Abstract
Fluorination in enhancing photoactivated antibacterial activity of Ru(ii) complexes with photo-labile ligands was studied. Ru(ii) polypyridine complexes containing a di-fluorinated dppz (dipyrido[3,2-a:2′,3′-c]phenazine) or mono-trifluoromethylated dppz bidentate ligand and four pyridine monodentate ligands (complexes 3 and 4) were found to show potent photoactivated antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), and Escherichia coli (E. coli) in both normoxic and hypoxic conditions. The bactericidal effect of complexes 3 and 4 under hypoxic conditions may stem from the fluorine-containing Ru(ii) aqua species after photo-induced pyridine dissociation, and DNA may be the potential antibacterial target. Photosensitized singlet oxygen may also account for their antibacterial activity under normoxic conditions. Moreover, negligible hemolysis rates as well as low dark- and photo-cytotoxicity toward human normal liver cells (L-O2) were also observed for both complexes. Our work may provide new insights into the development of novel and efficient Ru(ii) complex based photoactivatable antibacterial agents against antibiotic-resistant bacteria.
Fluorination in the dppz ligand efficiently enhanced the photoactivated antibacterial activity of Ru(ii) complexes with photo-labile ligands against antibiotic-resistant bacteria both under normoxic and hypoxic conditions.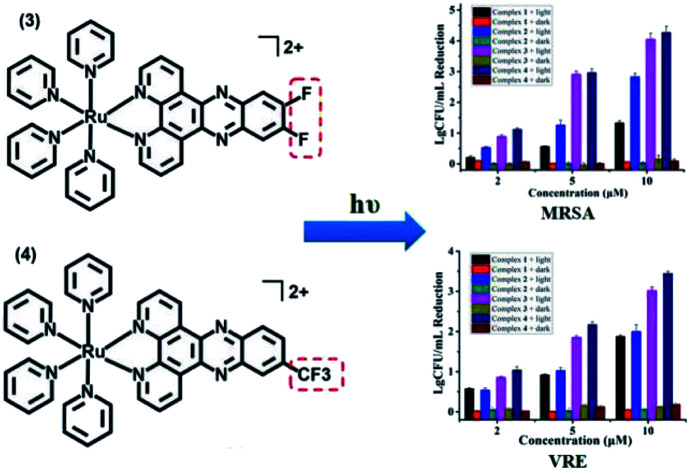
Introduction
Antibiotic resistance is an enormous threat to public health globally.1 In recent years, various types of drug-resistant bacteria have been reported, such as Methicillin-resistant Staphylococcus aureus (MRSA),2 vancomycin-resistant Enterococcus (VRE),3etc., leading to an urgent call for totally new antibacterial agents as well as novel antibacterial strategies. Metal complexes, such as Ru(ii) polypyridyl complexes with various structures and functions to interact with biomolecules, may act with different antibacterial mechanisms compared with the currently long used organic antibiotics.4 The antibacterial properties of inert lipophilic Ru(ii) polypyridyl complexes were first reported over 60 years ago,5 and were more extensively studied in recent years due to increasing antibiotic resistance.6
In addition to seeking for new antibacterial agents, development of novel antibacterial strategies which will not induce or slow down drug resistance is also important. Among them, photoactivation strategy is particularly attractive,7 and Ru(ii) complexes are also good candidates because of their rich photophysical and photochemical properties.8 According to the different antibacterial mechanisms upon irradiation, the reported Ru(ii) complexes for photoactivation antibacterial therapy can be mainly divided into two kinds. One kind of Ru(ii) complexes can generate reactive oxygen species (ROS, mainly singlet oxygen 1O2) to kill bacteria upon irradiation,9 which is well known as antimicrobial photodynamic therapy (aPDT). Bacteria can not generate resistance to aPDT due to the non-specific damage by highly toxic ROS. An oxygenated microenvironment is necessary for aPDT, however, this is not the case for all the infected sites. Thus aPDT may lost its efficacy against anaerobes or facultative anaerobes in hypoxic conditions. The other kind of Ru(ii) complexes contain antibacterial agents as the photolabile ligands,10 which can be released upon irradiation. The spatial and temporal release of antibacterial agents by light irradiation can avoid off-target side effects, and also slow down the development of resistance. Moreover, the oxygen-independent manner renders these complexes applicable even in a hypoxic environment. However, only antibacterial agents with coordinate groups can be applied in this strategy, which is not the case for most currently used antibiotics.
