Abstract
The emergence of machine learning-based in silico tools has enabled rapid and high-quality predictions in the biomedical field. In the COVID-19 pandemic, machine learning methods have been used in many topics such as predicting the death of patients, modeling the spread of infection, determining future effects, diagnosis with medical image analysis, and forecasting the vaccination rate. However, there is a gap in the literature regarding identifying epitopes that can be used in fast, useful, and effective vaccine design using machine learning methods and bioinformatics tools. Machine learning methods can give medical biotechnologists an advantage in designing a faster and more successful vaccine. The motivation of this study is to propose a successful hybrid machine learning method for SARS-CoV-2 epitope prediction and to identify nonallergen, nontoxic, antigen peptides that can be used in vaccine design from the predicted epitopes with bioinformatics tools. The identified epitopes will be effective not only in the design of the COVID-19 vaccine but also against viruses from the SARS family that may be encountered in the future. For this purpose, epitope prediction performances of random forest, support vector machine, logistic regression, bagging with decision tree, k-nearest neighbor and decision tree methods were examined. In the SARS-CoV and B-cell datasets used for education in the study, epitope estimation was performed again after the datasets were balanced with the synthetic minority oversampling technique (SMOTE) method since the epitope class samples were in the minority compared to the nonepitope class. The experimental results obtained were compared and the most successful predictions were obtained with the random forest (RF) method. The epitope prediction performance in balanced datasets was found to be higher than that in the original datasets (94.0% AUC and 94.4% PRC for the SMOTE-SARS-CoV dataset; 95.6% AUC and 95.3% PRC for the SMOTE-B-cell dataset). In this study, 252 peptides out of 20312 peptides were determined to be epitopes with the SMOTE-RF-SVM hybrid method proposed for SARS-CoV-2 epitope prediction. Determined epitopes were analyzed with AllerTOP 2.0, VaxiJen 2.0 and ToxinPred tools, and allergic, nonantigen, and toxic epitopes were eliminated. As a result, 11 possible nonallergic, high antigen and nontoxic epitope candidates were proposed that could be used in protein-based COVID-19 vaccine design (“VGGNYNY”, “VNFNFNGLTG”, “RQIAPGQTGKI”, “QIAPGQTGKIA”, “SYECDIPIGAGI”, “STFKCYGVSPTKL”, “GVVFLHVTYVPAQ”, “KNHTSPDVDLGDI”, “NHTSPDVDLGDIS”, “AGAAAYYVGYLQPR”, “KKSTNLVKNKCVNF”). It is predicted that the few epitopes determined by machine learning-based in silico methods will help biotechnologists design fast and accurate vaccines by reducing the number of trials in the laboratory environment.
Keywords: SARS-CoV-2, SARS-CoV, B-cell, Machine learning, In silico, Vaccine design
Graphical Abstract
1. Introduction
SARS-CoV-2 is a new type of coronavirus that presents with influenza-like symptoms in humans. Coronaviruses are viruses that typically have spikes in the surface region (Guo et al., 2020, Rabi et al., 2020). These pointed structures allow the virus to attach to the target cell. The coronavirus family is classified into 4 groups according to its genetic structure: alpha, beta, gamma and delta. Alpha and beta strains can infect mammalian species.
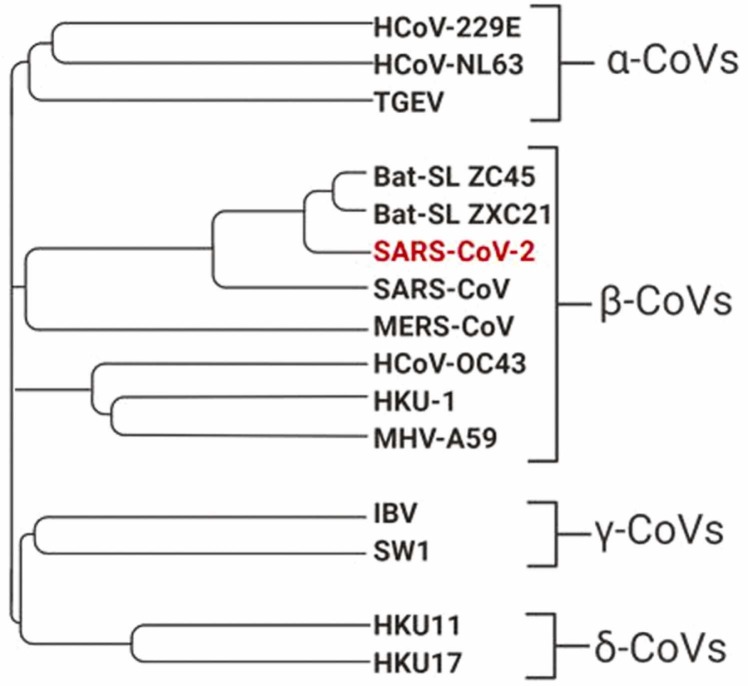
The genetic information of the nCoV-19 virus was identified and uploaded to GenBank (Zhu et al., 2020). SARS-CoV (severe acute respiratory syndrome) and MERS-CoV (Middle East respiratory syndrome) are also deadly coronaviruses that have emerged in recent years. The phylogenetic tree of the known coronavirus family is given in Fig. 1. It is clear that SARS-CoV, SARS-CoV-2, and MERS-CoV descended from the same ancestor (Misbah et al., 2020). SARS-CoV is the coronavirus most similar to SARS-CoV-2. The genome similarity of the two viruses has been reported to be 70% (Misbah et al., 2020).
