Abstract
Radiology reports are unstructured and contain the imaging findings and corresponding diagnoses transcribed by radiologists which include clinical facts and negated and/or uncertain statements. Extracting pathologic findings and diagnoses from radiology reports is important for quality control, population health, and monitoring of disease progress. Existing works, primarily rely either on rule-based systems or transformer-based pre-trained model fine-tuning, but could not take the factual and uncertain information into consideration, and therefore generate false positive outputs. In this work, we introduce three sedulous augmentation techniques which retain factual and critical information while generating augmentations for contrastive learning. We introduce RadBERT-CL, which fuses these information into BlueBert via a self-supervised contrastive loss. Our experiments on MIMIC-CXR show superior performance of RadBERT-CL on fine-tuning for multi-class, multi-label report classification. We illustrate that when few labeled data are available, RadBERT-CL outperforms conventional SOTA transformers (BERT/BlueBert) by significantly larger margins (6–11%). We also show that the representations learned by RadBERT-CL can capture critical medical information in the latent space.
Keywords: Thoracic Disorder, Contrastive Learning, Radiology Reports, Chest-Xray, Classification
1. Introduction
Chest radiography is a critical medical imaging technique used for diagnosis, screening, and treatment of many perilous diseases. Radiology reports are documented by radiologists after examining a patient’s medical history and diagnostic imaging, and represent complex anatomical and medical terms written for healthcare providers, along with indications of the presence or absence of any disease. Classifying radiology reports according to their description of abnormal findings is important for quality assurance and can mitigate the risks of diagnostic radiation exposure in children [24]. Additionally, the Precision Medicine Initiative (PMI) initiated by NIH and multiple research centers has highlighted the importance of text mining techniques to enable cohort pheno-typing of patients for population health (Shin et al., 2017). Classifying radiology reports can help to identify patient cohorts and enable precision medicine on a large scale. Labeling radiology reports with disease types can also assist in the development of deep learning applications for automated-diagnosis (Rajpurkar et al., 2017; Han et al., 2020; Yao et al., 2018).
ChestX-ray14 (Wang et al., 2017), MIMIC-CXR (Johnson et al., 2019), and OpenI (Demner-Fushman et al., 2016) are some of the largest radiology datasets available, and many classification algorithms have been developed based on the training sets provided by these datasets to classify reports into diseases. CheXpert (Irvin et al., 2019) is an automated rule-based labeler consisting of three stages: mention extraction, mention classification, and mention aggregation, to extract observations from the free text radiology reports to be used as structured labels for the images. CheXBert (Smit et al., 2020) uses the labels extracted by CheXpert to fine-tune BERT transformer along with ~ 1000 manually annotated reports to classify radiology reports. While these methods have shown great advancements, they cannot capture many critical and factual information (especially negated statements). Negated statements in a radiology report can lead to false positive classifications and therefore should be treated with caution. Also negated statements provide rich information that should be captured and integrated into the classification algorithms.
Motivated by the success of contrastive learning in computer vision (Chen et al., 2020a; He et al., 2020; Chen et al., 2020b; Grill et al., 2020; Robinson et al., 2020) to improve on the learning of feature representation in latent space, we propose to pre-train transformers using contrastive learning before the end-to-end fine-tuning for classification of radiology reports. Medical reports contain many critical and factual information such as the presence/absence of a disease (see Table 1 for more details). This information is central for making a classification decision, and many other downstream tasks such as Report Generation (Zhang et al., 2020a), Report Summarization (Zhang et al., 2020c), etc. Most existing approaches do not handle uncertainty/negation information explicitly, and depend on the deep learning models to capture them. We identified that the SOTA transformers such as Bert (Devlin et al., 2019), BlueBert (Peng et al., 2019), do not perform well at capturing uncertainity/negation information in latent space. Considering the significance of these critical information for both interpretability and performance improvement of deep learning models, we introduce RadBERT-CL, a pre-trained model using contrastive learning which can capture critical medical and factual nuances of radiology reports. It trains BlueBert (Peng et al., 2019) with the radiology report dataset and captures its fine-grained properties, in order to improve performance of report classification task at the fine-tuning stage. We introduce three novel data augmentation techniques at the sentence and document level, which can retain the critical medical concepts and factual information present in radiology reports while generating positive and negative pairs for contrastive learning.
Table 1:
Examples from the set of rules in our Info-Preservation Module for Negation and Uncertainty Detection and their corresponding matching sentences.
| BACKGROUND: Radiographic examination of the chest. clinical history: 80 years of age, male. PA AND LATERAL CHEST, ___ |
|
FINDINGS:
Heart size and mediastinal contours are normal. The right hilum is asymmetrically enlarged compared to the left hilum but has a similar size and configuration compared to a baseline radiograph___ ___. A chest CT performed in ___ demonstrated no evidence of a right hilum mass, and the observed asymmetry is probably due to a combination of a slight rotation related to mild scoliosis and a prominent pulmonary vascularity. Lungs are slightly hyperexpanded but grossly clear of pleural effusions. |
| IMPRESSION: No radiographic evidence of pneumonia. |
RadBERT-CL outperforms the previous best reported CheXbert labeler (Smit et al., 2020) with 0.5% improvement on F1-score without any need for hight quality manual annotation during training (note that the baseline (Smit et al., 2020) has claimed their results very close to human-level performance). We evaluated our system using 687 expert-annotated reports, same as CheXbert (Smit et al., 2020). We find that representations learned by RadBERT-CL are more informative, can capture and distinguish critical information present in the radiology reports. The improvements on F1-measure are more significant if few manually annotated data are available. This is particularly important since obtaining manually annotated data in medicine is extremely difficult and costly. In this case, our algorithm can achieve 6–11% improvements on disease classification. The highlights of our contributions are:
We propose two novel data augmentation techniques which retain factual and critical medical concepts, identified by our semi-rule based Info-Preservation Module, while generating positive and negative keys for contrastive learning.
