Permit number: ZA/LP/88394 (issued date: 26.04.2018) for Limpopo; Permit number: MPB. 1371 (Issued date: 16.03.2018) for Mpumalanga both from South Africa. Permit number: RPIV00312018 (Issued date: 29.10.2018) for Namibia. Samples from Tanzania were received from the Herbarium, Department of Botany, University of Dar es Salam, Tanzania through collaboration (Reference number: BOT/H.3/2017; Issued date: 08.06.2017). Leaf material from six populations sampled in Ethiopia, Sudan, Mali, Mauritania, Burkina Faso and Senegal were received from Odee et al. 2012 (NERC Centre for Ecology and Hydrology, Bush Estate, Penicuik Midlothian EH26 0QB Edinburgh, UK). Populations from Nigeria, Republic of Niger and Ghana were sampled under the coverage of the Economic Community of West African States (ECOWAS) protocol, in favor of the first author.
Supplementary Information
Acknowledgements
We are thankful to P. Otto (Leipzig) for facilitating fieldwork. We are also grateful to D. Odee (UK/Kenya), A. Favre, B. Pospichal (both Germany), S. Akpalu (Ghana), T. Abasse, M. Massaoudu (both Republic of Niger), B. Gado (Nigeria), H. Suleiman (Tanzania), F. Chase, L. Nanyeni (both Namibia) and the National Herbarium of Namibia (WIND) for assisting with sample collection. We specially thank T. Couvreur (Ecuador), for his comments on earlier version of the manuscript. The German Academic Exchange Service (DAAD grant no. 91562729) provided financial support to P.T. Lyam. Field and lab work were additionally supported by funds from the German Science Foundation (projects MU 2934/2-1 and MU 2934/3-1), the German Federal Ministry of Education and Research (BMBF Grant No. 16GW0120K) to A. Muellner-Riehl, and by Leipzig University. Finally, we acknowledge support from Leipzig University for Open Access Publishing.
Author contributions
P.T.L. conceived and designed the study. P.T.L., H.N., M.G. and A.M conducted fieldwork and sample collection. P.T.L., W.D and J.D performed the analysis and generated data. P.T.L. and W.D. interpreted the data. W.D and A.N.M-R., J.D., J.S., and F.H. contributed ideas and feedback on analysis. P.T.L. drafted the manuscript with support from W.D. and J.D. P.T.L., W.D., A.N.M-R., J.D., J.S., F.H., H.N. and M.G. discussed the results and contributed critically to the final revisions of the manuscript. W.D., J.S. and A.N.M-R supervised the research.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Genotype data of nSSR and cpSSR loci for all samples are available as supplementary file, as are the occurrence records used for SDM.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-11182-z.
References
- 1.D’Amen M, Zimmermann NE, Pearman PB. Conservation of phylogeographic lineages under climate change. Glob. Ecol. Biogeogr. 2013;22:93–104. doi: 10.1111/j.1466-8238.2012.00774.x. [DOI] [Google Scholar]
- 2.Espíndola A, et al. Predicting present and future intra-specific genetic structure through niche hindcasting across 24 millennia. Ecol. Lett. 2012;15:649–657. doi: 10.1111/j.1461-0248.2012.01779.x. [DOI] [PubMed] [Google Scholar]
- 3.Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Tr. Ecol. Evolut. 2003;18:189–197. doi: 10.1016/S0169-5347(03)00008-9. [DOI] [Google Scholar]
- 4.Fontaine C, Lovett P, Sanou H, Maley J, Bouvet JM. Genetic diversity of the shea tree (Vitellaria paradoxa CF Gaertn), detected by RAPD and chloroplast microsatellite markers. Heredity. 2004;93:639. doi: 10.1038/sj.hdy.6800591. [DOI] [PubMed] [Google Scholar]
- 5.Hampe A, El Masri L, Petit RJ. Origin of spatial genetic structure in an expanding oak population. Mol. Ecol. 2010;19:459–471. doi: 10.1111/j.1365-294X.2009.04492.x. [DOI] [PubMed] [Google Scholar]
