Abstract
Cadmium (Cd) pollution poses a serious risk to human health and ecological security. Bioremediation can be a promising and effective remediation technology for treating Cd contaminated soils. In this study, seven heterotrophic strains were isolated from Cd contaminated soil and 7 autotrophic strains were isolated from acid mine drainage. Cd removal efficiencies were compared after leaching with autotrophic bacteria (Att-sys), heterotrophic isolates (Htt-sys) and cooperative leaching systems (Co-sys) in laboratory agitating reactors. The results indicated that Cd removal efficiency of Co-sys (32.09%) was significantly higher than that of Att-sys (23.24%) and Htt-sys (0.74%). By analyzing the soil microbial community in different bioleaching systems, we found that the addition of heterotrophic isolates significantly promoted the growth of some heavy metal resistant inhabitants (Massilia, Alicyclobacillus, Micromonospora, etc.), and Co-sys had a minor effect on the growth of soil indigenous microbes. In Co-sys, the content of the four Cd fractions all decreased compared with other leaching systems. The analysis of soil physicochemical parameters during the leaching process showed that pH and ORP (oxidation reduction potential) were not the only determinants for Cd removal efficiency in Co-sys, synergistic metabolic activities of autotrophic and heterotrophic strains may be other determinants. This study demonstrated that cooperative bioremediation may prove to be a safe and efficient technique for field application in heavy metal soil pollution.
Bioremediation can be a promising and effective remediation technology for treating Cd contaminated soils. Cooperative bioremediation using heterotrophic and autotrophic mixtures proved to be an efficient, short-term bioremediation strategy for heavy metal contaminated soil.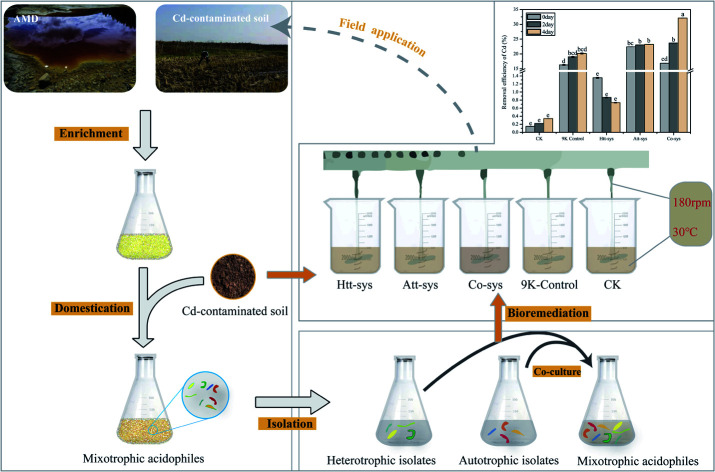
1. Introduction
Cadmium (Cd), as a toxic, cancerogenic, and teratogenic heavy metal, in soil can pose threats to biodiversity, agricultural productivity, food safety, and human health when it is transmitted via the food chain.1–4 Heavy metal pollution is an important factor of soil contamination.5 As a result of anthropogenic activities such as mining, smelting, electroplating, irrigation with wastewater and fertilizer abuse, paddy soil degradation has become a major global concern, particularly in China.6–8 Thus, it is urgent to find cost-effective and environmentally friendly techniques for Cd-polluted paddy soil remediation and resumption. To address these issues, a variety of methods have been explored to remediate heavy metal contaminated soils in recent years such as in-site immobilization with amendments, electro-kinetic, phytoremediation and chemical washing.9–12 However, these techniques are costly, labor intensive, time-consuming and may bring about secondary pollution.11 In particular, in-site immobilization efficiency is susceptible to environmental influences such as pH and redox potential and might result in releasing heavy metal back to soils.13,14
The bioleaching remediation can be a promising alternative remediation technology for heavy metal polluted soils due to simplicity of operation, low cost and eco-friendliness. For instance, Li isolated a heavy metal tolerant strain Providencia sp. LLDRA6 and demonstrated that the strain LLDRA6 has high bioremediation ability to remove heavy metals from contaminated soils.15 Khan found that indigenous isolates could be used to remediate soils contaminated with Pb and Hg.16 Yang studied bioleaching remediation of heavy metal-contaminated soils through a biosurfactant-producing strain Burkholderia sp. Z-90, and it showed that the removal efficiency of Zn, Pd, Mn, Cd, Cu and As in soils could be reached up to 44.0%, 32.5%, 52.2%, 37.7%, 24.1% and 31.6%, respectively.17 Deng used indigenous bacteria Penicillium chrysogenum strain F1 to bioleach heavy metals from a polluted soil and the bioavailability and toxicity of heavy metals were reduced significantly since the transformation of heavy metal speciation.18
For the bioleaching method, selection of the microbial agent is critical to remove heavy metals in soil.19 The autotrophic acidophilic microbes with Fe/S oxidizing metabolism can remove total and available heavy metals through bioleaching from sewage sludge, dredged sediments, sulphide ores and spent batteries, which has received considerable attention in recent years.20–23 In particular, the strains isolated from acid mine drainages (AMD) were commonly regarded as available microbial agents for bioleaching. In a previous study, we discovered more than 23 strains of bioleaching microbial species isolated from AMD from Dexing copper mine (Jiangxi, China), Zijinshan Copper Mine (Fujian, China) and Zambia's Luanshya Copper Mine, such as Acidithiobacillus ferrooxidans, Leptospirillum ferriphilum and Acidithiobacillus thiooxidans.24–27 It demonstrated that these acidophilic microbes could bioleach heavy metals from marmatite and chalcopyrite. For instance, Ma studied the bioleaching of chalcopyrite with different initial microbial proportions and presumed that the appropriate proportions of acidophilic microbes could accelerate the chalcopyrite dissolution rate.28 Xiao used an artificial microbial community with four acidophilic isolates obtained from AMD and inferred that the composition of the raw ores might be the key to determinate chalcopyrite bioleaching.26 Nevertheless, less attention has been paid to examine the bioleaching performance of autotrophic acidophilic microbe isolated from AMD on heavy metal contaminated soil. In addition, heterotrophic microbes have also attracted much attention in bioleaching due to the utilization of organic acid.29 Usually, organic acids such as formic, acetic, propionic and hexanoic, would disturb the ferrous and sulfur oxidation ability of the autotrophic acidophilic species.30 Besides, some researches have revealed that the mixotrophic acidophiles containing autotrophic and heterotrophic species are able to perform more complicated tasks in natural soils than pure species due to their better environmental adaptability.31–33 Therefore, in our recent research,25 we enriched the mixotrophic acidophiles that can effectively remove Cd from paddy soil, but the key strains and functions are not clear.
In order to confirm the speculation whether adding heterotrophic species to autotrophic acidophilic species forming mixotrophic acidophiles could enhance the bioleaching performance and clarify the critical strains and their functions in removing cadmium from paddy soil, we isolated 7 heterotrophic strains from the mixotrophic acidophiles at first, mixed them with 7 autotrophic leaching strains isolated previously from AMD, and analyzed the leaching effect of different cultures. The intermittent stirring test was designed to simulate field treatment, soil samples were collected to determine the Cd removal efficiency, soil physicochemical properties and microbial community structure. The specific aims of this research were to (i) investigate whether heterotrophic acidophilic can enhance the Cd removal efficiency and evaluate the effects of mixotrophic acidophiles on total Cd removal; (ii) reveal the major contributors to the high Cd removal efficiency of cooperative bioleaching system; (iii) analyze the effects of cooperative bioleaching system on soil indigenous microbial communities and explore the composition and diversity changes of microbial communities in different leaching systems. In summary, the possible mechanism of bioaugmentation of heterotrophic isolates was discussed. This study can lay a theoretical foundation for the Cd removal treatment in the paddy field by microbial agents.
2. Materials and methods
2.1. Sample collection and analysis
Soil samples from the plow layers (0–20 cm) of three adjacent Cd-contaminated paddy fields (S1, S2 and S3) were collected from Xiangtan County (N 27°21′12′′, E 112°03′18′′), Hunan Province. This contaminated area is only a few kilometers away from heavy industrial factories closed for more than ten years due to serious pollution to paddy fields and irrigation water. The main crop in the sampling area used to be rice. However, the contaminated area was fallow in recent years due to heavy metal pollution. We split the soil samples into three portions: (a) one portion of soil samples was stored at 4 °C for strains isolation; (b) the second portion of soil samples was air-dried in the absence of light and triturate to pass through a 2 mm sieve for soil property analysis; (c) the last portion of soil samples were stored at room temperature for bioremediation test.