We show that our model RadBERT-CL is able to learn and distinguish fine-grained medical concepts in latent space, which cannot be captured by SOTA pre-trained models like BERT, and BlueBert.
We apply contrastive learning for radiology report classification task and show improvements on the state-of-the-art methods. We use weakly-labeled data during our training and evaluate our system using 687 high-quality reports manually labelled by radiologists.
Lastly, we evaluate our model performance when a few data labels are available for training and show that our model outperforms significantly by 6–11% improvements in disease classification task.
2. Related Work
Contrastive Learning:
Contrastive learning (CL) seeks to learn effective representations by maximizing the agreement between two augmentations from one example and minimizing the agreement of augmentations from different instances. CL has been recently explored in computer vision and graph Neural Network due to its success in self-supervised representation learning. However, CL still receives limited interest in the NLP domain. The main reason is the discrete nature of text and it is hard to define and construct effective positive pairs. Several works have explored ways to perform augmentations. (Fang and Xie, 2020) back-translated source sentences to create sentence-level positive augmentations, which maintain semantic meaning of the source sentence. (Wu et al., 2020a) integrated four sentence-level augmentation techniques, namely word and span deletion, reordering and synonym substitution, to increase models’ robustness.
Factual Correctness and Consistency:
Factual correctness and factual consistency are key requirements for medical reports. Keeping factual information and avoiding hallucinations could support medical decision-making process. These requirements have been recently explored in NLP tasks, especially in abstractive text summarization. (Zhang et al., 2020b) directly took factual correctness as a training objective in their system via reinforcement learning. On the other hand, (Falke et al., 2019) and, (Goyal and Durrett, 2020) used textual entailment to detect factual inconsistency based on the assumption that summary should be entailed by the source document. (Zhu et al., 2021) built a knowledge graph containing all the facts in the text, and then fused it into the summarization process.
3. Methods
3.1. Problem Formulation
Radiology report classification is a multi-class multi-label classification problem, which classifies radiology reports into different disease observations (e.g., cardiomegaly, effusion, mass, edema). Following (Smit et al., 2020), we label each report rk in MIMIC-CXR dataset with a 14-dim vector y = [y1, y2, y3, …, y14] of observations, where observations y1 − y13 can take any value from the following 4 classes : blank, positive, negative, and uncertain. For y14, which corresponds to No Finding (no pathology is found in the scope of any of 13 disease observations), the classifier takes value from only 2 classes: blank, and uncertain.
3.2. Data Augmentation
In computer vision, it has been verified that contrastive learning benefits from strong data augmentation techniques like random cropping, rotation, blurring, color distortion, etc. However, in NLP, generating data augmentation is comparatively difficult due to the discrete representation of words, and it is unknown what kind of augmentation will benefit noise-invariant representational learning. (Fang et al., 2020) used back-translation to perform sentence augmentation while (Wu et al., 2020b) explored four different basic augmentation techniques: word and span deletion, reordering, and substitution. While these methods have shown improvements on some SentEval and GLUE benchmarks, they cannot be directly applied to generating augmentations for radiology reports. Radiology reports contain critical and factual information and that need to be preserved while generate augmentations. Table 1 presents an example of radiology report in which we have highlighted the information such as chest, left hilum, pulmonary vascularity, clear of, no evidence, pneumonia, etc.
Through augmentation, it is likely that (Wu et al., 2020b) dropped critical words or phrases which can lead to a completely different diagnosis. For example, dropping negation words, such as No, can lead to a diagnosis suggesting the presence of pleural effusion, and it can have negative consequences during our downstream task of disease classification. Also, as suggested by (Fang et al., 2020), back-translation cannot provide satisfactory results for the medical data because back-translation models have limited the cross-language translation ability for domain specific texts.
In order to ensure that critical and factual information is preserved while generating augmentations, we define an Info-Preservation module, which identifies and preserves facts during augmentation generation. We propose sentence-level and document-level augmentation techniques, to effectively pre-train our RadBERT-CL architecture.
3.2.1. Info-Preservation Module
Radiology reports consist of many important radiology concepts such as diseases, body parts, etc. In order to preserve them during augmentation, we develop a rule-based tool similar to Dynamic-LCS (Raj et al., 2020) to greedily match concepts in RadLex ontology (Langlotz, 2006) on sequences of the lemmatized tokens in the reports (longer matches are returned when possible). For capturing the presence of negation of any concept, we manually create a dictionary of 30 negation indicator keywords such as: not, without, clear of, ruled out, free of, disappearance of, without evidence of, no evidence of, absent, miss. Following (Chen et al., 2018), we create a dictionary of uncertainty keywords with a wide range of uncertain types, from speculations to inconsistencies present in the reports. We design a set of pattern matching rules following (Wang et al., 2017) for identifying sentences containing negation or uncertainty. Appendix Table 10 presents some examples of our rules and the matched sentences from the radiology reports. While generating augmentations, we make sure that any identified radiology concept or word from our negation and uncertainty list is not dropped.
3.2.2. Sentence-Level Augmentation
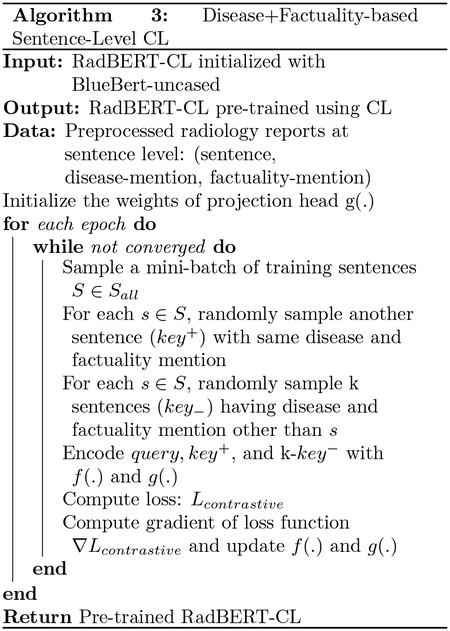
Sentence-level augmentations are generated by first splitting radiology reports into sentences and then applying random word and phrase dropping (Wu et al., 2020b), while preserving critical and factual information identified in Info - Preservation module. We propose two different augmentation techniques by associating each sentence with a disease concept from Radlex and a boolean variable indicating presence/absence of any negation or uncertainty phrase. Sentences without any mention of disease concepts are discarded.
