Abstract
With the emergence of Delta and Omicron variants, many other important variants of SARS-CoV-2, which cause Coronavirus disease-2019, including A.30, are reported to increase the concern created by the global pandemic. The A.30 variant, reported in Tanzania and other countries, harbors spike gene mutations that help this strain to bind more robustly and to escape neutralizing antibodies. The present study uses molecular modelling and simulation-based approaches to investigate the key features of this strain that result in greater infectivity. The protein-protein docking results for the spike protein demonstrated that additional interactions, particularly two salt-bridges formed by the mutated residue Lys484, increase binding affinity, while the loss of key residues at the N terminal domain (NTD) result in a change to binding conformation with monoclonal antibodies, thus escaping their neutralizing effects. Moreover, we deeply studied the atomic features of these binding complexes through molecular simulation, which revealed differential dynamics when compared to wild type. Analysis of the binding free energy using MM/GBSA revealed that the total binding free energy (TBE) for the wild type receptor-binding domain (RBD) complex was −58.25 kcal/mol in contrast to the A.30 RBD complex, which reported −65.59 kcal/mol. The higher TBE for the A.30 RBD complex signifies a more robust interaction between A.30 variant RBD with ACE2 than the wild type, allowing the variant to bind and spread more promptly. The BFE for the wild type NTD complex was calculated to be −65.76 kcal/mol, while the A.30 NTD complex was estimated to be −49.35 kcal/mol. This shows the impact of the reported substitutions and deletions in the NTD of A.30 variant, which consequently reduce the binding of mAb, allowing it to evade the immune response of the host. The reported results will aid the development of cross-protective drugs against SARS-CoV-2 and its variants.
Keywords: A.30 variant, Protein-protein docking, Dissociation constant, Simulation, Free energy
1. Introduction
COVID-19 is a disease instigated by a newly emerged, vastly infectious coronavirus, SARS-CoV-2, first reported in the city of Wuhan, China. The common symptoms of SARS-CoV-2 are fever, headache, shortness of breath, and cough. The spike protein acts as an entrance factor for the SARS-COV-2 infection in humans. There are six open reading frames in the coronavirus genome which encode for proteins, ordered from 5′ to 3′, M (membrane), E (envelope), S (spike), and N (nucleocapsid), as well as nonstructural proteins like protease and RNA-dependent RNA polymerase (RDRP) [1,2].
The coronavirus infection in cells begins when the spike protein binds to the host angiotensin-converting enzyme 2 (ACE2). Human ACE2 (hACE2) is mostly expressed in the lungs, kidneys, and small intestine, which might result in significant sickness [3]. After binding with ACE2, the host cell proteases split the SARS-COV-2 S-protein into the S1-ectodomain and S2 membrane-anchored domain, which are located at the N-terminal and C-terminal respectively. The S1 subunit aids in the recognition of cell surface receptors as well as facilitating virus entry into the cell [[4], [5], [6]]. The Mac-I domain of the SARS-COV-2 NSP3 protein has been identified as a promising therapeutic target due to its function in altering the innate immune response and increasing virulent qualities [7]. The Mac-1 domain of NSP3 was shown to enhance the response of interferons for viral neutralization and to play a critical role in viral infectivity [8]. Further studies revealed Mac-1 hijacking of the host immune response, which is accomplished by deactivation of the IFN pathway and suppression of STAT1 [9,10].
The SARS-CoV-2 are evolutionary gamblers that lacks a proofreading capabilities thus prone to enormous mutation frequency (million times > DNA-containing cells). Many variants of the SARS-CoV-2 virus have now been detected, as part of the ongoing worldwide pandemic, and have been associated with rapid transmissibility, evasion of immune responses, high occurrence of indisposition, and recurrence [11]. There are several variants of concern (VOC) including the alpha (B.1.1.7) variant, with 40–80% enhanced transmission, as well as beta (B.1.1.351), gamma, delta, and B.1.1.529 variants [12,13]. K417T, E484K, and N501Y mutations were detected in the receptor binding domain (RBD) of the P.1 variant reported in early 2021 in Brazil, which showed enhanced lethality, by 50%, and transmission, by 38% [14]. Another strain with significantly increased lethality was discovered in India in late 2020, officially known as B.1.617.2, which exhibited transmissibility enhanced by 87% [15].
The recently emerged B.1.1.529 strain of SARS-CoV-2 arose in November in South Africa and has spread to other countries, posing a serious public health threat. The spike protein of the B.1.1.529 strain has 30 different alterations including two deletions and seven substitutions found in the N-terminal domain (NTD), and fifteen substitutions found in the RBD. Several other strains, including new variants, have been identified to have the B.1.1.529 spike protein, which might affect SARS-CoV-2 transmission and infection. Recently, using structural modelling and simulation approaches, it has been reported that the binding affinity of B.1.1.529 spike protein RBD is enhanced for ACE2 and reduced for mAb [16].
Strain A.VOI.V2, also known as the A.30 variant, was first reported in several patients in Sweden and Angola in 2021, and reportedly originates from Tanzania. According to a recently published report on the cellular and molecular immunology of the A.30 variant, it evades vaccine-induced antibodies and has enhanced extra pulmonary spread, which is a great concern for public health response. The A.30 variant has 5 deletion and 10 substitution mutations in the N terminal domain of the S1 protein, which acts as an antigenic epitope and can be bound by neutralizing antibodies [17]. Furthermore, three mutations were discovered in the RBD, which interacts with the human ACE2 receptor and is a primary target of antibodies for neutralization. T478R and E484K are two of these mutations, occurring at the ACE2 binding site; however, the letter is responsible to develop resistance to antibody-mediated neutralization [18].
The application of molecular modelling and simulations in deciphering the molecular mechanism of pathogenesis for different variants of SARS-CoV-2, and therapeutics development has been invaluable. Here, we have also employed bio-molecular and simulation tools to comprehend the impact of these mutations on the binding of RBD to the host receptor ACE2, and of monoclonal antibodies to the NTD of A.30 variant. Protein coupling, simulation, and post-simulation investigations demonstrated the binding and dynamic features of this variant in contrast to the wild type. We explored the binding variations between the wild type and A.30 variant RBD and NTD interaction with the host receptor ACE2 and mAb. The current study will aid the development of prophylactic therapeutics using these key features for structure-based and rational drug design.
