Abstract
Environmental risks continue to grow due to heavy metal contamination caused by anthropogenic activities. Accumulation of harmful quantities of lead poses a threat to aquatic organisms, plants, and human beings. Whole-cell biosensors, which can proliferate independently, can detect the bioavailable fraction to assess the effect of target heavy metal on the environmental ecosystem. In this study, the biosynthesis pathway of violacein was heterogeneously constructed under the control of the T7 lac promoter in E. coli. A dose–response relationship existed between the inducer and the production of violacein. The biosynthesis pathway of violacein was finally engineered under the regulation of Pb(ii)-dependent metalloregulator PbrR to assemble Pb(ii)-inducible whole-cell biosensor. It permitted specific biosensing of Pb(ii) with extraordinary selectivity, and could resist the interferences from various metal ions. Color change by the intracellular accumulation of violacein could be recognized with the naked eye directly with high concentration of lead exposure, and quantified by determining the absorbance at 490 nm after butanol extraction. A good linear range for Pb(ii) concentrations of 0.1875–1.5 μM was obtained. The novel pigment-based whole-cell biosensor could detect concentrations as low as 0.1875 μM Pb(ii) based on in vitro quantification of violacein extracted by butanol, which is significantly lower than reported fluorescent protein-based PbrR-regulated biosensors based on direct measurement of whole cell fluorescence. These results indicate that genetically controlled violacein biosynthesis can enable a sensitive, visual, and qualitative biosensor for monitoring the presence of bioavailable Pb(ii) in lead-contaminated water.
Genetically controlled violacein biosynthesis can enable a sensitive, visual, and qualitative biosensor for monitoring the presence of bioavailable lead.
Introduction
Heavy metal pollution has become a major concern to the environment and public health in many rapidly growing regions in developing countries. Despite enormous efforts that have been made, heavy metal contamination was still reported in water, soil, sediment samples, and food in recent years mainly due to increasing anthropogenic activities.1–3
Significantly enhanced risks and increased morbidity are often correlated with increased bioavailability of heavy metals. When attempting to assess the distribution and the implication of heavy metal pollution, it is usually important to quantify both the total content of heavy metals and their bioavailable fraction.4 To adapt to environmental pollution, microorganisms encode a panel of metal-responsive transcriptional regulators that govern the expression of a series of detoxification-related genes that allows an organism to quickly adapt to maintain cellular homeostasis of heavy metals, such as Hg, Cd, Pb, and Cr, which do not have a biological role and usually cause extreme toxicity.5 Genetically engineered microorganisms, tailored to respond to the target metal ions using quantifiable fluorescent and enzymatic signals, may serves as potential tools for bioavailability assessment of heavy metals.6 Microbial cell-based sensors have great potential to detect various bioavailable heavy metal contaminants in an aqueous environment.7 Due to the extraordinary sensitivity and selectivity of metalloregulators towards target metal ions, many whole-cell biosensors using them as sensory elements have been successfully developed to cope with prevalent heavy metal contamination.8,9
The MerR family is a major metalloregulatory protein family, which is very large and has members that sense not only a wide range of biologically required metals but also heavy metal toxicants.10 The members of this family function as both transcription repressors with no target metal ions exposure and transcription activators with target metal ions exposure.11 The important characteristics of the MerR family are its extremely high selectivity and sensitivity towards target metal ions. Based on MerR-type regulators, many whole-cell biosensors towards heavy metal were developed using fluorescent and enzymatic reporters.9,12–14 Due to insufficient biosensing performance, many developed whole-cell sensors could not meet the practical requirements. The weak input signal could be transformed into a larger, easily detectable output signal in an engineered bacterial cell using a genetic amplifier.15 This strategy has been used to construct ultrasensitive bacterial sensors for arsenic and mercury.16 In addition, the development of small molecule reporter peptides,12 optimization of sensor receptor concentration,17 and genetic logic integration18 have been demonstrated to be beneficial to the improvement of the detection limit of biosensors.
Fluorescent proteins, luciferase, and beta-galactosidase are widely used for signal output of whole-cell and cell-free transcription-translation biosensors. These traditional genetic reporters have shown their own advantages, and have met the requirements of different applications.6 With the rapid development of synthetic biology, a few novel biosensors with customizable performance have been successfully constructed to meet the requirements for practical application.7 A modular cell-based biosensor could be engineered that can detect multiple environmental pollutants19 or integrate multiple-signal output modules into an artificial operon.9 As more and more genes for pigment metabolisms were elucidated, some natural pigments including carotenoid, lycopene, and pyocyanin have been employed as reporters for the development of whole-cell biosensors to enable visible signal output.20–23 Violacein-producing organisms, with a charming purple hue, have undoubtedly piqued imagination and curiosity of the researchers since their discovery. As a bisindole, violacein is a purple-colored pigment, which is known to have diverse biological activities, including being a broad-spectrum antibacterial agent and being an anticancer agent.24,25 The genes involved in the biosynthesis of violacein, and the regulatory mechanisms employed have been characterized within a small number of bacteria.26–28 Based on the natural pbr operon,29 lead whole-cell biosensors using fluorescent and enzymatic reporters as signal output elements were successfully developed.12,30 In this study, the genes vioA, vioB, vioC, vioD and vioE, which are the key biosynthetic enzymes involved in the biosynthesis of violacein, are successively assembled following the T7 lac promoter. The violacein was successfully expressed in E. coli by IPTG induction. The colorimetric biosensor for Pb(ii) was finally constructed by employing violacein production as a visual and measurable output signal. Compared with the traditional whole-cell signal output biosensors, the sensitivity of a novel biosensor specific towards Pb(ii) based on in vitro quantification of violacein was significantly enhanced.