Disease-based augmentation: In this technique, we discard all sentences which consist of any mention from our negation or uncertainty dictionary. For a given anchor sentence with disease concept d, any other sentence from any report mentioning d can be taken as positive example. Negative samples can be sentences which mention any disease concept except d. Refer Table 3 for the example.
Disease + Factuality based augmentation: In this technique, we consider any mention from our negation or uncertainty dictionary along with disease concept while generating augmentation pairs. For a given anchor sentence with disease concept d and negation or uncertainty present, any other sentence from any report mentioning d and negation or uncertainty present can be taken as positive example. Negative samples can be sentences which mention same disease d, but negation or uncertainty absent. Refer Table 3 for the example.
Table 3:
Examples highlighting the selection of positive and negative keys for a given anchor sentence using two different approaches for Sentence-Level Contrastive Learning. For Disease-based Augmentation, a given anchor sentence with disease concept d, any other sentence from any report mentioning d can be taken as positive example. In Disease + Factuality Based Augmentation, we incorporate mentions from our negation or uncertainty dictionary along with disease concept while generating augmentation pairs.
| a. Disease-based augmentation |
|---|
| Anchor/Query : definite focal consolidation is seen in left side of lungs |
| Positive Key : there is a focal consolidation at the left lung base adjacent to the lateral hemidiaphragm |
| Negative Key : there are low lung volumes and mild bibasilar atelectasis |
| b. Disease + Factuality based augmentation |
| Anchor/Query : definite focal consolidation is seen in left side of lungs |
| Positive Key : there is a focal consolidation at the left lung base adjacent to the lateral hemidiaphragm |
| Negative Key : the lungs are clear of any focal consolidation |

3.2.3. Document-Level Augmentation
Document-level augmentations are generated at the report-level, where each report is first pre-processed with removing extra spaces, newlines, and unwanted tokens. For a given report rk, we apply four types of augmentations (word deletion, span deletion, sentence reordering, and synonym substitution with probability 0.2) mentioned in (Wu et al., 2020b) while preserving critical and factual information identified in Info-Preservation module, to generate positive key. Negative keys can be any report not from the same patient.
3.3. Model Architecture
Our proposed model RadBERT-CL is a two-staged training process: pre-training and fine-tuning (Figure 1(a) and (b)). For pre-training, we follow Sim-CLR (Chen et al., 2020a) framework closely, and use BlueBert architecture as the encoder. Radiology reports are processed by Info-Preservation module and augmentations are generated using techniques proposed in Section 3.2. The augmentations are passed through the encoder f(.) and we take the CLS output of encoder and further pass it through the projection head g(.). Our projection heads consist of two MLP layers of size 768, along with non-linearity RELU and BatchNorm Layer. After pre-training we discard the projection head and use our pre-trained encoder for fine-tuning.
Figure 1:
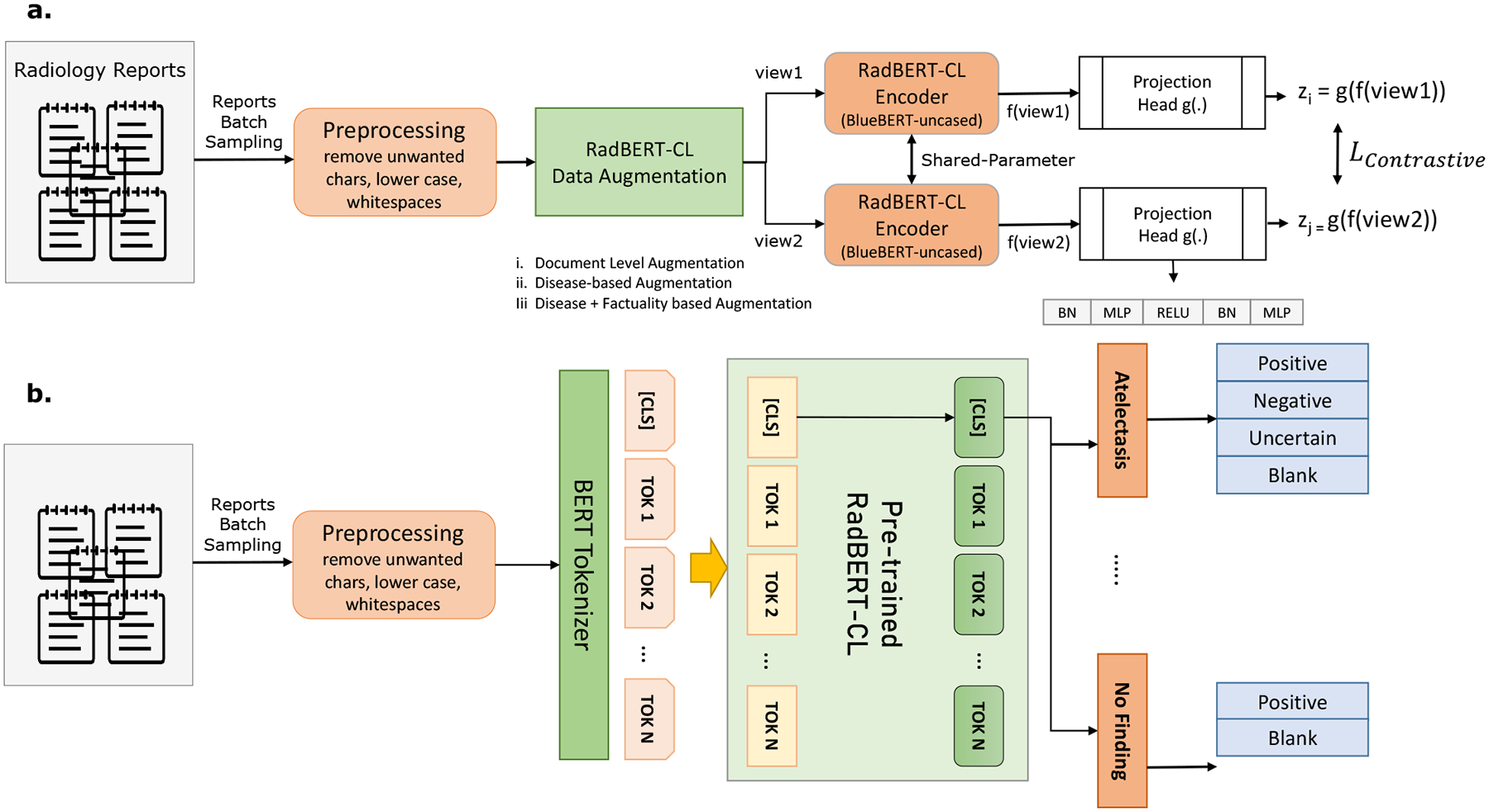
(a) Pre-training architecure of RadBERT-CL using contrastive learning. Two separate data augmentation views are generated using the augmentation techniques described in Section 3.2. Both views (query and key) are passed through RadBERT-CL, which is a transformer-based encoder f(.), and a projection head g(.). RadBERT-CL is trained to maximize agreement between the two augmented views using contrastive loss. (b) Fine-tuning Model architecture of RadBERT-CL. The model consists of 14 linear heads corresponding to 14 disease concepts. Among them, 13 linear heads can predict 4 outputs, while linear head corresponding to “No Finding” can predict 2 outputs.
