Key Points
Question
How does the diagnostic performance of home antigen tests change during the course of SARS-CoV-2 infection?
Findings
In this prospective cohort study of 225 adults and children with reverse transcription–polymerase chain reaction (RT-PCR)–confirmed SARS-CoV-2 infection, antigen test sensitivity was 64% and 84% when compared with same-day RT-PCR and viral culture, respectively. Antigen test sensitivity peaked 4 days after illness onset (77%); a second test 1 to 2 days later showed improved sensitivity (81%-85%).
Meaning
The study results suggest that symptomatic individuals with an initial negative home antigen test result for SARS-CoV-2 infection should test again 1 to 2 days later because test sensitivity seems to peak several days after illness onset.
Abstract
Importance
As self-collected home antigen tests become widely available, a better understanding of their performance during the course of SARS-CoV-2 infection is needed.
Objective
To evaluate the diagnostic performance of home antigen tests compared with reverse transcription–polymerase chain reaction (RT-PCR) and viral culture by days from illness onset, as well as user acceptability.
Design, Setting, and Participants
This prospective cohort study was conducted from January to May 2021 in San Diego County, California, and metropolitan Denver, Colorado. The convenience sample included adults and children with RT-PCR–confirmed infection who used self-collected home antigen tests for 15 days and underwent at least 1 nasopharyngeal swab for RT-PCR, viral culture, and sequencing.
Exposures
SARS-CoV-2 infection.
Main Outcomes and Measures
The primary outcome was the daily sensitivity of home antigen tests to detect RT-PCR–confirmed cases. Secondary outcomes included the daily percentage of antigen test, RT-PCR, and viral culture results that were positive, and antigen test sensitivity compared with same-day RT-PCR and cultures. Antigen test use errors and acceptability were assessed for a subset of participants.
Results
This study enrolled 225 persons with RT-PCR–confirmed infection (median [range] age, 29 [1-83] years; 117 female participants [52%]; 10 [4%] Asian, 6 [3%] Black or African American, 50 [22%] Hispanic or Latino, 3 [1%] Native Hawaiian or Other Pacific Islander, 145 [64%] White, and 11 [5%] multiracial individuals) who completed 3044 antigen tests and 642 nasopharyngeal swabs. Antigen test sensitivity was 50% (95% CI, 45%-55%) during the infectious period, 64% (95% CI, 56%-70%) compared with same-day RT-PCR, and 84% (95% CI, 75%-90%) compared with same-day cultures. Antigen test sensitivity peaked 4 days after illness onset at 77% (95% CI, 69%-83%). Antigen test sensitivity improved with a second antigen test 1 to 2 days later, particularly early in the infection. Six days after illness onset, antigen test result positivity was 61% (95% CI, 53%-68%). Almost all (216 [96%]) surveyed individuals reported that they would be more likely to get tested for SARS-CoV-2 infection if home antigen tests were available over the counter.
Conclusions and Relevance
The results of this cohort study of home antigen tests suggest that sensitivity for SARS-CoV-2 was moderate compared with RT-PCR and high compared with viral culture. The results also suggest that symptomatic individuals with an initial negative home antigen test result for SARS-CoV-2 infection should test again 1 to 2 days later because test sensitivity peaked several days after illness onset and improved with repeated testing.
This cohort study examines changes in the diagnostic performance of home antigen tests during the course of SARS-CoV-2 infection.
Introduction
Antigen tests for SARS-CoV-2 provide rapid, low-cost results and are approved for use outside of clinical settings. They may improve the availability, acceptability, and timeliness of SARS-CoV-2 diagnostic testing. Multiple studies have evaluated the sensitivity and specificity of antigen tests compared with real-time reverse transcription–polymerase chain reaction (RT-PCR).1,2 However, to our knowledge, few studies have examined how antigen test performance varies during the course of infection.3 To address this question, we evaluated daily use of a self-collected home antigen test compared with RT-PCR and viral culture in adults and children enrolled in a household transmission investigation.
Methods
Study Design and Oversight
Working with local and state health departments, the US Centers for Disease Control and Prevention (CDC) conducted a prospective household transmission investigation in San Diego County, California (January 18, 2021, to April 14, 2021) and metropolitan Denver, Colorado (March 22, 2021, to April 30, 2021), as previously described.4 This investigation was reviewed by CDC and was conducted according to applicable federal law and CDC policy (eg, 45 CFR part 46, 21 CFR part 565; 42 USC §241(d); 5 USC §552a; and 44 USC §3501 et seq6). The CDC determined that this investigation was a public health emergency response; as such, institutional review board review and informed consent were not required. The study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline for cohort studies.7
We recruited a convenience sample of individuals with recent RT-PCR–confirmed SARS-CoV-2 infection and their household members of all ages. Household members were defined as any individual who stayed 1 night or longer in the same residence during the infectious period (2 days before illness onset through 10 days afterward)8 of the earliest case in the household. Illness onset was defined as the symptom onset date or, if asymptomatic, the sample collection date of the first positive RT-PCR test result.8 Households were enrolled within 10 days of illness onset of the earliest case in the household.
Enrolled households were followed for 15 days. With questionnaires, we collected demographic information, including self-reported sex, race, and ethnicity, medical history, and vaccination history for each participant. Following federal government standards, participants self-reported Hispanic or Latino ethnicity and 1 or more races, and investigators categorized responses into the following categories: American Indian or Alaska Native, Asian, Black or African American, Hispanic or Latino, Native Hawaiian or Other Pacific Islander, White, or multiracial.9 All participants recorded daily symptoms during their 15-day enrollment period using a standardized form. Caregivers assisted young children with questionnaires and forms.