- 6.Omondi SF, Odee DW, Ongamo GO, Kanya JI, Khasa DP. Genetic consequences of anthropogenic disturbances and population fragmentation in Acacia senegal. Conserv. Genet. 2016;17:1235–1244. doi: 10.1007/s10592-016-0854-1. [DOI] [Google Scholar]
- 7.Hewitt G. Postglacial recolonization of European biota. Biol. J. Lin. Soc. 1999;68:87–112. doi: 10.1111/j.1095-8312.1999.tb01160.x. [DOI] [Google Scholar]
- 8.Donkpegan ASL, et al. Population genomics of the widespread African savannah trees Afzelia africana and Afzelia quanzensis reveals no significant past fragmentation of their distribution ranges. Am. J. Bot. 2020;107:498–509. doi: 10.1002/ajb2.1449. [DOI] [PubMed] [Google Scholar]
- 9.Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- 10.Holderegger R, Wagner H. Landscape genetics. Bioscience. 2008;58:199–207. doi: 10.1641/B580306. [DOI] [Google Scholar]
- 11.Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- 12.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 13.Pauls SU, Nowak C, Bálint M, Pfenninger M. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 2013;22:925–946. doi: 10.1111/mec.12152. [DOI] [PubMed] [Google Scholar]
- 14.Arnell NW, Lloyd-Hughes B. The global-scale impacts of climate change on water resources and flooding under new climate and socio-economic scenarios. Climatic Ch. 2014;122:127–140. doi: 10.1007/s10584-013-0948-4. [DOI] [Google Scholar]
- 15.Moss RH, et al. The next generation of scenarios for climate change research and assessment. Nature. 2010;463:747. doi: 10.1038/nature08823. [DOI] [PubMed] [Google Scholar]
- 16.van Vuuren DP, et al. The representative concentration pathways: an overview. Climatic Ch. 2011;109:5–31. doi: 10.1007/s10584-011-0148-z. [DOI] [Google Scholar]
- 17.Prather, M. et al. Annex II: climate system scenario tables. Climate Ch. 1395–1445 (2013).
- 18.Pachauri, R. K. et al. Climate change 2014: synthesis report. Contribution of Working Groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change. Synthesis report (Intergovernmental Panel on Climate Change, Geneva, Switzerland, 2014).
- 19.Müller, C. Climate change impact on Sub-Saharan Africa. An overview and analysis of scenarios and models (Dt. Inst. für Entwicklungspolitik, Bonn, 2009).
- 20.Serdeczny O, et al. Climate change impacts in Sub-Saharan Africa: From physical changes to their social repercussions. Reg. Environ. Ch. 2016;17:1585–1600. doi: 10.1007/s10113-015-0910-2. [DOI] [Google Scholar]
- 21.Linder HP, et al. The partitioning of Africa: Statistically defined biogeographical regions in sub-Saharan Africa. J. Biogeogr. 2012;39:1189–1205. doi: 10.1111/j.1365-2699.2012.02728.x. [DOI] [Google Scholar]
- 22.Sexton GJ, et al. Influence of putative forest refugia and biogeographic barriers on the level and distribution of genetic variation in an African savannah tree, Khaya senegalensis (Desr.) A. Juss. Tree Genet. Genomes. 2015 doi: 10.1007/s11295-015-0933-3. [DOI] [Google Scholar]
- 23.Linder HP, et al. Numerical re-evaluation of the sub-Saharan phytopchoria of mainland Africa. Biologiske Skrifter. 2005;55:229–252. [Google Scholar]
- 24.Ruiz Guajardo JC, et al. Landscape genetics of the key African acacia species Senegalia mellifera (Vahl)- the importance of the Kenyan Rift Valley. Mol. Ecol. 2010;19:5126–5139. doi: 10.1111/j.1365-294X.2010.04833.x. [DOI] [PubMed] [Google Scholar]
- 25.Kebede M, Enrich D, Taberlet P, Nemomissa S, Brochmann C. Phylogeography and conservation genetics of a giant lobelia (Lobelia giberroa) in Ethiopian and Tropical East African mountains. Mol. Ecol. 2007;16:1233–1243. doi: 10.1111/j.1365-294x.2007.03232.x. [DOI] [PubMed] [Google Scholar]