3.4. Dataset
For the disease labelling task, we use MIMIC-CXR dataset (Johnson et al., 2019) which consists of 377, 110 chest-Xray images of 227, 827 patients along with their corresponding de-identified radiology reports. The dataset is pseudo-labeled using automatic labeler (Irvin et al., 2019) for the intended set of 14 observations using the entire body of the report.
In our study, we apply the contrastive pre-training by using the radiology reports from the entire MIMIC-CXR dataset for generating positive and negative augmentations. We divide our dataset into two parts for the fine-tuning stage after removing the duplicate reports of same patient: 80% for training, 20% for validation. Note that there is no patient overlap between the training and validation split. Additionally, we have a set of 687 reports belonging to 687 unique patients, similar to (Smit et al., 2020), which has been manually annotated by radiologists for the same 14 observations, and we evaluate our RadBERT-CL on this dataset.
3.5. Contrastive Pre-training
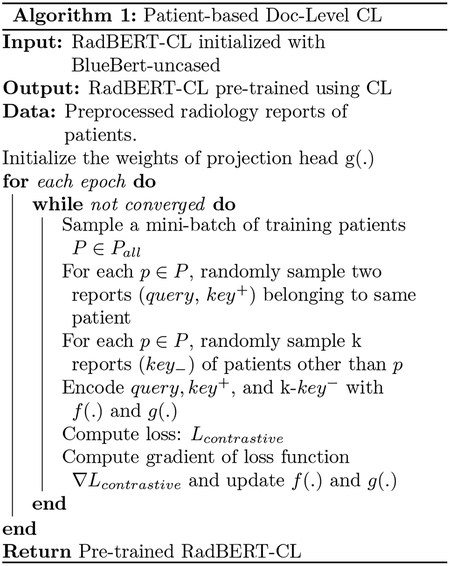
RadBERT-CL uses a transformer architecture similar to (Peng et al., 2019) and pre-trains it using contrastive self-supervised learning similar to (Chen et al., 2020a) on MIMIC-CXR dataset. Note that RadBERT-CL can be used on top of other language representation models and is not specific to (Peng et al., 2019). We propose three novel contrastive learning algorithms 1,2,3 with the help of augmentation techniques proposed in 3.2, which help RadBERT-CL to learn discriminative features across different medical concepts as well as factual cues. As shown in Figure 1(a), the augmentation views generated using techniques in 3.2, are passed through the our encoder RadBERT-CL f(.) and non-linear projection head g(.) to generate two 768-dimensional vectors zi = g(f(view1)) and zj = g(f(view2)). RadBERT-CL is pre-trained by maximizing the agreement between zi and zj using the contrastive loss similar to normalized temperature scaled cross-entropy loss (NT-Xent) (Chen et al., 2020a) defined as:
| (1) |
| (2) |
where τ is a temperature parameter, and num is the number of negative views. We calculate the loss for each sample in our mini-batch and sum them to estimate LContrastive. We calculate the gradient ∇Lcontrastive and back-propagate it to update our encoder f(.) and g(.). Contrastive learning benefits from training for larger epochs (He et al., 2020; Chen et al., 2020a; Grill et al., 2020), so we trained RadBERT-CL for 100 epochs using SGD optimizer. Note that after pre-training, we discard the project head g(.) and only use our encoder f(.) for fine-tuing on downstream task.
3.6. Supervised Fine-Tuning
In order to use the pre-trained RadBERT-CL model for our downstream task of report classification, we further fine-tune f(.) on the pseudo-labels of radiology report classification task as shown in Figure 1(b). Our disease is multi-class classification problem and We use cross-entropy loss as our supervised classification loss, defined as:
| (3) |
| (4) |
where, i denotes i − th training example, l denotes class label (Edema, Cardiomegaly, etc.), k ∈ {Positive, Negative, Uncertain, Blank}. We calculate the gradient ∇Lclassification and back-propagate it to update our encoder f(.).
4. Evaluation and Results
4.1. Evaluation
Following (Smit et al., 2020), we evaluate our system based on its average performance on three retrieval tasks: positive extraction, negative extraction, and uncertainty extraction. For each of the 14 observations, we compute a weighted average of the F1 scores on each of the above three tasks, weighted by the support for each class of interest, which we call the weighted-F1 metric. Table 4 presents the weighted-F1 score of RadBERT-CL using our three different variants of contrastive learning and their comparisons with SOTA methods. We have also presented the detailed evaluation score of our best RadBERT-CL variant (Algorithm 3) for all three retrieval tasks in Appendix Table 9.