Fig. 1.
Phylogenetic tree of SARS-CoV-2 (Misbah et al., 2020).
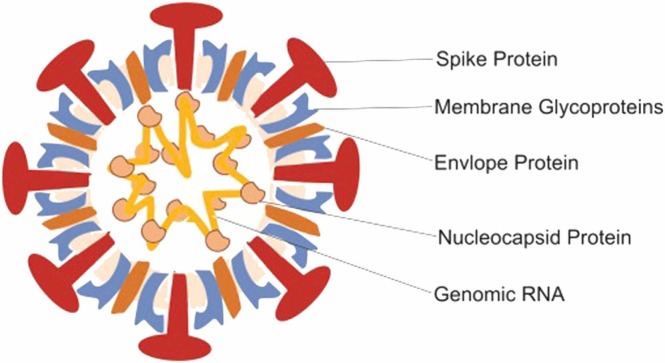
Similar to SARS-CoV, SARS-CoV-2 uses the antigen-converting enzyme 2 receptor, which is located in the lower respiratory tract of humans and allows human-to-human spread, to enter the target cell (Zhou et al., 2020, Gorbalenya et al., 2020). SARS-CoV-2 is a 29.9 kb, single-stranded RNA virus (Zhu et al., 2020). Similar to other coronaviruses, SARS-CoV-2 contains open reading frames in its genome. Approximately one-third of the entire virus genome encodes 4 basic structural proteins. These proteins include nucleocapsid, spike, envelope and membrane proteins (Mousavizadeh and Ghasemi, 2020). It is the nucleocapsid protein that holds the genome of the virus. As Fig. 2 shows, spike proteins are located on the outer surface of the virus. This protein, which is effective in identifying the host cell, allows the virus to attach to the membrane of the target cell. After the virus binds to the host cell, proteases present in that cell open the spike protein of the virus, revealing a fusion peptide. Thus, the RNA of the virus disperses into the cell and allows it to spread to more cells by replicating itself (Hoffmann et al., 2020). This whole process shows that the spike protein plays an important role in the entry of the virus into the cell. Therefore, vaccine studies have focused on the spike protein.
Fig. 2.
Structure of SARS-CoV-2 (Hosseini et al., 2020).
Since SARS-CoV-2 is a new virus and the vaccine and treatment methods are unknown, many people have died due to the virus. When the course of the disease is followed, it is seen that elderly individuals and people with weak immune systems have a more severe disease and are more likely to die. A weak immune system causes cells to be less able to fight and repair themselves (Yang et al., 2020).
The immune system is the body's defense mechanism against all external factors such as viruses, germs or harmful substances. Although immune system cells are spread throughout the body, they are more concentrated in immune system organs such as the spleen, thymus, lymph node and bone marrow. When a foreign substance such as a virus enters the body or a cancer cell develops, the immune system begins to produce substances called antibodies to destroy them. Foreign substances are targeted and fight antibodies until they are destroyed. Since it has a kind of memory, the immune system uses every experience in the next fight (Delves and Roitt, 2000).
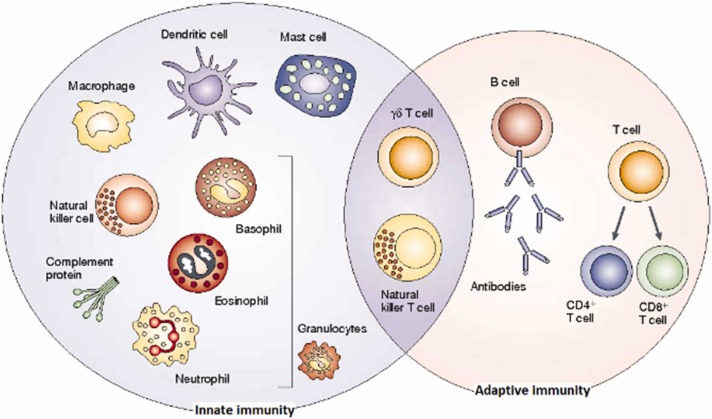
As shown in Fig. 3, there are 2 separate response mechanisms in the immune system, innate and adaptive. When innate immunity encounters microbes and viruses, it quickly steps in and creates the first immune responses. This response recognizes specific molecules carried by microorganisms, but is not agent specific. Since the innate immune system has no memory, it exhibits the same reaction in every encounter (Medzhitov and Janeway, 2000). Adaptive immunity, on the other hand, is acquired in various ways, such as disease or externally administered vaccines and serum. Adaptive immune systems have memories so they can remember pathogens they have encountered before. Therefore, adaptive immune systems produce antigen-specific responses. Lymphocytes, a type of white blood cell produced in the bone marrow, can recognize and destroy disease agents (Cooper and Alder, 2006).
