Abstract
Purpose
The effect of noncoding variants is often unknown in the absence of functional assays. Here, we characterized an ABCA4 intron 7 variant, c.859-25A>G, identified in Palestinian probands with Stargardt disease (STGD) or cone-rod dystrophy (CRD). We investigated the effect of this variant on the ABCA4 mRNA and retinal phenotype, and its prevalence in Palestine.
Methods
The ABCA4 gene was sequenced completely or partially in 1998 cases with STGD or CRD. The effect of c.859-25A>G on splicing was investigated in silico using SpliceAI and in vitro using splice assays. Homozygosity mapping was performed for 16 affected individuals homozygous for c.859-25A>G. The clinical phenotype was assessed using functional and structural analyses including visual acuity, full-field electroretinography, and multimodal imaging.
Results
The smMIPs-based ABCA4 sequencing revealed c.859-25A>G in 10 Palestinian probands from Hebron and Jerusalem. SpliceAI predicted a significant effect of this putative branchpoint-inactivating variant on the nearby intron 7 splice acceptor site. Splice assays revealed exon 8 skipping and two partial inclusions of intron 7, each having a deleterious effect. Additional genotyping revealed another 46 affected homozygous or compound heterozygous individuals carrying variant c.859-25A>G. Homozygotes shared a genomic segment of 59.6 to 87.9 kb and showed severe retinal defects on ophthalmoscopic evaluation.
Conclusions
The ABCA4 variant c.859-25A>G disrupts a predicted branchpoint, resulting in protein truncation because of different splice defects, and is associated with early-onset STGD1 when present in homozygosity. This variant was found in 25/525 Palestinian inherited retinal dystrophy probands, representing one of the most frequent inherited retinal disease-causing variants in West-Bank Palestine.
Keywords: ABCA4, Stargardt disease, SpliceAI, splice defect, founder mutation
Stargardt disease (STGD1) is the most frequent inherited maculopathy with an estimated prevalence of one in 10000.1 It is caused by biallelic variants in the gene encoding the transmembrane ATP-binding cassette transporter type A4 (ABCA4).2 More than 2200 unique variants have been reported for ABCA4 (http://www.lovd.nl/ABCA4),3,4 which have been linked to a spectrum of autosomal recessive maculopathies, such as classical STGD1, fundus flavimaculatus, cone-rod dystrophy (CRD), and atypical retinitis pigmentosa (RP).2,5–8
The observed clinical heterogeneity has led to a genotype-phenotype correlation model in which the severity of retinal dystrophy correlates with the residual activity of the ABCA4 protein because of the effect of the two causal alleles.9 In this model, variants can be categorized as deleterious (or severe), moderately severe, mild or hypomorphic.8,10,11 The identification of two pathogenic alleles is important to provide an accurate clinical prognosis and for the application of possible future gene- or variant-specific therapies.8,12
A conspicuously high proportion of reported ABCA4 variants (∼25%) affect RNA splicing,13 either resulting in exon skipping or intron inclusion when they are located at exon-intron junctions,11,14 or resulting in pseudo-exon inclusion in the case of deep-intronic variants.12,13,15–18 Notably, previous studies using whole ABCA4 gene (consisting of 50 exons spanning 128 kb) sequencing has uncovered pathogenic non-coding variants in ∼21% of STGD1 probands and in vitro splice assays have allowed a detailed characterization of putative splice variants.12,17,19,20
The prediction of the presence of pseudo-exons in ABCA4 was first shown by Braun et al.,21 who performed deep mRNA sequencing. In this way, low-abundant transcripts that contained putative pseudo-exons were revealed. Sequence analysis of these pseudo-exons in genetically unexplained STGD1 cases revealed the first causal deep-intronic variants in ABCA4. These low-abundant transcripts, in the absence of splice-site strengthening or creating variants in normal individuals, are often not present. In those cases, the discovery and selection of putative splice variants relies on the use of in silico prediction tools. Splicing defects occur when a variant alters one of the key splicing signals. These include the consensus sequences at the splice acceptor site (SAS) or splice donor site (SDS), the branchpoint site, as well as exonic or intronic cis regulators that act as enhancers or silencers.22,23 Several in silico tools are available to predict canonical and cryptic splice sites based on these key elements. The approaches utilized by these tools vary from position weight matrix, Markov model, and maximum entropy principle24–29 to more complex methods based on artificial neural networks and machine learning.30,31 Nevertheless, the majority of prediction tools are lacking accuracy in predicting the effect of novel deep-intronic variants outside of the known constitutive splice sites. There are several aspects of splicing not considered by prediction tools, such as the concerted role of different splicing signals and splice-site interdependency,32 chromatin organization33 and tissue-specific splicing events.16,34,35 Some of these shortcomings can be addressed by using recently developed tools based on deep residual neural networks such as SpliceAI,36 which is self-trained on pre-mRNA sequences to predict the likelihood that a certain position will be used as a splice site, and does not rely on a priori knowledge, allowing the incorporation of splicing determinants that are still not well understood.
In the context of a large-scale single molecule molecular inversion probes (smMIPs)-based ABCA4 sequencing study of STGD and STGD-like probands, we found, through the deep learning prediction tool SpliceAI,36 a putative pathogenic variant just upstream of exon 8, which potentially could result in a splice defect. Its frequent presence in other Palestinian STGD1 cases prompted us to investigate its effect on ABCA4 splicing, its associated ophthalmological characteristics, its founder characteristics, and its prevalence in the Palestinian population.
Materials and Methods
Patients and Genotyping Methods
The “discovery” cohort (cohort 1) consisted of 876 probands with a diagnosis of STGD, CRD, or macular dystrophy (MD) based on the clinical phenotype. They were ascertained in 21 centers from all over the world, and include 175 cases that originate from Palestine. For these probands, the entire 128-kb ABCA4 gene was sequenced using smMIPs, as previously described.18 In brief, series of ∼210 probands were sequenced using the Illumina NextSeq 500 platform, followed by variant calling and annotation through an in-house bioinformatics pipeline. The second cohort in this study (cohort 2) was described previously17 and originally consisted of 1,054 probands with genetically unsolved STGD, CRD and MD cases. The smMIPs-based ABCA4 sequencing genetically explained 448 probands,17 whereas 606 probands remained genetically unsolved, of which 38 probands were submitted by the St. John of Jerusalem Eye Hospital Group and Hadassah Medical Center, Jerusalem, Palestine. Whole exome sequencing (WES) data (M. Salameh, A. AlTalbishi, D. Sharon, unpublished data) and MIPs-based sequencing data (M. Salameh, A. AlTalbishi, D. Sharon, F.P.M. Cremers, unpublished data) from another 312 Palestinian inherited retinal dystrophy (IRD) probands (cohort 3) were re-analyzed for the presence of the c.859-25A>G variant. Similarly, 201 Palestinian IRD probands and affected siblings were analyzed through Sanger sequencing of ABCA4 exon 8 and flanking sequences (cohort 4). Three additional individuals from the National Eye Institute, National Institutes of Health in the United States, were identified to carry the variant by assessment of custom capture next-generation sequencing data from commercial clinical genetic testing panels for STGD, MD or retinal dystrophy. For allele frequency (AF) calculations, an overall cohort of 525 Palestinian probands was considered. This final cohort includes the Palestinian probands present in cohorts 1 and 2 (175 and 38, respectively) and all 312 probands in cohort 3. Because cohort 4 is largely composed of affected siblings and relatives, it was not included in the calculation of the AF. A group of 1400 non-IRD Palestinian probands, for which WES data was available, were used as controls. Statistical analysis of the results was performed using a one-tailed Fisher's exact test.
