Abstract
Genomic rearrangements in the 5′ part of the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) have been involved in multidrug resistance to nucleoside RT inhibitors (NRTI). We carried out a retrospective, multicenter study to investigate the prevalence, variability, and phenotypic consequences of such rearrangements. Data concerning the HIV-1 RT genotype and the biological and clinical characteristics of NRTI-treated patients were collected from 10 virology laboratories. Sensitivities of the different HIV-1 variants to RT inhibitors were analyzed in a single-cycle recombinant virus assay. Fifty-two of 2,152 (2.4%) RT sequences had a rearrangement in the 5′ part of the RT, with an extensive molecular variation. The number of codons inserted between positions 68 and 69 ranged from 1 (3 samples) or 2 (41 samples) to 5 and 11 in one case each. In four cases, codon 67 was deleted. High levels of phenotypic resistance to zidovudine (AZT), lamivudine (3TC), stavudine (d4T), abacavir (ABC), and didanosine (ddI) were found in 95, 92, 72, 62, and 15% of the 40 samples analyzed, respectively. Resistance to AZT, d4T, and ABC could be found in the absence of the T215Y/F mutations. Resistance to 3TC could develop in the absence of specific mutations. Low-level resistance to ddI was noticed in 40% of the patients. The deletions of codon 67 seemed to have little effect on NRTI sensitivity. Most of the rearrangements were shown to contribute to cross-resistance to NRTI. The results regarding susceptibility to ddI raise the question of the interpretation of the phenotypic data concerning this drug.
The efficacy of antiretroviral treatment can be impaired by several factors, including poor compliance with treatment regimens, suboptimal antiviral potency and drug concentrations, and selection of antiretroviral drug-resistant human immunodeficiency virus (HIV) quasispecies (11). The nucleoside analogue reverse transcriptase inhibitors (NRTI) were historically the first group of compounds used in anti-HIV therapy. They selected HIV type 1 (HIV-1) variants with mutations in the RT encoding region of the pol gene, which affected resistance to individual drugs (23). Moreover, two different pathways for the selection of HIV-1 showing multidrug resistance (MDR) to NRTI were identified. The first involves the Q151M mutation and four additional mutations (A62V, F77L, V75I, and F116Y) (12, 24). More recently, nucleotide rearrangements coding for different 1-amino-acid (aa) or 2-aa insertions, following position 68 or 69 of HIV-1 RT, have been reported to confer NRTI MDR (7, 8, 17, 19, 21, 22, 26, 29). Deletions at codon 67 associated with a T69G substitution have also been recently described (30; T. Imamichi, H. Imamichi, J. C. Lopez, J. Metcalf, J. Falloon, and H. C. Lane, Abstr. 7th Conf. Retrovir. Opportunistic Infect., abstr. 738, 2000). All these rearrangements involve the structure of the β3-β4 hairpin loop of HIV-1 RT and are likely to interact with the nucleotide binding process. Because previous reports concerning these rearrangements have been based on few cases, little is known about their prevalence, variability, molecular epidemiology, and clinical significance. We decided to carry out a multicenter, retrospective study in order to document these rearrangements emerging in the context of failure of antiretroviral therapy.
(This research was presented in part at the 3rd and 4th international workshops on HIV drug resistance and treatment strategies, June 1999, San Diego, Calif., and June 2000, Sitges, Spain.)
MATERIALS AND METHODS
RT genotype study.
A questionnaire regarding the molecular epidemiology of rearrangements in HIV-1 RT was sent to the virology laboratories belonging to the French Agence Nationale de Recherches sur le SIDA (ANRS) Antiretroviral Resistance study group. Data concerning the amino acid sequence between residues 60 to 71, additional resistance mutations, the treatment history, and the virological and immunological status of the patients were collected each time that a rearrangement was noticed in the RT encoding region. The number of patients with an RT rearrangement was compared, for each laboratory, to the total number of RTI-treated patients who had a documented HIV-1 genotype during the same period.
RT genotypic resistance studies were performed on viral plasma RNA using the ANRS consensus technique. Viral RNA was extracted using a standard guanidium isothiocyanate protocol and amplified in a one-step procedure using the RT-Titan kit (Boehringer) and the primers MJ3 and MJ4 (13). Amplified products were submitted to nested PCR using primers A-35 and Nel-35 (16). Direct population sequencing was performed on purified PCR products using the primers A-20 and Nel-20 (16) with the ABI PRISM DYE termination cycle sequencing Ready Reaction kit with AmpliTaq DNA polymerase (Perkin-Elmer) on an automated DNA sequencer (ABI Model 377; Applied Biosystems; Foster City, Calif.). Sequence alignement was performed with Sequence Navigator software (Perkin-Elmer).
Phenotypic analysis: recombinant virus RTI sensitivity assay.
Drug sensitivity assays were performed using a single-cycle recombinant virus assay (RVA) (20).