SARS-CoV-2 Testing
At enrollment, participants were offered home antigen tests that detect the SARS-CoV-2 nucleocapsid (N) protein using a lateral flow immunoassay (QuickVue At-Home OTC COVID-19 Test; Quidel Corporation), which received emergency use authorization from the US Food and Drug Administration on March 31, 2021, during the investigation. Participants were asked to perform 1 antigen test daily, regardless of symptoms. Participants were given 15 antigen test kits and printed instructions from the manufacturer (available in English and Spanish).10 Participants reviewed the instructions, self-collected anterior nasal swabs, and interpreted the antigen test results without guidance from investigators. At the discretion of caregivers, children either self-collected or were assisted in collecting nasal swabs and testing. Each day, immediately after testing, participants emailed a photograph of the antigen test strip and their interpretation of the result to investigators, who recorded their own interpretation. For a subset of households in Colorado, investigators observed participants perform their first antigen test and documented errors using a standardized form (eTable 1 in Supplement 1). All Colorado households were asked to complete a short survey on the last day that assessed the acceptability of home antigen testing (eTable 1 in Supplement 1).
Nasopharygneal (NP) swabs for RT-PCR and viral culture were collected by trained health care professionals from all participants at enrollment and 14 days later, regardless of symptom status. A subset of participants consented to undergo additional daily NP swabs for 7 days after enrollment. When a previously uninfected household member became symptomatic or had a newly positive home antigen test result, an additional NP swab was collected from all participating household members. All NP swabs were tested at a public health laboratory for SARS-CoV-2 ribonucleic acid by RT-PCR (TaqPath COVID-19 Combo Kit11 in Colorado; PerkinElmer New Coronavirus Nucleic Acid Detection Kit12 in California). Participants were notified of their RT-PCR results within 1 to 2 days of collection. Genome sequencing was performed for NP specimens with a positive RT-PCR result and N gene cycle threshold (Ct) value of less than 35. California specimens were sequenced at CDC,13 and Colorado specimens were sequenced at Colorado Department of Public Health and Environment as previously described.14 The SARS-CoV-2 lineages were assigned using pangolin (https://github.com/cov-lineages/pangolin). The NP specimens that were RT-PCR positive with an N gene Ct value of less than 32 were cultured as described previously.15,16 The NP specimens with an RT-PCR Ct value of 32 or greater were presumed to be culture-negative based on previous studies.17,18
Data Analysis
All participants who completed at least 1 home antigen test were included in analyses. We defined a case as a person with RT-PCR–confirmed SARS-CoV-2 infection (confirmatory laboratory criteria for COVID-19 in the Council of State and Territorial Epidemiologists case definition19) and illness onset within 10 days before or during the enrollment period. We defined symptomatic cases as individuals who reported symptoms consistent with the Council of State and Territorial Epidemiologists clinical criteria for COVID-19 at any point during their illness. We defined a noncase as a person who only had negative RT-PCR results during the investigation. When sequencing of a specimen collected from a participant with RT-PCR–confirmed infection was not successful, we assumed that the individual was infected by their household member and shared the same SARS-CoV-2 lineage. For all analyses, we excluded invalid, indeterminant, and missing test results. Partial data contributed by participants who were lost to follow-up were included.
Among cases, we calculated the daily percentage of positive home antigen tests, RT-PCR tests, and viral cultures by days from illness onset. The percentage of positivity for viral culture was defined as the number of positive cultures divided by the total number of NP specimens collected. The daily percentage of positive antigen tests was also examined by symptom status and vaccination status.
We calculated the sensitivity of the antigen test compared with 3 reference standards: (1) positive case status; (2) a positive RT-PCR test result collected the same day; and (3) a positive viral culture collected the same day. We repeated these calculations for subgroups defined by age, symptom status, vaccination status, and SARS-CoV-2 lineage. In addition, we calculated the overall specificity of home antigen tests among noncases.
To determine if serial antigen testing was associated with increased sensitivity, we compared the sensitivity of 3 antigen testing protocols: a single test (protocol 1), 2 tests on consecutive days (protocol 2), and 2 tests spaced 2 days apart (protocol 3). For these calculations, the reference standard was positive case status.
For the percentage of positivity, sensitivity, and specificity estimates, we calculated 95% confidence intervals with Wilson score intervals, a standard method for estimating confidence intervals for binomial proportions.20 For analyses that pooled repeated tests from the same participant, we adjusted confidence intervals for potential intraparticipant correlation using cluster-robust standard errors.
We calculated the concordance between the participant and investigator interpretations of the home antigen result, as well as the Cohen κ coefficient to account for concordance by chance. We calculated frequencies of observed user errors and responses to questions about the acceptability of home antigen tests. We compared the concordance of antigen and RT-PCR test results between households with and without observed errors using the Pearson χ2 test. Statistical analyses were performed with SAS, version 9.4 (SAS Institute), and Stata, version 16.1 (StataCorp). P < .05 was considered statistically significant.
Results
Among 552 individuals from 151 households enrolled in the household transmission investigation, 225 individuals (41%) from 107 households had RT-PCR–confirmed SARS-CoV-2 infection and completed at least 1 home antigen test (eFigure 1 in Supplement 1). Among these 225 cases, the median age was 29 years (range, 1-83 years); 117 (52%) were female and 205 (91%) were symptomatic (Table). Four of the 225 cases (2%) were hospitalized during their illness. Of the 225 cases, 194 (86%) had never received a COVID-19 vaccine. The most common SARS-CoV-2 lineages detected were Alpha (B.1.1.7) (126 of 225 cases [56%]), Epsilon (B.1.427/B.1.429) (35 of 225 [16%]), and Gamma (P.1) (8 of 225 [4%]); Alpha, Epsilon, and Gamma were the only variants of concern detected. Lineage was unknown for 17 of 225 cases (8%).