Variant Selection by in Silico Analysis
Variant selection from smMIPs sequencing data started with quality filtering on the overall coverage of the variants (>20 reads, given an average of 340 reads per nucleotide) and the percentage of variation at the position (variant present in ≥25% of all reads). Further variant prioritization was performed considering the AF, being <0.005 in the dbSNP database and <0.01 in general population databases, such as the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org/). In silico analysis was performed using the deep-learning tool SpliceAI to identify putative variants causing splice defects.36 SpliceAI predictions were considered for each nucleotide position in ABCA4, and the script was set to return the five highest delta scores for acceptor gain, acceptor loss, donor gain, and donor loss, in a window of 5000 nucleotides (nt) up- and downstream from the selected position. Likely pathogenic deep-intronic or noncanonical splice site (NCSS) variants were selected if the delta score was >0.1 (range 0–1). Variants were considered to be part of the NCSS when located between −14 to −3 upstream of an exon and +3 to +6 downstream of an exon, or positioned within the first and last two nucleotides of an exon. Alamut Visual 2.13 software and its splice defect prediction tools (SpliceSiteFinder-like,37 MaxEntScan,26 NNSPLICE30 and GeneSplicer27) were used as a visual aid to identify the position in which SpliceAI delta scores were predicted and to characterize the genomic context around the variant.
Midigene-Based Splice Assay
The splicing effect of c.859-25A>G was assessed using in vitro splice assays, based on a previously established wild-type midigene (BA7) containing ABCA4 exons 7 to 11.14 A mutant construct was generated for the variant through site-directed mutagenesis followed by Gateway Cloning. Subsequently, wild-type and mutant constructs were transfected separately in Human Embryonic Kidney (HEK293T, ATCC# CRL-3216) cells. HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% sodium pyruvate at 37°C and 5% CO2. For transfection, cells were seeded in a six-well plate and transfected at 70% confluency with 600 nanograms (ng) of plasmid using FuGENE HD reagent (Promega, Madison, WI, USA), as specified in the manufacturers protocol. Transfection of the mutant construct was performed in duplicate. After 48 hours, RNA was obtained from the cells using the Nucleospin RNA kit (Machery-Nagel, Düren, Germany), and cDNA was synthesized from 1000 ng of RNA through the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA). Reverse-transcription–polymerase chain reaction (RT-PCR), agarose gel analysis and Sanger sequencing were performed to assess the nature of splicing defects. RT-PCR conditions were as follows: 94°C for two minutes, followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and five minutes at 72°C, with a final extension step of two minutes at 72°C. Details on the primers used for mutagenesis, PCR, and Sanger sequencing can be found in Supplementary Table S1. Ratios between different RNA products were assessed by semi-quantification using Fiji software38 after agarose gel electrophoresis.
WES and Targeted Sanger Sequencing
WES was performed using 3billion Inc. (Seoul, South Korea) and Variantyx Inc. (Framingham, MA, USA) sequencing services for 250 and 63 additional samples, respectively. Sanger sequencing (Macrogen Europe B.V., Amsterdam, the Netherlands; analysis using Chromas software) was performed to confirm the presence of the c.859-25A>G variant in this cohort and for 200 additional Palestinian IRD probands. PCR was performed using the following conditions: 95°C for two minutes; 40 cycles of 15 seconds at 95°C, 15 seconds at 60°C, and 45 seconds at 72°C; final extension at 72°C for five minutes. Details on the primers used for PCR and Sanger sequencing can be found in Supplementary Table S1.
Haplotype Analysis for the ABCA4 Locus
Haplotype analysis was performed on WES data for 16 probands using Automap tool (https://automap.iob.ch/) to show homozygous regions in chromosome 1. For haplotype identification within these regions, BAM files (generated using https://usegalaxy.org/) were uploaded into Integrative Genomics Viewer software (IGV v.2.9.4)39,40 and were manually screened for each genomic location within the homozygous regions defined by Automap. The ABCA4 locus was screened manually. Variant calling format files (vcf) provided by 3billion were used to generate annotated genetic variants spreadsheets using ANNOVAR online tool (https://wannovar.wglab.org/). Within the ABCA4 region, haplotype information for 23 single nucleotide polymorphisms spanning over 125 kb was obtained.
Clinical Phenotyping
All individuals were examined by an experienced ophthalmologist for clinical analysis. The phenotype data collected included the following: anamnestic information on disease onset, symptoms and progression, age at last examination, best-corrected Early Treatment of Diabetic Retinopathy Study (ETDRS) chart visual acuity converted to logMAR (31/33 probands), clinical ocular exam by slit lamp biomicroscopy, kinetic and/or static perimetry, fundus photos, fundus autofluorescence (FAF) and optical coherence tomography (OCT). When available, age at electroretinography (ERG) and ERG respecting International Society for Clinical Electrophysiology of Vision standards were collected for both probands and affected relatives (20/56).41 For most patients, information on a specific age of onset was not available; thus age at last examination was reported instead. In most cases first examination took place in early childhood when the first symptoms appeared.
ERG for patients from the St John of Jerusalem Eye Hospital Group was performed using the Metrovision machine. The Topcon Triton 3D was used for OCT, fundus photos and FAF. The recently recruited patients had FAF and fundus photos taken using the Zeiss Clarus camera (Zeiss, Germany). In the National Eye Institute, ERG was performed using an LKC console and Burian-Allen contact lens electrodes. In addition, Cirrus-HD-OCT scans were performed as well as color fundus photography and fundus autofluorescence imaging. Full-field ERG (ffERG) information from Hadassah Medical Center was obtained using corneal electrodes and a computerized system (UTAS 3,000, LKC, MD). Briefly, in the dark-adapted state, a rod response to a dim blue flash and a mixed cone-rod response to a white flash were acquired. Cone responses to 30-Hz flashes of white light were acquired under a background light of 21 cd/m2. All responses were filtered at 0.3–500 Hz and signal averaging was used. OCT was performed using the Heidelberg Spectralis system, while color, infrared and FAF imaging were assessed using a Zeiss and/or Optos fundus camera and the Heidelberg Spectralis system. At Sickkids in Toronto phenotyping included ERG testing using the Diagnosys system whereas OCT imaging used the Heidelberg Spectralis system.
Results
Identification of ABCA4 Intron 7 Variant c.859-25A>G and In Silico Splice Defect Predictions
Incorporation of SpliceAI predictions in the annotation of smMIPs sequencing data led to the identification of the c.859-25A>G (chr1(GRCh37):g.94,546,299T>C) variant in 10 patients of Palestinian descent in the 876 STGD and STGD-like probands of cohort 1 (Table 1). This novel variant is absent in control populations in gnomAD. The position of the variant outside the SAS consensus sequence, but still close to the intron-exon junction, suggested a putative effect on other splicing elements, such as the branchpoint sequence (BPS). Therefore, an in-depth in silico analysis of the variant was performed using modified settings for the deep learning tool SpliceAI. Using a “scanning” window of 5000 nt upstream and 5000 nt downstream of c.859-25A>G, the strongest prediction entailed the skipping of exon 8 (SAS loss with a delta score of 0.39), together with a new SAS gain 138 nt upstream of exon 8 (delta score 0.31) (Supplementary Table S2). Interestingly, a donor loss with a delta score of 0.09 was predicted at the SDS of intron 7. Although this delta score did not reach the threshold of significance, it might be a contributing factor to exon 8 skipping (Supplementary Fig. S1A).
Table 1.