Table 4:
The weighted F1 scores for fine-tuned RadBERT-CL variants using Model architecture in Figure 1 (a) and (b). We compare RadBERT-CL variants with SOTA models for reports classification CheXpert (Irvin et al., 2019), and CheXbert (Smit et al., 2020) trained on MIMIC-CXR. Reported F1-scores are calculated on the test set of 687 manually labelled reports, similar to (Smit et al., 2020). Note that our method does not require any expensive labeled reports during training. Our contrastive pre-training helps RadBERT-CL to outperform the baselines.
| Category | CheXpert | Previous SOTA CheXbert | Algorithm 1 RadBERT-CL | Algorithm 2 RadBERT-CL | Algorithm 3 RadBERT-CL |
|---|---|---|---|---|---|
| Enlarged Cardiom. | 0.613 | 0.713 | 0.692 | 0.717 | 0.690 |
| Cardiomegaly | 0.764 | 0.815 | 0.808 | 0.806 | 0.817 |
| Lung Opacity | 0.763 | 0.741 | 0.761 | 0.747 | 0.746 |
| Lung Lesion | 0.683 | 0.664 | 0.732 | 0.685 | 0.701 |
| Edema | 0.864 | 0.881 | 0.885 | 0.889 | 0.891 |
| Consolidation | 0.772 | 0.877 | 0.876 | 0.886 | 0.885 |
| Pneumonia | 0.684 | 0.835 | 0.838 | 0.846 | 0.847 |
| Atelectasis | 0.917 | 0.940 | 0.926 | 0.936 | 0.931 |
| Pneumothorax | 0.882 | 0.928 | 0.950 | 0.933 | 0.943 |
| Pleural Effuison | 0.905 | 0.919 | 0.920 | 0.926 | 0.913 |
| Pleural Other | 0.478 | 0.534 | 0.541 | 0.577 | 0.581 |
| Fracture | 0.671 | 0.791 | 0.791 | 0.796 | 0.791 |
| Supported Devices | 0.867 | 0.888 | 0.888 | 0.884 | 0.889 |
| No Finding | 0.543 | 0.640 | 0.580 | 0.588 | 0.615 |
| Average | 0.743 | 0.798 | 0.799 | 0.801 | 0.804 |
To demonstrate the effectiveness of RadBERT-CL performance when only a few labeled data is available, we evaluated RadBERT-CL performance in two different training scenarios: (a) pre-train RadBERT-CL using Algorithm 3 on 687 high-quality annotated dataset (no manually annotated label is used), fine-tune on randomly selected 400 high-quality annotated dataset, and test it on remaining 287 high-quality annotated dataset. (b) pre-train RadBERT-CL using Algorithm 3 on entire MIMIC CXR, fine-tune on randomly selected 400 high-quality annotated dataset, and test it on remaining 287 high-quality annotated dataset.
4.2. Results
We observe that our RadBERT-CL model pre-trained using Algorithm 3 outperforms previous state-of-the-art model CheXbert in 7 out of 14 findings after fine-tuning. Table 4 presents the weighted F1 scores of RadBERT-CL varients and previous SOTA systems CheXpert and CheXbert. Our model variants combined together outperform CheXbert in 11 out of 14 findings. Note that CheXbert training is calibrated under the supervision of ~ 1000 manually annotated reports by radiologists while our system is trained using weakly labeled reports. With the help of the guided-supervision of expert-level annotated data as proposed in CheXbert (Smit et al., 2020), we believe that our system will show more significant improvements.
In our analysis using Algorithm 1,2,3, we found that RadBERT-CL is very successful in capturing the factual information present in radiology reports. We calculated the cosine similarity between CLS embeddings generated by two factually different report snippets as shown in Table 6, by BERT, BlueBert and RadBERT-CL. RadBERT-CL is able to distinguish between the factual nuances of medical reports which are not captured in the representations generated by BERT and BlueBert.
Table 6:
Cosine Similarity between the normalized-[CLS] embeddings of report snippets generated by RadBERT-CL after contrastive pre-training. Our RadBERT-CL embeddings are capable of distinguishing between the factual nuances of medical reports which cannot be captured by the embeddings generated by BERT, and BlueBert. Our model is able to capture fine-grained differences among diseases, negation, and uncertainty in the latent representations.
| Report Segment | BERT | BlueBert | Algorithm 3 RadBERT-CL |
|---|---|---|---|
| … definite focal consolidation is seen in left side of lungs… | 0.9411 | 0.9223 | −0.8266 |
| … the lungs are clear of any focal consolidation … | |||
| … subtle opacity at the right base could represent infection … | 0.9120 | 0.9038 | 0.4332 |
| … patchy left base opacity represent severe infection … | |||
| … pleural effusion is obserevd … | 0.9752 | 0.8931 | 0.3836 |
| … pleural edema is seen … |
While deep learning methods often require expert-annotated high-quality data for training, getting sufficiently annotated data in the medical domain is very costly due to the limited availability of human experts. However, we have enough unlabelled data which can be used to improve our DL models with the supervision of few high-quality annotated data. Table 5 illustrates our RadBERT-CL performance in such scenario. Clearly, our model outperforms conventional fine-tuning using BERT/BlueBert for the classification task, by huge margins of 0.06 to 0.11 on weighted F1-metric. Better performance in Linear evaluation settings indicates that the representations learned by RadBERT-CL in pre-training stage are significantly better than BERT/BlueBert. Our experiments confirm that using largely available unsupervised data to pre-train transformers using contrastive learning provide significant improvement in fine-tuning tasks when few labelled data is available.