For the second kind of Ru(ii) complexes mentioned above, the Ru(ii) moieties only served as drug carriers, and the bactericidal effect stemmed exclusively from the released ligands rather than the Ru(ii) aqua species generated after photo-induced ligand dissociation. The antibacterial activity of the Ru(ii) aqua species may be underestimated, especially considering the well-known fact that they can covalently bind with biomolecules, such as DNA.11 One possible reason for the lack of antibacterial activity of these common Ru(ii) aqua species may lie in their poor uptake by bacteria, and/or weak interaction with targeting biomolecules. Thus through proper structure design and modification on the non-labile ligands, the resulted Ru(ii) complexes themselves may also have good antibacterial property. The related studies may lead to new Ru(ii) based photoactivatable antibacterial agents that are not dependent on the known antibiotics, as well as ideal platforms for realizing multidrug combination therapy against antibiotic-resistance bacteria. However, to the best of our knowledge, no such an exploration has been reported yet.
Having the strongest electronegativity, the fluorine atom displays a variety of marvelous properties. Incorporation of fluorine atoms into a structure often gives rise to enhanced chemical and thermal stability and additional hydrogen bond interactions. These potentials make the fluorine atom an important pharmacophore in boosting activities of drugs ranging from antitumor to antibacterial.12 Very recently, we reported on three fluorinated dppz ligand coordinated Ru(ii) complexes (complexes 2–4 in Scheme 1, dppz = dipyrido[3,2-a:2′,3′-c]phenazine) containing four monodentate pyridine ligands.13 They exhibited much diminished cytotoxicity than their parent un-fluorinated complex (complex 1) in the dark, but much improved cytotoxicity against human cervical cancer (HeLa) cells and human ovary cancer (SKOV-3) cells rather than human normal liver cells (L-O2). The promising results prompt us to explore their possibility as new photoactivatable antibacterial agents. The studies herein show that these fluorinated Ru(ii) complexes, especially for complexes 3 and 4, can effectively photoinactivate MRSA and VRE, two kinds of resistant bacteria that have caused increasing incidence of infection with unsuccessful treatment and high fatality rates.2,3 The bactericidal effect of complexes 3 and 4 mainly stems from the fluorine-containing Ru(ii) aqua species after photo-induced pyridine dissociation, and DNA may be the potential antibacterial target. Their photoactivated bactericidal activities remained well in hypoxic conditions, indicating their all-weather capability.
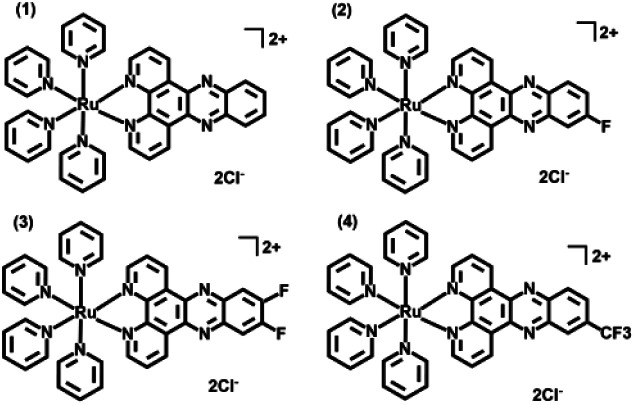
Scheme 1. Chemical structures of complexes 1–4.
Experimental section
CFU counting
Suspensions (1 mL) of bacteria (∼108/mL) at exponential phase were dispersed in PBS in transparent plastic centrifugal tubes. Then, the bacteria was treated with gradient concentrations of complexes 1–4 in the dark for 30 min. Afterwards, the light groups were exposed to LED irradiation (470 nm, 22.5 mW cm−2) for 20 min, while the dark groups were remained in the dark. The treated samples were spread on 3M Petrifilm Aerobic Count Plates after diluted into the proper concentration, and continually incubated in the incubator for 24 h.
As for the experiments in hypoxic condition, the transparent plastic centrifugal tubes were immediately transported and sealed in AnaeroGen™ 2.5L bio-bags produced by MITSUBISHI GAS CHEMICAL CO. to generate anaerobic conditions (O2% < 0.1%) for 30 min. The light groups were irradiated in hypoxia for 20 min, and spread to count plates as described. Each experiment was performed at least three independent times.
MIC/MBC measurements
Serial two-fold dilutions of each drug (128–0.5 μM or 0.25–0.002 μM) were made with the LB media in 96-well plates containing ∼105 bacteria per well. Cultured with complexes 1–4 for 30 min in the dark, the samples were exposed to LED irradiation (470 nm, 22.5 mW cm−2) for 20 min. Samples with methicillin or vancomycin were incubated in the dark all along. Incubating for 24 h, MIC and MBC were obtained by measuring the optical density of each sample at 570 nm on a microplate reader or by resazurin tests, respectively. The percentage of bacterial cells survival was calculated from the equation: cell growth% = (AT − AB)/(AC − AB), AT stands for the optical density (OD570) of each well in the presence of tested compounds, AB stands for the OD of the wells with only the LB medium, and AC stands for the OD of the wells with bacteria cells in the LB medium. MIC90 values were used in our manuscript, and were the lowest concentrations at which cell growth was inhibited to about 10%. MBC values were estimated by resazurin tests. The color of resazurin will be transformed from blue into pink in the presence of live bacteria, and MBC values are the lowest concentrations of tested compounds at which the color of resazurin is still blue.