Table. Characteristics of Participants With RT-PCR–Confirmed SARS-CoV-2 Infection Who Completed at Least 1 Home Antigen Test.
| Characteristic | Participants, No. (%)a |
|---|---|
| No. | 225 |
| Age, median (range), y | 29 (1-83) |
| Age group, y | |
| <12 | 39 (17) |
| 12-17 | 41 (18) |
| 18-49 | 119 (53) |
| ≥50 | 26 (12) |
| Sex | |
| Female | 117 (52) |
| Male | 108 (48) |
| Race and ethnicityb | |
| Hispanic or Latino, any race | 50 (22) |
| Non-Hispanic | |
| American Indian or Alaska Native | 0 |
| Asian | 10 (4) |
| Black or African American | 6 (3) |
| Native Hawaiian or Other Pacific Islander | 3 (1) |
| White | 145 (64) |
| Multiracial | 11 (5) |
| Symptomaticc | 205 (91) |
| COVID-19 vaccination statusd | |
| Not vaccinated | 194 (86) |
| Received at least 1 vaccine dose | 31 (14) |
| SARS-CoV-2 lineagee | |
| B.1.1.7 (Alpha) | 126 (56) |
| B.1.427 or B.1.429 (Epsilon) | 35 (16) |
| P.1 (Gamma) | 8 (4) |
| Other lineages | 39 (17) |
| Unknown | 17 (8) |
Abbreviations: NP, nasopharyngeal; RT-PCR, reverse transcription-polymerase chain reaction.
Percentages may not add to 100% owing to rounding.
Self-reported by participants; responses were categorized by investigators following federal government standards.9
Participants were considered symptomatic if they reported symptoms that fulfilled the clinical criteria for COVID-19 adopted by the Council of State and Territorial Epidemiologists on August 5, 2020 (https://ndc.services.cdc.gov/case-definitions/coronavirus-disease-2019-2020-08-05/). Symptoms were captured via the enrollment questionnaire and daily symptom questionnaires during the 15-day enrollment period.
Vaccination status was assigned at the start of the infectious period of the case. If the case was symptomatic, the start of the infectious period was 2 days before symptom onset; if asymptomatic, it was the collection date of the first positive SARS-CoV-2 RT-PCR test result.
Forty-two individuals did not have an NP specimen that was able to be sequenced. Of these 42 individuals, 25 (60%) lived with a household member who had an NP specimen that was successfully sequenced; we assumed that these individuals were infected with the same SARS-CoV-2 lineage as their household members. Seventeen individuals (40%) did not have a household contact with a successfully sequenced specimen and were categorized as unknown. No enrolled households had more than 1 lineage detected.
The 225 enrolled cases contributed 3044 home antigen tests and 642 NP swabs, including 593 pairs of antigen tests and NP swabs that were performed on the same date (eTable 2 in Supplement 1). Cases self-collected a median of 15 home antigen tests (IQR, 14-15; range, 1-17). We collected a median of 2 NP swabs (IQR, 2-3; range, 1-10) from each case, and 23 cases (10%) underwent daily NP swabs for 7 additional days following enrollment.
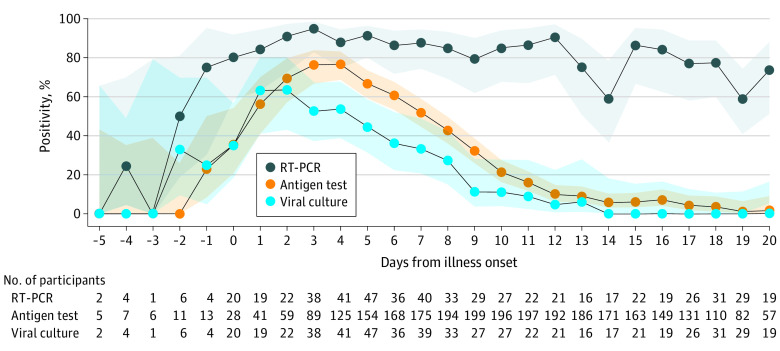
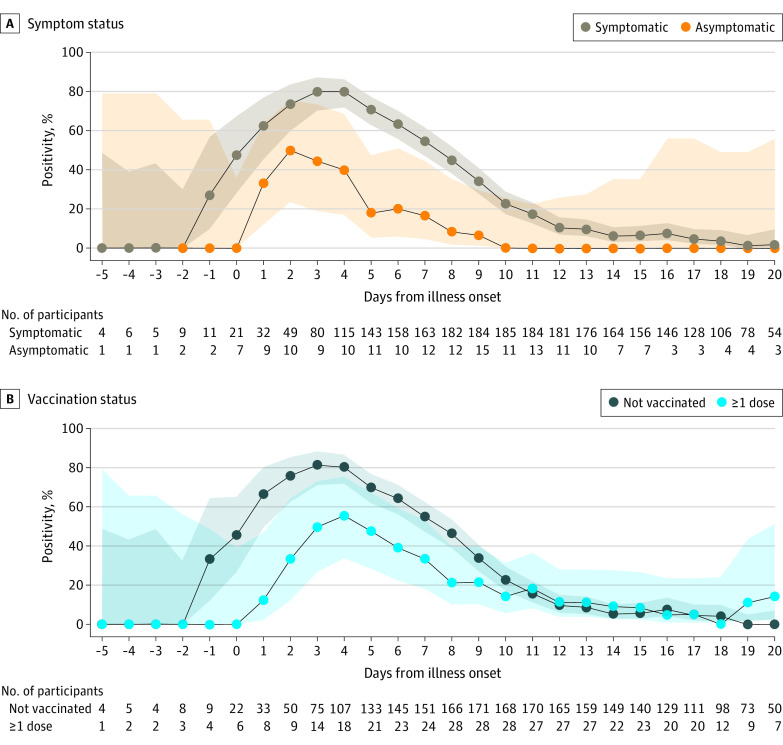
