Abstract
Cancers are caused by accumulated DNA mutations. This recognition of the central role of mutations in cancer and recent advances in next-generation sequencing, has initiated the massive screening of clinical samples and the identification of 1000s of cancer-associated gene mutations. However, proteomic analysis of the expressed mutation products lags far behind genomic (transcriptomic) analysis. With comprehensive global proteomics analysis, only a small percentage of single nucleotide variants detected by DNA and RNA sequencing have been observed as single amino acid variants due to current technical limitations. Proteomic analysis of mutations is important with the potential to advance cancer biomarker development and the discovery of new therapeutic targets for more effective disease treatment. Targeted proteomics using selected reaction monitoring (also known as multiple reaction monitoring) and parallel reaction monitoring, has emerged as a powerful tool with significant advantages over global proteomics for analysis of protein mutations in terms of detection sensitivity, quantitation accuracy and overall practicality (e.g., reliable identification and the scale of quantification). Herein we review recent advances in the targeted proteomics technology for enhancing detection sensitivity and multiplexing capability and highlight its broad biomedical applications for analysis of protein mutations in human bodily fluids, tissues, and cell lines. Furthermore, we review recent applications of top-down proteomics for analysis of protein mutations. Unlike the commonly used bottom-up proteomics which requires digestion of proteins into peptides, top-down proteomics directly analyzes intact proteins for more precise characterization of mutation isoforms. Finally, general perspectives on the potential of achieving both high sensitivity and high sample throughput for large-scale targeted detection and quantification of important protein mutations are discussed.
Keywords: Targeted proteomics, protein mutation, single amino acid variant (SAAV), single amino acid polymorphism (SAP), alternative splicing variant (ASV), fusion mutation, disease biomarker, top-down proteomics
1. INTRODUCTION
Cancers are caused by accumulated DNA mutations (Bignell et al., 2010; Cairns, 1975; Hanahan and Weinberg, 2000; Hanahan and Weinberg, 2011), which can contribute to cancer by activating protein function (oncogenes) or inactivating protein function (tumor suppressors). The recent advances in next-generation sequencing applied to large numbers of clinical samples has already led to the identification of 1000s of mutations (Shu et al., 2017). They can be divided into neutral (passenger) and disease-associated (driver) mutations. The passenger mutations occur spontaneously in somatic cells and have little or no apparent functional consequence on tumor progression, while the driver mutations play a key role in the malignant transformation and tumor progression (Kumar et al., 2019). However, it is challenging to sort out the cancer-associated mutations with functional significance (Manolio et al., 2009).
Functions encoded in the genome are generally executed by proteins; >2/3 of FDA-approved drugs directly target proteins (Santos et al., 2017). Mutation can affect protein folding and stability, protein function, subcellular localization, and protein-protein interactions (Reva et al., 2011; Sheynkman et al., 2016; Sheynkman et al., 2014; Smith et al., 2013). Protein mutations are not simply associated with tumor cells but actually responsible for altered cellular function and aberrant signaling pathways. Thus, they play a major role in cancer development for tumorigenesis, progression and therapy resistance (Reva et al., 2011; Smith et al., 2013; Tan et al., 2020). Furthermore, mutations at the genome level may not be readily reflected at the protein level. Many studies have revealed that mRNA abundance is poorly correlated with protein abundance across different types of tumor tissues (Lorentzian et al., 2019; Rodland et al., 2018; Sinha et al., 2019; Zhang et al., 2019). This strongly suggests that protein translation and degradation are tightly regulated and play a critical role in determining gene function, and mRNA may lack a direct link to protein activity and gene function. In addition, protein mutations produced only by tumor cells can also provide a unique opportunity for cancer biomarker development (Jensen et al., 1998; Nadal et al., 2014; Nishikawa et al., 2009; Wang et al., 2011). Therefore, direct analysis of protein mutations is critically important for discovery of functionally important mutations to improve our understanding of tumorigenesis, progression and therapy resistance (Reva et al., 2011) and to provide new therapeutic targets (Lima et al., 2019).
With recent technological advances in sample preparation and liquid chromatography–mass spectrometry (LC-MS) instrumentation, MS-based proteomics has emerged as a powerful tool for quantitative genome-scale profiling of proteome and precise comprehensive characterization of proteoforms with posttranslational modifications (PTMs) (e.g., phosphorylation, acetylation, and glycosylation) (Mertins et al., 2018; Rodland et al., 2018; Rodriguez and Pennington, 2018; Sheynkman et al., 2016; Zhang et al., 2019). The deep proteomic analysis is complementary to available genomic (and transcriptomic) analysis by providing additional biological insights that would have been difficult or impossible to obtain solely through genomics approaches (e.g., protein PTM analysis) (Kiseleva et al., 2018; Latysheva and Babu, 2016; Nadal et al., 2014; Smith et al., 2013). With continuous contributions by the NCI CPTAC (Clinical Proteomic Tumor Analysis Consortium) program, integrated analysis of proteomic and genomic data (i.e., proteogenomic analysis) has recently become an emerging trend with many high-impact papers for characterization of various cancer types (Chen et al., 2020; Dou et al., 2020; Gao et al., 2019; Gillette et al., 2020; Jiang et al., 2019; Mertins et al., 2016; Vasaikar et al., 2019; Wang et al., 2021; Zhang et al., 2014; Zhang et al., 2016). Proteogenomic analysis has been demonstrated to provide a more comprehensive understanding of disease biology and new insights into the functional regulation, which underscores the importance of comprehensive proteome and PTM characterization (Dou et al., 2020; Mertins et al., 2016; Vasaikar et al., 2019; Wang et al., 2021; Wisniewski et al., 2009; Zhang et al., 2014; Zhang et al., 2016). Nevertheless, the use of unbiased “discovery” MS-based proteomics platforms for large-scale analysis of mutation proteoforms has not been very effective thus far.
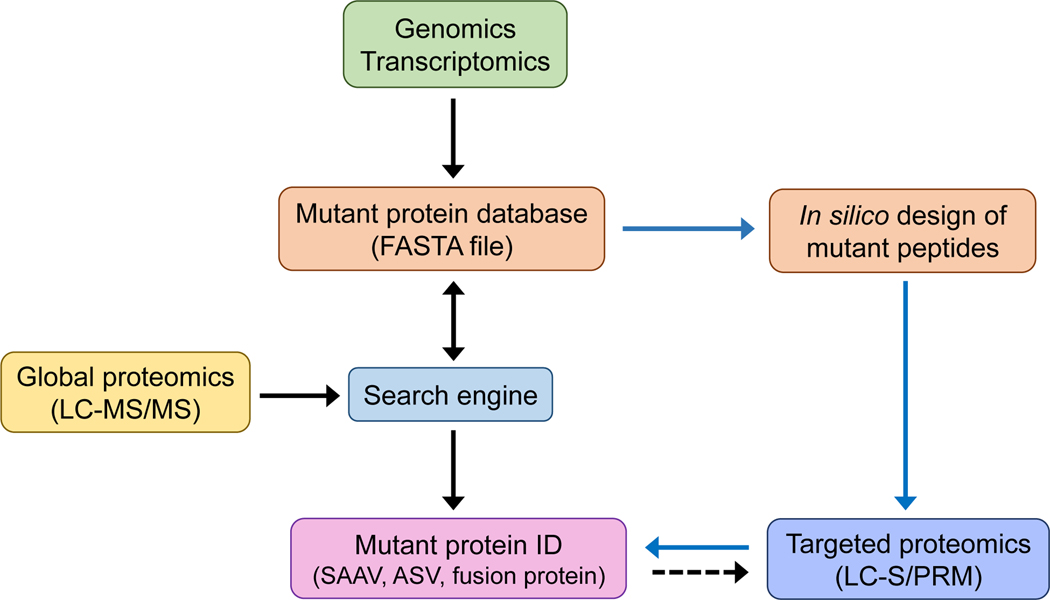
There are three common types of protein mutations encoded by mutated genes (Kiseleva et al., 2018; Latysheva and Babu, 2016; Reva et al., 2011; Vegvari, 2016; Wang et al., 2011): single nucleotide variants (SNVs), alternative splicing variants (ASVs), and gene fusion products. Coding SNVs at the protein level can be classified into three types: synonymous (sense) with normal function, which does not change the corresponding amino acid; nonsense with truncated form, which introduces a premature stop codon; nonsynonymous (missense) with mutation, which changes the corresponding amino acid (i.e., single amino acid variant (SAAV) or single amino acid polymorphism (SAP) (Vegvari, 2016). Mass spectrometry (MS)-based global proteomics has been attempted for deep profiling of protein mutations. However, only ~4–10% of single nucleotide variants detected by both DNA and RNA sequencing were observed as SAAVs (Rodland et al., 2018; Ruggles et al., 2016; Zhang et al., 2019). Very recently, Lubman’s group has applied three different global proteomics strategies for deep profiling of mutant peptides in PANC-1 cell line (Tan et al., 2020). Only ~540 SAAVs have been identified and which SAAVs are functionally important is largely unknown. Nevertheless, this study represents the most comprehensive discovery of SAAVs in single cell line. In addition, the same group has performed global proteomics analysis of subpopulations of breast cancer stem cells to study SAAVs and their relation to breast cancer (Tan et al., 2017). 374 unique SAAVs were identified in total, where 27 SAAVs are cancer-related SAAVs and 135 SAAVs were only found in the cancer stem cell population when compared to mature luminal cells. The low success rate can be attributed to lacking generic databases for sequence search, higher false discovery rates from an enlarged sequence search space, and mutant peptides often present at a low abundance resulting in escaping MS/MS sequencing (Rodland et al., 2018; Sheynkman et al., 2016; Zhang et al., 2019). Targeted proteomics has been broadly used for validation of mutations from global discovery (Lichti et al., 2015; Mostovenko et al., 2018; Tan et al., 2020; Vegvari, 2016) and for accurate quantification of mutations which are cancer-specific markers (Lesur et al., 2015; Wang et al., 2011) because it has higher sensitivity than global proteomics (Shi et al., 2012) and avoids complex database search (Figure 1).
FIGURE 1.
Schematic diagram for large-scale discovery of protein mutations by bottom-up MS-based proteomics (black arrow: global proteomics for analysis of protein mutations; blue arrow: targeted proteomics for analysis of protein mutations; dash black arrow: validation of global discovery with targeted proteomics).
Two types of MS-based global proteomics are used for global discovery: the commonly used data-dependent acquisition (DDA) and the more recently implemented data-independent acquisition (DIA) (Aebersold and Mann, 2016). However, it remains a challenge for reproducible detection and quantification of surrogate peptides from protein mutations because they often have low expression levels and happen at low frequency rates. For example, in DDA the MS randomly samples peptides for fragmentation and is biased to frequently pick the peptides with higher abundance, then leading to a bias against low abundance mutant peptides. Finally, the enzymatic digestion for shotgun proteomics also presents an issue in terms of sequence coverage since mutation containing peptides based on enzymatic cleavage patterns are sometimes simply too short or too long to be identifiable.
In contrast, MS-based targeted proteomics has the potential for precise broad targeted discovery of functionally important protein mutations because it selects the predefined targeted peptides without bias selection, and it has higher detection sensitivity, quantitation accuracy (accurate or absolute quantification), and reproducibility (≤10% CV) (Shi et al., 2016; Shi et al., 2012) with the same level of detection specificity (easy to distinguish SAAVs (Lesur et al., 2015; Lichti et al., 2015; Tan et al., 2020; Wang et al., 2011) as global proteomics (Figure 1). Furthermore, the shortcoming of targeted proteomics, lower multiplexing for target analytes, has recently been addressed effectively with enabling simultaneous quantification of ~1,000 analytes in a single analysis (Stopfer et al., 2021).
In this review we provide an overview of recent advances in bottom-up targeted proteomics and highlight its broad applications for identification and quantification of protein mutations in human bodily fluids, tissues, and cell lines. Furthermore, we briefly review recent applications of top-down targeted proteomics for analysis of protein mutations due to its unique ability for precise characterization of intact protein mutation isoforms. Finally, future perspectives in targeted proteomics for large-scale targeted discovery of functionally important protein mutations as biomarkers and therapeutic targets are discussed.
2. RECENT TECHNOLOGICAL ADVANCES IN TARGETED PROTEOMICS
Targeted proteomics can be divided into two groups: selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM), and parallel reaction monitoring (PRM). SRM is a classic targeted proteomics approach performed on a triple quadrupole mass spectrometer (QqQ MS). It exploits the unique features of QqQ with two levels of mass selection (i.e., Q1 and Q3 isolation of precursor ions and their product ions, respectively) and a relatively long dwell time (in general 10 ms per pair of precursor/product ions) over a narrow m/z window (± 0.02 m/z), which result in significantly improved selectivity and sensitivity, at least one to two orders of magnitude higher than full scan global proteomics analysis (Lange et al., 2008; Shi et al., 2016; Shi et al., 2012). The high specificity, quantitation accuracy (accurate or absolute quantification), and reproducibility make it ideally suitable for studying protein mutations.