Morphology studies
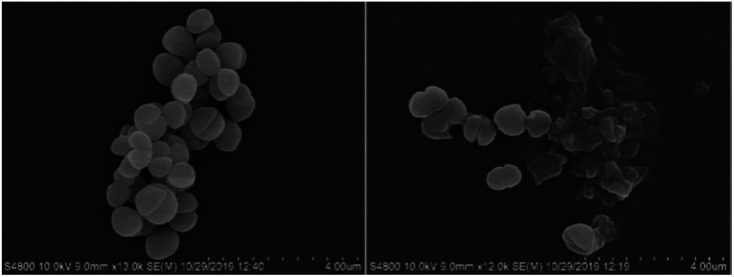
Scanning electronic microscopy (SEM) was employed to observe the morphology of bacterial cells with or without treatment by the complexes. S. aureus cells were harvested at exponential phase, washed by PBS and divided into 2 parallel groups. The treated group was incubated with 5 μM of complex 4 for 30 min, followed by 20 min of irradiation (470 nm, 22.5 mW cm−2). Control group was left untreated all along. Then, the cell pallets were obtained and subjected to fixation with 2% glutaraldehyde at 4 °C for 12 h. After washed by PBS, samples were dehydrated for 15 min in a graded ethanol series (30%, 50%, 70%, 90% and 95%) and 15 min in 100% ethanol. Finally the dried specimens were coated with gold and observed by SEM.
Co-culture experiments
L-O2 and S. aureus cells were employed in co-culture experiments to test the selectivity of complex 4. ∼4 × 105 L-O2 cells were incubated in 35 mm culture dishes (Corning) for 24 h. The medium was replaced by 1 mL of S. aureus (109 cells per mL) in PBS suspension, and 5 μM of complex 4 was added to the system. After incubation for 30 min, the sample was exposed to irradiation (470 nm, 22.5 mW cm−2) for 20 min. Then the bacteria and L-O2 cells were washed by PBS, before cultured in fresh DMEM for another 4 h. Washed by PBS, the samples were stained with PI and Hoechst and observed with Nikon N-C2-SIM inverted fluorescent microscope.
Results and discussion
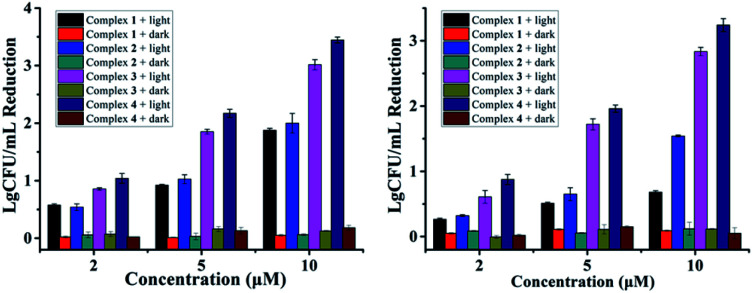
Antibacterial performance of complexes 1–4 against S. aureus and MRSA are presented in Fig. 1, neither the irradiation alone (Fig. S1†) nor complexes 1–4 in the dark did not inhibit the growth of bacteria. In contrast, concentration-dependent bactericidal abilities were observed upon light irradiation at 470 nm for 20 min (LED, 22.5 mW cm−2). Complexes 3 and 4 led to a colony forming unit (CFU) reduction larger than 4 log units against both S. aureus and MRSA at 10 μM, much higher than complexes 1 and 2 in the same conditions, indicating the important role of the F atoms. Minimum inhibitory concentrations (MIC) and minimum bactericidal concentrations (MBC) of 1–4 were also measured upon 470 nm irradiation, using methicillin and vancomycin as controls (Table 1). The MIC and MBC values of 3 and 4 against planktonic S. aureus and MRSA are 1–2 μM, much better than methicillin especially toward MRSA, and comparable to vancomycin, an antibiotic regarded as the last resort for MRSA.3
Fig. 1. Antibacterial activity of 1–4 against S. aureus (left) and MRSA (right) in the dark or under light irradiation (470 nm LED, 22.5 mW cm−2, 20 min).
