Abstract
SARS-CoV-2 continued circulation results in mutations and the emergence of various variants. Until now, whenever a new, dominant, variant appeared, it overpowered its predecessor after a short parallel period. The latest variant of concern, Omicron, is spreading swiftly around the world with record morbidity reports. Unlike the Delta variant, previously considered to be the main variant of concern in most countries, including Israel, the dynamics of the Omicron variant showed different characteristics. To enable quick assessment of the spread of this variant we developed an RT-qPCR primers-probe set for the direct detection of Omicron variant. Characterized as highly specific and sensitive, the new Omicron detection set was deployed on clinical and wastewater samples. In contrast to the expected dynamics whereupon the Delta variant diminishes as Omicron variant increases, representative results received from wastewater detection indicated a cryptic circulation of the Delta variant even with the increased levels of Omicron variant. Resulting wastewater data illustrated the very initial Delta-Omicron dynamics occurring in real time. Despite this, the future development and dynamics of the two variants side-by-side is still mainly unknown. Based on the initial results, a double susceptible-infected-recovered model was developed for the Delta and Omicron variants. According to the developed model, it can be expected that the Omicron levels will decrease until eliminated, while Delta variant will maintain its cryptic circulation. If this comes to pass, the mentioned cryptic circulation may result in the reemergence of a Delta morbidity wave or in the possible generation of a new threatening variant. In conclusion, the deployment of wastewater-based epidemiology is recommended as a convenient and representative tool for pandemic containment.
Keywords: SARS-CoV-2, Variants of concern, Reverse transcriptase quantitative polymerase chain reaction, Wastewater-based epidemiology, Delta, Omicron, Susceptible-infected-recovered model
Graphical abstract
1. Introduction
SARS-CoV-2 virus is circulating worldwide and is undergoing mutations resulting in the continuous emergence of new variants (Tao et al., 2021). Different variants could differ in various clinical parameters, including symptom severity, infection rate and immune response (Hoffmann et al., 2021a, Hoffmann et al., 2021b; Jewell, 2021). Therefore, when a new variant emerges, even in highly vaccinated populations, immunity might not be absolute. This may occur through vaccination deterioration over time, or through mutated targeting regions for neutralizing antibodies (generated by vaccine or previous variant recovery). One of the latest variants to materialize is the Omicron variant of concern (B.1.1.529) (Karim and Karim, 2021). This variant demonstrates numerous mutations especially in the spike protein gene (S gene) when compared to the original SARS-CoV-2 or other variants of concern (Venkatakrishnan et al., 2021). Therefore, despite vaccination efforts in Israel, with a large portion of the population being vaccinated between the 1st to 4th dose of vaccine and despite high infection rates by previous variants, the Omicron variant had now rooted itself in Israel (Israeli Ministry of Health Dashboard, n.d.; Leshem et al., 2022).
One methodology for assessment of viral infection is through wastewater-based epidemiology, a convenient tool that can provide one of the most accurate representation of the extent of virus' prevalence in a population (Collivignarelli et al., 2020; La Rosa et al., 2020a, La Rosa et al., 2020b; Martin et al., 2020; Westhaus et al., 2021; Yaniv et al., 2021a). Using such a tool can offer an early warning system that can provide the impetus for developing a vast clinical examination infrastructure. Using wastewater surveillance, it is possible to receive significant amount of information without having to depend on community cooperation and clinical testing resources. Previous reports from our group (Yaniv et al., 2021a, Yaniv et al., 2021b), continuously monitored SARS-CoV-2 prevalence in wastewater of a single city in Israel (the fourth in size) utilizing reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). Targeting variants of concern, we detected the shifts between the original SARS-CoV-2 and the Alpha variant (Yaniv et al., 2021b), and between the Alpha and Delta variants (Yaniv et al., 2021a). Considering clinical reports regarding the Omicron variant infection rate, it was therefore expected that as occurred in previous infections, this variant would appear in the wastewater and “overpower” its predecessor, the Delta variant.
The variant-population relations are dependent on many factors, some are circumstantial and others are specific to each variant. Infections caused by Delta or Omicron in the current morbidity wave might be prevented due to various levels of vaccination or through cross-immunization by the two variants. The cross-immunization may be symmetric, i.e., a recovered individual from a Delta infection is protected against infection by Omicron at the same level that a recovered individual from an Omicron infection is protected against Delta, or may be asymmetric (i.e., non-equal protections). In fact, recent studies (Laurie et al., 2022; Suryawanshi et al., 2022) suggest that the asymmetric case is more likely, but the extent of this asymmetry has not yet been quantified.
Here we present, the development of an RT-qPCR primers-probe set specifically meant for detection and quantification of the Omicron variant. Following full characterization, the developed set was utilized on clinical samples, as well as on wastewater samples. Wastewater from the Beer-Sheva site were simultaneously tested using a previously published set for Delta variant detection (Yaniv et al., 2021a), and the relations between the two variants of concern in wastewater were documented over a period of 7 weeks during December 2021 and January 2022. Wastewater results combined with model predictions may provide important tools for predicting the dynamics between the SARS-CoV-2 variants, Delta and Omicron, in the near future.
2. Methodology
2.1. Primers and probes design
The original sequence of SARS-CoV-2 (NC_045512.2) was taken from NCBI database. The Omicron variant (B.1.1.529, EPI_ISL_6794907) sequence was taken from GISAID database (Shu and McCauley, 2017). The new probe design focused on the S gene 22,083–22,181 bp location that includes 3 nucleotides deletion, changing N211 and L212 into Isoleucine, as well as a 9 nucleotides insertion at the 214 residue. All primers and probes were manufactured by Integrated DNA Technologies (IDT). The probes contain a ZEN Quencher as a second, internal quencher in qPCR 5′-nuclease assay. Probes were assigned with a 6-carboxy-fluorescein (FAM) fluorophore. N gene detection and Delta variant detection were performed utilizing previously developed sets (N2 by the CDC, Improved N3 and SΔ157), published by our group (Yaniv et al., 2021a, Yaniv et al., 2021b). All for primers and probes used in this study are listed in Table S1.