The organic matter (OM) content of soil was analyzed by loss-on-ignition at 600 °C.34 Soil pH and oxidation reduction potential (ORP) were measured in deionized water extract (soil-to-water 1 : 2.5, w/v) by a digital pH meter (BPH-220, Bell Instrument Equipment Co. Ltd., Dalian, China). 0.400–0.500 g sieved soil samples were digested by an acid mixture (HNO3, HF and HClO4: 2 : 2 : 1, v/v/v) on an electric heating plate (XJS20-42, Laboratory Instrument Equipment Co. Ltd., Tianjin, China) according to the method reported by Hao and Deng, following the temperature procedure. 110 °C for 30 min, 140 °C for 30 min, and finally at 180 °C until sample was digested completely. (a) Then, the clarified sample solution was extracted by 2 mL nitric acid, and cooled down to room temperature, and diluted with deionized water to 25 mL.25,34 The three-step sequential extraction procedure was carried out according to the modified BCR, by which the metals was divided into four fractions: weak acid soluble fraction, reducible fraction, oxidizable fraction, residual fraction.34 Then total Cd and the fractions of Cd contents were determined by ICP-OES (Perkin Elmer Analyst Optima 5300 DV, USA).35
2.2. Microorganisms
2.2.1. Consortium of autotrophic strains
A mixed consortium of A. caldus DX, A. thiooxidans DX, A. thiooxidans ZJ, A. thiooxidans AO1, F. acidiphilum DX, A. caldus S1 and L. ferriphilum DX isolated in the stable mixotrophic acidophiles was provided by Key Laboratory of Biometallurgy of Ministry of Education, Central South University. This mixed culture was composed of autotrophic bioleaching strains, able to oxidize Fe(ii) or S0 in extremely acidic conditions. The strains were routinely cultured in 9K liquid medium with ferrous (4.47%) and sulfur (1‰) (Table S1 of ESI†). The 9K basal medium consisted of (NH4)2SO4 (3 g L−1), KCl (0.1 g L−1), K2HPO4 (0.5 g L−1), MgSO4·7H2O (0.5 g L−1), Ca(NO3)2 (0.1 g L−1), deionized water as solvent, and this medium was generally used to cultivate Acidithiobacillus ferrooxidans.
2.2.2. Isolation and characterizations of heterotrophic isolates
Previous studies showed that the mixotrophic acidophiles had the capability of removing 34% of total Cd and increasing Cd uptake in plant.25 Besides, a comprehensive risk assessment of heavy metals was conducted previously in this experimental paddy fields and it turned out that Cd and Zn were the potential risks.34 The mixotrophic acidophiles were domesticated by adding the Cd-contaminated paddy soils (10 g) into the mixotrophic acidophiles (100 mL) and then inoculating them into 900 mL modified 9K medium at an initial pH of 3.5. After the solution pH dropped down to 2.5, the next subculturing was conducted via inoculating the culture (100 mL) into fresh 900 mL modified 9K medium at the same initial pH value. Subculturing was carried out when the pH of the solution was below 2.5. After approximately 50 generations of domestication, the incubation period of each subculture was consistent (4–5 days). Then the stable mixotrophic acidophiles were considered to be obtained. The modified 9K medium contained (NH4)2SO4 (3 g L−1), KCl (0.1 g L−1), K2HPO4 (0.5 g L−1), MgSO4·7H2O (0.5 g L−1), Ca(NO3)2 (0.1 g L−1), Glucose (0.7 g L−1), yeast (0.3 g L−1) extract and sulfur (1 g L−1).
For isolation of heterotrophic strains, 10 mL stable mixotrophic acidophiles were inoculated into 90 mL modified PDA medium containing Glucose (2%), MgSO4·7H2O (0.05%), K2HPO4 (0.1%) and yeast extract (0.2%) in 250 mL flasks. All flasks were cultured at 30 °C and 170 rpm in the incubator shaker for 5 days to obtain enrichment cultures.36,37 The heterotrophic strains were isolated through gradient dilution separation method.38 The diluted cultures were separated and purified on solid PDA plates by spreading plate and streaking plate methods. Solid PDA plates containing 2% agar powder and 0.01% bromocresol green were used to screen out acid-producing strains. Consequently, the acid-producing strains were considered to be obtained when the growth surrounding the colony turned yellow from green. Each target colony was streaked repeatedly to get the purified strain. Subsequently, 7 fungal strains (named as isolate F1, F2, F3, F4, zp1, zp2, and zp3, respectively) were isolated and routinely incubated in PDA medium on a rotary shaker (170 rpm) at 30 °C.
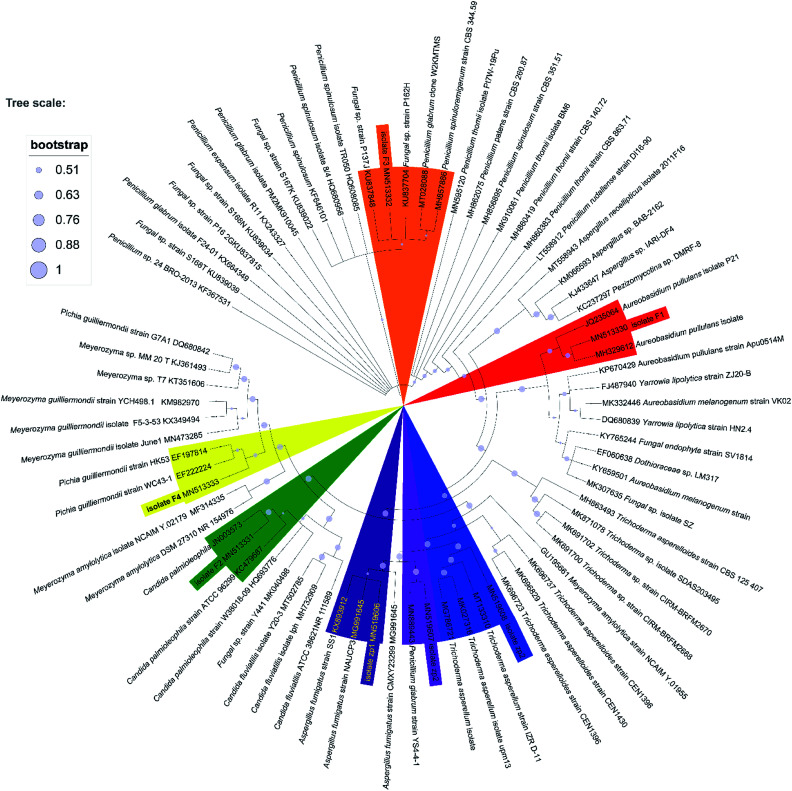
Preliminary characterizations of the 7 fungal isolates were based on their morphological characteristics, such as microscopic observation of cell dimensions and some physiological and biochemical tests.39 ITS rRNA identification was further used to analyze the phylogenetic relationships of these 7 isolates. ITS rRNA gene was amplified using the universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), and the PCR products were sequenced by Sangon Biotech (Shanghai, China). Specific procedure was shown in ESI.† The obtained sequences were assembled and submitted to NCBI Ribosomal Database (http://www.ncbi.nlm.nih.gov/BLAST/) for alignment and phylogenetic analysis. Sequences with a percentage identity of 97% or higher were considered to represent the closest species. More than 70 other strains selected from GenBank were used for constructing the phylogenetic tree of the 7 isolates. The phylogenetic trees were constructed using MEGA X version 10.1.8 (http://www.megasoftware.net). The robustness of the phylogenetic tree topology was confirmed using bootstrap analysis with 1000 bootstrap replications. Evolutionary distances were calculated using the p-distance method, and the neighbor-joining algorithm was used to generate the initial tree. Then the NWK file exported from MEGA was submitted to ITOL (https://itol.embl.de/tree) to embellish the phylogenetic tree.
2.2.3. Co-culture of autotrophic and heterotrophic isolates
In order to reveal the bioremediation mechanism of the mixotrophic acidophiles and to analyze how autotrophic and heterotrophic isolates removed Cd from soil. We performed the bioleaching experiments using three groups according to their different trophic-types. More specifically, 7 autotrophic bioleaching strains, A. caldus DX, A. thiooxidans DX, A. thiooxidans ZJ, A. thiooxidans AO1, F. acidiphilum DX, A. caldus S1 and L. ferriphilum DX were employed as a autotrophic microbial group according to their high heavy metal tolerance and special habitat with low pH during leaching process.26,27,40 Meanwhile, seven heterotrophic isolates were employed and cultured as a heterotrophic microbial group in modified PDA medium. The mixotrophic microbial group were co-cultured by autotrophic leaching strains and heterotrophic isolates proportionally. Before this, the suspension of the strains were centrifugated at 12 000g for 20 minutes. Then the centrifuged sediments of 7 isolates were combined and inoculated into 100 mL PDA medium in 250 mL glass flask. The mass ratio of strains in three treatments were exhibited as follow: autotrophic microbial group: A. caldus DX, A. thiooxidans DX, A. thiooxidans ZJ, A. thiooxidans AO1, F. acidiphilum DX, A. caldus S1 and L. ferriphilum DX (1 : 1 : 1 : 1 : 1 : 1 : 1); heterotrophic microbial group: F1, F2, F3, F4, zp1, zp2 and zp3 (1 : 1 : 1 : 1 : 1 : 1 : 1); mixotrophic microbial group: autotrophic microbial group and heterotrophic microbial group (1 : 1). The growth medium for mixotrophic microbial group was modified with 9K and ferrous (4.47%) and sulfur (1‰). Then, three microbial groups were incubated at 30 °C in a rotary platform at 170 rpm for 5 days.