Geographic Origin and Genotypic Details of Probands Carrying Variant c.859-25A>G
| Family | Pedigree | District/ | Clinical | Allele 1 | Allele 2 | ||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Number | Reference | Village | Diagnosis | cDNA | Protein | cDNA | Protein | Cohort |
| 1 | SJH0003 | II-1 | Bethlehem/Beit Fajjar | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 3 | SJH0010 | II-1 | Hebron/Beit Ommar | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 4 | SJH0021 | II-1 | Hebron/Sureef | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 6 | SJH0048 | II-1 | Bethlehem/Aida Camp | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.6816+2T>A | p.(?) | 2 |
| 7 | SJH0100 | II-1 | Hebron/Dura | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 9 | SJH0101 | II-2 | Hebron/Dura | RP | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 3 |
| 13 | SJH0102 | III-1 | Bethlehem/Husan | CRD/RP | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 14 | SJH0103 | II-1 | Hebron/Sureef | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.4979C>T | p.(Pro1660Leu) | 2 |
| 18 | SJH0125 | II-1 | Hebron/Sureef | CRD/STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 19 | SJH0133 | II-1 | Bethlehem/Tqou' | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 2 |
| 20 | SJH0195 | II-1 | Hebron/Sureef | CD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.5882G>C | p.(Gly1961Ala) | 4 |
| 23 | SJH0216 | II-1 | Bethlehem/Ertas | RP | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 3 |
| 24 | SJH0228 | II-1 | Hebron/Beit Ommar | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 3 |
| 25 | SJH0243 | II-1 | Hebron/Sureef | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.4979C>T | p.(Pro1660Leu) | 4 |
| 26 | SJH0255 | II-1 | Hebron/Dura | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 4 |
| 27 | SJH0265 | II-1 | Hebron/Dura | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 3 |
| 28 | SJH0268 | II-1 | Bethlehem/Aida Camp | CD/CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 3 |
| 29 | SJH0297 | II-1 | Hebron/Dura | AV-MD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.5882G>A | p.(Gly1961Glu) | 3 |
| 30 | SJH0351 | II-1 | Hebron/Sureef | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 32 | SJH0385 | II-1 | Hebron/Dura | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.5882G>A | p.(Gly1961Glu) | 1 |
| 33 | SJH0459 | II-1 | Hebron/Dura | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 4 |
| 34 | SJH0461 | I-1 | Hebron/Sureef | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 4 |
| 36 | MOL0003 | II-1 | Hebron | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 3 |
| 37 | MOL0136 | II-1 | Jerusalem | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 39 | MOL0419 | II-1 | Hebron/Dura | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 40 | MOL0497 | II-1 | Hebron/Dura | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.5882G>A | p.(Gly1961Glu) | 1 |
| 41 | MOL0844 | II-1 | Jerusalem | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 43 | MOL1066 | III-1 | Hebron/Dura | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 46 | MOL1216 | III-2 | Hebron/Beit Ommar | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 51 | MOL1417 | II-1 | Jerusalem | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 1 |
| 53 | NEI1 | III-1 | Washington DC | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | 5 |
| 55 | NEI2 | II-1 | Washington DC | CRD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.4979C>T | p.(Pro1660Leu) | 5 |
| 56 | 072537 | II-1 | Toronto | STGD | c.859-25A>G | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | c.3262C>T | p.(Pro1088Ser) | 1 |
AV-MD, Adult vitelliform macular dystrophy; STGD, Stargardt disease; CD, cone dystrophy; CRD, cone-rod dystrophy; RP, retinitis pigmentosa.
Cohort 1: consisting of 876 probands sequenced by smMIPs; cohort 2: consisting of 606 unsolved probands sequenced by single molecule Molecular Inversion Probes for which data re-analysis was performed; cohort 3: consisting of 312 probands sequenced by whole exome sequencing or Molecular Inversion Probes for which data re-analysis was performed; cohort 4: consisting of 201 probands and affected relatives sequenced by Sanger; cohort 5: consisting of probands and affected relatives sequenced with Next Generation Sequencing.
Considering that previous smMIPs sequencing datasets were not analyzed with the aid of SpliceAI, we re-examined the sequencing results of 606 genetically unsolved STGD or STGD-like probands (cohort 2)17 and identified c.859-25A>G in nine additional cases (Table 1). Similarly, analysis of cohorts 3 and 4 lead to the identification of seven and five additional probands carrying this variant, respectively. Additionally, 22 affected relatives were found to carry this variant (Supplementary Table S3). Finally, two additional probands and one affected relative carrying the variant were independently identified in the National Eye Institute in the United States (Cohort 5). In total, we found 56 cases carrying c.859-25G>A, comprising 41 homozygotes and 15 compound heterozygotes, belonging to 33 families. Most of the cases carrying c.859-25A>G reside in or in the vicinity of Hebron with the following distribution: Hebron Governorate (n = 39 cases), Bethlehem (n = 7), Jerusalem (n = 6) (Supplementary Fig. S2). The four cases who carry this variant and reside in the US or Canada originated from Jerusalem and Bethlehem (Table 1 and Supplementary Figs. S3 and S4).
Allele Frequency Comparison Between Patient and Control Cohort
An overall cohort of 525 Palestinian probands was considered to calculate the AF of c.859-25A>G in the patient population. As 20 homozygous and 5 compound heterozygous cases were identified, we calculated an AF of 0.04. Screening of the WES control cohort (1400 non-IRD probands) returned an AF of 0.003, since the variant was identified in only 9 alleles. The difference between the two AF was assessed for statistical significance with a one-sided Fisher's exact test, that returned a p-value of 0.0001. Additionally, an odds ratio (OR) of 13.89 (confidence interval: 6.76–28.51) was obtained, which is in line with the OR>5 value ACMG guidelines report as a strong indicator of pathogenicity.42 These results show that c.859-25A>G is significantly enriched in the patient population.
In Vitro Splicing Assay
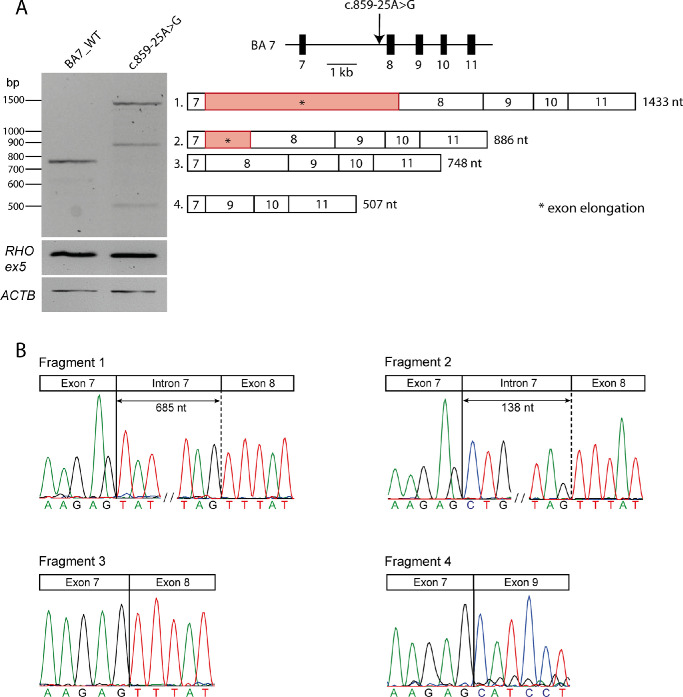
The effect of c.859-25A>G was assessed using in vitro splice assays in HEK293T cells (Fig. 1). The cells were transfected either with a wild-type midigene construct (spanning ABCA4 exon 7 to exon 11) or a mutant construct carrying c.859-25A>G. Sanger sequencing analysis of RT-PCR products revealed three splicing defects and no remaining wild-type product in HEK293T cells. Skipping of exon 8 was observed, which resulted in a shift of the reading frame and the introduction of a premature stop codon (p.Phe287Hisfs*7). In addition, two distinct partial intron inclusions were observed among the RT-PCR products obtained from HEK293T cells transfected with the previously described midigene construct. These are likely due to the abolition of the BPS and lead to the activation of two cryptic SASs in intron 7. A partial intron inclusion of 138 nt at the 5ʹ end of exon 8 corresponds with the activation of a cryptic SAS at position c.859-138, as predicted by SpliceAI (Supplementary Table S2 and Fig. S1A), and results in a frameshift leading to the introduction of a premature stop codon (p.Phe287Leufs*3). A second partial intron inclusion of 685 nt is the result of the use of a second cryptic SAS at position c.859-685, also leading to a frameshift and a premature stop codon (p.Phe287Tyrfs*33). Interestingly, this partial inclusion was predicted by the second strongest delta score, which yielded a SAS gain with a delta score of 0.06 at position c.859-685 (Supplementary Table S2 and Supplementary Fig. S1B). Considering that all three mutated products lead to a nonfunctional ABCA4 protein, c.858-25A>G can be considered to be deleterious and will act as a severe allele. The protein notation of this defect, based on semi-quantification of the cDNA products (Supplementary Table S4), is expected to be p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3].
Figure 1.