The daily positivity during the infectious period of cases peaked at 95% for RT-PCR tests (3 days after illness onset), 77% for antigen tests (4 days after illness onset), and 64% for viral cultures (2 days after illness onset) (Figure 1). The daily positivity decreased more quickly for the antigen test and culture compared with RT-PCR. Six days after illness onset, when people with mild or asymptomatic SARS-CoV-2 infection may discontinue isolation according to current CDC guidance,21 RT-PCR positivity was 86%, antigen test positivity was 61%, and culture positivity was 36%. At 11 days after illness onset, when most individuals are no longer considered infectious, RT-PCR positivity remained high (86%), while antigen test positivity and culture positivity were low (16% and 9%, respectively). Of 76 NP specimens obtained 11 to 14 days after illness onset, only 1 (1%) was culture positive. Home antigen test positivity peaked 3 days after illness onset at 80% for symptomatic cases and 50% for asymptomatic cases (Figure 2A). Home antigen test positivity was consistently higher for cases who were not vaccinated compared with those who received at least 1 vaccine dose before infection (Figure 2B).
Figure 1. Daily Percentage of Positive SARS-CoV-2 Tests in Participants With Reverse Transcription–Polymerase Chain Reaction (RT-PCR)–Confirmed Infection.
Daily percentage of positive SARS-CoV-2 tests (lines) and 95% CIs (shaded areas) of RT-PCR tests, home antigen tests, and viral culture among 225 participants with RT-PCR–confirmed SARS-CoV-2 infection. If the participant was symptomatic, illness onset was defined as the symptom onset date; if asymptomatic, illness onset was the collection date of the first positive RT-PCR test result. Confidence intervals were calculated by the Wilson score interval method.
Figure 2. Daily Percentage of Positive Home Antigen Tests by Symptom Status and Vaccination Status.
Daily percentage of positive SARS-CoV-2 tests (lines) and 95% CIs (shaded areas) of home antigen tests among 225 participants with reverse transcription–polymerase chain reaction (RT-PCR)–confirmed SARS-CoV-2 infection by symptom status (A) and vaccination status (B). Participants were considered symptomatic if they reported symptoms that fulfilled the clinical criteria for COVID-19 adopted by the Council of State and Territorial Epidemiologists on August 5, 2020 (https://ndc.services.cdc.gov/case-definitions/coronavirus-disease-2019-2020-08-05/). Symptoms were captured via the enrollment questionnaire and daily symptom questionnaires during the 15-day enrollment period. Confidence intervals were calculated using the Wilson score interval method.
Overall sensitivity of home antigen tests for detecting cases was 50% (95% CI, 45%-55%) (Figure 3), whereas specificity was 97% (95% CI, 95%-98%). Sensitivity was higher for symptomatic cases (53%; 95% CI, 48%-57%) compared with asymptomatic cases (20%; 95% CI, 10%-35%) and varied by lineage (Epsilon, 70%; Alpha, 49%; Gamma, 42%; other, 51%) (Figure 3). The sensitivity of antigen tests was 64% (95% CI, 56%-70%) compared with RT-PCR tests collected on the same day and 84% (95% CI, 75%-90%) compared with viral cultures collected on the same day. The sensitivity of antigen tests compared with same-day cultures was 85% for symptomatic cases, 87% for unvaccinated cases, and between 81% to 90% for all identified SARS-CoV-2 lineages. For asymptomatic cases, the sensitivity of antigen tests compared with same-day cultures was 33% (95% CI, 6%-80%). An increase in RT-PCR Ct values from same-day NP specimens was associated with a decrease in antigen test sensitivity (eFigure 2 in Supplement 1).
Figure 3. Sensitivity of Home Antigen Tests Compared With 3 Reference Standards.