The HIV RT encoding region was amplified from patient plasma samples by nested RT PCR using the outer primers MJ3 (5′AGT AGG ACC TAC ACC TGT CA 3′) and RT-EXT (5′TTC CCA ATG CAT ATT GTG AG 3′) with inner primers A35 (5′ TTG GTT GCA TAA ATT TTC CCA TTA GTC CTA TT 3′) and RT-IN (5′ TTC CCA ATG CAT ATT GTG AG 3′). The resultant 1,530-bp fragment, extending between codon 93 of the protease-encoding region and codon 503 of RT-encoding region of pol, was purified on QiaAmp columns.
The plasmid with a deletion of RT (pSRT) was constructed from pNLenv, a previously described pNL4-3-derived plasmid carrying a near-full deletion of the env gene (bp 6343 to 7611) (5). Site-directed mutagenesis was used to alter the sequence to give the unique restriction sites SnaB1 at position 3872 and NruI at position 3892. BalI and SnaBI were used to remove the RT encoding region (between positions 2618 and 3872), and linearization was achieved using NruI.
Subconfluent 293T cells in T25 flasks were transfected by the calcium phosphate precipitation method with 8 μg of NruI-linearized pSRT, 0.1 μg of VSV-G plasmid (encoding for the vesicular stomitis virus envelope protein), and between 0.5 and 1 μg of the HIV reverse transcriptase PCR product.
The transfection precipitate was washed off the cells after 18 h of incubation, and fresh growth medium was added. After a further 24 h of culture, supernatant was clarified by centrifugation (500 × g, 15 min) and transferred to P4 indicator cells (3, 31) that had been preincubated with serial dilutions of RTI, in triplicate wells, for 4 h. The range of drug concentrations used varied between compounds. The level of expression of β-galactosidase in the P4 cell lysates was measured using a colorimetric assay based on the cleavage of chlorophenol red-β-d-galactopyranoside by β-galactosidase. The 50% inhibitory concentration (IC50) was calculated using the median effect equation (4). An RVA index was calculated as the ratio of the IC50 for the patient sample to the IC50 obtained for pNL4-3 wild-type HIV-1 tested in parallel.
Nucleotide sequence accession numbers.
The sequences reported in this study have been assigned the GenBank accession numbers AF315232 to AF315274, AF311177, AF311157, AF311187, AF311159, AF311179, AF311162, and AF311173.
RESULTS
Prevalence of RT rearrangements.
In the 10 laboratories participating in the study, 52 rearrangements were reported in the 5′ part (aa 20 to 240) of the RT encoding region from 52 different patients. In the same period (January 1998 to June 1999), 2,152 RT sequences from RTI-treated patients had been documented (prevalence of RT rearrangements, 2.4%). The prevalence in the different centers ranged from 0.8 to 4.5%; this variation was not significant (P = 0.22).
Molecular variability of the RT rearrangements.
The amino acid variability deduced from the nucleotide sequence of codons 60 to 71 of the RT encoding region is shown in Table 1. The number of inserted residues was 1 (3 samples), 2 (41 samples), 5 (1 sample) or 11 (1 sample). In all cases, insertions were located between aa 68 and 69. In four cases, a deletion of codon 67 was noted. The presence of an 11-aa insertion in one case could be confirmed by sequencing a later sample (data not shown).
TABLE 1.
Genotypic and phenotypic patterns of the HIV-1 variants in the studya
| Patient no. | HIV-1 RNA (copies/ml) | No. CD4+ (cells/μl) | Therapy
|
RT mutation(s) | Amino acid sequence
|
||
|---|---|---|---|---|---|---|---|
| RTI | PI(s) | 60–66 | 67/68 | ||||
| VFAIKKK | DS | ||||||
| 1 | 18,700 | 18 | d4T 3TC | RTV SQV | 1181 215Y | VFAIKKK | ES |
| 2 | 94,326 | 144 | d4T 3TC | RTV | 1841 215F 219Q | VFAINKK | GS |
| 3 | 49,400 | 298 | AZT 3TC NVP | 69D 7DR 181C 184I 219Q | VFAINKK | GY | |
| 4 | 40,421 | 548 | d4T ddI | IDV | 41L 215Y | VFAIKKK | A/DS |
| 5 | 26,045 | 311 | AZT ddI | NFV | 41L 103N 181C 215Y | VFAIKKK | ES |