2.2. Clinical samples RNA extraction, RT-qPCR and analysis
Previously detected SARS-CoV-2 positive samples in either UTM (COPAN Italia S.p.A, Brescia, Italy, or NOVAMES Ltd., Jerusalem, Israel) or VTM (BI, Biological Industries, Beit Haemek, Israel) were subjected to Viral RNA extraction using QIAamp® 96 Viral RNA kit (QAIGEN, Hilden, Germany) following manufacturer instructions on a FREEDOM EVO 200 (Tecan Group Ltd., Männedorf, Switzerland). The extracted RNA was subjected to three different RT-qPCR assays using 1. The ALLPLEX™, SARS-CoV-2 Variants I Assay (Seegene, Seoul, South Korea) and 2. Our newly designed Omicron assay (this paper) and 3. Our previously published Delta assay (Yaniv et al., 2021a). All assays were performed on a Bio-Rad CFX 96 PCR instrument (Bio-Rad, Hercules, CA, USA) following manufacturer instructions or as described in the RT-qPCR section using the same RNA. Result for the ALLPLEX™, SARS-CoV-2 Variants I Assay were analyzed using Seegene SARS-CoV-2 Viewer V3, and the Bio-Rad Maestro 2.0 for the Omicron and Delta assays.
For analysis purposes, a few criteria were chosen. Samples considered “Invalid”, “out of range” or “Unique” were not taken for analysis. Samples deemed as “Invalid” were samples without a detection signal for the RdRP gene (83/376). Samples deemed as “Out of range” were samples with a detection signal for Delta or Omicron larger than Ct 40 (17/376). Samples deemed as “Unique” were samples with a detected signal for RdRP gene, yet no detection signal for neither the Delta nor the Omicron detection (9/376).
2.3. Wastewater RNA extraction
Wastewater samples were collected from the wastewater treatment plant of the city of Beer-Sheva, Israel. Samples were collected twice a week for 24 h (composite sewage samples). The samples were transferred under chilled conditions to the lab and were kept at 4 °C until processed. Samples underwent a duplicate RNA extraction directly from the wastewater sample, according to Zymo Environmental Water RNA (Zymo Research R2042) manufacture protocol, including enrichment step starting from 5 mL sample. MS2 phage exist in non-detectable levels in Israel's wastewater, therefore spiking wastewater samples with high concentration of the phage allows its usage as external control. We added 105 copies of MS2 phage to 5 mL of each sample prior to the initial lysis step in the RNA extraction protocol. The final elution volume was 35 μL of RNase free water. RNA samples were kept at –80 °C. Wastewater samples resulted in negative detection of a variant, underwent a concentration protocol using MCE electronegative membrane procedure as was previously described (Yaniv et al., 2021a). However, none of the concentrated samples manifested a new signal, meaning there was a full correlation between the Zymo kit results and the concentrated results.
2.4. Wastewater samples RT-qPCR
The Wastewater's RNA were subjected to an RT-qPCRs assays as previously described (Yaniv et al., 2021a, Yaniv et al., 2021b). Briefly, reaction final volume is 20 μL with primers and probes concentration of 0.5 μM and 0.2 μM respectively. Each reaction mixture was added with 5 μL of RNA sample. Assay steps were preformed according to manufacture recommended protocol (One Step PrimeScript III RT-qPCR mix RR600 TAKARA, Japan) using Applied Biosystems Thermocycler (Thermo Scientific). RNA copy number per Liter of wastewater for raw sample was calculated using an equation available in the SI file (page 17). Each RT-qPCR assay run included relevant quality controls (such as: Non template control (NTC) and MS2 phage detection for wastewater RNA sample (Dreier et al., 2005)).
2.5. Calibration curves and limit of detection determination
Calibration curve for the Omicron detection assay was performed on a known-positive dsDNA gene block. A double stranded linear DNA (produced by IDT Technologies) containing S gene sequence matching the reported 211–212 modification together with the 214 insertion (Table S2). Negative controls were carried out in two ways. First a negative control was by a DNA gene block matching the original SARS-CoV-2 sequence relevant for the tested primers-probe set (Table S2). Further negative controls were performed using Wastewater samples confirmed as positive for the original variant, Alpha variant of concern or Delta variant of concern. Serial dilutions for the relevant gene block were prepared based on copy number calculations. The resulting Ct values (Y axis) were plotted against the log copy number of the DNA gene block template (X axis). While not being able to represent the cDNA synthesis step, DNA standards are acceptable and of high use (Ahmed et al., 2020, Ahmed et al., 2021; Bivins et al., 2021; Gerrity et al., 2021; Medema et al., 2020; Philo et al., 2021). In accordance, copy number calculations were accounted for the use of a double stranded DNA instead of single strand RNA. Each concentration was examined by ten repetitions and a standard deviation was calculated. In addition, linear regression was employed between the log copy number and the Ct values from the RT-qPCR results.
2.6. Complex matrix detection
In order to assess the designed Omicron RT-qPCR assay's sensitivity in a wastewater matrix, we used RNA extracted from pre-determined SARS-CoV-2-negative wastewater sample. The predetermined negative RNA sample was examined using our Omicron detection set. We added known concentration of the gene block used as positive control for calibration (0/100/101/102) to the negative RNA sample (1:10 v/v). Each spiking condition was measured by eight repetitions. Ct values were plotted to correspond to the new probes limit of detection in a complex wastewater environment.