2. Methodology
2.1. Structure retrieval and mutant modelling
The spike protein, which is required for viral interaction with the host cell, was reported to have mutations in emerging strains. The recently submitted SARS-CoV-2 amino acid sequence was retrieved from the UniProt database (accession number P0DTC2) for identification of mutation loci in newly evolved variants [19]. The wild type protein structure of SARS-CoV-2 spike protein (6M0J) was retrieved from the PDB database and AlphFold2 was used to model the predicted mutations in the wild type protein structure [20,21]. The NTD and mAb structures were also collected from RCSB using 7C2L accession number [20].
2.2. Binding network and dissociation constant (KD) determination
The HADDOCK (high ambiguity-driven protein-protein docking) online server was used to investigate the binding differences between the wild type and novel strain A.30 RBD and NTD [22,23]. A guru interface was used for the docking where all the available features are executed for the best docking. A restraint docking by defining the interface residues i.e. 21, 24, 27, 28, 30, 35, 38, 79, 80, 82, 83, and 353 for ACE2, 449, 453, 455, 456, 486, 487, 489, 493, 496, 498, 500, 501, 502, and 505 for RBD, 25–32, 25–32, 51–58, and 100–116 for mAbs, while a region between 145 and 150 was defined for NTD. This server generate different docking conformations, which are then clustered according to the related orientations, and the final conformation is based on higher number of cluster size and lowest Z score. The PDBsum online server (https://www.ebi.ac.uk/) was used to determine the different binding networks (hydrogen bonds, salt bridge and non-bonded contacts) between the wild type and mutant proteins [24]. Subsequently, to provide more accurate information about the strength of the binding complex between the spike protein and ACE2 receptor of both mutant and wild type strains, we used the online server PROtein binDIng enerGY prediction (PRODIGY) to calculate the dissociation constant (KD) and the binding affinity [25].
2.3. Molecular dynamics simulation
The molecular dynamics behavior of both wild type and mutant complexes was analyzed with the AMBER20 package, which uses the FF19SB force field [[26], [27], [28]]. The system was neutralized by 23 sodium ions added to each system and the system solvation was performed by using the OPC (optimal point charge) water box model (9180 water molecules). Afterward, the system energy was minimized for the removal of bad clashes. 3000 cycles of conjugate gradient algorithm and 6000 cycles of steepest descent algorithm were used and, after heating to 300 K, 1 atm of constant pressure with weak restraint was used for system equilibration [29,30]. Finally, the 300 ns production was run [31]. A time step 0.02 was used for the simulation. The CPPTRAJ package of AMBER20 was used to analyze trajectories and CUDA was used to run the simulation [32]. For structural stability, root mean square deviation (RMSD) analysis, as a function of time, was performed using the following equation:
| (1) |
Where:
di = position difference between atoms,
-
i
= reference and superimposed structure.
The residues flexibility was indexed by estimating the root mean square fluctuation (RMSF) employing B-factor [33], which is the most imperative constraint for calculating the flexibility of all residues in a protein. Numerically, the flexibility can be calculated with the given equation:
| (2) |
The radius of gyration (Rg) measures an overall size of the protein during simulations. For the calculation of radius of gyration, the following equation was used.
| (3) |
Where.
mi = mass of the atom i,
ri = distance of atom i.
2.4. Analysis of total binding free energies
MM/GBSA method, which is reported to be the most accepted protocol for the estimation of the complex free energy in molecular interactions. The protocol calculated the BFE for the WT and mutant RBD in association with hACE2 [[33], [34], [35], [36], [37]].
| (4) |
Different contributing components of total binding energy were calculated by the following equation:
| (5) |
It has a wide range of applications i.e. used to estimate the binding energy for proteins in different studies including SARS-CoV-2 and neurological disorders [[38], [39], [40], [41], [42], [43]].
3. Results and discussion
3.1. Structural modelling and analysis
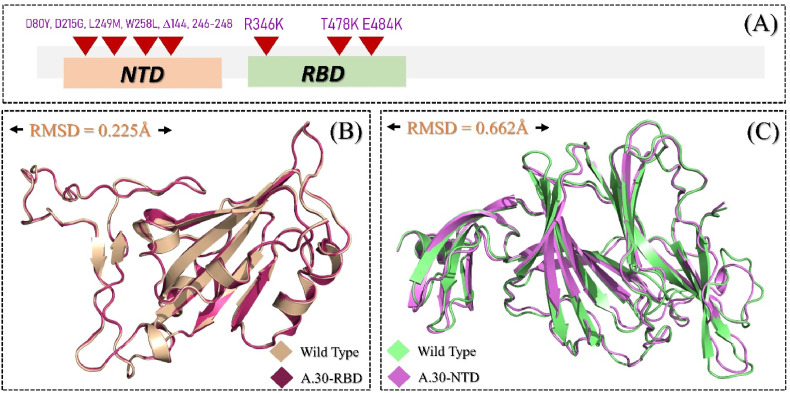
The emergence of more lethal variants of SARS-CoV-2, that can also have increased transmissibility, chance of re-infection, and evasion of the immune response, has been ongoing since the global pandemic was declared in March 2020 and unceasing reports of emerging variants has aggravated global public health. Many variants, including B.1.1.7, B.1.1.529, P.1, B.1.351, have been reported in different geographic locations around the world with a diverse set of mutations that help the virus to improve its survivability. Recently, a heavily mutated strain of SARS-CoV-2, formally known as A.30, has been reported to have significantly increased transmissibility and immune evasion. This variant has R346K, T478K and E484K mutations in the RBD of the spike protein that help the virus to increase binding affinity and, therefore, infectivity. Additionally, D80Y, D215G, L249 M, and W258L mutations, and deletions at loci 144, 246, 247 and 248, have been reported in the NTD. The distribution of these mutations in the spike protein is shown in Fig. 1 A.
Fig. 1.
(A) Mutational landscape of A.30 variant RBD and NTD. (B) Superimposed structure of the wild type RBD and A.30 RBD with R346K, T478K and E484K mutations. (C) Demonstrates the superimposed structures of the wild type NTD and A.30 NTD.
Although previous studies have classified A.30 as an antibody escaping strain, however, atomic-level insights are needed to understand the key differences between the wild type and A.30 variants to guide structure-based drug design. In this study, we investigated the effect of these unique mutations on the structure, function, and binding of the RBD to ACE2, and of mAbs to the spike protein NTD, in great detail. To illustrate the interaction variations and associate them with the infection rates of the A.30 variant, the present work combines integrative protein-protein modelling and molecular simulation techniques. Prior to protein-protein docking, AlphFold2 was used to model the 3D structures of A.30 RBD and NTD for comparison with wild type. A RMSD difference of 0.225 Å was seen between the wild type and A.30 RBDs, while a 0.662 Å RMSD difference was recorded between NTDs. This indicates that the variants have undergone secondary structural reconfiguration, resulting in a new strategy for binding and infection. The reference and modelled structures were superimposed (wild type-RBD, A.30-RBD, wild type-NTD, and A.30-NTD) as shown in Fig. 1B and C.