| 6 | 221,500 | 81 | ABC ddI | NFV SQV | 98C 210W 215Y | IFAIKKK | EN/S |
| 7 | 47,500 | 60 | d4T 3TC | IDV | 41L 44D 118I 184I 215Y | VFVIKKK | SS |
| 8 | 5,100 | 172 | d4T 3TC | IDV | 41L 44D 118I 184I 210W 215Y | VFVIKKK | SS |
| 9 | 58,600 | 18 | d4T | NFV | 41L 74V 118I 190Q 210L 215Y 219Q | VFAIKKK | DS/NS |
| 10 | 202,653 | 1 | d4T ddI ABC | IDV | 41L 181C 215I | VFAIKKK | ES |
| 11 | 58,800 | 323 | Stop ARV | Stop ARV | 41L 181C 210W 215Y 219K/E | VFAIKKK | DS |
| 12 | 2,009,600 | 103 | d4T 3TC | NFV | 41L 210W 215Y | VFAIKKK | ES |
| 13 | 20,060 | 114 | d4T ddI | 41L44D 184I 210W 215Y | VFAIRKK | DS | |
| 14 | 46,650 | 54 | d4T ddI H. Urea | 41L 74V 75M 184V 210W 215Y | VFAIKKK | SL | |
| 15 | 23,680 | 166 | AZT 3TC | RTV | 41L 210W 215Y | VFAIRKK | DS |
| 16 | 50,000 | 34 | d4T ddI | NFV SQV | 98G 210W 215Y | VFAIKKK | DS |
| 17 | 272,015 | 20 | d4T ddI NVP | IDV | 41L 103N 181C 210W 215Y | VFAIRKK | DS |
| 18 | 416,096 | ND | 3TC ABC | 108I 184I 210W 215Y | VFAIRKK | QS | |
| 19 | 89,000 | 157 | d4T ABC EFV | APV | 41L 74V 181C 103N 190A 210W215Y | VFAIKKK | DS |
| 20 | 50,000 | 459 | d4T 3TC | IDV | 7OR 118I 184V 215F | VFVIKKK | DS |
| 21 | 247,000 | 10 | ABC NVP | NFV | 101Q 190A 210W 215Y | VFA/VIKKK | ES |
| 22 | 35,000 | 45 | ddI EFV | APV | 41L44D 74V 98G 103N 108I 184V 190A 210W 215Y | VFVIKKK | DS |
| 23 | 652,535 | ND | AZT 3TC NVP | 41L 44D 74I 103N 184V 190A 210W 215Y | VFVIKKK | DS | |
| 24 | 230,760 | 50 | Stop ARV | Stop ARV | 41L 184I 210W 215Y | VFVIRKKK | ES |
| 25 | 319,000 | 185 | AZT ddI DLV | 210W 215F | IFAIKKK | ES | |
| 26 | 22,000 | 494 | AZT ddC | 41L 210W 215Y | VFAIKKK | DS | |
| 27 | 530,000 | 325 | AZT | RTV SQV | 41L 103N 210W 215Y | VFAIRKK | ES |
| 28 | 110,000 | ND | d4T 3TC | NFV | 98G 215Y | VFAIKKK | ES |
| 29 | 3,230 | ND | d4T 3TC | RTV | 184V 215Y | VFA/VIRKK | DS |
| 30 | 4,800 | ND | ABC | SQV RTV | None | VFAIKKK | GS |
| 31 | 512,131 | 238 | AZT 3TC | T215Y | VFAIKKK | V/AS | |
| 32 | 410,876 | 9 | AZT 3TC ddI | 41L 74V 118I184V 215F 219Q 103N | VFAINKK | KG | |
| 33 | 200,000 | 30 | d4T ABC | 184I | VFAIKKK | GS | |
| 34 | 50,000 | 98 | ABC EFV | RTV SQV | 100I 103N 215Y 41L | VFAIKKK | ET |
| 35 | 800,000 | 40 | ABC EFV | NFV | 62V 74V 75I 103N 181C 215F 219E | VFVIKKK | ES |
| 36 | 380,000 | ND | AZT 3TC NVP | IDV | 41L 62V 98S 184I 210W 215Y | VFVIKKK | ES |
| 37 | 850 | 1,190 (child) | d4T 3TC | RTV | 41L 62V 215Y | VFVIKKK | D S/T |
| 38 | 9,291 | 550 | d4T 3TC | NFV | 62V 184V | VFVIKKK | SS |
| 39 | 128,800 | 197 | d4T 3TC | SQV | 41L 62V 98S 184I 210W 215Y | VTVIKKK | ES |
| 40 | 50,000 | 25 | AZT 3TC EFV | RTV APV | 103N 190A 210W 215Y | VFAIKKK | GS |
| 41 | 50,000 | 7 | ddI ABC NVP | 188C 215Y/F | VFAIKKK | DS | |
| 42 | 3,000 | 495 | d4T ddI | 215Y | VFAIKKK | GS | |
| 43 | 200,000 | 14 | d4T ddI EFV | RTV | 41L 103N 210W 215Y | IFAIKKK | DS |
| 44 | 55,000 | 430 | d4T 3TC | NFV SQV | 62V 184I 215Y | VFVIKKK | ES |
| 45 | 30,203 | 456 | d4T 3TC | NFV | 62V 70R 184V 215F | VFVIKKK | DS |
| 46 | 4,280 | 231 | d4T ddI | 41L 62V 70R | VFVIKKK | DS | |
| 47 | 59,280 | 376 | d4T 3TC | RTV | 41L 67N 75L 98S 215Y 219E | VFAIKKK | NN |
| 48 | 108,840 | 168 | d4T ddI NVP | NFV | 41L 74I 103N 118I 181C 190A 210W 215Y | VFAIKKK | DS |
| 49 | 15,000 | 310 | d4T EFV | SQV | 41L 70R 103N 184V 215Y | VFAIKKK | S |
| 50 | 221 | 478 | AZT 3TC | RTV | 41L 184V 215Y 219E | VFAIKKK | S |
| 51 | 36,320 | 80 | d4T ddI | 210W 215F 219E | VFAIKKK | S | |
| 52 | 26,000 | 232 | d4T 3TC | NFV | 184V | VFAIKKK | S |
| Amino acid sequence
|
RVA index for drug
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 69 | 70/71 | Comment | AZT | 3TC | d4T | ddI | ABC | EFV | NVP |
| T | KW | Wild type | |||||||
| SG | KW | Insertion 1 aa | 183 | 15 | 9.0 | 2.8 | 10.1 | 0.6 | 1.0 |
| TT | RW | Insertion 1 aa | 9 | >339 | 5.3 | 4.4 | 18.3 | 0.7 | 0.7 |
| SD | RW | Insertion 1 aa | ND | ND | ND | ND | ND | ND | ND |
| SSG | KW | Insertion 2 aa | >1,805 | >245 | 55.1 | 3.4 | >9 | 0.5 | 1.7 |