2.3. Bioremediation experiments in laboratory simulations
The cell concentrations of three microbial groups were exceeded 2 × 108 CFU mL−1 after 5 days incubation. The pH values of microbial groups were 4.20, 1.15 and 1.40 for heterotrophic microbial group, autotrophic microbial group and mixotrophic microbial group, respectively. Besides, deionized water and abiotic 9K medium were used as control groups with their pH value adjusted to 6.5 and 2.0 with sulfuric acid. Five agitating bioleaching systems were conducted to evaluate the performance of cooperative bioleaching. All leaching experiments were implemented with different acidophilic microbial solutions (500 mL) into Cd-contaminated paddy soils (500 g): the autotrophic leaching system (Att-sys), the heterotrophic leaching system (Htt-sys), the cooperative leaching system (Co-sys) and equal volume of 9K medium and deionized water were employed as 9K control and CK.
The five leaching systems were placed on a six-unit electric agitator platform (JJ-3A, Jinyi Instrument Technology Co., Ltd, Jiangsu, China) at 30 °C. Each unit was equipped with a heating plate and an agitating paddle, and its impeller blades extended into four fifths of the beaker in depth (Fig. S1 of ESI†). All bioleaching experiments were carried out in triplicate. In order to simulate the field treatment of bioremediation, the intermittent agitating experiment were carried out in 4 days. More specifically, every single leaching system was agitated twice a day, and each time was agitated for 10 minutes with 180 rpm.
During the bioleaching process, 5 mL soil slurry of five leaching systems at 0, 2, 4 day were collected for microbial community analysis and procedures are shown in ESI.†
2.4. Analytical methods
The physicochemical parameters were measured during intermittent agitating bioleaching. The 5 mL of soil slurry supernatant after each agitation was withdrawn and the concentration of leached Cd was determined using ICP-OES. Meanwhile, pH and ORP of the slurry supernatant were also measured by a digital pH meter. Residual soil sample after 4 days intermittent agitating experiment was collected and dried at room temperature (25–30 °C), and then ground to pass 2 mm sieves for digestion. Determination of total Cd concentration in digesting solution was performed using ICP-OES as mentioned in sample analysis. Fractional Cd distribution in soils were extracted and detected according to the methods reported by Gleyzes.41
2.5. Data analysis
Operational taxonomic unit (OTU) clustering was analyzed by reported methods.42 Nonmetric Multidimensional scaling (NMDS) and Alpha-diversity indexes including Shannon index, Simpson index, OTU abundance and evenness were calculated through the R statistical platform. All graphs and charts were generated by SigmaPlot 14.0. Permutational multivariate analysis of variance (PerMANOVA) was used to identify the difference in soil microbial diversity after various treatments. The Fisher's least significant difference test (LSD) was used to compare the differences between different leaching systems.
The reads data were deposited into the NCBI Sequence Read Archive (SRA) database, in BioProject PRJNA576993.
3. Results
3.1. Paddy physicochemical properties
The physicochemical properties of paddy soils including pH, OM and Cd content is summarized in Table 1. As shown, the total Cd concentrations of the paddy soil varied from 9.09 to 10.03 mg kg−1 which were 30.3 to 33.4 times higher than the reference background values (0.3 mg kg−1) in Hunan province.34 Microbial isolates from Cd-contaminated soil are generally more resistant to high concentrations of Cd than microorganisms isolated from the pristine soil.37 The pH values of soil 1, soil 2 and soil 3 were 5.96, 5.89 and 6.05, respectively. pH is one of the most important factors affecting the form of heavy metals in the soil and heavy metals generally are easier to migrate in acidic soils with low pH.35 OM contents in the three studied soil samples showed no significant difference ranging from 3.79% to 4.66%. The average level of total N, P and K in the three experimental soils were 2.26 g kg−1, 0.56 g kg−1 and 14.06 g kg−1, respectively, while the available N, P and K were 224.22 mg kg−1, 2.07 mg kg−1 and 109.89 mg kg−1, respectively.
Physiochemical properties of experimental soils, mean ± standard deviation (n = 3).
| Characteristics | Soil 1 | Soil 2 | Soil 3 |
|---|---|---|---|
| Soil pH | 5.96 ± 0.23 | 5.89 ± 1.05 | 6.05 ± 0.27 |
| Soil ORP | 290.30 ± 21.40 | 322.30 ± 20.60 | 250.80 ± 18.10 |
| Available N (mg kg−1) | 234.67 ± 60.48 | 214.33 ± 54.05 | 223.67 ± 36.75 |
| Available P (mg kg−1) | 0.64 ± 0.35 | 4.25 ± 3.15 | 1.32 ± 1.38 |
| Available K (mg kg−1) | 108.33 ± 17.90 | 101.67 ± 9.24 | 119.67 ± 19.22 |
| Total N (g kg−1) | 2.38 ± 0.33 | 2.12 ± 0.31 | 2.28 ± 0.14 |
| Total P (g kg−1) | 0.48 ± 0.02 | 0.66 ± 0.18 | 0.54 ± 0.02 |
| Total K (g kg−1) | 13.7 ± 0.20 | 14.7 ± 0.78 | 13.77 ± 0.71 |
| OM (%) | 4.66 ± 0.90 | 3.79 ± 0.39 | 4.26 ± 0.49 |
| Total Cd (mg kg−1) | 9.09 ± 0.44 | 10.03 ± 0.45 | 9.73 ± 1.62 |
3.2. Characterizations of heterotrophic isolates
Seven heterotrophic acidophilic isolates formed obvious yellow halos on modified PDA medium. The seven fungal isolates distinguished by their morphological characters were named as F1, F2, F3, F4, zp1, zp2 and zp3, respectively (Fig. S2 of ESI†). Then the proofread sequences and 20 other strains selected from GenBank were used for constructing the phylogenetic tree of the 7 isolates as reflected in Fig. 1.43 Among them, F1 showed a 99% sequence homology with the strain Aureobasidium pullulans isolate LABBIO17. Thus, F1 was identified as Aureobasidium pullulans isolate F1 (MN513330).44 Accordingly, the isolate F2, F3, F4, zp1, zp2 and zp3 were identified as Candida palmioleophila isolate F2 (MN513331), Fungal sp. isolate F3 (MN513332), Meyerozyma guilliermondii isolate F4 (MN513333), Aspergillus fumigatus isolate zp1 (MN519606), Penicillium glabrum isolate zp2 (MN519607) and Trichoderma asperellum isolate zp3 (MN519608), respectively.
Fig. 1. Phylogenetic tree based on ITS gene sequence of F1, F2, F3, F4, zp1, zp2 and zp3, derived from Neighbour Joining method using MEGA X software.
