Abstract
The vitamin D endocrine system is essential for calcium metabolism and skeletal integrity. 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] regulates bone mineral homeostasis and acts directly on osteoblasts. In the present study we characterized the transcriptional regulation of the class 3 semaphorin (Sema3) gene family by 1,25(OH)2D3 in osteoblastic cells. Class 3 semaphorins are secreted proteins that regulate cell growth, morphology and migration, and were recently shown to be involved in bone homeostasis. In ST2, MC3T3-E1 and primary calvarial osteoblast cell cultures we found that all members of the Sema3 gene family were expressed, and that Sema3e and Sema3f were the most strongly induced 1,25(OH)2D3 target genes among the studied cell types. In addition, transcription of Sema3b and Sema3c was upregulated, whereas Sema3d and Sema3g was downregulated by 1,25(OH)2D3 in different osteoblastic cells. Chromatin immunoprecipitation analysis linked to DNA sequencing (ChIP-seq analysis) revealed the presence of the vitamin D receptor at multiple genomic loci in the proximity of Sema3 genes, demonstrating that the genes are primary 1,25(OH)2D3 targets. Furthermore, we showed that recombinant SEMA3E and SEMA3F protein were able to inhibit osteoblast proliferation. However, recombinant SEMA3s did not affect ST2 cell migration. The expression of class 3 semaphorins in osteoblasts together with their regulation by 1,25(OH)2D3 suggests that these genes, involved in the regulation of bone homeostasis, are additional mediators for 1,25(OH)2D3 signaling in osteoblasts.
Keywords: Vitamin D, Vitamin d receptor, Semaphorin, Osteoblast, Transcription, Gene expression regulation
1. Introduction
The semaphorin (SEMA) superfamily consists of a large number of cell surface and secreted glycoproteins that are involved in axonal growth cone guidance, cell migration, cardiogenesis, angiogenesis, tumor growth and immune responses [1–5]. The class 3 semaphorin (SEMA3) family consists of 7 members (SEMA3A through 3G) and are the only secreted semaphorins in vertebrates [1]. Except for SEMA3E, which directly binds the plexin (PLXN) D1 receptor [6], SEMA3 proteins first bind to neuropilin (NRP) receptors. After NRP binding, PLXNA co-receptors are recruited that serve as the primary receptors that transduce the signal to the cytoplasm, while NRPs stabilize the receptor complex [7]. The composition of the NRP/PLXN receptor complex determines the binding affinity of the SEMA3 ligands to the NRP receptors and may depend on cell or tissue type [8], allowing cell-type specific signaling.
Lately, semaphorins have gained attention for their involvement in normal bone homeostasis and bone pathology [9–11]. Semaphorin molecules of different classes are widely expressed in bone cells and SEMA3s may either promote (SEMA3B) or inhibit (SEMA3A, SEMA3E) osteoclast formation [12]. Moreover, SEMA3A is also able to promote osteoblastic bone formation [9]. These studies suggest that semaphorin signaling participates in the complex cross-talk between osteoblasts and osteoclasts, which indicates that they may have therapeutic potential for the treatment of bone disorders.
The classical role of 1α,25-dihydroxyvitamin D3 [1,25(OH)2D3], the most biologically active vitamin D3 metabolite, is to maintain adequate serum levels of calcium and phosphate by stimulating calcium absorption in the intestine, by increasing reabsorption of calcium and phosphate in the kidney, and by releasing calcium and phosphate from skeletal stores [13]. Although 1,25(OH)2D3 has direct effects on bone formation and osteoblasts, its exact role in bone cells has yet to be completely unraveled [14]. 1,25(OH)2D3 exerts its functions through the vitamin D receptor (VDR), a ligand-activated transcription factor that belongs to the nuclear receptor superfamily. To directly regulate the transcription of primary 1,25 (OH)2D3 target genes, ligand-activated VDR forms a heterodimer with the retinoid X receptor (RXR) and binds DNA at specific vitamin D responsive elements (VDREs), preferentially the DR3-type VDREs that are formed by direct repeats of two hexameric core binding sequences spaced by three nucleotides [15,16]. Interestingly, different studies have illustrated that 1,25(OH)2D3 induces the expression of several class 3 semaphorins. Indeed, Sema3b as well as Sema3e are transcriptionally upregulated by 1,25 (OH)2D3 in osteoblasts [17,18]. Moreover, 1,25(OH)2D3 upregulates Sema3b in prostate cancer cells [19], and induces SEMA3B and SEMA3F protein levels in malignant endometrial cancer cells [20]. However, none of these studies deciphered whether these Sema3 genes are primary targets of 1,25(OH)2D3.
The findings that 1,25(OH)2D3 induces expression levels of different Sema3 ligands prompted us to investigate the transcriptional regulation of all seven members of the Sema3 family genes in osteoblastic cell cultures after treatment with 1,25(OH)2D3. In addition, we demonstrated by chromatin immunoprecipitation sequencing (ChIP-seq) analyses a number of active VDR binding sites across the genomic loci for the Sema3 genes. Finally, we attempted to decipher possible roles of recombinant SEMA3s in osteoblast proliferation and migration.
2. Materials and methods
2.1. Cell culture
MC3T3-E1 pre-osteoblast and bone marrow-derived stromal ST2 cells (both from Riken Cell Bank, Japan) were maintained in Minimum Essential Medium alpha with GlutaMAX (α-MEM, Gibco) supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. Primary calvarial osteoblasts were isolated from newborn C57BL/6 mice (Harlan Laboratories) as described previously [21]. The day after seeding, treatment with vehicle (PBS or ethanol), with recombinant SEMA3A, 3B, 3C, 3E, or 3F (R&D Systems), or with 10−8 M 1,25(OH)2D3 (Sigma-Aldrich) was started. The final concentration of ethanol in the medium was lower than 0.001%.
Gene expression during in vitro osteoblastic mineralization was assessed by culturing calvarial osteoblasts until confluency, and supplementing the culture medium with 50 μg/ml ascorbic acid (Sigma-Aldrich) and 10 mM β-glycerophosphate (Sigma-Aldrich). At days 13 and 20 after the switch to osteogenic medium, cultures were incubated with vehicle or 10−8 M 1,25(OH)2D3 for 24 h.
To measure cellular proliferation, ST2 and MC3T3-E1 cells were incubated for 72 h with recombinant SEMA3A, 3B, 3C, 3E, or 3F (R&D Systems) and 1 μCi [3H]thymidine (specific activity of 2 Ci/mmol, PerkinElmer) was added. After an additional 4 h incubation period [3H]thymidine incorporation into cells was measured as described previously [22].
2.2. RNA extraction, cDNA synthesis and qPCR
Total RNA was isolated, cDNA prepared with Oligo d(T)16 primers (Invitrogen), and quantitative real-time PCR (qPCR) reactions performed using 1/10 diluted cDNA templates with the Fast SYBR Green Master Mix (Applied Biosystems) or the TaqMan Fast Universal PCR Master Mix (Applied Biosystems) as described previously [22]. To obtain relative gene expression levels, the 2−ΔCt method was used and the reference gene β-actin was used to normalize the gene expression.