| SSG | KW | Insertion 2 aa | >1,805 | >245 | 66.7 | 8.0 | >9 | 6.8 | >463 |
| SSC | KW | Insertion 2 aa | >1805 | >245 | 67.8 | 3.7 | >9 | 3.9 | 9.0 |
| SCT | KW | Insertion 2 aa | 297 | >245 | 68.1 | 3.0 | >9 | 0.7 | 0.9 |
| SCT | KW | Insertion 2 aa | 131 | >245 | 27.4 | 5.6 | 6.3 | 0.2 | 1.0 |
| SCA | KW | Insertion 2 aa | >301 | >25 | >28 | 6.2 | 21.1 | >68 | >947 |
| SVG | KW | Insertion 2 aa | >301 | >25 | >28 | 7.4 | >24 | 23.9 | >947 |
| SSG | KW | Insertion 2 aa | >301 | >25 | 8.9 | 5.1 | >24 | 9.1 | 20.4 |
| SSG | KW | Insertion 2 aa | >301 | >25 | 20.6 | 5.4 | >24 | 1.5 | 5.8 |
| SSG | KW | Insertion 2 aa | >301 | >25 | >28 | 4.7 | 15.2 | 0.2 | 0.4 |
| AVG | KW | Insertion 2 aa | >30.1 | >25 | >28 | 10.7 | >24 | 1.2 | 2.5 |
| STT | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SSG | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SSA | KW | Insertion 2 aa | >1805 | >245 | 67.7 | 2.5 | >9 | 91.0 | >1,463 |
| SSA | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SST | KW | Insertion 2 aa | 169 | 4 | 1.8 | 2.1 | 7.2 | >209 | >2,894 |
| SSS | RW | Insertion 2 aa | 36 | >339 | 1.3 | 1.3 | 12.2 | 0.3 | 0.4 |
| SES | KW | Insertion 2 aa | 1,999 | >339 | 32.3 | 11.7 | 32.4 | 43.5 | >2,894 |
| SSK | KW | Insertion 2 aa | 3,745 | >339 | 9.7 | 2.7 | >48 | 1.2 | 13.0 |
| SSG | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SST | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SMT | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SST | KW | Insertion 2 aa | >1,805 | 46 | 7.3 | 2.5 | 7.5 | 0.4 | 0.4 |
| SSA | KW | Insertion 2 aa | >1,805 | >245 | 20.3 | 3.1 | >9 | 0.8 | 1.6 |
| SSG | KW | Insertion 2 aa | >1,805 | >245 | 17.1 | 5.1 | >9 | 0.7 | 0.9 |
| SSS | KW | Insertion 2 aa | 168 | >245 | 11.9 | 2.3 | 7.2 | 0.5 | 1.1 |
| SSG | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SSG | RW | Insertion 2 aa | >1,805 | >245 | 26.8 | 4.3 | 5.6 | 0.6 | 1.4 |
| STT | RW | Insertion 2 aa | >1,805 | >245 | 32.4 | 5.1 | 8.1 | >137 | >1,463 |
| SVG | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SVT | KW | Insertion 2 aa | >3,397 | >339 | 53.9 | 10.8 | >48 | >209 | >2,894 |
| SSG | KW | Insertion 2 aa | >1,805 | >245 | 35.3 | 2.6 | 5.7 | >137 | >1,463 |
| SVT | KW | Insertion 2 aa | 758 | >245 | 203.2 | 5.9 | 7.7 | 1.0 | 0.1 |
| SSA | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SSG | KW | Insertion 2 aa | 1,214 | >245 | 11.2 | 1.6 | 5.0 | 1.1 | 2.1 |
| SVT | KW | Insertion 2 aa | 1,129 | >245 | 37.8 | 3.2 | 6.5 | 0.6 | 0.7 |
| SSG | KW | Insertion 2 aa | >3,397 | >339 | 19.6 | 4.4 | >48 | 76.1 | 262.5 |
| SSG | KW | Insertion 2 aa | ND | ND | ND | ND | ND | ND | ND |
| SIG | KW | Insertion 2 aa | >3,397 | >339 | 10.5 | 2.4 | 17.9 | 0.3 | 0.3 |
| SSG | KW | Insertion 2 aa | >586 | >619 | 155.8 | 18.4 | >19 | 1.1 | 2.5 |
| SSG | KW | Insertion 2 aa | >586 | >619 | >382 | 22.6 | >19 | 1.0 | 0.8 |
| SVS | RW | Insertion 2 aa | >586 | >619 | 321.7 | 40.2 | >19 | 0.4 | 0.7 |
| SVA | RW | Insertion 2 aa | >3,688 | >346 | 14.8 | 6.1 | 9.2 | 1.0 | 1.0 |
| ITT | RVMGK | Insertion 5 aa | >301 | >25 | 24.7 | 5.8 | 18.3 | 1.0 | 3.8 |
| TTE | GKKDSTRWRKI | Insertion 11 aa | 168 | 2 | 2.8 | 1.3 | 4.1 | 38,3 | >947 |
| G | RW | Deletion | 15 | >619 | 2.1 | 6.8 | 0.8 | >229 | 5,147.0 |
| G | RW | Deletion | ND | ND | ND | ND | ND | ND | ND |
| G | RW | Deletion | 1,029 | 9 | 0.9 | 0.8 | 5.1 | 0.2 | 0.2 |
| G | KW | Deletion | 4 | >339 | 1.5 | 2.3 | 2.7 | 0.5 | 0.3 |
RTI and PI, antiretroviral therapy at the time of detection of an RT rearrangement; 60 to 71, amino acid sequence of the HIV-1 variants between positions 60 and 71 in the RT. Phenotypic results are expressed as the RVA index for each drug tested. ND, not determined; PI, protease inhibitor; ddC, zalcitabine; APV, amprenavir; ARV, antiretroviral therapy; IDV, indinavir; NFV, nelfinavir; RTV, ritonavir; SQV, saquinavir; DLV, delavirdine; H. Urea, hydroxyurea.