3.2. Protein-protein docking of the wild type and A.30 variant RBDs with human ACE2
Prediction of protein complexes for bound ACE2 and RBD was carried out using HADDOCK web docking server. The binding interfaces for RBD-ACE2 and NTD-mAb were defined for constraint docking and accuracy. Nine residues of hACE2 and 13 residues of the RBD directly interacted with each other. The regions 140–150 were defined for the NTD and the three essential regions, CDR1-3 (25–32), (51–58), and (100–116), from mAb were selected as the binding interface residues for mAb and NTD. Previously these residues have been used to study the interaction of RBD from different variants such as Alpha, delta, delta+, and Omicron variants [20,44,45].
For the wild type RBD-ACE2 complex, the docking score was predicted to be −111.8 ± 1.5 kcal/mol, which has been reported previously by other studies [16,35,46,47]. In contrast, the docking prediction for the A.30 RBD-ACE2 complex reported a value of −129.6 ± 1.8 kcal/mol ( Table 1 ). The variations in the docking scores demonstrate the impact these substitutions have on the function, with a higher binding affinity seen for the A.30 RBD towards ACE2. This report strongly aligns with the already published data on other SARS-CoV-2 variants, including B.1.1.7, B.1.351, P.1, B.1.617, and B.1.618, where the spike mutations, particularly in the RBD, decreased the docking score compared to wild type [35,44,[47], [48], [49]].
Table 1.
Estimated docking affinities and KD (dissociation constant) for wild type RBD, wild type NTD, A.30 RBD and A.30 NTD.
| Parameters | Wild Type-RBD | A.30-RBD | Wild Type-NTD | A.30-NTD |
|---|---|---|---|---|
| HADDOCK score | −111.8 ± 1.5 | −129.6 ± 1.8 | −63.6 ± 5.2 | −51.4 ± 1.3 |
| Cluster size | 51 | 160 | 9 | 8 |
| RMSD (Å) | 14.6 ± 0.2 | 14.5 ± 0.4 | 13.0 ± 0.3 | 21.6 ± 0.3 |
| Van der Waals energy | −48.1 ± 1.3 | −54.0 ± 4.1 | −55.2 ± 4.6 | −34.8 ± 5.6 |
| Electrostatic energy | −169.7 ± 13.2 | −248.1 ± 31.6 | −131.8 ± 17.0 | −165.5 ± 23.1 |
| Desolvation energy | −30.0 ± 3.4 | −26.3 ± 5.2 | −1.6 ± 1.7 | −4.8 ± 2.8 |
| Restraint's violation energy | 1.9 ± 1.1 | 3.1 ± 4.0 | 195.5 ± 40.9 | 26.6 ± 24.7 |
| Buried surface area (A2) | 1661.1 ± 57.7 | 1829 ± 72.9 | 1341.8 ± 21.7 | 1105.2 ± 80.7 |
| Z-score | −1.6 | −1.7 | −1.5 | 0.7 |
| KD (dissociation constant) | 3.2E−09 | 1.3E−10 | 1.9E−09 | 5.4E−08 |
| Ref | [16] | [16] |
We further compared the van der Waals (vdW) and electrostatic energies for both complexes, where wild type reported a vdW of −48.1 ± −1.3 kcal/mol, while the A.30 complex demonstrated a vdW of −54.0 ± −4.1 kcal/mol. When determining the electrostatic energy, the wild type complex reported −169.7 ± −13.2 kcal/mol and for the A.30 complex, −248.1 ± −31.6 kcal/mol. This suggests that the higher binding affinity of the A.30 variant is due to the enhanced contribution by vdW and electrostatic energy. Increased electrostatic energy has been seen in other variants and is regarded as the main cause of stronger interactions in these complexes [16,35,44,47,49].
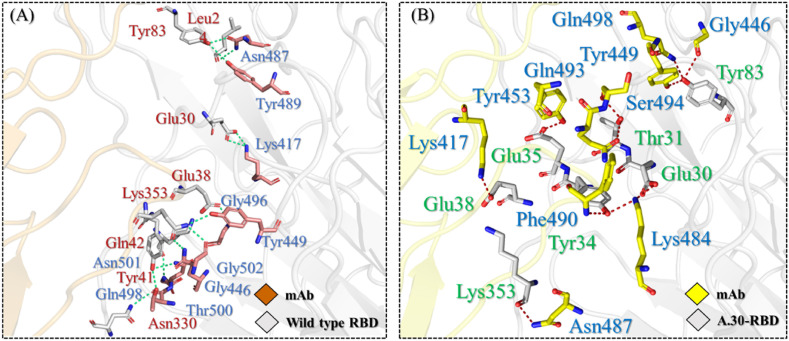
Further study of these complexes showed both had 11 hydrogen bonds, however, wild type RBD-ACE2 had only one salt bridge with hACE2, while the A.30 variant complex was reported to have three. Both the complexes reported some conserved interactions, while additional interactions were also seen in the A.30 RBD-ACE2 complex only. As given in Fig. 2 A, the interaction Tyr83-As487 was seen in the wild type but altered in the A.30 complex where Tyr83 was shown to interact with Gln498 and Glu496. The interaction of Lys417 in the RBD for both wild type and A.30 variant was strongly conserved for its interaction with Glu30. A single salt bridge between Glu30-Lys417 was seen in both complexes, while two additional salt bridges between Glu38-Lys484 were reported in the A.30 complex. Additionally, the interaction Glu35-Arg493 was only reported in the A.30 variant and has been shown to be accountable for the anchor locking and correct orientation of the RBD when binding to ACE2 [35,50]. Likewise, the hydrogen bond established by Lys353 in each complex determines the role of these residues to recognize and bind to ACE2 [35,50]. The conserved salt bridges indicate the functional significance of Glu38 and the substitution Lys484 that essentially increase the binding of wild type and various variants [35,47,49]. The binding variations displayed by both the complexes, and particularly for the A.30 variant, shows the importance of key substitutions and consequent binding alteration, forming an altered approach towards binding and host cell entrance. The 3D-interactions of wild type RBD and A.30 RBD complexes are shown in Fig. 2A and B. The docking scores for each complex are given in Table 1.