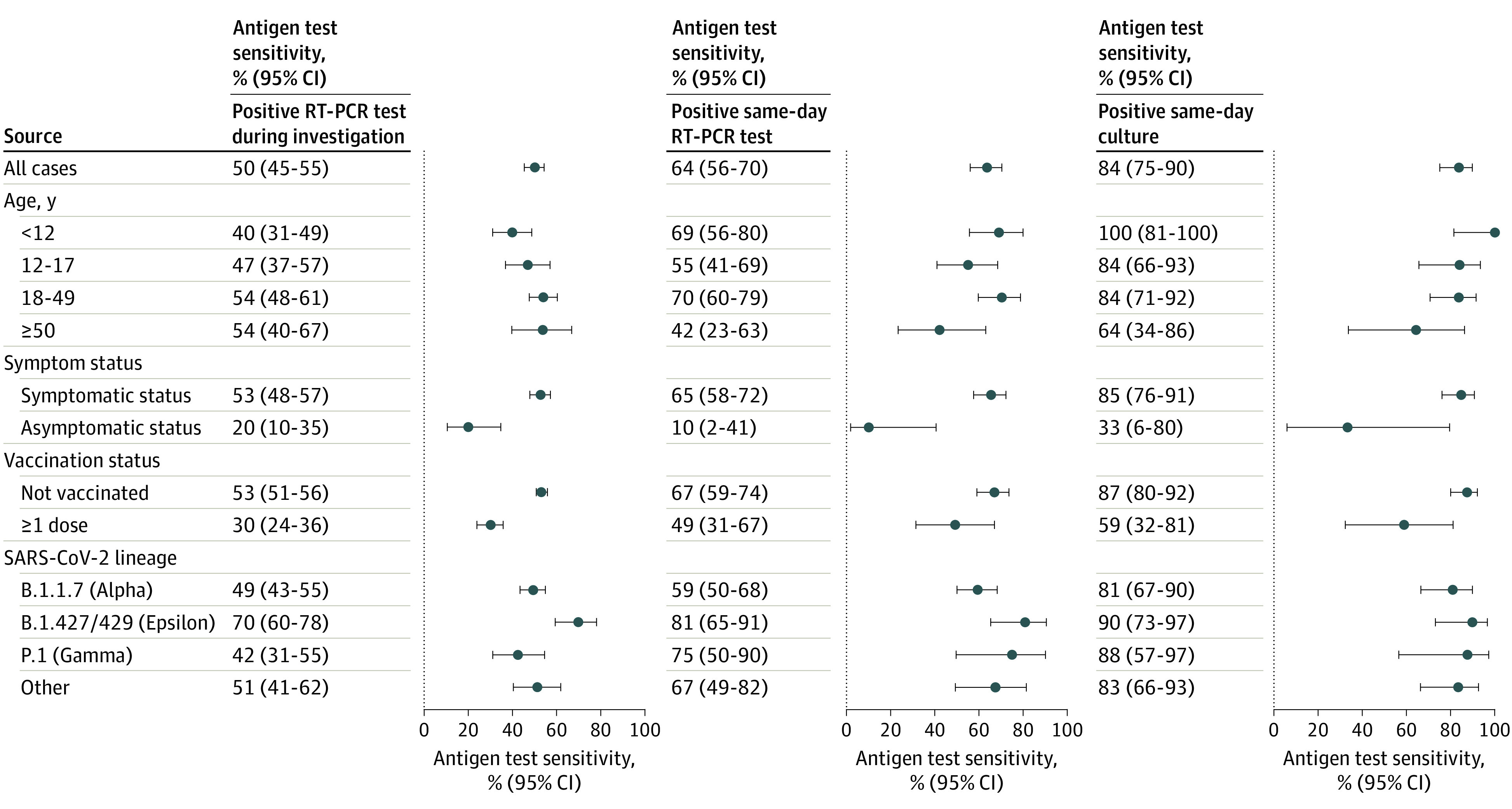
Sensitivity (dots) and 95% CIs (error bars) of home antigen tests compared with 3 reference standards: a positive reverse transcription–polymerase chain reaction (RT-PCR) test at any time during the investigation, a positive RT-PCR test collected the same day as the antigen test, and a positive viral culture collected the same day as the antigen test. Results are displayed for all cases and subgroups as defined by age, symptom status, vaccination status, and SARS-CoV-2 lineage. Participants were considered symptomatic if they reported symptoms that fulfilled clinical criteria for COVID-19 adopted by the Council of State and Territorial Epidemiologists on August 5, 2020 (https://ndc.services.cdc.gov/case-definitions/coronavirus-disease-2019-2020-08-05/). Symptoms were captured via the enrollment questionnaire and daily symptom questionnaires during the 15-day enrollment period. Vaccination status was defined at the time of enrollment. Confidence intervals were calculated using Wilson score intervals with cluster-robust standard errors accounting for correlation of tests performed by the same participant. For participants younger than 12 years, the 95% CI for antigen test sensitivity compared with a positive same-day viral culture was calculated without cluster adjustment as all tests were contributed by unique participants.
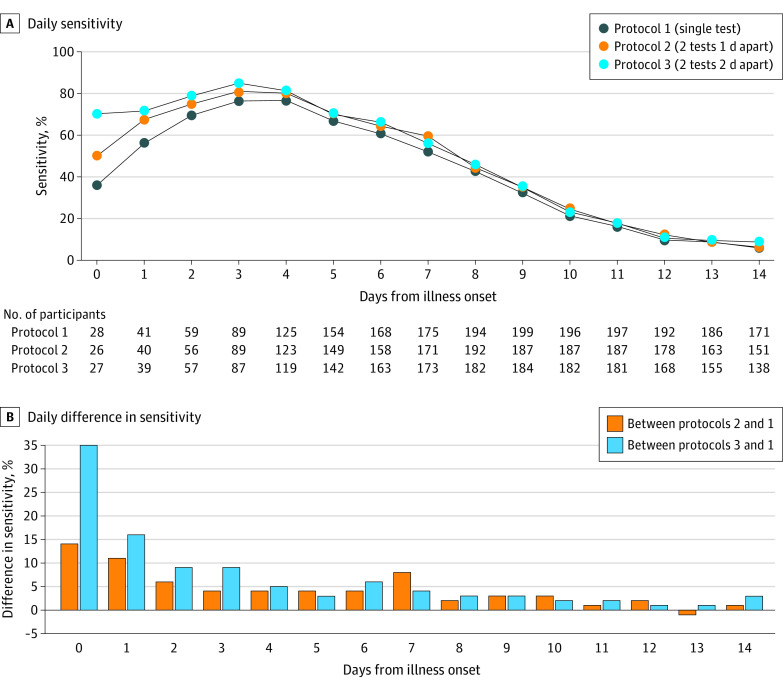
The frequency and timing of home antigen testing affected the sensitivity to detect cases. During the first 3 days after illness onset, performing 2 antigen tests 2 days apart (protocol 3) was more sensitive than administering 2 tests on consecutive days (protocol 2) and a single test (protocol 1) (Figure 4). Both serial testing protocols (protocols 2 and 3) remained more sensitive than a single test (protocol 1) throughout the 14 days after illness onset, with the largest differences in the first 3 days. Protocol 3 saw the highest peak sensitivity (85%) compared with protocols 2 (81%) and 1 (77%).