MIC and MBC values (μM) of 1–4 upon irradiation (470 nm LED, 22.5 mW cm−2, 20 min) against E. coli, S. aureus, MRSA, and VRE.
| E. coli | S. aureus | MRSA | VRE | |||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| 1 | 16 | 32 | 8 | 16 | 16 | 32 | 16 | 32 |
| 2 | 16 | 32 | 4 | 4 | 4 | 8 | 8 | 8 |
| 3 | 8 | 8 | 2 | 2 | 2 | 2 | 2 | 4 |
| 4 | 4 | 8 | 1 | 1 | 1 | 2 | 2 | 4 |
| Methicillin | >128 | >128 | 4 | 8 | >128 | >128 | >128 | >128 |
| Vancomycin | 128 | >128 | 2 | 2 | 2 | 4 | >128 | >128 |
Morphology studies were also conducted along with the CFU measurements by SEM (Fig. 2). Complex 4 without irradiation caused little effect on the morphology of S. aureus, consistent with its little antibacterial activity in the dark. Upon 470 nm LED irradiation for 20 min, most of S. aureus cells turned deformed, and the leaking intracellular components may indicate the death of the bacteria.
Fig. 2. SEM images of S. aureus cells treated by 4 (5 μM) without (left) or with (right) irradiation at 470 nm (LED, 22.5 mW cm−2) for 20 min.
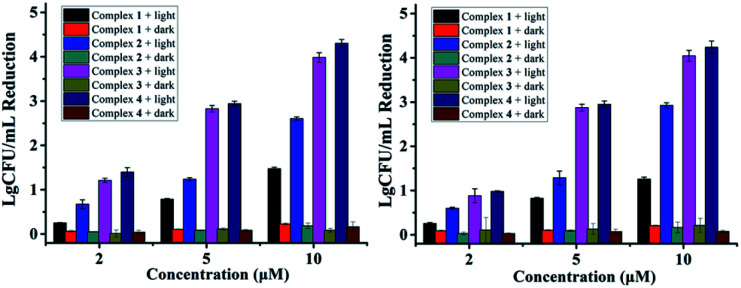
Additionally, VRE, a bacterial strain highly resistant against both vancomycin and methicillin (Table 1), had little resistance against complex 4. A CFU reduction of 3.45 log units was obtained by 10 μM of 4 under irradiation condition (Fig. 3). Besides Gram-positive bacteria, complexes 3 and 4 can photoinactivate Gram-negative ones with a similar potency. As shown in Fig. 3, E. Coli cells had a CFU reduction of 3.24 log units by 4 at 10 μM upon light irradiation.
Fig. 3. Antibacterial activity of 1–4 against VRE (left) and E. coli (right) in the dark or under light irradiation (470 nm, LED, 22.5 mW cm−2, 20 min).
As reported in our previous research, complexes 1–4 containing monodentate pyridine ligands can undergo photo-induced one pyridine dissociation, and the resulted Ru(ii) aqua complexes are toxic towards cancer cells.11 For the antibacterial experiments here, the resulted Ru(ii) aqua complexes upon irradiation may also be the active species, which can kill bacteria in an O2-independent pathway. As shown in Fig. 4, the antibacterial performance of complexes 1–4 against S. aureus and MRSA in hypoxic conditions (O2 < 0.1%) was not compromised, confirming the O2-independent antibacterial mechanism. Moreover, we also examined the antibacterial activity of the photo-product of complex 4 in the dark (Fig. S2†), the results of which were comparable with that upon photoactivation (Fig. 1 and 3). Free pyridine (10 μM) did not obviously inhibit bacterial growth, confirming the important role of Ru(ii) aqua compounds. Ru(ii) complexes with weak ligand fields usually have quite low singlet oxygen generation ability, and complex 1 is such an example (Fig. S3†). Interestingly, fluorination increased the ability of complexes 2–4, thus singlet oxygen may also partially account for the antibacterial activity in normoxic conditions.
Fig. 4. Antibacterial activity of 1–4 against S. aureus (left) and MRSA (right) in hypoxic conditions (O2 < 0.1%).
The steady enhanced bactericidal ability from 1 to 4 may be mainly ascribed to the gradually increased bacterial cell uptake. As displayed in Fig. 5, the uptake levels by S. aureus measured by ICP-MS gradually increased from 1–4 and show a positive correlation with the F atom numbers (Fig. 5), indicating the important role of F atoms. Generally, the uptake levels of drugs are mainly related with their lipophilicity. However, complexes 1–4 display very similar hydrophobicity (Table 2), hinting that other factors, such as enhanced hydrogen bond interactions by F atoms, may facilitate cell uptake.