3.3. Bioleaching performance
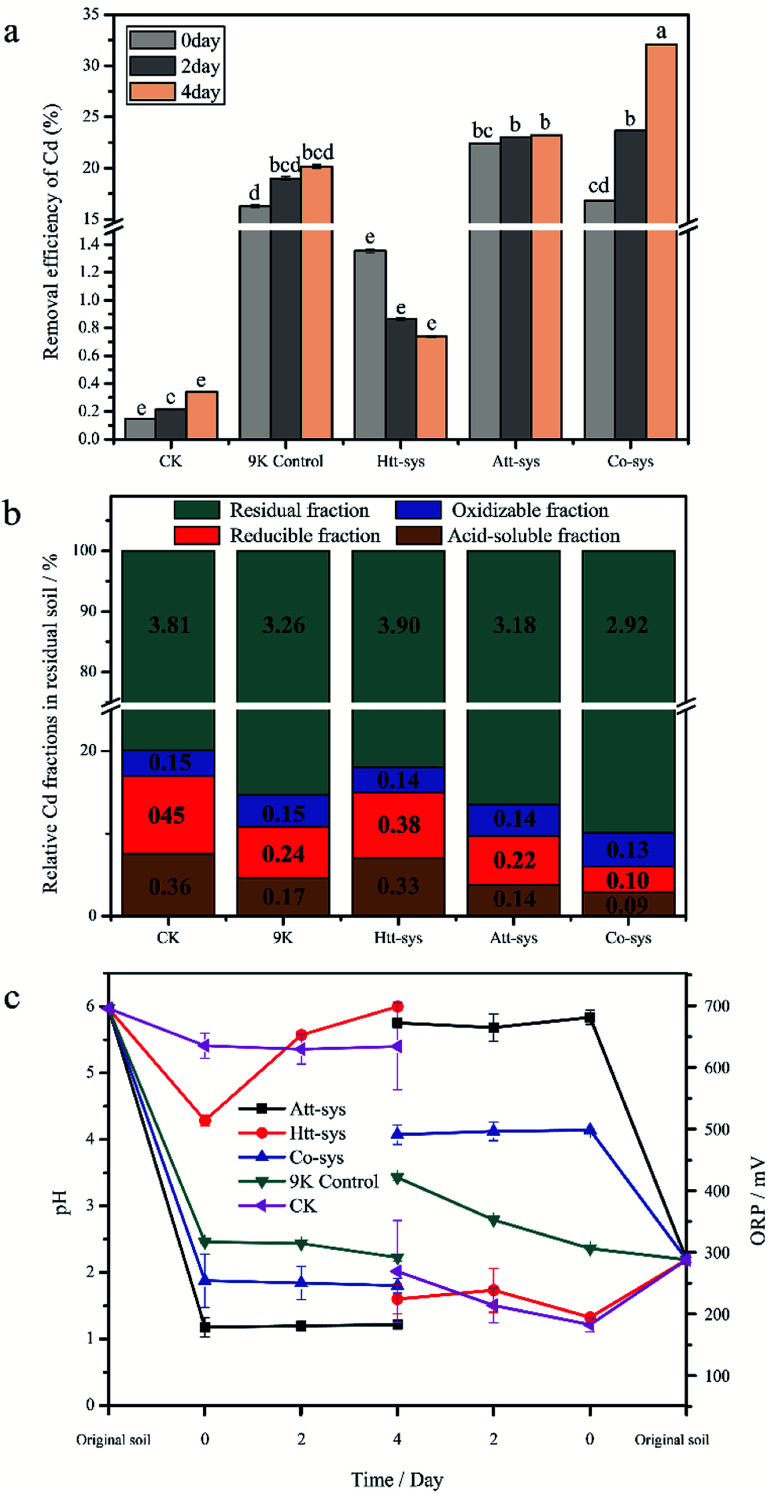
The removal efficiency of total Cd in the bioremediation process is shown in Fig. 2a. It could be clearly observed that there is no significant differences between the Htt-sys and CK (p > 0.05), which means that the effect of Htt-sys on Cd removal was not significant in the presence of pure heterotrophic mixture. Specifically, the Cd removal efficiencies of CK and Htt-sys on fourth day were 0.73% and 0.34%, respectively. Unlike the rising trend of removal percentages of other groups, the removal efficiency of Htt-sys decreased as time proceeded. The decrease of heavy metal content in leaching solution may be attributed to the adsorption of fungi in the Htt-sys. The Cd removal efficiencies of 9K control, Att-sys and Co-sys were significantly higher than that of CK and Htt-sys (p < 0.05). Prominent Cd removal efficiencies were observed in the Co-sys (23.66%), Att-sys (23.03%) and 9K control (18.99%) at the first 2 days, and there was no significant (p > 0.05) difference among these three leaching systems. Especially, the leaching efficiency of Co-sys at the second day was higher than that of the Att-sys at the fourth day. Compared with CK group, 9K control showed a higher leaching efficiency which might attribute to due to its lower pH value (Fig. 2c). However, the removal efficiency of the Co-sys (32.09%) was significantly (p < 0.05) higher than that of 9K control (20.15%) and Att-sys (23.24%) in next 2 days.
Fig. 2. (a) Removal efficiency of Cd in three leaching systems (means ± SD). The different small letters represented the significantly different according to the Fisher's LSD test. (b) Relative Cd fractions in residual soils of five leaching systems, figures represent the contents of each Cd fraction in soils. (c) Variations of pH and ORP in three leaching systems (means ± SD).
The content of four fractions (acid-soluble fraction, reducible fraction, oxidizable fraction and residual fraction) of Cd in residual soils using five leaching systems were shown in Fig. 2b. In five leaching systems, residual fractions (79.9% to 90.0%) were the dominant phase and the results were coincided with the Cd fraction distribution of previous study.25 In CK group, the oxidizable fraction (3.1%) only accounted for a small proportion, while the residual fraction (79.9%) was the dominant phase and followed by reducible fraction (9.4%) and acid-soluble fraction (7.6%). Compared with CK group, the similar distribution was observed in Htt-sys. Conversely, the distinct Cd partitioning were observed in 9K group, Att-sys and Co-sys. The acid-soluble fraction and reducible fraction in the three leaching systems all reduced to varying degrees compared to the CK group. As for the acid-soluble fraction and reducible fraction, their distribution in 9K (4.6% and 6.3%) were closed to that in Att-sys (3.8% and 5.6%). However, the acid-soluble fraction and reducible fraction in Co-sys (2.8% and 3.2%) were significantly (p < 0.05) different from that in other four leaching systems. The percentages of residual fraction and oxidizable fraction (90.0% and 4.0%) in Co-sys were increased compared with other leaching systems. In Co-sys, the content of the four Cd fractions all decreased compared with other four leaching systems.
The variations of physicochemical parameters during the leaching process exhibited that five leaching systems with different initial microbial compositions promoted the leaching process to different degrees (Fig. 2c). The pH decreasing extents of different leaching systems could be ranked as order as following, Att-sys > Co-sys > 9K control > Htt-sys > CK. In the Co-sys, Att-sys and 9K control, pH decreased rapidly when three different fresh solutions were mixed with the contaminated soils. The leachate pH of Att-sys and Co-sys were declined suddenly and then maintained at about 1.20, and 2.20 respectively, which could be attributed to the excess protons in the soil of the two bioleaching systems. However, the pH in the Htt-sys gradually increases to about 6.0 during the leaching process. Notably, the pH of Co-sys was higher than that of Att-sys during the whole leaching process. As shown in Fig. 2c, a similar ORP trend as pH values was observed. Specifically, the Att-sys and Co-sys showed the highest ORP, while the 9K control, Htt-sys and CK showed lower ORP. As expected, the Att-sys and Co-sys both exhibited high removal efficiencies of Cd.
3.4. Changes of microbial community structure
3.4.1. Microbial community composition and relative abundance
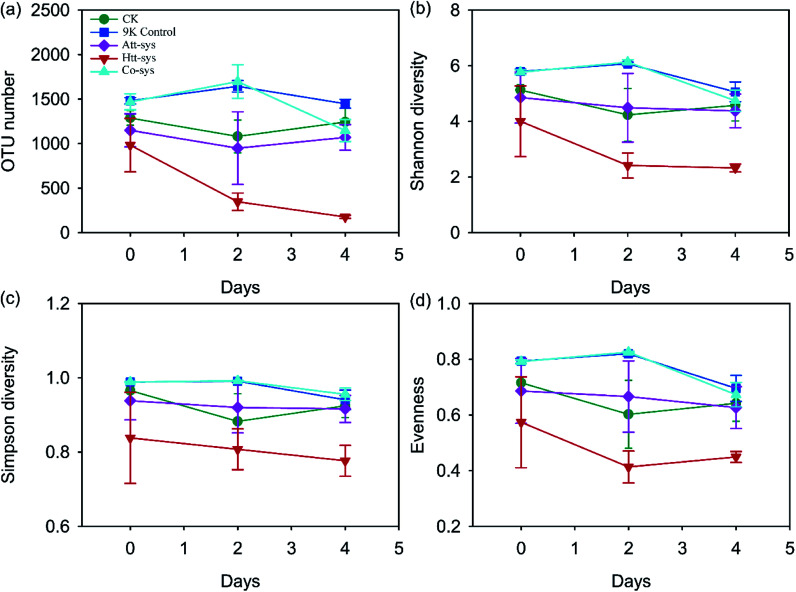
Alpha-diversity indexes (OTU number, Shannon index, Simpson index and evenness) in original soil, CK, 9K control, Att-sys and Co-sys were all significantly higher than that in Htt-sys (p < 0.05) (Table S2 of ESI†). The OTU number of leached residues in Co-sys and Att-sys were close to that of CK, and no significant differences (p > 0.05) were observed between the Att-sys and Co-sys. The Co-sys performed the highest Cd removal efficiency (Fig. 2a), which showed that the survived functional species in Co-sys would have a higher Cd bioleaching performance due to interactions between heterotrophic isolates and autotrophic leaching bacteria. The changes of soil microbial diversity during the leaching process are shown in Fig. 3. No significant change was observed in CK group during the whole leaching process, which corroborated that heterotrophic isolates can significantly reduce soil microbial diversity. Compared with 9K control and CK, the observed OTU number, Shannon diversity, Simpson diversity and evenness of Htt-sys decreased sharply in the first two days, but the decreasing trend became inconspicuous in next two days. It also could be observed that the Att-sys had lower microbial diversity compared with Co-sys and 9K control.