2.3. Chromatin immunoprecipitation (ChIP) and ChIP-seq analysis
ChIP-seq analyses were previously performed in MC3T3-E1 cells (GSE51515 and GSE41955) [23,24]. Tag density for each factor was normalized to 107 tags and data normalized to input were displayed using the UCSC genome browser (http://genome.ucsc.edu/) [25], and is based on the Build 37 assembly (mm9) by the National Center for Biotechnology Information (NCBI) and the Mouse Genome Sequencing Consortium. ChIP-qPCR experiments using anti-VDR (sc-1008, Santa Cruz), anti-RXR (sc-774, Santa Cruz) and anti-tetra-acetylated histone H4 (06–866, Millipore) were performed with MC3T3-E1 cells treated for 3 h with 10−7 M 1,25(OH)2D3 as described previously [23]. Cell lysates used in the ChIP-qPCR experiments were from three independent experiments and results were normalized with respect to input samples. The specificity of receptor binding data by ChIP was confirmed by unspecific negative control region, where no VDR/RXR binding was observed according to ChIP-seq.
2.4. Transient transfection assays
Genomic DNA fragments (mm9) of Sema3b (chr9: 107,507,703–107,508,041; 339 bp in size) and Sema3f (chr9: 107,605,724–107,606,022; 299 bp in size) were cloned in a pGL3-Basic luciferase reporter vector (Promega). Vectors, in which 4 nucleotides from the DR3-type VDREs present in these genomic fragments were mutated, were prepared with the QuickChange II Site-Directed Mutagenesis kit (Stratagene). Exponentially growing MC3T3-E1 cells were transfected, stimulated following day with 1,25(OH)2D3 (10−8 M) or vehicle, and subjected to luciferase activity assessment as described previously [26].
2.5. Scratch wound cell migration assay
Cell migration was analyzed in ST2 cells with the IncuCyte ZOOM Scratch Wound assay (Essen BioScience). Cells were grown overnight to form sub-confluent monolayers and treated for 2 h with recombinant SEMA3A, 3B, 3C, 3E, or 3F before 700–800 μm wide scratch wounds were prepared for each well with a 96-pin WoundMaker (Essen BioScience). The wound recovery was monitored every two hours by the IncuCyte ZOOM live cell imaging system and the data were analyzed with software version 2015A. A minimum of 12 replicate treatments per experiment was performed.
Data for each experiment were further analyzed as relative wound densities (RWD) over time. RWD (%) is a measure of the percentage change in relative spatial cell density in the initial wound area over the cell density outside the wound at every time point. RWD values as a function of time were plotted in a line chart, and area under the curve (AUC) was determined for each individual wound separately to obtain a comparable measure for cell migration kinetics. AUC was determined from RWD data obtained during 36 h of wound healing, as illustrated in Fig. S1, where approximately 75–85% RWD was reached and RWD still increased linearly over time (Fig. S1B).
2.6. Statistical analysis
The results are expressed as the mean and standard deviation. Two-tailed Student’s t-tests of sample means, and one sample t-tests using log-transformed values of fold change data, were carried out to detect significant differences. The applied statistical tests are indicated in the figure legends. p < 0.05 was accepted as significant.
3. Results
3.1. 1,25(OH)2D3 induced the transcription of different semaphorin 3 family members
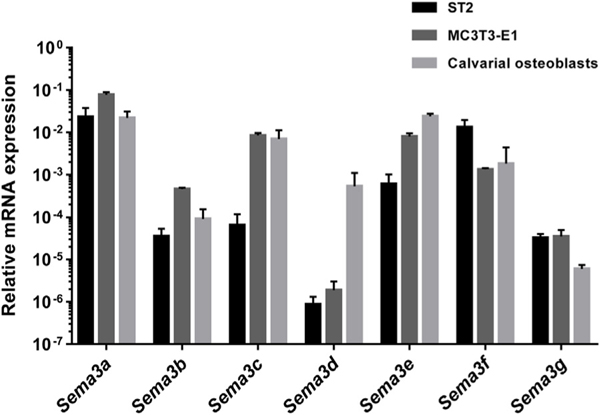
In order to get an overview of osteoblastic gene expression of the different members of class 3 semaphorins, relative basal mRNA expression levels were analyzed by qPCR in the ST2 bone marrow-derived stromal cell line, in the MC3T3-E1 mouse pre-osteoblast cell line, and in cultures of primary calvarial osteoblasts (Fig. 1). Expression profiling revealed that all 7 genes were expressed in the studied cell types with Sema3a, 3c, 3e and 3f being the most abundantly expressed in all investigated cell types. The expression pattern of the Sema3 genes was rather similar among the studied cell types, with the exception of Sema3d, which was only weakly expressed in ST2 and MC3T3-E1 cells, but rather abundantly expressed in primary calvarial osteoblasts.
Fig. 1.
Basal mRNA expression levels of semaphorin 3 family members in osteoblastic cells. qPCR was performed to determine the relative basal mRNA expression of Sema3a, 3b, 3c, 3e, 3f and 3g in ST2 cells (black bars), MC3T3-E1 cells (dark grey bars), and in primary calvarial osteoblasts (light grey bars). Gene expression was analyzed by qPCR in non-treated cells and normalized to the reference gene β-actin. The mean and standard deviation of at least three independent experiments are shown.