Figure 4. Daily Sensitivity of 3 Home Antigen Testing Protocols.
A, Daily sensitivity of 3 home antigen testing protocols; the reference standard was a positive reverse transcription–polymerase chain reaction (RT-PCR) test at any time during the investigation. B, Daily difference in testing protocol sensitivities. For protocols 2 and 3, sensitivity is displayed by day of the first test collected.
Among 2808 antigen tests for which the participant and investigator recorded an interpretation (positive, negative, or invalid), 2761 interpretations (98%) were concordant (Cohen κ, 0.96; 95% CI, 0.95-0.97) (eTable 4 in Supplement 1). Of 48 households that were observed, 24 (50%) had at least 1 household member who did not use the antigen test according to manufacturer instructions when collecting their first antigen test at enrollment. Errors observed included not placing the swab or test strip in the provided solution for the full time indicated (n = 11), contamination of the nasal swab before use (n = 9), and improper swabbing technique (n = 8). Concordance between the antigen test and RT-PCR results at enrollment from households with observed errors was similar to households without observed errors (72% vs 66%; P = .54). Almost all participants surveyed reported no difficulties collecting the nasal swabs (264 of 277 [95%]) and that they would be more likely to get tested for SARS-CoV-2 infection if the home antigen test were available over the counter (261 of 271 [96%]). No adverse events were reported during home antigen testing or collection of NP specimens.
Discussion
In this prospective cohort study of 225 adults and children with RT-PCR–confirmed SARS-CoV-2 infection who were observed for 15 days, home antigen test sensitivity peaked 4 days after illness onset. Sensitivity improved when a second antigen test was performed 1 to 2 days later, particularly early in the illness course. More than half of those who performed antigen tests on day 6 of illness had positive test results, but by day 11, fewer than one-fifth had positive test results. Home antigen tests were moderately sensitive compared with RT-PCR but highly sensitive compared with viral culture. With the widespread availability, ease of use, and rapid turnaround time, home antigen tests may increase testing in populations with barriers to testing in other settings and facilitate identification and isolation of cases.
The overall sensitivity of the home antigen test during the infectious period was 50%. This sensitivity was within the 34% to 88% range that was reported in a Cochrane review of other rapid, point-of-care SARS-CoV-2 antigen tests,1 but it was less than the 80% target set by the World Health Organization for point-of-care tests.22 Self-collection did not have a negative association with the overall sensitivity of the home antigen tests. Although antigen test use errors were commonly observed, the errors were not associated with reduced test accuracy. Similarly, another study found that even with errors in self-testing, the sensitivity of self-collected antigen tests was comparable with professionally collected antigen tests.23 Rather, the low overall sensitivity in this study may be associated with daily testing over a long period; many antigen test results that contributed to the overall sensitivity calculation were obtained late in illness, when the infection may have cleared. Compared with a positive same-day RT-PCR and same-day viral culture, antigen test sensitivity was higher.
Similar to other studies on antigen test performance,3,24,25,26 we found that the antigen test sensitivity was higher among symptomatic persons and earlier during the illness course. Antigen test positivity was lower before and on the day of illness onset. This is notable, as SARS-CoV-2 transmission often occurs before and during the first few days following symptom onset.27,28,29 Serial antigen tests spaced 1 to 2 days apart were associated with greatly improved sensitivity, particularly if the first test was performed around the time of illness onset. As with any diagnostic test, negative antigen test results should be interpreted within the context of estimated pretest probability, which should incorporate symptoms, known exposures, and community incidence of SARS-CoV-2. In particular, individuals with a high pretest probability of SARS-CoV-2 infection and initial negative home antigen test result should consider repeating an antigen test in 1 to 2 days or obtaining a confirmatory RT-PCR.
In addition to expanding diagnostic capabilities, antigen tests could also help optimize the duration of isolation. Reverse transcription–polymerase chain reaction detects small amounts of SARS-CoV-2 nucleic acid fragments and can remain positive well after illness recovery.30,31,32,33 For this reason, CDC no longer recommends an RT-PCR test–based strategy to end isolation for most patients.21 At present, CDC guidance allows for antigen tests to be used toward the end of the 5-day isolation period for individuals with infection; if positive, isolation should be continued for 10 days.34 In the present study, more than half of individuals with infection who tested on day 6 still had positive results on home antigen tests and would be recommended to remain isolated; however, by day 11, fewer than one-fifth had positive test results. These findings support the current CDC recommendation for strict use of face masks in settings with other people and continued isolation from unvaccinated or immunocompromised individuals through 10 days after illness onset.
These findings suggest that antigen test performance may differ in vaccinated vs unvaccinated individuals and between SARS-CoV-2 lineages. Viral load dynamics differ by viral lineage,35,36,37,38 and fully vaccinated persons with infection demonstrate accelerated viral clearance.39 Although this investigation occurred before the emergence of the Omicron (B.1.1.529) lineage, a recent study with 731 participants found that antigen tests continue to perform well with Omicron infections.40 However, as most of the US population has now received a COVID-19 vaccine or been infected with SARS-CoV-2,41 further studies should better assess the performance of antigen tests in vaccinated persons and those with natural immunity.
