Abstract
Adenocarcinoma of the pancreas (PAAD) is a cancerous growth that deteriorates rapidly and has a poor prognosis. Researchers are investigating autophagy in PAAD to identify a new biomarker and treatment target. An autophagy-related gene (ARG) model for overall survival (OS) was constructed using multivariate Cox regression analyses. A cohort of the Cancer Genome Atlas (TCGA)-PAAD was used as the training group as a basis for model construction. This prediction model was validated with several external datasets. To evaluate model performance, the analysis with receiver operating characteristic curves (ROC) was performed. The Human Protein Atlas (HPA) and Cancer Cell Line Encyclopedia (CCLE) were investigated to validate the effects of ARGs expression on cancer cells. Comparing the levels of immune infiltration between high-risk and low-risk groups was finished through the use of CIBERSORT. The differentially expressed genes (DEGs) between the low-/high-risk groups were analyzed further via Gene Ontology biological process (GO-BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, which were used to identify potential small-molecule compounds in Connectivity Map (CMap), followed by half-maximal inhibitory concentration (IC50) examination with PANC-1 cells. The risk score was finally calculated as follows: BAK1 × 0.34 + ITGA3 × 0.38 + BAG3 × 0.35 + APOL1 × 0.26–RAB24 × 0.67519. ITGA3 and RAB24 both emerged as independent prognostic factors in multivariate Cox regression. Each PAAD cohort had a significantly shorter OS in the high-risk group than in the low-risk group. The high-risk group exhibited infiltration of several immune cell types, including naive B cells (p = 0.003), plasma cells (p = 0.044), and CD8 T cells (nearly significant, p = 0.080). Higher infiltration levels of NK cells (p = 0.025), resting macrophages (p = 0.020), and mast cells (p = 0.007) were found in the high-risk group than the low-risk group. The in vitro and in vivo expression of signature ARGs was consistent in the CCLE and HPA databases. The top 3 enriched Gene Ontology biological processes (GO-BPs) were signal release, regulation of transsynaptic signaling, and modulation of chemical synaptic transmission, and the top 3 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were MAPK, cAMP, and cell adhesion molecules. Four potential small-molecule compounds (piperacetazine, vinburnine, withaferin A and hecogenin) that target ARGs were also identified. Taking the results together, our research shows that the ARG signature may serve as a useful prognostic indicator and reveal potential therapeutic targets in patients with PAAD.
Subject terms: Cancer genetics, Gastrointestinal cancer, Tumour biomarkers
Introduction
Adenocarcinoma of the pancreas (PAAD) is a cancerous growth that deteriorates rapidly and has a poor prognosis. Due to its high mortality rate, PAAD has a low incidence rate and an almost identical mortality rate1. According to some studies, pancreatic cancer may overtake lung cancer by 2020 as the fourth most common cause of death from cancer in developed countries2,3. A significant proportion of deaths due to pancreatic cancer is caused by the lack of evident clinical symptoms in the early stages of the disease, which delays treatment4. It is estimated that only 20% of patients diagnosed with pancreatic cancer can undergo surgery, and the 5-year survival rate increases by only 20–30% after surgery5,6. Considering that the prognosis of pancreatic cancer patients is poor, it is imperative that a prognosis prediction model be developed and then treatment plans based upon it developed.
The autophagic process is a highly conserved system of degrading nonessential components within cells. The autophagy process is activated during times of metabolic stress for the purpose of providing alternative energy sources, including autophagosome formation, nucleation, double-membrane growth and closure, and fellowed by lysosomal fusion; this process helps maintain homeostasis and viability7. Various diseases have been linked to abnormal autophagy, including malignant tumors8,9. Cancer treatments such as chemotherapy10, targeted therapy11 or immunotherapy12 have most commonly been described as resistant to autophagy as a mechanism of resistance. Autophagy, however, appears to work in both directions to regulate tumorigenesis. Low autophagy levels promote cancer initiation in early-stage cancers, while high autophagy levels promote survival of tumor cells in nutrient-deficient environments13,14. It has been demonstrated that several human pancreatic ductal carcinoma cell lines exhibit high levels of autophagy, whereas no autophagy is evident in the pancreatic ducts of normal individuals15–17. Despite this, several reports have found that impaired autophagy promotes cancer since chronic stress, which contributes to PDAC in patients, impairs the levels of autophagy in the pancreas18. Therefore, it is unclear what role autophagy plays in pancreatic cancer19,20.
On the basis of gene expression signatures based on autophagy-related genes (ARGs), several cancers such as colon cancer21, breast cancer22, ovarian cancer23, and non-small-cell lung cancer24 have been associated with gene expression signatures. An ARG-based prognostic model for pancreatic cancer was reported in a recent study; however, the results were not confirmed with external databases25. To identify differentially expressed genes (DEGs), we integrated data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. In the development of our prognostic model, ARGs were examined, and their performance was evaluated using multiple external datasets (GSE57495, GSE78229), GDS4336, GSE85916, and ICGC-PACA-AU), all of which are associated with pancreatic cancer. According to our research, the expression of ARGs is associated with immune infiltration of tumors, and we developed a series of small-molecule compounds that target ARGs. These results suggest that the ARG signature may offer patients with PAAD prognostic information and provide potential drug targets.
Results
Characteristics of the patients included in the datasets
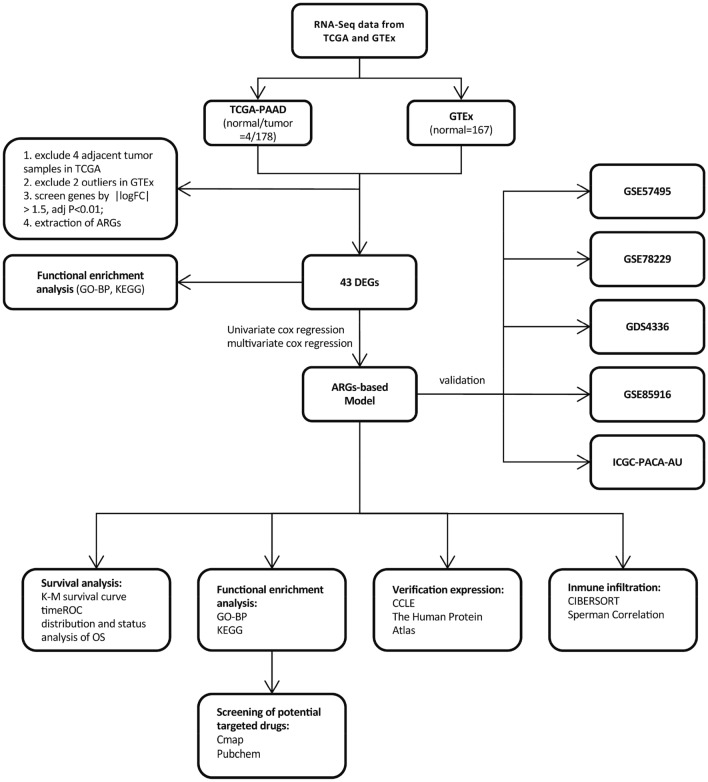
The sequencing information of pancreatic cancer patients from the TCGA-PAAD database, which contains 176 tumor samples and 4 adjacent samples, was downloaded. Subsequently, we used only the 176 tumor samples because there were a small number of adjacent tumor samples. As a control group, we downloaded sequence data from the GTEx database. Following filtering, 2 tissue samples with extremely low sequencing data were excluded, and finally, 165 normal pancreatic tissue samples were used to identify DEGs. As external data sources, we used the GEO and ICGC websites to examine the clinical applicability of the signature in various databases. Table 1 shows the characteristics of each dataset (Fig. 1).
Table 1.
| TCGA-PAAD | GDS4336 | GSE57495 | GSE78229 | GSE85916 | ICGC-PACA-AU | |
|---|---|---|---|---|---|---|
| Age (year) | ||||||
| < 65 | 81 | – | – | – | – | 33 |
| ≥ 65 | 95 | – | – | – | – | 46 |
| Unknown | 0 | – | – | – | – | 1 |
| Gender | ||||||
| Female | 80 | – | – | – | – | 40 |
| Male | 96 | – | – | – | – | 40 |
| Survival status | ||||||
| Alive | 88 | 13 | 21 | 14 | 22 | 32 |
| Dead | 88 | 29 | 42 | 35 | 57 | 48 |
| Grade | ||||||
| Grade 1 | 30 | – | – | 2 | – | – |
| Grade 2 | 94 | – | – | 24 | – | – |
| Grade 3 | 48 | – | – | 22 | – | – |
| Grade 4 | 2 | – | – | 1 | – | – |
| unknown | 2 | – | – | 0 | – | – |
| Stage | ||||||
| Stage I | 21 | – | 13 | 4 | – | – |
| Stage II | 145 | – | 50 | 45 | – | – |
| Stage III | 3 | – | 0 | 0 | – | – |
| Stage IV | 4 | – | 0 | 0 | – | – |
| Unknown | 3 | – | 0 | 0 | – | – |
| T | ||||||
| T1 | 7 | – | – | – | – | – |
| T2 | 24 | – | – | – | – | – |
| T3 | 140 | – | – | – | – | – |
| T4 | 3 | – | – | – | – | – |
| Unknown | 2 | – | – | – | – | – |
| M | ||||||
| M0 | 79 | – | – | – | – | – |
| M1 | 4 | – | – | – | – | – |
| Unknown | 93 | – | – | – | – | – |
| N | ||||||
| N0 | 49 | – | – | – | – | – |
| N1 | 122 | – | – | – | – | – |
| Unknown | 5 | – | – | – | – | – |
Figure 1.
Diagram of the Overall Study Design.