PRM is conceptually similar to SRM (Shi et al., 2016). The major difference between PRM and SRM is that in PRM full MS/MS spectra are acquired for each precursor in the high-resolution accurate-mass Orbitrap mass analyzer, while for SRM the predefined product ions are monitored by the low-resolution quadrupole mass analyzer. The principle and performance of SRM and PRM have been well documented with systematic performance comparison (Kockmann et al., 2016; Ronsein et al., 2015; Shi et al., 2016). Compared to antibody-based approaches, SRM/PRM has higher specificity for site-specific detection and quantification of protein mutations. Recent advances in targeted proteomics detection sensitivity and analyte multiplexing capability (e.g., simultaneous quantification of 100s of proteins (Lee et al., 2020; Shi et al., 2012) and phosphorylation sites (Stopfer et al., 2021) enable researchers to perform discovery and validation of 100s-1000s of low-abundance protein mutations.
2.1. Detection sensitivity
Direct LC-S/PRM can provide detection sensitivity for reproducible quantification of target proteins in the concentration range of 4–5 orders of magnitude. However, such sensitivity is insufficient to cover the entire proteome (e.g., protein mutations at the low expression level) with up to 7 and 11 orders of magnitude for mammalian cell lines and human plasma/serum, respectively. Therefore, different front-end sample processing strategies have been explored to greatly enhance detection sensitivity: target analyte enrichment with affinity reagents and sample fractionation to enrich target analytes and reduce complexity (Figures 2 and 3).
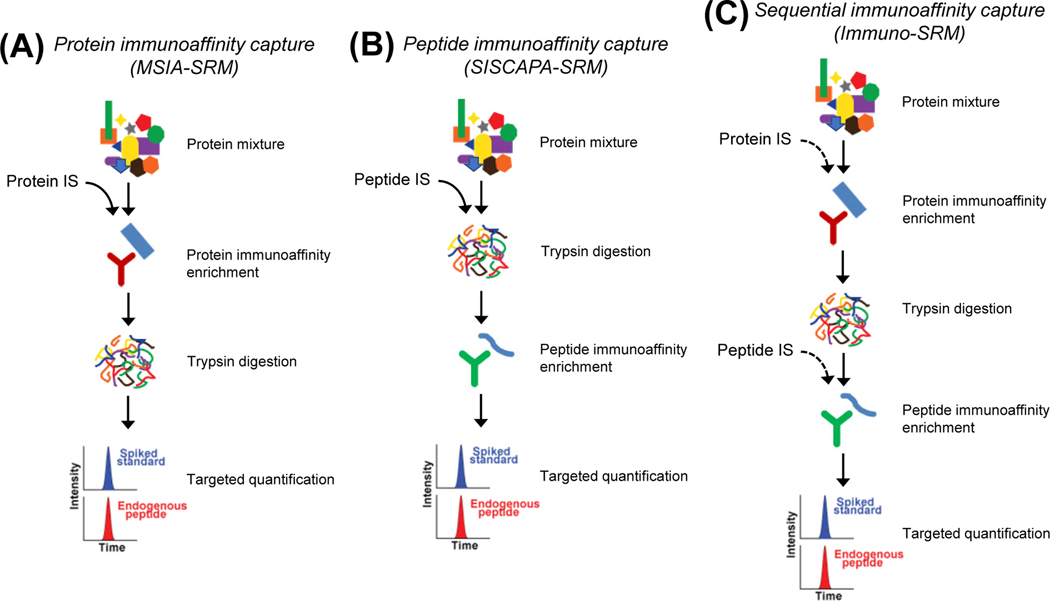
FIGURE 2.
Antibody-based affinity enrichment for improving targeted proteomics detection sensitivity: (A) Protein immunoaffinity capture (MSIA-SRM); (B) Peptide immunoaffinity capture (SISCAPA-SRM); (C) Sequential immunoaffinity capture (Immuno-SRM).
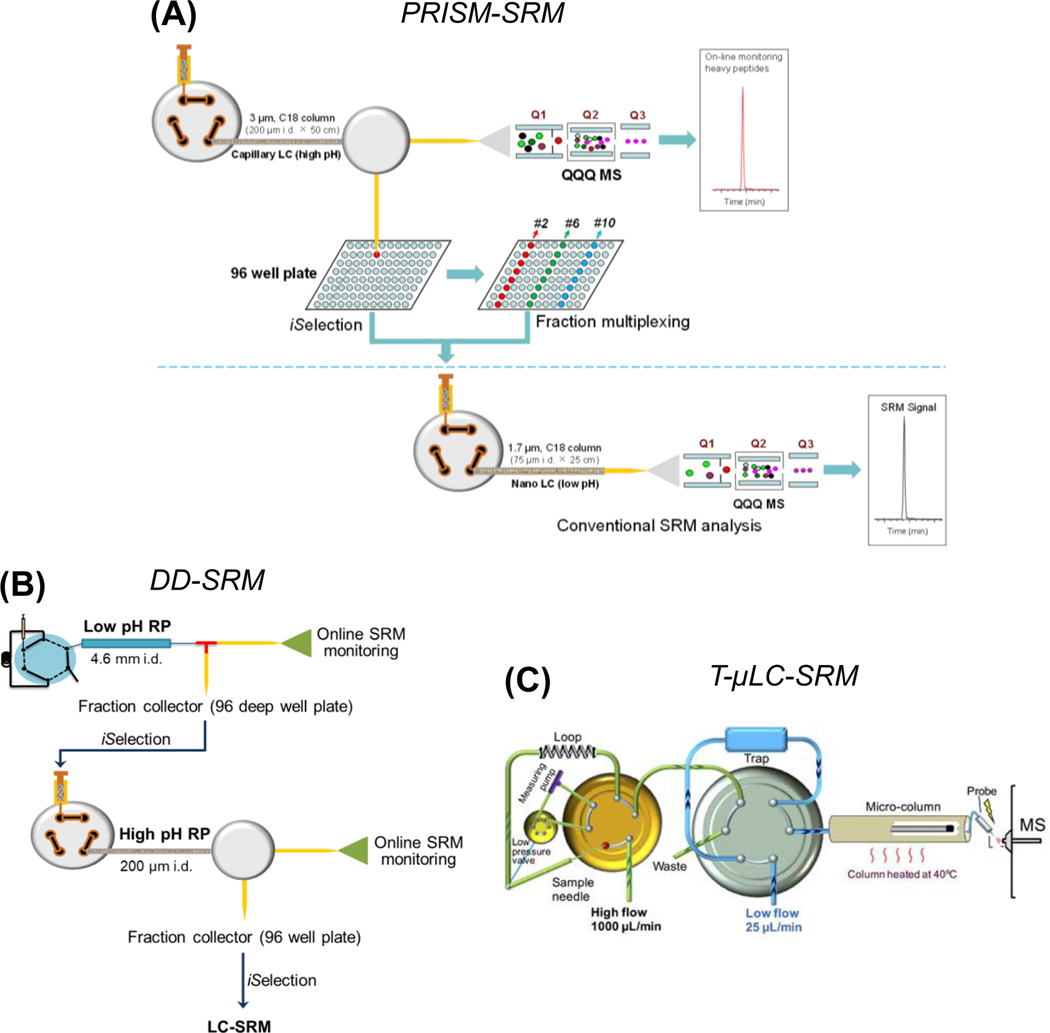
FIGURE 3.
High-resolution LC-based enrichment for improving targeted proteomics detection sensitivity: (A) PRISM-SRM; (B) DD-SRM; (C) T-μLC-SRM. Reprinted with permission from Zhang et al. Anal Chem 2018, 90(3):1870–1880 for Figure 3C. Copyright 2018 American Chemical Society
2.1.1. Antibody-based affinity enrichment
A. Protein immunoaffinity capture
With antibody-based immunoaffinity (IA) enrichment of target proteins or peptides, immuno-SRM can be used for sensitive reproducible quantification of target proteins from complex biological samples. There are three types of immuno-SRM assays: protein IA capture, peptide IA capture, and sequential IA capture (Figure 2). Among them, the protein IA capture is the most commonly used method for identification and quantification of protein mutations because the common shared epitopes recognized by commercially available antibodies that are originally raised against the wild type proteins can be used for enrichment of their corresponding mutant proteins. For example, an anti-SPINK1 antibody was used for effective enrichment of SPINK1 mutant proteins (N34S and P55S), followed by LC-SRM analysis. Immuno-SRM enabled to detect SPINK1 mutations at the limit of detection (LOD) of 0.5 ng/mL in patient sera at different disease conditions (Ravela et al., 2018) (Figure 2A).
B. Peptide immunoaffinity capture
The peptide IA capture is commonly referred as stable isotope standards and capture by antipeptide antibodies (SISCAPA) (Anderson et al., 2004; Anderson et al., 2009; Whiteaker et al., 2010; Whiteaker et al., 2012) (Figure 2B). When compared to the protein IA capture, SISCAPA has an advantage to reduce background interference more significantly with selective enrichment of target peptides of interest rather than all the peptides from target proteins. It can provide an average of ~100-fold enhancement in detection sensitivity of targeted proteomics (Anderson et al., 2004). Potential clinical applications of SISCAPA-SRM have been illustrated by implementation of SISCAPA in a clinical laboratory environment for quantification of low-abundance plasma biomarker proteins (Whiteaker and Paulovich, 2011). Another advantage over the protein IA capture is that harsh sample processing conditions can be used for SISCAPA without the need to maintain native protein epitopes for protein enrichment. SISCAPA-SRM was applied for targeted quantification of membrane proteins in tissue homogenates (Fan and Neubert, 2016) and plasma biomarkers in the presence of autoantibodies (Hoofnagle et al., 2008; Kushnir et al., 2013). In addition, Katafuchi et al., developed a highly sensitive SISCAPA-SRM assay for enabling detection of FGF15 at LOD of 0.1 ng/mL in mouse plasma (Katafuchi et al., 2015).
C. Sequential immunoaffinity capture
To further increase immuno-SRM detection sensitivity, sequential IA enrichment, first targeting protein and then surrogate peptide for enrichment, can be developed for detection and quantification of extremely low-abundance proteins (Figure 2C). One example is to measure β-nerve growth factor (β-NGF) in human serum. β-NGF protein was first isolated from patient serum using an anti-protein antibody. Subsequently, the peptide IA capture was performed to further reduce background interference using an antibody against the surrogate peptide for the β-NGF protein. As a result, reliable SRM quantification of β-NGF at the levels of ~7–450 pg/mL in human serum can be achieved with the inter-assay CV of <15% (Neubert et al., 2013). The sequential enrichment was demonstrated to have additional >10-fold improvement in detection sensitivity when compared to single-step immuno-SRM analysis.
2.1.2. High-resolution LC-based enrichment
A. PRISM-SRM
Antibody-based immuno-SRM is attractive for enhanced detection sensitivity, however it shares similar limitations as other antibody-based approaches (e.g., unavailability of antibodies for new proteins and the long lead time and high cost for the development of high-quality antibodies). To address this issue, we have developed an antibody-independent high-resolution LC-based approach termed PRISM (high-pressure, high-resolution separations coupled with intelligent selection and multiplexing) for highly sensitive SRM quantification of target proteins in complex biological samples (Figure 3A) (Shi et al., 2012). PRISM-SRM capitalizes on high-resolution reversed-phase liquid chromatographic separations for analyte enrichment, intelligent selection of target fractions via on-line SRM monitoring of heavy internal standards, and the partial orthogonality between high pH and low pH RPLC separations. The accurate elution profiles of the internal standards allow precise determination of the locations of target peptides in the 96-well plate, thus allowing the selection of the most informative target fractions for downstream LC-SRM analysis. It is also practically feasible to use other alternative strategies to pinpoint the target fractions, such as off-line determination of the locations of target fractions and relying on the reproducibility of HPLC to locate target fractions from subsequent experiments. Furthermore, a limited number of target fractions eluted at different times during the first-dimension separation can be multiplexed to enhance the overall sample throughput (Figure 3A). Fractions eluted at early, middle, and late retention times have little overlap in their elution profiles of the second-dimension LC separation, and thus can be effectively combined before LC-SRM analysis.
In contrast to standard LC-SRM, PRISM-SRM can provide ~200-fold improvement in detection sensitivity (Shi et al., 2012; Shi and Qian, 2013). PRISM-SRM allows for accurate and reproducible quantification of plasma proteins at the 50–100 pg/mL range in human blood with IgY14 immunoaffinity depletion (Shi et al., 2012) and the sub-ng/mL to low ng/mL range without immunoaffinity depletion (Shi et al., 2013). When applied to human cell lines it can reliably quantify proteins at 10–100 copies per cell (Shi et al., 2016). Such sensitivity is comparable to or even better than most analytically validated ELISAs (Shi et al., 2012; Shi et al., 2014). PRISM-SRM enabled the detection and quantification of all the selected EGFR pathway proteins including extremely low-abundant negative feedback regulators (e.g., DUSP4,6 and SPRY4) and the low expression EGFR in MCF7 that are rarely detected by other sensitive MS platforms or ELISAs (Catenacci et al., 2012). Very recently, we have applied PRISM-SRM to evaluate 52 candidate biomarkers in prostate cancer primary tumors (Gao et al., 2020). A 5-protein classifier demonstrated significant improvement over the standard of care base models using only the clinical and pathological variables in predicting distant metastasis and patient stratification.