Limitations
This study had limitations. The findings are limited to the SARS-CoV-2 lineages that were circulating at the time of the investigation. Participants were primarily non-Hispanic White, younger, and unvaccinated; thus, they are not representative of the entire US population. Most participants were symptomatic and household contacts of a known COVID-19 case. Therefore, results may not be generalizable for use of home antigen tests to screen individuals who are asymptomatic or without a known exposure to SARS-CoV-2. Almost all symptomatic cases experienced mild disease, so diagnostic performance for severe disease could not be assessed. Not all specimens were sent for culture; some specimens that were assumed to be culture negative based on Ct values and prior studies may have been misclassified. As we did not collect daily NP specimens for all participants, we had fewer data for RT-PCR and viral cultures; the confidence intervals for calculations involving these tests are thus wider than those that involved only home antigen tests.
Conclusions
In this cohort study of 225 adults and children with RT-PCR–confirmed SARS-CoV-2 infection, home antigen test sensitivity for SARS-CoV-2 was moderate compared with RT-PCR and high compared with viral culture. Sensitivity peaked several days after illness onset and improved with repeated testing. These findings suggest that symptomatic individuals with an initial negative home antigen test result should test again 1 to 2 days later.
eTable 1. Forms Used to Observe Home Antigen Test Use and Acceptability
eTable 2. Home Antigen Tests and Nasopharyngeal Swabs Completed by Participants with RT-PCR–confirmed SARS-CoV-2 Infection, by Days from Illness Onset
eTable 3. Home Antigen Test Results by Participant Case Status
eTable 4. Participant and Investigator Interpretation of Home Antigen Test Results
eFigure 1. Flow Diagram for Participants with RT-PCR–confirmed SARS-CoV-2 Infection and At Least One Home Antigen Test Result
eFigure 2. Sensitivity of Home Antigen Tests by RT-PCR Cycle Threshold Value
Nonauthor collaborators
References
- 1.Dinnes J, Deeks JJ, Berhane S, et al. ; Cochrane COVID-19 Diagnostic Test Accuracy Group . Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2021;3(3):CD013705. doi: 10.1002/14651858.CD013705.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson KE, Altayar O, Caliendo AM, et al. The Infectious Diseases Society of America guidelines on the diagnosis of COVID-19: antigen testing. Clin Infect Dis. Published online June 23, 2021. doi: 10.1093/cid/ciab557 [DOI] [PubMed] [Google Scholar]
- 3.Smith RL, Gibson LL, Martinez PP, et al. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. J Infect Dis. 2021;224(6):976-982. doi: 10.1093/infdis/jiab337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly MAP, Chuey MR, Soto R, et al. ; COVID-19 Household Transmission Team . Household transmission of SARS-CoV-2 Alpha variant—United States, 2021. Clin Infect Dis. Published online February 11, 2022. doi: 10.1093/cid/ciac125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Archives . Code of Federal Regulations. Accessed January 15, 2021. https://www.ecfr.gov/
- 6.US House of Representatives . US Code. Accessed January 15, 2021. https://uscode.house.gov/
- 7.von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP; STROBE Initiative . The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370(9596):1453-1457. doi: 10.1016/S0140-6736(07)61602-X [DOI] [PubMed] [Google Scholar]
- 8.US Centers for Disease Control and Prevention . Case investigation & contact tracing guidance: appendices. Accessed June 16, 2021. https://www.cdc.gov/coronavirus/2019-ncov/php/contact-tracing/contact-tracing-plan/appendix.html#contact
- 9.Office of Management and Budget . Revisions to the standards for the classification of federal data on race and ethnicity. Accessed April 1, 2021. https://www.govinfo.gov/content/pkg/FR-1997-10-30/pdf/97-28653.pdf
- 10.Quidel . QuickVue At-Home COVID-19 Test: user instructions. Accessed June 16, 2021. https://www.fda.gov/media/146313/download
- 11.ThermoFisher Scientific . TaqPath COVID-19 Combo Kit and TaqPath COVID-19 Combo Kit Advanced: instructions for use. Accessed June 16, 2021. https://www.fda.gov/media/136112/download
- 12.PerkinElmer . Instructions for PerkinElmer New Coronavirus Nucleic Acid Detection Kit v 8.0. Accessed June 16, 2021. https://www.fda.gov/media/136410/download
- 13.Paden CR, Tao Y, Queen K, et al. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(10):2401-2405. doi: 10.3201/eid2610.201800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin Webb L, Matzinger S, Grano C, et al. Identification of and surveillance for the SARS-CoV-2 variants B.1.427 and B.1.429—Colorado, January-March 2021. MMWR Morb Mortal Wkly Rep. 2021;70(19):717-718. doi: 10.15585/mmwr.mm7019e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harcourt J, Tamin A, Lu X, et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis. 2020;26(6):1266-1273. doi: 10.3201/eid2606.200516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Killerby ME, Ata Ur Rasheed M, Tamin A, et al. Shedding of culturable virus, seroconversion, and 6-month follow-up antibody responses in the first 14 confirmed cases of coronavirus disease 2019 in the United States. J Infect Dis. 2021;224(5):771-776. doi: 10.1093/infdis/jiab125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C-G, Lee K-M, Hsiao M-J, et al. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J Clin Microbiol. 2020;58(8):e01068-e20. doi: 10.1128/JCM.01068-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basile K, McPhie K, Carter I, et al. Cell-based culture informs infectivity and safe de-isolation assessments in patients with coronavirus disease 2019. Clin Infect Dis. 2021;73(9):e2952-e2959. doi: 10.1093/cid/ciaa1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.US Centers for Disease Control and Prevention . Coronavirus disease 2019 (COVID-19): 2020. interim case definition, approved August 5, 2020. Accessed September 19, 2020. https://ndc.services.cdc.gov/case-definitions/coronavirus-disease-2019-2020-08-05/