Fig. 2.
Interaction profiling of wild type and A.30 RBD-ACE2 complexes. (A) Shows the binding mode of wild type-RBD with ACE2. (B) Shows the binding mode of A.30-RBD with ACE2.
3.3. Protein-protein docking of the wild type and A.30 NTD variant with mAb
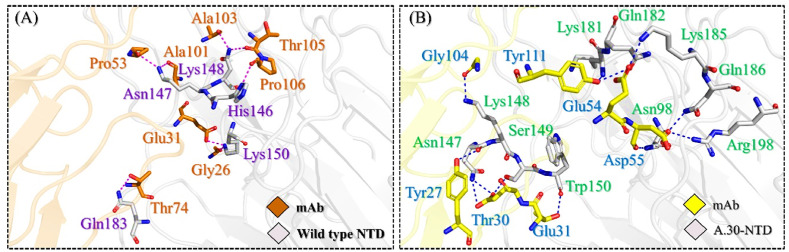
Evaluation of the binding variations between the wild type and A.30 NTDs with mAb was also performed using the HADDOCK docking approach. The wild type NTD-mAb complex demonstrated a docking score of −63.6 ± 5.2 kcal/mol, while the A.30 complex reported −51.4 ± 1.3 kcal/mol. This variation in the HADDOCK docking scores demonstrated conformational changes persuaded by the deletions and replacement of key amino acids, which consequently, altered binding. Investigation into the binding differences discovered the impact of the mutations and deletions on the changes in binding conformations. Comparative investigation of the wild type and A.30 NTD-ACE2 complexes show that the key interactions required for recognition and processing by mAbs are lost in the A.30 complex. The variations in the A.30 NTD induced different binding dynamics in the CDR regions of mAb compared to wild type. In the key binding regions of NTD (amino acids 140–150) and CDR 1–3 of the mAbs, A.30 was shown to have lost interactions that are sustained in the wild type. Earlier studies have shown that the deletion of two key residues, Tyr145 and, to a greater extent, His146, reduces the identification and targeting of the B.1.618 variant, corroborate the hypothesis that the deletion of key residues helps the virus to evade neutralization by escaping the antibody response [47]. The interaction patterns of the wild type and the A.30 NTD in complex with mAb are shown in Fig. 3 A and B. The docking scores for each complex are given in Table 1.
Fig. 3.
The comparative binding analysis of wild type and A.30 NTD to the mAb. (A) Shows the binding mode of wild type NTD with mAb while (B) demonstrates the binding of A.30 NTD with mAb.
3.4. Determining the dissociation constant (KD) for RBD and NTD
The binding strength of the two A.30 complexes, RBD-ACE2 and NTD-mAb, was estimated through KD prediction which has been previously used for other variants such as B.1.1.7, B.1.351, P.1, B.1.617, and B.1.618 [35,47,49]. This study, which employed the same approach as these previous reports, revealed a KD of 1.3 E−10 for the A.30 RBD-ACE2 complex and 1.3 E−10 and 5.4 E−08 for the NTD-mAb complex. When compared, the RBD of A.30 binds more strongly than the wild type, while the NTD of the wild type binds more strongly than that of A.30. The results strongly corroborate the docking score analysis as well as earlier studies based on other variants [35,47,49]. The docking scores and KD values for the wild type RBD and NTD, and the A.30 RBD and NTD complexes are given in Table 1.
4. Conformational dynamics of the wild type and A.30 RBD
4.1. Dynamics stability analysis of RBD
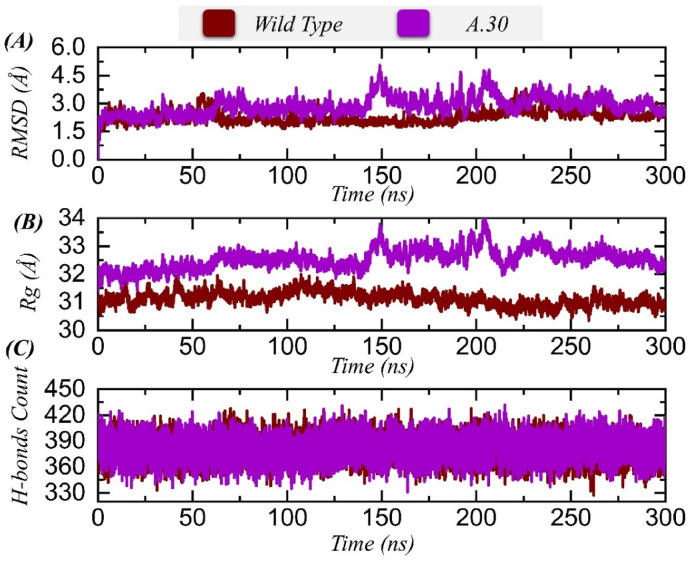
The conformational dynamics of wild type and A.30 variant were investigated through root mean square deviation (RMSD) based on carbon alpha atoms. This analysis is fundamental in understanding the binding affinity of wild and variant spike proteins to the ACE2 receptor and is vital for explaining the effects of mutations on the structure, function, and overall binding characteristics of the spike protein. This is also important in deciphering the virus infection ability in terms of attachment, replication, and transmission. The RMSD was estimated for a 300 ns (ns) period and is presented in Fig. 4 A. As shown in Fig. 4A, the wild type displayed constant equilibrium and revealed stable dynamics with a net RMSD of ∼1 Å (Å) throughout the simulation time. The maximum RMSD reported for the wild type spike system was ∼1.5 nm at 150 ns? This suggests that the intermolecular interactions were strong, keeping the binding conformation stable. For the A.30 RBD, similar dynamics were seen, however, a short period of instability occurred between 150 and 200 ns? These variations corresponded to structural adjustments to achieve better binding conformation between the molecules and involved the binding/unbinding of chemical interactions. However, good binding stability between the variant RBD and ACE2 was seen towards the end of the period, showing stable dynamics for the system.
Fig. 4.
Structural and dynamic stability analysis of wild type/A.30 RBD-ACE2 predicted by RMSD analysis. (A) shows the RMSD of wild type/A.30 complexes, (B) Rg plot for wild type/A.30 RBD variants, (C) hydrogen bonds analysis of the wild type and variant.