2.7. Susceptible-infected-recovered modeling
We present here a double susceptible-infected-recovered (SIR) model with cross-variant immunization and time-dependent waning immunity. We define the following time-dependent variables:
-
(i)
fD and f O are the fractions of actively infected populations in Delta and Omicron, respectively.
-
(ii)
sD and s O are the effective fractions of susceptible populations to Delta and Omicron infections, respectively, henceforth “susceptibilities”. These variables present an average over the diverse immunity presented in the population, although in the original SIR model they simply present the fraction of population that is neither actively infected nor recovered.
-
(iii)
rD and r O are the fractions of recovered population from Delta and Omicron infections, respectively, in the present outbreak of infection. The contribution of recovered individuals from previous outbreaks is accounted for in the initial conditions for s D and s O.
In addition, we use the following (time-independent) parameters:
-
(i)
τD and τ O are the infection time-period of Delta and Omicron, respectively.
-
(ii)
R0(D) and R 0 (O) are the basic reproduction numbers of Delta and Omicron, respectively.
We note in passing that the actual (time-dependent) reproduction numbers of Delta and Omicron (respectively) are given by R D = R 0 (D) s D and R O = R 0 (O) s O, making the actual reproduction numbers much below the basic ones.
The model equations are as follows:
| (1) |
| (2) |
| (3) |
| (4) |
where s D and s O take the following expressions, accounting for both cross-variant immunization and waning immunity that is linear in time,
| (5) |
| (6) |
where s D0 and s O0 are the initial susceptibilities (t = 0) that were gained from past pandemic waves and vaccination (using the relation s O0 = 1 − (1 − s D0) q OD), a is the rate at which the relative immunity decreases, q DO represents the relative mean protection against Delta infection that a newly recovered individual from Omicron gained, and similarly q OD represents the relative mean protection against Omicron infection that a newly recovered individual from Delta gained. Asymmetric cross-immunization, supported by recent immunology studies (Laurie et al., 2022; Suryawanshi et al., 2022), implies q DO < q OD, i.e., the protection against Omicron due to Delta past infection is higher than the protection against Delta due to Omicron past infection. Note that the model allows double infections at the same time (i.e., Eq. (5) does not depend on f O and Eq. (6) does not depend on f D), which is possible during the first week of infection but unlikely later, however, fine-tuning of this issue is out of the scope here.
Due to the (5), (6), Eqs. (1), (2), (3), (4) form a set of non-linear differential equations and are therefore solved numerically. Further description of the model choice of parameters is elaborated in the SI.
3. Results and discussion
3.1. Direct RT-qPCR Omicron variant of concern (B.1.1.529) detection
Whenever a new variant of concern emerges, there is an obvious need for rapid and direct detection of variant positive cases in the population. In most countries, the employed strategy is to sequence all COVID-19 positive patients entering the country along with random sequencing of COVID-19 positive patients in the population for the detection of new variants. Next generation sequencing (NGS) retrieves the most data and therefore applies to all variants. While NGS can reveal the appearance of a new variant, once revealed it is costly and time-consuming to keep using sequencing methods for the detection of any previously detected variants. The current infrastructures around the world cannot provide rapid NGS analysis in terms of large-scale sampling and produce immediate results. It is for these reasons that current NGS analyses are performed on a significantly smaller sub-group of samples and may result in overlooking variants distribution. When focusing on specific, known variants, RT-qPCR can overcome such limitations. Therefore, a specifically designed RT-qPCR assay for variant detection provides an important tool for broad, existing variants survey in the population with immediate results.
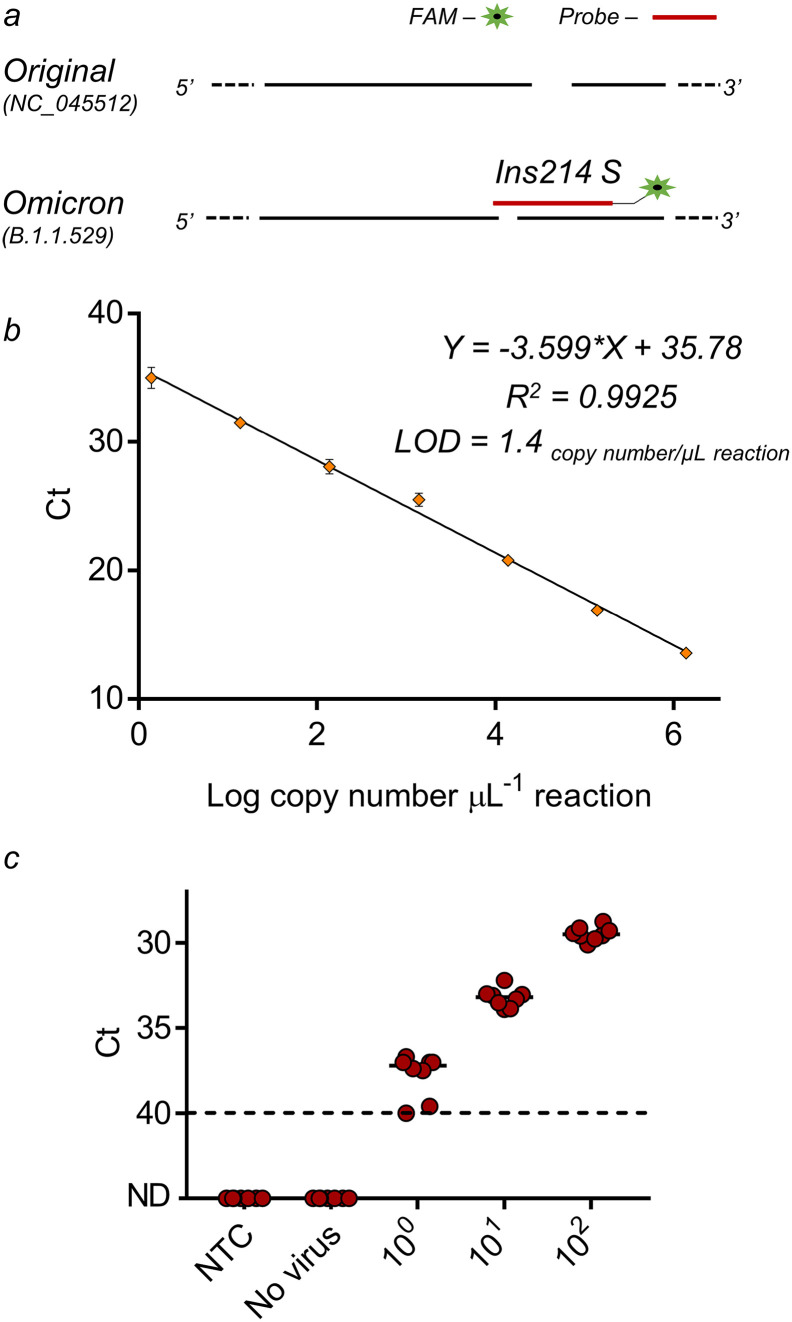