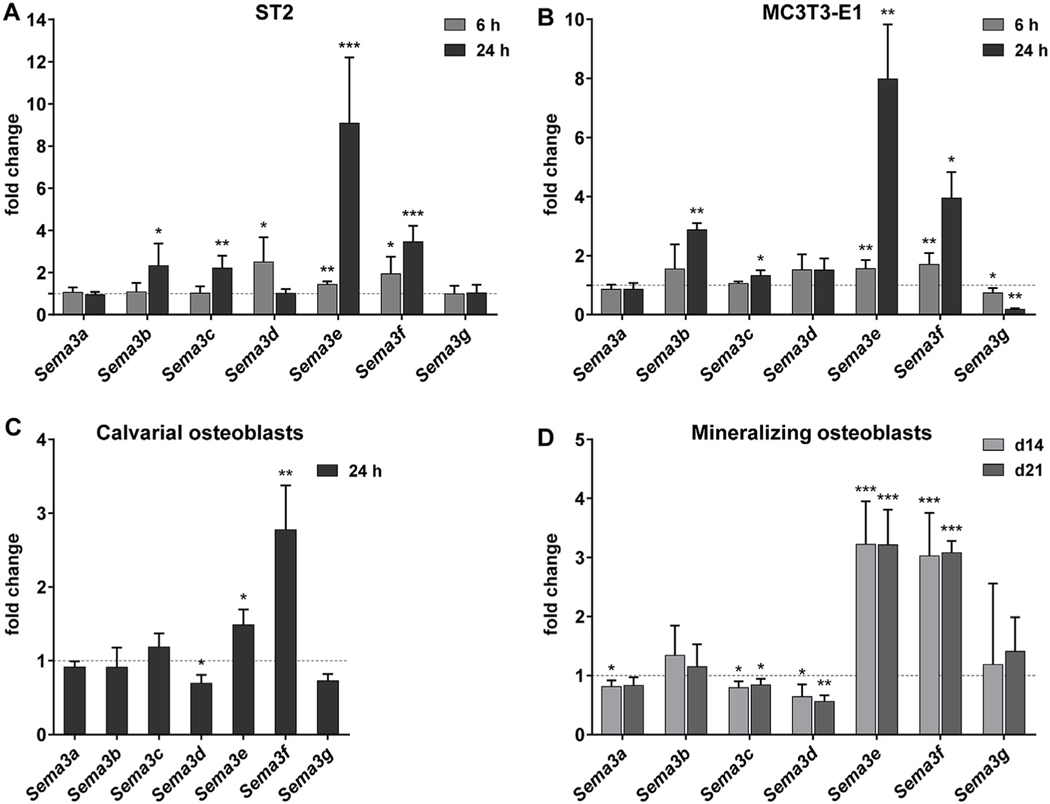
When these cells were stimulated with 1,25(OH)2D3 for 6 and 24 h, we observed that transcript levels of four semaphorin 3 family members, Sema3b, 3c, 3e and 3f, were significantly induced in at least 2 cell types (Fig. 2). Transcriptional upregulation of Sema3e and 3f by 1,25(OH)2D3 was apparent after a 6 h incubation period in ST2 and MC3T3-E1 cells (Fig. 2A and B), and was observed at 24 h after stimulation in all investigated cell cultures (Fig. 2A, B, C, and D). Interestingly, even though their basal expression levels were high, Sema3e and 3f were the most inducible semaphorin 3 family genes in response to a 24 h treatment with 1,25(OH)2D3 in all cell types, showing 9.1 and 3.5, 8.0 and 4.0, and 1.5 and 2.8 −fold inductions in ST2, MC3T3-E1 and calvarial osteoblast cells, respectively. Additionally, the induction of the genes was sustained for at least 72 h after stimulation (data not shown). Furthermore, the genes Sema3b and 3c were induced in both ST2 and MC3T3-E1 cells after a 24 h incubation period with 1,25(OH)2D3 demonstrating significant, but weaker and delayed upregulation compared to that of Sema3e and 3f (Fig. 2A and B). Additionally, expression of Sema3g and Sema3d was regulated in a cell type-specific manner. Indeed, Sema3g expression was significantly downregulated in MC3T3-E1 cells at both measured time points (Fig. 2B), whereas Sema3d showed prominent downregulation in calvarial osteoblasts (Fig. 2C).
Fig. 2.
Transcript levels of class semaphorin 3 family members were regulated by 1,25(OH)2D3 in osteoblastic cells and during osteoblast mineralization. ST2 (A) and MC3T3-E1 (B) cells were treated with 1,25(OH)2D3 (10−8 M) or vehicle for 6 and 24 h. Primary calvarial osteoblasts (C) were treated for 24 h with 1,25(OH)2D3 (10−8 M) or vehicle. Cultures of primary calvarial osteoblasts (D) were mineralized 14 or 21 days, and prior to RNA isolation mineralizing cells were treated with 1,25(OH)2D3 (10−8 M) or vehicle for 24 h mRNA was isolated and transcript levels of all class 3 semaphorins were analyzed by qPCR. Changes in expression levels by 1,25(OH)2D3 treatment relative to vehicle-treated controls are shown. The mean and standard deviation of at least three independent experiments (A to C), or of merged data from two independent experiments performed in triplicate cultures (D), are shown. One samples t-tests were performed to determine the significance of the mRNA induction by 1,25(OH)2D3 (* p < 0.05, ** p < 0.01, *** p < 0.001).
In summary, all semaphorin 3 family members were expressed in the osteoblastic cells we examined. The Sema3e and 3f genes were strongly upregulated 1,25(OH)2D3 targets in osteoblastic cells, while the Sema3b and 3c genes responded to 1,25(OH)2D3 only in ST2 and MC3T3-E1 cells, and Sema3d only in primary osteoblasts.
3.2. Sema3d, 3e and 3f were transcriptionally regulated by 1,25 (OH)2D3 in primary mineralizing osteoblast cultures
Subsequently, the effect of 1,25(OH)2D3 on the expression of Sema3 family members was investigated in vitro during osteogenic differentiation of primary calvarial osteoblasts. mRNA was isolated during early and late mineralization, days 14 and 21, respectively. In both studied time points, transcription of both Sema3e and 3f showed prominent induction by 1,25(OH)2D3 when mineralizing cultures were subjected to a short 24 h incubation period with 1,25 (OH)2D3 (Fig. 2D). The magnitude of the inductions was similar at both mineralization phases (3–3.2 −fold for both genes), which was slightly stronger for Sema3e than that observed in non-mineralized osteoblasts (Fig. 2C). In addition, transcript levels of Sema3d were significantly reduced after treatment with 1,25 (OH)2D3, which was also observed before mineralization. No induction of Sema3b was observed by 1,25(OH)2D3 during the mineralization process (Fig. 2D) whereas a modest downregulation of Sema3c expression was demonstrated.
Taken together, the upregulation of the Sema3e and Sema3f genes and the downregulation of Sema3d by 1,25(OH)2D3 was observed during mineralization of primary calvarial osteoblasts.