Overview of splice defects caused by variant c.859-25A>G in HEK293T cells. (A) Wild-type and mutant midigenes assay results. Rhodopsin exon 5 (RHO ex5) RT-PCR was used as a control for transfection efficiency. The housekeeping gene Beta-Actin (ACTB) was selected as a control for transcription. To the right, schematic representation of WT midigene (BA7_WT), in which the position of the variant is indicated with an arrow. Beneath, schematic representation of the four RT-PCR products identified in panel (A). c.859-25A>G leads to exon 8 skipping (Fragment 4), a partial intron 7 inclusion of 138-nt 5ʹ (Fragment 2) and a partial intron 7 inclusion of 685-nt 5ʹ elongation of exon 8 (Fragment 1). WT product (Fragment 3) was not detected in the mutant construct. (B) The chromatograms show the exact exonic and intronic breakpoints in the four fragments as confirmed by Sanger sequencing.
c.859-25A>G Resides on a Founder Haplotype
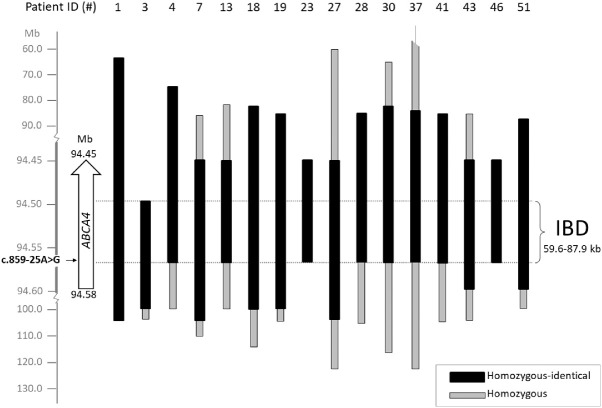
Homozygosity mapping was performed using WES data from 16 c.859-25G>A homozygous probands. Homozygous regions in probands carrying c.859-25G>A varied in size from 85 kb to 108 Mb. The identity-by-descent (IBD) region starts at g.94,546,299 (c.859-25G>A) and ends at position g.94,486,667 (rs933073), resulting in a shared homozygous region of 59.6 kb (Fig. 2 and Supplementary Table S5). Because WES data does not cover the ABCA4 locus completely, the exact boundaries of the IBD region cannot be determined. Intervals of 18.2 kb and 10.1 kb lacking SNP data to assess the haplotype are present at the 5ʹ (telomeric) and 3ʹ (centromeric) side, respectively. Thus the IBD region spans between 59.6 and 87.9 kb. In the 5ʹ region of the ABCA4 gene, four haplotypes can be distinguished (Supplementary Table S5), suggesting that at least four different recombination events occurred in the ancestors of the STGD1 cases between variant c.859-25G>A at position g.94,546,299 and the most 5’ (centromeric) ABCA4 SNP analyzed (rs3789451; g.94,586,328). Of these, haplotype 1 was the most prevalent in the analyzed cases (8/16), and proband 1 carrying this haplotype was thus considered as a reference to determine shared homozygosity regions between probands (Fig. 2 and Supplementary Table S5). At the 3ʹ end of the ABCA4 gene, all probands except proband 3 are homozygous and carry the same haplotype.
Figure 2.
Founder haplotype analysis for c.859-25A>G in 16 homozygous probands. On the left, the genomic positions on chromosome 1 and at the top patient ID numbers are reported. The haplotype of patient 1 was taken as a reference for comparison. Black boxes represent identical homozygous segments between individuals, while grey boxes represent regions of homozygosity in single individuals. IBD: identity-by-descent, encompassing between 59.6 kb and 87.9 kb. The homozygous region in 37 ends at 12.8 Mb.
Ophthalmic Characteristics of Biallelic c.859-25G>A Cases
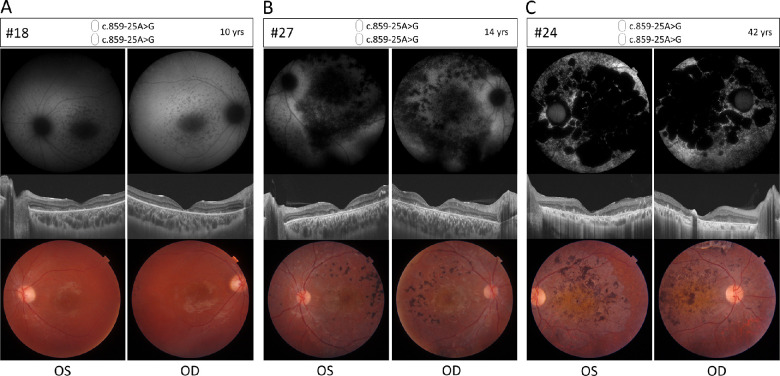
Phenotypic details for a representative selection of homozygous and compound heterozygous patients harboring c.859-25A>G are provided in Table 2 and Figures 3 and 4. The mean visual acuity of homozygous c.859-25G>A patients was 1.1 logMAR, indicating legal blindness (at mean age of 12 years). Examples of FAF, OCT, and fundus images are depicted for cases aged 10 years, 14 years and 42 years (Figs. 3A–C), which show different stages of STGD1. All cases showed foveal photoreceptor atrophy. None exhibit the yellow flecks that are typically seen in intermediate STGD1 cases. The 10-year-old presents with bull's eye maculopathy and the 14-year-old shows severe macular retinal pigment epithelium (RPE) and photoreceptor (PR) atrophy with localized hyperpigmentation deposits. The 42-year-old displays diffuse retinal atrophy and hypo-autofluorescence involving the whole retinal posterior pole and mid periphery, as observed in fundus images and FAF. OCT shows advanced atrophy of RPE and outer retinal layers of the macula. ERG information was available for 15 homozygous individuals. All of these showed abnormal cone responses and 13/15 showed abnormal rod responses suggesting a diagnosis of CRD (Supplementary Table S6).
Table 2.
Clinical Characteristics of Selected ABCA4-Retinopathy Patients With c.859-25A>G
| BCVA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID (no.) | Second ABCA4 Allele | Sex | Age at Last Examination (yrs) | OD | OS | Fishman Classification* | Foveal Photo-Receptors | FAF Abnormalities | Initial Clinical Diagnosis |
| 18 | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | F | 10 | 0.9 | 0.8 | 3 | Atrophy | Beyond | CRD/STGD |
| 27 | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | F | 14 | 1.1 | 1.1 | 4 | Atrophy | Beyond | CRD |
| 24 | p.[Phe287Hisfs*7,Phe287Tyrfs*33,Phe287Leufs*3] | M | 42 | 1.0 | 1.3 | 4 | Atrophy | Beyond | CRD |
| 6 | c.6816+2T>A | F | 12 | 0.8 | 0.7 | 3 | Atrophy | Normal | STGD |
| 20 | p.(Gly1961Ala) | M | 8 | 0.8 | 0.7 | 2 | Atrophy | Minimal | CD |
| 32 | p.(Gly1961Glu) | F | 18 | / | / | 2 | Atrophy | Beyond | STGD |
| 25 | p.(Pro1660Leu) | F | 51 | 1.5 | 1.7 | 4 | Atrophy | Beyond | Adv. CRD |
| 55 | p.(Pro1660Leu) | M | 39 | NLP | 1.3 | 4 | Atrophy | Beyond | CRD |
| 56 | p.(Pro1088Ser) | M | 18 | 1.1 | 1.0 | 3 | Atrophy | Beyond | CRD |
Adv. CRD, Advanced cone-rod dystrophy; BCVA, best corrected visual acuity; CD, cone dystrophy; F, female; M, male; NLP, no light perception; OD, right eye; OS, left eye; STGD, stargardt disease.
In all cases reported in the table, the first allele is c.859-25A>G. For sample 6 the cDNA notation is reported for the second allele since no assay was performed to determine the protein effect of the variant affecting the canonical splice site sequence. Best corrected visual acuity is reported in logMAR. Fundus autofluorescence reports the extent of abnormalities with regards to the vascular arcades.
*Fishman classification: 1 – flecks limited to within the vascular arcades, 2 – fleck-like lesions anterior to the vascular arcades and/or nasal to the optic disc, 3 – most diffuse flecks resorbed leaving diffuse RPE atrophy, and 4 – not only diffusely resorbed fundus flecks and atrophy of the retinal pigment epithelium but also diffuse choriocapillaris atrophy (Rotenstreich et al. Ophthalmology. 2003;100:1151–1158).
Figure 3.