The rearrangements also frequently included mutations at positions 67, 68, and 69 in 63, 17, and 78% of the cases, respectively. As previously reported, the molecular changes frequently created serine stretches, with the pattern SSG present at a frequency of 35%.
Associated RTI resistance mutation.
The different mutations coding for HIV resistance to NRTI or nonnucleoside RTI (NNRTI) observed in the 52 sequences are shown in Table 1. The major mutations T215Y or T215F (T215Y/F), M184V/I, and K103N were observed in 86, 40, and 25% of the sequences, respectively.
Antiretroviral therapies and virological evolution of the patients.
All patients were RTI experienced at the time of sampling for genotype, with a median duration of therapy of 50.5 months; most of them had undergone multiple therapy changes, with a median use of eight drugs and five NRTI. At the time of sampling, the RTI most frequently in use were stavudine (d4T) (55% of the patients), lamivudine (3TC) (44%), and didanosine (ddI) (33%) (Table 1). The antiretroviral therapy comprised at least one protease inhibitor for 69% of the patients. Two patients had stopped antiretroviral therapy at the time of sampling.
At the time of sampling for genotype, the median CD4+ cell count was 124.4/μl (range, 1 to 550) and the median amount of plasma HIV-1 RNA was 4.06 log 10 copies/ml (range, 2.34 to 6.30).
Individual follow-up data concerning 20 patients are shown in Table 2. For some patients, significant decreases of the plasma viral load were observed; this corresponded to the change of therapies, including new protease inhibitors, or to intensification of treatment. In one case, the viral load decreased without any change in treatment, with conservation of an insertion of 11 aa in the RT.
TABLE 2.
Follow-up of 20 patients of the studya
| Patient no. | Pattern 69, baseline | Antiretroviral therapy | Time of follow-up (weeks) | Δ log10 HIV-1 RNA | Pattern 69, follow-up |
|---|---|---|---|---|---|
| 3 | S-D | AZT 3TC APV | 8 | 0.25 | ND |
| 6 | S-SG | ABC NVP NFV SQV | 36 | −0.82 | S-ST/S |
| 9 | S-CA | d4T 3TC RTV SQV EFV | 40 | 0.36 | S-CA |
| 10 | S-VG | Megahaart | 10 | −2.26 | ND |
| 12 | S-SG | 2 months wash-out | 35 | −0.47 | S |
| 13 | S-SG | 3 months wash-out | 13 | −0.26 | ND |
| 15 | S-TT | ddI EFV NFV SQV | 64 | −2.06 | ND |
| 16 | S-SG | AZT 3TC EFV RTV SQV | 8 | 0.11 | ND |
| 17 | S-SA | d4T 3TC APV | 24 | 0.22 | ND |
| 18 | S-SS | d4T 3TC NFV | 4 | −1.01 | ND |
| 21 | S-ES | ABC NFV NVP | 16 | 0.48 | ND |
| 23 | S-SG | AZT 3TC ABC NVP SQV | 24 | −0.32 | ND |
| 25 | S-MT | d4T 3TC IDV | 28 | −3.19 | ND |
| 26 | S-ST | d4T 3TC RTV | 8 | −2.03 | ND |
| 29 | S-SS | d4T 3TC IDV | 28 | 0.76 | ND |
| 30 | S-SG | ABC SQV RTV | 22 | 1.68 | ND |
| 31 | S-SG | AZT 3TC RTV | 36 | −0.93 | S-SG |
| 47 | Insertion 5 aa | ddI ABC EFV | 24 | 0.12 | ND |
| 48 | Insertion 11 aa | d4T ddI NVP NFV | 24 | −0.74 | Insertion 11 aa |
| 52 | Deletion | d4T 3TC NFV | 24 | 0.07 | ND |
Δ log10 HIV-1 RNA, changes of the HIV plasma viral load from the baseline. Antiretroviral therapy, the drugs which were absent at baseline appear in bold (can be recycled from previous regimens). Megahaart, 8 antiretroviral drugs combined with hydroxyurea. Pattern 69, genotype at codon 69 and rearrangements. ND, genotype not determined. RTV, ritonavir; SQV, saquinavir; IDV, indinavir; NFV, nelfinavir.