Besides sensitive quantification of low-abundance proteins, we have also applied PRISM-SRM for site-specific quantification of stoichiometric protein phosphorylation (Shi et al., 2015), SAAVs (Vegvari, 2016; Wang et al., 2017), and gene fusion products (He et al., 2014). PRISM-SRM was demonstrated to enable direct quantification of ERK phosphorylation isoforms (pT, pY, pTpY) in human mammary epithelial cells (HMECs) and their phosphorylation dynamics in HMEC treated by different doses of EGF using as little as 25 μg tryptic peptides from whole cell lysates (Shi et al., 2015). When compared to immobilized metal affinity chromatography (IMAC), PRISM provided ~10-fold higher signal intensities, presumably due to the better peptide recovery of PRISM. The description of PRISM-SRM quantification of SAAVs and gene fusion products can be found in the following Section 3.
B. DD-SRM
To further improve targeted proteomics detection sensitivity, we have developed another “deep-dive” SRM (DD-SRM) approach that capitalizes on multidimensional high-resolution RPLC separation for target peptide separation and enrichment combined with precise selection of target peptide fractions (Nie et al., 2017) (Figure 3B). The concept of DD-SRM was built on three features of multidimensional RPLC separations: 1) the partial orthogonality between low- and high-pH RPLC separations that provides high peak capacity, and thus the combination of low-high-low pH RPLC separations would significantly reduce matrix background interference, 2) the consecutive implementation of three-dimensional RPLC separations (high-flow, microflow, and nanoflow LC) that allows for exceptionally high sample loading (e.g., ~4 mg for DD-SRM, ~25–50 μg for PRISM-SRM, and ~0.2–0.5 μg for LC-SRM), and 3) precise selection of target peptide fractions of interest by online SRM monitoring due to the compatibility of RPLC separation buffers with electrospray ionization. RPLC separation provides higher resolution and reproducibility and does not need additional sample cleanup to minimize sample handling for improved peptide recovery. All these features contribute to the ability of DD-SRM to detect extremely low abundance proteins in complex biological samples.
DD-SRM was demonstrated to enable detection and quantification of target proteins at ≤10 pg/mL levels in nondepleted human blood or <10 copies per cell in human tissue. Such levels of sensitivity are better than those of many analytically validated immunoassays. In contrast to conventional LC-SRM at detection sensitivity of ~100 ng/mL in human blood, DD-SRM improves SRM detection sensitivity by >4 orders of magnitude due to its greatly increased sample loading and reduced background interference. When compared to PRISM-SRM, DD-SRM provides ~100-fold improvement in detection sensitivity due to significantly enhanced sample loading as well as ultrahigh resolving power from serial orthogonal low-high-low pH RPLC separations. Furthermore, unlike immuno-SRM methods which require approximately 1 mL of human blood to achieve detection sensitivity of ~100 pg/mL, DD-SRM has ~10-fold higher sensitivity with ~5-fold less starting material (i.e., only ~200 μL of human blood).
C. T-μLC-SRM
To improve robustness and sample throughput, Zhang et al., has recently developed a novel trapping-micro-LC-SRM (T-μLC-SRM) method for sensitive quantification of target proteins (Zhang et al., 2018). It employs a dual-flow system for high-capacity loading and sensitive μLC-SRM analysis (Figure 3C). T-μLC-SRM consists of two synchronized LC units where high-flow LC at high pH is used for online trapping of high amounts of biological samples on a large-capacity trapping column, and low-flow μLC-MS at low pH is used for separation and detection. It utilizes the partial orthogonality between high and low pH RPLC for sample loading and analysis and selective-trapping and delivery as well as narrow-window-isolation SRM, which significantly reduce background noise and thus improve SRM detection sensitivity and selectivity. In addition, unique column configuration and peak compression were introduced to enable large-capacity, rapid, and quantitative loading while maintaining excellent chromatographic resolution. When compared to conventional LC-SRM, T-μLC-SRM improves detection sensitivity by up to 25-fold with exceptional robustness. No appreciable peak deterioration or loss of sensitivity was observed after >1,500 injections of tissue or plasma samples. It can be readily implemented for multiplexed quantification of many low-abundance proteins and coupled to other affinity-based enrichment methods to further improve detection sensitivity.
2.1.3. Other enrichment strategies
Besides the above methods, the use of nanoparticles (NPs) has recently emerged as an alternative method for highly sensitive, specific enrichment of proteoforms. NPs have similar size and diffusion kinetics as proteins, allowing effective penetration through complex biological mixtures, and high surface-to-volume ratios to enhance protein interaction. Furthermore, they are versatile scaffolds with the ability to couple diverse affinity ligands for effective protein binding and capture (Kelley et al., 2014; You et al., 2007). Tiambeng et al. have recently designed an organosilane surface functionalization molecule bearing a cysteine (Cys)-thiol reactive handle for synthesizing peptide-functionalized superparamagnetic NPs to capture and enrich low-abundance proteins from human serum (Tiambeng et al., 2020). The newly synthesized NPs enabled highly specific reproducible enrichment of cTnI (<1 ng/mL) by efficient depletion of highly abundant proteins (e.g., serum albumin at >1010 higher concentration than cTnI). Following the enrichment, top-down targeted proteomics was used to reveal different proteoforms of cTnI from human blood (Tiambeng et al., 2020).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is another convenient method for enrichment of target proteins based on their molecular weight. When combined with LC-SRM (termed GeLC-SRM), SDS-PAGE has been used for detection and quantification of protein mutations. Halvey et al., applied GeLC-SRM for reliable quantification of wide-type (WT) and mutant KRAS in relatively low protein inputs (5–50 μg) (Halvey et al., 2012). It allows one to detect KRAS mutant variants (G12D, G13D, G12V, G12S) in a panel of cancer cell lines. They also measured KRAS mutant peptides in fluid from benign pancreatic cysts and pancreatic cancers at concentrations from 0.08–1.1 fmol/μg of protein. This result demonstrated that GeLC-SRM provides a robust, sensitive approach for targeted quantification of mutant proteins in complex biological samples.
2.2. Analyte multiplexing capability
Recent advances in QqQ MS allow for simultaneous quantification of up to 30,000 transitions per analysis in the retention time (RT) scheduling mode. This can be translated into ~5,000 target analytes assuming 6 transitions for each pair of heavy and light analytes. Thus, with the well-established conditions the most advanced QqQ MS (e.g., Thermo TSQ Altis) has the potential for multiplexed quantification of 1,000s of protein mutations in a single LC-SRM analysis. We have recently applied TSQ Altis for reliable measurements of ~150 HNSCC protein markers with 2–3 surrogate peptides per protein (a total of ~300 target peptides) across ~1,000 serum samples (Lee et al., 2020). SureQuant operated in Thermo Orbitrap Exploris, Eclipse, or Lumos MS instrument, is another high-multiplexing method for enabling highly sensitive, multiplexed, reliable, and accurate quantification of ~100s-1000s of target analytes. This method has been built upon traditional internal standard (IS)-PRM (Gallien et al., 2015) with leveraging heavy isotope-labeled IS trigger peptides to efficiently guide MS acquisition in real-time analysis. The use of trigger peptides eliminates the need for RT scheduling to greatly expand multiplexing capability for highly multiplexed analysis of target proteins or peptides. Very recently, SureQuant has been used for reliable quantification of 385 commonly dysregulated pTyr targets with high quantitative accuracy, enhances target detection success rates, and improves the robustness and usability of targeted acquisition (Stopfer et al., 2021). It has also been used for multiplexed quantification of 804 surrogate peptides for 582 plasma proteins with extending the dynamic range by >1 order of magnitude in contrast to conventional SRM.
3. TARGETED PROTEOMICS FOR ANALYSIS OF PROTEIN MUTATIONS
In recent years S/PRM-based targeted proteomics has been demonstrated to have significant advantages over traditional antibody-based assays for reliable quantification of protein mutations in different diseases (Table 1). The protein mutation candidates are generated from either genomic studies or proteogenomic analysis (Figure 1). Furthermore, it has been recognized that novel mutant peptides from global proteomics discovery should be reported with extreme caution because of frequent false positive identification (Li et al., 2011; Nesvizhskii, 2014). Validation of these mutant peptides with targeted proteomics is often required to avoid artificial assignment (Bunger et al., 2007; Chernobrovkin et al., 2015). For example, proteogenomic analysis has recently been used for identification of protein mutations, p53R273C for breast cancer (Dimitrakopoulos et al., 2017), CAPN2D22E for prostate cancer (Kwon et al., 2019), and HSPA12AE365G, AHNAK2P1072S, and GAAV220L for neurodegenerative disease for neurodegenerative disease (Wingo et al., 2017). These protein mutations have been further validated by LC-S/PRM to confirm their existence. In this section, we will review recent studies for detection and quantification of protein mutations using targeted proteomics, including SAAVs, alternative splicing variants, and gene fusion products.
TABLE 1.
Discovery and validation of protein mutations using targeted proteomics
| Bottom-up targeted proteomics | |||||||
|---|---|---|---|---|---|---|---|
| Mutation type | Disease type | Protein | Mutation site | Specimen | Assay type | Clinical application | Ref. |
| SAAVs | Pancreatic cancer | Serotransferrin | I448V | Serum samples | LC-SRM | Diagnosis biomarker | Nie et al., 2017 |
| Pancreatic and colorectal cancer | KRAS | G12V | Cell lines, tissue and cyst fluids | Immuno-SRM | Diagnosis biomarker | Wang et al., 2011 | |
| G12D | |||||||
| Pancreatic and colorectal cancer | KRAS | G12D | Tissues | Immuno-SRM | Diagnosis biomarker | Ruppen-Cañás et al., 2012 | |
| Pancreatic cancer | KRAS | G12D | Cell Lines | PRISM-SRM | Therapeutic target | Tan et al., 2020 | |
| p53 | R273H | ||||||
| SLC37A4 | G88D | ||||||
| Pancreatic disease | KRAS | G12D | Cell lines and tissues | SDS-PAGE-based MRM (GeLC-MRM) | Diagnosis biomarker | Halvey et al., 2012 | |
| G13D | |||||||
| G12V | |||||||
| G12S | |||||||
| Pancreatitis | SPINK1 | N34S | Serum samples | Immuno-pseudo-MRM | Prediction of chronic pancreatitis | Ravela et al., 2018 | |
| P55S | |||||||
| Prostate cancer | PSA | L132I | Plasma and seminal samples | Immuno-SRM (plasma depletion) | Diagnosis biomarker | Vegvari et al., 2013 | |
| Prostate Cancer | CAPN2 | D22E | Cell lines and tissues | PRM | Prognosis and prediction of advanced prostate cancer | Kwon et al., 2019 | |
| Prostate cancer | SPOP | Y87N | Cell lines | PRISM-SRM | Diagnosis biomarker | Wang et al., 2017 | |
| F102C | |||||||
| F133V | |||||||
| Neurodegenerative disease | Heat shock 70 kDa protein 12A (HSPA12A) | E365G | Brain tissues | PRM | Diagnosis biomarker | Wingo et al., 2017 | |
| AHNAK nucleoprotein 2 (AHNAK2) | P1072S | ||||||
| lysosomal alpha-glucosidase (GAA) | V220L | ||||||
| Obesity and diabetes | Complement component C7 | P587T | Plasma samples | LC-SRM | Diagnosis biomarker | Su et al., 2011 | |
| Complement factor H | V62I | ||||||
| Complement component C5 | D966Y | ||||||
| Age-related macular degeneration | Complement factor H (CFH) | Y402H | Plasma samples | LC-SRM | Diagnosis biomarker | Zhang et al., 2017 | |
| I62V | |||||||
| Brain disease | ALDH5A1 | H180Y | Brain tissues | LC-SRM | Diagnosis biomarker | Su et al., 2014 | |
| HADH | P215T | ||||||
| RBP1 | M50V | ||||||
| GRIA1 | N768S | ||||||
| CDC42 | K163R | ||||||
| Glioblastoma | Cytoplasmic C-1-tetrahydrofolate synthase (MTHFD1) | R653Q | Stem cells | LC-PRM | Therapeutic target | Mostovenko et al., 2018 | |
| Neurological and developmental disorder | HSP10 | L73F | Primary cells | LC-SRM | Diagnosis biomarker | Bie et al., 2016 | |
| Breast cancer | p53 | R273C | Cell lines and tissues | LC-SRM and PRM | Diagnosis biomarker | Dimitrakopoulos et al., 2017 | |
| Dileucineopathies | GLUT1 | P485L | Cell lines and primary cells | LC-PRM | Diagnosis biomarker | Meyer et al., 2018 | |
| ITPR1 | P1059L | ||||||
| CACNA1H | P648L | ||||||
| Infertility | TEX101 | G99V | Spermatozoa | Immuno-PRM (anti-TEX101) |
Diagnosis biomarker | Schiza et al., 2019 | |
| Colon Cancer | KRAS | G13D | Cell lines | LC-SRM | Monitor of disease progress | Demory Beckler et al., 2013 | |
| Colorectal carcinoma | BRAF | V600E | Tissues | Immuno-SRM | Therapeutic target | Chen et al., 2016 | |
| Colorectal adenocarcinoma | Cell lines | Immuno-SRM | Diagnosis biomarker | Lin et al., 2019 | |||
| Lung disease | Pulmonary surfactant protein A (SP-A) | Q223K | Bronchoalveolar lavage | SDS-PAGE-based MRM (GeLC-MRM) | Genotyping validation | Foster et al., 2014 | |
| Congenital adrenal hyperplasia | CYP21A2 | L388R | Plasma samples | SDS-PAGE-based MRM (GeLC-MRM) | Diagnosis biomarker | Brønstad et al., 2014 | |
| E140K | |||||||
| P45L | |||||||
| V211M | |||||||
| V281L | |||||||
| Alexander disease | GFAP | R79C | Tissues | SDS-PAGE-based MRM (GeLC-MRM) | Diagnosis biomarker | Heaven rt al., 2019 | |
| R239H | |||||||
| R416W | |||||||
| ASVs | PKM1/2 | iPSC lines and tissues | LC-PRM | Splicing isoform function | Lau et al., 2019 | ||
| MYOM1 | |||||||
| NDUA5 | |||||||
| TENV | |||||||
| SVIL | |||||||
| RTR2 | |||||||
| MYBPC3 | |||||||
| Neuronal disease | NRX1 | Alternative splicing 3, 4 and 6 | Brain tissues | LC-SRM | Cell recognition processes | Schreiner et al., 2014 | |
| NRX2 | Alternative splicing 3 and 6 | ||||||
| NRX3 | Alternative splicing 3, 4 and 6 | ||||||
| Alzheimer’s disease | Tau | 0N3R | Cerebrospinal fluids | Combination of IP and GeLC-MRM | Diagnosis biomarkers and therapeutic targets | Xu et al., 2021 | |
| 1N3R | |||||||
| 2N3R | |||||||
| 0N4R | |||||||
| 1N4R | |||||||
| 2N4R | |||||||
| Premature aging disorders | Lamin A/C | 78–89 | Cell lines | LC-SRM | Diagnosis biomarker | Al-Qahtani et al., 2019 | |
| 547–572 | |||||||
| 529–553 | |||||||
| 529–615 | |||||||
| Heart disease | TPM1 | 189–212 | Tissues | LC-PRM | Discovering functionally relevant isoforms in the heart. | Han et al., 2021 | |
| Histone-deregulated disease | H2A | 22 H2A variants | Mouse testes | LC-MRM/PRM | Histone variant and isoform identification | El Kennani et al., 2018 | |
| H2B | 3 H2B variants | ||||||
| Non-small cell lung cancer (NSCLC) | Osteopontin (OPN) | 16–81 | Plasma samples | Immuno-SRM | Diagnosis biomarker | Wu et al., 2012 | |
| 16–58_72–81 | |||||||
| 16–30_58–81 | |||||||
| Gene fusion product | Prostate cancer | TMPRSS2-ERG | Cell line and tissues | PRISM-SRM | Diagnosis biomarker | He et al., 2014 | |
| Cell lines | Immuno-SRM | Fu et al., 2021 | |||||
| Top-down targeted proteomics | |||||||
| Mutation type | Disease type | Protein | Mutation site | Specimen | Assay type | Clinical application | Ref. |
| SAAVs | Hemoglobinopathies | Hemoglobin A | E6V | whole blood samples | ETD and pseudo-SRM |
Diagnosis biomarker | Coelho Graça et al., 2012 |
| E6K | |||||||
| Muscle-related diseases | Tropomyosin (Tpm) | R38Q | Skeletal muscles | Offline multi-step purification | Diagnosis biomarker | Jin et al., 2016 | |
| P64L | |||||||
| Colorectal cancer | KRAS4b | WT | Cell line and tissues | Immunoaffinity enrichment | Therapeutic targets | Ntai et al., 2018 | |
| G13D | |||||||
3.1. SAAVs
SAAV, also known as SAP, is the most common protein mutation and often occurs on tumor-related proteins. SAAVs may lead to significant alteration in protein function and are directly associated with human diseases, and thus reliable quantification of SAAV mutations is crucial to understand molecular mechanism in disease (or cancer) initiation and progression with the potential of developing highly specific protein mutation markers for diagnosis and prognosis and revealing therapeutic targets for better treatment.