Experimental section
Bacterial strains, plasmids, and agents
The bacterial strain and plasmids involved in this study are listed in Table S1.†E. coli TOP10 was used for all the cloning steps and for the engineered heavy metal biosensors. BL21(DE3)plysS was used for the production of violacein under the control of the T7 lac promoter. All the bacterial strains were cultured in Luria–Bertani (LB) broth (10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl) supplemented with 50 μg mL−1 ampicillin, as necessary. Stock solutions of HgCl2, CaCl2, MgCl2, FeSO4, MnSO4, NiSO4, CuSO4, ZnSO4, CdCl2, and Pb(NO3)2 were freshly prepared with analytical grade chemicals and distilled water. Isopropyl-β-d-thiogalactopyranoside (IPTG) was obtained from Sangon Biotech (Shanghai, China). All oligonucleotide primers and DNA fragments were synthesized by Sangon Biotech (Shanghai, China).
Cloning and construct assembly
The natural vioABCDE operon derived from Chromobacterium violaceum was previously characterized.26 To assemble the artificial violacein biosynthetic pathway, a fragment of 7430 bp encoding five open reading frames designate vioA (GenBank: MH029788.1), vioB (GenBank: MH029788.1), vioC (GenBank: KX461961.1), vioD (GenBank: KX461962.1) and vioE (GenBank: MH029788.1), was synthesized after codon preference optimization (Fig. S1†). The synthetic fragment was cloned into the NdeI and SacI sites of pET21a to generate pET-vio. The genetic element including the pbrR gene and its divergent pbr promoter was amplified from pT-Ppbr by PCR using primer pairs F-pbr and R-pbr (Table S1†). Then, the amplified DNA fragment was inserted into the BglII–XbaI site of the pET-vio to yield pPpbr-vio.
IPTG-induced assays
Freshly transformed BL21(DE3)plysS colonies were inoculated in triplicate into 3 mL LB medium, and then cultured overnight at 37 °C. 30 μL of each culture was inoculated into 3 mL fresh LB medium. The culture was grown at 37 °C for 3 h with rotation at 250 rpm, and then induced with different concentrations of IPTG at 30 °C. OD600 and violacein content were determined after an 8 h induction.
Heavy metal-induced assays
Freshly transformed TOP10 colonies were inoculated in triplicate in 3 mL LB medium, and then cultured overnight at 37 °C. 30 μL of each culture was inoculated into 3 mL fresh LB medium. The culture was grown at 37 °C for 3 h with rotation at 250 rpm. For the evaluation of detection specificity towards target metal, a final concentration of 5 μM Pb(ii), Cd(ii), Ca(ii), Mg(ii), Fe(ii), Mn(ii), Ni(ii), Cu(ii), Zn(ii), or Hg(ii) was added to the medium, followed by culturing at 37 °C for 3 h before OD600 and violacein content were determined. For the competitive assays, the mixture of 5 μM Pb(ii) with other different metal ions at the concentration of 5 μM was added to the medium. Following up to 3 h of incubation at 37 °C, the biosensor cells were harvested for analysis. For the sensitivity detections, different concentrations of Pb(ii) was added to the culture, followed by culturing at 37 °C, OD600 and violacein content were determined after a 3 h induction. For the environmental samples detection, 2.7 mL of exponential phase LB-cultures of TOP10/pPpbr-vio were mixed with either distilled water, tap water or lake water in 300 μL aliquots spiked with 1.5, 0.75, 0.375, 0 μM Pb(ii), respectively. The tap water and lake water used was collected from the laboratory tap and a local lake in Shenzhen, China. The culture continued to be cultivated for 3 h at 37 °C before detections.
Determination of the violacein biosynthesis
Violacein was extracted and assayed using a method previously described with a slight modification.31 1 mL induced culture was centrifuged at 5000 g for 5 min, and the supernatant was removed. The captured bacterial cells were lysed by adding 200 μL of 2% SDS, followed by violent vortexing for 10 min with a vortex mixer. Violacein was finally extracted from the cell lysate by adding 400 μL butanol, followed by vortexing for 10 min, and centrifuging at 8000g for 2 min. The organic phase (upper) containing the violacein was transferred, and aliquots of 150 μL were analysed at 490 nm using a Bio-Rad iMark spectrophotometer. The absorbance at 490 nm was finally normalized by the bacterial density (optical density at 600 nm) of the same culture.