Following South Africa's declaration regarding the appearance of Omicron B.1.1.529 variant towards the end of November 2021, we designed a new primers-probe detection set specific to the Omicron variant (Table S1). Other groups also published sequences for Omicron detection, yet they differ from the design presented here (Erster et al., 2021; Lee et al., 2022). The designed set in this paper focused on a unique region in the S gene (located between 22,083–22,181 bp), containing a deletion of 3 nucleotides together with an insertion of 9 nucleotides (Fig. 1a). At a later stage a new sub-variant emerged, BA.2, while due to sequence differences, the developed set does not match BA.2 detection. However, the BA.2 sub-variant emerged in a later stage than the presented time-period of this publication and was therefore irrelevant for the desired analysis. The new set was named “Ins214 S" based on the insertion and underwent full characterization, including limit of detection (LOD) determination in different environments.
Fig. 1.
RT-q-PCR primers-probe set designed for the detection of SARS-CoV-2 Omicron variant. a, Detection primers-probe set designed for specific identification of the Omicron variant (B.1.1.529). Design was based on S gene unique features of the Omicron variant compared to other known variants. b, Calibration curve for Omicron set detection. Resulted Ct value plotted against the tested Log copy number. Error bars present standard deviation for ten replicates. c, Ins214 S primers–probe set in wastewater matrix. RNA extracted from negative detection wastewater sample (No virus) spiked with known concentrations of positive template (100–102 S gene template copies per μL) and Non-Template Control (NTC, water). ND - not detected. Solid lines indicate the median and dashed lines indicate the detection limit as decided by clinical guidelines.
A calibration curve was performed using Linear DNA gene block possessing the same unique features as the targeted Omicron variant. Serial dilutions of the gene block were tested between 1.4*106 and 1.4*100 copies per μL reaction (Fig. 1b). LOD was determined as the lowest dilution tested in which a Ct value of <40 was received for >95% of the measurements and was found to be 1.4*100 copies per μL reaction. When translated to wastewater monitoring and detection using the equation appearing in the SI file (page 17), the new set LOD is 4.7*102 RNA copies per L wastewater (Ct = 40). Apart from being found as sensitive, the newly designed Omicron set did not have cross-reactivity with any of the tested negative controls. Thus, if original variant gene block showed positive signals in wastewater samples (to the original/Alpha/Delta variants), a signal from the Omicron set did not manifest, proving this set to be highly specific.
We further evaluated the sensitivity and the stability of the designed Omicron set in wastewater matrix. Wastewater contains many factors that may inhibit the RT-qPCR assay due to the complexity of the environment, therefore it was crucial to evaluate the designed set functionality in such an environment if it is aimed for wastewater detection. Predetermined negative for SARS-CoV-2 wastewater samples, were spiked with Omicron positive gene block at different concentrations (Fig. 1c). As can be seen in Fig. 1c, despite the wastewater's difficult environment, the sensitivity of the Omicron set was not compromised and the LOD remained as before (1.4*100 copies per μL reaction and 4.7*102 RNA copies per L wastewater).
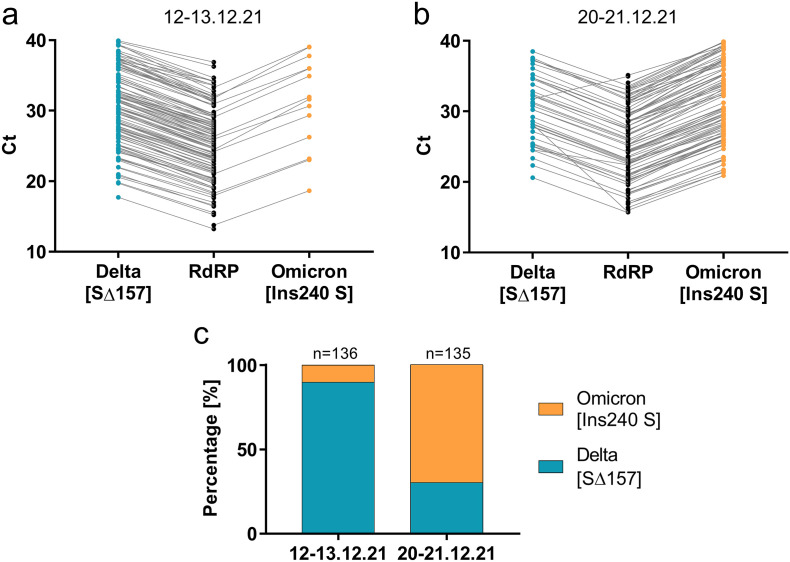
Subsequent to the Omicron primers-probe set characterization, it was validated and tested on clinical samples. Taking 271 clinical samples predetermined as positive to SARS-CoV-2 Variants I by the Seegene variants commercial kit, the newly designed Omicron set was employed, together with the Delta set previously published by our group (Yaniv et al., 2021a) (Fig. 2 , raw data available in Table S3). In all samples, a signal manifested for one of the sets, meaning either Delta or Omicron, apart from a single sample positive for both. Such a case could be explained by double variants infection or by a new variant possessing the two sets targets, regardless this is very rare (<0.5%).