3.3. ChIP-seq experiments revealed direct binding of VDR and RXR to the genomic regions of the Sema3 genes
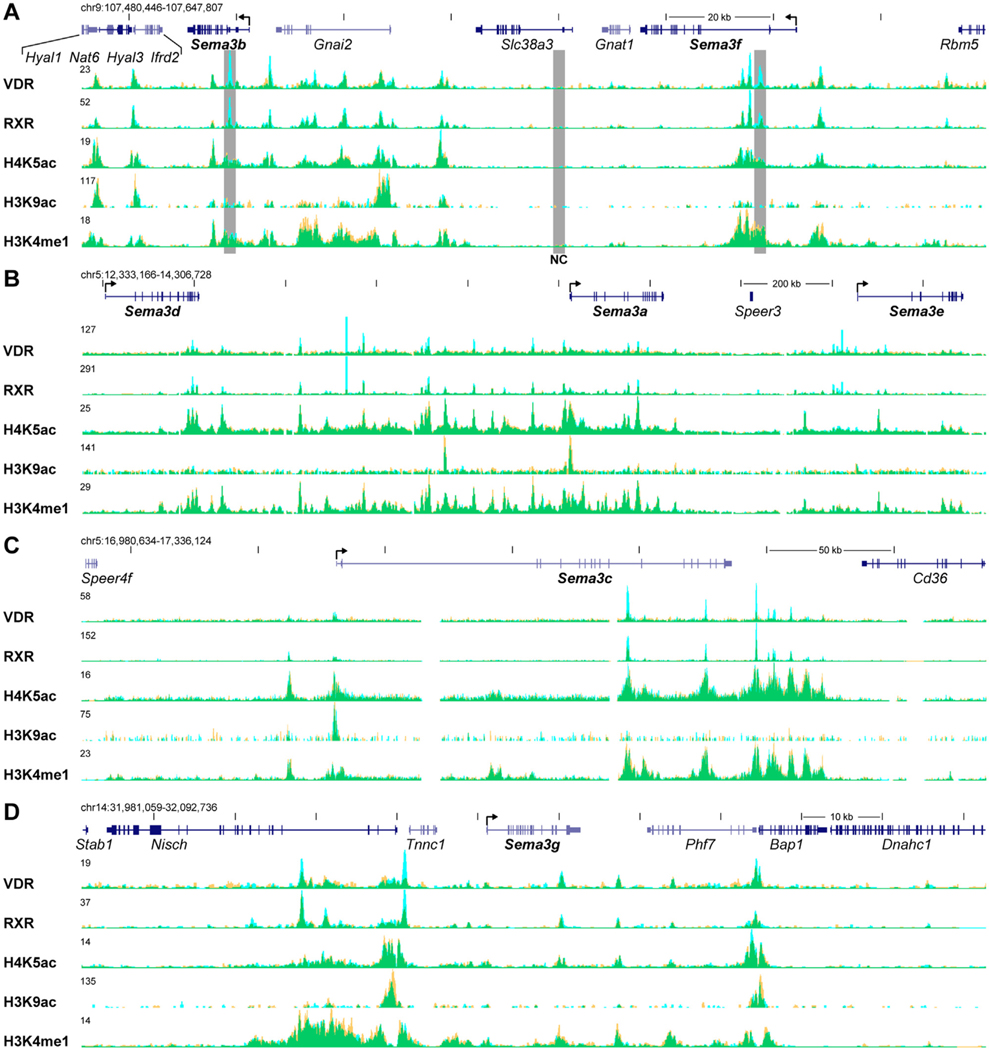
Because the induction of Sema3 genes by 1,25(OH)2D3 was observed in osteoblastic cell cultures, we used ChIP-seq analyses of VDR and RXR binding and selected histone modifications in MC3T3-E1 cells to demonstrate the basis of Sema3 regulation by VDR in more detail. All observed VDR peaks co-localized with RXR peaks demonstrating the presence of VDR/RXR heterodimers at these sites (Fig. 3). Intronic VDR/RXR peaks were present at two and three sites of the Sema3b and 3f genes, respectively (Fig. 3A). In the larger genomic region spanning 20 kb downstream from Sema3b to 35 kb upstream from Sema3f genes, which contains eight intervening and adjacent genes, the presence of at least 14 VDR/RXR peaks was demonstrated with the strongest and most inducible peaks near the Sema3b and 3f transcription start site (TSS). In contrast, a ten-fold larger genomic region covering the genes Sema3d, Sema3a and Sema3e contains only one intervening gene, but demonstrated at least 15 peaks for VDR and RXR binding (Fig. 3B). Interestingly in this genomic locus, VDR peaks focused between the Sema3d and Sema3e genes, since no VDR/RXR peaks were observed within 500 kb down- or upstream from the displayed region (data not shown). Similarly, ChIP-seq data revealed clustered VDR/RXR binding around the Sema3c (Fig. 3C) and Sema3g (Fig. 3D) genes, both genes being 1,25 (OH)2D3 targets in MC3T3-E1 cells.
Fig. 3.
VDR and RXR showed direct 1,25(OH)2D3-inducible binding to the genomic regions of Sema3b and 3f genes. Representative ChIP-seq tag density tracks for VDR and RXR binding, and modified histones in vehicle- and 1,25(OH)2D3-treated samples in MC3T3-E1 cells are shown (Veh, yellow; 1,25(OH)2D3, blue; overlap, green). Genomic locations and scales are indicated. Maximum height of tag density for the data track was set according the highest peak of each track and is indicated on the tracks. Genomic regions containing the genes Sema3b and 3f (A), Sema3d, 3a, 3e (B), Sema3c (C) and Sema3g (D) are shown. Transcriptional directions of Sema3 genes are indicated by an arrow at the TSS. Regions that were further analyzed by ChIP-qPCR are highlighted by gray boxes (NC, negative control).
The specific histone modifications, H4K5 and H3K9 acetylation (H4K5ac and H3K9ac, enriched at active chromatin [27]), and H3K4 mono-methylation (H3K4me1, enriched across enhancers [27]) showed a remarkably similar landscape amongst most of the VDR/RXR binding sites (Fig. 3). With no exceptions, VDR/RXR binding sites were aligned with H4K5ac peaks that, in general, demonstrate accessible chromatin structures. In addition, H4K5ac levels showed some response to 1,25(OH)2D3 treatment displaying both increase and decrease of acetylation at different VDR binding sites. H3K9ac peaks aligned with H4K5ac peaks. However, the magnitude of H3K9ac peaks showed great peak-to-peak variation. With few exceptions all VDR/RXR binding sites aligned with strong H3K4me1 levels demonstrating enhancer activity around the VDR/RXR heterodimer. Moreover, VDR binding sites at all studied regions generally aligned within valleys of distinct peak-valley-peak patterns created by histone modification levels (For detailed view, the reader is referred to the original ChIP-seq data tracks). This pattern highlights nucleosome free regions that are accessible for additional regulatory factors, and are enriched within enhancers in active state [28].
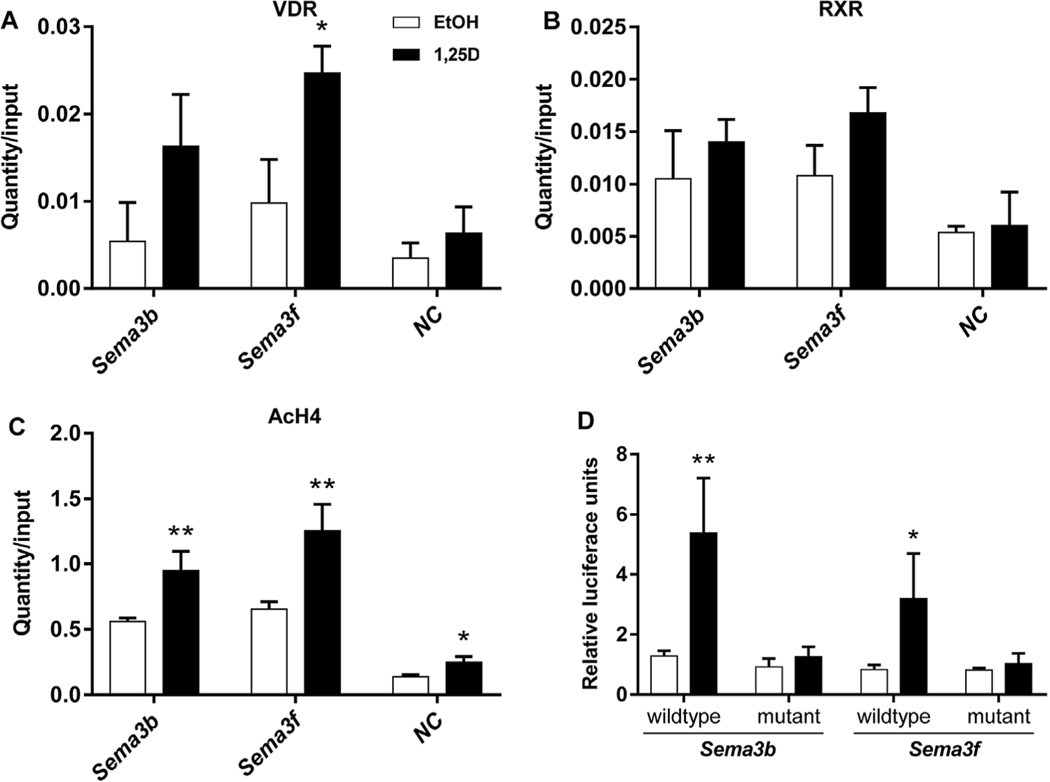
Subsequently, we performed ChIP-qPCR experiments with selected VDR binding sites at the Sema3b and Sema3f loci based upon their highly induced VDR binding and proximity of the respective gene TSS (locations indicated at Fig. 3). For both sites, we confirmed the increase in VDR and RXR binding after incubation with 1,25(OH)2D3 (Fig. 4A and B). Both VDR binding sites carried a DR3-type VDRE sequence for direct VDR binding and demonstrated a significant increase in acetylation levels (AcH4) (Fig. 4C). The specificity of receptor binding was confirmed by qPCR of a negative control region that is located at the second intron of the Slc38a3 gene between the genes Sema3b and Sema3f (Fig. 3, 4A and 4B). No VDR or RXR binding was seen on the negative region. In order to further confirm the functionality of the studied VDR binding sites of Sema3b and Sema3f, both DR3-type VDREs were cloned in luciferase reporter vectors and used in transient transfection experiments (Fig. 4D). Luciferase activities of reporter vectors with the genomic fragments of Sema3b and 3f were significantly induced when transfected MC3T3-E1 cells were incubated for 24 h with 1,25(OH)2D3 (4.2 and 3.8 −fold, respectively). Subsequently, the DR3-type binding sites were mutated before transfection, to prevent VDR binding, and luciferase activity was evaluated in 1,25 (OH)2D3-or vehicle-treated cells. Interestingly, the induction of luciferase activity by 1,25(OH)2D3 was completely abolished with the mutated reporter constructs, indicating that the suspected VDR binding sites were responsible for the transactivation of the reporter constructs.