Fig. 3.
The innate and adaptive immune response (Dranoff, 2004).
There are 2 types of adaptive immune responses, humoral and cellular. The humoral response is elicited by proteins called antibodies formed by B lymphocytes. When B lymphocytes encounter an antigen, they produce antibodies that can target the antigen. Antibodies are a type of chemical substance called immunoglobulins. They are responsible for the elimination of extracellular pathogens(Pathak and Palan, 2005).
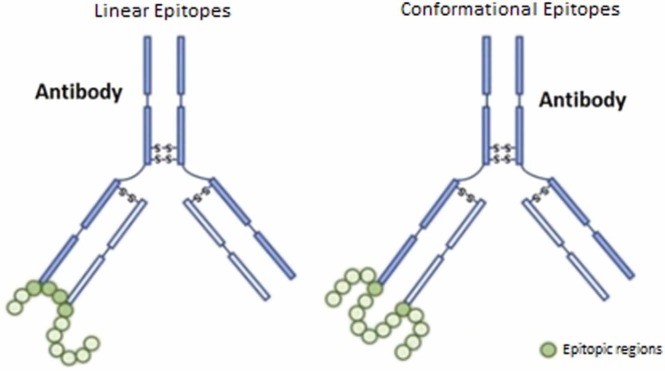
B-cells are responsible for the synthesis of antibody molecules called immunoglobulins and have a Y-shaped structure. B-cells have specific antigen receptors called B-cell receptors (BCR) on their surface. These receptors bind pathogens with immunoglobin molecules. A region of antigen recognized by a particular antibody or B-cell is called a B-cell epitope (Ansari and Raghava, 2010). As shown in Fig. 4, there are 2 types of B-cell epitopes: continuous (linear) and discontinuous. Linear epitopes consist of continuous residues found in the antigen protein sequence. That is why it is also called a continues epitope. Discontinuous or conformational epitopes, in contrast, consist of noncontiguous residues in the antigen sequence (Sanchez-Trincado et al., 2017). Both epitopes play an important role in peptide-based vaccine studies. However, linear B-cell epitope estimation is performed within the scope of the study, since linear B-cell epitopes consist of peptides that can be used more easily to replace antigens for immunity and antibody production.
Fig. 4.
Linear and conformational epitopes (Melo et al., 2018).
The Immune Epitope Database (IEDB) is a publicly available database containing experimentally validated T- and B-cell epitope data presented in the literatüre (Vita et al., 2019). It also includes epitopes identified for specific viruses such as SARS-CoV and MERS. Since SARS-CoV-2 is a new virus, limited information is available for vaccine and drug studies. However, when the structural proteins of SARS-CoV and SARS-CoV-2 are compared, as shown in Fig. 5, it is seen that the spike and nucleocapsid proteins are largely preserved for both viruses. This similarity shows that SARS-CoV data can be used in peptide-based vaccine studies (Chen et al., 2020). Considering this similarity, epitope prediction was made for SARS-CoV-2, the spike protein. SARS-CoV and linear B-cell epitope information from IEDB were used to create the model.
Fig. 5.
Phylogenetic tree for structural proteins (Chen et al., 2020).
Although epitope data are not yet available for SARS-CoV-2, since the gene and protein sequence information is known, the characteristics of the virus and the epitopes in the pathogen can be predicted by machine learning-based in silico methods (Tahir ul Qamar et al., 2019). There are many studies in the literature on predicting the death of patients diagnosed with SARS-CoV-2 using machine learning/artificial intelligence algorithms, modeling the spread of infection (Ceylan, 2020, Cihan, 2022), diagnosis with medical image analysis (Saygılı, 2021), and forecasting the COVID-19 vaccination rate (Cihan, 2021, Zhou and Li, 2022).