Our results are consistent with previous research that shows that RBD global stability leads to a greater ACE2 binding capacity [51]. Furthermore, earlier research has shown a close link between the RBD structural stability and binding affinity, with mutations that improve structural stability and stiffness accompanied by increases in binding affinity [52,53]. Other investigations have shown that the mutation C432D in the RBD reduced ACE2 mediated entrance through destabilization of the RBD structure [51]. Recent research on the variants B.1.1.7, B.1.351, P.1, B.1.617, and B.1.618 demonstrated an improvement in structural stability that is significantly linked to variant stability and stronger binding affinity in these variants [35,[47], [48], [49]]. These data substantially support our findings, which demonstrated that A.30 structural stability has increased along with its binding affinity, following the global pattern of greater stability and binding affinity shown in other variants.
The change in the dynamics of A.30 compared to that of wild type might be an adaptation for evading neutralizing antibodies. This is supported by a very recent study demonstrating that humoral antibody response is impaired due to A.30 mutations [54]. These dynamics may also be helpful for the A.30 virus in retaining high transmissibility due to the presence of P681H and D614G mutations. Previously, said mutations were shown to enhance the strain's ability to infect and transmit more aggressively than the parent strain [54]. The A.30 mutations are further reported to allow close interactions between the HR1 and HR2 domains of the virus spike, resulting in the virus fusion with the host cells and initiation of a strong infection cycle [55].
4.2. Structural compactness analysis of RBD
To validate the RMSD findings, radius of gyration (Rg) analysis was performed to examine the system's compactness over time (Fig. 4B). As with RMSD, Rg illustrated that the wild type system is more compact and did not experience any major relaxation event in the structure during the simulation time. The mean and maximum Rg of the wild type spike-ACE2 complex was 30.8 Å, while for the A.30 variant; the complex underwent deviation at the same points seen in RMSD. Again, these changes were due to unstable binding in the start and middle period of the simulation time but binding stability was gained later. The mean and maximum Rg of the variant spike-ACE2 complex was 32.5 Å.
4.3. Hydrogen bonding analysis of RBD
The protein-protein complexes are holded by hydrogen and hydrophobic bonds. of the interface is always surrounded by water molecules that also compete for hydrogen bonds [56,57]. Considering the higher importance of hydrogen bonds in protein coupling, it is worthy to desipher hydrogen bonding landscape of protein interfaces. For instance, the H-bonds understanding has been previously implemented for SARS-CoV-2 variants to reveal the binding differences. Hydrogen bonding is a very important stabilizing factor in biological systems. These forces are formed when a hydrogen atom is shared between strong electronegative molecules. Fig. 4C shows the number of hydrogen bonds over time, with the average the number of hydrogen bonds throughout reported to be 390 for the A.30 variant. Throughout the simulation time, both studied systems showed the maximum number of hydrogen bonding. This reveals that the interactions between wild type and A.30 variant spike proteins and ACE2 are enriched with strong and close distance hydrogen bonds, keeping both system dynamics stable. Post-simulation hydrogen bonding occupancy for each complex is given in Table 2 .
Table 2.
Post-simulation hydrogen bonding occupancy analysis for each complex.
| Index | Interaction | Wild type RBD % | A.30 RBD% | |
|---|---|---|---|---|
| 1. | LEU19-ALA475 | 34% | 39% | |
| 2. | ASP355-THR500 | 8% | 3% | |
| 3. | TYR83-ASN487 | 66% | 69% | |
| 4. | LYS353-GLY502 | 71% | 52% | |
| 5. | GLU38-GLY496 | 26% | 41% | |
| 6. | Tyr41-THR500 | 37% | 2% | |
| 7. | LYS353-TYR493 | 64% | 12% | |
| 8. | LYS353-TYR446 | 47% | 39% | |
| 9. | GLU38-LYS417 | 42% | 78% | |
| 10. | TYR83-GLY496 | 4% | 64% | |
| 11. |
TYR83-GLN498 |
12% |
51% |
|
| Index | Interactions | Wild type NTD % | Interactions | A.30 NTD% |
| 1. | GLY56-LEU249 | 28% | TYR27-ASN147 | 29% |
| 2. | PRO53-LYS147 | 36% | ASP55-ASN98 | 32% |
| 3. | ALA101-LYS147 | 54% | GLU31-ASN147 | 46% |
| 4. | ALA103-ASN148 | 69% | GLY104-LYS148 | 20% |
| 5. | THR105-ASN148 | 33% | ASP55-ASP176 | 33% |
| 6. | GLU31-LYS150 | 57% | GLY56-ASP176 | 15% |
| 7. | GLU31-LYS150 | 42% | TYR111-LYS180 | 31% |
| 8. | GLU54-LYS185 | 12% | ||
| 9. | ASP55-ASN186 | 17% | ||
4.4. Residue flexibility analysis of RBD
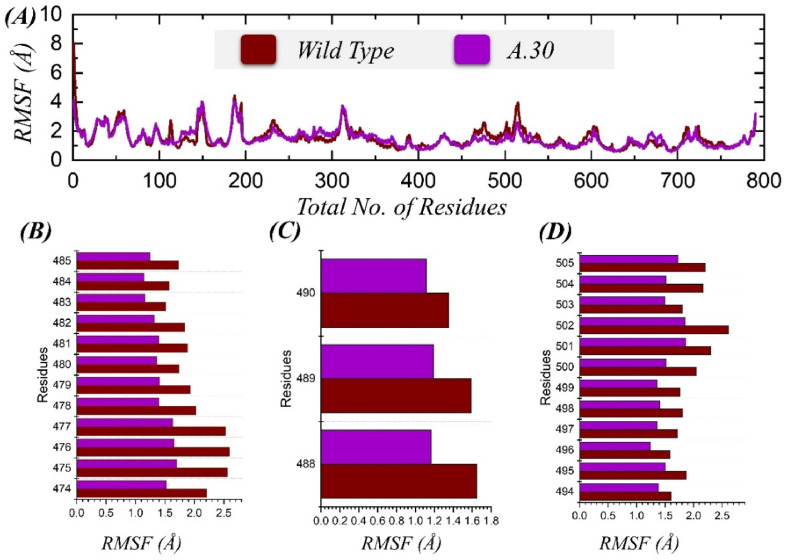
For understanding flexibility on the residue level, root mean square fluctuation (RMSF) was applied to the simulation trajectories of the systems. It is crucial to examine the deviations of a system's residues to conclude which residues are vital for holding the interaction between spike and ACE2 and providing stabilization to the overall complex. As shown in Fig. 5 A, the mean RMSF for the wild type system and A.30 system were 2.20 and 2.26 Å, respectively. Similarly, for the wild type spike-ACE2 complex, the maximum RMSF observed was at residues 100–200, 300–340, and 450–550, while for the A.30 complex, highly flexible residues also included the region of 650–680, alongside those seen for wild type. Additionally, we calculated the residue flexibility index for the three important loops in the RBD of wild type and A.30 variants, which come in direct contact with ACE2. As given in Fig. 5C-D, the three loops demonstrated differential values for residue flexibility index in each complex. These loops (residues 484–505) were more flexible in the wild type, with minimal fluctuation in the A.30 variant. This shows the A.30 loops flexibility is stabilized by binding with ACE2 and by the mutation induced variation in conformational dynamics. Hence, A.30 shows greater stability in binding ACE2 than the wild type.