Fig. 5. Uptake levels of 1–4 (5 μM) by S. aureus measured by ICP-MS.
Zeta potential values of S. aureus, oil/water partition coefficients and hemolysis behaviors of 1–4.
| S. aureus | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Zeta potential (mV) | 12.10 ± 0.57 | 10.35 ± 0.50 | 9.74 ± 0.18 | 8.80 ± 0.13 | 8.44 ± 0.14 |
| Log Pq/wa | — | −1.09 ± 0.05 | −1.07 ± 0.02 | −1.15 ± 0.05 | −1.14 ± 0.03 |
| Hemolysis percentage at 20 μM (%) | — | 0.64 ± 0.01 | 0.65 ± 0.01 | 1.05 ± 0.04 | 1.19 ± 0.01 |
| Hemolysis percentage at 5 μM (%) | — | 0.07 ± 0.01 | 0.06 ± 0.01 | 0.24 ± 0.02 | 0.37 ± 0.08 |
Logarithmic values of n-octanol/water partition coefficients.
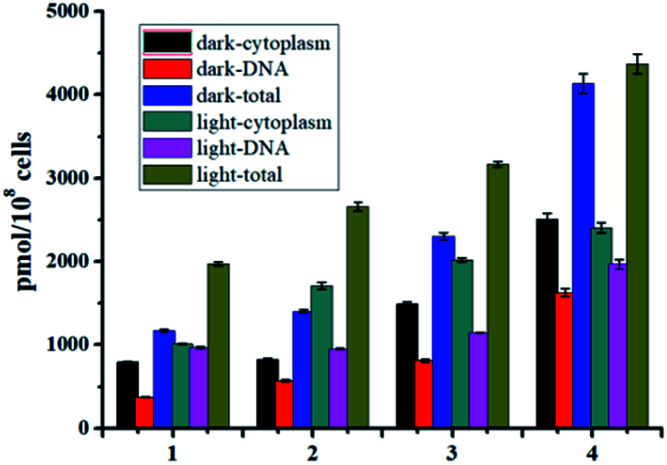
As well studied,11 the resulted Ru(ii) aqua complexes after photo-induced ligand dissociation can covalently bind with DNA like cisplatin, which is the general anticancer mechanism for these Ru(ii) complexes. Bacterial DNAs are mostly nakedly locates in cytoplasm without the protection of nuclear membrane, which is more convenient for drug access. Moreover, the well-known DNA intercalation dppz ligand in complexes 1–4 will facilitate DNA targeting.14 Thus DNA may also be the potential target of these Ru(ii) complexes based antibacterial agents. The DNA targeting ability of complexes 1–4 with and without irradiation were studied by separating DNA from cytoplasm using commercial kits. As shown in Fig. 5, the accumulation of complexes 1–4 onto DNA is obvious, and about 40% of complexes 3 and 4 can bind with DNA with or without irradiation. In the dark, complexes 1–4 may bind to DNA through dppz-based noncovalent intercalation interaction. Upon light irradiation and dissociation of a pyridine ligand, the resultant Ru(ii) aqua complexes may bind DNA covalently, leading to irreversible DNA damage, and ultimate bacteria death. The total binding/uptake of bacterial cells and inner DNA binding levels upon irradiation increased with different extent for complexes 1–4, possibly due to the enhanced permeability of bacterial cells and formation of DNA covalent binding after photo-induced ligand dissociation.
ζ-potential detection results of S. aureus are also in line with the binding/uptake measurements. The original ζ-potential value of S. aureus was −12.10 mV due to the highly negatively charged cell walls. After incubation with the examined Ru(ii) complexes, the measured ζ-potentials all experienced a positive shift as shown in Table 2. Complex 4 showed the biggest shift, indicative of its highest cellular binding/uptake further. The cationic complexes 1–4 may bind with the cell walls by electrostatic interaction, and we also examined the possible uptake pathways by comparing the binding/uptake levels of complex 4 at 37 °C and 4 °C (Table S1†). No obvious difference was observed, indicating that complex 4 may enter bacterial cells in an energy-independent passive diffusion way.