Fig. 4.
ChIP-qPCR and transient transfection assay. ChIP-qPCR results confirmed the ligand-inducible association of VDR (A) and RXR (B), and demonstrated enhanced histone H4 acetylation (C) in response to treatment at the two studied VDR binding sites at genes Sema3b and Sema3f. ChIP was performed with chromatin samples from MC3T3-E1 cells treated with 1,25(OH)2D3 (10−7 M) or vehicle for 3 h. Results show receptor associations and histone H4 acetylation at both binding sites and at NC. The DR3-type VDR binding sites of Sema3b and 3f were further confirmed to be responsive to 1,25(OH)2D3 in transient transfection experiments (D). A DNA fragment, containing either the wildtype or a mutated (non-functional) VDR binding site, was cloned and inserted into the pGL3-Basic vector. Luciferase reporter gene assays were performed to evaluate the functionality of the identified VDR binding sites in exponentially growing MC3T3-E1 cells treated with 1,25(OH)2D3 (10−8 M) or vehicle for 24 h. The results were normalized to that of the empty pGL3-Basic luciferase reporter vectors and expressed as means and standard deviations of at least three independent experiments. Two-tailed Student’s t-tests were performed to determine the significant differences between 1,25(OH)2D3-treated cells and vehicle-treated controls in the receptor association or chromatin acetylation (A-C), and in the promoter activity (D) (* p < 0.05, ** p < 0.01).
In summary, ChIP experiments revealed around all seven Sema3 genes numerous intronic and intergenic VDR/RXR heterodimer binding sites that were co-localized with histone markers for active chromatin structure and enhancer activity. Ligand-inducible VDR and RXR binding was confirmed and a prominent increase in histone H4 acetylation was shown for both studied VDR sites in ChIP-qPCR experiments. In addition, the functionality of the VDR binding sites was confirmed in transient transfection assays.
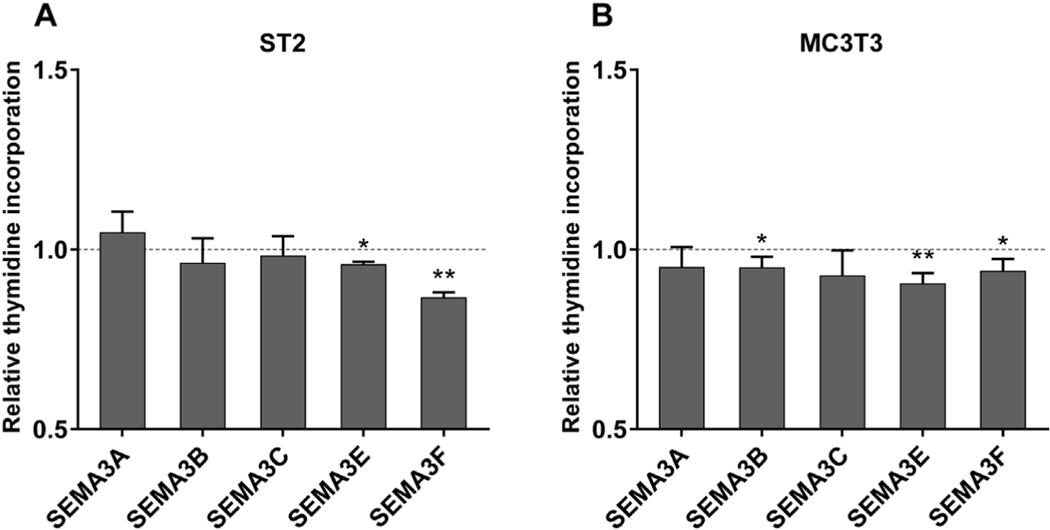
3.4. Recombinant SEMA3E and SEMA3F decreased cell proliferation
In order to evaluate the possible function of the VDR-regulated class 3 semaphorins, we investigated whether recombinant semaphorin 3 proteins affected cell proliferation. ST2 and MC3T3-E1 cells were cultured for 3 days in the presence of recombinant SEMA3 proteins and proliferation was measured by a [3H]thymidine incorporation assay. Incubation with recombinant SEMA3E as well as SEMA3F led to a small, but significant, reduction of cell proliferation in both cell lines (Fig. 5).
Fig. 5.
ST2 and MC3T3-E1 cell proliferation was reduced by SEMA3E and 3F. ST2 (A) and MC3T3-E1 (B) cells were treated with 640 ng/ml recombinant SEMA3A, 3B, 3C, 3E or 3F protein for 72 h and cell proliferation was evaluated by [3H]thymidine incorporation assay. Changes in thymidine incorporation in SEMA3-treated cells relative to vehicle-treated controls are shown. Data are presented as means and standard deviations of at least three independent experiments. One samples t-tests were performed to determine the significance of the inhibition by the treatment (*p < 0.05, **p < 0.01).