Fig. 5.
(A) Residue flexibility of wild type and A.30 variants calculated as RMSF. (B–D) represent the flexibility index for the three important loops from residues 484–505.
5. Conformational dynamic of the wild type and A.30 NTD
5.1. Dynamics stability analysis of NTD
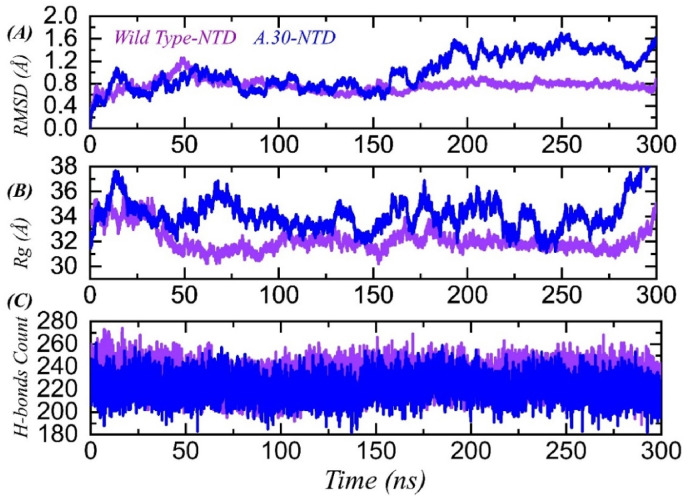
We further assessed the stability variations between the wild type and A.30 NTD in complex with mAb. As given in Fig. 6 A, the RMSD of both complexes remained comparable until 150 ns, where an increase was seen for the A.30 NTD-mAb complex. For wild type, the RMSD remained ∼0.85 Å during the first 150 ns, while during this period the RMSD for A.30 was also reported to be over 0.80 Å. After 150 ns, the wild type NTD gained more stability and demonstrated a uniform RMSD for the remaining period. The average RMSD for the wild type NTD-mAb complex was reported to be 0.9 Å. The RMSD for the A.30 NTD-mAb complex continued to increase gradually but reported more structural perturbation than the wild type. The average RMSD increased during the last 150 ns and was calculated to be 1.10 Å. Previously, similar findings were reported for other variants, such as B.1.1.7, B.1.1.617, B.1.1.618, and B.1.1.529, suggesting that the mutations and deletions which change the protein dynamics help the A.30 variant to escape the neutralizing antibodies [16,35,45,46].
Fig. 6.
Structural and dynamic stability analysis of wild type/A.30 NTD variant complexes with mAb, predicted by RMSD analysis. (A) shows the RMSD of wild type/A.30 NTD complexes, (B) Rg plot for wild type/A.30 NTD variants, (C) hydrogen bonds analysis of the wild type and A.30 variant NTD.
5.2. Structural compactness analysis of NTD
The radius of gyration for both the complexes reported a strong agreement with the RMSD results. As can be seen in Fig. 6B, the Rg graphs for the wild type and A.30 variant reported a similar pattern to RMSD. Wild type and A.30 complexes demonstrated comparable Rg during the first 150ns which then increased during the last 150ns for the A.30 complex only. This shows that the A.30 variant reported significant binding and unbinding events, thus destabilizing the neutralizing antibody, and consequently helping the virus to escape the immune response. These findings strongly align with the previous reports of the other variants [16,35,45,46]. The average Rg for the wild type complex was calculated to be 32.80 Å while for A.30 it was calculated to be 34.2 Å.
5.3. Hydrogen bonding analysis of NTD-mAb
Estimation of hydrogen bonding during the simulation provided information about the binding stability incurred by the hydrogen bonds. To calculate the total number of hydrogen bonds in the simulation trajectory, hydrogen bonding analysis was performed. As given in Fig. 6C, the wild type reported more hydrogen bonds than the A.30 NTD-mAb complex. In the wild type complex, the average number of hydrogen bonds was reported to be 248 while in the A.30 complex the average number of hydrogen bonds was reported to be 234. This shows that the mutations and deletions in the NTD of A.30 induce conformational changes that enable the virus to lose important contact with the mAb. Post-simulation hydrogen bonding occupancy for each complex is given in Table 2.
5.4. Residue flexibility analysis of NTD
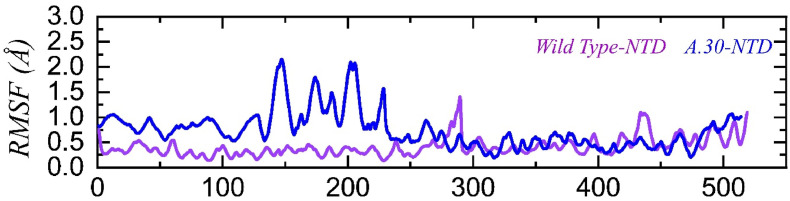
We also predicted the local level residue flexibility for the NTD of wild type and A.30 variants in complex with mAb. As shown in Fig. 7 , the wild type NTD is comparatively more stable than the A.30 NTD. In particular, the A.30 NTD demonstrated higher fluctuation between residues 1–240, while a less substantial fluctuation was displayed by the wild type NTD between residues 420–450. The difference in the flexibility index shows how conformational optimization by these mutations affects the change in binding strength. The RMSF (flexibility) graph for the wild type and A.30 NTD in complex with mAb are given in Fig. 7.
Fig. 7.
Residual flexibility of wild type and A.30 NTDs in complex with mAb, calculated as RMSF.