Fig. 3. Changes of OTU number (a), Shannon diversity (b), Simpson diversity (c) and Evenness (d) of five leaching systems during the whole leaching process.
The Mantel test listed in Table 2 showed that the whole environmental factors were significantly (r = 0.366, p = 0.001) correlated with the change of microbial community structure. The total Cd content of the paddy soil, pH and ORP were crucial factors affecting the microbial community composition (p < 0.01).
Mantel test of different environmental factors and the change of microbial community structure. The r value represents the correlation between different factors, and the p value indicates the correlation is significant.
| r | p | |
|---|---|---|
| Total factors | 0.366 | 0.001 |
| pH | 0.447 | 0.001 |
| ORP | 0.163 | 0.006 |
| Total Cd | 0.357 | 0.001 |
3.4.2. Taxonomic complexity of microbial community
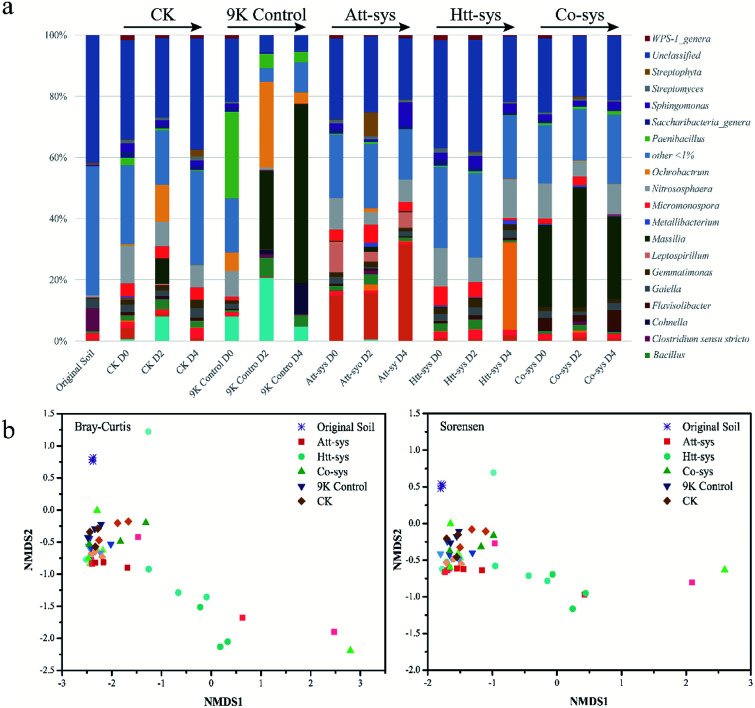
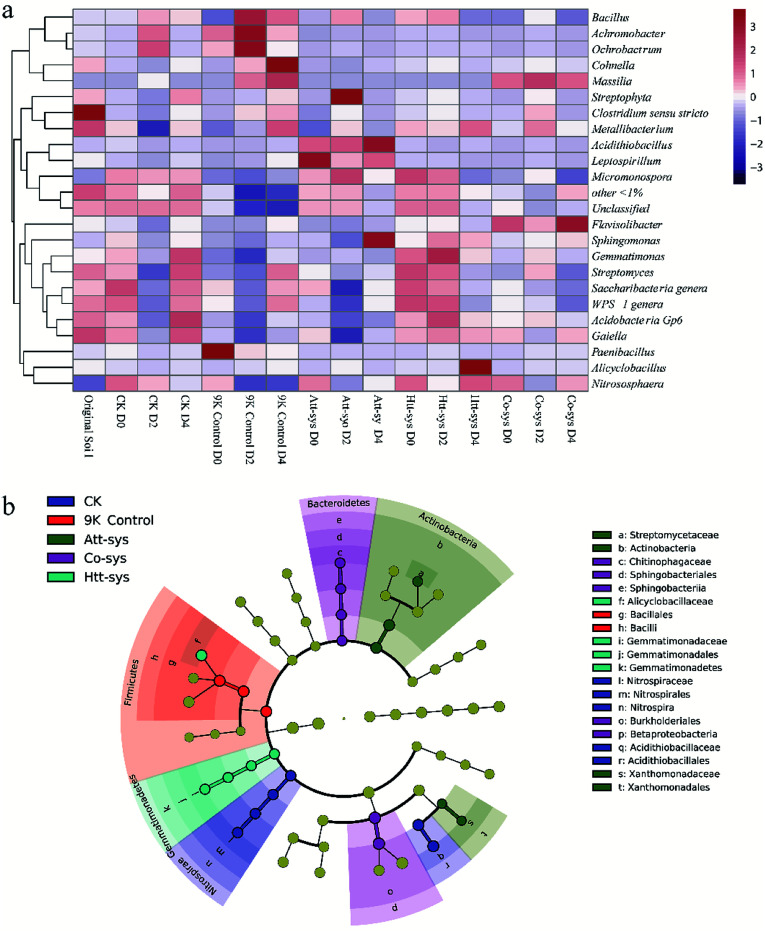
The shifts of microbial community compositions in different leaching systems are crucial to evaluate the bioremediation effects and soil microenvironmental changes.8 Since the Co-sys showed the highest Cd removal efficiency compare to single leaching system, further investigations were conducted to analyze the microbial community composition and dynamics by Illumina Sequencing. Microbial community compositions in different leaching systems were shown in Fig. 4a. It also could be observed that the microbial community changed significantly at the genus level in different leaching systems. The relative abundance of Massilia in 9K control increased sharply from 0.1% to 59% after 4 days leaching. Similarly, the relative abundance of Achromobacter and Ochrobactrum also increased in 9K control especially achieved about 20.7% and 28% after 2 days leaching. However, the existence of Achromobacter was hardly observed in bioleaching groups. The composition of soil microbial community of Att-sys was significantly different from that of CK and 9K control. As shown in Fig. 4a, the relative abundance of Acidithiobacillus increased sharply from initial 15% to 32%, and gradually became the dominant microbe. However, a considerable proportion of indigenous microorganisms survived from Att-sys, specially the high abundance unclassified genera decreased from 27% to 20% and low abundance genera (<1%) decreased from 21% to 16%. Furthermore, the soil microbial community composition also shifted significantly in Htt-sys. The relative abundance of acidophilic Alicyclobacillus increased to 29% after 4 days leaching while the proportion of Micromonospora were gradually decreased. In Co-sys, the Massilia kept a high relative abundance during the whole leaching process and increased from 27% to 39%. Unexpectedly, the low relative abundance (0.6%–1.2%) of the Alicyclobacillus was observed in Co-sys. Yet even so, highest Cd leaching efficiency was obtained in Co-sys (pH 2.23), followed by Att-sys (pH 1.22) and 9K group (pH 1.80), and this might be related to the high abundance of Massilia. It can be dramatically observed that the Acidithiobacillus had low percentages in the Co-sys while the Flavisolibacter had high percentages (6.6%). We speculated that in Co-sys, the heterotrophic isolates increased the pH of the leaching solution, leading to the growth of some pH-sensitive indigenous microorganisms in the soil, which might improve the removal efficiency of Cd by the Co-sys.
Fig. 4. (a) Relative abundance of microbial community structure using different treatments during the leaching process (Genus level). (b) Changes in microbial community structure after various treatments based on NMDS, the same symbol represents the same leaching group, with light colors for 0 days, darker for 2 days, and darkest for 4 days.
The changes of microbial community structure in different leaching systems were analyzed based on NMDS as shown in Fig. 4b. Clearly, all of the samples in treated soils from 0 to 4 day were not clustered together and separated from soil samples of 9K control, CK and Co-sys. Further, the Htt-sys showed a transformation gradient trend during the leaching process, indicated that the effect of heterotrophic isolates on indigenous microorganisms was a continuous process. Besides, the soil samples in Att-sys at the initial bioleaching were clustered together with CK but separated subsequently. This result might indicate that microbial composition changed in autotrophic bioleaching process. However, 9K, CK and Co-sys all showed no significant differences along with the treat time. The results showed that soil microbial communities changed indistinctly in the three leaching systems, and Co-sys had less impact on indigenous microorganisms. PerMANOVA (Permutational multivariate analysis of variance) (Fig. S3 of ESI†) illustrated that the microbial community in the Co-sys was closer to that in original soil.