Producing vaccines against infectious diseases by conventional methods has proven time-consuming and very expensive. Vaccine candidates have been effectively identified in previous viruses (HPV, Ebola, Zika, and MERS) using in silico methods (Yazdani et al., 2020). However, there is a gap in the literature on determining epitopes that can be used in vaccine design by using machine learning-based in silico methods and bioinformatics tools. Designing a useful and effective vaccine against new mutant viruses which escape COVID-19 vaccines or different viruses that may emerge in the future will be one of the biggest challenges scientists can face. As such, the determination of vaccine candidates with the traditional method in silico predictions is very important because of limited time and resources (Yazdani et al., 2020).
Sohail et al. (2021) used two in silico methods for T-cell epitope prediction. These are prediction methods based on SARS-CoV immunological data and peptide-HLA binding prediction methods due to genetic similarity. Quadeer et al. (2020) presented positive T-cell immune responses against epitopes containing COVID-19 proteins from blood samples of COVID-19 patients. The authors compared the epitopes obtained by in vitro methods with the predicted epitopes and found that the methods using SARS-CoV immunological data were more in line with the experimental results in general.
Due to genetic similarity in our study, we aimed to predict the B-cell epitope for SARS-CoV-2 from SARS-CoV immunological data. In this context, there are a limited number of studies in the literature. While some researchers have performed epitope prediction on protein sequences, others have tried to identify candidate epitopes using protein and epitope sequence features. Grifoni et al. (2020) utilized bioinformatics tools for B- and T-cell epitope prediction for SARS-CoV-2. The BepiPred 2.0 tool (Jespersen et al., 2017) for linear B-cell epitopes and the Discotope 2.0 tool (Kringelum et al., 2012) for conformational B-cell epitope prediction were used. Chen et al. (2020) aimed to predict B-cell epitopes in the spike protein of SARS-CoV-2 and T-cell epitopes in the nucleocapsid protein. In the study, they determined the conserved regions of the virus by aligning the SARS-CoV-2 protein sequences obtained from the NCBI database with Clustal Omega. BepiPred and ABCPred (Saha and Raghava, 2006) tools were used for linear B-cell epitope prediction. Estimation of T-cell epitopes in the nucleocapsid protein was made with the free online tool provided by IEDB. Shoukat et al. (2021) proposed a method to classify T-cell responses by analyzing TCR beta information from people infected and uninfected with COVID-19. The proposed method aimed to detect protective immunity acquired through natural infection or vaccine-induced immunity. PCA and hierarchical clustering were applied to the sequence data separated into K-mers. Since the number of samples in the used dataset is small, the dataset is divided with hold one out. Accordingly, an accuracy value of 96% was obtained in the training data and 92.9% in the test data. SARS-CoV B-cell linear epitope prediction was performed using the Bayesian neural network classification method by Ghoshal et al. (2021). In this study, 85% prediction accuracy was obtained for the SARS-CoV dataset. Aleatoric and epistemic uncertainty methods were used to measure the uncertainty in epitope estimation. In this study, only SARS-CoV epitope prediction was carried out, and no prediction was made for SARS-CoV-2. Noumi et al. (2021) used the attentional mechanism LSTM network for epitope prediction. The results obtained were compared with the epitope sequences predicted by BepiPred 2.0 for the same protein sequences. In this study, the epitope peptide length was limited to 8–14 amino acids. The highest accuracy value was obtained as 0.79 for the case where the peptide length was 12. Jain et al. (2021) made epitope predictions for SARS-CoV by using various machine learning methods and epitope and peptide properties. In this study, the dataset containing B-cell epitopes was used to develop the model, and the SARS-CoV dataset was used for testing. The most successful result was obtained with the ensemble learning model with an accuracy value of 87%.
The limited number of studies available on this topic is based on either analysis of protein sequences with bioinformatics tools or prediction using sequence features. Higher accuracy estimates are needed for the proposed epitope regions to be used as vaccine candidates. The motivation of this study is to propose a new and successful hybrid machine learning approach for SARS-CoV-2 by using physico-chemical and sequence-based features in proven datasets for SARS-CoV in combination with feature engineering and data preprocessing. We aimed to determine nonallergen, high antigen and nontoxic epitopes among them by performing bioinformatic analyses for the predicted epitopes. This study presents the following contributions:
-
•
To examine and compare the epitope prediction performances of machine learning methods in SARS-CoV and B-cell datasets.
-
•
To compare the prediction performance of the methods for the original SARS-CoV and B-cell datasets vs. the dataset balanced by the SMOTE method.
-
•
Identifying epitopes in the SARS-CoV-2 spike protein dataset with the proposed SMOTE-RF-SVM method.
-
•
To analyze epitopes determined by machine learning methods using AllerTop, VaxiJen and ToxinPred bioinformatics tools.