Identification of differentially expressed ARGs
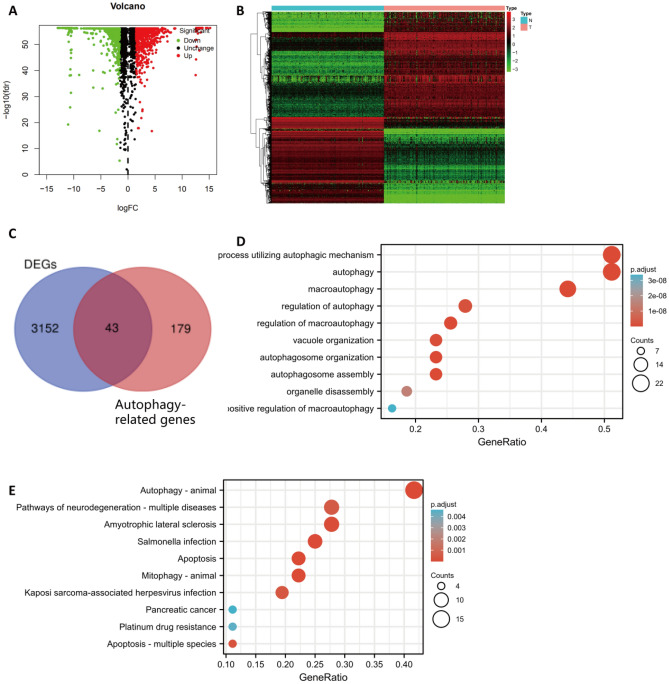
Using the normalizeBetweenArrays function in R software, we combined the TCGA-PAAD dataset with the GTEx normal pancreatic tissue dataset. Based on the criteria, a total of 3195 genes were found to be differentially expressed (Supplementary Table 1). Figure 2A–B shows the volcano and heatmaps. By overlapping the differentially expressed ARGs with the 232 ARGs from the HADb website, a total of 43 differentially expressed ARGs were selected (Fig. 2C and Supplementary Table 2).
Figure 2.
Identification and Enrichment Analysis of the Autophagy-Related DEGs. (A) Using the GTEx cohort data with the PAAD cohort data from the TCGA, the following volcano plot is derived. Red dots represent genes which are upregulated, green dots represent genes which are downregulated, and black dots indicate genes which are neither up- nor downregulated. (B) A heatmap illustrating the expression levels of DEGs. (C) Overlapping genes between the DEGs and ARGs. A bubble chart showing the enriched GO-BPs and KEGG pathways (D–E).
Numerous enriched GO-BPs and KEGG pathways were examined in the functional enrichment analysis (Fig. 2D–E). Based on GO-BP analysis, the ARGs mainly functioned to regulate autophagy and to utilize autophagic mechanisms. Based on KEGG analysis, these genes were indicated to be mainly involved in signaling pathways related to autophagy-animal. Moreover, we added unbiased GO-BP and KEGG analyses to Supplementary Fig. 1 by using all the DEGs.
Establishment of a prognostic signature with 5 ARGs
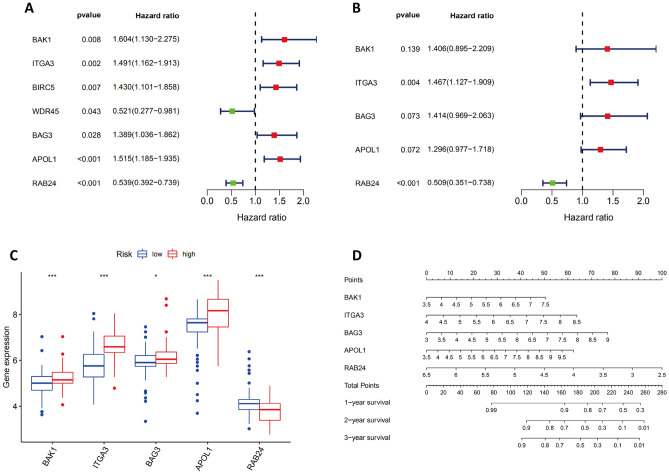
Using univariate Cox regression, a forest map was generated showing seven ARGs associated with pancreatic cancer prognosis: BAK1, ITGA3, BIRC5, WDR45, BAG3, APOL1, and RAB24 (Fig. 3A). Two of these 7 ARGs were protective, whereas 5 were associated with an increased risk. Then, we constructed a multivariate regression equation of these 7 genes, and we included 5 of them in the risk signature model. The final risk score was defined as [Expression level of BAK1 × (0.34085)] + [Expression level of ITGA3 × (0.38309)] + [Expression level of BAG3 × (0.34635)] + [Expression level of APOL1 × (0.25892)] + [Expression level of RAB24 × (−0.67519)]. Based on a multivariable Cox regression analysis, ITGA3 was identified as an independent high-risk ARG (hazard ratio [HR] = 1.47, 95% confidence interval [CI] = 1.13–1.91); RAB24 was an independent low-risk ARG (HR = 0.51, 95% CI = 0.35–0.74) (Fig. 3B).
Figure 3.
Establishment of a Prognostic Signature with 5 ARGs. Cox regression analyses of the ARG hazard ratios (HRs) in PAAD according to univariate (A) and multivariate (B) analyses. Red boxes indicate higher-risk ARGs with HRs > 1; green boxes indicate lower-risk ARGs with HRs < 1. (C) The levels of the 5 ARGs in the low/ high-risk groups. (D) A nomogram was generated to predict the survival rates of pancreatic cancer patients after one, two, and three years of therapy. A symbol of *, **, or *** denotes a p < 0.05, p < 0.01, or p < 0.001, respectively.
Based upon the median risk scores of the TCGA-PAAD cohort, the patients in the TCGA dataset, and a series of external datasets (GSE57495, GSE78229, GDS4336, GSE85916, and ICGC-PACA-AU) were divided into high- and low-risk groups.
In the TCGA cohort, we examined the expression of the 5 ARGs in the two risk groups. One of the five identified ARGs (RAB24) is downregulated in the high-risk group, whereas four of them are upregulated in the high-risk group (BAK1, ITGA3, BAG3, and APOL1) (Fig. 3C). To assess the ARG risk score's ability to accurately predict patients' 1-, 2-, and 3-year survival rates we generated nomograms which were used to make this evaluation (Fig. 3D).
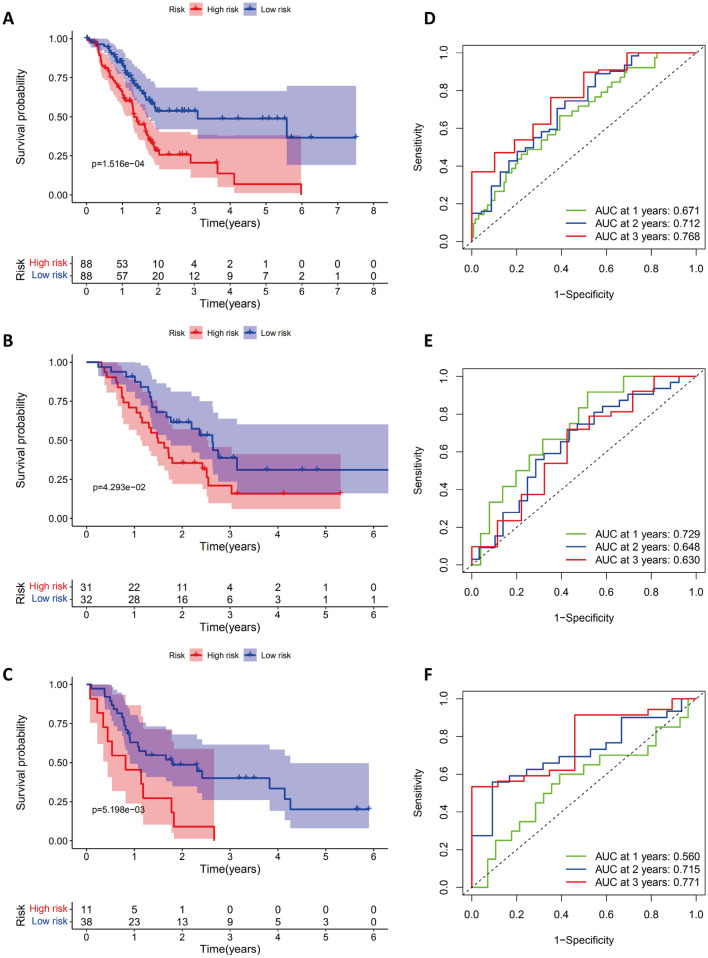
The TCGA and GEO cohorts were analyzed for clinicopathologic characteristics. Kaplan–Meier analysis was performed on the survival curves of the low-risk and high-risk patient groups. In the high-risk group, the probability of survival after one year was significantly lower (p = 0.001 in TCGA, p = 0.043 in GSE57495, p = 0.005 in GSE78229, p = 0.017 in GDS4336, p = 0.031 in GSE85916, and p = 0.003 in ICGC-PACA-AU) (Fig. 4A–C and Supplementary Fig. 12). The areas under the receiver operating characteristic curves (AUCs) for 1-year, 2-year and 3-year OS were 0.671, 0.712 and 0.768 in the TCGA-PAAD dataset (Fig. 4D); 0.729, 0.648 and 0.630 in the GSE57495 dataset (Fig. 4E); and 0.560, 0.715, and 0.771 in the GSE78229 dataset (Fig. 4F).
Figure 4.
Survival Analysis of the PAAD Cohort. Kaplan–Meier analysis of PAAD patients in the TCGA (A), GSE57495 (B), and GSE78229 cohorts (C) by high-risk and low-risk groups. A comparison of the ROC curves for the risk scores of PAAD patients for the TCGA (D), GSE57495 (E), and GSE78229 cohorts (F).