3.1.1. Pancreatic cancer
Pancreatic cancer is the 4th most common cancer for both men and women and 11th most common cause of death globally (Ilic and Ilic, 2016). CA19–9 glycoprotein is used for clinical diagnosis, but it lacks sufficient sensitivity and specificity. To identify alternative biomarkers for this lethal cancer, Lubman’s group has applied global proteomics for deep profiling of SAAV peptides in PANC-1 cell line and human serum, and then validated the SAAV candidate biomarkers of interest by targeted proteomics (Nie et al., 2014; Tan et al., 2019; Tan et al., 2020). One mutant peptide from serotransferrin (I448V) was measured in a cohort of serum samples from pancreatic cancer patients. When combined with α−1-antichymotrypsin (AACT) and thrombospondin-1 (THBS1), the mutant peptide has shown an excellent diagnostic performance for significant differentiation of pancreatic cancer patient from healthy controls and pancreatitis (Nie et al., 2014).
Oncogenic KRAS gene mutations are known to be one of characteristics of pancreatic cancer (Waddell et al., 2015). For sensitive detection of KRAS protein mutations, a pan-KRAS antibody which recognizes the common epitope on both the WT and mutant KRAS (G12V and G12D) was used for efficient enrichment of KRAS protein mutations. The enriched samples were analyzed by LC-SRM with detection of both KRAS mutations (Wang et al., 2011). With the use of a more sensitive targeted proteomics platform, immuno-SRM enabled for quantification of both WT KRAS and KRASG12D as low as 12 amol (0.25 pg) in tumor tissue samples from pancreatic and colorectal cancer patients (Ruppen-Canas et al., 2012). Besides antibody-based enrichment, Halvey et al. employed SDS-PAGE for selectively enriching KRAS proteins. Four KRAS variants (G12D, G13D, G12V and G12S) were identified by GeLC-SRM from a panel of cancer cell lines and fluid from benign pancreatic cysts and pancreatic cancer patients (Halvey et al., 2012). GeLC-SRM was used for reliable quantification of their corresponding mutant peptides at concentrations of ~0.08–1.1 fmol/μg, which makes it possible to use KRAS protein mutations as specific diagnosis biomarkers for pancreatic cancer.
In addition, SPINK1 mutation (N34S) was reported to be associated with earlier onset and pancreatic insufficiencies (Witt et al., 2000). Both wild type and mutant (N34S and P55S) SPINK1 can be effectively enriched using an anti-SPINK1 antibody followed by LC-SRM analysis. Immuno-SRM assays have been demonstrated to enable detection of SPINK1 mutations at the LOD of 0.5 ng/mL in patient sera at different disease conditions (Ravela et al., 2018).
3.1.2. Prostate cancer
Prostate cancer (PCa) is the second most common cancer among men worldwide (Jemal et al., 2011). Prostate-specific antigen (PSA) is routinely used for clinical diagnosis and frequently chosen as a model protein for the S/PRM method development (Fortin et al., 2009; Li et al., 2011; Liu et al., 2012). With immuno-SRM assays, the PSAL132I mutation was identified in the PCa patient plasma, and PSAL132I was estimated to be present in 10% of the human population (Vegvari et al., 2013). However, its value for PCa screening needs further evaluation in large cohorts of clinical samples. We have recently used targeted proteomics for analysis of SPOP mutations (up to 15% of PCa patients) which represent one of the most specific biomarkers that define distinct molecular subtypes of PCa. Based on genomic data we selected 11 surrogate peptides for SPOP SAAVs (Y87C/N, F102C, K129C, D130H, W131C/G, F133L/V/S and K134N) (Wang et al., 2017). We applied a tiered approach (first LC-SRM then PRISM-SRM) for quantifying them in PCa cell lines. PRISM-SRM enable confident detection of three most frequent SPOP mutations (Y87N, F102C, and F133V) in the mutation positive cell lines but not in the negative cell lines (Wang et al., 2017). Lower expression of the F133V mutation and WT SPOP was observed when compared to that of other two F102C and Y87N mutations. This result suggests that SRM-based targeted proteomics is a promising tool for multiplexed targeted discovery of protein mutations predicted from genomic data.
3.1.3. Aging- or neurodegeneration-related diseases
Point mutations of single amino acids were proved to cause functional change of proteins in age-related diseases (e.g., β-amyloid precursor protein in Alzheimer’s disease (Di Fede et al., 2009)). SRM-based targeted proteomics was used to detect and quantify SAAVs (e.g., complement component C7P587T, ALDH5A1H180Y, HADHP215T, RBP1M50V, GRIA1N768S, CDC42K163Rand MTHFD1R653Q) which were reported to be significantly associated with obesity, diabetes (Su et al., 2011), age-related macular degeneration (Zhang et al., 2017), brain disease (Su et al., 2014), and glioblastoma (Mostovenko et al., 2018). Targeted proteomics analysis of plasma SAAVs provides a new way for assessing physiological or pathological traits. For functional study, Bie et al., applied SRM assays to measure the steady state levels of the WT and L73F mutant HSP10 in fibroblasts of a patient with a complex disease history (e.g., infantile spasms, hypotonia, developmental delay, a slightly enlarged liver, macrocephaly, and mild non-specific dysmorphic features) (Bie et al., 2016). Based on SRM quantification, they discovered that mutant HSP10L73F not only decreased the formation of HSP60/HSP10 chaperonin complex but also reduced the expression of SOD2. This result suggests that mutant HSP10L73F acts as a strong contributing factor for the disorder in the affected patient.
3.1.4. Other diseases
Targeted proteomics has also been used for quantification of mutant proteins in other diseases, such as GLUT1P485L, ITPR1P1059L, and CACNA1HP648L for in dileucineopathies (Meyer et al., 2018), TEX101G99V in infertility (Schiza et al., 2019), and KRASG13D and BRAFV600E in colon cancer (Chen et al., 2016; Demory Beckler et al., 2013; Lin et al., 2019). With gel electrophoresis fractionation, SRM-based targeted proteomics was applied for detection and quantification of SP-A variants (Gln223 and Lys223) from bronchoalveolar lavage (BAL)(Foster et al., 2014), GFAP variants (R79C, R239H and R416W) for Alexander disease(Heaven et al., 2019), and CYP21A2 variants (L388R, E140K, P45L, V211M and V281L) from congenital adrenal hyperplasia (CAH) patient plasma(Bronstad et al., 2014). The mutations of L388R and E140K for CYP21A2 were demonstrated to affect the enzyme activity for steroid synthesis in the adrenal cortex(Bronstad et al., 2014).
3.2. ASVs
Large-scale RNA-seq sequencing has shown that there are over 100,000 alternative splicing transcripts in the human genome (Pan et al., 2008; Wang et al., 2008). However, whether their corresponding protein isoforms are produced remains largely unknown due to the lack of suitable proteomics tools. Recent technological advances in bottom-up MS proteomics have greatly increased the ASV identification, but current bottom-up proteomics still lacks the depth to cover the majority of the ASV proteome and the sufficient resolution to discern 10s-100s of closely related protein isoforms that differ by only a single peptide (Nesvizhskii and Aebersold, 2005). One promising tool to tackle these challenges is targeted proteomics, as demonstrated by SRM-based measurement of alternative splicing forms of neuronal neurexin receptors (Schreiner et al., 2015).
3.2.1. Neuronal disease
Neurexins are a class of synaptic adhesion molecules that play an important role in the synapse formation and function (Dean et al., 2003). In mammals, three neurexin genes (Nrxn1–3) are transcribed into long alpha-neurexin and short beta-neurexin transcripts with >1,000 unique isoforms in the adult brain due to the presence of six alternatively spliced segments (AS1–6) and extensive alternative splicing (Baudouin and Scheiffele, 2010; Schreiner et al., 2014). The diversity of neurexin molecules may serve as synaptic recognition events which control neuronal wiring and function (Reissner et al., 2013). To detect endogenous neurexin splicing protein variants, Schreiner et al. developed SRM assays for quantification of surrogate peptides which are unique to segments AS3, 4, and 6 (Schreiner et al., 2015). With these SRM assays, 17 unique protein isoforms can be reliably detected and quantified. For the panel of SRM assays, pan-neurexin peptides and peptides unique to either the alpha or beta form of neurexin were also included to measure the total neurexin and neurexin-alpha/beta, respectively. Such SRM assays not only allow them to answer the fundamental question about which splicing variants are translated to proteins, but also provide a proteomics tool for highly specific quantification of protein splicing variants.
Another example is targeted quantification of ASVs from Alzheimer’s disease (AD), one of the most common causes of dementia (Kukull and Bowen, 2002). Tau protein was demonstrated to promote microtubule assembly, stabilize microtubules, and maintain normal morphology of neurons (Weingarten et al., 1975). It has at least six isoforms and each isoform has differential physiological roles (Andreadis, 2005). Quantification of individual Tau isoform may be helpful to better understand the role of Tau in AD and to improve AD diagnostic and treatment accuracy. Very recently, Xu et al. developed multidimensional separations (i.e., the combined immunoprecipitation and SDS-PAGE) coupled with LC-SRM for detection and quantification of all the six Tau isoforms with ~250-fold improvement in detection sensitivity (Xu et al., 2021). The multidimensional targeted proteomics platform was then applied for simultaneous quantification of these isoforms in CSF of AD patients.