Table 5:
Transfer learning performance (F1-score) of RadBERT-CL, BERT, and BlueBERT when few labeled data is available. Fine-Tuning is done using randomly selected 400 reports and F1-score is reported on the remaining 287 reports of 687 high-quality manually annotated reports. Reported results are the mean F1-score of the 10 random training experiments and rounded to 3 decimal places. We identify significant improvements by RadBERT-CL in both Linear Evaluation setting (freeze encoder f(.) parameters and train the classifier layer), and Full-network Evaluation setting (train encoder f(.) and classifier layer end-to-end).
| Model | Linear Evaluation | Full-Network Evaluation |
|---|---|---|
| BERT-uncased | 0.137 ±0.012 | 0.477 ±0.009 |
| BlueBERT-uncased | 0.153 ±0.005 | 0.480 ±0.007 |
|
Algorithm 3 RadBERT-CL (pre-trained using 687 test reports) |
0.258 ±0.015 | 0.543 ±0.021 |
|
Algorithm 3 RadBERT-CL (pre-trained using Full MIMIC-CXR unlabelled data) |
0.282 ±0.011 | 0.591 ±0.019 |
5. Conclusion
In this work, we present novel data augmentation techniques for contrastive learning to capture factual nuances of medical domain. Our method involves pre-training transformers using abundance of unsupervised data to capture fine-grained domain knowledge before fine-tuning it for downstream tasks such as disease classification. We further show that such training strategy improves the performance in downstream tasks significantly in limited data settings. We hope that this work can draw community attention towards the ability of contrastive learning to capture discriminative properties in the medical domain.
Table 2:
Explanation of class value predicted by RadBERT-CL for disease observations
| Blank | observation not mentioned in the report |
| Positive | observation mentioned and its presence is confirmed eg. definite focal consolidation is seen in lungs |
| Negation | observation mentioned and its absence is confirmed eg. the lungs are clear of any focal consolidation |
| Uncertain | observation mentioned with uncertainty eg. signs of parenchymal changes suggesting pneumonia |
6. Appendix
Table 7:
Training details for RadBERT-CL Pretraining and Finetuning Stages.
| Hyperparameter | Pretraining | Finetuning |
|---|---|---|
| batch-size | 128 | 32 |
| learning-rate | 0.1 | 2e-5 |
| optimizer | SGD | Adam |
| temperature (CL) | 0.4 | - |
| n_epochs | 100 | 10 |
| beta | - | [0.9, 0.99] |
| Aug. Probability | 0.2 | - |
Figure 2:
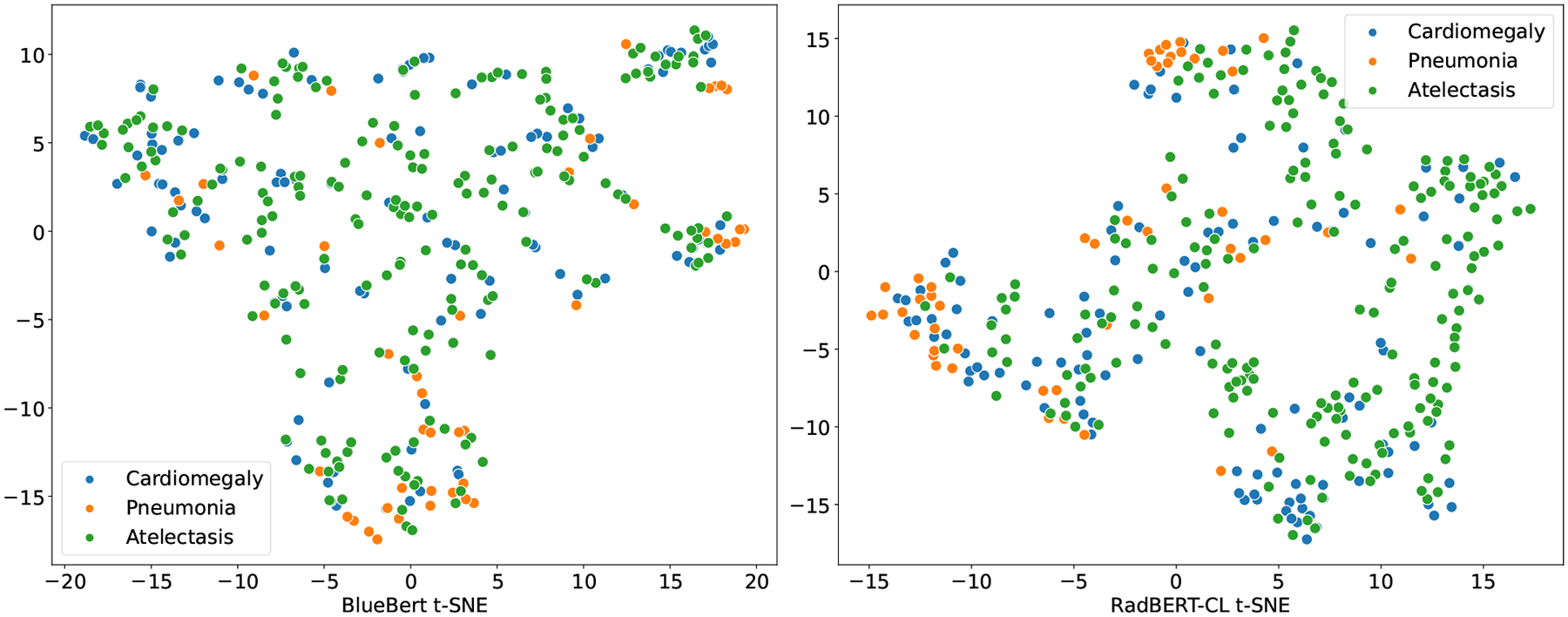
t-SNE visualization of BlueBert and RadBERT-CL(Algorithm 3) for radiology reports annotated positive for three major diseases (Cardiomegaly, Pneumonia, and Atelectasis). Note that the reports used for generating the t-SNE plot are sampled from 687 radiologists annotated test set which are not used in RadBERT-CL pre-training. From the figure, it is evident that embeddings generated after pre-training RadBERT-CL with contrastive learning, is more informative compared to BlueBert on unseen data.