-
•
To determine probable nonallergenic, highly antigenic and nontoxic epitopes that can be used in vaccine design against SARS-CoV-2.
It is anticipated that the findings obtained from this study can be used to design a fast, reliable and cost-effective vaccine, especially against SARS-CoV-2 and other viruses in the SARS family.
2. Materials and methods
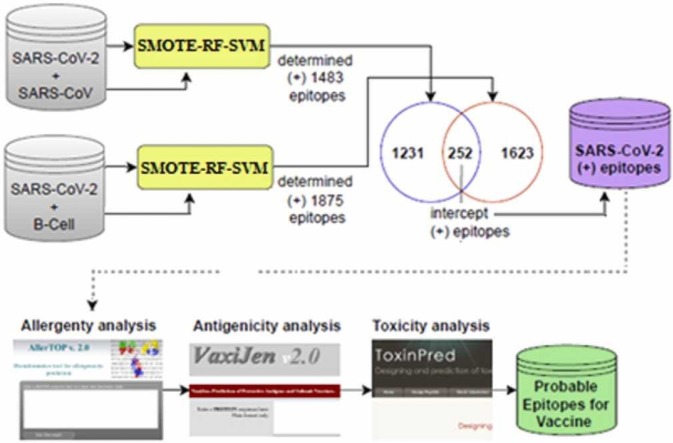
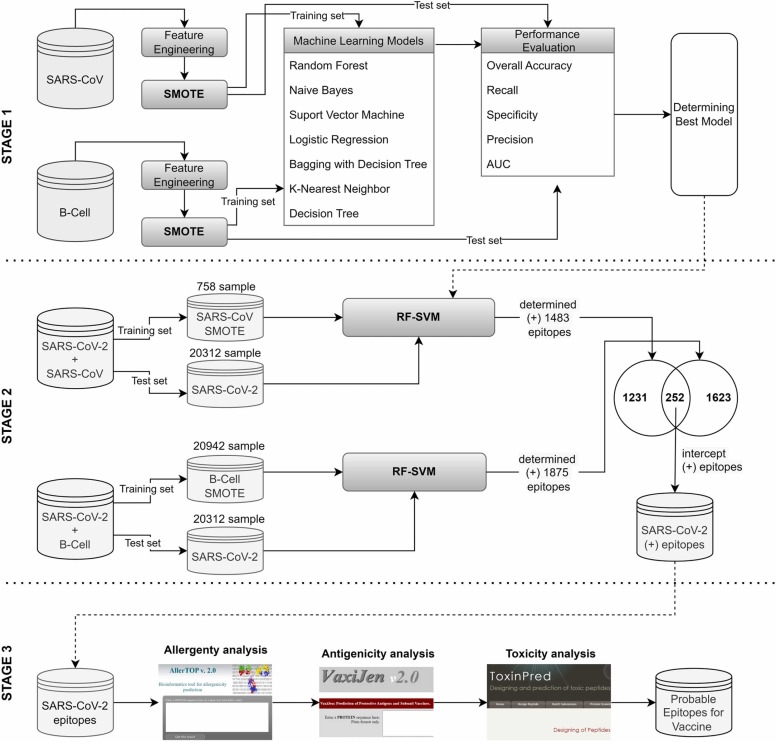
The flowchart followed in this study to identify epitopes that can be used in vaccine design is illustrated in Fig. 6. In the first stage, the performances of different machine learning methods for determining epitopes in the original datasets (SARS-CoV, B-cell) and SMOTE datasets are examined and compared. In the second step, the SARS-CoV-2 epitopes were predicted after the proposed hybrid method was trained with SARS-CoV and B-cell datasets. In the third stage, epitopes determined by machine learning methods are analyzed with bioinformatic tools (AllerTop, VaxiJen, ToxinPred), and probable nonallergen, antigen, and nontoxic epitopes are selected that can be used in the vaccine. Within the scope of our study, analyses and algorithms were developed using R programming.
Fig. 6.
Flowchart of determining SARS-CoV-2 epitopes in this study.
2.1. Datasets used in the study
The datasets used in this study are publicly available and provided by the Kaggle database (Kaggle, 2021). The database contains three datasets namely SARS-CoV, B-cell, and SARS-CoV-2. Details on these datasets are presented below.