3.2.2. Other diseases
Lamin A/C mutations have been linked to numerous heritable disorders including premature aging syndrome (Worman, 2018). SRM assays have recently been developed for highly specific quantification of four splicing variants (lamin A, lamin C, laminAΔ10, and laminAΔ50) for the Lamin A/C proteins in MCF7 and U937 cancer cell lines(Al-Qahtani et al., 2019). Except for laminAΔ50, three out of four Lamin A/C variants have been detected with direct LC-SRM. The performance of splicing variant SRM assays (e.g., sensitivity, reproducibility, and reliability) has further been evaluated and compared with that of qRT-PCR for quantification of splicing variants at the mRNA level.
To improve the number and the confidence of ASVs identification, Lau et al. developed an isoform-inferred approach in which RNA-seq data are used to guide the discovery of splicing transcripts harboring junction pairs (Lau et al., 2019). They identified >1,000 isoform sequences which were not documented previously. They further validated selected junction sequences by performing PRM-based targeted quantification. Prior to LC-PRM analysis, normal human heart tissue lysates were digested and separated into 10 fractions with high-pH RPLC to improve PRM sensitivity. Targeted quantification not only validated of known junction peptides, but also identified six protein isoforms. For easy method adoption, the same group has recently published the detailed workflow for the computation-assisted targeted proteomics method (Han et al., 2021). Targeted proteomics analysis of histone variants is another example because of the existence of many variants for histone proteins (El Kennani et al., 2017). Kennani et al. demonstrated the power of the targeted proteomics for precise quantification of histone variants and isoforms with high sequence similarity (El Kennani et al., 2018). These assays can be easily applied to study human histone variants whose abundance may be dysregulated in various diseases.
In addition, Wu et al. developed immuno-SRM assays for quantification of three osteopontin splicing variants (OPNa, b, and c) in human plasma (Wu et al., 2012). All OPN isoforms were immunocaptured to reduce sample complexity using the pan-OPN antibody and SRM assays were used for targeted quantification of isoform-specific signature peptides. For the non-small cell lung cancer (NSCLC) patients, the concentrations of OPNa were found to be substantially elevated when compared to normal controls.
3.3. Protein fusion products
Gene fusions are generated through structural rearrangements (e.g., inversions, interstitial deletion, or translocations of two independent genes). They can lead to the dysregulation of gene expression, the formation of novel fusion proteins, and the truncation of protein products. Recent studies have revealed that fusion genes can act as driver mutations across diverse cancer types (Watson et al., 2013). Fusion between the androgen-responsive gene TMPRSS2 and the transcription factor ERG (TMPRSS2-ERG) is a common driver mutation in nearly 50% of PCa (Nam et al., 2007) and has been used as drug target (Shao et al., 2020) as well as diagnostic (Yang et al., 2016) and prognostic biomarkers (Hagglof et al., 2014). Due to the lack of high-quality antibodies suitable for quantitative analysis, studies of TMPRSS2-ERG gene fusions have seldom been performed at the protein level.
With the development of the antibody-independent, highly sensitive and specific PRISM-SRM platform, we have applied PRISM-SRM assays for reliable quantification of the TMPRSS2-ERG gene fusion protein products in PCa cell lines and prostate tumor tissues (He et al., 2014). At least two distinct ERG protein isoforms were observed to be simultaneously expressed in TMPRSS2-ERG gene fusion positive samples as evidenced by the concomitant detection of two mutually exclusive peptides in two patient tumors and the VCaP cell line. Three peptides, shared across almost all fusion protein products, were determined to be the most abundant peptides, providing “signature” peptide markers for detection of ERG over-expression resulting from TMPRSS2-ERG gene fusion. With its demonstrated analytical performance, PRISM-SRM is a valuable tool for studying gene fusion protein products without the need of specific affinity reagents.
Very recently, Fu et al. developed immuno-SRM assays for quantification of a low-abundance T1E4 TMPRSS2-ERG fusion protein and its isoforms in VCaP cells (Fu et al., 2021). With accurate quantification of the total ERG and its four unique isoforms, immuno-SRM revealed that the T1E4-ERG isoform accounted for ~50% of the total ERG protein in VCaP cells and formalin-fixed paraffin-embedded PCa tissues. With immuno-SRM assay, the amount of total ERG was estimated as 2.2 fg per VCaP cell, consistent with the level of 1.8 fg per cell from our PRISM-SRM assay (He et al., 2014). This suggests that both targeted proteomics methods are reliable for quantification of the TMPRSS2-ERG gene fusion products. Immuno-SRM assays present an alternative tool for quantifying ERG and its isoforms in clinical samples, thus paving the way for development of more accurate PCa diagnostics.
4. TOP-DOWN PROTEOMICS FOR ANALYSIS OF PROTEIN MUTATIONS
Unlike bottom-up proteomics that requires the digestion of proteins into peptides, top-down proteomics directly analyzes intact proteins, allowing precise and comprehensive characterization of various proteoforms such as protein mutations from genetic polymorphisms and RNA splice variants as well as PTMs (Aebersold et al., 2018; Schaffer et al., 2019; Smith and Kelleher, 2018). Since it directly analyzes intact proteins, top-down proteomics can potentially provide the richest data for both reliable protein identification and precise localization of mutation and PTM sites for a complete view of proteoforms (Smith and Kelleher, 2018). Similar to bottom-up proteomics, top-down proteomics can also be performed in either a shotgun fashion (i.e., global profiling) or a targeted manner (Chen et al., 2018). However, there are many technical challenges for current top-down proteomics technologies (e.g., difficulties in characterization of high molecular weight and low-abundance proteins, and bioinformatics tools for analysis of complex proteoform MS data) (Schaffer et al., 2019), which severely hinder its broad applications with technical limitations in depth and throughput.
4.1. Global top-down proteomics
In top-down proteomics, intact proteins extracted from cells are typically fractionated by LC, electrophoresis, or serial size exclusion chromatography (sSEC) followed by RPLC-MS/MS analysis. Current state-of-the-art global top-down proteomics workflows enable to identify ~3,000–6,000 proteoforms corresponding to ~800–1,200 proteins. Using a multiple dimensional separation system (electrophoresis, solution isoelectric focusing (sIEF), and gel-eluted liquid fraction entrapment electrophoresis (GELFrEE)), ~1,000 proteins were identified from HeLa cells with >3,000 proteoforms (Tran et al., 2011) and ~1,200 proteins from H1299 cells with >5,000 proteoforms (Catherman et al., 2013). Cai et al. developed a sSEC strategy for high-resolution size-based fractionation of 10–223 kDa intact proteins from human heart tissues. The cSEC fractions were further separated by RPLC followed by MS analysis. In contrast to one dimensional RPLC separation, >4,000 unique proteoforms were identified with a 15-fold increase in the detection of proteins above 60kDa (Cai et al., 2017). Using an orthogonal multidimensional separation platform coupling SEC and RPLC based protein prefractionation to capillary zone electrophoresis (CZE)-MS/MS with ~4,000 of peak capacity, ~5,700 proteoforms were identified from the E. coli proteome (McCool et al., 2018), representing the largest bacterial top-down proteomics data set. A 10-fold improvement in the number of identified proteoforms was achieved when compared with previous CZE-MS/MS studies. Toby et al. applied the GELFrEE fractionation and RPLC-MS/MS for global analysis of human blood and ~2,900 proteoforms were identified from human peripheral blood mononuclear cells (Toby et al., 2019). Using quantitative top-down proteomics analysis of specific regions of the female mouse brain, several proteoform changes in abundance were observed after mice were trained for cocaine-conditioned place preference. These observations provide insight into estrogen signaling in the brain with potential new approaches to treat women with cocaine use disorder (Park et al., 2019). However, most proteoforms were caused by the events of various PTMs (e.g., methylation, acetylation, and phosphorylation) and truncations at the protein termini. Few protein mutations were identified by global profiling presumably due to their low frequency rate and/or the low- or moderate-abundance of their corresponding proteins.
4.2. Targeted top-down proteomics
To improve detection sensitivity for analysis of protein mutations, targeted top-down proteomics were used for detection and quantification of proteoforms from specific proteins of interest. With the combination of electron transfer dissociation and pseudo-SRM, targeted top-down proteomics was used for analysis of hemoglobin A and hemoglobin variants where glutamic acid at position 6 of the β globin chain is replaced by either valine (hemoglobin S) or lysine (hemoglobin C) in clinical samples (Coelho Graca et al., 2012). Subtle difference in the β globin chain can be unambiguously identified from the whole blood by direct injection. With the use of a top-down LC/MS strategy for antibody-independent purification and offline top-down MS analysis, tropomyosin (Tpm) isoforms associated with muscle diseases were reliably identified and quantified across different skeletal muscles from multiple species, including swine, rat, and human (Jin et al., 2016). Among these isoforms Tpm1.1 was confirmed to have two amino acid polymorphisms R38Q and P64L and N-terminal acetylation. This study demonstrates the utility of targeted top-down MS for precise characterization of proteoforms from single protein.
Another example for targeted top-down MS is to study how genetically encoded mutations affect PTMs on the same protein molecule by detection and quantification of mutation-specific consequences (Ntai et al., 2018). Immunoaffinity enrichment combined with top-down MS was used to discover and quantify KRAS proteoforms caused by the KRASG13D mutation. Analysis of isogenic CRC cell lines has shown a direct link between the knockout of the mutant G13D allele and the complete nitrosylation of cysteine 118 of the remaining WT KRAS4b. Application of the top-down MS workflow to other cancer cell lines with three mutations at Gly12 along with primary colorectal tumor samples enabled to quantify mutant versus WT KRAS4b expression ratios to reveal the major difference in the levels of C-terminal carboxymethylation, a modification critical for membrane association. This study highlights the importance of targeted top-down proteomics for discovery of dynamic PTMs underlying key regulatory mechanisms with the potential link to disease stage and chance of survival.
5. FUTURE PERSPECTIVES
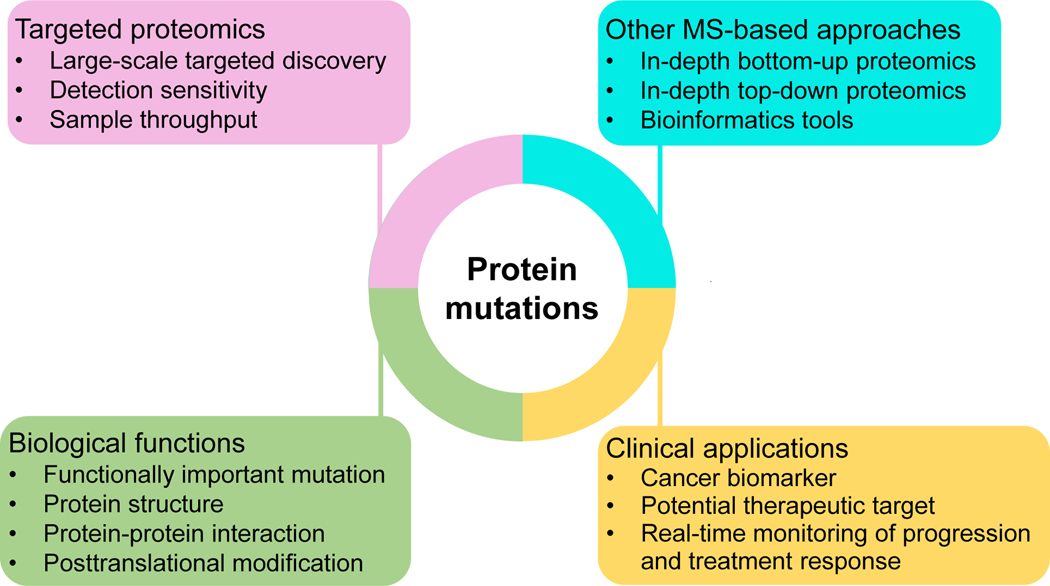
The central dogma of biology is the flow of genetic information from DNA to RNA to protein. Recent technological advances allow for cost-effective whole genome (transcriptome) analysis of genetic mutations, resulting in the identification of 1000s of cancer-associated mutations (Reva et al., 2011). However, proteomics technologies for comprehensive analysis of protein mutations are lagging far behind genomics technologies, which severely prevents a more comprehensive view of cancer-associated mutations through integrated protegenomic analysis and the identification of functionally important mutations to connect disease genotype to phenotype. More importantly, protein mutations identified in tumor cells not only confirm genetic mutations but actually are responsible for tumorigenesis and altered cellular functions (Wang et al., 2011). Therefore, large-scale analysis of protein mutations promises a unique opportunity for cancer biomarker development and discovery of new therapeutic targets for more effective disease treatment (Figure 4).
FIGURE 4.
Future developments for large-scale discovery of protein mutations and their applications to biomedical research.