Fig. 2.
RT-qPCR of SARS-CoV-2 Omicron variant vs. Delta variant in clinical samples. Ct results for 3 genes for COVID-19 positive cases in December 12-13th 2021 (a) and in December 20-21st 2021 (b). RdRP is relevant for all known SARS-CoV-2 variants, while the other two targets are specific to the Delta or Omicron variants. c, Summary of the Delta and Omicron percentage in the tested clinical cases.
In Israel, on December 12-13th 89.7% of the samples were positive to the Delta variant and only 10.3% positive for Omicron variant. Yet, approximately one week later, the “tides shifted” and on December 20-21st 70.4% were positive for Omicron and only 29.6% were positive for Delta (Fig. 2c). Clinical and epidemiological background was not disclosed and therefore the results presented here could not help in concluding the true Omicron-Delta dynamics. However, the high infection rate of the Omicron variant that was reported by other countries, seemed to also affect reported morbidity in Israel.
The use of RT-qPCR corresponding to the most relevant variants of concern on a regular basis, may serve as a good indicator for emergence of other, new variants. Meaning, any clinical sample which is positive to SARS-CoV-2 in general (usually through N gene or RdRP gene detection), yet negative for specific prevalent variants detection, as resulted for 9 clinical samples by our assays, can reveal a new variant. Moreover, clinical samples may result in a positive signal for several variants at the same time, as resulted for 2 clinical samples by our assays, consequence of either a single new variant or multiple infections. These explanatory samples are therefore high-risk targets worth of sequencing, more than random survey sequencing and can help refine sequencing efforts. It is therefore recommended to embrace an approach of RT-qPCR assays development for circulating variants and utilizing these assays on routine testing, thus discovering infections suspected as new variants that should be sequenced.
3.2. Wastewater portrait of Omicron and Delta variants interactions
Our group continuously monitors Israel's 4th largest city's (Beer-Sheva) wastewater since the beginning of the COVID-19 pandemic. We monitor general SARS-CoV-2 levels using N gene detection by CDC's N2 set together with our previously published set (Yaniv et al., 2021b). In addition, a more specific variant detection is being employed in accordance with the most relevant variants of concern distribution in the world and specifically in Israel. Utilizing such a monitoring system, it was possible to observe the change from the original variant to the Alpha variant around February 2021 (Yaniv et al., 2021b) and from the Alpha variant to the Delta variant around June 2021 (Yaniv et al., 2021a). It would seem that the rise of Omicron took place globally around the end of November 2021, therefore the developed Omicron detection set in this study was employed on wastewater samples from the same time-point and onwards. The Delta variant detection set was also employed as this variant still appears in Israel's wastewater. It is important to note that N gene detection levels are higher than S gene detection levels, most likely due to transcriptional level differences during the SARS-CoV-2 “lifespan” (Emanuel et al., 2021; Kim et al., 2020).
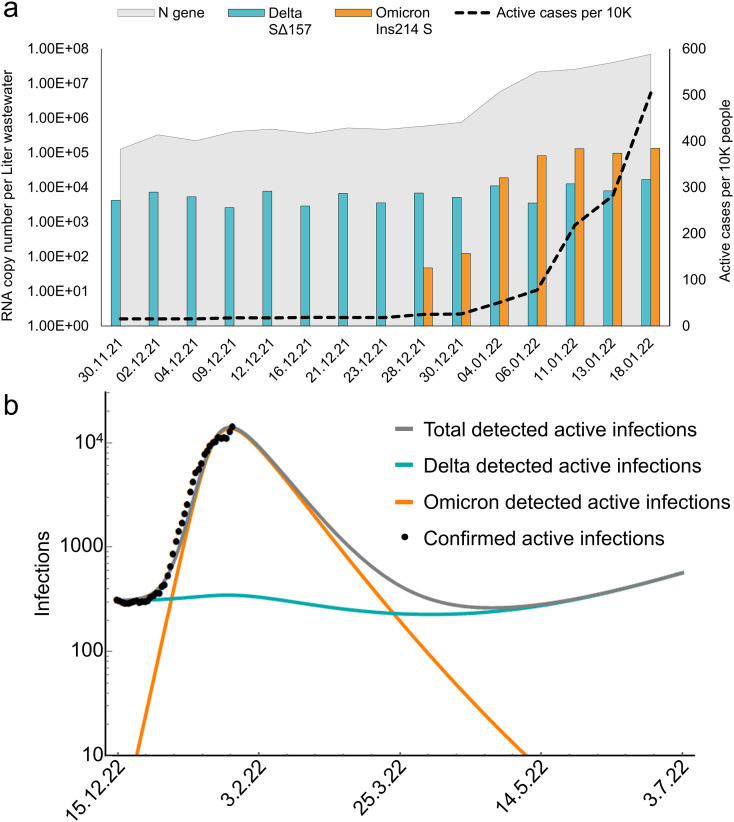
Fig. 3a demonstrates detected levels of N gene, Omicron's S gene, Delta's S gene and the reported morbidity cases per 10 K people. Delta variant's morbidity peaked in around September 2021 and then declined steadily (Yaniv et al., 2021a). Since then, the Delta variant detection levels in the wastewater reached a relatively constant level of 103–104 RNA copies per Liter wastewater. The constant levels observed are unlike the occurrences of the Alpha variant in Israel, where levels declined until full eradication of Alpha variant traces in wastewater (Yaniv et al., 2021b). The relatively low levels of Delta variant observed in our samples may indicate cryptic circulation of this variant throughout the population despite many recovered cases and national vaccination efforts. The Delta variant behavior is considered cryptic, as it was expected to diminish and vanish fully by the over-powering Omicron variant, as has happened for previous variants shifts. Such cryptic circulation allows the variant preservation in the population without increasing morbidity levels (With an average of 17 active cases per 10 K people, Fig. 3a).