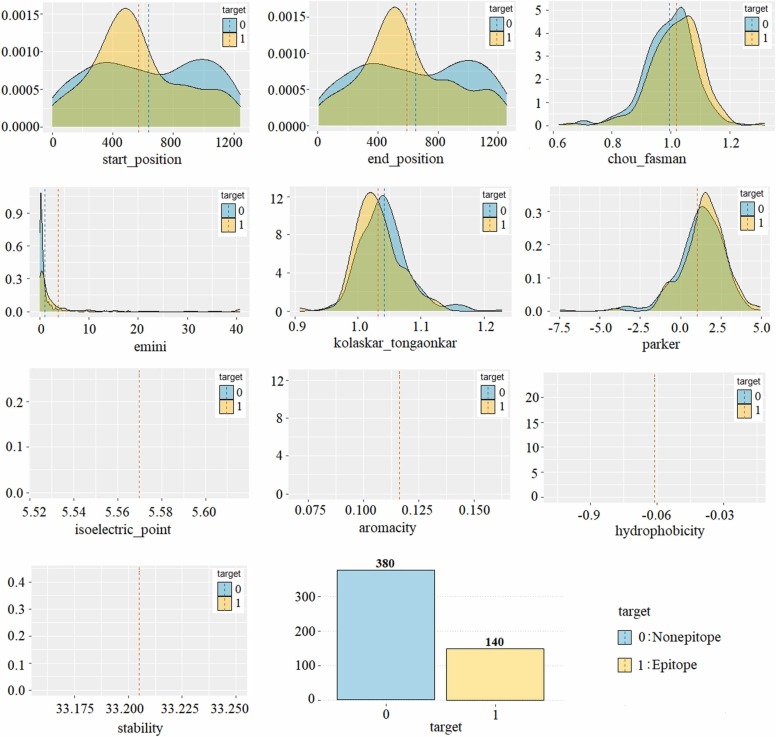
The SARS-CoV dataset is labeled and consists of 520 samples (peptides). In this study, the SARS-CoV dataset was used for model training. The dataset contains 380 nonepitopes and 140 epitopes. Since the dataset is imbalanced, it has been balanced using by synthetic minority oversampling technique (SMOTE) method. Thus, epitope prediction performances in the original and balanced SARS-CoV datasets were compared. The information about the variables in the dataset and the minimum-maximum values of the features according to the label information is shown in Table 1, and the density of the variables according to the target variable is shown in Fig. 7.
Table 1.
The variables in the SARS-CoV dataset and the minimum and maximum values of these variables according to the target variable ([minimum, maximum]).
| Variable | Type | Target: 0 (non-epitope) | Target: 1 (epitope) |
|---|---|---|---|
| parent_protein_id | Categoric | – | – |
| protein_seq | Categoric | – | – |
| start_position | Integer | [1,1241] | [ 1, 1236] |
| end_position | Integer | [10,1255] | [33,1255] |
| peptide_seq | Categoric | – | – |
| chou_fasman | Numeric | [ 0.62, 1.29] | [0.66, 1.32] |
| Emini | Numeric | [0.00, 17.97] | [0.00, 40.61] |
| kolaskar_tongaonkar | Numeric | [0.94, 1.23] | [0.91, 1.13] |
| Parker | Numeric | [− 7.47, 4.91] | [− 4.02, 4.76] |
| isoelectric_point | Numeric | [5.57, 5.57] | [5.57, 5.57] |
| Aromacity | Numeric | [0.12, 0.12] | [0.12, 0.12] |
| Hydrophobicity | Numeric | [− 0.06, − 0.06] | [− 0.06, − 0.06] |
| Stability | Numeric | [33.21, 33.21] | [33.21, 33.21] |
| Target | Binary | N = 380 | N = 140 |
Fig. 7.
Density plot of the variables in the SARS-CoV dataset by target (epitope/nonepitope).
In the study, some variables were removed from the dataset by feature engineering, also some variables were added to the dataset. The protein id was eliminated from the dataset because it does not represent epitope information. Protein sequence and peptide sequence categorical variables were converted into numerical variables namely protein length and peptide length by taking their lengths. Since the protein length is the same for all samples (peptides), it was not used in this study. Because all peptides in the SARS-CoV dataset were identified from the same protein sequence of the SARS virus. The values of isoelectric point, aromaticity, hydrophobicity, and stability are the same in all peptides as they are properties dependent on the protein sequence. Finally, the start position and end position variables were removed from the dataset as they were sufficiently representative of the dataset and were 100% related to each other. The position variable has been added to the SARS-CoV dataset. This variable was obtained from (end_position - start_position)+ 1 formula.
When Fig. 7 is examined, it is seen that 140 of the 520 peptides are epitopes and 380 are nonepitopes. This shows that the positive class is in the minority and the dataset is imbalanced distributed or unevenly. When the input variables in the dataset are examined, it is seen that the values of the epitope class samples are higher than the nonepitopes class in all variables. Furthermore, it is seen that the dataset has a normal distribution.