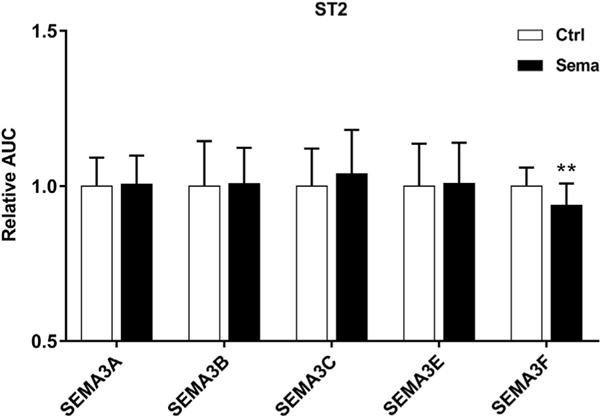
3.5. Recombinant semaphorin 3 proteins did not affect ST2 cell migration
Class 3 semaphorins are known modulator of cell migration in different tissues [29]. Therefore, we performed scratch wound assays in ST2 cells that were treated with recombinant SEMA3 proteins prior to scratching the wound. A minor, but significant, inhibition of ST2 cell migration after incubation with SEMA3F was observed (Fig. 6). The other class 3 semaphorin family members did not alter the wound healing speed.
Fig. 6.
SEMA3 treatment did not affect ST2 cell migration in scratch wound assays. Treatment of sub-confluent ST2 cell cultures with recombinant SEMA3A, 3B, 3C, 3E, or 3F protein was started 2 h prior to the introduction of the scratch wound and was continued during the complete experiment. Cell migration was monitored using the IncuCyte ZOOM Scratch Wound assay and an AUC-value was derived for each individual wound as a measure of cell migration (as demonstrated in Fig. S1). Representative experiments performed with 600 ng/ml of SEMA3A, 3B, 3C, 3E, or 3F treatment are shown with results normalized to the mean of the AUC of control wells. Data are presented as means and standard deviations of at least 12 wells. Two-tailed Student’s t-tests were performed in order to determine the significant differences in cell migration between Sema3-treated cells and vehicle-treated controls (** p < 0.01).
In order to further investigate the role of SEMA3 in pre-osteoblasts, we treated exponentially growing ST2 cells for 72 h with recombinant SEMA3 proteins (600 ng/ml), and measured the mRNA inductions of osteoblast lineage genes (runt-related transcription factor 2, osteopontin and osteocalcin) and of osteoclastic marker receptor activator of nuclear factor κB ligand (RANKL, encoded by the Tnfsf11 gene). We saw no changes in the expression levels of the studied genes in response to the treatment (data not shown).
4. Discussion
Semaphorins were first identified as molecules involved in axonal guidance during the development of the nervous system but are now recognized as crucial regulators of homeostasis over a wide range of organ systems. SEMA3s in particular affect cell growth, survival and migration in neuronal as well as non-neuronal tissues [3,5], and there is an increasing body of evidence that SEMA3 signaling is present in bone and may affect bone homeostasis [12]. In order to investigate the effect of 1,25(OH)2D3 on Sema3 gene expression at distinct stages of osteoblast differentiation, we used ST2 cells derived from mouse bone marrow, which have stem cell characteristics and can be differentiated into osteoblasts [30], MC3T3-E1 pre-osteoblasts, and both unmineralized and mineralizing primary calvarial osteoblasts. Our study demonstrated that all members of the Sema3 family were expressed in the three osteoblastic cell types and that the basal expression patterns of Sema3 genes in these cells were remarkably similar. We showed that the expression of six different Sema3 family members (Sema3b, 3c, 3d, 3e, 3f and 3g) was altered by 1,25(OH)2D3 in at least one osteoblastic cell type, suggesting that these molecules are additional mediators for the effects of 1,25(OH)2D3 on osteoblasts. Of interest, transcript levels of Sema3e and 3f were rapidly and robustly induced by 1,25(OH)2D3 (within 6 h after treatment), not only in osteoblastic cell lines but also in both unmineralized and mineralizing primary calvarial osteoblasts, suggesting that Sema3e and Sema3f are conserved 1,25 (OH)2D3 targets over the different osteoblastic cell types. Since these strongly inducible genes were also highly expressed in these cells, Sema3e and Sema3f may be important mediators of the 1,25 (OH)2D3 signaling in osteoblasts. Consistent with our results, the induction of Sema3e in response to 1,25(OH)2D3 was previously reported in non-differentiated calvarial osteoblasts [18]. Furthermore, transcript levels of Sema3b and Sema3c were increased after 1,25(OH)2D3 treatment, but only in the ST2 and MC3T3-E1 cell lines and not in calvarial osteoblasts. The induction of Sema3b in response to 1,25(OH)2D3 in ST2 cells was previously reported by Sutton et al. [17]. However, in their study a modest induction of Sema3b by 1,25(OH)2D3 was observed also in mineralizing primary calvarial osteoblasts. Downregulation of transcript levels of Sema3g by 1,25(OH)2D3 was only observed in MC3T3-E1 cells whereas Sema3d mRNA levels were only downregulated in calvarial osteoblasts, both in unmineralized and mineralizing cultures. The latter corresponds with a study performed in mouse calvarial osteoblasts, where a trend for reduction in Sema3d transcripts in response to 1,25(OH)2D3 is observed [18]. While we demonstrated that all other members of the Sema3 gene family were regulated by 1,25(OH)2D3 in osteoblastic cells, we did not observe any evidence that Sema3a was a 1,25(OH)2D3 target gene in these cells, which is consistent with a previous report in osteoblasts [18].
To investigate whether the 1,25(OH)2D3-induced Sema3 genes were directly regulated by VDR, we utilized ChIP-seq data from MC3T3-E1 cells to demonstrate VDR/RXR binding and presence of histone modifications indicating active enhancers at the genomic regions around the Sema3 genes. We observed the presence of strongly inducible VDR/RXR heterodimers accompanied by accessible chromatin architecture (H4K5ac and H3K9ac) and enhancer activity (H3K4me1) at several intronic and intergenic locations of all transcriptionally upregulated Sema3 genes (Sema3b, 3f, 3e and 3c) and the downregulated Sema3g gene. Our observations suggest that these genes are indeed primary 1,25(OH)2D3 targets and that each gene is most likely regulated by multiple VDR binding sites in osteoblastic cells. For example, the Sema3c locus demonstrated a group of ligand-induced VDR peaks positioned around the end of the Sema3c gene. Accordingly, the VDR peaks aligned with highly induced RXR peaks and showed open chromatin structure in combination with enhancer activity as defined by the histone modifications, which suggest that these VDR binding sites are activated by 1,25(OH)2D3 stimulation and responsible for the Sema3c transactivation.
Interestingly, non-responsive Sema3d and Sema3a in this cell line also showed numerous prominent VDR/RXR binding sites clustered around these two genes. Although the histone modification landscape was generally similar amongst the VDR binding sites, the underlying mechanisms for the observed ineffective VDR binding may involve the small differences observed in histone modification responses to 1,25(OH)2D3 treatment. For example, all the H4K5ac peaks upstream of Sema3b TSS showed some degree of induction after 1,25(OH)2D3 treatment. In contrast, at many VDR sites of non-responsive Sema3d and Sema3a acetylation levels were unchanged or reduced. In accordance, a very strong VDR peak between the genes lacked an H3K4me1 peak. We speculate that this may suggest inactivity of the VDR site. It is also possible that the lack of response to 1,25(OH)2D3 may be due to the presence of silencing marks such as H3K27me3 or H3K9me3 not evaluated here or perhaps the absence of additional unknown transcription factors. Nevertheless, these data suggest that the epigenetic histone status of the enhancers to which the VDR is bound likely contributes to the receptor’s transactivation ability at these genes, and may also explain why the non-responding Sema3d gene in MC3T3-E1 cell line can respond to treatment in primary osteoblasts. The similarity of Sema3d and Sema3a chromatin landscape with multiple VDR/RXR binding sites raises the question as to whether Sema3a would respond to 1,25(OH)2D3 in osteoblastic cells in an alternative epigenetic state.