Fig. 3.
SARS-CoV-2 Delta variant of concern and Omicron variant of concern dynamics. a, RT-qPCR detection of SARS-CoV-2 in the city of Beer-Sheva's wastewater during late November 2021 until late January 2022. Overall SARS-CoV-2 variants detected by N gene sets (gray area), Delta variant of concern detected by SΔ157 set (teal columns) and Omicron variant of concern detected by Ins214 S set (orange columns). Reported population's morbidity was extracted from the Ministry of Health database (black dashed line). b, Simulation results for Omicron (orange line), Delta (teal line), and total number (gray line) of detected actively infected people in the city of Beer-Sheva according to a moderately asymmetric cross-immunity, qDO = qOD/4 (see SI for the choice of parameters). Black dots represent confirmed active infections obtained from the Ministry of Health dashboard (irrespective of variants) up to January 25th, 2022. Time t = 0 corresponds to the date December 15th, 2021.
Usually, wastewater detection precedes virus' presence in the population between a few days to several weeks prior to clinical reports (Chavarria-Miró et al., 2021; La Rosa et al., 2021; Róka et al., 2021; Wu et al., 2022). In the case of Omicron detection in the wastewater, variant's presence preceded population's morbidity increase by approximately a week prior to clinical reports (Fig. 3a). This was somewhat expected in the case of the Omicron and reflected the variant's high infection rate compared to previous variants. Moreover, the previous variant (Delta) reached a peak of 9.31*104 copies per Liter wastewater, multiplying by two orders of magnitude in 2.5 months since first being detected (Yaniv et al., 2021a). The Omicron however, multiplied by two orders of magnitude in just a week of first being detected. Overall, at the end of December 2021 the Delta variant composed >99% and Omicron variant <1% of the detected SARS-CoV-2 in Beer-Sheva's wastewater. Within a short period of 9 days the SARS-CoV-2 composition changed with the Delta variant being <5% and Omicron variant >95% (exact viral numbers available in the SI). Additional data should be collected in the near future, in order to understand if the Omicron has reached its peak in Beer-Sheva or if higher levels of infection should be expected.
While the reported wastewater results supply a real-time image of virus' prevalence in the population, it is worthy to better understand and predict the epidemiological behavior. To do so, we present here a double susceptible-infected-recovered (Espinosa et al., 2020) (SIR) model with cross-immunization after recovery and time-dependent waning immunity. Adapting possible model parameters based on initial data received from wastewater, together with information from the Ministry of Health database, and a variety of other sources (see SI), it is possible to shed light on the recent parallel spread of Omicron and Delta and to predict its future trends.
The (homogeneous) SIR model, and higher-level multi-compartment models, are highly suited to describe quantitatively epidemic spread when human mobility is high, as the spread of the infection remains approximately homogeneous. For COVID-19, one may include the short exposure stage – yielding the SEIR model (Mwalili et al., 2020), and even the pre-symptomatic-infecting stage – yielding the SEPIR model (Tsori and Granek, 2021a, Tsori and Granek, 2021). Since the exposed (and non-infecting) stage is relatively short, about two days for the parent virus and most of its variants, and probably down to one day for Omicron, it was neglected here. The pre-symptomatic-infecting stage is about 3 days long for most variants (between days 3–5 from infection) (He et al., 2020; Lauer et al., 2020), and is likely to be around 1–2 days long for Omicron (between days 3–4 from infection) (European Centre for Disease Prevention and Control, 2022), and the difference in the level of infectiousness during the symptomatic stage is obviously present. For simplicity, we used here only the SIR model for both variants, introducing the differences between them in the choice of model parameters. In addition, we do not include here the shedding processes of Delta and Omicron virions to wastewater (Proverbio et al., 2022), which could allow better comparison with wastewater data, as this will require the use of yet unknown parameters.
We relied on an asymmetric cross-immunization, where on the average, considering the status of vaccination and past infections of the population, a person recovering from Omicron gained 4 times less protection against a Delta infection, than a person recovering from Delta gained protection against an Omicron infection (further explained in the SI). Moreover, to receive a better correlation with the steady, cryptic circulation of the Delta observed in wastewater, a linear in time waning of immunity was considered. The resulting model predicts a significant decrease in Omicron levels within the near future, diminishing until fully disappearing (Fig. 3b). Furthermore, the Delta is also expected to decrease, but at a much lower pace, thereby still maintaining a low (yet non-vanishing) level of cryptic circulation in the population (Fig. 3b), before starting to rise again due to the waning immunity. These conclusions remain qualitatively true when two other plausible choices of asymmetric cross-immunization are considered (Fig. S1).