Prognosis value of the ARG-based risk model in the TCGA-PAAD dataset
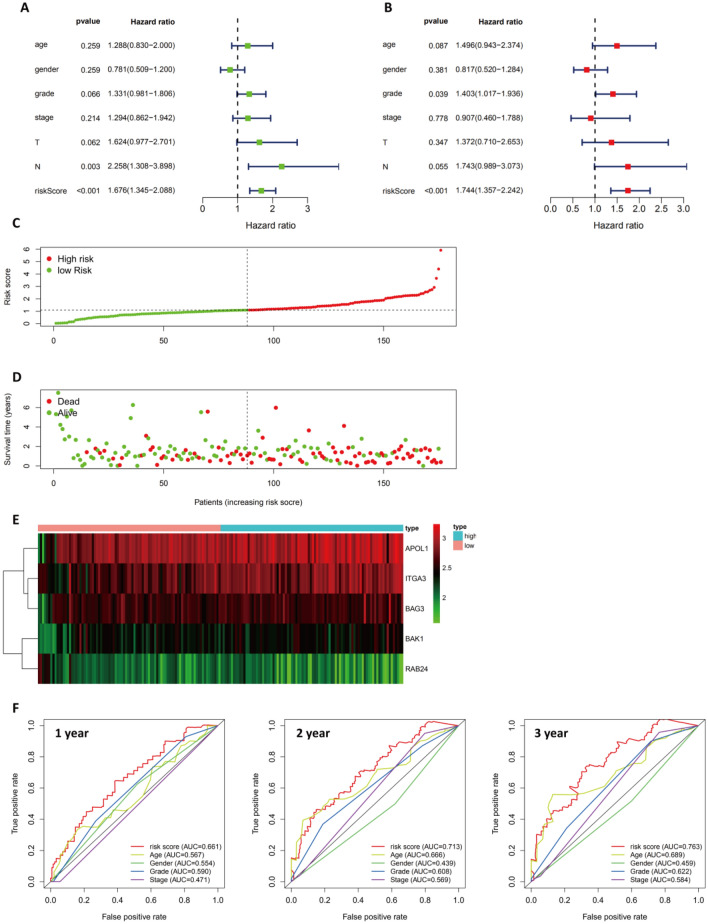
We conducted univariate and multivariate analyses to identify factors that might be associated with a poor/better prognosis in pancreatic cancer patients registered in TCGA (Fig. 5A–B). Multivariate analysis confirmed the significance of the grade and risk score in the forest map. Thus, risk scores based on the five ARGs were independently associated with patient outcomes (HR = 1.744, 95% CI = 1.36–2.24).
Figure 5.
Examination of the Prognostic ARG Signature in the TCGA-PAAD Dataset. In PAAD, the forest plot of univariate and multivariate Cox regression analyses is shown in (A–B). (C) Prognostic index distribution. (D) Survival data for patients in the low- and high-risk groups. (E) An interactive heatmap of gene expression profiles for the included ARGs. (F) The AUC values of the risk score at 1, 2, and 3 years, as well as the clinicopathological characteristics.
In the next step, a ranking of the risk scores was used to analyze the distribution of risk scores and survival status (Fig. 5C–D). Based on the results of the study, patients with higher risk scores were more likely to die than those with lower risk scores. The differential expression profiles of the 5 risk score-associated ARGs between the low-risk group and the high-risk group are shown in the heatmap of Fig. 5E. As a means of further examining the diagnostic efficiency of the five risk score-associated ARGs and the clinical characteristics, 1-, 2-, and 3-year ROC curves have been generated for the risk score and the clinical characteristics, as shown in Fig. 5F.
Investigation of the ARG-based risk model in the testing group
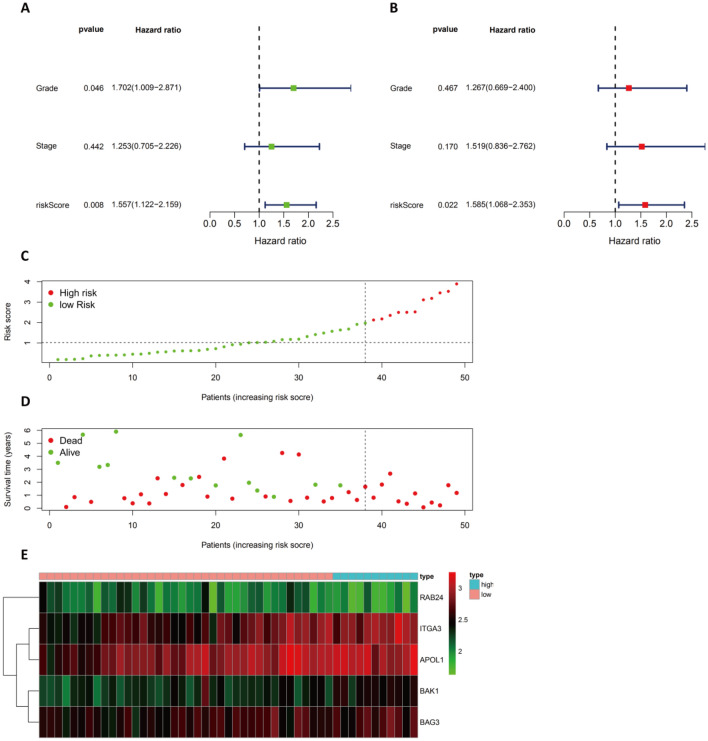
Following the analysis of the data from the TCGA datasets of pancreatic cancer cohorts, univariate and multivariate Cox regression analyses were conducted to examine the impact of the ARG signature on the prognosis of pancreatic cancer patients in external datasets. In GSE78229, both univariate and multivariate Cox regression analyses revealed that the risk score remained significant after multivariate analysis (Fig. 6A–B). This finding indicates that the risk score is independently related to the prognosis of patients (HR of 1.585, 95% CI of 1.08–2.35). From the GSE78229 dataset, groups of high-risk and low-risk patients were ranked according to risk scores to analyze the distribution of risk scores and survival status and the expression profiles of risk-associated ARGs (Fig. 6C–E). Therefore, ARG-based models were confirmed to be accurate in the independent validation pancreatic cancer cohorts.
Figure 6.
Evaluation of the Prognostic ARG Signature in the GSE78229 Dataset. (A–B) Forest plot illustrating the univariate and multivariate Cox regression analyses in PAAD. (C) Distribution of prognostic index. (D) Survival status of patients in the low- and high-risk groups. (E) The expression profile of the ARGs included in the heatmap.
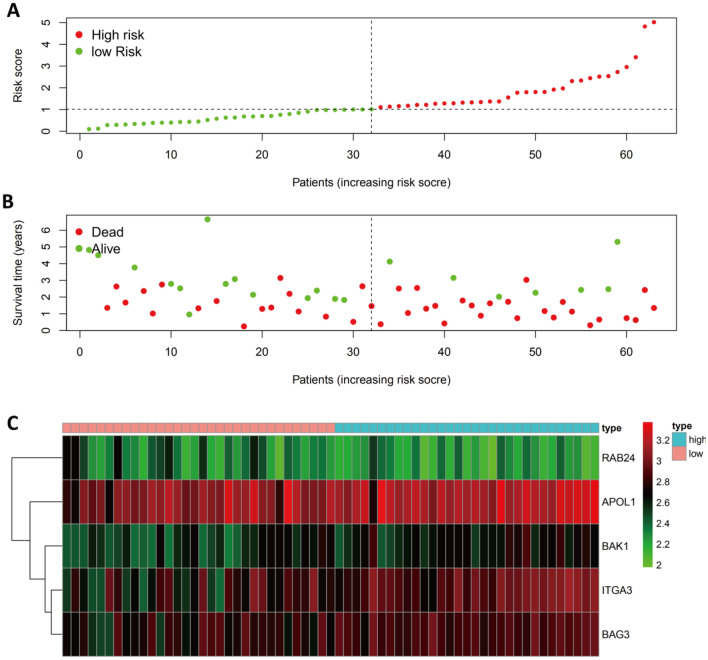
As the GSE57495 dataset does not contain any clinical information, it was not considered as part of the univariate or multivariate Cox regression analysis. However, we evaluated the distribution of risk scores and survival variables and the expression profiles of the risk-associated ARGs for the high-risk and low-risk pancreatic cancer patient groups based on their risk scores in the GSE57495 dataset (Fig. 7A–C). Supplementary Figs. 3–5 list the results from other external datasets (GDS4336, GSE85916, and ICGC-PACA-AU).
Figure 7.
Verification of the Prognostic ARG Signature in the GSE57495 Dataset. (A) Graph illustrating the distribution of the prognostic index. (B) The survival status of patients in low-risk and high-risk groups. (C) A heatmap showing the expression profiles of included ARGs.
Differences between high-risk and low-risk groups in terms of immune cell infiltration
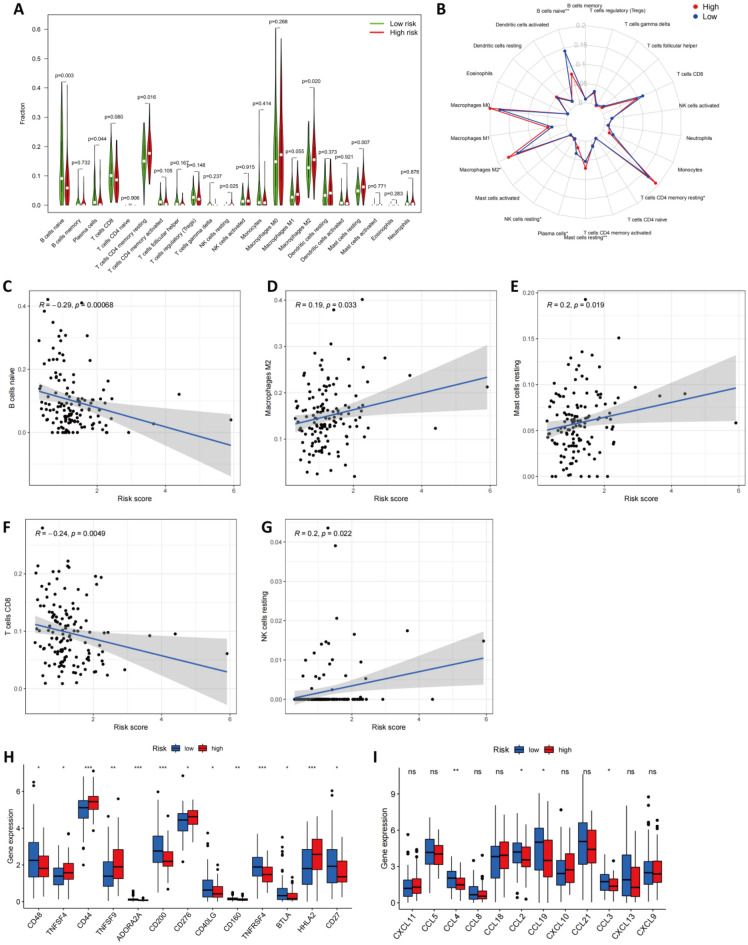
In our next analysis, we examined how ARGs impacted the prognosis of patients with pancreatic cancer. As autophagy is correlated with immune cell infiltration and immune cell infiltration has been linked to cancer development and prognosis, we used CIBERSORT to compare the abundance of 22 immune cell types between the high- and low-risk groups in TCGA-PAAD (Fig. 8A–B). A total of six immune cells were found to show varying degrees of infiltration between the two groups with respect to the 22 types that were studied: the infiltration levels of naive B cells (p = 0.003) and plasma cells (p = 0.044) were higher in the low-risk group, and the infiltration levels of memory CD4 T cells (p = 0.016), resting NK cells (p = 0.025), M2 macrophages (p = 0.020) and mast cells (p = 0.007) were higher in the high-risk group. In addition, the CD8 T-cell infiltration level was nearly significantly lower in the high-risk group (p = 0.080). The immune infiltration landscape is summarized in a radar plot in Fig. 8B. Additionally, the correlation between the risk score and the infiltration of the seven immune cells was investigated by Spearman correlation analysis. According to our results, the infiltration levels of naive B cells (r = −0.29, p < 0.001, Fig. 8C), M2 macrophages (r = 0.19, p = 0.033, Fig. 8D), mast cells (r = 0.20, p = 0.019, Fig. 8E), CD8 T cells (r = -0.24, p < 0.001, Fig. 8F) and NK resting cells (r = 0.20, p = 0.022, Fig. 8G) were statistically correlated with the ARG risk scores.