5.1. Detection sensitivity and sample throughput
Future developments will focus on significant improvements in detection sensitivity and sample throughput for rapid deep profiling of protein mutations across 100s of clinical samples. Enhancing detection sensitivity could be achieved by effective integration of ultralow-flow LC (Shen et al., 2003; Shen et al., 2004; Shen et al., 2002) or capillary electrophoresis (CE) (Sun et al., 2013) and a high-efficiency ion source/ion transmission interface (Cox et al., 2014; Marginean et al., 2010; Page et al., 2008; Tang et al., 2011) with the most advanced MS platform for sensitive detection of low-abundance protein mutations. Sample throughput could be increased by using ultrafast high-resolution ion mobility-based gas-phase separation (e.g., timsTOF (Meier et al., 2018), FAIMS (Hebert et al., 2018) and SLIM IMS (Ibrahim et al., 2017)) to replace current slow liquid-phase (LC or CE) separations, and effective integration of liquid- and gas-phase separations for greatly reducing separation time (i.e., improved sample throughput) but without trading off separation resolving power. For example, the combined low-resolution FAIMS and high-resolution LC separations enabled analysis of up to 2,000 peptides per minute and more than 5,000 protein groups in 20 mins (Bekker-Jensen et al., 2020). All these advancements could significantly improve the performance of both bottom-up and top-down MS platforms for in-depth profiling of protein mutations in a reasonable time frame. They will also help close the gap between proteomics and transcriptomics or genomics technologies for more effective proteogenomic analysis of mutations. For confident detection of protein mutations by global proteomics profiling, an accurate and more complete mutation database is a prerequisite as the identification relies on the quality of the database (Schaffer et al., 2019; Sheynkman et al., 2016). In addition, new bioinformatics tools are also needed to better control the FDR (Schaffer et al., 2019; Sheynkman et al., 2016).
5.2. Large-scale targeted discovery of protein mutations
MS-based bottom-up global proteomics has been used for deep profiling of protein mutations. However, only low percentage of SNVs detected by either DNA or RNA sequencing had been validated as SAAVs at the protein level (Rodland et al., 2018; Ruggles et al., 2016; Zhang et al., 2019). Unlike global proteomics that need database searching for mutant protein identification and may fail to detect important mutations at low expression levels, targeted proteomics can be used to exclusively analyze 1000s of presumably important mutations obtained from curated literature, mutation database, and prediction tools. For example, targeted proteomics has been used to validate mutations from global discovery (Lichti et al., 2015; Mostovenko et al., 2018; Tan et al., 2020; Vegvari, 2016) and to accurately quantify cancer-specific mutations as markers (Lesur et al., 2015; Wang et al., 2011). With recently demonstrated exceptionally high multiplexing capability (e.g., ~1000s of target analytes in single LC-PRM analysis operated in the SureQuant mode (Stopfer et al., 2021)) and constant improvement in MS detection sensitivity, targeted proteomics is well suited for precise and comprehensive targeted discovery (identification) and quantification of functionally important protein mutations.
To further improve the success rate of targeted discovery, the use of multiple enzymes in a complementary fashion needs to be considered to improve sequence coverage (Sheynkman et al., 2016; Trevisiol et al., 2016). This is critically important for large-scale profiling of protein mutations because of an uneven distribution of their cleavage sites. If only trypsin (lysine and arginine as the cleavage sites) is used, some tryptic peptides may be too long or too short to be effectively detected and sequenced by MS. In the case of no suitable tryptic peptide available to cover the mutation site, a different proteolytic enzyme may be used to generate peptides covering the targeted mutation site for S/PRM. This was evidenced in our study for quantification of SPOP mutations using the enzyme of Arg-C rather than trypsin for digestion because there are many lysine residues in the SPOP mutation region (Wang et al., 2017).
5.3. Discovery of disease biomarkers and therapeutic targets
Protein mutation markers are supposed to provide complementary diagnostic and prognostic values to genetic mutation markers because the correlation between protein and mRNA abundance is typically low to moderate (Rodland et al., 2018; Sinha et al., 2019; Zhang et al., 2019). Proteogenomic analysis of the same mutation markers across the same cohort of clinical samples is expected to fully reveal disease biology for accurate diagnosis, prognosis, and better monitoring of the response to therapy, contributing to better opportunities toward precision medicine. It has also been recognized that comprehensive proteogenomic analysis is powerful for differentiating functionally important mutations (e.g., driver mutations) from passenger mutations and for revealing the connection between genotype and phenotype (Dou et al., 2020; Mertins et al., 2016; Rodland et al., 2018; Ruggles et al., 2016; Sheynkman et al., 2016; Vasaikar et al., 2019; Wang et al., 2021; Zhang et al., 2016). With constant improvement in MS-based proteomics technologies, more functionally important protein mutations are expected to be detected and quantified in clinical samples. Since these protein mutations are expressed only in tumor cells and responsible for tumorigenesis and progression, the expression level of these mutations can more accurately reflect cellular phenotype and regulatory processes than genetic mutations (Vegvari, 2016; Wang et al., 2011). Thus, they have the potential to be novel disease markers with high specificity for early diagnosis and prognosis and monitoring disease progression.
Cancers are caused by the accumulation of mutations (Hanahan and Weinberg, 2000). An individual mutation may slightly increase the risk of cancer development, but the combination of multiple mutations can result in a significant level of risk. Therefore, multiplexed targeted proteomics technologies are important for quantification of 100s of functionally important mutations across 100–1000s of clinical samples to prioritize the most promising disease biomarkers. Furthermore, the stoichiometries of protein mutations (i.e., the abundance ratio of mutant over the total mutant and WT proteins) can be accurately determined by multiplexed targeted proteomics. When compared to the abundance of protein mutations alone, the threshold of their stoichiometries may be more critical to define the onset of tumorigenesis and tumor progression, which could facilitate biomarker discovery and improving our understanding of the biology of complex diseases.
Besides the above, functionally important protein mutations may also alter protein structures, which could affect protein association and dissociation, leading to aberrant protein-protein interactions and signaling transduction pathways (e.g., different dynamic signaling patterns of PTMs) (Reva et al., 2011). Therefore, high-resolution structural analysis of disease-associated functional mutations allows for discovery of new therapeutic targets for targeted therapy. In addition, aberrant signaling pathways caused by disease-associated mutations are another attractive therapeutic target because activation of signaling pathways contributes to tumor growth and therapeutic resistance, and constitutively active signaling is commonly observed in cancer (Vegvari, 2016). Another important yet difficult to access research area is the gain or loss of PTMs caused by SAAVs. This is particularly important for protein glycosylation because the large glycan group will alter protein folding, resulting in changing protein function. Limited studies on glycosylation were conducted by Lubman’s and Goldman’s research groups (Fan et al., 2018; Mazumder et al., 2012; Tan et al., 2019). Future studies for simultaneous targeted quantification of PTMs and SAAVs are needed to help better understand the functional impact of SAAVs on human diseases.
6. CONCLUSIONS
Proteomic analysis of mutations is still at the infancy stage. With the recent advances in LC and MS instrumentation, MS-based global proteomics allows genome-scale proteome profiling and proteomic data are being increasingly integrated with genomic data for proteogenomic analysis, which is expected to significantly improve our understanding of the biology of the diseases and the potential of moving toward precision medicine. However, from the genome-scale proteome profiling data, less than 10% single nucleotide variants were detected as SAAVs at the protein level. The low success rate can be attributed to lacking generic databases for sequence search and low abundance mutant peptides as well as high false discovery. Targeted proteomics is a powerful tool for targeted discovery and validation of protein mutations because there is no need for database search, and it has high specificity, multiplexing capability, and reproducibility. When combined with the front-end enrichment of target analytes to enhance detection sensitivity, targeted proteomics has been broadly used for multiplexed detection and quantification of low abundance protein mutations and identification of functionally important mutations. These mutations are responsible for tumorigenesis and tumor progression, and thus they can be used as disease markers for diagnosis and monitoring disease progression due to their high specificity to tumor cells. Moreover, functional studies of these mutations have the potential for discovery of new therapeutic targets. Targeted top-down proteomics has also been used for analysis of protein mutations with precise localization of mutation sites in the intact proteins. Still, there are many technical challenges for current top-down proteomics, which greatly limits its broad applications at present. Further improvement in detection sensitivity and sample throughput for targeted proteomics is needed for cost effective, rapid, sensitive quantification of many important protein mutations in large clinical cohorts for translation of mutation markers into clinical use.
ACKNOWLEDGEMENTS
This paper is dedicated to Professor David M. Lubman in honor of his outstanding contributions to the field of mass spectrometry and his pioneer achievements in protein mutation analysis. Dr. Shi would like to thank David for his inspiration, guidance, and support during their long-term collaboration. Portions of the research were supported by NIH UG3CA256967 (to T.S.), R01GM139858 (to T.S.), RF1MH128885 (to T.S.), R21CA223715 (to T.S.), P41GM103493 (to R.D.S.), U01DK124020 (to W.J.Q.), U54DA049116 (to W.J.Q.), U24CA210955 (to T.L. and R.D.S.), and NCI EDRN Interagency Agreement ACN20007-001 (to T.L.). The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, a national scientific user facility sponsored by the United States of America Department of Energy under Contract DE-AC05-76RL0 1830.
Abbreviations:
- AACT
α-1-antichymotrypsin
- AD
Alzheimer’s disease
- ASV
alternative splicing variant
- AS
alternative splicing
- BAL
bronchoalveolar lavage
- β-NGF
β-nerve growth factor
- CAH
congenital adrenal hyperplasia
- CE
capillary electrophoresis
- cTnI
cardiac troponin I
- Cys
cysteine
- CZE
capillary zone electrophoresis
- DD-SRM
deep-dive selected reaction monitoring
- DDA
data-dependent acquisition
- DIA
data-independent acquisition
- FDR
false-discovery rate
- GeLC-SRM
gel electrophoresis-LC-SRM
- GELFrEE
gel-eluted liquid fraction entrapment electrophoresis
- HMEC
human mammary epithelial cell
- HNSCC
head and neck squamous cell carcinoma
- IA
immunoaffinity
- IMAC
immobilized metal affinity chromatography
- IS
internal standard
- LC-MS
liquid chromatography–mass spectrometry
- LOD
limit of detection
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- NCI CPTAC
National Cancer Institute Clinical Proteomic Tumor Analysis Consortium
- NPs
nanoparticles
- Nrxn
neurexin gene
- NSCLC
non-small cell lung cancer
- OPN
osteopontin
- PCa
prostate cancer
- PRISM
high-pressure, high-resolution separations coupled with intelligent selection and multiplexing
- PRM
parallel reaction monitoring
- PSA
prostate-specific antigen
- PTM
posttranslational modification
- QqQ
triple quadrupole mass spectrometer
- RPLC
reversed-phase LC
- RT
retention time
- SAAV
single amino acid variant
- SAP
single amino acid polymorphism
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- sIEF
solution isoelectric focusing
- SISCAPA
stable isotope standards and capture by antipeptide antibodies
- SNV
single nucleotide variant
- SOD2
superoxide dismutase 2
- SPINK1
pancreatic secretory trypsin inhibitor Kazal type 1
- SPOP
Speckle-type POZ protein
- SRM
selected reaction monitoring
- sSEC
serial size exclusion chromatogram
- THBS1
thrombospondin-1
- Tpm
tropomyosin
- T-μLC-MS
trapping-micro-LC-MS
- WT
wide type
Biography

Tai-Tu Lin obtained his PhD degree form National Taiwan university in 2017, studying membrane proteomics and glycobiology under the guidance of Professors Yu-Ju Chen and Chi-Huey Wong. He is currently working as a postdoctoral fellow in the Integrative Omics Group, Biological Sciences Division, Pacific Northwest National Lab under Dr. Wei-Jun Qian, focusing on developing mass spectrometric assays for precisely monitoring the pathophysiology of disease and applying the state-of-the-art mass spectrometry method for biomarker discovery.

Tong Zhang obtained his PhD degree from the University of Florida in 2016, studying biochemistry and proteomics under Professors Sixue Chen and Alice C. Harmon. He worked as a postdoctoral fellow in the Integrative Omics Group, Biological Sciences Division, Pacific Northwest National Laboratory under Dr. Wei-Jun Qian, focusing on developing proteomic strategies to characterize biological systems. He is currently working as a research scientist in the Seattle Children’s Research Institute. His research interest includes highly sensitive proteomic detection of mutations that cause human diseases, large-scale identification of post-translational modifications in the proteomes, and mechanistic studies for understanding how redox modifications relay cell signaling.

Reta B. Kitata obtained his PhD degree from National Tsing Hua University in 2017, studying membrane proteomics and cancer biology under the guidance of Professor Yu-Ju Chen. Later he worked as postdoctoral fellow in Academia Sinica, Taiwan. He is currently working as research associate in Integrative Omics Group, Biological Sciences Division, Pacific Northwest National Laboratory under Dr. Tujin Shi focusing on spatial phosphoproteome mapping of human tissues.

Tao Liu obtained his PhD in Biochemistry and Molecular Biology in 2001 from Shanghai Institute of Biochemistry, Chinese Academy of Sciences. He is a Senior Staff Scientist and Team Leader in the Integrative Omics group in the Biological Sciences Division at the Pacific Northwest National Laboratory. His research includes development and application of advanced MS technologies for protein post-translational modifications, single-cell, and multiomic characterization of cancers, leading to improved understanding of aberrant regulatory and signal transduction networks underlying cancers, as well as cancer biomarker discovery and verification.