- 26.Kadu C, et al. Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands. Mol. Ecol. 2011;20:165–178. doi: 10.1111/j.1365-294X.2010.04931.x. [DOI] [PubMed] [Google Scholar]
- 27.Lyam PT, Duque-Lazo J, Schnitzler J, Hauenschild F, Müllner-Riehl AN. Testing the forest refuge hypothesis in sub-Saharan Africa using species distribution modeling for a key savannah tree species, Senegalia senegal (L.) Britton. Front. Biogeogr. 2020 doi: 10.21425/F5FBG48689. [DOI] [Google Scholar]
- 28.Logossa ZA, et al. Molecular data reveal isolation by distance and past population expansion for the shea tree (Vitellaria paradoxa C.F. Gaertn) in West Africa. Mol. Ecol. 2011;20:4009–4027. doi: 10.1111/j.1365-294X.2011.05249.x. [DOI] [PubMed] [Google Scholar]
- 29.Lompo D, Vinceti B, Konrad H, Gaisberger H, Geburek T. Phylogeography of African locust bean (Parkia biglobosa) reveals genetic divergence and spatially structured populations in west and central Africa. J. Heredity. 2018;109:811–824. doi: 10.1093/jhered/esy047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong Pock Tsy J-M, et al. Chloroplast DNA phylogeography suggests a West African centre of origin for the baobab, Adansonia digitata L. (Bombacoideae, Malvaceae) Mol. Ecol. 2009;18:1707–1715. doi: 10.1111/j.1365-294X.2009.04144.x. [DOI] [PubMed] [Google Scholar]
- 31.Allal F, et al. Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity. 2011;107:174–186. doi: 10.1038/hdy.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fagg CW, Allison GE. Acacia Senegal and the gum arabic trade: monograph and annotated bibliography. United Kingdom: University of Oxford; 2004. [Google Scholar]
- 33.Lézine AM. Late Quaternary vegetation and climate of the Sahel. Quatern. Res. 1989;32:317–334. doi: 10.1016/0033-5894(89)90098-7. [DOI] [Google Scholar]
- 34.Steele, T. Vertebrate records: Late Pleistocene of Africa. In Encyclopedia of Quaternary Science, edited by S. Elias. (Elsevier, Oxford, 2007), 3139–3150.
- 35.Raddad E, Salih A, Fadl M, Kaarakka V, Luukkanen O. Symbiotic nitrogen fixation in eight Acacia senegal provenances in dryland clays of the Blue Nile Sudan estimated by the 15N natural abundance method. Plant Soil. 2005;275:261–269. doi: 10.1007/s11104-005-2152-4. [DOI] [Google Scholar]
- 36.Gray A, et al. Does geographic origin dictate ecological strategies in Acacia senegal (L.) Willd? Evidence from carbon and nitrogen stable isotopes. Plant Soil. 2013;369:479–496. doi: 10.1007/s11104-013-1593-4. [DOI] [Google Scholar]
- 37.Ross, J. H. A conspectus of African acacia species (1979).
- 38.Odee DW, Telford A, Wilson J, Gaye A, Cavers S. Plio-Pleistocene history and phylogeography of Acacia senegal in dry woodlands and savannahs of sub-Saharan tropical Africa: evidence of early colonisation and recent range expansion. Heredity. 2012;109:372–382. doi: 10.1038/hdy.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyam P, et al. Genetic diversity and distribution of Senegalia senegal (L.) Britton under climate change scenarios in West Africa. PLoS ONE. 2018;13:e0194726. doi: 10.1371/journal.pone.0194726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicotra, A. B. et al. Plant phenotypic plasticity in a changing climate. Trends in Plant Science15, 684–692; 10.1016/j.tplants.2010.09.008 (2010). [DOI] [PubMed]
- 41.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 42.ESRI. ArcGIS Desktop: Release 10.5. Redlands, CA: Environmental Systems Research Institute (2020).