Results and discussion
Design of Pb(ii) responsive, violacein-producing, bacterial biosensor
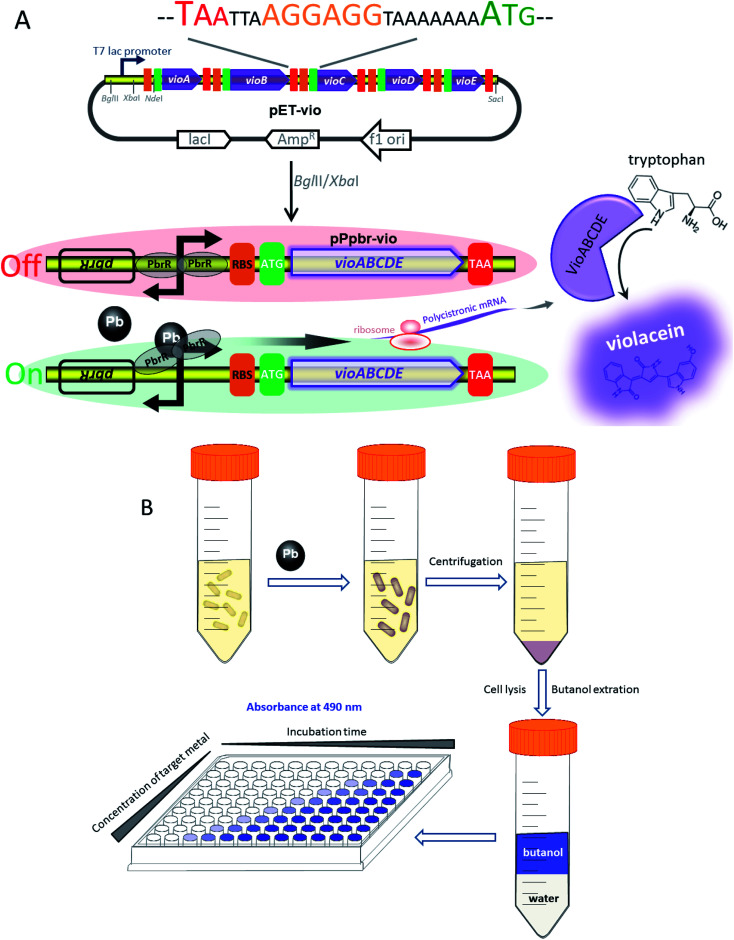
The strategy used for the assembly of Pb(ii)-responsive biosensor with a pigment-based signal output is summarized in Fig. 1A. All five violacein biosynthetic pathway genes were placed individually under the control of the strong T7 lac promoter in the high-copy-number plasmid pET21a. A high-copy-number plasmid was chosen to enhance the transcription and translation levels that could improve the sensitivity in heavy metal biosensing. The first RBS (AAGGAG) was derived from the cloning plasmid pET21a, and a reinitiation RBS (AGGAGG) located after an individual ORF was present 17 bp from the start codon of the downstream gene. The DNA sequence of pPpbr-vio involved in this study is shown in Fig. S2.† The pbr operon originating from Cupriavidus metallidurans has been previously characterized.29 Inspired by the specific regulation mechanism of MerR-family metalloregulators, the Pb(ii)-specific biosensor was designed by substituting the detoxification genes with the violacein biosynthesis module. The bisindole violacein is formed by the conversion of two molecules of endogenous l-tryptophan through the action of five proteins, which are transcribed and translated as a single polycistronic unit under the induction of Pb(ii) (Fig. 1A). For qualitative analysis, violacein-producing biosensor cells with striking purple hues were conveniently recognized in centrifuged bacterial cell pellets by the naked eye. For quantification analysis, the violacein was butanol-extracted from the cell lysate and determined by measuring the absorption at 490 nm using a conventional visible light spectrometer (Fig. 1B).
Fig. 1. Assembly of the violacein expression cassette and its application in Pb(ii) biosensing. (A) The design of the pigment-based whole-cell biosensors. Circuit diagram and schematic depicting the assembly of the violacein biosynthetic module vioABCDE under the control of T7 lac promoter (top). The DNA sequence containing the stop codon of the upstream gene, an extra RBS, and the start codon of the downstream gene is shown. Genetic organization of the lead (bottom) inductive violacein expression constructs. The Pb(ii) sensing element (the pbrR gene and its divergent promoter) was inserted in front of the vioABCDE to generate the lead biosensing module. (B) A schematic of the workflow envisioned for the implementation of the colorimetric biosensors towards Pb(ii).