Phenotypic analysis and correlation with RT genotype.
Additional frozen plasma samples were available for phenotypic analysis using the single-cycle RVA for 40 of the samples showing a rearrangement in the HIV-1 RT. The RVA index was obtained for five NRTI, zidovudine (AZT), 3TC, d4T, ddI, and abacavir (ABC); and for two NNRTI, nevirapine (NVP) and efavirenz (EFV). HIV-1 variants with an RVA index of <4 were considered sensitive; an RVA index of >10 indicated high-level resistance, while an RVA index between 4 and 10 was considered low-level resistance.
Sensitivity to NRTI.
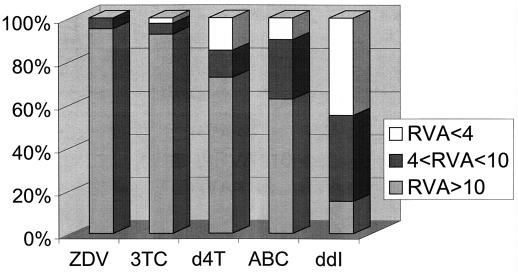
The HIV-1 variants exhibited high-level resistance to a mean of 3.4 ± 1.2 NRTI (median of 4). The pattern of phenotypic sensitivity to the different NRTIs is shown in Fig. 1. The frequency of high-level resistance was greatest for AZT (95%). High-level resistance to d4T was slightly less frequent at 72% (29 of 40). Resistance to d4T was more often observed in samples with a 2-aa insertion than in samples with other rearrangements. In three patients with a 2-aa insertion (patients 10, 38, and 46), high-level resistance to AZT and d4T was detected in the absence of the T215Y/F mutations, which were originally described as crucial for the appearance of a significant degree of resistance to AZT (2, 14, 15); however two of these three samples also contained other thymidine analogue mutations.
FIG. 1.
Phenotypic sensitivities to NRTIs of 40 HIV-1 variants with an RT rearrangement. Phenotypic results are expressed for each drug as percentages of variants divided into three groups: RVA of <4, phenotypic sensitivity; RVA of >4 and <10, low-level resistance; RVA of >10, high-level resistance. ZDV, zidovudine.
High-level resistance to 3TC was seen in 37 (92%) of the samples, including one with an insertion of 11 aa, one with an insertion of two codons, and one with a deletion. Not all high-level resistance to 3TC could be related to the presence of M184V/I mutations, which were present in only 16 (43%) of the variants with an RVA index of >10 for 3TC. Another mutational pattern which has been described as involved in 3TC resistance (E44D/A and/or V118I mutations with M41L and T215Y changes) (10) was present in seven cases in conjunction with 184V/I mutations and in three cases with the 184M wild-type genotype with RVA indices for 3TC of 15 (1-aa insertion), >25 (2-aa insertion), and 2 (11-aa insertion). High-level resistance to ABC was noted in 25 of 40 patients (62%). In these 25 samples, the M184V/I mutations were present in only 7 cases and fewer than three NRTI resistance mutations were detected in 11 cases.
ddI phenotypic sensitivity displayed a different pattern, with only 15% (6 of 40) of samples showing high-level resistance and 40% (16 of 40) showing low-level (median increase, sevenfold) resistance. The mutation L74V, usually associated with resistance to ddI (25), was present in six samples, three of which remained sensitive to ddI.
Sensitivity to NNRTI.
Fourteen patients had a phenotypic assay with high-level resistance to NVP and/or EFV, explained in 13 cases by the presence of mutations related to NNRTI resistance in the RT encoding region (K103N, Y181C, or G190A). In one case (patient 9), no explicative mutation was found, but the mutation G190Q was detected. The remaining 26 (65%) samples were sensitive to NNRTI.
DISCUSSION
This study enabled us to describe HIV-1 RT rearrangements in a large cohort of HIV-1-infected, NRTI-treated patients. The rearrangements were present in 2.4% of the patients. We confirm the results of previously published studies, based on smaller numbers of patients, which reported prevalences of RT rearrangements of between 0.5 and 2.7% (1, 27, 28). Despite this low prevalence, future studies will be necessary to assess the evolution of the incidence of such rearrangements. A genotypic study on 400 antiretroviral therapy-naive patients included during the same period in France did not detect any RT rearrangement (9), suggesting that RTI are responsible for the selection of these mutational patterns.