Richard D. Smith received his PhD degree in physical chemistry from the University of Utah. He is currently Director of Proteomics Research, a Battelle Fellow, as well as Chief Scientist for the Biological Sciences Division at Pacific Northwest National Laboratory. He is the author or co-author of more than 1100 peer reviewed publications, holds 75 US patents, and his recognition has included the 2003 American Chemical Society Award for Analytical Chemistry, the 2009 HUPO Discovery Award in Proteomics Sciences and the 2013 ASMS Distinguished Contribution in Mass Spectrometry Award. His early research included the development of the combinations of both capillary supercritical fluid chromatography and capillary electrophoresis separations with mass spectrometry. He also led in the early development and use of high-resolution capillary LC combined with high resolution mass spectrometry for applications in proteomics. More recently his research has included contributions to ion mobility spectrometry (IMS) development, including Structures for Lossless Ion Manipulations (SLIM) IMS for achieving much higher resolution ion mobility separations with mass spectrometry, for increasing the speed, sensitivity and utility of measurements.

Wei-Jun Qian obtained his Ph.D. in Bioanalytical Chemistry from the Chemistry Department at the University of Florida in 2002. He is currently a Laboratory Fellow at the Pacific Northwest National Laboratory and a member of the Integrative Omics group within the Biological Sciences Division. His research involves the development of chemical proteomic approaches for posttranslational modifications, particularly cysteine thiol-based redox modifications, and targeted quantification with applications in metabolic and oxidative stress-related disease areas.

Tujin Shi obtained his PhD degree from York University in 2007, working under the guidance of Professors K. W. Michael Siu and Alan C. Hopkinson. He is a senior scientist in the Integrative Omics Group, Biological Sciences Division, Pacific Northwest National Laboratory. His research interest encompasses targeted proteomics, single-cell proteomics, liquid biopsy proteomics, and spatial tissue mapping as well as gas-phase ion chemistry. His team members develop mass spectrometry-based proteomics approaches and then apply them to address current biomedical need in combination with other advanced technologies.
REFERENCES
- Aebersold R, Agar JN, Amster IJ, et al. How many human proteoforms are there? Nat Chem Biol. 2018;14(3):206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537(7620):347–355. [DOI] [PubMed] [Google Scholar]
- Al-Qahtani WS, Abduljabbar M, AlSuhaibani ES, Abel Rahman A, Aljada A. Quantification of the Lamin A/C Transcript Variants in Cancer Cell Lines by Targeted Absolute Quantitative Proteomics and Correlation with mRNA Expression. Int J Mol Sci. 2019;20(8):1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J Proteome Res. 2004;3(2):235–244. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Jackson A, Smith D, Hardie D, Borchers C, Pearson TW. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol Cell Proteomics. 2009;8(5):995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta. 2005;1739(2–3):91–103. [DOI] [PubMed] [Google Scholar]
- Baudouin S, Scheiffele P. SnapShot: Neuroligin-neurexin complexes. Cell. 2010;141(5):908. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen DB, Martinez-Val A, Steigerwald S, et al. A Compact Quadrupole-Orbitrap Mass Spectrometer with FAIMS Interface Improves Proteome Coverage in Short LC Gradients. Mol Cell Proteomics. 2020;19(4):716–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bie AS, Fernandez-Guerra P, Birkler RI, et al. Effects of a Mutation in the HSPE1 Gene Encoding the Mitochondrial Co-chaperonin HSP10 and Its Potential Association with a Neurological and Developmental Disorder. Front Mol Biosci. 2016;3:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstad I, Breivik L, Methlie P, et al. Functional studies of novel CYP21A2 mutations detected in Norwegian patients with congenital adrenal hyperplasia. Endocr Connect. 2014;3(2):67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Cargile BJ, Sevinsky JR, et al. Detection and validation of non-synonymous coding SNPs from orthogonal analysis of shotgun proteomics data. J Proteome Res. 2007;6(6):2331–2340. [DOI] [PubMed] [Google Scholar]
- Cai W, Tucholski T, Chen B, et al. Top-Down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy. Anal Chem. 2017;89(10):5467–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255(5505):197–200. [DOI] [PubMed] [Google Scholar]
- Catenacci D, Xu P, Henderson L, Liao W, Burrows J, Hembrough T. Development of a Quantitative Gastroesophageal Cancer Selected Reaction Monitoring Mass Spectrometric Multiplex Assay for Use in FFPE Tumor Tissues. Eur J Cancer. 2012;48:172-172. [Google Scholar]
- Catherman AD, Durbin KR, Ahlf DR, et al. Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol Cell Proteomics. 2013;12(12):3465–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BF, Brown KA, Lin ZQ, Ge Y. Top-Down Proteomics: Ready for Prime Time? Anal Chem. 2018;90(1):110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hsiao YC, Chiang SF, et al. Quantitative analysis of wild-type and V600E mutant BRAF proteins in colorectal carcinoma using immunoenrichment and targeted mass spectrometry. Anal Chim Acta. 2016;933:144–155. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Roumeliotis TI, Chang YH, et al. Proteogenomics of Non-smoking Lung Cancer in East Asia Delineates Molecular Signatures of Pathogenesis and Progression. Cell. 2020;182(1):226–244. [DOI] [PubMed] [Google Scholar]
- Chernobrovkin AL, Kopylov AT, Zgoda VG, et al. Methionine to isothreonine conversion as a source of false discovery identifications of genetically encoded variants in proteogenomics. J Proteomics. 2015;120:169–178. [DOI] [PubMed] [Google Scholar]
- Coelho Graca D, Lescuyer P, Clerici L, et al. Electron transfer dissociation mass spectrometry of hemoglobin on clinical samples. J Am Soc Mass Spectrom. 2012;23(10):1750–1756. [DOI] [PubMed] [Google Scholar]
- Cox JT, Kronewitter SR, Shukla AK, Moore RJ, Smith RD, Tang K. High sensitivity combined with extended structural coverage of labile compounds via nanoelectrospray ionization at subambient pressures. Anal Chem. 2014;86(19):9504–9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, et al. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6(7):708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler M, Higginbotham JN, Franklin JL, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12(2):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fede G, Catania M, Morbin M, et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323(5920):1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrakopoulos L, Prassas I, Berns E, Foekens JA, Diamandis EP, Charames GS. Variant peptide detection utilizing mass spectrometry: laying the foundations for proteogenomic identification and validation. Clin Chem Lab Med. 2017;55(9):1291–1304. [DOI] [PubMed] [Google Scholar]
- Dou Y, Kawaler EA, Cui Zhou D, et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell. 2020;180(4):729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kennani S, Adrait A, Permiakova O, et al. Systematic quantitative analysis of H2A and H2B variants by targeted proteomics. Epigenetics Chromatin. 2018;11(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kennani S, Adrait A, Shaytan AK, et al. MS_HistoneDB, a manually curated resource for proteomic analysis of human and mouse histones. Epigenetics Chromatin. 2017;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Hu Y, Yan C, et al. Loss and gain of N-linked glycosylation sequons due to single-nucleotide variation in cancer. Sci Rep. 2018;8(1):4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YY, Neubert H. Quantitative Analysis of Human Neonatal Fc Receptor (FcRn) Tissue Expression in Transgenic Mice by Online Peptide Immuno-Affinity LC-HRMS. Anal Chem. 2016;88(8):4239–4247. [DOI] [PubMed] [Google Scholar]
- Fortin T, Salvador A, Charrier JP, et al. Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol Cell Proteomics. 2009;8(5):1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Thompson JW, Ledford JG, et al. Identification and Quantitation of Coding Variants and Isoforms of Pulmonary Surfactant Protein A. J Proteome Res. 2014;13(8):3722–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Rais Y, Bismar TA, Hyndman ME, Le XC, Drabovich AP. Mapping Isoform Abundance and Interactome of the Endogenous TMPRSS2-ERG Fusion Protein by Orthogonal Immunoprecipitation-Mass Spectrometry Assays. Mol Cell Proteomics. 2021;20:100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S, Kim SY, Domon B. Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM). Mol Cell Proteomics. 2015;14(6):1630–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhu H, Dong L, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179(5):1240. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang YT, Chen Y, et al. Proteomic Tissue-Based Classifier for Early Prediction of Prostate Cancer Progression. Cancers (Basel). 2020;12(5):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette MA, Satpathy S, Cao S, et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell. 2020;182(1):200–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglof C, Hammarsten P, Stromvall K, et al. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS One. 2014;9(2):e86824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvey PJ, Ferrone CR, Liebler DC. GeLC-MRM quantitation of mutant KRAS oncoprotein in complex biological samples. J Proteome Res. 2012;11(7):3908–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wood SD, Wright JM, Dostal V, Lau E, Lam MPY. Computation-assisted targeted proteomics of alternative splicing protein isoforms in the human heart. J Mol Cell Cardiol. 2021;154:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- He J, Sun X, Shi T, et al. Antibody-independent targeted quantification of TMPRSS2-ERG fusion protein products in prostate cancer. Mol Oncol. 2014;8(7):1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaven MR, Wilson L, Barnes S, Brenner M. Relative stabilities of wild-type and mutant glial fibrillary acidic protein in patients with Alexander disease. J Biol Chem. 2019;294(43):15604–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Prasad S, Belford MW, et al. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal Chem. 2018;90(15):9529–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54(11):1796–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim YM, Hamid AM, Deng LL, et al. New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst. 2017;142(7):1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22(44):9694–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16(9):1097–112. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Sun A, Zhao Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257–261. [DOI] [PubMed] [Google Scholar]
- Jin Y, Peng Y, Lin Z, et al. Comprehensive analysis of tropomyosin isoforms in skeletal muscles by top-down proteomics. J Muscle Res Cell Motil. 2016;37(1–2):41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katafuchi T, Esterhazy D, Lemoff A, et al. Detection of FGF15 in plasma by stable isotope standards and capture by anti-peptide antibodies and targeted mass spectrometry. Cell Metabolism. 2015;21(6):898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat Nanotechnol. 2014;9(12):969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva OI, Lisitsa AV, Poverennaya EV. Proteoforms: Methods of Analysis and Clinical Prospects. Mol Biol (Mosk). 2018;52(3):394–410. [DOI] [PubMed] [Google Scholar]
- Kockmann T, Trachsel C, Panse C, et al. Targeted proteomics coming of age - SRM, PRM and DIA performance evaluated from a core facility perspective. Proteomics. 2016;16(15–16):2183–2192. [DOI] [PubMed] [Google Scholar]
- Kukull WA, Bowen JD. Dementia epidemiology. Med Clin North Am. 2002;86(3):573–90. [DOI] [PubMed] [Google Scholar]
- Kumar S, Clarke D, Gerstein MB. Leveraging protein dynamics to identify cancer mutational hotspots using 3D structures. Proc Natl Acad Sci USA. 2019;116(38):18962–18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir MM, Rockwood AL, Roberts WL, Abraham D, Hoofnagle AN, Meikle AW. Measurement of thyroglobulin by liquid chromatography-tandem mass spectrometry in serum and plasma in the presence of antithyroglobulin autoantibodies. Clin Chem. 2013;59(6):982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OK, Ha YS, Lee JN, et al. Comparative Proteome Profiling and Mutant Protein Identification in Metastatic Prostate Cancer Cells by Quantitative Mass Spectrometry-based Proteogenomics. Cancer Genomics Proteomics. 2019;16(4):273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latysheva NS, Babu MM. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016;44(10):4487–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E, Han Y, Williams DR, et al. Splice-Junction-Based Mapping of Alternative Isoforms in the Human Proteome. Cell Rep. 2019;29(11):3751–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Shi T, Petyuk VA, et al. Detection of Head and Neck Cancer Based on Longitudinal Changes in Serum Protein Abundance. Cancer Epidemiol Biomarkers Prev. 2020;29(8):1665–1672. [DOI] [PubMed] [Google Scholar]
- Lesur A, Ancheva L, Kim YJ, Berchem G, van Oostrum J, Domon B. Screening protein isoforms predictive for cancer using immunoaffinity capture and fast LC-MS in PRM mode. Proteomics. Clinical applications. 2015;9(7–8):695–705. [DOI] [PubMed] [Google Scholar]
- Li J, Su Z, Ma ZQ, et al. A bioinformatics workflow for variant peptide detection in shotgun proteomics. Mol Cell Proteomics. 2011;10(5):M110 006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tian Y, Rezai T, et al. Simultaneous analysis of glycosylated and sialylated prostate-specific antigen revealing differential distribution of glycosylated prostate-specific antigen isoforms in prostate cancer tissues. Anal Chem. 2011;83(1):240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti CF, Mostovenko E, Wadsworth PA, et al. Systematic identification of single amino acid variants in glioma stem-cell-derived chromosome 19 proteins. J Proteome Res. 2015;14(2):778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019; 12(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Chang HY, Wu CC, Wu CW, Chang KP, Yu JS. BRAF protein immunoprecipitation, elution, and digestion from cell extract using a microfluidic mixer for mutant BRAF protein quantification by mass spectrometry. Anal Bioanal Chem. 2019;411(5):1085–1094. [DOI] [PubMed] [Google Scholar]