Ophthalmic features of homozygous retinopathy cases carrying c.859-25A>G. Fundus autofluorescence (upper panels), optical coherence tomography (OCT) (middle panels) and color fundus (lower panels) for left (OS) and right (OD) eyes of three homozygous cases. The pictures are ordered by age to show different stages of STGD1 phenotypic characteristics. (A) Proband 18, 10 years of age. FAF images show enlarged foveal hypo-autofluorescence with hypo-autofluorescent spots in the posterior pole and mid periphery. OCT images show central macular atrophy of the RPE, photoreceptors and the outer retinal layers with preserved parafoveal retina, fundus images show bull's eye maculopathy (beaten-bronze pattern) with no clear flecks and few pigmentary clumps in the temporal retina. (B) Proband 27, 14 years of age. FAF images show patchy and pinpoint hypo-autofluorescence in the posterior pole and mid periphery, OCT images show diffused RPE and photoreceptor atrophy with outer retinal layers disruption. Fundus images show beaten-bronze foveal appearance with orange hue and pigment clumps. (C) Proband 24, 42 years of age. FAF images show a geographic pattern of diffuse hypo-autofluorescence involving the whole retinal posterior pole and mid-periphery. OCT show advanced atrophy of the RPE and outer retinal layers of the whole macula. Fundus images show diffuse retinal atrophy in the posterior pole with pigment clumps and orange macular hue.
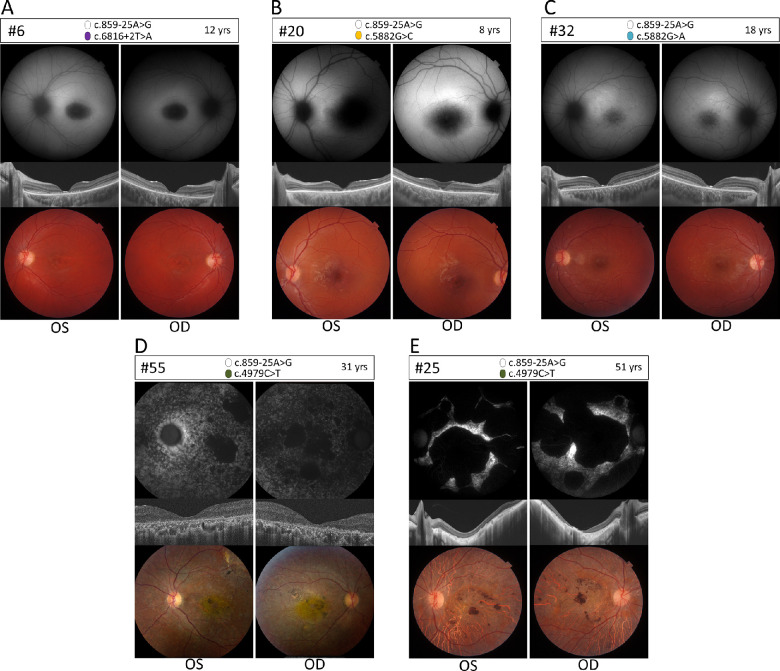
Figure 4.
Ophthalmic features of compound heterozygous retinopathy cases carrying c.859-25A>G. Fundus autofluorescence (upper panels), OCT (middle panels), and color fundus (lower panels) for left (OS) and right (OD) eyes of five compound heterozygous cases. (A) Proband 6, carrying c.6816+2T>A p.(?) as the second allele. FAF images show enlarged foveal hypo-autofluorescence, OCTs show central macular atrophy of RPE, photoreceptors and the outer retinal layers with preserved parafoveal retina. Fundus images show beaten-bronze foveal atrophy pattern with no flecks. (B) Proband 20, carrying c.5882G>C p.(Gly1961Ala) as the second allele. FAF images show enlarged foveal hypo-autofluorescence, OCTs show central macular atrophy of RPE and photoreceptors with preserved parafoveal retina. Fundus images show beaten-bronze foveal atrophy pattern with no flecks. (C) Proband 32, carrying c.5882G>A p.(Gly1961Glu) as the second allele. FAF images abnormal foveal hypo-autofluorescence signal with hypo-hyper autofluorescence flecks in the posterior pole of the retina, OCTs show images with a narrow area of foveal RPE and photoreceptor atrophy. Fundus images show blunt foveal reflex with posterior pole yellowish flecks. (D–E) Probands 55 and 25, both carrying c.4979C>T p.(Pro1660Leu) as the second allele. (D) FAF images show patchy hypo-autofluorescence islands in the posterior pole with diffused pinpoint hypo-autofluorescence involving the posterior pole and mid-periphery, OCT images show diffused RPE and outer retinal layers atrophy involving the whole macula and fundus images show diffused atrophy of the posterior pole with pigment clumps and yellow hue of the macula. (E) FAF images show large islands of complete hypo-autofluorescence with remaining small island of hyper-autofluorescence, OCT images show diffused atrophy of the outer retina, RPE and choroid with posterior staphyloma appearance. Fundus images show diffused posterior pole chorioretinal atrophy with pigment clumps.
The compound heterozygous case carrying c.859-25A>G and the splice site variant c.6816+2T>A (no. 6; Fig. 4A) shows increased foveal hypo-autofluorescence in FAF images. Additionally, OCT shows central macular atrophy of RPE, photoreceptors and the outer retinal layers with preserved parafoveal retina. Fundus images show a beaten-bronze foveal atrophy pattern with no flecks. The variant c.6816+2T>A disrupts the exon 49 SDS and, in the absence of the activation of a novel or wild-type cryptic SDS in exon 49 or intron 49, is predicted to result in exon 49 skipping. The predicted mutant protein (p.(Val2244_Gln2272del)) would lack the C-terminus of the protein.
Compound heterozygous cases carrying c.5882G>C;p.(Gly1961Ala) as a second allele have a mean visual acuity of 0.9 logMAR. Fundus photos and FAF in most cases show diffuse patchy retinal atrophy of the posterior pole with pigmentary changes and diffuse macular atrophy and retinal thinning in OCT (no. 20; Fig. 4B). Based on our data, we classified variant c.5882G>C as moderately severe. Patient 32 (Fig. 4C) carries c.5882G>A;p.(Gly1961Glu) as a second variant. Fundus and FAF images show an abnormal foveal hypo-autofluorescence signal with some hypo-autofluorescent and hyper-autofluorescent flecks in the posterior pole of the retina and blunt foveal reflex with posterior pole yellowish flecks. OCT images show a narrow area of foveal RPE and photoreceptor atrophy. The retinal images show defects that are confined to the vascular arcade, which is typical for STGD1 cases carrying p.(Gly1961Glu) in trans with any deleterious allele.43
Six patients from three separate families (Table 1 and Supplementary Fig. S4) carry c.4979C>T, p.(Pro1660Leu) as the second allele. This is a frequent pathogenic ABCA4 variant in the Israeli population.44 The two unrelated probands carrying p.(Pro1660Leu) in Figure 4D and 4E show a severe CRD phenotype with visual acuities ranging from 1.3 logMAR to no light perception, with an average of 1.5 logMAR. In patient 55 (Fig. 4D), living in the United States, color fundus pictures at 31 years have a beaten-bronze macular atrophic appearance with a few flecks extending beyond the nasal aspect of the optic nerve head. FAF imaging shows a combination of macular hyper-autofluorescence (corresponding to flecks) and some hypo-autofluorescent lesions (RPE atrophy), extending beyond the nasal edge of the optic nerve head. OCT confirms the macular atrophy with the affected area showing loss of ellipsoid zone and retinal pigment epithelium. ERG for the patient showed amplitude reductions in both scotopic and photopic responses, measuring at around 50% of the lower range amplitude value compared to normal, with asymmetric responses (worse in the right eye) and notable delay in photopic response implicit times. The abnormal cone and rod responses are suggestive of a CRD diagnosis. In patient 25 (Fig. 4E) FAF and OCT at 51 years show large islands of complete hypo-autofluorescence with remaining small island of hyper-autofluorescence, an diffuse atrophy of the outer retina, RPE and choroid with posterior staphyloma appearance. Fundus images show diffuse posterior pole chorioretinal atrophy with pigment clumps, however no ERG data was available for this patient. The suggested diagnosis for the patient is CRD. In the third family carrying c.4979C>T as second allele, ERG results were available for patient 14 who had a clinical diagnosis of STGD and presented with normal ERG values for both cones and rods.