To further analyze the observations from ChIP-seq data, we focused on the genomic region containing the Sema3b and Sema3f genes as an example, and selected two highly inducible VDR peaks at the proximity of the respective gene TSS for our ChIP-qPCR and transfection assays. ChIP-qPCR results confirmed the 1,25(OH)2D3-inducible VDR/RXR binding to both sites and demonstrated prominent chromatin opening during VDR activation. In addition, Sema3f region showed in both absence and presence of ligand stronger VDR binding than that at Sema3b, which is in line with the earlier and more prominent mRNA inductions of Sema3f. Subsequent in silico screening revealed the presence of putative DR3-type VDREs in VDR/RXR-bound regions. Transient transfection experiments combined with site directed mutagenesis demonstrated that the in silico identified DR3-type VDREs were indeed functional. Interestingly, increased protein levels of SEMA3B and SEMA3F in response to 1,25(OH)2D3 treatment is observed in endometrial cancer cells [20], which demonstrates that these primary target genes can be regulated by 1,25(OH)2D3 in other tissues.
Although the definite physiological outcome of the 1,25 (OH)2D3-inducible Sema3 gene expression remains obscure, these proteins are suggested to be involved in the communication between osteoblasts and osteoclasts [18,31]. Expression of Sema3f, but also that of Sema3a and Sema6b, was reduced in ST2 cells in which Runx2, a master regulator of osteoblast differentiation, is overexpressed [31]. Overexpression of Runx2 in ST2 cells results in an enhanced differentiation of co-cultured pre-osteoclasts and the authors propose that the downregulation of Sema3f, Sema3a and Sema6b is involved in Runx2-driven osteoclastogenesis [31]. In addition, loss of Nrp2, the main receptor for SEMA3F, leads to bone loss that is accompanied by an enhanced osteoclast number and a reduced osteoblast count [32]. Interestingly, SEMA3E is also suggested to inhibit osteoclastogenesis, as an inhibitory effect of recombinant SEMA3E on osteoclast formation is reported in mouse marrow hematopoietic cells that were cultured in the presence of macrophage colony stimulating factor and RANKL [18]. These studies suggest that upregulation of Sema3e and Sema3f may inhibit osteoclast formation. In contrast, osteoblast-derived SEMA3B is demonstrated to promote in vivo osteoclastogenesis, leading to osteopenia when overexpressed [17]. However, recent observations that osteoblastic SEMA3B is suppressed in osteoporotic mice and that SEMA3B may promote in vitro osteoblast differentiation [33], suggest a more complex role of this protein in bone homeostasis. Furthermore, reduced expression of Sema3d in cortical bone in a mouse model for hypophosphatemic rickets is reported suggesting that SEMA3D may be associated with mineralization defects observed in the osteoblasts of these mice [34]. However, our observation that SEMA3 treated ST2 cells did not change the expression of early osteoblastic markers or Tnfsf11 gene suggests that SEMA3s are not involved in osteoblast differentiation or in the regulation of RANKL.
SEMA3s are demonstrated to modulate cell migration and chemotaxis in a variety of tissues by their capabilities to influence cytoskeletal organization and cell adhesion [29,35,36]. However, we did not observe major effects of recombinant semaphorins on cell migration in ST2 cells. Our findings are inconsistent with those reported in mouse calvarial osteoblasts, where an inhibitory effect of recombinant SEMA3E protein on osteoblast migration is observed [18]. In our cellular model, the faint reduction in wound healing after SEMA3F treatment may be explained by a reduction in cell proliferation. Indeed, we demonstrated that recombinant SEMA3E and SEMA3F were able to inhibit osteoblast proliferation, which may suggest that Sema3e and Sema3f are involved in the antiproliferative effects of 1,25(OH)2D3.
In conclusion, we showed that 1,25(OH)2D3 directly controls the transcriptional regulation of members of the Sema3 gene family. In addition, we showed that Sema3b, Sema3c, Sema3e and Sema3f are upregulated and Sema3d and sema3g are downregulated 1,25 (OH)2D3 target genes within osteoblastic cells, providing more evidence for the involvement of 1,25(OH)2D3 in the osteoblastic signaling.
Supplementary Material
Acknowledgements
We thank Ine Beullens, Biauw Keng Tan and Mark Van Camp for excellent technical assistance. This work was supported by grants from the Fund for Scientific Research [FWO G.OA17.14N]; the KU Leuven [GOA/14/010]; the Orion Research Foundation (to J.R.); the Osk. Huttunen Foundation (to J.R.); and, in part, by the National Institutes of Health grant R01-DK072281 (to J.W.P.).
Abbreviations:
- 1,25(OH)2D3
1,25 dihydroxyvitamin D3
- AUC
area under the curve
- ChIP
chromatin immunoprecipitation
- ChIP-seq
ChIP sequencing
- NRP
neuropilin
- PLXN
plexin
- RWD
relative wound density
- RXR
retinoid X receptor
- Sema3
class 3 semaphorin
- TSS
transcription start site
- VDR
vitamin D receptor
- VDRE
vitamin D responsive element
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsbmb.2017.02.005.