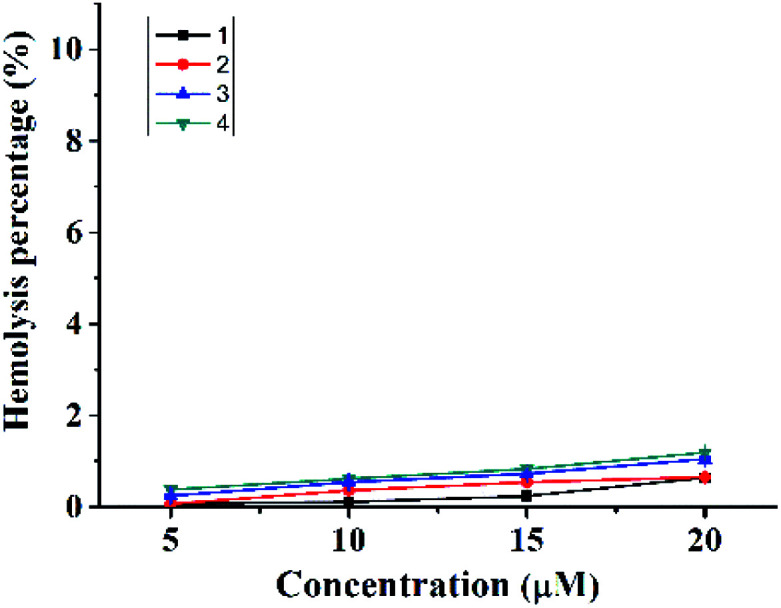
As an antibacterial agent, negligible hemolysis is one of the most critical prerequisites. The hemolysis rates toward rabbit red blood cells of complexes 1–4 were measured. As shown in Fig. 6 and Table 2, their hemolysis rates are all lower than 1.2% at the concentration of 20 μM. For complexes 3 and 4, the hemolysis rates may be negligible at the concentration (around 5 μM as shown in Fig. 1) where the bactericidal effect (CFU reduction over 3 log units) may be observed.
Fig. 6. Hemolysis percentage of 1–4 toward rabbit RBCs.
Low cytotoxicity toward normal human cells is another important requirement for an antibacterial agent. Our previous work has shown that complexes 3 and 4 had negligible dark cytotoxicity toward human normal liver cells (L-O2) with IC50 values over 200 μM.13 The phototoxicity of 3 and 4 were also low with IC50 values of 30.0 ± 2.1 μM and 29.3 ± 1.5 μM, respectively. In the presence of 5 μM of 3 or 4 and upon irradiation, 70% and 90% L-O2 cells still survived after incubation for 24 h.
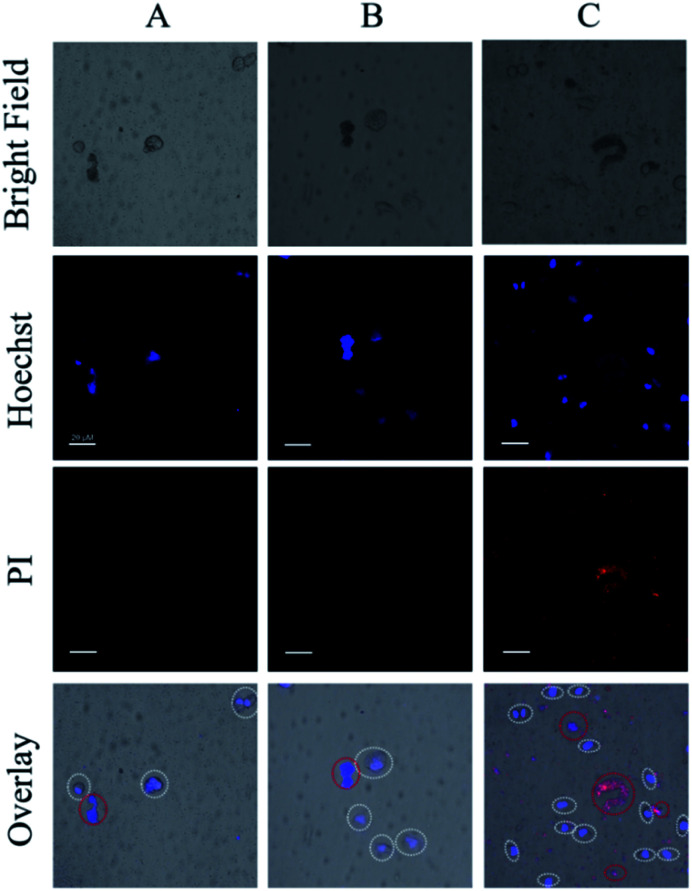
To ensure the safety and selectivity of the examined Ru(ii) complexes, co-culture experiments were conducted using 4 as an example. Mixtures of L-O2 and S. aureus cells were incubated with complex 4 (5 μM) for 30 min, then were exposed to irradiation (470 nm, 22.5 mW cm−2) for 20 min. After further culture in new medium for 4 h, the mixtures of L-O2 and S. aureus cells were stained with propidium iodide (PI) and Hoechst 33342, and imaged with confocal laser scanning microscope (CLSM). PI can stain the dead L-O2 and S. aureus cells with red fluorescence, and Hoechst 33342 can help to see L-O2 more clearly. As shown in Fig. 7C, only S. aureus cells were stained with PI, indicating a selective photoinactivation toward bacteria by complex 4.