Figure 8.
Correlation between the ARG-Based Model and the Infiltration of Immune Cells. The difference in the infiltration of immune cells between the high‐ and low‐risk groups are shown in (A) and summarized in (B). (C–G) An analysis of Spearman correlations of risk scores with immune cell infiltration levels was performed, and results with statistical differences are illustrated. (H) The level of checkpoint genes was determined in the low- and high-risk groups. (I) The expression of TLS-associated genes was determined in the low- and high-risk groups. An analysis of Spearman correlations of risk scores with immune cell infiltration levels was performed.
Multiple immunotherapies produce therapeutic effects by expressing immune checkpoint genes. Therefore, we next compared the expression of checkpoint genes between the two groups (Fig. 8H). It was statistically significant that there was a difference between the two groups in terms of all the checkpoint genes. The expression of CD48, ADORA2A, CD200, CD40LG, CD160, TNFRSF4, BTLA, and CD27 was obviously lower in the high-risk group, and that of TNFSF4, CD44, TNFSF9, CD276, and HHLA2 was higher in the high-risk group than in the low-risk group. In addition, we investigated the level of tumor tertiary lymphoid structure (TLS)-related genes between the low/high-risk group according to previous reports26, and the results indicated that the level of CCL2, CCL3, CCL4, as well as CCL19 was lower in the high-risk group than in the low-risk group (Fig. 8I).
Verification of ARG expression in tumor samples and cell lines
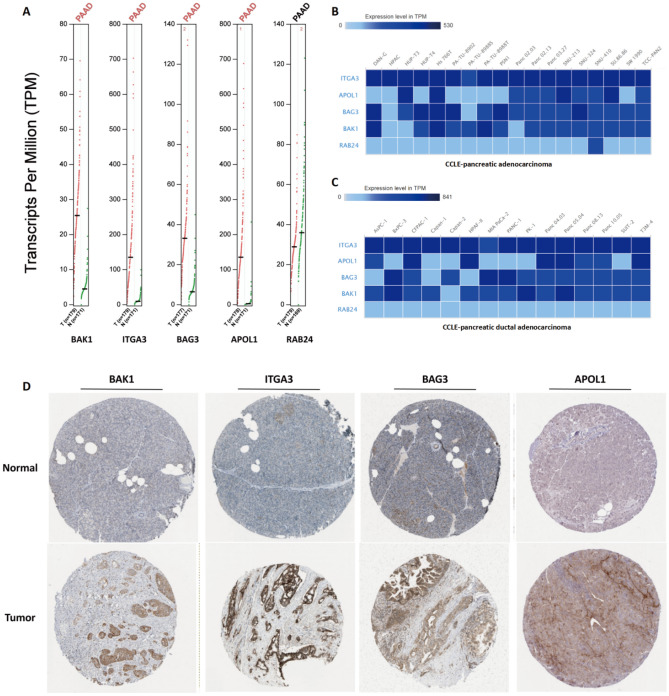
We collected data from pancreatic cancer patients and cell lines to verify the expression of ARGs. GEPIA was used to investigate clinical sequencing information from the TCGA and GTEx databases. The expression levels of BAK1, ITGA3, BAG3, and APOL1 were statistically increased in pancreatic cancer samples versus normal samples, while those of RAB24 was decreased (Fig. 9A). These results were confirmed with the CCLE database. BAK1, ITGA3, BAG3 and APOL1 were highly expressed in most pancreatic cancer and pancreatic ductal adenocarcinoma cell lines (Fig. 9B–C), while RAB24 was poorly expressed.
Figure 9.
Differential Expression of the 5 Signature ARGs in PAAD In Vitro and In Vivo. (A) ARG expression in pancreatic cancer tumor tissues and normal tissues according to the TCGA database. (B–C) The level of BAK1, ITGA3, BAG3 and APOL1 in most pancreatic cancer cell lines and pancreatic ductal adenocarcinoma cell lines based on the CCLE database. (D) Immunohistochemical staining of BAK1, ITGA3, BAG3 and APOL1 in pancreatic cancer tumor tissues and normal tissues. These data were obtained from the HPA database.
Using the HPA database, we conducted a further analysis of ARG expression. Compared with those from the normal controls, immunohistochemical results from the pancreatic tissues revealed statistical increases in the expression of BAK1, ITGA3, BAG3, and APOL1 in pancreatic cancer patients; no immunohistochemical data were available for RAB24 (Fig. 9D).
Functional enrichment analysis and small molecule drug screening using DEGs
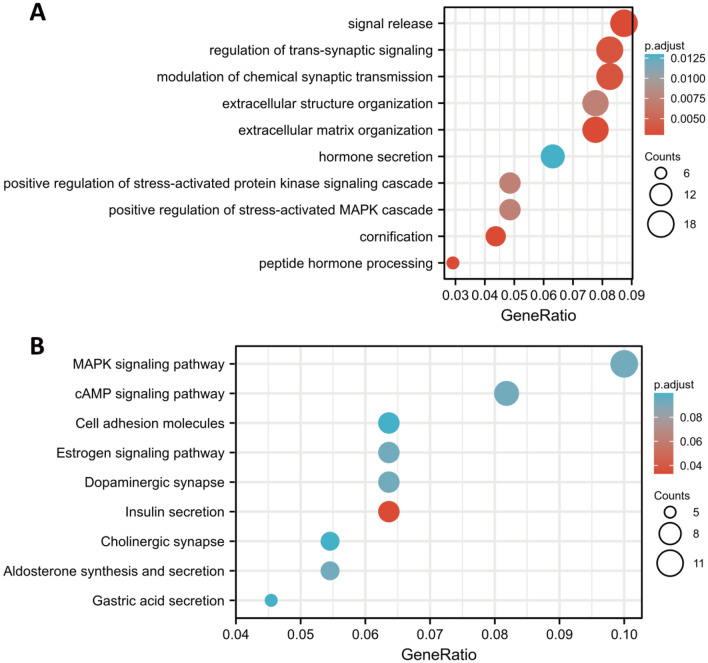
Further GO-BP and KEGG analyses were conducted on DEGs between the high-risk and low-risk groups. According to our analysis, the top three BPs pathways were signal release, transsynaptic signaling, and chemical synaptic transmission (Fig. 10A), and the top three KEGG pathways were MAPK, cAMP, and cell adhesion molecules (Fig. 10B).
Figure 10.
An analysis of functional enrichment between the low- and high-risk groups (A) GO-BP analysis and (B) KEGG pathway analysis.
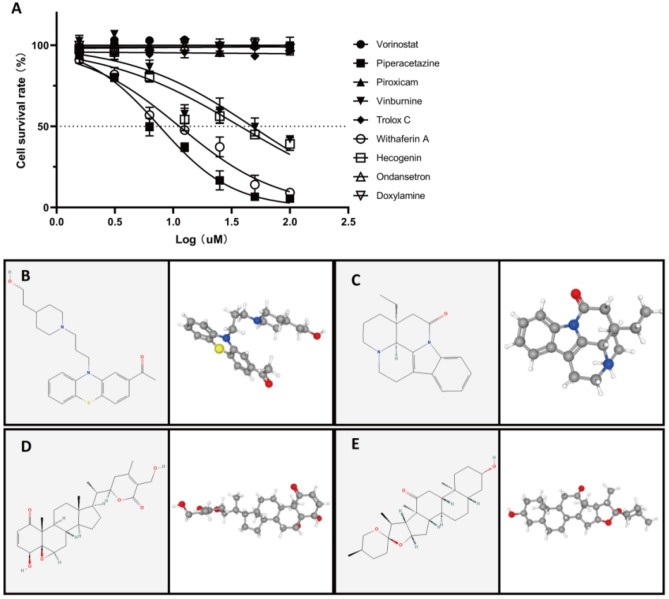
Our research matched upregulated and downregulated DEGs with small-molecule therapies using the Connectivity Map (CMap) website to identify potential drugs for PAAD. Table 2 lists the 9 most significant small-molecule drugs and their similarity scores. The IC50 values of the drugs were further examined in PANC-1 cells. We identified 4 drugs, namely, piperacetazine (IC50 = 7.627 μM), vinburnine (IC50 = 47.28 μM), withaferin A (IC50 = 11.26 μM), and hecogenin (IC50 = 37.45 μM) as potential drugs for improving the prognosis of patients with pancreatic cancer (Fig. 11A). Utilizing the PubChem website, their 2/3D spatial structure were visualized (Fig. 11B–E). These potent small-molecule drugs could reverse autophagy-induced gene expression, which provides a framework for developing targeted drugs for the treatment of PAAD. There are still many more studies to be conducted to investigate the usefulness of these candidate drugs for PAAD treatment.
Table 2.