IPTG inducible violacein production and evaluation of color change
Under the induction of increasing concentrations of IPTG, the violacein biosynthetic gene cluster on the pET-vio regulated by T7 lac promoter, showed elevated production of blue-purple pigment in the culture (Fig. 2). The results demonstrated that violacein could be heterogeneously expressed by engineered E. coli in a dose–response relationship between inducer and pigment accumulation. Violacein has been demonstrated to have antibacterial properties, particularly towards Gram-positive bacteria.25,32 Many Gram-negative bacteria, including E. coli, carry an outer membrane which makes them insensitive to violacein.24 Although violacein was heterogeneously overexpressed in the E. coli cytoplasm with a high concentration of IPTG in this study, the overexpressed violacein did not exert significant influences on the growth of the engineered cells.
Fig. 2. The response of BL21(DE3)plysS/pET-vio with IPTG induction. Exponential phase LB-cultures of BL21(DE3)plysS harboring pET-vio was incubated with increased concentration of IPTG at 30 °C for 8 h. Absorbance measurements of the butanol phase were taken at 490 nm after extraction. All the data are the means of three independent experiments. Error bars indicate standard deviation.
Selectivity of violacein-based biosensor towards target metal
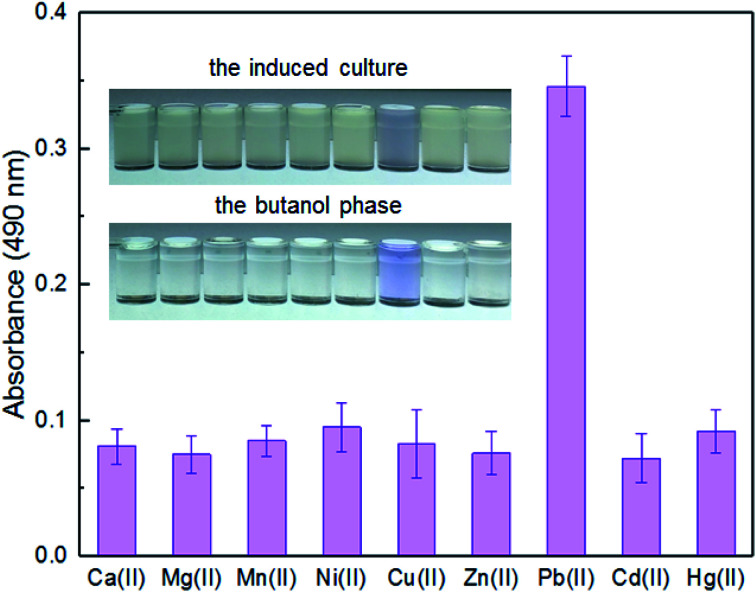
Enzymatic, fluorescent, and bioluminescent reporters have been used for heavy metal biosensing, yet special substrates, agents, or expensive equipment were usually needed for target signal detection.9,11,30 The development of a novel biosensor using natural pigment as the output signal makes it possible to recognize signal by the naked eye.20,33 A whole-cell biosensor was assembled by transforming TOP10 with pPpbr-vio. To evaluate the specificity of an engineered biosensor, the exponential phase of biosensor cells was exposed to various metal ions, individually, at the concentration of 5 μM. The color of the culture exposed with Pb(ii) changed to blue-purple, which could be directly distinguished by the naked eye. The absorbance at 490 nm of its butanol phase was subsequently demonstrated to be significantly higher than that of other groups with the various metal ions exposure (Fig. 3). The Pb(ii) specificity of the pigment-based biosensor was demonstrated, which was expected due to highly specific sensing characteristics of MerR-family metalloregulators.11
Fig. 3. The response of TOP10/pPpbr-vio towards different metal ions. TOP10/pPpbr-vio at the exponential growth phase was cultured in LB media with 5 μM of various metal ions at 37 °C for 3 h. The signals of the whole-cell biosensor towards various metal ions were determined at 490 nm after butanol extraction. All the data are the means of three independent experiments. Error bars indicate standard deviation.