- 43.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Res. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elhadji SD, et al. Exploring genetic diversity and structure of Acacia senegal (L.) Willd to improve its conservation in Niger. African J. Biotechnol. 2017;16:1650–1659. doi: 10.5897/AJB2016.15846. [DOI] [Google Scholar]
- 45.Muriira NG, Muchugi A, Yu A, Xu J, Liu A. Genetic Diversity Analysis Reveals Genetic Differentiation and Strong Population Structure in Calotropis Plants. Sci. Rep. 2018;8:7832. doi: 10.1038/s41598-018-26275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conord C, Gurevitch J, Fady B. Large-scale longitudinal gradients of genetic diversity: a meta-analysis across six phyla in the Mediterranean basin. Ecol. Evol. 2012;2:2600–2614. doi: 10.1002/ece3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omondi SF, et al. Genetic diversity and population structure of Acacia senegal (L) Willd Kenya. Trop. Plant Biol. 2010;3:59–70. doi: 10.1007/s12042-009-9037-2. [DOI] [Google Scholar]
- 48.Marko PB, Hart MW. The complex analytical landscape of gene flow inference. Trends Ecol. Evol. 2011;26:448–456. doi: 10.1016/j.tree.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Goncalves AL, García MV, Heuertz M, González-Martínez SC. Demographic history and spatial genetic structure in a remnant population of the subtropical tree Anadenanthera colubrina var cebil (Griseb.) Altschul (Fabaceae) Ann. Forest Sci. 2019 doi: 10.1007/s13595-019-0797-z. [DOI] [Google Scholar]
- 50.Rosenzweig, M. L. Species diversity in space and time (Cambridge university press, 1995).
- 51.Vellend M, Geber MA. Connections between species diversity and genetic diversity. Ecol. Lett. 2005;8:767–781. doi: 10.1111/j.1461-0248.2005.00775.x. [DOI] [Google Scholar]
- 52.Ackerly DD, et al. The geography of climate change: implications for conservation biogeography. Divers. Distrib. 2010;16:476–487. doi: 10.1111/j.1472-4642.2010.00654.x. [DOI] [Google Scholar]
- 53.Waldvogel A-M, et al. Evolutionary genomics can improve prediction of species' responses to climate change. Evol. Lett. 2020;4:4–18. doi: 10.1002/evl3.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutchison DW, Templeton AR. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evol.; Int. J. Org. Evol. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- 55.Shi MM, Michalski SG, Welk E, Chen XY, Durka W. Phylogeography of a widespread Asian subtropical tree: genetic east-west differentiation and climate envelope modelling suggest multiple glacial refugia. J. Biogeogr. 2014;41:1710–1720. doi: 10.1111/jbi.12322. [DOI] [Google Scholar]
- 56.Voss N, Eckstein RL, Durka W. Range expansion of a selfing polyploid plant despite widespread genetic uniformity. Ann. Botany. 2012;110:585–593. doi: 10.1093/aob/mcs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiorini CF, et al. Phylogeography of the specialist plant Mandirola hirsuta (Gesneriaceae) suggests ancient habitat fragmentation due to savanna expansion. Flora. 2020;262:151522. doi: 10.1016/j.flora.2019.151522. [DOI] [Google Scholar]
- 58.Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution. 2014;68:1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- 59.Wang IJ, Bradburd GS. Isolation by environment. Mol. Ecol. 2014;23:5649–5662. doi: 10.1111/mec.12938. [DOI] [PubMed] [Google Scholar]
- 60.Nosil P, Vines TH, Funk DJ. Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evol.; Int. J. Org. Evol. 2005;59:705–719. [PubMed] [Google Scholar]
- 61.Wang IJ, Summers K. Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Mol. Ecol. 2010;19:447–458. doi: 10.1111/j.1365-294X.2009.04465.x. [DOI] [PubMed] [Google Scholar]
- 62.Xu B, et al. Population genetic structure is shaped by historical, geographic, and environmental factors in the leguminous shrub Caragana microphylla on the Inner Mongolia Plateau of China. BMC Plant Biol. 2017;17:200. doi: 10.1186/s12870-017-1147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendry AP, Day T. Population structure attributable to reproductive time: isolation by time and adaptation by time. Mol. Ecol. 2005;14:901–916. doi: 10.1111/j.1365-294X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- 64.Solomon, S., Manning, M., Marquis, M. & Qin, D. Climate change 2007-the physical science basis: Working group I contribution to the fourth assessment report of the IPCC (Cambridge university press, 2007).