As shown in Fig. 5a, it can be observed the distribution of different genera in different leaching systems. In Co-sys, the transformation of dominant microbes at the genus level was more moderate compared to 9K treatment and Att-sys. In CK group, the distribution of indigenous microbes was closed to that in original soils, such as genera Gaiella, Acidobacteria Gp6, Saccharibacteria, Streptomyces, etc. However, in 9K control and Att-sys, the microbial transformation at the genus level was sharper compared to CK group, Htt-sys and Co-sys. LEfSe (Linear discriminant analysis Effect Size) is a reliable method to detect biomarkers in microbiome.45,46 Various taxa were enriched in different leaching systems. For example, Sphingobacteriia (Class), Sphingobacteriales (Order), Chitinophagaceae (Family), Betaproteobacteria (Class) and Burkholderiales (Order) were enriched in the Co-sys (Fig. 5b). Xanthomonadales (Order), Xanthomonadaceae (Family), Actinobacteria (Class) and Streptomycetaceae (Family) were enriched in Att-sys. LEfSe analysis also indicated that Gemmatimonadaceae (Family) and Alicyclobacillaceae (Family) acted as the biomarkers in Htt-sys.
Fig. 5. (a) A heatmap showing the relationship between relative abundances (>1.0% of total OTU abundance) and different leaching groups. Colour codes: Red rectangles = high relative abundance, purple rectangles = low relative abundance and white rectangles = intermediate abundance. Groups are clustered in columns and OTUs in rows. (b) LEfSe analysis of microbial community in Cd-contaminated soils of five leaching systems (Yellow circles represent non-significant (p > 0.05) differences in abundance among five leaching system, other colors represent biomarkers with significant (p < 0.05) differences in different leaching groups).
4. Discussion
The Cd removal efficiency of the Co-sys was significantly higher than that of the Att-sys or Htt-sys within four days leaching. It suggested that Cd removal efficiency of Co-sys would be much higher than that of the Att-sys if given sufficient leaching time. Many changes of oxidizable fraction and residual fraction between different leaching systems were observed. The Cd fractions with high activity (e.g. reducible fraction and acid-soluble fraction) could lead to potentially environmental risks.47 It lays a fact that, in Co-sys, the increase of these forms means a decrease in the toxicity of Cd to the environment. The bioremediation of heavy metals contaminated solid wastes is generally based on bioleaching by Acidithiobacillus spp. which are autotrophic microbes.48 In fact, the bioleaching treatments were difficult to extensively carry out for paddy soils bioremediation due to its dependence on acidic conditions.49
However, the protons produced by heterotrophic isolates in the soil that were insufficient to dissolve heavy metals from the paddy soil. This confirmed previous speculations that heterotrophic isolates could not only dissolve heavy metals, could also be used in biomineralization and detoxification of heavy metals. To a certain extent, 7 heterotrophic isolates (Aureobasidium pullulans F1, Candida palmioleophila F2, Fungal sp. F3, Meyerozyma guilliermondii F4, Aspergillus fumigatus isolate zp1, Penicillium glabrum isolate zp2 and Penicillium glabrum isolate zp3) were played important roles in detoxification during the bioleaching process. We speculated that the inhibition of heavy metals on indigenous microbes will be alleviated. The lower pH values of Att-sys, Co-sys and 9K control might explain the higher Cd removal efficiencies compared with the Htt-sys and CK. pH was a crucial factor affecting the form of heavy metals in soils and low pH not only improved heavy metals mobility but also prevents heavy metals from re-stabilizing.50,51 Low pH will cause the dissolution of organic matter, while heterotrophic isolates can reduce the toxicity of dissolved organic matter to acidophilic autotrophic bacteria.52 Unexpectedly, higher pH values were observed in Co-sys which may be determined by the initial pH value of fresh leaching solutions.53 Yet, it cannot be excluded that heterotrophic isolates participate in the role of pH in the Co-sys. Seidel believed that the carbon produced by heterotrophic microbes in the bioleaching process far exceeds the carbon demand of autotrophic bacteria.54 That, the supplemented carbon can promote the growth of certain A. ferrooxidans. Dramatically, the Co-sys which had heterotrophic isolates attained lower acidification efficiency but almost reached the bioleaching end point 2 days earlier compared to Att-sys. In fact, the dissolved organic matter hampered the utilization of substrates by functional microorganisms, thus prolongated the bioleaching process.55 The highest Cd removal efficiency observed might suggest that pH and ORP were not sole determinants for cooperative bioremediation. The heterotrophic isolates might play important roles in cooperative bioremediation.56,57 Conclusively, the dependence of cooperative bioleaching on acidic conditions were abated. The bioremediation by Co-sys was a complex process and the co-existence of heterotrophic and autotrophic acidophiles could determine a higher Cd removal efficiency due to a synergistic metabolism.
The microbial diversity of soil after treated by different leaching systems might suggest that the metabolites of autotrophic microbes can inhibit the growth of soil microbes, but the inhibition was alleviated in the Co-sys, and similar phenomena was described on several occasions by Johnson.58 All the observations of Alpha-diversity indexes demonstrated that the inoculated heterotrophic isolates and autotrophic mixture could reduce soil microbial diversity while Co-sys alleviated the impact of inoculating microbes on altering soil microbial community. It can be observed that the distribution of the dominant genera in the Co-sys was similar with the original soil and CK group (Fig. 5a). It confirmed that inhibition of heavy metals on indigenous microbes will be alleviated and the impact of exogenous microorganisms on indigenous microorganisms was moderate in Co-sys.
As shown in Fig. 5b, we found that Order Burkholderiales, Family Alicyclobacillaceae, genera Massilia and genera Flavisolibacte played major roles in Co-sys. The Order Burkholderiales was a biomarker in Co-sys, it meant the Order Burkholderiales was mportant in Co-sys during the bioleaching process. Yang isolated a strain of Burkholderiales (Burkholderia sp. Z-90) and this strain was a biosurfactant producer.17 The formation of metal complexes of heavy metals and biosurfactants is the key pathway to remove heavy metals. It was verified that the inhibition of heavy metals on indigenous microbes will be alleviated when inoculated heterotrophic isolates, and it can explain the bioaugmentation mechanism of heterotrophic isolates to autotrophic bioleaching. Significant growth of the Massilia in the 9K control might be resulted with high salinity and low pH could especially promote the survival of some species of Massilia, but the acidic habitat in 9K control inhibited the growth of some indigenous microorganisms. The highest Cd removal efficiency was obtained in Co-sys, followed by Att-sys. However, in Co-sys, genus Massilia kept a high relative abundance during the whole leaching process. This result might imply that the Massilia may play an important role in the bioremediation performance, thus resulting in high Cd removal efficiency in Co-sys. Zhang isolated four strains of the Massilia from heavy metal contaminated soil and preliminarily considered that the Massilia was related to heavy metal pollution.59 45 arsenic-resistant bacteria of Massilia were isolated by Du from mining soil.60 Based on this, we speculated that the genus Massilia existed extensively in Cd contaminated soils in this study. For example, LEfSe analysis (Fig. 4b) showed that the genus Massilia of Order Burkholderiales were biomarkers in Co-sys. However, some species of Massilia were widely existed in 9K but LEfSe analysis indicated that only Bacillales (Order) and Bacilli (Class) were biomarkers in 9K. The results suggested the genus Massilia had strong correlation with high removal efficiency. The emergence of Acidithiobacillus was expected in Att-sys and proved that the Acidithiobacillus was a key factor influencing bioleaching performance of Att-sys. However, a considerable proportion of indigenous microorganisms survived from Att-sys might also play an important role in bioleaching performance. Interestingly, the genus Micromonospora only exists in a large proportion of Htt-sys while existed in a small proportion of other bioleaching systems. Ali found that the Micromonospora were predominant heavy metals resistant inhabitants in soils.61 Furthermore, the increase of acidophilic Alicyclobacillus in Htt-sys might suggested that the inoculation of heterotrophic isolates promoted the growth of some heavy metals resistant inhabitants.62–64 It was found that Alicyclobacillus and Sulfobacillus had similar habitats and a synergistic effect, in which Sulfobacillus oxidize Fe2+ to Fe3+ and the metabolites of Sulfobacillus can be consumed by Alicyclobacillus.63 The increase of acidophilic heterotrophs promoted the Cd leaching since they could decrease the content of reducible Cd.48 Unexpectedly, though the Family Alicyclobacillaceae which contains genus Alicyclobacillus was a biomarker in Htt-sys, the low relative abundance the Alicyclobacillus was observed in Co-sys. Some studies demonstrated that all of Alicyclobacillus genus can coexist with autotrophic bioleaching bacteria by utilizing the Fe(ii) and sulfur.63,65,66 Further, Gurung reported that the Alicyclobacillus genus could alleviate the inhibition of dissolved organic matter on the growth of acidophilic Acidiphilium by consuming these organics.67 This might infer that certain indigenous microbes have become dominant genera in Co-sys. It is worth exploring the emergence of Flavisolibacter in the Co-sys. The Flavisolibacter was identified by Kasemodel in highly contaminated metal soils.68 Furthermore, LEfSe also illustrated that the Family Chitinophagaceae which contains genus Flavisolibacter was a biomarker in Co-sys (Fig. 5b). However, the indigenous Flavisolibacter had low percentage in Att-sys, we can draw a conclusion that the Co-sys can promote the growth of some heavy metal-resistant bacteria in indigenous microorganisms. PerMANOVA reflected by Fig. S3 of ESI† was corroborated that the impact of Co-sys on indigenous soil microbes was more moderate than that of single leaching system. The results demonstrated that Co-sys could reduce the effects on indigenous microbial community composition while avoid soil acidification due to the colonization of acidophilic microorganisms.