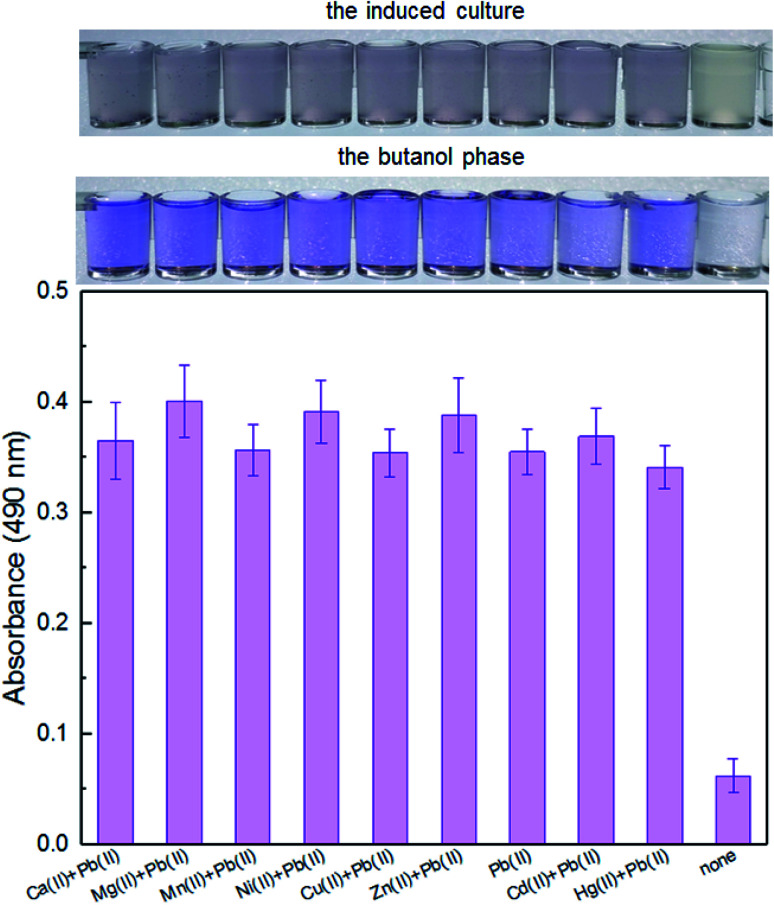
It is well-known that environmental contamination is often complex and accompanied by many heavy metal ions and microorganisms. The anti-interference ability of an engineered biosensor will be especially important when real environmental samples are tested. In the presence of various interfering metal ions, the violacein-based biosensor still showed intense pigment output which could be comparable to the signal output of the biosensor cells with Pb(ii) exposure alone (Fig. 4).
Fig. 4. The response of TOP10/pPpbr-vio towards Pb(ii) under the influence of different metal ions. TOP10/pPpbr-vio at the exponential growth phase was cultured in LB media with various metal ions exposure at 37 °C for 3 h. The signals of the whole-cell biosensor towards 5 μM Pb(ii) in the presence of 5 μM Ca(ii), Mg(ii), Mn(ii), Ni(ii), Cu(ii), Zn(ii), Cd(ii), and Hg(ii) were determined at 490 nm after butanol extraction. All the data are the means of three independent experiments. Error bars indicate standard deviation.
Lead sensitivity detection with the colorimetric biosensor
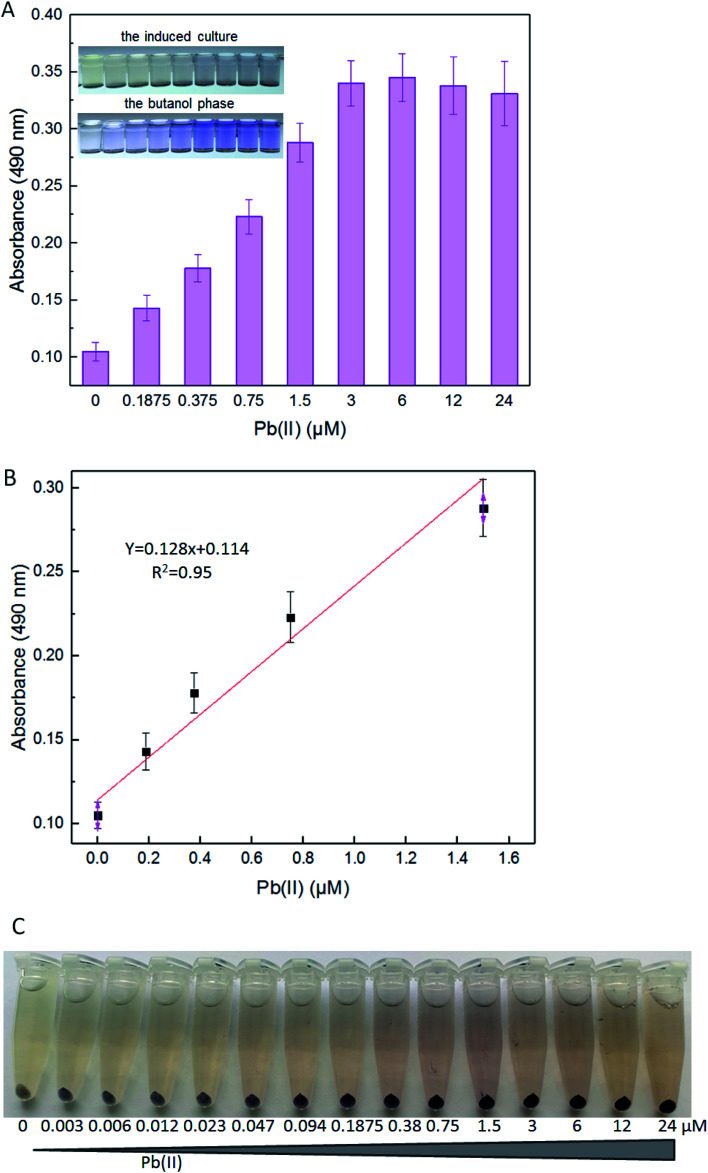
The detection limits of lead E. coli whole-cell biosensor previously constructed were above 10 μM Pb(ii) with the fluorescent reporter, and above 1 μM Pb(ii) with the enzymatic reporter, respectively.9,30 The improved lead sensitivity is expected to be based on the violacein reporter. Even a low-level translation of the violacein biosynthesis enzymes can, over time, biosynthesize enough pigment to produce a detectable signal. As expected, the violacein increased as the concentration of Pb(ii) was increased in the media. Even when the concentration of Pb(ii) was as low as 0.1875 μM, violacein accumulation was still detected with a visible light spectrophotometer (Fig. 5A). A good linear relationship between pigment response and concentration of Pb(ii) was observed within 0.1875–1.5 μM (Fig. 5B).