Fig. 7. CLSM images of the mixed solutions of S. aureus and L-O2, (A) light irradiation only, (B) addition of 4 (5 μM) only, (C) addition of 4 (5 μM) and light irradiation. For clarity, the L-O2 cells were marked with white circles, red circles indicated the aggregated S. aureus cells (Scale bar: 20 μm).
Conclusions
In summary, fluorinated Ru(ii) complexes 3 and 4 show potent photoactivated antibacterial activity against MRSA, VRE, and E. coli. at a concentration as low as 5 μM. The bactericidal effect stems from both of singlet oxygen and in situ generated Ru(ii) fraction after photo-induced ligand dissociation, and the latter may still remain active in a hypoxic condition due to its oxygen-independent manner. To the best of our knowledge, the results here first disclosed that Ru(ii) complexes with photo-labile ligands can be developed as potent antibacterial agents against antibiotic-resistant bacteria. Moreover, both 3 and 4 may serve as ideal platforms for further introduction of photolabile antibacterial ligands to realize more efficient combination therapy to combat intractable antibiotic-resistant bacteria.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the National Key R&D Program of China (2018YFC1602204), the NSFC (21390400, 21571181, and 21773277), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB17000000).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra01806f
Notes and references
- (a) Brown E. D. Wright G. D. Nature. 2016;529:336. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]; (b) Lakemeyer M. Zhao W. Mandl F. A. Hammann P. Sieber S. A. Angew. Chem., Int. Ed. 2018;57:14440. doi: 10.1002/anie.201804971. [DOI] [PubMed] [Google Scholar]
- (a) Wang Y. Zhou Q. Wang Y. Ren J. Zhao H. Wu S. Yang J. Zhen J. Luo Y. Wang X. Gu Y. Photochem. Photobiol. 2015;91:124. doi: 10.1111/php.12378. [DOI] [PubMed] [Google Scholar]; (b) Lam P. L. Lu G. L. Hon K. M. Lee K. W. Ho C. L. Wang X. Tang J. C. O. Lam K. H. Wong R. S. M. Kok S. H. L. Bian Z. X. Li H. Lee K. K. H. Gambari R. Chui C. H. Wong W. Y. Dalton Trans. 2014;43:3949. doi: 10.1039/C3DT52879K. [DOI] [PubMed] [Google Scholar]
- Dhanda G. Sarkar P. Samaddar S. Haldar J. J. Med. Chem. 2019;62:3184. doi: 10.1021/acs.jmedchem.8b01093. [DOI] [PubMed] [Google Scholar]
- (a) Li F. Collins J. G. Keene F. R. Chem. Soc. Rev. 2015;44:2529. doi: 10.1039/C4CS00343H. [DOI] [PubMed] [Google Scholar]; (b) Yang Y. Liao G. Fu C. Polymers. 2018;10:650. doi: 10.3390/polym10060650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer F. P. Gyarfas E. C. Rogers W. P. Koch J. H. Nature. 1952;170:190. doi: 10.1038/170190a0. [DOI] [PubMed] [Google Scholar]
- (a) Li F. Feterl M. Warner J. M. Keene F. R. Collins J. G. J. Antimicrob. Chemother. 2013;68:2825. doi: 10.1093/jac/dkt279. [DOI] [PubMed] [Google Scholar]; (b) Kumar S. V. Scottwell S. O. Waugh E. McAdam C. J. R Hanton L. Brooks H. J. Crowley J. D. Inorg. Chem. 2016;55:9767. doi: 10.1021/acs.inorgchem.6b01574. [DOI] [PubMed] [Google Scholar]; (c) Smitten K. L. Fairbanks S. D. Robertson C. C. Serna J. B. Foster S. J. Thomas J. A. Chem. Sci. 2020;11:70. doi: 10.1039/C9SC04710G. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Li F. Feterl M. Mulyana Y. Warner J. M. Collins J. G. Keene F. R. J. Antimicrob. Chemother. 2012;67:2686. doi: 10.1093/jac/dks291. [DOI] [PubMed] [Google Scholar]; (e) Li F. Mulyana Y. Feterl M. Warner J. M. Collins J. G. Keene F. R. Dalton Trans. 2011;40:5032. doi: 10.1039/C1DT10250H. [DOI] [PubMed] [Google Scholar]