The Screened Drugs for PAAD Treatment by CMAP.
| Rank | cmap name | Mean | n | Enrichment | p | Specificity |
|---|---|---|---|---|---|---|
| 1 | Vorinostat | −0.458 | 12 | −0.534 | 0.00108 | 0.3274 |
| 2 | Piperacetazine | −0.336 | 4 | −0.77 | 0.00567 | 0.0146 |
| 3 | Piroxicam | −0.581 | 4 | −0.71 | 0.01438 | 0.0292 |
| 4 | Vinburnine | −0.372 | 4 | −0.705 | 0.01566 | 0.0213 |
| 5 | Trolox C | −0.454 | 4 | −0.669 | 0.02697 | 0.0159 |
| 6 | Withaferin A | −0.418 | 4 | −0.661 | 0.02972 | 0.1475 |
| 7 | Hecogenin | −0.319 | 4 | −0.637 | 0.04201 | 0.0939 |
| 8 | Ondansetron | −0.1 | 4 | −0.627 | 0.0476 | 0.1053 |
| 9 | Doxylamine | −0.243 | 5 | −0.532 | 0.07332 | 0.2032 |
Figure 11.
Examination of Potential Small-Molecule Compounds. CMap has been used to screen for small-molecule compounds based on DEGs uploaded to the site. The IC50 values of the drugs were further examined in PANC-1 cells (A). Furthermore, for each of the four compounds that had been screened, the PubChem website was used to visualize the results. The 2/3D spatial structure of piperazine (B), vinburnine (C), withaferin A (C), and hecogenin (D).
Discussion
Pancreatic cancer is an aggressive malignancy with a low prognosis; the survival rate for patients suffering from pancreatic cancer is less than 10%1. Even though rapid advances have been made in the diagnostic and therapeutic treatment of malignant tumors, a lack of progress has actually been made for PAAD. According to a recent study, ARGs play an important role in PAAD25; however, less external datasets were used to verify these results. It is especially pertinent for PAAD, since sample sizes in each database are relatively small20. In this study, we evaluated the performance of an ARG-based prediction model in multiple datasets (GSE57495, GSE78229, GDS4336, GSE85916, and ICGC-PACA-AU). Considering the significance of autophagy in PAAD, in this study, we systematically constructed an ARG-based signature for PAAD to identify potential biomarkers for diagnosis and treatment.
First, we identified differentially expressed ARGs by combining the GTEx and TCGA datasets due to a lack of control samples in the TCGA database; then, we utilized GO-BP analysis and KEGG analysis to verify the role of ARGs in PAAD. ARG-based models were further established by univariate and multivariate Cox analyses. A final risk score was calculated as [Expression level of BAK1 × (0.34085)] + [Expression level of ITGA3 × (0.38309)] + [Expression level of BAG3 × (0.34635)] + [Expression level of APOL1 × (0.25892)] + [Expression level of RAB24 × (−0.67519)]. Evidence from the TCGA cohort and multiple external data sets indicate that ARGs play an important role in the prognosis of PAAD patients, even though the size of each cohort is relatively small. Overall, the ARG-based model in this study exhibits good universality and prognostic value.
In multivariate Cox analysis of the five ARGs, only ITGA3 (HR = 1.47, 95% CI = 1.13–1.99) and RAB24 (HR = 0.51, 95% CI = 0.35–0.74) were identified as independent ARGs. Integrin alpha 3 (ITGA3) belongs to the family of integrins, which are heterodimeric integral membrane proteins that function as cell surface adhesion molecules. This gene has been reported in several autophagy-related prognostic models for various tumors, indicating its important role in the progression of tumors27–32. Mechanistically, it has been reported that miRNA-524-5p inhibits the progression of papillary thyroid carcinoma cells by targeting FOXE1 and ITGA3 in cell autophagy and cycling pathways33. In addition, the IκB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells, resulting from decreased ITGA3-ITGB1 function, and these phenomena are associated with cancer progression and metastasis34. However, the related literature is still limited, and the role of ITGA3 should be further studied. As the only protective factor, high RAB24 expression was associated with a good prognosis. RAB24 is a small GTPase belonging to the Rab subfamily of Ras-related proteins; it regulates intracellular protein trafficking and has been shown to be important in autophagosome maturation35–39. RAB24 was the first protein to be shown to be necessary in the very late stages of basal autophagy40 and has been linked to various diseases, including carotid atherosclerosis41 and multiple sclerosis42. In studies in tumors, RAB24 was identified as a direct target of miR-615-5p in hepatocellular carcinoma (HCC). Research has shown that the downregulation of miR-615-5p expression and the upregulation of RAB24 expression promotes epithelial-mesenchymal transition, adhesion and vasculogenic mimicry in HCC cells, which enhance metastasis43. This finding is somewhat similar to that in a study of an ARG-based model for prostate cancer44, which also identified RAB24 as a protective factor. One possible explanation is that changes in RAB24 expression are a result of autophagy-associated physiological phenomena rather than a cause of tumorigenesis.
Although BAK1, BAG3 and APOL1 were not identified as independent risk factors, they are still underlying biomarkers and targets in PAAD treatment because the small sample size reduced the statistical power. BAK1 (known as BCL2 antagonist/killer 1) is an inhibitor of the BCL2 protein, which is the central player in the mitochondrion-dependent apoptotic program. As a result of multiple apoptotic events, mitochondrial outer membrane permeabilization (MOMP) has been shown to result in the release of cytochrome c into the cytosol to activate caspases, and BAK is a key effector involved in MOMP. However, BAK1 is also reported to be involved in autophagy45. Beclin-1 is an important regulatory hub to which proautophagic and antiautophagic proteins can bind. Bcl-2 family members are widely reported to modulate Beclin-1-dependent autophagy, which can be inhibited by BAK146–49. Further, BCL2 and BCL2L1/BCL-XL are thought to inhibit autophagy indirectly through an interaction with the proapoptotic members of the BCL2 family, BAX and BAK150,51. It is known as BCL2-associated athanogene 3 (BAG3), which is a multifunctional HSP70 cochaperone and antiapoptotic protein that interacts with the ATPase domain of HSP70 through its C-terminal BAG domain, and plays a crucial role in maintaining cellular proteostasis52. Along with HSP70 and LC3, BAG3 can also target polyubiquitinated client proteins for degradation by autophagy. BAG3 therefore plays a key physiological role in the regulation of both proteasomal degradation and autophagy, which are major cellular pathways for protein degradation53–55. Aberrant expression of BAG3 has been linked to different cancer entities56–61. Due to its ability to promote cell survival signaling by interacting with distinct client proteins in complex with HSP70, BAG3 overexpression contributes to the development of apoptosis resistance in various types of tumors62. APOL1 (apolipoprotein L 1) is a secreted high-density lipoprotein63, which has been demonstrated to participate in the progression of hyperlipidemia, obesity and atherosclerosis64–67. APOL1 overexpression induces autophagy and autophagy-associated cell death in a variety of cancer cell types68,69.
Understanding the cellular and molecular mechanisms underlying autophagy-related immune modulation is a prerequisite for the development of immunotherapy-based targeted approaches for this deadly malignancy. Via immune infiltration analysis, we found that the high-risk group exhibited increased levels of memory CD4 T cells, NK cells, M2 macrophages, and mast cell infiltration. Conversely, this group exhibited lower levels of naive B-cell and plasma cell infiltration than the low-risk group. The infiltration of another CD8 T-cell population was almost significantly decreased in the high-risk group compared with the low-risk group (p = 0.08). Tertiary lymphoid structures (TLSs) are composed of complex aggregates of cytotoxic lymphocytes, B lymphocytes (including plasma cells) and dendritic cells70. For patients with solid tumors, the presence of TLSs has been associated with a favorable outcome71. Studies have shown that cytotoxic CD8+ T cells are important effector cells that contribute to adaptive immunity by specifically recognizing and wiping out tumor cells, and they are thereby associated with improved survival in cancer patients20,72. In previous studies, it was found that infiltration of CD20+ B cells in ovarian, non-small lung and cervical cancers was associated with improved survival and decreased relapse rates20,72,73. According to two recent prognostic models of PAAD, low numbers of B cells (plasma cells) and CD8+ T cells were associated with a poor prognosis in the disease20,74,75. Monocytes can differentiate into M1 and M2 macrophages, and M2 macrophages exhibit immunosuppressive and tumor-promoting roles76. Therefore, high M2 macrophage infiltration levels may limit the antitumor response. In terms of CD4 + memory T cells, two recent studies reported that a high level of CD4 + naive/CD4 + memory cell infiltration predicted improved OS in PAAD and non-small-cell lung cancer77,78, which partly supports our results. In conclusion, the overall effect of the ARGs in this signature on the immune microenvironment of PAAD involves a comprehensive and complex process. We hypothesized that the risk score ultimately reflects the degree of antitumor response suppression because the high-risk group had worse survival outcomes than the low-risk group in multiple databases. It should be noted that the investigation of the expression of checkpoint genes and TLS-associated genes also revealed obvious differences between the two groups, indicating that the ARG-based model may provide potential targets for PAAD treatments, which is very similar to the results of another research model we built20. Interestingly, several checkpoints were more highly expressed in the high-risk group, indicating a worse prognosis in the PAAD cohort. Among the genes, several genes (CD4479, TNFSF980, and CD27681) have been reported and demonstrated previously; however, the high expression of HHLA2 is inconsistent with the results in a newly published study82. Exploration of the phenomena and mechanisms may require more experiments.