Fig. 5. The response of TOP10/pPpbr-vio induced with different concentrations of Pb(ii). LB-incubated exponential-phase cultures of TOP10/pPpbr-vio were induced with increased concentration of Pb(ii) at 37 °C for 3 h. Whole-cell biosensor dose–response (A) and linear response (B) to Pb(ii). All the data are the means of three independent experiments. Error bars indicate standard deviation. (C) The centrifuged biosensor cell pellets induced with 0, 0.003, 0.006, 0.012, 0.023, 0.047, 0.094, 0.1875, 0.38, 0.75, 1.5, 3, 6, 12, and 24 μM Pb(ii). The experiment was repeated three times, and a representative image is shown.
Violacein cannot be secreted from the cells into the medium due to its liposoluble property.25 For this reason, although the color change of the induced culture is less tangible at the low concentration of Pb(ii), the color change in the butanol phase can be detected by the colorimetric method (Fig. 5B). In particular, this is because the produced pigment mostly accumulated in the bacterial cells. The color change of the supernatant of the cultures was not obvious because of the poor water solubility of violacein. The color of this biosensor's cell pellets changed from grey-white to blue-purple, and then to black-purple, notably after increased concentration of Pb(ii) exposure (Fig. 5C). The color of cell pellets decreased with the decrease of lead exposure concentration, and the uniformity of purple in the cell pellets also gradually decreased. More and more colorless cells (with no obvious pigment accumulation) were found in the cell pellets with less than 0.38 μM Pb(ii) exposure (Fig. 5C). The saturation of Pb(ii) binding sites in dimeric PbrR with high concentration of Pb(ii) exposure might result in the overproduction of violacein in all engineered cells. However, the Pb(ii) binding sites in dimeric PbrR were unsaturated with inadequate Pb(ii) exposure, which would lead to the failure of violacein accumulation in some engineered cells. Generally, The color change of cell pellets is notably visible to the naked eye even at less than 0.1875 μM Pb(ii) induction. However, this difference cannot be distinguished using the colorimetric method.
In summary, compared with reported whole-cell based signal output biosensors, the color change of violacein-based biosensor cultures could be recognized by the naked eye, but only when engineered cells were exposed to a high concentration of Pb(ii). Furthermore, additional processing steps, such as centrifugation, butanol extraction, and colorimetric detection, were required for the quantification of violacein produced by bacterial cells. All of these extra procedures led to increased testing time and cost.
Environmental samples detection with the colorimetric biosensor
To determine the effectiveness of the violacein-based biosensor to detect bioavailable Pb(ii) in environmental samples, the exponential phase of engineered cells were exposed to 0, 0.375, 0.75, and 1.5 μM Pb(ii) dissolved in either distilled water, tap water or lake water. It was noted that an increase in the absorbance at 490 nm correlated with an increase in Pb(ii) concentration among three groups, and the cell density slightly decreased due to the toxicity of Pb(ii). In particular, no significant difference (p > 0.05) was observed when comparing the violacein signals of the butanol phases with the same concentration of Pb(ii) exposure in three different water samples. Our findings suggest that the violacein-based biosensor has the potential for ecotoxicological assessment of lead-polluted environmental water sample (Fig. 6).
Fig. 6. The response of TOP10/pPpbr-vio after exposure to varying concentrations of Pb(ii) in the environmental water samples. LB-incubated exponential-phase cultures of whole-cell biosensor TOP10/pPpbr-vio was exposed to varying concentrations (0, 0.375, 0.75, and 1.5 μM) of Pb(ii) in the following water samples: sterile distilled water, unsterilized tap water and unsterilized lake water. After culturing at 37 °C for 3 h, the bacterial density (OD600) was first determined (point line diagram, right-Y scale), and then the violacein was extracted with butanol and quantified at 490 nm (bar chart, left-Y scale).