Table 8:
Examples where RadBERT-CL incorrectly assign or misses label while making prediction. We include speculative reasoning for the classification errors.
| Report Snippet: … apparent new small right pleural edema manifested by posterior blunting of right costophrenic sulcus … |
| Prediction: Pleural Other |
| Ground Truth: Edema |
| Reasoning: the presence of pleural keyword along with edema may have confused the model to classify it as Pleural Other. |
| Report Snippet: … new area of pleural abnormality has developed in right side of lungs, and the heart and mediastinal structures and bony structures remain normal in appearance … |
| Prediction: Pleural Effusion |
| Ground Truth: Pleural Other |
| Reasoning: we found in reports that many pleural disorders share similar context which possibly make it difficult to classify them correctly. This can also explain the low F1-score of Pleural Other category. |
| Report Snippet: … mild interstitial edema and small right pleural effusion are new since ___… |
| Prediction: Pleural Effusion |
| Ground Truth: Pleural Effusion, Edema |
| Reasoning: the model misses to identify edema and only identified Pleural Effusion possibly because majority of times, edema is mentioned as Pleural Edema in reports. |
Table 9:
Detailed F1-evaluation of RadBERT-CL variant (Algorithm 3) for the classification tasks of positive extraction, negation extraction, uncertainty extraction, and blank for each of our 14 observations. Note that for ”Blank”, we have f1-scores related to positive extraction and blank, while the other two are set to zero.
| Category | Positive F1 | Negation F1 | Uncertain F1 | Blank F1 |
|---|---|---|---|---|
| Enlarged Cardiomediastinum | 0.579 | 0.786 | 0.831 | 0.965 |
| Cardiomegaly | 0.870 | 0.862 | 0.433 | 0.978 |
| Lung Opacity | 0.820 | 0.200 | 0.512 | 0.910 |
| Lung Lesion | 0.777 | 0.571 | 0.211 | 0.983 |
| Edema | 0.913 | 0.901 | 0.745 | 0.993 |
| Consolidation | 0.909 | 0.824 | 0.876 | 0.997 |
| Pneumonia | 0.786 | 0.916 | 0.807 | 0.991 |
| Atelectasis | 0.962 | 0.444 | 0.874 | 0.999 |
| Pneumothorax | 0.850 | 0.971 | 0.526 | 0.996 |
| Pleural Effuison | 0.938 | 0.957 | 0.596 | 0.985 |
| Pleural Other | 0.623 | 0.234 | 0.114 | 0.981 |
| Fracture | 0.894 | 0.333 | 0.667 | 0.993 |
| Supported Devices | 0.902 | 0.100 | 0.000 | 0.942 |
| No Finding | 0.592 | 0.000 | 0.000 | 0.978 |
Table 10:
Examples from the set of rules in our Info-Preservation Module for Negation and Uncertainty Detection and their corresponding matching sentences.
| a. Negation Detection |
|---|
| RULE: * + clear/free/disappearance + <prep_of> + * + DISEASE_CONCEPT |
| 1. the left lung is free of consolidations or pneumothorax |
| 2. the lungs are clear of any focal consolidation |
| 3. pleural sinuses are free of any fluid accumulation |
| RULE: * + no/not + evidence/ * + <prep_[of |for] > + * + DISEASE_CONCEPT |
| 1. within the remaining well-ventilated lung, there is no evidence of pneumonia |
| 2. there is not evidence for pulmonary edema |
| 3. there are no evidences of acute pneumothorax |
| b. Uncertainty Detection |
| RULE: * + couldbe/maybe/… + * + DISEASE_CONCEPT |
| 1. there are bibasilar opacities which could be due to atelectasis given low lung volumes |
| 2. perihilar opacity could be due to asymmetrical edema |
| 3. left base opacity may be due to atelectasis |
| RULE: * + suggest/suspect/[−ing| − ed] + * + DISEASE_CONCEPT |
| 1. signs of parenchymal changes suggesting pneumonia |
| 2. the left heart border is silhouetted, with a suspected left basilar opacity |
| 3. prominence of the central pulmonary vasculature suggesting mild pulmonary edema |
References
- Chen Chaomei, Song Min, and Heo Go Eun. A scalable and adaptive method for finding semantically equivalent cue words of uncertainty. Journal of Informetrics, 12, Feb 2018. ISSN 1751–1577. doi: 10.1016/j.joi.2017.12.004. [DOI] [Google Scholar]
- Chen Ting, Kornblith Simon, Norouzi Mohammad, and Hinton Geoffrey. A simple framework for contrastive learning of visual representations, 2020a.
- Chen Xinlei, Fan Haoqi, Girshick Ross, and He Kaiming. Improved baselines with momentum contrastive learning, 2020b.
- Demner-Fushman Dina, Kohli M, Rosenman M, Shooshan SE, Rodriguez Laritza, Antani S, Thoma G, and McDonald C. Preparing a collection of radiology examinations for distribution and retrieval. Journal of the American Medical Informatics Association : JAMIA, 23 2:304–10, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. Bert: Pre-training of deep bidirectional transformers for language understanding, 2019. [Google Scholar]
- Falke Tobias, Ribeiro Leonardo F. R., Prasetya Ajie Utama Ido Dagan, and Gurevych Iryna. Ranking generated summaries by correctness: An interesting but challenging application for natural language inference. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics, 2019. doi: 10.18653/v1/p19-1213. URL 10.18653/v1/p19-1213. [DOI] [Google Scholar]
- Fang Hongchao and Xie Pengtao. Cert: Contrastive self-supervised learning for language understanding. CoRR, abs/2005.12766, 2020. URL https://arxiv.org/abs/2005.12766. [Google Scholar]
- Fang Hongchao, Wang Sicheng, Zhou Meng, Ding Jiayuan, and Xie Pengtao. Cert: Contrastive self-supervised learning for language understanding, 2020.