The DEGs were then analyzed for GO-BP and KEGG enrichment. A GO analysis revealed that signal release, regulation of transsynaptic signaling, and modulation of chemical synaptic transmission, all of which were associated with hormone secretion. Other evidence from both CMap (Table 2) and IC50 (Fig. 11A) examination showed that piperacetazine serves as a dopamine receptor antagonist and has the potential to be used to treat PAAD. Recently, the dopamine D2 receptor (D2R) family was demonstrated to be upregulated in many cancers and tied to stemness83. Interestingly, the expression of D2R and its associated G protein Gai2 has been reported to be obviously upregulated in pancreatic ductal adenocarcinoma tissue samples84. In addition, there is increasing evidence that autophagy is closely related to the activity of dopamine receptors85. Whether autophagy can regulate the activity of dopamine receptors and affect the occurrence and development of pancreatic cancer must be explored. Comparing the high-risk and low-risk groups, KEGG analysis revealed that DEGs were mainly enriched in MAPK and cAMP pathways, which are crucial for the activation and regulation of autophagy86–89. Interestingly, potential treatments targeting autophagy via MAPK90,91 and cAMP92 in PAAD have been recently reported. Further investigation was conducted on potential small-molecule drugs with significant negative fractions. Although several of these drugs are not clinically used, comparing the functions of different drugs targeting differentially expressed ARGs may provide putative biomarkers for further validation as changes in gene expression in cancer can influence treatment outcomes20,93. In our analysis, the DEGs were uploaded to the CMap website, and the IC50 values of the predicted drugs were further examined in PANC-1 cells. In addition to piperacetazine described above (IC50 = 7.627 μM), withaferin A also showed a strong killing effect on pancreatic cancer cells (IC50 = 11.26 μM). According to research, withaferin A, a natural compound derived from the ashwagandha plant Withania somnifera94, has anti-diabetic properties95, protects the liver from injury caused by acetaminophen96, and triggers apoptosis in various cancers, including breast97, prostate98, colorectal99, non-small-cell lung100 and pancreatic cancer. In addition, withaferin A also induces incomplete autophagy by suppressing the fusion of autophagosomes and lysosomes in human pancreatic cancer cells101.
To conclude, the identified risk-associated ARGs may provide a basis for developing PAAD treatments involving autophagy. Importantly, the predictive value of the ARGs-based signature was confirmed by external cohort of PAAD, indicating that this model is able to benefit formulation of precise treatment plans. In spite of this, additional prospective experiments are necessary to determine the clinical relevance of this model in defining the optimal personalized targeted treatments and to explore treatments that target ARGs in PAAD20.
Methods
The authors declare that all the methods in this article were performed in accordance with the relevant guidelines and regulations in the editorial and publishing policies of Scientific Reports (https://www.nature.com/srep/journal-policies/ editorial-policies#experimental-subjects).
Study design and data collection
Figure 1 illustrates the experimental design and data analysis flow. Transcription profiles and clinical data of TCGA-PAAD patients and normal controls from the GTEx database were obtained from the UCSC Xena website (http://xenabrowser.net/datapages/), and these data were used as a training group. Microarray datasets (GSE57495, GSE78229, GDS4336, and GSE85916) were downloaded from the Gene Expression Omnibus (GEO) portal (https://www.ncbi.nlm.nih.gov/geo/). RNA-seq data from the PACA-AU cohort were downloaded from the ICGC Data Portal (https://dcc.icgc.org/) for further validation of the signature and used as an external test group. The ARGs were obtained from the Human Autophagy Database (HADb) (http://www.autophagy.lu/clustering/) (listed in Supplementary Table 3).
Identification of DEGs and extraction of ARGs
The normalized gene expression was calculated using fragments per kilobase of transcript per million mapped reads (FPKM) and log2-based transformation. Following this, the "sva" package of R was used to normalize RNA expression profiles and to remove batch effects. Using the limma package, we identified the DEGs with |logFC|> 1.5 and adjusted p < 0.01 by the limma package. We then extracted the ARGs from the combined PAAD cohort data.
Construction of the prognostic risk model
Utilizing the TCGA-PAAD database, a univariate Cox regression analysis was conducted to identify differentially expressed ARGs with prognostic significance. To construct a potential independent prognostic ARG model for pancreatic cancer, the identified prognostic ARGs were further included in a multivariate Cox regression calculation. After establishing a formula for the risk score, we calculated the risk score in each case as follows:
Coefi represents the correlation coefficient of each ARG and X represents gene expression. Using the median risk score of the TCGA-PAAD cohort as a cutoff value, the external datasets patients were divided into high- and low-risk groups in accordance with the cutoff value.
Evaluation of the prognostic capacity of the ARG model
Both the TCGA cohort and external datasets (GSE57495, GSE78229, GDS4336, GSE85916 and ICGC-PACA-AU) were analyzed using R software. A Kaplan–Meier analysis was performed on the survival data based on ARGs for pancreatic cancer patients. Univariate and multivariate Cox regression analyses were used to identify the independent risk factors. The receiver operating characteristic (ROC) curves were generated by the timeROC package. The prognostic efficiency of the model was measured by the area under the ROC curve (AUC).
Estimation of immune cell infiltration
The levels of 22 cancer-infiltrating immune cell subgroups were quantified using CIBERSORT to evaluate immune cell infiltration. Using Spearman correlation analysis, further relationships were explored between immunocellular subgroups infiltrating tumors and ARG expression. The expression of potential immune checkpoint genes102–111 and TLS-related genes26 was also investigated according to previous literature.
Functional and pathway enrichment analysis
The DEGs between groups were analyzed with the “limma” and “clusterProfiler” packages to perform Gene Ontology biological process (GO-BP) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG)112–114 pathway enrichment analysis, as described previously20,115.
Verification of ARG expression
To verify the expression of ARGs, the expression in cells from the Cancer Cell Line Encyclopedia (CCLE) database was visualized by Expression Atlas (https://www.ebi.ac.uk/gxa/home), and immunohistochemistry data for clinical samples were obtained from the Human Protein Atlas (HPA) website (https://www.proteinatlas.org/). The baseline immunohistochemical image data are provided in Supplementary Fig. 6.
Identification of potential compounds
DEGs based on the ARG signature were divided into up- and downregulated gene groups and uploaded to the Connectivity Map website (https://portals.broadinstitute.org/cmap/), as our described previously116.
CCK-8 assay and IC50 examination
The cytotoxicity of the predicted drugs against pancreatic cancer cells (PANC-1) was assessed by using a Cell Counting Kit-8. All cells were seeded in 96-well plates at a density of 5 × 103 cells/well at 37 °C. Each well was treated with different drug concentrations (100/50/25/12.5/6.25/3.125/1.5625 nM) or 1% DMSO for 48 h. Then, 10 µL of CCK-8 solution was added, and the cells were incubated for another 1 h. A microplate reader was used to determine absorbance at 450 nm and 650 nm (SpectraMax M5, Molecular Devices, USA). The assay was performed in triplicate. The IC50 was determined according to the previous literature. The 2D/3D structures of the screened drugs were further investigated by the PubChem website (pubchem.ncbi.nlm.nih.gov).
Statistical analysis
Data were analyzed by R software version 4.0.4. Data following a normal or nonnormal distribution were compared using unpaired Student's t test and the Wilcoxon test, respectively, and the statistical significance threshold was set at p < 0.05116.
Supplementary Information
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Grant No. 81902055) and the National Science and Technology Major Project of China (Grant No. 2018ZX10302205).
Author contributions
J.D. collected the papers, analyzed the data, and drafted the manuscript. N.Z., Q.Z. and L.L. reviewed the data. P.M. and Y.Z. contributed to writing. Y.Z. presented the idea of this manuscript, analyzed the conclusions, and revised the manuscript. Contributions from all authors were included in the article and the final version was approved.
Data availability
The datasets analyzed during the current research are all available in The Cancer Genome Atlas (http://cancergenome.nih.gov) repositories, Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), and International Cancer Genome Consortium Pancreatic Cancer Australian [ICGC-PACA-AU]) (https://dcc.icgc.org/). The data used to support the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-11050-w.