The United States Environmental Protection Agency (USEPA) recommends the criteria for maximum concentration for lead in freshwater to be 82 μg L−1 (0.396 μM).34 The World Health Organization (WHO) guidelines recommend the guideline value for lead in drinking water to be 10 μg L−1 (0.048 μM).35 The engineered violacein-based biosensor developed in this study could detect below the USEPA criteria of maximum concentration to prevent acute toxicity in aquatic organisms. However, the quantitative detection limit of the colorimetry method is 0.1875 μM, which is higher than the WHO criteria for lead in drinking water. The accumulation of violacein in the biosensor cell pellets could actually be recognized with less than 0.1875 μM Pb(ii) exposure (Fig. 5C), but the color difference failed to be distinguished using the colorimetry method. More sensitive methods for violacein detection and quantification, such as high performance liquid chromatography, may be helpful to improve the detection limit.
Environmental lead exists in several forms including elemental, organic and inorganic.36 Atomic absorption spectroscopy, inductively coupled plasma optical emission spectrometry and inductively coupled plasma mass spectrometry have been widely used for the determination of the total lead in environmental samples. The great advantage of a whole-cell biosensor is that it can detect and quantify the bioavailable Pb(ii), which is harmful for aquatic lives, directly.37 There are many factors that influence the bioavailability of lead, such as the environmental matrix, the physicochemical characteristics of the lead compounds, and the exposure organisms.30 This kind of biosensing method for bioavailable Pb(ii) detection is always a good supplement to the traditional instrumental methods. Cell-based sensors offer diversified, portable and cheap methods for the detection of heavy metal but have yet to be truly adopted commercially.7 The state-of-the-art tools offered by synthetic biology enable us to develop more novel biosensors with customizable performance to meet the requirements for practical application.
Conclusions
Few attempts have been made to assemble colorimetric whole-cell biosensors using the natural pigment synthetic pathway. In this study, a sensitive, visual, and qualitative pigment-based bacterial biosensor, in which the output signal is generated based on genetical control of violacein biosynthesis, was developed for preliminary estimation of the bioavailability of toxic lead. Pb(ii)-sensing regulator PbrR was employed in this novel whole-cell biosensor, which was demonstrated to respond specifically to Pb(ii), and to be resistant to the interferences from other different metal ions. Color change by the intracellular accumulation of violacein could be recognized directly with the naked eye after the biosensor cells were induced with a high concentration of Pb(ii), and quantified by determining the absorbance at 490 nm after butanol extraction. The linear range of Pb(ii) detection is 0.1875–1.5 μM. The developed violacein-based biosensor for Pb(ii) quantification had much higher sensitivity than reported fluorescent protein-based Pb(ii) biosensor, while it possessed merits such as the stability of output violacein, the removal of contaminated Gram-positive bacteria from environmental samples due to extensive anti-bacterial activities of violacein, and the simple spectrophotometric detection procedure. The idea of assembling a whole-cell biosensor using brightly-colored violacein as the signal output may have potential for developing novel biosensors for the detection of other heavy metals.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by Shenzhen Key Medical Discipline Construction Fund, the Science and Technology Program of Shenzhen (JCYJ20180306170237563 and JCYJ20190808175205480) and the Natural Science Foundation of Guangdong Province (2019A1515011989).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04815a
References
- Williams P. N. Islam M. R. Adomako E. E. Raab A. Hossain S. A. Zhu Y. G. Feldmann J. Meharg A. A. Environ. Sci. Technol. 2006;40:4903–4908. doi: 10.1021/es060222i. [DOI] [PubMed] [Google Scholar]
- Williams P. N. Lei M. Sun G. Huang Q. Lu Y. Deacon C. Meharg A. A. Zhu Y. G. Environ. Sci. Technol. 2009;43:637–642. doi: 10.1021/es802412r. [DOI] [PubMed] [Google Scholar]
- Vareda J. P. Valente A. J. M. Duraes L. J. Environ. Manage. 2019;246:101–118. doi: 10.1016/j.jenvman.2019.05.126. [DOI] [PubMed] [Google Scholar]