5. Conclusions
In the present study, 7 heterotrophic isolates, Aureobasidium pullulans F1 (MN513330), Candida palmioleophila F2 (MN513331), Fungal sp. F3 (MN513332), Meyerozyma guilliermondii F4 (MN513333), Aspergillus fumigatus isolate zp1 (MN519606), Penicillium glabrum isolate zp2 (MN519607) and Penicillium glabrum isolate zp3 (MN519608) were isolated from the mixotrophic acidophiles which could remove Cd from the paddy soil. The results of bioremediation by three leaching systems clearly demonstrated that the Co-sys had the highest Cd removal efficiency (32.09%) from Cd-contaminated soils compared with Att-sys (23.24%) and Htt-sys (0.74%). The pH and ORP were the major contributors to the high Cd removal efficiency in the bioleaching process. The inoculation of heterotrophic isolates promoted the growth of some heavy metal resistant inhabitants (Massilia, Alicyclobacillus, Micromonospora, etc.). It was found that, the cooperative bioremediation had minor effects on soil indigenous microbial community and could ameliorate the habitat. The cooperative bioremediation by using heterotrophic and autotrophic mixtures proved to be an efficient and short-term bioremediation strategy for heavy metal contaminated soil without disturbing the habitat.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grants No. 2018YFC1800400), Major Science and Technology Programs of Changsha (No. kq1902047) and the National Natural Science Foundation of China (41907032).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03935g
Notes and references
- Li X. Peng W. Jia Y. Lu L. Fan W. Chemosphere. 2016;156:228–235. doi: 10.1016/j.chemosphere.2016.04.098. [DOI] [PubMed] [Google Scholar]
- Wu C. Shi L. Xue S. Li W. Jiang X. Rajendran M. Qian Z. Sci. Total Environ. 2019;647:1158–1168. doi: 10.1016/j.scitotenv.2018.08.087. [DOI] [PubMed] [Google Scholar]
- Kang C.-H. Kwon Y.-J. So J.-S. Ecol. Eng. 2016;89:64–69. doi: 10.1016/j.ecoleng.2016.01.023. [DOI] [Google Scholar]
- Peng W. Li X. Song J. Jiang W. Liu Y. Fan W. Chemosphere. 2018;197:33–41. doi: 10.1016/j.chemosphere.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Wang D. Bai J. Wang W. Zhang G. Cui B. Liu X. Li X. Land Degrad. Dev. 2018;29:3783–3794. doi: 10.1002/ldr.3086. [DOI] [Google Scholar]
- Wu G. Kang H. Zhang X. Shao H. Chu L. Ruan C. J. Hazard. Mater. 2010;174:1–8. doi: 10.1016/j.jhazmat.2009.09.113. [DOI] [PubMed] [Google Scholar]
- Luptakova A. Kusnierova M. Hydrometallurgy. 2005;77:97–102. doi: 10.1016/j.hydromet.2004.10.019. [DOI] [Google Scholar]
- Cui Z. Zhang X. Yang H. Sun L. J. Environ. Chem. Eng. 2017;5:3616–3621. doi: 10.1016/j.jece.2017.07.021. [DOI] [Google Scholar]
- Wang T. Sun H. Mao H. Zhang Y. Wang C. Zhang Z. Wang B. Sun L. J. Hazard. Mater. 2014;278:483–490. doi: 10.1016/j.jhazmat.2014.06.028. [DOI] [PubMed] [Google Scholar]
- Fan W. Jia Y. Li X. Jiang W. Lu L. Chemosphere. 2012;88:751–756. doi: 10.1016/j.chemosphere.2012.04.047. [DOI] [PubMed] [Google Scholar]
- Geebelen W. Adriano D. C. Lelie D. V. D. Mench M. Carleer R. Clijsters H. Vangronsveld J. Plant Soil. 2003;249:217–228. doi: 10.1023/A:1022534524063. [DOI] [Google Scholar]
- Mench M. J. Manceau A. Vangronsveld J. Clijsters H. Mocquot B. Agronomie. 2000;20:383–397. doi: 10.1051/agro:2000135. [DOI] [Google Scholar]
- Gray C. W. Dunham S. J. Dennis P. G. Zhao F. J. McGrath S. P. Environ. Pollut. 2006;142:530–539. doi: 10.1016/j.envpol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Sun Y. Zhou Q. Wang L. Liu W. J. Hazard. Mater. 2009;161:808–814. doi: 10.1016/j.jhazmat.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Li D. Li R. Ding Z. Ruan X. Luo J. Chen J. Zheng J. Tang J. Chemosphere. 2019;241:125039. doi: 10.1016/j.chemosphere.2019.125039. [DOI] [PubMed] [Google Scholar]
- Khan I. Ali M. Aftab M. Shakir S. Qayyum S. Haleem K. S. Tauseef I. Environ. Monit. Assess. 2019;191:622. doi: 10.1007/s10661-019-7781-9. [DOI] [PubMed] [Google Scholar]
- Yang Z. Zhang Z. Chai L. Wang Y. Liu Y. Xiao R. J. Hazard. Mater. 2016;301:145–152. doi: 10.1016/j.jhazmat.2015.08.047. [DOI] [PubMed] [Google Scholar]
- Deng X. Chai L. Yang Z. Tang C. Wang Y. Shi Y. J. Hazard. Mater. 2013;248–249:107–114. doi: 10.1016/j.jhazmat.2012.12.051. [DOI] [PubMed] [Google Scholar]
- Jang M. Hwang J. S. Choi S. I. Chemosphere. 2007;66:8–17. doi: 10.1016/j.chemosphere.2006.05.056. [DOI] [PubMed] [Google Scholar]
- Beolchini F. Fonti V. Rocchetti L. Saraceni G. Pietrangeli B. Dell'Anno A. Chem. Ecol. 2013;29:415–426. doi: 10.1080/02757540.2013.776547. [DOI] [Google Scholar]
- Fonti V. Dell'Anno A. Beolchini F. Water Res. 2013;47:5139–5152. doi: 10.1016/j.watres.2013.05.052. [DOI] [PubMed] [Google Scholar]
- Camargo F. P. Prado P. F. D. Tonello P. S. Dos Santos A. C. A. Duarte I. C. S. J. Environ. Manage. 2018;211:28–35. doi: 10.1016/j.jenvman.2018.01.045. [DOI] [PubMed] [Google Scholar]
- Fang D. Zhang R. Zhou L. Li J. J. Hazard. Mater. 2011;192:226–233. doi: 10.1016/j.jhazmat.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Ma L. Wang X. Tao J. Xue F. Kai Z. Xiao Y. Liang Y. Yin H. Liu X. Hydrometallurgy. 2016;169:41–46. doi: 10.1016/j.hydromet.2016.12.007. [DOI] [Google Scholar]
- Hao X. Zhu P. Zhang H. Liang Y. Yin H. Liu X. Bai L. Liu H. Jiang H. Sci. Total Environ. 2019;655:347–355. doi: 10.1016/j.scitotenv.2018.11.221. [DOI] [PubMed] [Google Scholar]
- Xiao Y. Liu X. Dong W. Liang Y. Niu J. Gu Y. Ma L. Hao X. Zhang X. Xu Z. Yin H. Arch. Microbiol. 2017;199:757–766. doi: 10.1007/s00203-017-1342-9. [DOI] [PubMed] [Google Scholar]
- Ma L. Wang H. Wu J. Wang Y. Zhang D. Liu X. Bioresour. Technol. 2019;280:9–17. doi: 10.1016/j.biortech.2019.01.117. [DOI] [PubMed] [Google Scholar]
- Ma L. Wang X. Feng X. Liang Y. Xiao Y. Hao X. Yin H. Liu H. Liu X. Bioresour. Technol. 2017;223:121–130. doi: 10.1016/j.biortech.2016.10.056. [DOI] [PubMed] [Google Scholar]