- Goyal Tanya and Durrett Greg. Evaluating factuality in generation with dependency-level entailment. In Findings of the Association for Computational Linguistics: EMNLP 2020. Association for Computational Linguistics, 2020. doi: 10.18653/v1/2020.findings-emnlp.322. URL 10.18653/v1/2020.findings-emnlp.322. [DOI] [Google Scholar]
- Grill Jean-Bastien, Strub Florian, Altché Florent, Tallec Corentin, Richemond Pierre H, Buchatskaya Elena, Doersch Carl, Pires Bernardo Avila, Guo Zhaohan Daniel, Azar Mohammad Gheshlaghi, et al. Bootstrap your own latent: A new approach to self-supervised learning. arXiv preprint arXiv:2006.07733, 2020. [Google Scholar]
- Han Yan, Chen Chongyan, Tang Liyan, Lin Mingquan, Jaiswal Ajay, Wang Song, Tewfik Ahmed, Shih George, Ding Ying, and Peng Yifan. Using radiomics as prior knowledge for thorax disease classification and localization in chest x-rays. arXiv preprint arXiv:2011.12506, 2020. [PMC free article] [PubMed] [Google Scholar]
- He Kaiming, Fan Haoqi, Wu Yuxin, Xie Saining, and Girshick Ross. Momentum contrast for unsupervised visual representation learning, 2020.
- Irvin Jeremy, Rajpurkar Pranav, Ko Michael, Yu Yifan, Silviana Ciurea-Ilcus Chris Chute, Marklund Henrik, Haghgoo Behzad, Ball Robyn, Shpanskaya Katie, Seekins Jayne, Mong David A., Halabi Safwan S., Sandberg Jesse K., Jones Ricky, Larson David B., Langlotz Curtis P., Patel Bhavik N., Lungren Matthew P., and Ng Andrew Y.. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison, 2019.
- Johnson Alistair E. W., Pollard Tom J., Greenbaum Nathaniel R., Lungren Matthew P., Chih ying Deng Yifan Peng, Lu Zhiyong, Mark Roger G., Berkowitz Seth J., and Horng Steven. Mimic-cxr-jpg, a large publicly available database of labeled chest radiographs, 2019. [DOI] [PMC free article] [PubMed]
- Langlotz C. Radlex: a new method for indexing online educational materials. Radiographics : a review publication of the Radiological Society of North America, Inc, 26 6:1595–7, 2006. [DOI] [PubMed] [Google Scholar]
- Peng Yifan, Yan Shankai, and Lu Zhiyong. Transfer learning in biomedical natural language processing: An evaluation of bert and elmo on ten benchmarking datasets, 2019.
- Raj Mayank, Jaiswal Ajay, Rohit RR, Gupta Ankita, Sahoo Sudeep Kumar, Srivastava Vertika, and Kim Yeon Hyang. Solomon at semeval-2020 task 11: Ensemble architecture for fine-tuned propaganda detection in news articles. In Proceedings of the Fourteenth Workshop on Semantic Evaluation, pages 1802–1807, 2020. [Google Scholar]
- Rajpurkar Pranav, Irvin Jeremy, Zhu Kaylie, Yang Brandon, Mehta Hershel, Duan Tony, Ding Daisy, Bagul Aarti, Langlotz Curtis, Shpanskaya Katie, Lungren Matthew P., and Ng Andrew Y.. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning, 2017.
- Robinson Joshua, Chuang Ching-Yao, Sra Suvrit, and Jegelka Stefanie. Contrastive learning with hard negative samples. arXiv preprint arXiv:2010.04592, 2020. [Google Scholar]
- Bonggun Shin, Chokshi F, Lee Timothy, and Choi Jinho D.. Classification of radiology reports using neural attention models. 2017 International Joint Conference on Neural Networks (IJCNN), pages 4363–4370, 2017. [Google Scholar]
- Smit Akshay, Jain Saahil, Rajpurkar Pranav, Pareek Anuj, Ng Andrew Y., and Lungren Matthew P.. Chexbert: Combining automatic labelers and expert annotations for accurate radiology report labeling using bert, 2020.
- Wang Xiaosong, Peng Yifan, Lu Le, Lu Zhiyong, Bagheri Mohammadhadi, and Summers Ronald M.. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Jul 2017. doi: 10.1109/cvpr.2017.369. URL . [DOI] [Google Scholar]
- Wu Zhuofeng, Wang Sinong, Gu Jiatao, Khabsa Madian, Sun Fei, and Ma Hao. Clear: Contrastive learning for sentence representation, 12 2020a.
- Wu Zhuofeng, Wang Sinong, Gu Jiatao, Khabsa Madian, Sun Fei, and Ma Hao. Clear: Contrastive learning for sentence representation, 2020b.
- Yao Li, Poblenz Eric, Dagunts Dmitry, Covington Ben, Bernard Devon, and Lyman Kevin. Learning to diagnose from scratch by exploiting dependencies among labels, 2018.
- Zhang Yixiao, Wang Xiaosong, Xu Ziyue, Yu Qihang, Yuille Alan, and Xu Daguang. When radiology report generation meets knowledge graph, 2020a.
- Zhang Yuhao, Merck Derek, Tsai Emily, Manning Christopher D., and Langlotz Curtis. Optimizing the factual correctness of a summary: A study of summarizing radiology reports. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics, 2020b. doi: 10.18653/v1/2020.acl-main.458. URL 10.18653/v1/2020.acl-main.458. [DOI] [Google Scholar]
- Zhang Yuhao, Merck Derek, Emily Tsai, Manning Christopher D., and Langlotz Curtis P.. Optimizing the factual correctness of a summary: A study of summarizing radiology reports, 2020c.
- Zhu Chenguang, Hinthorn William, Xu Ruochen, Zeng Qingkai, Zeng Michael, Huang Xuedong, and Jiang Meng. Enhancing factual consistency of abstractive summarization. North American Chapter of the Association for Computational Linguistics (NAACL) 2021, June 2021. [Google Scholar]