- 20.Brown LD, Cai TT, DasGupta A. Interval estimation for a binomial proportion. Stat Sci. 2001;16(2):101-133. doi: 10.1214/ss/1009213286 [DOI] [Google Scholar]
- 21.US Centers for Disease Control and Prevention . Ending isolation and precautions for people with COVID-19: interim guidance. Accessed April 1, 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html
- 22.World Health Organization . COVID-19 target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0. Accessed November 1, 2021. https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1
- 23.Lindner AK, Nikolai O, Rohardt C, et al. Diagnostic accuracy and feasibility of patient self-testing with a SARS-CoV-2 antigen-detecting rapid test. J Clin Virol. 2021;141:104874. doi: 10.1016/j.jcv.2021.104874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah MM, Salvatore PP, Ford L, et al. Performance of repeat BinaxNOW severe acute respiratory syndrome coronavirus 2 antigen testing in a community setting, Wisconsin, November 2020-December 2020. Clin Infect Dis. 2021;73(suppl 1):S54-S57. doi: 10.1093/cid/ciab309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pray IW, Ford L, Cole D, et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses—Wisconsin, September-October 2020. MMWR Morb Mortal Wkly Rep. 2021;69(5152):1642-1647. doi: 10.15585/mmwr.mm695152a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prince-Guerra JL, Almendares O, Nolen LD, et al. Evaluation of Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection at two community-based testing sites—Pima County, Arizona, November 3-17, 2020. MMWR Morb Mortal Wkly Rep. 2021;70(3):100-105. doi: 10.15585/mmwr.mm7003e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2(1):e13-e22. doi: 10.1016/S2666-5247(20)30172-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672-675. doi: 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 29.Cheng H-Y, Jian S-W, Liu D-P, Ng T-C, Huang W-T, Lin H-H; Taiwan COVID-19 Outbreak Investigation Team . Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med. 2020;180(9):1156-1163. doi: 10.1001/jamainternmed.2020.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surie D, Huang JY, Brown AC, et al. Infectious period of severe acute respiratory syndrome coronavirus 2 in 17 nursing home residents—Arkansas. Open Forum Infect Dis. 2021;8(3):ofab048. doi: 10.1093/ofid/ofab048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li N, Wang X, Lv T. Prolonged SARS-CoV-2 RNA shedding: not a rare phenomenon. J Med Virol. 2020;92(11):2286-2287. doi: 10.1002/jmv.25952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W-D, Chang S-Y, Wang J-T, et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81(2):318-356. doi: 10.1016/j.jinf.2020.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mallett S, Allen AJ, Graziadio S, et al. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? a systematic review of individual participant data. BMC Med. 2020;18(1):346. doi: 10.1186/s12916-020-01810-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.US Centers for Disease Control and Prevention . COVID-19 quarantine and isolation. Accessed April 1, 2022. https://www.cdc.gov/coronavirus/2019-ncov/your-health/quarantine-isolation.html
- 35.Kidd M, Richter A, Best A, et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis. 2021;223(10):1666-1670. doi: 10.1093/infdis/jiab082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calistri P, Amato L, Puglia I, et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int J Infect Dis. 2021;105:753-755. doi: 10.1016/j.ijid.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373(6551):eabi5273. doi: 10.1126/science.abi5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyngse FP, Mølbak K, Skov RL, et al. ; Danish Covid-19 Genome Consortium . Increased transmissibility of SARS-CoV-2 lineage B.1.1.7 by age and viral load. Nat Commun. 2021;12(1):7251. doi: 10.1038/s41467-021-27202-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singanayagam A, Hakki S, Dunning J, et al. ; ATACCC Study Investigators . Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2022;22(2):183-195. doi: 10.1016/S1473-3099(21)00648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrom J, Marquez C, Pilarowski G, et al. Comparison of SARS-CoV-2 reverse transcriptase polymerase chain reaction and BinaxNOW rapid antigen tests at a community site during an Omicron surge: a cross-sectional study. Ann Intern Med. Published online March 15, 2022. doi: 10.7326/M22-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.US Centers for Disease Control and Prevention . COVID data tracker: nationwide COVID-19 infection- and vaccination-induced antibody seroprevalence (blood donations). Accessed April 1, 2022. https://covid.cdc.gov/covid-data-tracker/#nationwide-blood-donor-seroprevalence
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Forms Used to Observe Home Antigen Test Use and Acceptability
eTable 2. Home Antigen Tests and Nasopharyngeal Swabs Completed by Participants with RT-PCR–confirmed SARS-CoV-2 Infection, by Days from Illness Onset
eTable 3. Home Antigen Test Results by Participant Case Status
eTable 4. Participant and Investigator Interpretation of Home Antigen Test Results
eFigure 1. Flow Diagram for Participants with RT-PCR–confirmed SARS-CoV-2 Infection and At Least One Home Antigen Test Result
eFigure 2. Sensitivity of Home Antigen Tests by RT-PCR Cycle Threshold Value
Nonauthor collaborators