Discussion
In this study, we identified a novel Palestinian founder mutation, c.859-25A>G, in ABCA4. The variant affects the putative branchpoint of intron 7, leading to a complex in vitro splice defect in which exon 8 is skipped and two partial inclusions of intron 7 occur. All splice defects are likely to induce nonsense-mediated RNA decay and result in the absence of ABCA4 activity. Additionally, no other pathogenic variants were identified in the probands in the shared homozygous region, offering c.859-25A>G as a compelling pathogenic variant. Patients homozygous for the variant show early-onset STGD1 and a phenotype that strongly points to a deleterious effect of the variant. This is further supported by the genotype-phenotype correlation observed in compound heterozygous probands carrying p.(Gly1961Glu) as second allele, which is in line with previously reported cases of p.(Gly1961Glu) being present in trans with a deleterious allele.43
Genotype-phenotype correlations in our study cohort also suggests the classification of variant c.5882G>C;p.(Gly1961Ala) as moderately severe. Regarding c.4979C>T, different phenotypes were observed in the three families carrying the variant in trans with c.859-25A>G. In particular, two families showed a more severe phenotype with a proposed diagnosis of CRD, suggesting a classification severe, whereas in the third family, all affected individuals presented with a milder phenotype associated to classical STGD, pointing to a moderately severe effect. No other pathogenic variants in ABCA4 that would explain the different phenotypes were identified. It is possible that currently unknown modifiers, either in ABCA4 itself or in other genes, play a role in the difference of severity observed. From the data collected in this study, we propose a classification of moderately severe or severe. Finally, phenotypic assessment of the patient carrying c.6816+2T>A suggests a mild or moderately severe effect of the variant. To better classify the effect, further analysis is required to assess the percentage of correct residual ABCA4 transcript.
ABCA4 founder mutations have been extensively described in literature45–48 and are known to widely differ between ethnic groups and geographic regions. The Palestinian population has a particularly high incidence of IRD-associated founder mutations, due to the presence of geographically and culturally isolated settlements that originate from small founder populations. Consequently, many of these villages have high consanguinity rates, which results in an enrichment of haplotype homozygosity and a high number of founder mutations.44,49 Statistical analysis of the AFs calculated for the patient and control cohorts showed a significantly higher frequency of the variant in probands compared to non-IRD individuals, supporting the pathogenicity of the founder variant c.859-25A>G. Considering that we found 27 homozygous c.859-25A>G STGD1 cases in the Hebron Governorate that has approximately 1,000,000 inhabitants, and that not all IRD cases from this area were investigated for the presence of this variant, we can deduce a minimal AF of 0.0052 for c.859-25A>G, and a minimal carrier rate of 0.0103 for this variant in healthy individuals (i.e., 1/97 individuals). This is likely an overestimate because of the high consanguinity levels observed in the population. Nevertheless, knowledge of the carriership of c.859-25A>G is relevant for genetic counseling in Palestinian families living in Hebron.
It is of interest that despite its high frequency the variant was not previously detected, which is likely due to the position of the variant in a BPS. Although the recognition of the BPS by the splicing machinery is critical in RNA splicing, because it leads to the formation of the lariat structure that precedes intron excision from pre-mRNA, BPS variants are scarce in literature compared to other variants affecting splicing. Nevertheless, several examples of variants abolishing the BP have been reported in hereditary diseases, such as LCAT-associated fish-eye disease,50 Ehlers-Danlos syndrome due to COL5A1 variants,51 hypophosphatasia due to variants in ALPL52 and in hereditary cancer-associated genes, such as XPC mutated in patients with xeroderma pigmentosum,53 and RB1 mutated in patients with retinoblastoma.54 Earlier this year, the first BPS variant associated with non-syndromic retinitis pigmentosa was described for BBS1.55 The rarity of published pathogenic BPS variants is likely due to the difficulties in identifying the BPS sites. Correct identification is a challenge for two main reasons: the conserved motif and the localization of BPS. Firstly, the consensus motif has been identified as the extremely short and degenerate sequence “yUnAy,”56 where y = a pyrimidine and n = any nucleotide. In this sequence, only the A residue (position 0, corresponding to the point of formation of the lariat structure) and the U residue (at position −2) are highly conserved. Mutations of these two residues cause BPS abolishment, leading to splice defects, such as exon skipping or pseudo-exon inclusion.50,57 Second, the majority of BPS have been identified in a window of 18 to 44 nt upstream of the SAS, although BPSs as far away as 400 nt are known.58
The sparse number of experimentally proven BPSs, both wild-type and mutated, has resulted in additional challenges in the development of effective tools to predict the impact of variants upstream of SASs. The currently available prediction tools perform poorly, and are mostly focused on the recognition of wild-type BPSs.59 In this study we show that SpliceAI, given the right parameters, was able to correctly predict the effect of the BPS variants on splicing. These results suggest that SpliceAI is one of the best tools for putative splice affecting variant prioritization, because it is able to flag variants independently of the biological cause of the expected splicing disruption. Our results are in line with the recently reported performance assessment of SpliceAI for intronic ABCA4 splicing variants.60 Further confirmation and implementation of visualization tools to check the genetic context around the predicted effects is still advisable. Nevertheless, use of SpliceAI allows one to bypass more lengthy and spurious research of variants, which would otherwise entail the use of multiple tools focused on predicting single aspects of splicing, such as effects on NCSS, BPS, enhancer and silencer motifs separately. As for analysis parameters, the window of flanking sequences considered by the program has a significant impact on the predicted splicing defects of a variant, thus should be carefully taken into account. Both partial intron inclusions observed in the in vitro splice assay were predicted by SpliceAI, but a small window of analysis (e.g., ±100 nt) would not have identified either of these inclusions as they are located 113 and 660 nt upstream of the variant. Notably, the considerable distance of the cryptic SASs also prevented their detection in all other programs used in this study. In particular, analysis of the effect of c.859-25A>G using Alamut Visual prediction tools resulted in a reduction of 1.9 in the predicted intron 7 SAS strength (wild-type, 3.7; mutant 1.8 [range 0–21]) on GeneSplicer, which on its own would not be considered sufficient to select the variant for further testing.
In this study, the effect of the variant was assessed at the RNA level using an in vitro splice assay using HEK293T cells. There is ample evidence that this in vitro model is able to correctly recapitulate splice defects affecting the consensus splice site sequences at the junctions of exons and introns.61 This is also true for almost all variants that create new splice sites in introns or strengthen cryptic splice sites in introns, resulting in the recognition by the splicing machinery and pseudo-exon inclusion, exon elongation or intron retention.12,13,17,62 Nevertheless, considering the absence of retina-specific factors and the artificial nature of the midigene system, it remains interesting to further validate the results obtained in HEK293T cells in an alternative model. The use of induced pluripotent stem cells to obtain patient-derived photoreceptor precursor cells or retinal organoids, allows observation of the variant effect in a context that is more similar to that of the patient. Retina-specific pseudo-exon recognition was found for intronic variants in ABCA4, creating new or strengthening existing exonic splice enhancer motifs.16 The most frequent variant associated with Leber congenital amaurosis, an intron 26 variant in CEP290, c.2991+1655A>G, was shown to enhance a pseudo-exon RNA insertion in retinal organoids compared to fibroblasts or EBV-immortalized B lymphocytes.34,35
Intronic variants resulting in aberrant splicing are good targets for antisense oligonucleotide (AON)-based therapeutic strategies. The use of AONs to modulate splicing and obtain correct transcripts has been proven to be effective in ABCA4.13,16,63–65 Regarding the treatment of the effects of c.859-25A>G, the use of AONs might prove challenging, since binding to the region upstream of the canonical SAS comes with the risk of disrupting regulatory motifs and the binding of auxiliary splice proteins. Considering the involvement of two cryptic splice sites upstream of the BPS variant, a potential rescue approach would be to target the new SASs of the partial intron retention with AONs, which are unlikely to affect exon 8 recognition by the splicing machinery. On the other hand, it remains to be seen whether blocking these new SASs would lead to a correct transcript. We hypothesize, partially based on SpliceAI predictions, that this approach may lead to more exon 8 skipping. Experimental testing of potential AONs is needed to investigate this further.