References
- [1].Castellani V, Rougon G, Control of semaphorin signaling, Curr. Opin. Neurobiol. 12 (5) (2002) 532–541. [DOI] [PubMed]
- [2].Kruger RP, Aurandt J, Guan KL, Semaphorins command cells to move, Nat. Rev. Mol. Cell Biol. 6 (10) (2005) 789–800. [DOI] [PubMed] [Google Scholar]
- [3].Gaur P, Bielenberg DR, Samuel S, Bose D, Zhou Y, Gray MJ, Dallas NA, Fan F, Xia L, Lu J, Ellis LM, Role of class 3 semaphorins and their receptors in tumor growth and angiogenesis, Clin. Cancer Res. 15 (22) (2009) 6763–6770. [DOI] [PubMed] [Google Scholar]
- [4].Takamatsu H, Okuno T, Kumanogoh A, Regulation of immune cell responses by semaphorins and their receptors, Cell Mol. Immunol. 7 (2) (2010) 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Epstein JA, Aghajanian H, Singh MK, Semaphorin signaling in cardiovascular development, Cell Metab. 21 (2) (2015) 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oh WJ, Gu C, The role and mechanism-of-action of Sema3E and Plexin-D1 in vascular and neural development, Semin. Cell Dev. Biol. 24 (3) (2013) 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Janssen BJ, Malinauskas T, Weir GA, Cader MZ, Siebold C, Jones EY, Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex, Nat. Struct. Mol. Biol. 19 (12) (2012) 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sharma A, Verhaagen J, Harvey AR, Receptor complexes for each of the Class 3 Semaphorins, Front. Cell. Neurosci. 6 (2012) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hayashi M, Nakashima T, Taniguchi M, Kodama T, Kumanogoh A, Takayanagi H, Osteoprotection by semaphorin 3A, Nature 485 (7396) (2012) 69–74. [DOI] [PubMed] [Google Scholar]
- [10].Kang S, Kumanogoh A, Semaphorins in bone development, homeostasis, and disease, Semin. Cell Dev. Biol. 24 (3) (2013) 163–171. [DOI] [PubMed] [Google Scholar]
- [11].Fukuda T, Takeda S, Xu R, Ochi H, Sunamura S, Sato T, Shibata S, Yoshida Y, Gu Z, Kimura A, Ma C, Xu C, Bando W, Fujita K, Shinomiya K, Hirai T, Asou Y, Enomoto M, Okano H, Okawa A, Itoh H, Sema3A regulates bone-mass accrual through sensory innervations, Nature 497 (7450) (2013) 490–493. [DOI] [PubMed] [Google Scholar]
- [12].Verlinden L, Vanderschueren D, Verstuyf A, Semaphorin signaling in bone, Mol. Cell. Endocrinol. (2015). [DOI] [PubMed]
- [13].Eisman JA, Bouillon R, Vitamin D: direct effects of vitamin D metabolites on bone: lessons from genetically modified mice, Bonekey. Rep. 3 (2014) 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].van de Peppel J, van Leeuwen JP, Vitamin D and gene networks in human osteoblasts, Front. Physiol. 5 (2014) 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Umesono K, Murakami KK, Thompson CC, Evans RM, Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors, Cell 65 (7) (1991) 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C, Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy, Nucleic Acids Res. 39 (21) (2011) 9181–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sutton AL, Zhang X, Dowd DR, Kharode YP, Komm BS, Macdonald PN, Semaphorin 3 B is a 1 25-Dihydroxyvitamin D3-induced gene in osteoblasts that promotes osteoclastogenesis and induces osteopenia in mice, Mol. Endocrinol. 22 (6) (2008) 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hughes A, Kleine-Albers J, Helfrich MH, Ralston SH, Rogers MJ, A class III semaphorin (Sema3e) inhibits mouse osteoblast migration and decreases osteoclast formation in vitro, Calcif. Tissue Int. 90 (2) (2012) 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Munetsuna E, Kawanami R, Nishikawa M, Ikeda S, Nakabayashi S, Yasuda K, Ohta M, Kamakura M, Ikushiro S, Sakaki T, Anti-proliferative activity of 25-hydroxyvitamin D3 in human prostate cells, Mol. Cell. Endocrinol. 382 (2) (2014) 960–970. [DOI] [PubMed] [Google Scholar]
- [20].Nguyen H, Ivanova VS, Kavandi L, Rodriguez GC, Maxwell GL, Syed V, Progesterone and 1,25-dihydroxyvitamin D3 inhibit endometrial cancer cell growth by upregulating semaphorin 3 B and semaphorin 3F, Mol. Cancer Res. 9 (11) (2011) 1479–1492. [DOI] [PubMed] [Google Scholar]
- [21].Daci E, Udagawa N, Martin TJ, Bouillon R, Carmeliet G, The role of the plasminogen system in bone resorption in vitro, J. Bone Miner. Res. 14 (6) (1999) 946–952. [DOI] [PubMed] [Google Scholar]
- [22].Leyssens C, Marien E, Verlinden L, Derua R, Waelkens E, Swinnen JV, Verstuyf A, Remodeling of phospholipid composition in colon cancer cells by 1alpha,25(OH)2D3 and its analogs, J. Steroid Biochem. Mol. Biol. 148 (2015) 172–178. [DOI] [PubMed] [Google Scholar]
- [23].Meyer MB, Benkusky NA, Lee CH, Pike JW, Genomic determinants of gene regulation by 1 25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation, J. Biol. Chem. 289 (28) (2014) 19539–19554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Meyer MB, Benkusky NA, Pike JW, The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression, J. Biol. Chem. 289 (23) (2014) 16016–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, The human genome browser at UCSC, Genome Res. 12 (6) (2002) 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eelen G, Verlinden L, van Camp M, van Hummelen P, Marchal K, de Moor B, Mathieu C, Carmeliet G, Bouillon R, Verstuyf A, The effects of 1alpha 25-dihydroxyvitamin D3 on the expression of DNA replication genes, J. Bone Miner. Res. 19 (1) (2004) 133–146. [DOI] [PubMed] [Google Scholar]
- [27].Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE, Mapping and analysis of chromatin state dynamics in nine human cell types, Nature 473 (7345) (2011) 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pundhir S, Bagger FO, Lauridsen FB, Rapin N, Porse BT, Peak-valley-peak pattern of histone modifications delineates active regulatory elements and their directionality, Nucleic Acids Res. 44 (9) (2016) 4037–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tran TS, Kolodkin AL, Bharadwaj R, Semaphorin regulation of cellular morphology, Annu. Rev. Cell Dev. Biol. 23 (2007) 263–292. [DOI] [PubMed] [Google Scholar]
- [30].Otsuka E, Yamaguchi A, Hirose S, Hagiwara H, Characterization of osteoblastic differentiation of stromal cell line ST2 that is induced by ascorbic acid, Am. J. Physiol. 277 (1 Pt 1) (1999) C132–138. [DOI] [PubMed] [Google Scholar]
- [31].Baniwal SK, Shah PK, Shi Y, Haduong JH, Declerck YA, Gabet Y, Frenkel B, Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells, Osteoporos. Int. 23 (4) (2012) 1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Verlinden L, Kriebitzsch C, Beullens I, Tan BK, Carmeliet G, Verstuyf A, Nrp2 deficiency leads to trabecular bone loss and is accompanied by enhanced osteoclast and reduced osteoblast numbers, Bone 55 (2) (2013) 465–475. [DOI] [PubMed] [Google Scholar]
- [33].Sang C, Zhang Y, Chen F, Huang P, Qi J, Wang P, Zhou Q, Kang H, Cao X, Guo L, Tumor necrosis factor alpha suppresses osteogenic differentiation of MSCs by inhibiting semaphorin 3 B via Wnt/b-catenin signaling in estrogen-deficiency induced osteoporosis, Bone 84 (2016) 78–87. [DOI] [PubMed] [Google Scholar]
- [34].Liu S, Tang W, Fang J, Ren J, Li H, Xiao Z, Quarles LD, Novel regulators of Fgf23 expression and mineralization in Hyp bone, Mol. Endocrinol. 23 (9) (2009) 1505–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nasarre P, Kusy S, Constantin B, Castellani V, Drabkin HA, Bagnard D, Roche J, Semaphorin SEMA3F has a repulsing activity on breast cancer cells and inhibits E-cadherin-mediated cell adhesion, Neoplasia 7 (2) (2005) 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mendes-da-Cruz DA, Brignier AC, Asnafi V, Baleydier F, Messias CV, Lepelletier Y, Bedjaoui N, Renand A, Smaniotto S, Canioni D, Milpied P, Balabanian K, Bousso P, Leprêtre S, Bertrand Y, Dombret H, Ifrah N, Dardenne M, Macintyre E, Savino W, Hermine O, Semaphorin 3F and neuropilin-2 control the migration of human T-cell precursors, PLoS One 9 (7) (2014) e103405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.