- 65.Thuiller W. Climate change and the ecologist. Nature. 2007;448:550–552. doi: 10.1038/448550a. [DOI] [PubMed] [Google Scholar]
- 66.Osland MJ, et al. Tropicalization of temperate ecosystems in North America: The northward range expansion of tropical organisms in response to warming winter temperatures. Global Ch. Biol. 2021;27:3009–3034. doi: 10.1111/gcb.15563. [DOI] [PubMed] [Google Scholar]
- 67.Higgins SI, Lavorel S, Revilla E. Estimating plant migration rates under habitat loss and fragmentation. Oikos. 2003;101:354–366. doi: 10.1034/j.1600-0706.2003.12141.x. [DOI] [Google Scholar]
- 68.Jump AS, Penuelas J. Running to stand still: adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005;8:1010–1020. doi: 10.1111/j.1461-0248.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- 69.Jump AS, Marchant R, Peñuelas J. Environmental change and the option value of genetic diversity. Trends Plant Sci. 2009;14:51–58. doi: 10.1016/j.tplants.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 70.Kirk H, Freeland JR. Applications and implications of neutral versus non-neutral markers in molecular ecology. Int. J. Mol. Sci. 2011;12:3966–3988. doi: 10.3390/ijms12063966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bucharova A, et al. Mix and match: regional admixture provenancing strikes a balance among different seed-sourcing strategies for ecological restoration. Conserv. Genet. 2019;20:7–17. doi: 10.1007/s10592-018-1067-6. [DOI] [Google Scholar]
- 72.Tong Y, et al. Ex situ conservation of Pinus koraiensis can preserve genetic diversity but homogenizes population structure. Forest Ecol. Manag. 2020;465:117820. doi: 10.1016/j.foreco.2019.117820. [DOI] [Google Scholar]
- 73.Vessella F, Simeone MC, Schirone B. Quercus suber range dynamics by ecological niche modelling: from the Last Interglacial to present time. Quat. Sci. Rev. 2015;119:85–93. doi: 10.1016/j.quascirev.2015.04.018. [DOI] [Google Scholar]
- 74.Lovejoy TE. Climate change and biodiversity. India: TERI Press; 2006. [Google Scholar]
- 75.Poczai, P., Varga, I., Bell N.E. & Hyvonen, J. The molecular basis of plant genetic diversity. In Genomics meets biodiversity: advances in molecular marker development and their applications in plant genetic diversity assessment. The molecular basis of plant genetic diversity, edited by M. Caliskan (InTech Open Access Publisher2012), 3–31.
- 76.Botermans M, Sosef MSM, Chatrou LW, Couvreur TLP. Revision of the African Genus Hexalobus (Annonaceae) Syst. Bot. 2011;36:33–48. doi: 10.1600/036364411X553108. [DOI] [Google Scholar]
- 77.Sosef M, et al. Exploring the floristic diversity of tropical Africa. BMC Biol. 2017;15:15. doi: 10.1186/s12915-017-0356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chapuis M-P, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 79.Escoffier L, Lische H. ARLEQUIN suite ver. 3.5. A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 80.Lewis PO, Zaykin D. Genetic data analysis: computer program for the analysis of allelic data. Mol. Ecol. 2002;11:1157–1164. doi: 10.1046/j.1365-294X.2002.01512.x. [DOI] [PubMed] [Google Scholar]
- 81.AComputer Program to Calculate F-Statistics Goudet, J. FSTAT (Version 1.2) J. Hered. 1995;6:245–246. [Google Scholar]
- 82.El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl. Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- 83.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity. 1995;86:248–249. doi: 10.1093/oxfordjournals.jhered.a111573. [DOI] [Google Scholar]
- 84.Pritchard, J., Stephens, M. & Donelly, P. Inference of Population Structure Using Multilocus Genotype Data, 945–959 (2000). [DOI] [PMC free article] [PubMed]
- 85.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Earl DA, von Holdt BM. STRUCTURE HARVESTER A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 87.Pritchard, J. K., Wen, W. & Falush, D. Documentation for STRUCTURE software: Version 2.3. University of Chicago, Chicago, IL, 1–37 (2010).
- 88.Eliades, N. G. & Eliades, D. G. HAPLOTYPE ANALYSIS: software for analysis of haplotype data. Forest Goettingen (Germany): Genetics and Forest Tree Breeding, Georg-August University Goettingen (2009).
- 89.Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 90.Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Title PO, Bemmels JB. ENVIREM: an expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography. 2018;41:291–307. doi: 10.1111/ecog.02880. [DOI] [Google Scholar]
- 92.Hengl T, et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE. 2017;12:e0169748. doi: 10.1371/journal.pone.0169748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang IJ. Examining the full effects of landscape heterogeneity on spatial genetic variation: a multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution. 2013;67:3403–3411. doi: 10.1111/evo.12134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data of nSSR and cpSSR loci for all samples are available as supplementary file, as are the occurrence records used for SDM.