5.5. Binding free energy calculation for the wild type and A.30 complexes RBD-ACE2 & NTD-mAb
The MM/GBSA technique for computing the BFE of biological partners is a widely used method for examining the putative docking configuration. This method, which is less costly than the alchemical free energy methods, displays the binding stability of interacting key regions and the BFE. It is also regarded to be more precise than any rational scoring function. We employed the MM/GBSA technique because it allows us to see how the mutations in the spike RBD influence the binding with hACE2 and NTD with mAb. The BFE results are given in Table 3 .
Table 3.
MM/GBSA results, which show the binding free energy for each complex. All the energies are represented in kcal/mol.
5.5.1. Binding free energy for RBD-ACE2 complexes
As given in Table 3, the vdW for the wild type RBD-ACE2 complex was estimated to be −87.75 kcal/mol while for the A.30 complex the vdW was reported to be −99.23 kcal/mol. Moreover, for the wild type complex, the electrostatic energy was estimated to be −616.79 kcal/mol, whereas the A.30 complex was estimated to have electrostatic energy of −1168.78 kcal/mol. Previously, a higher electrostatic energy for the variant has also been reported and was considered the prime factor in its enhanced binding to ACE2 [35,47,49]. Finally, BFE for the wild type complex was −58.25 kcal/mol in contrast to the A.30 complex, which reported −65.59 kcal/mol. The higher BFE for the A.30 complex implies vigorous binding with the ACE2 than for wild type, allowing the variant to bind and spread more promptly.
5.5.2. Binding free energy for NTD-mAb complexes
The BFE for NTD-mAb complexes revealed contrasting results to the above RBD-ACE2 complexes. The BFE for the wild type NTD-mAb complex was computed to be −65.76 kcal/mol while for the A.30 complex it was −49.35 kcal/mol. This shows the impact of the reported substitutions and deletions in the NTD of A.30 variant which reduce the binding affinity of mAb towards NTD and helps in evading the immune response instigated by the host. Our findings corroborate with an earlier study that showed that mutations and deletions in the NTD of B.1.618 variants led to immune evasion [47].
6. Conclusions
In conclusion, the current study employed protein-protein coupling and molecular simulation approaches to decipher the key features required for stronger interaction with the ACE2 and escaping the neutralizing antibodies for the A.30 variant. We reported that the key substitutions altered the binding modes of RBD and NTD towards ACE2 and mAb. The only limitation of the current study is long run replicated simulations are missing which could further provide deep understanding on the reproducible results and basis for accurate interactions. The explored key features can be considered while designing novel therapeutics against this variant. These data will aid in the development of cross-protective drugs against SARS-CoV-2 and its variants.
Funding
Dong-Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2021ZD02).
Availability of data and material
All the data is available on RCSB, UniProt and any simulation data would be provided on reasonable demand. The accession numbers to access this data are given in the manuscript.
Ethics approval and consent to participate
N/A.
Consent for publication
N/A.
Conflicts of interest
Declared none.
Declaration of competing interest
Authors declare there is no declaration of interest.
Acknowledgments
The computations were partially performed at the PengCheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University.
References
- 1.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V.J.N.R.M. Coronavirus biology and replication: implications for. SARS-CoV-2. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontanet A., Autran B., Lina B., Kieny M.P., Karim S.S.A., Sridhar D.J.T.L. SARS-CoV-2 variants and ending the COVID-19 pandemic. 2021;397:952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus evolution. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain I., Pervaiz N., Khan A., Saleem S., Shireen H., Wei D.-Q., Labrie V., Bao Y., Abbasi A.A. Genes & Immunity; 2020. Evolutionary and Structural Analysis of SARS-CoV-2 Specific Evasion of Host Immunity; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosey C.A., Houl J.H., Katsonis P., Balapiti-Modarage L.P.F., Bommagani S., Arvai A., Moiani D., Bacolla A., Link T., Warden L.S., Lichtarge O., Jones D.E., Ahmed Z., Tainer J.A. Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors. Prog. Biophys. Mol. Biol. 2021;163:171–186. doi: 10.1016/j.pbiomolbio.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin M.-H., Chang S.-C., Chiu Y.-C., Jiang B.-C., Wu T.-H., Hsu C.-H. Structural, biophysical, and biochemical elucidation of the SARS-CoV-2 nonstructural protein 3 macro domain. ACS Infect. Dis. 2020;6:2970–2978. doi: 10.1021/acsinfecdis.0c00441. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12:360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duong D. Can Med Assoc; 2021. Alpha, Beta, Delta, Gamma: What's Important to Know about SARS-CoV-2 Variants of Concern? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng B., Kemp S.A., Papa G., Datir R., Ferreira I.A., Marelli S., Harvey W.T., Lytras S., Mohamed A., Gallo G. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B. 1.1. 7. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauring A.S., Malani P.N. Variants of SARS-CoV-2. JAMA. 2021;326:880. doi: 10.1001/jama.2021.14181. [DOI] [PubMed] [Google Scholar]
- 15.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A., Datir R., Collier D.A., Albecka A., Singh S., SARS-CoV-2 B. 1.617 2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan A., Waris H., Rafique M., Suleman M., Mohammad A., Ali S.S., Khan T., Waheed Y., Liao C., Wei D.-Q. The Omicron (B. 1.1. 529) variant of SARS-CoV-2 binds to the hACE2 receptor more strongly and escapes the antibody response: insights from structural and simulation data. Int. J. Biol. Macromol. 2022;200 doi: 10.1016/j.ijbiomac.2022.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., Zepeda S., di Iulio J., Bowen J.E., Montiel-Ruiz M., Zhou J., Rosen L.E., Bianchi S., Guarino B., Fregni C.S., Abdelnabi R., Foo S.C., Rothlauf P.W., Bloyet L.M., Benigni F., Cameroni E., Neyts J., Riva A., Snell G., Telenti A., Whelan S.P.J., Virgin H.W., Corti D., Pizzuto M.S., Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.028. 2332-2347.e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora P., Rocha C., Kempf A., Nehlmeier I., Graichen L., Winkler M.S., Lier M., Schulz S., Jäck H.-M., Cossmann A., Stankov M.V., Behrens G.M.N., Pöhlmann S., Hoffmann M. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 2021;18:2673–2675. doi: 10.1038/s41423-021-00779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magrane M. 2011. UniProt Knowledgebase: a Hub of Integrated Protein Data. 2011. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021:1. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein− protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 23.Koukos P.I., Reau M.F., Bonvin A.M. bioRxiv; 2021. Shape-restrained Modelling of Protein-Small Molecule Complexes with HADDOCK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 26.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., III, DeBolt S., Ferguson D., Seibel G., Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 28.Salomon‐Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- 29.Meza J.C. Steepest descent. Wiley Interdisciplinary Reviews: Comput. Stat. 2010;2:719–722. [Google Scholar]