References
- 1.McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018;24(43):4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019;10(1):10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Korc M. Pancreatic cancer-associated stroma production. Am. J. Surg. 2007;194(4 Suppl):S84–S86. doi: 10.1016/j.amjsurg.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H, Li T, Du Y, Li M. Pancreatic cancer: Challenges and opportunities. BMC Med. 2018;16(1):214. doi: 10.1186/s12916-018-1215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 7.Deretic V, Klionsky DJ. Autophagy and inflammation: A special review issue. Autophagy. 2018;14(2):179–180. doi: 10.1080/15548627.2017.1412229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124(16):3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy J, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sui X, Chen R, Wang Z, et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4(10):e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tracey N, Creedon H, Kemp AJ, et al. HO-1 drives autophagy as a mechanism of resistance against HER2-targeted therapies. Breast Cancer Res. Treat. 2020;179(3):543–555. doi: 10.1007/s10549-019-05489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark CA, Gupta HB, Curiel TJ. Tumor cell-intrinsic CD274/PD-L1: A novel metabolic balancing act with clinical potential. Autophagy. 2017;13(5):987–988. doi: 10.1080/15548627.2017.1280223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5(9):726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 14.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30(17):1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Wang X, Contino G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endo S, Nakata K, Ohuchida K, et al. Autophagy Is required for activation of pancreatic stellate cells, associated with pancreatic cancer progression and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;152(6):1492–1506.e24. doi: 10.1053/j.gastro.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todoric J, Antonucci L, Di Caro G, et al. Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell. 2017;32(6):824–839.e8. doi: 10.1016/j.ccell.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piffoux M, Eriau E, Cassier PA. Autophagy as a therapeutic target in pancreatic cancer. Br. J. Cancer. 2021;124(2):333–344. doi: 10.1038/s41416-020-01039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Lv L, Ma P, Zhang Y, Deng J, Zhang Y. Identification of an autophagy-related pair signature for predicting prognoses and immune activity in pancreatic adenocarcinoma. Front. Immunol. 2021;12:743938. doi: 10.3389/fimmu.2021.743938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Dai S, Yuan Y, Xiao Q, Ding K. A prognostic model for colon cancer patients based on eight signature autophagy genes. Front. Cell Dev. Biol. 2020;8:602174. doi: 10.3389/fcell.2020.602174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin QG, Liu W, Mo YZ, et al. Development of prognostic index based on autophagy-related genes analysis in breast cancer. Aging (Albany NY). 2020;12(2):1366–1376. doi: 10.18632/aging.102687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An Y, Bi F, You Y, Liu X, Yang Q. Development of a novel autophagy-related prognostic signature for serous ovarian cancer. J. Cancer. 2018;9(21):4058–4071. doi: 10.7150/jca.25587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Wu L, Ao H, et al. Prognostic implications of autophagy-associated gene signatures in non-small cell lung cancer. Aging (Albany NY). 2019;11(23):11440–11462. doi: 10.18632/aging.102544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue P, Zhu C, Gao Y, et al. Development of an autophagy-related signature in pancreatic adenocarcinoma. Biomed. Pharmacother. 2020;126:110080. doi: 10.1016/j.biopha.2020.110080. [DOI] [PubMed] [Google Scholar]
- 26.Lin Z, Huang L, Li S, Gu J, Cui X, Zhou Y. Pan-cancer analysis of genomic properties and clinical outcome associated with tumor tertiary lymphoid structure. Sci. Rep. 2020;10:21530. doi: 10.1038/s41598-020-78560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, C., Li, A.H., Liu, S., Sun, H. Identification of an 11-Autophagy-related-gene signature as promising prognostic biomarker for bladder cancer patients. Biology (Basel). 2021. 10(5). [DOI] [PMC free article] [PubMed]
- 28.Wang SS, Chen G, Li SH, et al. Identification and validation of an individualized autophagy-clinical prognostic index in bladder cancer patients. Onco. Targets Ther. 2019;12:3695–3712. doi: 10.2147/OTT.S197676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Gao L, Guo X, et al. Development and validation of a nomogram with an autophagy-related gene signature for predicting survival in patients with glioblastoma. Aging (Albany NY). 2019;11(24):12246–12269. doi: 10.18632/aging.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren Z, Zhang L, Ding W, et al. Development and validation of a novel survival model for head and neck squamous cell carcinoma based on autophagy-related genes. Genomics. 2021;113(1 Pt 2):1166–1175. doi: 10.1016/j.ygeno.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 31.Jin Y, Qin X. Development of a prognostic signature based on autophagy-related genes for head and neck squamous cell carcinoma. Arch. Med. Res. 2020;51(8):860–867. doi: 10.1016/j.arcmed.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Yang C, Mei H, Peng L, Jiang F, Xie B, Li J. Prognostic correlation of an autophagy-related gene signature in patients with head and neck squamous cell carcinoma. Comput. Math. Methods Med. 2020;2020:7397132. doi: 10.1155/2020/7397132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Chen X, Lin T, Chen X, Yan J, Jiang S. MicroRNA-524-5p suppresses the progression of papillary thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell autophagy and cycling pathways. J. Cell Physiol. 2019;234(10):18382–18391. doi: 10.1002/jcp.28472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen N, Debnath J. IκB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells independently of the PI3K-AKT-MTORC1 pathway. Autophagy. 2013;9(8):1214–1227. doi: 10.4161/auto.24870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olkkonen VM, Dupree P, Killisch I, Lütcke A, Zerial M, Simons K. Molecular cloning and subcellular localization of three GTP-binding proteins of the rab subfamily. J. Cell Sci. 1993;106(Pt 4):1249–1261. doi: 10.1242/jcs.106.4.1249. [DOI] [PubMed] [Google Scholar]
- 36.Munafó DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3(7):472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- 37.Gutierrez MG, Vázquez CL, Munafó DB, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7(7):981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 38.Wu M, Yin G, Zhao X, et al. Human RAB24, interestingly and predominantly distributed in the nuclei of COS-7 cells, is colocalized with cyclophilin A and GABARAP. Int. J. Mol. Med. 2006;17(5):749–754. [PubMed] [Google Scholar]
- 39.Tambe Y, Yamamoto A, Isono T, Chano T, Fukuda M, Inoue H. The drs tumor suppressor is involved in the maturation process of autophagy induced by low serum. Cancer Lett. 2009;283(1):74–83. doi: 10.1016/j.canlet.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 40.Ylä-Anttila P, Eskelinen EL. Roles for RAB24 in autophagy and disease. Small GTPases. 2018;9(1–2):57–65. doi: 10.1080/21541248.2017.1317699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaminathan B, Goikuria H, Vega R, et al. Autophagic marker MAP1LC3B expression levels are associated with carotid atherosclerosis symptomatology. PLoS ONE. 2014;9(12):e115176. doi: 10.1371/journal.pone.0115176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Igci M, Baysan M, Yigiter R, et al. Gene expression profiles of autophagy-related genes in multiple sclerosis. Gene. 2016;588(1):38–46. doi: 10.1016/j.gene.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 43.Chen Z, Wang X, Liu R, et al. KDM4B-mediated epigenetic silencing of miRNA-615-5p augments RAB24 to facilitate malignancy of hepatoma cells. Oncotarget. 2017;8(11):17712–17725. doi: 10.18632/oncotarget.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu D, Jiang L, Luo S, et al. Development of an autophagy-related gene expression signature for prognosis prediction in prostate cancer patients. J. Transl. Med. 2020;18(1):160. doi: 10.1186/s12967-020-02323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen HC, Kanai M, Inoue-Yamauchi A, et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat. Cell. Biol. 2015;17(10):1270–1281. doi: 10.1038/ncb3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J. Cell. Biol. 2010;190(5):881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molejon MI, Ropolo A, Re AL, Boggio V, Vaccaro MI. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 2013;3:1055. doi: 10.1038/srep01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedro JM, Wei Y, Sica V, et al. BAX and BAK1 are dispensable for ABT-737-induced dissociation of the BCL2-BECN1 complex and autophagy. Autophagy. 2015;11(3):452–459. doi: 10.1080/15548627.2015.1017191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akar U, Chaves-Reyez A, Barria M, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4(5):669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- 50.Lindqvist LM, Vaux DL. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy. 2014;10(8):1474–1475. doi: 10.4161/auto.29639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc. Natl. Acad. Sci. U S A. 2014;111(23):8512–8517. doi: 10.1073/pnas.1406425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kögel, D., Linder, B., Brunschweiger, A., Chines, S., Behl, C. At the crossroads of apoptosis and autophagy: Multiple roles of the co-chaperone BAG3 in stress and therapy resistance of cancer. Cells. 2020. 9(3). [DOI] [PMC free article] [PubMed]
- 53.Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: A multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2(4):e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Behl C. Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 2016;37(8):672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Stürner E, Behl C. The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front. Mol. Neurosci. 2017;10:177. doi: 10.3389/fnmol.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosati A, Bersani S, Tavano F, et al. Expression of the antiapoptotic protein BAG3 is a feature of pancreatic adenocarcinoma and its overexpression is associated with poorer survival. Am. J. Pathol. 2012;181(5):1524–1529. doi: 10.1016/j.ajpath.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Raffone A, Travaglino A, et al. BAG3 expression correlates with the grade of dysplasia in squamous intraepithelial lesions of the uterine cervix. Acta Obstet. Gynecol. Scand. 2020;99(1):99–104. doi: 10.1111/aogs.13716. [DOI] [PubMed] [Google Scholar]
- 58.Shi H, Chen W, Dong Y, Lu X, Zhang W, Wang L. BAG3 promotes chondrosarcoma progression by upregulating the expression of β-catenin. Mol. Med. Rep. 2018;17(4):5754–5763. doi: 10.3892/mmr.2018.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu H, Wu W, Fu Y, et al. Overexpressed BAG3 is a potential therapeutic target in chronic lymphocytic leukemia. Ann. Hematol. 2014;93(3):425–435. doi: 10.1007/s00277-013-1883-1. [DOI] [PubMed] [Google Scholar]
- 60.Xiao H, Cheng S, Tong R, et al. BAG3 regulates epithelial-mesenchymal transition and angiogenesis in human hepatocellular carcinoma. Lab Invest. 2014;94(3):252–261. doi: 10.1038/labinvest.2013.151. [DOI] [PubMed] [Google Scholar]
- 61.Yang D, Zhou J, Wang H, Wang Y, Yang G, Zhang Y. High expression of BAG3 predicts a poor prognosis in human medulloblastoma. Tumour Biol. 2016;37(10):13215–13224. doi: 10.1007/s13277-016-5197-5. [DOI] [PubMed] [Google Scholar]
- 62.De Marco M, Basile A, Iorio V, et al. Role of BAG3 in cancer progression: A therapeutic opportunity. Semin Cell Dev. Biol. 2018;78:85–92. doi: 10.1016/j.semcdb.2017.08.049. [DOI] [PubMed] [Google Scholar]