One major finding of our study is the extensive heterogeneity of the rearrangements. Because of this heterogeneity, we decided to document the phenotypic sensitivity to RTI of the different variants, using an RVA. Insertions of 1, 2 (majority in our study), and 5 aa after residue 69 were associated with high-level phenotypic resistance to AZT, 3TC, d4T, and ABC. This phenotype was, in some cases, found in the absence of the different mutations reported to be involved in high-level resistance to these four drugs. In contrast, the 11-codon insertion, found surprisingly in one patient, was not shown to be involved in resistance to NRTI; the later evolution of the viral load, decreasing without any treatment change, was in concord in this case with the phenotype. Similarly, none of the three deletions with an RVA available in our study were linked to phenotypic resistance to NRTI. Recent studies have used site-directed mutagenesis to determine the specific phenotypic impact of the rearrangements; Larder et al. (17) reported that the 69S-SS insertion (mutation T69S with an insertion of two serines) did not code for MDR by itself but contributed to MDR in the presence of AZT resistance mutations. Inversely, 69S-SA and 69S-SG patterns were found to code for MDR even in absence of other genotypic changes (29). In a recent report (30), Winters et al. described the frequent association of 3-bp deletions in the β3-β4 region of the RT gene with the Q151M mutation. Using site-directed mutagenesis, they showed that the deletions could code for MDR in the presence of the T215Y mutation. These findings are not confirmed by our RVA results; this can be explained by differences in the phenotypic assays used, as well by the genetic background of the viruses, which can be present in recombinant viruses but not in isolates obtained by site-directed mutagenesis. Other studies involving greater numbers of isolates will be necessary to clarify the role of deletions in MDR.
One intriguing finding of this study is the relatively low level of phenotypic resistance to ddI induced by the rearrangements. In previous studies, assays using peripheral blood mononuclear cell cocultured virus isolates, or recombinant viruses, have also failed to detect high-level phenotypic resistance to ddI, even in the presence of the L74 V mutation (6, 18). It is thus difficult to know whether these data represent a true sensitivity to ddI or an inadequacy of such in vitro tests to measure resistance to ddI. Standardization of the interpretation of RVAs, including the definition of the threshold values for sensitivity or resistance, seems to be particularly crucial for this drug.
Our study thus provides correlates between genotype and phenotype for a wide variety of RT rearrangements. This information is of particular importance at a time when genotypic assays are becoming more widely used in the management of antiretroviral treatment failure. MDR is potentially a major limitation to antiretroviral efficacy. Some individual follow-up in our study suggested that therapy intensification could have some impact on the viral load in plasma. However, 30% of all included patients were infected by HIV-1 quasispecies that were resistant to both NRTI and NNRTI, and most of the variants also displayed major resistance mutations for protease inhibitors (data not shown). In such patients with a probable resistance to most of the inhibitors presently available, genotype-guided treatments have few chances to be effective. Alternative strategies, including treatment interruption strategies and/or the use of new drugs targeting other stages of HIV replication, should be evaluated in this context.
ACKNOWLEDGMENTS
We thank Valérie Birac for excellent technical assistance. We thank Dominique Costagliola for help with the statistical analysis.
This work was supported by ANRS (Agence Nationale de Recherches sur le SIDA).
REFERENCES
- 1.Balotta C, Violin M, Monno L, Bagnarelli P, Riva C, Facchi G, Berlusconi A, Lippi M, Rusconi S, Clementi M, Galli M, Angarano G, Moroni M. Prevalence of multiple dideoxynucleoside analogue resistance (MddNR) in a multicenter cohort of HIV-1-infected Italian patients with virologic failure. J Acquir Immune Defic Syndr. 2000;24:232–240. doi: 10.1097/00126334-200007010-00007. [DOI] [PubMed] [Google Scholar]
- 2.Boucher C A B, O'Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M A, Goudsmit J, Larder B A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. HIV-1 reverse transcription: a termination step at the centre of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 4.Chou T-C. The median-effect principle and the combination index for quantitation of synergism and antagonism, P. 61–102. In: Chou T-C, Rideout J, editors. Synergism and antagonism in chemotherapy. New York, N.Y: Academic Press; 1991. [Google Scholar]
- 5.Clavel F, Hoggan M D, Willey R L, Strebel K, Martin M A, Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989;63:1455–1459. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colonno R J, Wachtfogel Y T, Hertogs K, Larder B A. Frequency of stavudine and didanosine resistance among a large number of randomly selected HIV-1 isolates. Antivir Ther. 2000;5:85. [Google Scholar]