- Magrisso S. Erel Y. Belkin S. Microb. Biotechnol. 2008;1:320–330. doi: 10.1111/j.1751-7915.2008.00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron K. J. Rutherford J. C. Ford D. Robinson N. J. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Lopreside A. Wan X. Michelini E. Roda A. Wang B. Anal. Chem. 2019;91:15284–15292. doi: 10.1021/acs.analchem.9b04444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks M. Bachmann T. T. Wang B. ChemPhysChem. 2020;21:131. doi: 10.1002/cphc.201901191. [DOI] [PubMed] [Google Scholar]
- Kumar S. Verma N. Singh A. K. Sens. Actuators, B. 2017;240:248–254. doi: 10.1016/j.snb.2016.08.160. [DOI] [Google Scholar]
- Hui C. Y. Guo Y. Liu L. Zheng H. Q. Gao C. X. Zhang W. PLoS One. 2020;15:e0228456. doi: 10.1371/journal.pone.0228456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L. Stoyanov J. V. Kidd S. P. Hobman J. L. FEMS Microbiol. Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Jung J. Lee S. J. J. Microbiol. Biotechnol. 2019;29:1522–1542. doi: 10.4014/jmb.1908.08002. [DOI] [PubMed] [Google Scholar]
- Guo Y. Hui C. Y. Liu L. Zheng H. Q. Wu H. M. Front. Microbiol. 2019;10:1454. doi: 10.3389/fmicb.2019.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. Liu X. Sun P. Wang X. Zhu H. Hong M. Mao Z. W. Zhao J. Environ. Sci. Technol. 2014;48:3363–3371. doi: 10.1021/es4046567. [DOI] [PubMed] [Google Scholar]
- Cayron J. Effantin G. Prudent E. Rodrigue A. Res. Microbiol. 2020;171:21–27. doi: 10.1016/j.resmic.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Wang B. Barahona M. Buck M. Nucleic Acids Res. 2014;42:9484–9492. doi: 10.1093/nar/gku593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X. Volpetti F. Petrova E. French C. Maerkl S. J. Wang B. Nat. Chem. Biol. 2019;15:540–548. doi: 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- Wang B. Barahona M. Buck M. Nucleic Acids Res. 2015;43:1955–1964. doi: 10.1093/nar/gku1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. Shen Y. Zou Y. Sun P. Wei W. Zhao J. Zhang C. Analyst. 2018;143:630–634. doi: 10.1039/C7AN00587C. [DOI] [PubMed] [Google Scholar]
- Wang B. Barahona M. Buck M. Biosens. Bioelectron. 2013;40:368–376. doi: 10.1016/j.bios.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Zheng Y. Fan X. Xu L. Pang T. Liu T. Liang L. Huang S. Xiao Q. J. Biosci. Bioeng. 2020;129:223–228. doi: 10.1016/j.jbiosc.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Watstein D. M. Styczynski M. P. ACS Synth. Biol. 2018;7:267–275. doi: 10.1021/acssynbio.7b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNerney M. P. Michel C. L. Kishore K. Standeven J. Styczynski M. P. Nat. Commun. 2019;10:5514. doi: 10.1038/s41467-019-13454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K. Inoue K. Takahashi Y. Ueda S. Isoda K. Yagi K. Maeda I. Appl. Environ. Microbiol. 2008;74:6730–6738. doi: 10.1128/AEM.00498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauz A. C. G. Carretero G. P. B. Saraiva G. K. V. Park P. Mortara L. Cuccovia I. M. Brocchi M. Gueiros-Filho F. J. ACS Infect. Dis. 2019;5:539–549. doi: 10.1021/acsinfecdis.8b00245. [DOI] [PubMed] [Google Scholar]
- Choi S. Y. Yoon K. H. Lee J. I. Mitchell R. J. BioMed Res. Int. 2015;2015:465056. doi: 10.1155/2015/465056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- August P. R. Grossman T. H. Minor C. Draper M. P. MacNeil I. A. Pemberton J. M. Call K. M. Holt D. Osburne M. S. J. Mol. Microbiol. Biotechnol. 2000;2:513–519. [PubMed] [Google Scholar]
- Rodrigues A. L. Trachtmann N. Becker J. Lohanatha A. F. Blotenberg J. Bolten C. J. Korneli C. de Souza Lima A. O. Porto L. M. Sprenger G. A. Wittmann C. Metab. Eng. 2013;20:29–41. doi: 10.1016/j.ymben.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Zhang X. Enomoto K. Appl. Microbiol. Biotechnol. 2011;90:1963–1971. doi: 10.1007/s00253-011-3203-9. [DOI] [PubMed] [Google Scholar]
- Borremans B. Hobman J. L. Provoost A. Brown N. L. van Der Lelie D. J. Bacteriol. 2001;183:5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereza-Malcolm L. Aracic S. Franks A. E. Sensors. 2016;16:2174. doi: 10.3390/s16122174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi T. Kato M. Fukamachi K. Kato N. Ikeda T. FEMS Microbiol. Lett. 2008;279:124–130. doi: 10.1111/j.1574-6968.2007.01016.x. [DOI] [PubMed] [Google Scholar]
- Aruldass C. A. Rubiyatno Venil C. K. Ahmad W. A. RSC Adv. 2015;5:51524–51536. doi: 10.1039/C5RA05765E. [DOI] [Google Scholar]
- Yong Y. C. Zhong J. J. Biosens. Bioelectron. 2009;25:41–47. doi: 10.1016/j.bios.2009.06.010. [DOI] [PubMed] [Google Scholar]
- USEPA, National Recommended Water Quality Criteria, 1984 [Google Scholar]
- WHO, Guidelines for Drinking-Water Quality, Geneva, Switzerland, 2011 [Google Scholar]
- de Souza I. D. de Andrade A. S. Dalmolin R. J. S. Crit. Rev. Toxicol. 2018;48:375–386. doi: 10.1080/10408444.2018.1429387. [DOI] [PubMed] [Google Scholar]
- Kim H. J. Jeong H. Lee S. J. Anal. Bioanal. Chem. 2018;410:1191–1203. doi: 10.1007/s00216-017-0751-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.