Outside of AON-based rescue, other therapeutic approaches that could be implemented are gene augmentation or gene editing. Gene augmentation therapy is successful for some retinal disorders (e.g. RPE65-associated Leber congenital amaurosis or early onset RP)66 but remains challenging for STGD1. The relatively large size of the ABCA4 transcript prevents delivery with a single adeno-associated virus vector, which has a maximum packaging capacity of about 5 kb. Multiple approaches are being developed to bypass this issue (e.g. the use of a dual vector system)67–70 and the implementation of nanoparticles, such as non-viral gene therapy delivery system.67–70 An attractive alternative to gene replacement for ABCA4 is RNA-editing, which allows the correction of point mutations at the transcript level with low risk of permanent off-target effects. Unfortunately, currently only A>G and C>U corrections can be obtained using engineered deaminase enzymes (such as ADAR and APOBEC), neither of which are viable options for our A>G variant.71–73
In conclusion, we identified a severe and frequent Palestinian founder variant, c.859-25A>G, leading to a complex splicing defect very likely due to the disruption of the BPS in intron 7 of ABCA4. The high frequency of the variant in the Palestinian population makes it highly relevant in terms of genetic counselling, in particular in the Hebron Governorate and in Jerusalem. Our study highlights the relevance of the use of correct prediction tools to identify elusive splice-altering variants, such as BPS variants, that have the potential to strongly impact genetic diagnosis and genetic counselling in patients affected by STGD1.
Supplementary Material
Acknowledgments
The authors thank Moien Kanaan for allele frequency data from 1,400 Palestinian persons who do not show inherited retinal dystrophies. We thank Bin Guan, Ehsan Ullah, and Chelsea Bender for technical assistance, and Stéphanie S. Cornelis for help with Hardy-Weinberg calculations.
Supported by the RetinaUK, grant no. GR591 (to FPMC), a Horizon 2020, Marie Sklodowska-Curie Innovative Training Network entitled European Training Network to Diagnose, Understand and Treat Stargardt Disease, a Frequent Inherited Blinding Disorder-StarT (813490) (to FPMC), the Foundation Fighting Blindness USA, grant no. PPA-0517-0717-RAD (to FPMC and SR), the Rotterdamse Stichting Blindenbelangen, the Stichting Blindenhulp, and the Stichting tot Verbetering van het Lot der Blinden (to FPMC), and by the Landelijke Stichting voor Blinden en Slechtzienden, the Macula Degeneratie fonds and the Stichting Blinden-Penning that contributed through Uitzicht 2016-12 (to FPMC). This work was also supported by the ProRetina foundation Germany, the Stichting Blindenhulp, the Stichting ter Verbetering van het Lot der Blinden, the Gelderse Blindenstichting, the Stichting voor Ooglijders, and the Oogfonds that contributed through Uitzicht 2020-17 (FPMC and SR). This work was also supported by EJPRD19-234 (Solve-RET) (FPMC and SR). This work was also supported by “Groupement de Coopération Sanitaire Interrégional G4 qui réunit les Centres Hospitaliers Universitaires Amiens, Caen, Lille et Rouen (GCS G4)” and by the Fondation Stargardt France (to CMD), the Foundation Fighting Blindness, Grant/Award Number: BR‐GE‐0214–0639‐TECH and BRGE‐0518–0734‐TECH TECH (to DS and EB), the Israel Science Foundation (grant No. 1778/20) within the Israel Precision Medicine Partnership program (DS and EB), the STEP-GTP fellowship (to MS), and EuropeAid (Peace of Sight project; DS, EB, and AAT). BB, WMZ and RBH were supported from the intramural research fund of the National Eye Institute, National Institutes of Health (NIH). The Henry Brent Chair in Innovative Pediatric Ophthalmology Research (EH).
Disclosure: Z. Corradi, None; M. Salameh, None; M. Khan, None; E. Héon, Novartis (C), Jansen (C), Atsena Therapeutics (E); K. Mishra, None; R.J. Hitti-Malin, None; Y. AlSwaiti, None; A. Aslanian, None; E. Banin, None; B.P. Brooks, None; W.M. Zein, None; R.B. Hufnagel, None; S. Roosing, None; C‐M. Dhaenens, None; D. Sharon, None; F.P.M. Cremers, None; A. AlTalbishi, None
References
- 1. Blacharski PA, Newsome DA.. Bilateral macular holes after Nd-Yag laser posterior capsulotomy. Am J Ophthalmol. 1988; 105: 417–418. [DOI] [PubMed] [Google Scholar]
- 2. Allikmets R, Singh N, Sun H, et al.. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997; 15: 236–246. [DOI] [PubMed] [Google Scholar]
- 3. Cornelis SS, Bax NM, Zernant J, et al.. In Silico Functional Meta-Analysis of 5,962 ABCA4 Variants in 3,928 Retinal Dystrophy Cases. Hum Mutat. 2017; 38: 400–408. [DOI] [PubMed] [Google Scholar]
- 4. Cornelis SS, Runhart EH, Bauwens M, et al.. Genetic risk estimates for offspring of patients with Stargardt disease. medRxiv. 2021.
- 5. Cremers FP, van de Pol DJ, van Driel M, et al.. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet. 1998; 7: 355–362. [DOI] [PubMed] [Google Scholar]
- 6. Rozet JM, Gerber S, Ghazi I, et al.. Mutations of the retinal specific ATP binding transporter gene (ABCR) in a single family segregating both autosomal recessive retinitis pigmentosa RP19 and Stargardt disease: evidence of clinical heterogeneity at this locus. J Med Genet. 1999; 36: 447–451. [PMC free article] [PubMed] [Google Scholar]
- 7. Maugeri A, Klevering BJ, Rohrschneider K, et al.. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000; 67: 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cremers FPM, Lee W, Collin RWJ, Allikmets R.. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog Retin Eye Res. 2020; 79: 100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maugeri A, van Driel MA, van de Pol DJ, et al.. The 2588G–>C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet. 1999; 64: 1024–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Runhart EH, Khan M, Cornelis SS, et al.. Association of sex with frequent and mild ABCA4 alleles in Stargardt disease. JAMA Ophthalmol. 2020; 138: 1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Runhart EH, Sangermano R, Cornelis SS, et al.. The common ABCA4 variant p.Asn1868Ile shows nonpenetrance and variable expression of Stargardt disease when present in trans with severe variants. Invest Ophthalmol Vis Sci. 2018; 59: 3220–3231. [DOI] [PubMed] [Google Scholar]
- 12. Sangermano R, Garanto A, Khan M, et al.. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet Med. 2019; 21: 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bauwens M, Garanto A, Sangermano R, et al.. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet Med. 2019; 21: 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sangermano R, Khan M, Cornelis SS, et al.. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018; 28: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fadaie Z, Khan M, Del Pzo-oValero M, et al.. Identification of splice defects due to noncanonical splice site or deep-intronic variants in ABCA4. Hum Mutat. 2019; 40: 2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Albert S, Garanto A, Sangermano R, et al.. Identification and rescue of splice defects caused by two neighboring deep-intronic ABCA4 mutations underlying Stargardt disease. Am J Hum Genet. 2018; 102: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khan M, Cornelis SS, Del Pozo-Valero MD, et al.. Resolving the dark matter of ABCA4 for 1054 Stargardt disease probands through integrated genomics and transcriptomics. Genet Med. 2020; 22: 1235–1246. [DOI] [PubMed] [Google Scholar]
- 18. Khan M, Cornelis SS, Khan MI, et al.. Cost-effective molecular inversion probe-based ABCA4 sequencing reveals deep-intronic variants in Stargardt disease. Hum Mutat. 2019; 40: 1749–1759. [DOI] [PubMed] [Google Scholar]
- 19. Bauwens M, Garanto A, Sangermano R, et al.. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet Med. 2019; 21: 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujinami K, Zernant J, Chana RK, et al.. Clinical and molecular characteristics of childhood-onset Stargardt disease. Ophthalmology. 2015; 122: 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braun TA, Mullins RF, Wagner AH, et al.. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013; 22: 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang ZF, Burge CB.. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008; 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frendewey D, Keller W.. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985; 42: 355–367. [DOI] [PubMed] [Google Scholar]
- 24. Shapiro MB, Senapathy P.. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987; 15: 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C.. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yeo G, Burge CB.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004; 11: 377–394. [DOI] [PubMed] [Google Scholar]