Some reservations are in order when relying on the model predictions, especially since many factors are likely to dynamically change as time unfolds, for example the appearance of a new variant in the coming weeks. Moreover, boosting or applying new vaccines can reduce the waning immunity thus keeping the Delta at a low level. However, if the essence of the model predictions presented in Fig. 3b (as well as the other scenarios depicted in Fig. S1) will be proven accurate, the Delta variant has the potential of becoming far more problematic than the Omicron. With the expected significant decline in morbidity from all the recovered Omicron cases, the Israeli government and Ministry of Health will most likely remove various restrictions (i.e., crowding). In the meantime, the Delta, that is still circulating in a population with waning immunity and under less restrictions, may reemerge in larger numbers or even produce a new, different variant to generate infections in Israel.
4. Conclusions
Continuous monitoring of Israel's 4th largest city (Beer-Sheva) was performed through wastewater analysis to determine SARS-CoV-2 levels in wastewater. With escalated global infection by the Omicron variant of concern, it was expected to appear in the wastewater and eradicate the presence of its predecessor, the Delta variant of concern. For the purpose of quick, efficient and direct monitoring of the Omicron variant, without relying on complex sequencing methodology, we developed a primers-probe set for detection utilizing RT-qPCR assay. The developed Omicron primers-probe set was found to be highly specific and sensitive with an LOD of 1.4 copies per μL reaction even in a complex wastewater environment. The detection set has proven differentiation ability both in clinical and environmental samples.
The detection set developed in this study, together with a previously developed Delta detection set (Yaniv et al., 2021a) were employed on a single city's wastewater from the end of November 2021 until mid-January 2022. Even before the emergence of the Omicron variant, the Delta levels remained present in the wastewater with a low, constant signal. Indications for the Delta variant cryptic circulation in the population were further observed when the Omicron suddenly emerged at the end of December 2021. Despite an extremely high infection rate and increasing Omicron virus' prevalence in the population, where detection signal was multiplied by a 100 in just a week, Delta variant signals remained without a significant change.
We modeled the Delta-Omicron dynamics using a SIR model to predict their possible future behavior. The results obtained from the wastewater analysis were taken as a reference point, considering it is the most representative image on virus' prevalence status. The resulting model predictions might not necessarily come to pass, especially if other significant factors will shift unexpectedly in the near future. However, if correct, it is possible that the Omicron variant will diminish and disappear in the next 2 months, yet the Delta will keep circulating with minor levels and present an unnoticed risk. In a case where morbidity will rapidly decline over Omicron disappearance, public restrictions may be removed and result in the Delta variant reemergence or the rise of a new variant.
The study presented here emphasizes that once a new variant is identified as a possible risk factor, the development of an RT-qPCR assay for that specific variant is crucial for a more effective pandemic containment and assessment. Moreover, wastewater-based epidemiology may serve as a very informative tool both in times of high and low virus' levels, for better risk assessment and managing health policy. Finally, gaining early, representative data can help generate more accurate prediction models to improve and support pandemic management.
Funding sources
We would like to acknowledge funding from Ben Gurion University through The Corona Challenge Covid-19 and funding from the Israeli Ministry of Health.
CRediT authorship contribution statement
Karin Yaniv: Conceptualization, Writing – original draft, Visualization, Formal analysis. Eden Ozer: Conceptualization, Writing – original draft, Formal analysis. Marilou Shagan: Visualization. Yossi Paitan: Formal analysis, Writing – original draft. Rony Granek: Formal analysis, Modeling, Writing – original draft. Ariel Kushmaro: Conceptualization, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We thank Electra Company, which holds WWTP of Beer- Sheva management, especially Nikolay Schipunov. We gratefully acknowledge GISAID database for access to SARS-CoV-2 variants sequences. We gratefully acknowledge Esti Kramarsky-Winter assistance for comments and scientific editing of the manuscript.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.155599.
Appendix A. Supplementary data
Include primers, probes and DNA templates used in this study, as well as raw data for clinical and wastewater samples and in-depth explanation for SIR model development.
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. J. 2020;728(138764) doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761(144216) doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Kaya D., Bibby K., Simpson S.L., Bustin S.A., Shanks O.C., Ahmed W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021;203(117516) doi: 10.1016/j.watres.2021.117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona, Spain. Appl. Environ. Microbiol. 2021;87:1–9. doi: 10.1128/AEM.02750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Prot. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier J., Störmer M., Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 2005;43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel W., Kirstin M., Vedran F., Asija D., Theresa G.L., Roberto A., Filippos K., David K., Katja H., Salah A., Christopher B., Karen H., Anja R., Ivano L., Andranik I., Tommaso M., Simone D.G., Jan P., Samantha P., Meyer Thomas F., Alexander M.M., Daniela N., Andreas H., Matthias S., Altuna A., Nikolaus R., Christian D., Markus L. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience. 2021:102151. doi: 10.1016/j.isci.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster O., Beth-Din A., Asraf H., Levy V., Kabat A., Mannasse B., Azar R., Shifman O., Lazar S., Mandelboim M., Fleishon S., Mendelson E., Zuckerman N.S. Specific Detection of SARS-COV-2 B.1.1.529 (Omicron) Variant by Four RT-qPCR Differential Assays. medRxiv; 2021. pp. 1–12. [DOI] [Google Scholar]