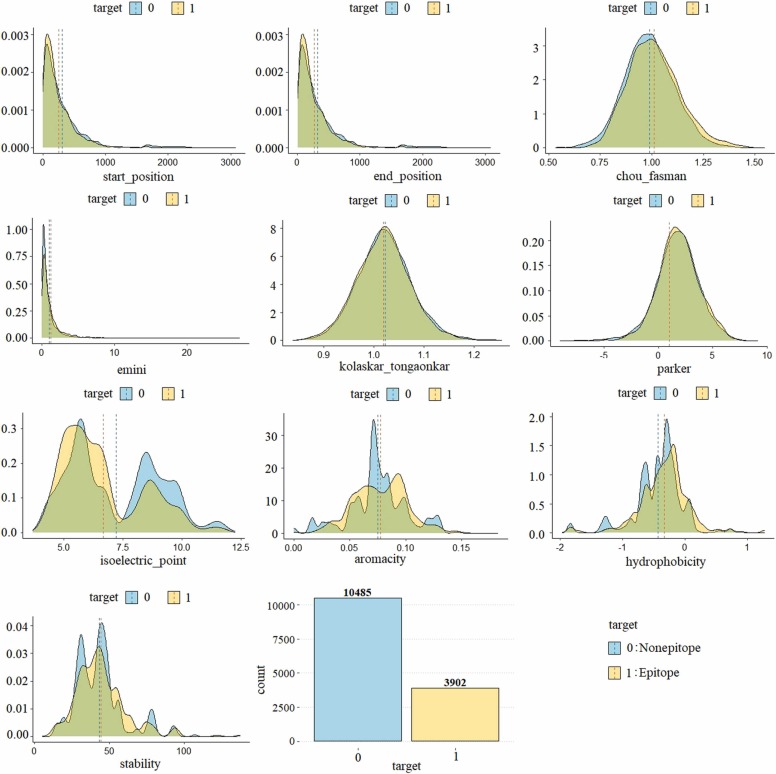
The B-cell dataset consists of 14732 peptide combinations identified from 757 different proteins. The variables of the dataset and the minimum and maximum values of these variables according to the target variable are given in Table 2. The density distribution of the variables in the dataset according to the class label is presented in Fig. 8.
Table 2.
The variables in the B-cell dataset and the minimum and maximum values of these variables according to the target variable ([minimum, maximum]).
| Variable | Type | Target: 0 (nonepitope) | Target: 1 (epitope) |
|---|---|---|---|
| parent_protein_id | Categoric | – | – |
| protein_seq | Categoric | – | – |
| start_position | Integer | [1,2757] | [ 1, 3079] |
| end_position | Integer | [6,2768] | [6,3086] |
| peptide_seq | Categoric | – | – |
| chou_fasman | Numeric | [ 0.53, 1.50] | [0.62, 1.55] |
| Emini | Numeric | [0.00, 27.19] | [0.00, 23.31] |
| kolaskar_tongaonkar | Numeric | [0.84, 1.26] | [0.85, 1.25] |
| Parker | Numeric | [− 9.03, 9.12] | [− 7.09, 7.81] |
| isoelectric_point | Numeric | [4.08, 2.23] | [3.69, 11.76] |
| Aromacity | Numeric | [0.00, 0.15] | [0.00, 0.18] |
| Hydrophobicity | Numeric | [− 1.84, 0.97] | [− 1.97, 1.27] |
| Stability | Numeric | [14.45, 137.05] | [5.45, 137.05] |
| Target | Binary | N = 10,485 | N = 3902 |
Fig. 8.
Density plot of the variables in the B-cell dataset by target (epitope/nonepitope).
The B-cell dataset contains the same variables as the SARS-CoV dataset. As with the SARS-CoV dataset, categorical variables were removed from the B-cell dataset. Position, protein length, and peptide length variables were added to the dataset. Position variable was obtained with (end_position - start_position)+ 1 formula, the protein length variable was obtained from the length of the protein sequence and the peptide length variable was obtained from the length of the peptide sequence variable.
When the density plot of the B-cell dataset is examined, it is seen that there is an unbalanced distribution with 10,485 nonepitope and 3902 epitope samples. Contrary to the SARS-CoV dataset, it is seen that the values of the epitope samples in the B-cell dataset are not always higher than the nonepitopes. It is seen that the input variables of the B-cell dataset are normally distributed.