- 30.Watowich S.J., Meyer E.S., Hagstrom R., Josephs R. A stable, rapidly converging conjugate gradient method for energy minimization. J. Comput. Chem. 1988;9:650–661. [Google Scholar]
- 31.Salomon-Ferrer R., Gotz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theor. Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 32.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 33.Khan A., Kaushik A.C., Ali S.S., Ahmad N., Wei D.-Q. Deep-learning-based target screening and similarity search for the predicted inhibitors of the pathways in Parkinson's disease. RSC Adv. 2019;9:10326–10339. doi: 10.1039/c9ra01007f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ali A., Khan M.T., Khan A., Ali S., Chinnasamy S., Akhtar K., Shafiq A., Wei D.-Q. Pyrazinamide resistance of novel mutations in pncA and their dynamic behavior. RSC Adv. 2020;10:35565–35573. doi: 10.1039/d0ra06072k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan A., Khan M.T., Saleem S., Junaid M., Ali A., Ali S.S., Khan M., Wei D.-Q. Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020;18 doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahir ul Qamar M., Ahmad S., Khan A., Mirza M.U., Ahmad S., Abro A., Chen L.-L., Almatroudi A., Wei D.-Q. Structural probing of HapR to identify potent phytochemicals to control Vibrio cholera through integrated computational approaches. Comput. Biol. Med. 2021 doi: 10.1016/j.compbiomed.2021.104929. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Khan A., Chandra Kaushik A., Junaid M., Zhang X., Wei D.-Q. The systematic modeling studies and free energy calculations of the phenazine compounds as anti-tuberculosis agents. J. Biomol. Struct. Dyn. 2019;37:4051–4069. doi: 10.1080/07391102.2018.1537896. [DOI] [PubMed] [Google Scholar]
- 38.Arshia A.H., Shadravan S., Solhjoo A., Sakhteman A., Sami A. De novo design of novel protease inhibitor candidates in the treatment of SARS-CoV-2 using deep learning, docking, and molecular dynamic simulations. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu T., Li F., Zhang Y., Yin J., Qiu W., Li X., Liu X., Xin W., Wang C., Yu L., Gao J., Zheng Q., Zeng S., Zhu F. Varidt 2.0: structural variability of drug transporter. Nucleic Acids Res. 2022;50:D1417–d1431. doi: 10.1093/nar/gkab1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu T., Zheng G., Tu G., Yang F., Chen Y., Yao X., Li X., Xue W., Zhu F. Exploring the binding mechanism of metabotropic glutamate receptor 5 negative allosteric modulators in clinical trials by molecular dynamics simulations. ACS Chem. Neurosci. 2018;9:1492–1502. doi: 10.1021/acschemneuro.8b00059. [DOI] [PubMed] [Google Scholar]
- 41.Jomhori M., Mosaddeghi H., Farzin H. Tracking the interaction between single-wall carbon nanotube and SARS-Cov-2 spike glycoprotein: a molecular dynamics simulations study. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue W., Yang F., Wang P., Zheng G., Chen Y., Yao X., Zhu F. What contributes to serotonin-norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018;9:1128–1140. doi: 10.1021/acschemneuro.7b00490. [DOI] [PubMed] [Google Scholar]
- 43.Xue W., Fu T., Deng S., Yang F., Yang J., Zhu F. Molecular mechanism for the allosteric inhibition of the human serotonin transporter by antidepressant escitalopram. ACS Chem. Neurosci. 2022;13:340–351. doi: 10.1021/acschemneuro.1c00694. [DOI] [PubMed] [Google Scholar]
- 44.Suleman M., Yousafi Q., Ali J., Ali S.S., Hussain Z., Ali S., Waseem M., Iqbal A., Ahmad S., Khan A. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z., Safdar M., Abubaker J., Uddin N.N., Cao L., Wei D.-Q., Mohammad A. The SARS-CoV-2 B.1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11:30132–30147. doi: 10.1039/d1ra04694b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236 doi: 10.1002/jcp.30367. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z. The SARS-CoV-2 B. 1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11:30132–30147. doi: 10.1039/d1ra04694b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celik I., Yadav R., Duzgun Z., Albogami S., El-Shehawi A.M., Fatimawali, Idroes R., Tallei T.E., Emran T.B. Interactions of the receptor binding domain of SARS-CoV-2 variants with hACE2: insights from molecular docking analysis and molecular dynamic simulation. Biology. 2021;10:880. doi: 10.3390/biology10090880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Andrade J., Gonçalves P.F.B., Netz P.A. Why does the novel coronavirus spike protein interact so strongly with the human ACE2? A thermodynamic answer. Chembiochem. 2021;22:865–875. doi: 10.1002/cbic.202000455. [DOI] [PubMed] [Google Scholar]
- 51.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davenport Thaddeus M., Gorman J., Joyce M.G., Zhou T., Soto C., Guttman M., Moquin S., Yang Y., Zhang B., Doria-Rose Nicole A., Hu S.-L., Mascola John R., Kwong Peter D., Lee Kelly K. Somatic hypermutation-induced changes in the structure and dynamics of HIV-1 broadly neutralizing antibodies. Structure. 2016;24:1346–1357. doi: 10.1016/j.str.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ovchinnikov V., Louveau J.E., Barton J.P., Karplus M., Chakraborty A.K. Role of framework mutations and antibody flexibility in the evolution of broadly neutralizing antibodies. Elife. 2018;7 doi: 10.7554/eLife.33038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arora P., Rocha C., Kempf A., Nehlmeier I., Graichen L., Winkler M.S., Lier M., Schulz S., Jäck H.-M., Cossmann A. The spike protein of SARS-CoV-2 variant A. 30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 2021;18:2673–2675. doi: 10.1038/s41423-021-00779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D., Oezguen N., Urvil P., Ferguson C., Dann S., Savidge T. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2(3) doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chodera J.D., Mobley D.L. Entropy-enthalpy compensation: role and ramifications in biomolecular ligand recognition and design. Annu. Rev. Biophys. 2013;42:121–142. doi: 10.1146/annurev-biophys-083012-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data is available on RCSB, UniProt and any simulation data would be provided on reasonable demand. The accession numbers to access this data are given in the manuscript.