- 7.De Antoni A, Foli J, Lisziewicz J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:899–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 8.de Jong J J, Goudsmit J, Lukashov V V, Hillebrand M E, Baan E, Huismans R, Danner S A, Ten Veen J H, De Wolf F, Jurrians S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 9.Descamps D, Costagliola D, Glaude G, Buffet-Janvresse C, Calvez V, Collin G, Cottalorda J, Ferchal F, Harzic M, Izopet J, Masquelier B, Ruffault A, Schmuck A, Tamalet C, Yvon A, Brun-Vézinet F the French ANRS Antiretroviral Resistance Study Group. Prevalence of resistance mutations in antiretroviral-naive patients: French National Study. Antivir Ther. 1999;4(S1):87. [Google Scholar]
- 10.Hertogs K, Bloor S, de Vroey V, Van Den Eynde C, Dehertogh P, Van Cauwenberge A, Stürmer M, Alcorn T, Wegner S, Van Houtte M, Miller V, Larder B A. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob Agents Chemother. 2000;44:568–573. doi: 10.1128/aac.44.3.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch M S, Brun-Vézinet F, D'Aquila R T, Hammer S M, Johnson V A, Kuritzkes D R, Loveday C, Mellors J W, Clotet B, Conway B, Demeter L M, Vella S, Jacobsen D M, Richman D D. Antiretroviral drug testing in adult HIV-1 infection. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 12.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung M, Agut H, Candotti D, Ingrand D, Katlama C, Huraux J M. Susceptibility of HIV-1 isolates to zidovudine: correlation between widely applicable culture test and PCR analysis. J Acquir Immune Defic Syndr Hum Retrovirol. 1992;5:359–364. [PubMed] [Google Scholar]
- 14.Kellam P, Boucher C A B, Larder B A. A novel fifth mutation in HIV-1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 16.Larder B A, Kellam P, Kemp S D. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masquelier B, Pellegrin I, Ruffault A, Ragnaud J-M, Morlat P, Michelet C, Doignon F, Biteau N, Fleury H J A. Genotypic evolution of HIV-1 isolates from patients after a switch of therapy from zidovudine to didanosine. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:330–334. [PubMed] [Google Scholar]
- 19.Masquelier B, Descamps D, Carriere I, Ferchal F, Collin G, Denayrolles M, Ruffault A, Chanzy B, Izopet J, Buffet-Janvresse C, Schmitt M P, Race E, Fleury H J A, Aboulker J P, Yeni P, Brun-Vézinet F. Zidovudine resensitization and dual HIV-1 resistance to zidovudine and lamivudine in the Delta lamivudine roll-over study. Antivir Ther. 1999;4:7–18. [PubMed] [Google Scholar]
- 20.Race E, Dam E, Obry V, Paulous S, Clavel F. Analysis of human immunodeficiency virus type 1 cross-resistance to protease inhibitors in patients failing on combination therapies using a rapid single-cycle recombinant virus assay. AIDS. 1999;13:2061–2068. doi: 10.1097/00002030-199910220-00008. [DOI] [PubMed] [Google Scholar]
- 21.Rakik A, Ait-Khaled M, Griffin P, Thomas D A, Tisdale M, Kleim J P for the abacavir CNA2007 International Study Group. A novel genotype encoding a single amino acid insertion and five other substitutions between residues 64 and 74 of the HIV-1 reverse transcriptase confers high-level cross-resistance to nucleoside reverse transcriptase inhibitors. J Aquir Immune Defic Syndr. 1999;22:139–145. doi: 10.1097/00126334-199910010-00005. [DOI] [PubMed] [Google Scholar]
- 22.Ross L, Johnson M, Graham N, Shaefer M, St. Clair M. The reverse transcriptase codon 69 insertion is observed in nucleoside reverse transcriptase inhibitor-experienced HIV-1 infected individuals, including those without prior or concurrent zidovudine therapy. J Hum Virol. 1999;2:290–295. [PubMed] [Google Scholar]
- 23.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance: 2000–2001 update. Int Antivir News. 2000;8:65–91. [Google Scholar]
- 24.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, et al. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 26.Tamalet C, Izopet J, Koch N, Fantini J, Yahi N. Stable rearrangements of the β3-β4 hairpin loop of HIV-1 reverse transcriptase in plasma viruses from patients receiving combination therapy. AIDS. 1998;12:F161–F166. doi: 10.1097/00002030-199814000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Tamalet C, Yahi N, Tourres C, Colson P, Quinson A M, Poizot-Martin I, Dhiver C, Fantini J. Multidrug resistance genotypes (insertions in the beta3-beta4 finger subdomain and MDR mutations) of HIV-1 reverse transcriptase from extensively treated patients: incidence and association with other resistance mutations. Virology. 2000;270:310–316. doi: 10.1006/viro.2000.0261. [DOI] [PubMed] [Google Scholar]
- 28.Van Vaerenbergh K, Van Laethem K, Albert J, Boucher C A, Clotet B, Floridia M, Gerstoft J, Hejdeman B, Nielsen C, Pannecouque C, Perrin L, Pirillo M F, Ruiz L, Schmit J C, Schneider F, Schoolmeester A, Schuurman R, Stellbrink H J, Stuyver L, Van Lunzen J, Van Remoortel B, Van Wijngaerden E, Vella S, Witvrouw M, Yerly S, De Clercq E, Destmyer J, Vandamme A M. Prevalence and characteristics of multinucleoside-resistant human immunodeficiency virus type 1 among European patients receiving combinations of nucleoside analogues. Antimicrob Agents Chemother. 2000;44:2109–2117. doi: 10.1128/aac.44.8.2109-2117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winters M A, Cooley K I, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winters M A, Coolley K L, Cheng P, Girard Y A, Hamdan H, Kovari L C, Merigan T C. Genotypic, phenotypic, and modeling studies of a deletion in the β3-β4 region of the human immunodeficiency virus type 1 reverse transcriptase gene that is associated with resistance to nucleoside reverse transcriptase inhibitors. J Virol. 2000;74:10707–10713. doi: 10.1128/jvi.74.22.10707-10713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]