- (a) Velema W. A. Berg J. P. Hansen M. J. Szymanski W. Driessen A. J. Feringa B. L. Nat. Chem. 2013;5:924. doi: 10.1038/nchem.1750. [DOI] [PubMed] [Google Scholar]; (b) Babii O. Afonin S. Berditsch M. Reibetaer S. Mykhailiuk P. K. Kubyshkin V. S. Steinbrecher T. Ulrich A. S. Komarov I. V. Angew. Chem., Int. Ed. 2014;53:3392. doi: 10.1002/anie.201310019. [DOI] [PubMed] [Google Scholar]; (c) Feng Y. Liu L. Zhang J. Aslan H. Dong M. J. Mater. Chem. B. 2017;5:8631. doi: 10.1039/C7TB01860F. [DOI] [PubMed] [Google Scholar]
- (a) Balzani V. Juris A. Coord. Chem. Rev. 2001;211:97. doi: 10.1016/S0010-8545(00)00274-5. [DOI] [Google Scholar]; (b) Juris A. Balzani V. Barigelletti F. Campagna S. Belser P. von Zelewsky A. Coord. Chem. Rev. 1988;84:85. doi: 10.1016/0010-8545(88)80032-8. [DOI] [Google Scholar]
- (a) Frei A. Rubbiani R. Tubafard S. Blacque O. Anstaett P. Felgenträger A. Maisch T. Spiccia L. Gasser G. J. Med. Chem. 2014;57:7280. doi: 10.1021/jm500566f. [DOI] [PubMed] [Google Scholar]; (b) Arenasa Y. Monrob S. Shib G. Mandela A. McFarland S. Lilgec L. Photodiagn. Photodyn. Ther. 2013;10:615. doi: 10.1016/j.pdpdt.2013.07.001. [DOI] [PubMed] [Google Scholar]; (c) Feng Y. Sun W. Wang X. Zhou Q. Chem.–Eur. J. 2019;25:13879. doi: 10.1002/chem.201903923. [DOI] [PubMed] [Google Scholar]
- (a) Smith N. A. Zhang P. Greenough S. E. Horbury M. D. Clarkson G. J. McFeely D. Habtemariam A. Salassa L. Stavros V. G. Dowson C. G. Sadler P. J. Chem. Sci. 2017;8:395. doi: 10.1039/C6SC03028A. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Garner R. N. Pierce C. G. Reed C. R. W Brennessel W. Inorg. Chim. Acta. 2017;461:261. doi: 10.1016/j.ica.2017.02.031. [DOI] [Google Scholar]
- (a) Tian N. Feng Y. Sun W. Lu J. Lu S. Yao Y. Li C. Wang X. Zhou Q. Dalton Trans. 2019;48:6492. doi: 10.1039/C9DT00441F. [DOI] [PubMed] [Google Scholar]; (b) Zhou Q. Zheng Y. Wang T. Chen Y. Li K. Zhang Y. Li C. Hou Y. Wang X. Chem. Commun. 2015;51:10684. doi: 10.1039/C5CC03291A. [DOI] [PubMed] [Google Scholar]; (c) Singh T. N. Turro C. Inorg. Chem. 2004;43:7260. doi: 10.1021/ic049075k. [DOI] [PubMed] [Google Scholar]; (d) Betanzos-Lara S. Salassa L. Habtemariam A. Sadler P. J. Chem. Commun. 2009:6622. doi: 10.1039/B914153G. [DOI] [PubMed] [Google Scholar]
- (a) Purser S. Moore P. R. Swallow S. Gouverneur V. Chem. Soc. Rev. 2008;37:320. doi: 10.1039/B610213C. [DOI] [PubMed] [Google Scholar]; (b) Gillis E. P. Eastman K. J. Hill M. D. Donnelly D. J. Meanwell N. A. J. Med. Chem. 2015;58:8315. doi: 10.1021/acs.jmedchem.5b00258. [DOI] [PubMed] [Google Scholar]; (c) Hagmann W. K. J. Med. Chem. 2008;51:4359. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]; (d) Asahina Y. Araya I. Iwase K. Iinuma F. Hosaka M. Ishizaki T. J. Med. Chem. 2005;48:3443. doi: 10.1021/jm040204g. [DOI] [PubMed] [Google Scholar]
- Boerhan R. Sun W. Tian N. Wang Y. Lu J. Li C. Cheng X. Wang X. Zhou Q. Dalton Trans. 2019;48:12177. doi: 10.1039/C9DT01594A. [DOI] [PubMed] [Google Scholar]
- (a) Li G. Sun L. Ji L. Chao H. Dalton Trans. 2016;45:13261. doi: 10.1039/C6DT01624C. [DOI] [PubMed] [Google Scholar]; (b) Erkkila K. E. Odom D. T. Barton J. K. Chem. Rev. 1999;99:2777. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.