- 63.Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586(7):947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duchateau PN, Movsesyan I, Yamashita S, et al. Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J. Lipid Res. 2000;41(8):1231–1236. doi: 10.1016/S0022-2275(20)33430-1. [DOI] [PubMed] [Google Scholar]
- 65.Albert TS, Duchateau PN, Deeb SS, et al. Apolipoprotein L-I is positively associated with hyperglycemia and plasma triglycerides in CAD patients with low HDL. J. Lipid Res. 2005;46(3):469–474. doi: 10.1194/jlr.M400304-JLR200. [DOI] [PubMed] [Google Scholar]
- 66.Li Q, Fan P, Bai H, et al. Distribution and effect of apoL-I genotype on plasma lipid and apolipoprotein levels in Chinese normalipidemic and endogenous hypertriglyceridemic subjects. Clin. Chim. Acta. 2009;403(1–2):152–155. doi: 10.1016/j.cca.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Horrevoets AJ, Fontijn RD, van Zonneveld AJ, de Vries CJ, ten Cate JW, Pannekoek H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood. 1999;93(10):3418–3431. doi: 10.1182/blood.V93.10.3418.410k23_3418_3431. [DOI] [PubMed] [Google Scholar]
- 68.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4(8):1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J. Biol. Chem. 2008;283(31):21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Engelhard VH, Rodriguez AB, Mauldin IS, Woods AN, Peske JD, Slingluff CL., Jr Immune cell infiltration and tertiary lymphoid structures as determinants of antitumor immunity. J. Immunol. 2018;200(2):432–442. doi: 10.4049/jimmunol.1701269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trajkovski G, Ognjenovic L, Karadzov Z, et al. Tertiary lymphoid structures in colorectal cancers and their prognostic value. Open Access Maced. J. Med. Sci. 2018;6(10):1824–1828. doi: 10.3889/oamjms.2018.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol. Immunol. 2017;14(8):662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varn FS, Tafe LJ, Amos CI, Cheng C. Computational immune profiling in lung adenocarcinoma reveals reproducible prognostic associations with implications for immunotherapy. Oncoimmunology. 2018;7(6):e1431084. doi: 10.1080/2162402X.2018.1431084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu F, Zhang Z, Yuan M, et al. M6A regulatory genes play an important role in the prognosis, progression and immune microenvironment of pancreatic adenocarcinoma. Cancer Invest. 2021;39(1):39–54. doi: 10.1080/07357907.2020.1834576. [DOI] [PubMed] [Google Scholar]
- 75.Zhuang H, Huang S, Zhou Z, et al. A four prognosis-associated lncRNAs (PALnc) based risk score system reflects immune cell infiltration and predicts patient survival in pancreatic cancer. Cancer Cell Int. 2020;20:493. doi: 10.1186/s12935-020-01588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee HW, Choi HJ, Ha SJ, Lee KT, Kwon YG. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochim. Biophys. Acta. 2013;1835(2):170–179. doi: 10.1016/j.bbcan.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 77.Hang J, Huang J, Zhou S, et al. The clinical implication of CD45RA(+) naïve T cells and CD45RO(+) memory T cells in advanced pancreatic cancer: A proxy for tumor biology and outcome prediction. Cancer Med. 2019;8(3):1326–1335. doi: 10.1002/cam4.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang P, Ma J, Yang X, Li W. Peripheral CD4+ naïve/memory ratio is an independent predictor of survival in non-small cell lung cancer. Oncotarget. 2017;8(48):83650–83659. doi: 10.18632/oncotarget.19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li XP, Zhang XW, Zheng LZ, Guo WJ. Expression of CD44 in pancreatic cancer and its significance. Int. J. Clin. Exp. Pathol. 2015;8:6724–6731. [PMC free article] [PubMed] [Google Scholar]
- 80.Wu J, Wang Y, Yang Y, Liu F, Chen J, Jiang Z, Jiang Z. TNFSF9 promotes metastasis of pancreatic cancer through Wnt/Snail signaling and M2 polarization of macrophages. Aging (Albany NY) 2021;13:21571–21586. doi: 10.18632/aging.203497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inamura, K. et al. Tumor B7-H3 (CD276) Expression and survival in pancreatic cancer. J Clin. Med.7, (2018). [DOI] [PMC free article] [PubMed]
- 82.Boor P, et al. HHLA2 is expressed in pancreatic and ampullary cancers and increased expression is associated with better post-surgical prognosis. Br. J. Cancer. 2020;122:1211–1218. doi: 10.1038/s41416-020-0755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weissenrieder JS, Neighbors JD, Mailman RB, Hohl RJ. Cancer and the dopamine D(2) receptor: A pharmacological perspective. J. Pharmacol. Exp. Ther. 2019;370:111–126. doi: 10.1124/jpet.119.256818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jandaghi P, et al. Expression of DRD2 Is increased in human pancreatic ductal adenocarcinoma and inhibitors slow tumor growth in mice. Gastroenterology. 2016;151:1218–1231. doi: 10.1053/j.gastro.2016.08.040. [DOI] [PubMed] [Google Scholar]
- 85.Wang, D., Ji, X., Liu, J., Li, Z. & Zhang, X. Dopamine receptor subtypes differentially regulate autophagy. Int. J. Mol. Sci.19, (2018). [DOI] [PMC free article] [PubMed]
- 86.Skah S, Richartz N, Duthil E, et al. cAMP-mediated autophagy inhibits DNA damage-induced death of leukemia cells independent of p53. Oncotarget. 2018;9(54):30434–30449. doi: 10.18632/oncotarget.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou, Y.Y., Li, Y., Jiang, W.Q., Zhou, L.F. MAPK/JNK signalling: A potential autophagy regulation pathway. Biosci Rep. 2015. 35(3). [DOI] [PMC free article] [PubMed]
- 88.Ko JH, Yoon SO, Lee HJ, Oh JY. Rapamycin regulates macrophage activation by inhibiting NLRP3 inflammasome-p38 MAPK-NFκB pathways in autophagy- and p62-dependent manners. Oncotarget. 2017;8(25):40817–40831. doi: 10.18632/oncotarget.17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiang H, Zhang J, Lin C, Zhang L, Liu B, Ouyang L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm. Sin B. 2020;10(4):569–581. doi: 10.1016/j.apsb.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bryant KL, Stalnecker CA, Zeitouni D, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang C, Xu J, Meng Q, et al. TGFB1-induced autophagy affects the pattern of pancreatic cancer progression in distinct ways depending on SMAD4 status. Autophagy. 2020;16(3):486–500. doi: 10.1080/15548627.2019.1628540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matrood, S., de Prisco, N., Wissniowski, T., et al. Modulation of pancreatic neuroendocrine neoplastic cell fate by autophagy mediated death. Neuroendocrinology. 2020. [DOI] [PubMed]
- 93.Wang Z, Lachmann A, Keenan AB, Ma’ayan A. L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics. 2018;34(12):2150–2152. doi: 10.1093/bioinformatics/bty060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tang Q, Ren L, Liu J, Li W, Zheng X, Wang J, Du G. Withaferin A triggers G2/M arrest and intrinsic apoptosis in glioblastoma cells via ATF4-ATF3-CHOP axis. Cell Prolif. 2020;53:e12706. doi: 10.1111/cpr.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee J, et al. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nat. Med. 2016;22:1023–1032. doi: 10.1038/nm.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jadeja RN, Urrunaga NH, Dash S, Khurana S, Saxena NK. Withaferin-A reduces acetaminophen-induced liver injury in mice. Biochem. Pharmacol. 2015;97:122–132. doi: 10.1016/j.bcp.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hahm ER, Singh SV. Withaferin A-induced apoptosis in human breast cancer cells is associated with suppression of inhibitor of apoptosis family protein expression. Cancer Lett. 2013;334:101–108. doi: 10.1016/j.canlet.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roy RV, Suman S, Das TP, Luevano JE, Damodaran C. Withaferin A, a steroidal lactone from Withania somnifera, induces mitotic catastrophe and growth arrest in prostate cancer cells. J. Nat. Prod. 2013;76:1909–1915. doi: 10.1021/np400441f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koduru S, Kumar R, Srinivasan S, Evers MB, Damodaran C. Notch-1 inhibition by Withaferin-A: A therapeutic target against colon carcinogenesis. Mol. Cancer Ther. 2010;9:202–210. doi: 10.1158/1535-7163.MCT-09-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X, Guo S, Liu X, Su L. Chaetocin induces endoplasmic reticulum stress response and leads to death receptor 5-dependent apoptosis in human non-small cell lung cancer cells. Apoptosis. 2015;20:1499–1507. doi: 10.1007/s10495-015-1167-4. [DOI] [PubMed] [Google Scholar]
- 101.Li X, et al. Simultaneous inhibition of the ubiquitin-proteasome system and autophagy enhances apoptosis induced by ER stress aggravators in human pancreatic cancer cells. Autophagy. 2016;12:1521–1537. doi: 10.1080/15548627.2016.1191722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xie Z, Li M, Hong H, Xu Q, He Z, Peng Z. Expression of N6-methyladenosine (m6A) regulators correlates with immune microenvironment characteristics and predicts prognosis in diffuse large cell lymphoma (DLBCL) Bioengineered. 2021;12:6115–6133. doi: 10.1080/21655979.2021.1972644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Janakiram M, Chinai JM, Zhao A, Sparano JA, Zang X. HHLA2 and TMIGD2: New immunotherapeutic targets of the B7 and CD28 families. Oncoimmunology. 2015;4:e1026534. doi: 10.1080/2162402X.2015.1026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jutz S, Hennig A, Paster W, Asrak Ö, Dijanovic D, Kellner F, et al. A cellular platform for the evaluation of immune checkpoint molecules. Oncotarget. 2017;8:64892–64906. doi: 10.18632/oncotarget.17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li B, Chan HL, Chen P. Immune checkpoint inhibitors: Basics and challenges. Curr. Med. Chem. 2019;26:3009–3025. doi: 10.2174/0929867324666170804143706. [DOI] [PubMed] [Google Scholar]
- 106.Young A, Ngiow SF, Gao Y, Patch AM, Barkauskas DS, Messaoudene M, et al. A2AR adenosine signaling suppresses natural killer cell maturation in the tumor microenvironment. Cancer Res. 2018;78:1003–1016. doi: 10.1158/0008-5472.CAN-17-2826. [DOI] [PubMed] [Google Scholar]
- 107.Liu JQ, Hu A, Zhu J, Yu J, Talebian F, Bai XF. CD200-CD200R pathway in the regulation of tumor immune microenvironment and immunotherapy. Adv. Exp. Med. Biol. 2020;1223:155–165. doi: 10.1007/978-3-030-35582-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Flies DB, Higuchi T, Harris JC, Jha V, Gimotty PA, Adams SF. Immune checkpoint blockade reveals the stimulatory capacity of tumor-associated CD103(+) dendritic cells in late-stage ovarian cancer. Oncoimmunology. 2016;5:e1185583. doi: 10.1080/2162402X.2016.1185583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–542e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Capalbo C, Scafetta G, Filetti M, Marchetti P, Bartolazzi A. Predictive biomarkers for checkpoint inhibitor-based immunotherapy: The Galectin-3 signature in NSCLCs. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harjunpää H, Guillerey C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020;200:108–119. doi: 10.1111/cei.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucl. Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen G, et al. Identification of key signaling pathways and genes in eosinophilic asthma and neutrophilic asthma by weighted gene co-expression network analysis. Front. Mol. Biosci. 2022;9:805570. doi: 10.3389/fmolb.2022.805570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Q, Lv L, Ma P, Zhang Y, Deng J, Zhang Y. Identification of an autophagy-related pair signature for predicting prognoses and immune activity in pancreatic adenocarcinoma. Front. Immunol. 2021;12:5073. doi: 10.3389/fimmu.2021.743938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current research are all available in The Cancer Genome Atlas (http://cancergenome.nih.gov) repositories, Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), and International Cancer Genome Consortium Pancreatic Cancer Australian [ICGC-PACA-AU]) (https://dcc.icgc.org/). The data used to support the findings of this study are available from the corresponding author upon request.