- Espinosa P., Quirola-Amores P., Teran E. Application of a susceptible, infectious, and/or recovered (SIR) model to the COVID-19 pandemic in Ecuador. Front. Appl. Math. Stat. 2020;6:1–12. doi: 10.3389/fams.2020.571544. [DOI] [Google Scholar]
- European Centre for Disease Prevention and Control Assessment of the further spread and potential impact of the SARS-CoV-2 Omicron variant of concern in the EU/EEA, 19th update [WWW Document] 2022. https://www.ecdc.europa.eu/sites/default/files/documents/RRA-19-update-27-jan-2022.pdf
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10(100086) doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., Winkler M.S., Schulz S., Jäck H.M., Jahrsdörfer B., Schrezenmeier H., Müller M., Kleger A., Münch J., Pöhlmann S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Arora P., Sidarovich A., Moldenhauer A.-S., Winkler M.S., Schulz S., Jäck H.-M., Stankov M.V., Behrens G.M.N., Pöhlmann S. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli Ministry of Health Dashboard, n.d.Israeli Ministry of Health Dashboard [WWW Document], n.d. URL https://datadashboard.health.gov.il/COVID-19/general?utm_source=go.gov.il&utm_medium=referral
- Jewell B.L. Monitoring differences between the SARS-CoV-2 B.1.1.7 variant and other lineages. Lancet Public Health. 2021;6:e267–e268. doi: 10.1016/S2468-2667(21)00073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/s0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since december 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie M.T., Liu J., Sunshine S., Peng J., Black D., Mitchell A.M., Mann S.A., Pilarowski G., Zorn K.C., Rubio L., Bravo S., Marquez C., Sabatino J.J., Mittl K., Petersen M., Havlir D., DeRisi J. SARS-CoV-2 variant exposures elicit antibody responses with differential cross-neutralization of established and emerging strains including Delta and Omicron. J. Infect. Dis. 2022;1–16 doi: 10.1093/infdis/jiab635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Gu X., Armas F., Wu F., Chandra F., Chen H., Xiao A., Leifels M., Chua F.J.D., Kwok G.W., Tay J.Y., Lim C.Y., Thompson J., Alm E.J. Quantitative Detection of SARS-CoV-2 Omicron Variant in Wastewater Through Allele-specific RT-qPCR. medRxiv; 2022. pp. 1–17. [DOI] [Google Scholar]
- Leshem E., Gonen T., Hoffman T., Barsisat A., Kreiss Y., Regev-Yochay G. Low rate of transmission to triple-vaccinated contacts of an imported case of SARS-CoV-2 omicron infection: a contact tracing study in Israel. J. Travel Med. 2022 doi: 10.1093/jtm/taac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Klapsa D., Wilton T., Zambon M., Bentley E., Bujaki E., Fritzsche M., Mate R., Majumdar M. Tracking SARS-CoV-2 in sewage: evidence of changes in virus variant predominance during COVID-19 pandemic. Viruses. 2020;12:1144. doi: 10.3390/v12101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus‑2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mwalili S., Kimathi M., Ojiambo V., Gathungu D., Mbogo R. SEIR model for COVID-19 dynamics incorporating the environment and social distancing. BMC Res. Notes. 2020;13:1–5. doi: 10.1186/s13104-020-05192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., Harrison J.C., Demeke B.A., Zhou N.A., Beck N.K., Shirai J.H., Meschke J.S. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. J. 2021;760(144215) doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proverbio D., Kemp F., Magni S., Ogorzaly L., Cauchie H.-M., Gonçalves J., Skupind A., Aalto A. Model-based assessment of COVID-19 epidemic dynamics by wastewater analysis. Sci. Total Environ. 2022;827 doi: 10.1016/j.scitotenv.2022.154235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róka E., Khayer B., Kis Z., Kovács L.B., Schuler E., Magyar N., Málnási T., Oravecz O., Pályi B., Pándics T., Vargha M. Ahead of the second wave: early warning for COVID-19 by wastewater surveillance in Hungary. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22:2–4. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi R.K., Chen I.P., Ma T., Syed A.M., Simoneau C.R., Ciling A., Khalid M.M., Sreekumar B., Chen P.-Y., George A.F., Kumar G.R., Montano M., Garcia-Knight M.A., Brazer N., Saldhi P., Sotomayor-Gonzalez A., Servellita V., Gliwa A., Nguyen J., Silva I., Milbes B., Kojima N., Hess V., Shacreaw M., Lopez L., Brobeck M., Turner F., Soveg F.W., Fang X., Maishan M., Matthay M., Morris M.K., Wadford D., Hanson C., Greene W.C., Andino R., Spraggon L., Roan N.R., Chiu C.Y., Dounda J., Ott M. Limited Cross-variant Immunity After Infection With the SARS-CoV-2 Omicron Variant Without Vaccination. medRxiv; 2022. pp. 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsori Y., Granek R. Epidemiological model for the inhomogeneous spatial spreading of COVID-19 and other diseases. PLoS One. 2021;1–25 doi: 10.1371/journal.pone.0246056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsori Y., Granek R. Comparison of the Inhomogeneous SEPIR Model and Data From the COVID-19 Outbreak in South Carolina. medRxiv; 2021. pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A.J., Anand P., Lenehan P.J., Suratekar R., Raghunathan B., Niesen M.J.M., Soundararajan V. Omicron variant of SARS-CoV-2 harbors a unique insertion mutation of putative viral or human genomic origin. OSF Prepr. 2021:1–16. doi: 10.31219/osf.io/f7txy. [DOI] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Lewis Y., Kushmaro A. RT-qPCR assays for SARS-CoV-2 variants of concern in wastewater reveals compromised vaccination-induced immunity. Water Res. 2021;207 doi: 10.1016/j.watres.2021.117808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Shagan M., Lakkakula S., Plotkin N., Bhandarkar N.S., Kushmaro A. Direct RT-qPCR assay for SARS-CoV-2 variants of concern (Alpha, B.1.1.7 and Beta, B.1.351) detection and quantification in wastewater. Environ. Res. 2021;201 doi: 10.1016/j.envres.2021.111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Include primers, probes and DNA templates used in this study, as well as raw data for clinical and wastewater samples and in-depth explanation for SIR model development.