- 27. Pertea M, Lin X, Salzberg SL.. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001; 29: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fairbrother WG, Yeo GW, Yeh R, et al.. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004; 32: W187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR.. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003; 31: 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reese MG, Eeckman FH, Kulp D, Haussler D.. Improved splice site detection in Genie. J Comput Biol. 1997; 4: 311–323. [DOI] [PubMed] [Google Scholar]
- 31. Mort M, Sterne-Weiler T, Li B, et al.. MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol. 2014; 15: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khan M, Cornelis SS, Sangermano R, et al.. In or out? New insights on exon recognition through splice-site interdependency. Int J Mol Sci. 2020; 21: 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matera AG, Wang Z.. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014; 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. den Hollander AI, Koenekoop RK, Yzer S, et al.. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006; 79: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parfitt DA, Lane A, Ramsden CM, et al.. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell. 2016; 18: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al.. Predicting splicing from primary sequence with deep learning. Cell. 2019; 176: 535–548 e524. [DOI] [PubMed] [Google Scholar]
- 37. Leman R, Gaildrat P, Le Gac G, et al.. Novel diagnostic tool for prediction of variant spliceogenicity derived from a set of 395 combined in silico/in vitro studies: an international collaborative effort. Nucleic Acids Res. 2018; 46: 7913–7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schindelin J, Arganda-Carreras I, Frise E, et al.. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinson JT, Thorvaldsdottir H, Wenger AM, Zehir A, Mesirov JP.. Variant review with the integrative genomics viewer. Cancer Res. 2017; 77: e31–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thorvaldsdottir H, Robinson JT, Mesirov JP.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013; 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCulloch DL, Marmor MF, Brigell MG, et al.. ISCEV Standard for full-field clinical electroretinography (2015 update). Doc Ophthalmol. 2015; 130: 1–12. [DOI] [PubMed] [Google Scholar]
- 42. Richards S, Aziz N, Bale S, et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zernant J, Lee W, Collison FT, et al.. Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J Med Genet. 2017; 54: 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharon D, Ben-Yosef T, Goldenberg-Cohen N, et al.. A nationwide genetic analysis of inherited retinal diseases in Israel as assessed by the Israeli inherited retinal disease consortium (IIRDC). Hum Mutat. 2020; 41: 140–149. [DOI] [PubMed] [Google Scholar]
- 45. Cremers FP, Maugeri A, den Hollander AI, Hoyng CB.. The expanding roles of ABCA4 and CRB1 in inherited blindness. Novartis Found Symp. 2004; 255: 68–79; discussion 79–84, 177–178. [DOI] [PubMed] [Google Scholar]
- 46. Burke TR, Fishman GA, Zernant J, et al.. Retinal phenotypes in patients homozygous for the G1961E mutation in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012; 53: 4458–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beit-Ya'acov A, Mizrahi-Meissonnier L, Obolensky A, et al.. Homozygosity for a novel ABCA4 founder splicing mutation is associated with progressive and severe Stargardt-like disease. Invest Ophthalmol Vis Sci. 2007; 48: 4308–4314. [DOI] [PubMed] [Google Scholar]
- 48. Chacon-Camacho OF, Granillo-Alvarez M, Ayala-Ramirez R, Zenteno JC.. ABCA4 mutational spectrum in Mexican patients with Stargardt disease: identification of 12 novel mutations and evidence of a founder effect for the common p.A1773V mutation. Exp Eye Res. 2013; 109: 77–82. [DOI] [PubMed] [Google Scholar]
- 49. Zlotogora J. Molecular basis of autosomal recessive diseases among the Palestinian Arabs. Am J Med Genet. 2002; 109: 176–182. [DOI] [PubMed] [Google Scholar]
- 50. Kuivenhoven JA, Weibusch H, Pritchard PH, et al.. An intronic mutation in a lariat branchpoint sequence is a direct cause of an inherited human disorder (fish-eye disease). J Clin Invest. 1996; 98: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burrows NP, Nicholls AC, Richards AJ, et al.. A point mutation in an intronic branch site results in aberrant splicing of COL5A1 and in Ehlers-Danlos syndrome type II in two British families. Am J Hum Genet. 1998; 63: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mentrup B, Girschick H, Jakob F, Hofmann C. A homozygous intronic branch-point deletion in the ALPL gene causes infantile hypophosphatasia. Bone. 2017; 94: 75–83. [DOI] [PubMed] [Google Scholar]
- 53. Khan SG, Yamanegi K, Zheng ZM, et al.. XPC branch-point sequence mutations disrupt U2 snRNP binding, resulting in abnormal pre-mRNA splicing in xeroderma pigmentosum patients. Hum Mutat. 2010; 31: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Houdayer C, Dehainault C, Mattler C, et al.. Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum Mutat. 2008; 29: 975–982. [DOI] [PubMed] [Google Scholar]
- 55. Fadaie Z, Whelan L, Dockery A, et al.. BBS1 branchpoint variant is associated with non-syndromic retinitis pigmentosa. J Med Genet. 2021. [DOI] [PubMed] [Google Scholar]
- 56. Gao K, Masuda A, Matsuura T, Ohno K.. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008; 36: 2257–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. De Klein A, Riegman PH, Bijlsma EK, et al.. A G–>A transition creates a branch point sequence and activation of a cryptic exon, resulting in the hereditary disorder neurofibromatosis 2. Hum Mol Genet. 1998; 7: 393–398. [DOI] [PubMed] [Google Scholar]
- 58. Leman R, Tubeuf H, Raad S, et al.. Assessment of branch point prediction tools to predict physiological branch points and their alteration by variants. BMC Genomics. 2020; 21: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Canson D, Glubb D, Spurdle AB.. Variant effect on splicing regulatory elements, branchpoint usage, and pseudoexonization: strategies to enhance bioinformatic prediction using hereditary cancer genes as exemplars. Hum Mutat. 2020; 41: 1705–1721. [DOI] [PubMed] [Google Scholar]
- 60. Riepe TV, Khan M, Roosing S, Cremers FPM, t Hoen PAC.. Benchmarking deep learning splice prediction tools using functional splice assays. Hum Mutat. 2021; 42: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sangermano R, Khan M, Cornelis SS, et al.. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018; 28: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khan M, Arno G, Fakin A, et al.. Detailed Phenotyping and Therapeutic Strategies for Intronic ABCA4 Variants in Stargardt Disease. Mol Ther Nucleic Acids. 2020; 21: 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sangermano R, Garanto A, Khan M, et al.. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genetics in Medicine. 2019; 21: 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Garanto A, Duijkers L, Tomkiewicz TZ, Collin RWJ.. Antisense oligonucleotide screening to optimize the rescue of the splicing defect caused by the recurrent deep-intronic ABCA4 variant c.4539+2001G>A in Stargardt disease. Genes (Basel). 2019; 10: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tomkiewicz TZ, Suarez-Herrera N, Cremers FPM, Collin RWJ, Garanto A.. Antisense oligonucleotide-based rescue of aberrant splicing defects caused by 15 pathogenic variants in ABCA4. Int J Mol Sci. 2021; 22(9): 4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Russell S, Bennett J, Wellman JA, et al.. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017; 390: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McClements ME, Barnard AR, Charbel Issa P, MacLaren RE. Assessment of AAV dual vector safety in the Abca4 (-/-) mouse model of Stargardt disease. Transl Vis Sci Technol. 2020; 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dyka FM, Molday LL, Chiodo VA, Molday RS, Hauswirth WW.. Dual ABCA4-AAV vector treatment reduces pathogenic retinal A2E accumulation in a mouse model of autosomal recessive Stargardt disease. Hum Gene Ther. 2019; 30: 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Trapani I. Dual AAV vectors for Stargardt disease. Methods Mol Biol. 2018; 1715: 153–175. [DOI] [PubMed] [Google Scholar]
- 70. Trapani I, Toriello E, de Simone S, et al.. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum Mol Genet. 2015; 24: 6811–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fry LE, Peddle CF, Barnard AR, McClements ME, MacLaren RE.. RNA editing as a therapeutic approach for retinal gene therapy requiring long coding sequences. Int J Mol Sci. 2020; 21(3): 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Abudayyeh OO, Gootenberg JS, Franklin B, et al.. A cytosine deaminase for programmable single-base RNA editing. Science. 2019; 365: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang X, Lv J, Li Y, et al.. Programmable C-to-U RNA editing using the human APOBEC3A deaminase. EMBO J. 2020; 39: e104741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.