- Wang S. Zheng G. Zhou L. Water Res. 2010;44:5423–5431. doi: 10.1016/j.watres.2010.06.055. [DOI] [PubMed] [Google Scholar]
- Gan M. Song Z. Zhu J. Liu X. Environ. Earth Sci. 2016;75:457. doi: 10.1007/s12665-016-5307-0. [DOI] [Google Scholar]
- Emenike C. U. Agamuthu P. Fauziah S. H. J. Geochem. Explor. 2017;182:275–278. doi: 10.1016/j.gexplo.2016.10.002. [DOI] [Google Scholar]
- Pathak A. Dastidar M. G. Sreekrishnan T. R. J. Environ. Manage. 2009;90:2343–2353. doi: 10.1016/j.jenvman.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Rodríguez Y. Hydrometallurgy. 2003;71:57–66. doi: 10.1016/S0304-386X(03)00174-9. [DOI] [Google Scholar]
- Deng Y. Jiang L. Xu L. Hao X. Zhang S. Xu M. Zhu P. Fu S. Liang Y. Yin H. Liu X. Bai L. Jiang H. Liu H. Ecotoxicol. Environ. Saf. 2019;171:281–289. doi: 10.1016/j.ecoenv.2018.12.060. [DOI] [PubMed] [Google Scholar]
- Liu B. L. Ma X. W. Ai S. W. Zhu S. Y. Zhang W. Y. Zhang Y. M. J. Soils Sediments. 2016;16:1547–1556. doi: 10.1007/s11368-016-1351-3. [DOI] [Google Scholar]
- Rajkumar M. Freitas H. Chemosphere. 2008;71:834–842. doi: 10.1016/j.chemosphere.2007.11.038. [DOI] [PubMed] [Google Scholar]
- Umrania V. V. Bioresour. Technol. 2006;97:1237–1242. doi: 10.1016/j.biortech.2005.04.048. [DOI] [PubMed] [Google Scholar]
- Zeng W.-m. Wu C.-b. Zhang R.-b. Hu P.-l. Qiu G.-z. Gu G.-h. Zhou H.-b. Trans. Nonferrous Metals Soc. China. 2009;19:222–227. doi: 10.1016/S1003-6326(08)60256-3. [DOI] [Google Scholar]
- Hu Q. Dou M.-n. Qi H.-y. Xie X.-m. Zhuang G.-q. Yang M. J. Environ. Sci. 2007;19:1114–1119. doi: 10.1016/S1001-0742(07)60181-8. [DOI] [Google Scholar]
- Zhang X. Liu X. Liang Y. Xiao Y. Ma L. Guo X. Miao B. Liu H. Peng D. Huang W. Yin H. Front. Microbiol. 2017;8:790. doi: 10.3389/fmicb.2017.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleyzes C. Tellier S. Astruc M. Trends Anal. Chem. 2002;21:451–467. doi: 10.1016/S0165-9936(02)00603-9. [DOI] [Google Scholar]
- Edgar R. C. Nat. Methods. 2013;10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Zloch M. Thiem D. Gadzala-Kopciuch R. Hrynkiewicz K. Chemosphere. 2016;156:312–325. doi: 10.1016/j.chemosphere.2016.04.130. [DOI] [PubMed] [Google Scholar]
- Qian X. Fang C. Huang M. Achal V. J. Clean. Prod. 2017;164:198–208. doi: 10.1016/j.jclepro.2017.06.195. [DOI] [Google Scholar]
- Segata N. Izard J. Waldron L. Gevers D. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao E. Krumins V. Xiao T. Dong Y. Tang S. Ning Z. Huang Z. Sun W. Environ. Pollut. 2017;221:244–255. doi: 10.1016/j.envpol.2016.11.071. [DOI] [PubMed] [Google Scholar]
- Kasemodel M. C. Sakamoto I. K. Varesche M. B. A. Rodrigues V. G. S. Sci. Total Environ. 2019;675:367–379. doi: 10.1016/j.scitotenv.2019.04.223. [DOI] [PubMed] [Google Scholar]
- Beolchini F. Dell'Anno A. De Propris L. Ubaldini S. Cerrone F. Danovaro R. Chemosphere. 2009;74:1321–1326. doi: 10.1016/j.chemosphere.2008.11.057. [DOI] [PubMed] [Google Scholar]
- Chen S. Y. Lin J. G. J. Hazard. Mater. 2001;82:77–89. doi: 10.1016/S0304-3894(00)00357-5. [DOI] [PubMed] [Google Scholar]
- Sauvé S. Hendershot W. Allen H. E. Environ. Sci. Technol. 2000;34:1125–1131. doi: 10.1021/es9907764. [DOI] [Google Scholar]
- Chen S. Y. Lin J. G. Chemosphere. 2001;44:1093–1102. doi: 10.1016/S0045-6535(00)00334-9. [DOI] [PubMed] [Google Scholar]
- Min G. Song Z. Zhu J. Liu X. Environ. Earth Sci. 2016;75:457. doi: 10.1007/s12665-016-5307-0. [DOI] [Google Scholar]
- Yu R.-l. Liu J. Tan J.-x. Zeng W.-m. Shi L.-j. Gu G.-h. Qin W.-q. Qiu G.-z. Int. J. Miner., Metall. Mater. 2014;21:311–316. doi: 10.1007/s12613-014-0910-0. [DOI] [Google Scholar]
- Seidel H. Ser C. L. Zehnsdorf A. Hoffmann P. Schmerold R. Environ. Sci. Technol. 2004;38:1582–1588. doi: 10.1021/es030075d. [DOI] [PubMed] [Google Scholar]
- Fang D. Zhou L. X. Chemosphere. 2007;69:303–310. doi: 10.1016/j.chemosphere.2007.03.059. [DOI] [PubMed] [Google Scholar]
- Tyagi R. D. and Couillard D., Biological Degradation of Wastes, 1997, pp. 307–322 [Google Scholar]
- Labana S. Singh O. V. Basu A. Pandey G. Jain R. K. Appl. Microbiol. Biotechnol. 2005;68:417–424. doi: 10.1007/s00253-005-1926-1. [DOI] [PubMed] [Google Scholar]
- Johnson D. B. FEMS Microbiol. Ecol. 1998;27:307–317. doi: 10.1111/j.1574-6941.1998.tb00547.x. [DOI] [Google Scholar]
- Zhang Y. Q. Li W. J. Zhang K. Y. Tian X. P. Jiang Y. Xu L. H. Jiang C. L. Lai R. Int. J. Syst. Evol. Microbiol. 2006;56:459–463. doi: 10.1099/ijs.0.64083-0. [DOI] [PubMed] [Google Scholar]
- Du Y. Yu X. Wang G. Int. J. Syst. Evol. Microbiol. 2012;62:2356–2362. doi: 10.1099/ijs.0.034306-0. [DOI] [PubMed] [Google Scholar]
- Ali N. Dashti N. Al-Mailem D. Eliyas M. Radwan S. Environ. Sci. Pollut. Res. Int. 2012;19:812–820. doi: 10.1007/s11356-011-0624-z. [DOI] [PubMed] [Google Scholar]
- Kaksonen A. H. Särkijärvi S. Puhakka J. A. Peuraniemi E. Junnikkala S. Tuovinen O. H. Hydrometallurgy. 2016;159:46–53. doi: 10.1016/j.hydromet.2015.10.032. [DOI] [Google Scholar]
- Wang S. Zheng Y. Yan W. Chen L. Dummi Mahadevan G. Zhao F. J. Hazard. Mater. 2016;320:393–400. doi: 10.1016/j.jhazmat.2016.08.054. [DOI] [PubMed] [Google Scholar]
- Mehrotra A. Sreekrishnan T. R. Environ. Technol. 2017;38:2709–2724. doi: 10.1080/09593330.2016.1275821. [DOI] [PubMed] [Google Scholar]
- Xu G. Xiao-Yan Y. Li-Jun L. Jia-Yue Z. Shuang-Jiang L. Cheng-Ying J. Int. J. Syst. Evol. Microbiol. 2009;59(0):2415–2420. doi: 10.1099/ijs.0.008870-0. [DOI] [PubMed] [Google Scholar]
- Cheng-Ying J. Ying L. Yan-Yang L. Xiao-Yan Y. Xu G. Shuang-Jiang L. Int. J. Syst. Evol. Microbiol. 2008;58:2898–2903. doi: 10.1099/ijs.0.2008/000562-0. [DOI] [PubMed] [Google Scholar]
- Gurung A. Chakraborty R. Can. J. Microbiol. 2009;55:1040–1048. doi: 10.1139/W09-062. [DOI] [PubMed] [Google Scholar]
- Kasemodel M. C. Sakamoto I. K. Varesche M. B. A. Rodrigues V. G. S. Sci. Total Environ. 2019;675:367–379. doi: 10.1016/j.scitotenv.2019.04.223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.