- Liu T, Hossain M, Schepmoes AA, et al. Analysis of serum total and free PSA using immunoaffinity depletion coupled to SRM: correlation with clinical immunoassay tests. J Proteomics. 2012;75(15):4747–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzian A, Uzozie A, Lange PF. Origins and clinical relevance of proteoforms in pediatric malignancies. Expert Rev Proteomics. 2019;16(3):185–200. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marginean I, Page JS, Tolmachev AV, Tang K, Smith RD. Achieving 50% ionization efficiency in subambient pressure ionization with nanoelectrospray. Anal Chem. 2010;82(22):9344–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R, Morampudi KS, Motwani M, Vasudevan S, Goldman R. Proteome-Wide Analysis of Single-Nucleotide Variations in the N-Glycosylation Sequon of Human Genes. Plos One. 2012;7(5):e36212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool EN, Lubeckyj RA, Shen X, et al. Deep Top-Down Proteomics Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry: Identification of 5700 Proteoforms from the Escherichia coli Proteome. Anal Chem. 2018;90(9):5529–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Brunner AD, Koch S, et al. Online Parallel Accumulation Serial Fragmentation (PASEF) with a Novel Trapped on Mobility Mass Spectrometer. Mol Cell Proteomics. 2018;17(12):2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534(7605):55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13(7):1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Kirchner M, Uyar B, et al. Mutations in Disordered Regions Can Cause Disease by Creating Dileucine Motifs. Cell. 2018;175(1):239–253. [DOI] [PubMed] [Google Scholar]
- Mostovenko E, Vegvari A, Rezeli M, et al. Large Scale Identification of Variant Proteins in Glioma Stem Cells. ACS Chem Neurosci. 2018;9(1):73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal E, Chen G, Prensner JR, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2014;9(10):1513–1522. [DOI] [PubMed] [Google Scholar]
- Nam RK, Sugar L, Yang W, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97(12):1690–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Methods. 2014;11(11):1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4(10):1419–1440. [DOI] [PubMed] [Google Scholar]
- Neubert H, Muirhead D, Kabir M, Grace C, Cleton A, Arends R. Sequential protein and peptide immunoaffinity capture for mass spectrometry-based quantification of total human beta-nerve growth factor. Anal Chem. 2013;85(3):1719–1726. [DOI] [PubMed] [Google Scholar]
- Nie S, Shi T, Fillmore TL, et al. Deep-Dive Targeted Quantification for Ultrasensitive Analysis of Proteins in Nondepleted Human Blood Plasma/Serum and Tissues. Anal Chem. 2017;89(17):9139–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Yin H, Tan Z, et al. Quantitative analysis of single amino acid variant peptides associated with pancreatic cancer in serum by an isobaric labeling quantitative method. J Proteome Res. 2014;13(12):6058–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Wu W, Koike A, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009;69(1):111–119. [DOI] [PubMed] [Google Scholar]
- Ntai I, Fornelli L, DeHart CJ, et al. Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc Natl Acad Sci U S A. 2018;115(16):4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page JS, Tang K, Kelly RT, Smith RD. Subambient pressure ionization with nanoelectrospray source and interface for improved sensitivity in mass spectrometry. Anal Chem. 2008;80(5):1800–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. [DOI] [PubMed] [Google Scholar]
- Park HM, Satta R, Davis RG, et al. Multidimensional Top-Down Proteomics of Brain-Region-Specific Mouse Brain Proteoforms Responsive to Cocaine and Estradiol. J Proteome Res. 2019;18(11):3999–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravela S, Valmu L, Domanskyy M, et al. An immunocapture-LC-MS-based assay for serum SPINK1 allows simultaneous quantification and detection of SPINK1 variants. Anal Bioanal Chem. 2018;410(6):1679–1688. [DOI] [PubMed] [Google Scholar]
- Reissner C, Runkel F, Missler M. Neurexins. Genome Biol. 2013;14(9):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodland KD, Piehowski P, Smith RD. Moonshot Objectives: Catalyze New Scientific Breakthroughs-Proteogenomics. Cancer J. 2018;24(3):121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez H, Pennington SR. Revolutionizing Precision Oncology through Collaborative Proteogenomics and Data Sharing. Cell. 2018;173(3):533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsein GE, Pamir N, von Haller PD, et al. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J Proteomics. 2015;113:388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggles KV, Tang Z, Wang X, et al. An Analysis of the Sensitivity of Proteogenomic Mapping of Somatic Mutations and Novel Splicing Events in Cancer. Mol Cell Proteomics. 2016;15(3):1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppen-Canas I, Lopez-Casas PP, Garcia F, et al. An improved quantitative mass spectrometry analysis of tumor specific mutant proteins at high sensitivity. Proteomics. 2012;12(9):1319–1327. [DOI] [PubMed] [Google Scholar]
- Santos R, Ursu O, Gaulton A, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16(1):19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer LV, Millikin RJ, Miller RM, et al. Identification and Quantification of Proteoforms by Mass Spectrometry. Proteomics. 2019;19(10):e1800361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiza C, Korbakis D, Jarvi K, Diamandis EP, Drabovich AP. Identification of TEX101-associated Proteins Through Proteomic Measurement of Human Spermatozoa Homozygous for the Missense Variant rs35033974. Mol Cell Proteomics. 2019;18(2):338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner D, Nguyen TM, Russo G, et al. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 2014;84(2):386–398. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Simicevic J, Ahrne E, Schmidt A, Scheiffele P. Quantitative isoform-profiling of highly diversified recognition molecules. Elife. 2015;4:e07794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Kahraman N, Yan G, Wang J, Ozpolat B, Ittmann M. Targeting the TMPRSS2/ERG fusion mRNA using liposomal nanovectors enhances docetaxel treatment in prostate cancer. Prostate. 2020;80(1):65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Moore RJ, Zhao R, et al. High-efficiency on-line solid-phase extraction coupling to 15–150-microm-i.d. column liquid chromatography for proteomic analysis. Anal Chem. 2003;75(14):3596–3605. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tolic N, Masselon C, et al. Ultrasensitive proteomics using high-efficiency on-line micro-SPE-NanoLC-NanoESI MS and MS/MS. Analytical Chemistry. 2004;76(1):144–154. [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74(16):4235–4249. [DOI] [PubMed] [Google Scholar]
- Sheynkman GM, Shortreed MR, Cesnik AJ, Smith LM. Proteogenomics: Integrating Next-Generation Sequencing and Mass Spectrometry to Characterize Human Proteomic Variation. Annu Rev Anal Chem (Palo Alto Calif). 2016;9(1):521–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheynkman GM, Shortreed MR, Frey BL, Scalf M, Smith LM. Large-scale mass spectrometric detection of variant peptides resulting from nonsynonymous nucleotide differences. J Proteome Res. 2014;13(1):228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Fillmore TL, Sun X, et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci U S A. 2012;109(38):15395–15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Gao Y, Gaffrey MJ, et al. Sensitive targeted quantification of ERK phosphorylation dynamics and stoichiometry in human cells without affinity enrichment. Anal Chem. 2015;87(2):1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Gao Y, Quek SI, et al. A highly sensitive targeted mass spectrometric assay for quantification of AGR2 protein in human urine and serum. J Proteome Res. 2014;13(2):875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Niepel M, McDermott JE, et al. Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway. Sci Signal. 2016;9(436):rs6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Qian WJ. Antibody-free PRISM-SRM for multiplexed protein quantification: is this the new competition for immunoassays in bioanalysis? Bioanalysis. 2013;5(3):267–269. [DOI] [PubMed] [Google Scholar]
- Shi T, Song E, Nie S, et al. Advances in targeted proteomics and applications to biomedical research. Proteomics. 2016;16(15–16):2160–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Su D, Liu T, et al. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12(8):1074–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Sun X, Gao Y, et al. Targeted quantification of low ng/mL level proteins in human serum without immunoaffinity depletion. J Proteome Res. 2013;12(7):3353–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Wu X, Tong X, et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Sci Rep. 2017;7(1):583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Huang V, Livingstone J, et al. The Proteogenomic Landscape of Curable Prostate Cancer. Cancer Cell. 2019;35(3):414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL. Proteoforms as the next proteomics currency. Science. 2018;359(6380):1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL, Consortium for Top Down P. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10(3):186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopfer LE, Flower CT, Gajadhar AS, et al. High-Density, Targeted Monitoring of Tyrosine Phosphorylation Reveals Activated Signaling Networks in Human Tumors. Cancer Res. 2021;81(9):2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su ZD, Sheng QH, Li QR, et al. De novo identification and quantification of single amino-acid variants in human brain. J Mol Cell Biol. 2014;6(5):421–433. [DOI] [PubMed] [Google Scholar]
- Su ZD, Sun L, Yu DX, et al. Quantitative detection of single amino acid polymorphisms by targeted proteomics. J Mol Cell Biol. 2011;3(5):309–315. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhu G, Zhao Y, Yan X, Mou S, Dovichi NJ. Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests. Angew Chem Int Ed Engl. 2013;52(51):13661–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Nie S, McDermott SP, Wicha MS, Lubman DM. Single Amino Acid Variant Profiles of Subpopulations in the MCF-7 Breast Cancer Cell Line. J Proteome Res. 2017;16(2):842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Yi X, Carruthers NJ, Stemmer PM, Lubman DM. Single Amino Acid Variant Discovery in Small Numbers of Cells. J Proteome Res. 2019;18(1):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Zhu J, Stemmer PM, et al. Comprehensive Detection of Single Amino Acid Variants and Evaluation of Their Deleterious Potential in a PANC-1 Cell Line. J Proteome Res. 2020;19(4):1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Page JS, Marginean I, Kelly RT, Smith RD. Improving liquid chromatography-mass spectrometry sensitivity using a subambient pressure ionization with nanoelectrospray (SPIN) interface. J Am Soc Mass Spectrom. 2011;22(8):1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiambeng TN, Roberts DS, Brown KA, et al. Nanoproteomics enables proteoform-resolved analysis of low-abundance proteins in human serum. Nat Commun. 2020;11(1):3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby TK, Fornelli L, Srzentic K, et al. A comprehensive pipeline for translational top-down proteomics from a single blood draw. Nat Protoc. 2019;14(1):119–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran JC, Zamdborg L, Ahlf DR, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480(7376):254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisiol S, Ayoub D, Lesur A, Ancheva L, Gallien S, Domon B. The use of proteases complementary to trypsin to probe isoforms and modifications. Proteomics. 2016;16(5):715–728. [DOI] [PubMed] [Google Scholar]
- Vasaikar S, Huang C, Wang X, et al. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell. 2019;177(4):1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegvari A. Mutant Proteogenomics. Adv Exp Med Biol. 2016;926:77–91. [DOI] [PubMed] [Google Scholar]
- Vegvari A, Sjodin K, Rezeli M, et al. Identification of a novel proteoform of prostate specific antigen (SNP-L132I) in clinical samples by multiple reaction monitoring. Mol Cell Proteomics. 2013;12(10):2761–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Barbieri CE, He J, et al. Quantification of mutant SPOP proteins in prostate cancer using mass spectrometry-based targeted proteomics. J Transl Med. 2017;15(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LB, Karpova A, Gritsenko MA, et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39(4):509–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chaerkady R, Wu J, et al. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci U S A. 2011;108(6):2444–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14(10):703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975;72(5):1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker JR, Paulovich AG. Peptide immunoaffinity enrichment coupled with mass spectrometry for peptide and protein quantification. Clin Lab Med. 2011;31(3):385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010;9(1):184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker JR, Zhao L, Lin C, Yan P, Wang P, Paulovich AG. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol Cell Proteomics. 2012;11(6):M111 015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo TS, Duong DM, Zhou M, et al. Integrating Next-Generation Genomic Sequencing and Mass Spectrometry To Estimate Allele-Specific Protein Abundance in Human Brain. J Proteome Res. 2017;16(9):3336–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. [DOI] [PubMed] [Google Scholar]
- Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25(2):213–216. [DOI] [PubMed] [Google Scholar]
- Worman HJ. Cell signaling abnormalities in cardiomyopathy caused by lamin A/C gene mutations. Biochem Soc Trans. 2018;46(1):37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Pungaliya P, Kraynov E, Bates B. Identification and quantification of osteopontin splice variants in the plasma of lung cancer patients using immunoaffinity capture and targeted mass spectrometry. Biomarkers. 2012;17(2):125–133. [DOI] [PubMed] [Google Scholar]
- Xu M, Lu M, Zhang W, Jin Q, Chen Y. Simultaneous Detection of Six Isoforms of Tau Protein in Human Cerebrospinal Fluid by Multidimensional Mass Spectrometry-Based Targeted Proteomics. J Proteome Res. 2021;20(5):2299–2307. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yu L, Wang Z. PCA3 and TMPRSS2-ERG gene fusions as diagnostic biomarkers for prostate cancer. Chin J Cancer Res. 2016;28(1):65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You CC, Miranda OR, Gider B, et al. Detection and identification of proteins using nanoparticle-fluorescent polymer ‘chemical nose’ sensors. Nat Nanotechnol. 2007;2(5):318–323. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Whiteaker JR, Hoofnagle AN, Baird GS, Rodland KD, Paulovich AG. Clinical potential of mass spectrometry-based proteogenomics. Nat Rev Clin Oncol. 2019;16(4):256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu T, Zhang Z, et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 2016;166(3):755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, An B, Qu Y, et al. Sensitive, High-Throughput, and Robust Trapping-Micro-LC-MS Strategy for the Quantification of Biomarkers and Antibody Biotherapeutics. Anal Chem. 2018;90(3):1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhu M, Geng-Spyropoulos M, et al. A novel, multiplexed targeted mass spectrometry assay for quantification of complement factor H (CFH) variants and CFH-related proteins 1–5 in human plasma. Proteomics. 2017;17(6):10.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]