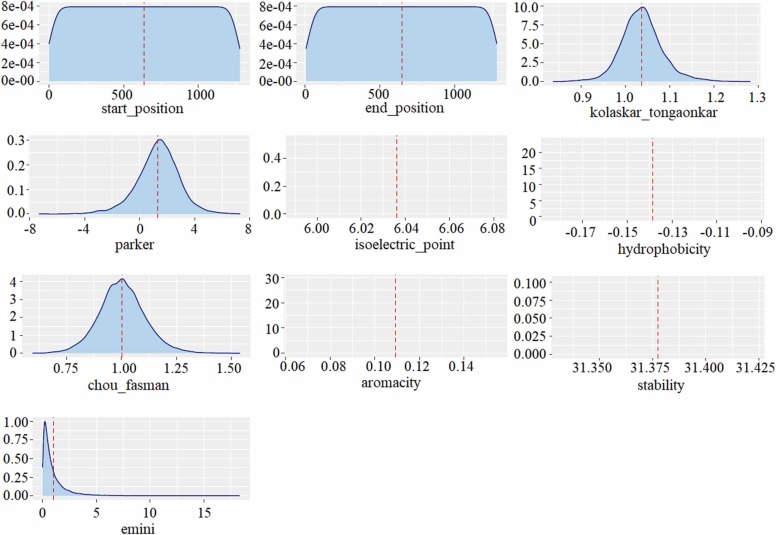
The SARS-CoV-2 dataset contains 20312 peptides obtained from the spike protein of the SARS-CoV-2 virus and there is no label information. Since the SARS-CoV-2 dataset was unlabeled, it was used as a test set. In other words, the SARS-CoV dataset was modeled with different algorithms, the method with a high success in modeling the data was determined, and the epitope estimation was made to use the SARS-CoV-2 dataset as the test set. Likewise, epitope prediction was performed using the B-cell dataset for the training set and the SARS-CoV-2 dataset for the test set. The obtained results were compared and the concurrence/intersection epitopes predicted by the models trained with both data (SARS-CoV, B-cell) were determined. The variables in the SARS-CoV-2 dataset are given in Table 3 and the density plot of the input variables is given in Fig. 9.
Table 3.
The variables in the SARS-CoV-2 dataset and the minimum, maximum and mean value of variables.
| Variable | Type | Minimum | Maximum | Mean |
|---|---|---|---|---|
| Parent_protein_id | Categoric | – | – | – |
| Protein_seq | Categoric | – | – | – |
| Start_position | Integer | 1 | 1277 | 635 |
| End_position | Integer | 5 | 1281 | 646 |
| Peptide_seq | Categoric | – | – | – |
| Chou_fasman | Numeric | 0.596 | 1.538 | 1.003 |
| Emini | Numeric | 0.003 | 18.298 | 1.000 |
| Kolaskar_tongaonkar | Numeric | 0.837 | 1.282 | 1.037 |
| Parker | Numeric | -7.317 | 7.300 | 1.335 |
| Isoelectric_point | Numeric | 6.036 | 6.036 | 6.036 |
| Aromacity | Numeric | 0.109 | 0.109 | 0.109 |
| Hydrophobicity | Numeric | -0.139 | -0.139 | -0.139 |
| Stability | Numeric | 31.380 | 31.380 | 31.380 |
Fig. 9.
Density plot of the variables in the SARS-CoV dataset.
2.2. Synthetic minority oversampling technique (SMOTE)
The imbalanced distribution between labels in a dataset negatively affects training and testing performance while developing a model (Cao et al., 2019). The imbalance in the dataset can be resolvable by different methods. Sampling methods aim to balance the class distribution in the training data by either repeating minority samples or generating new minority samples (oversampling) or removing samples from the majority class (undersampling) (Douzas et al., 2018). Various techniques have been proposed for oversampling and undersampling. Random subsampling is a non-intuitive method used to eliminate samples of a large number of classes to balance class distributions (Hundi and Shahsavari, 2020). The disadvantage of this method is the potential to destroy useful or important samples. Therefore, the information to be learned from the data is lost. On the other hand in the over-sampling method, the samples in the minority class are increased synthetically and they are brought closer to the number of samples in the majority class (Turlapati and Prusty, 2020). In this study, the synthetic minority oversampling technique (SMOTE), which is one of the over-sampling techniques was used to balance the data (Chawla et al., 2002).
SMOTE is one of the most frequently used resampling methods proposed by Chawla et al. (2002). SMOTE starts from existing minority samples and interpolates to create new artificial minority samples. The overtraining data is created by the rotation of the actual data. This method first finds the k-nearest neighbors of each minority sample, then randomly selects one of its nearest neighbors. Creates a new minority class instance that connects the minority class instance and its nearest neighbor. This procedure repeats until both classes have an equal number of elements (Chawla et al., 2002, Batista et al., 2004). In the study, 3 and 5 nearest neighbors were tried and 5-NN was used due to its success. The steps of the algorithm can be summarized as follows:
Step 1: The k nearest neighbors of each observation belonging to the minority class are searched,
Step 2: The difference between the observation belonging to the minority class and the observation with its k nearest neighbors (kNN) is taken,
Step 3: A random number (α) is chosen between (0,1), this number is multiplied by the difference found in Step 2,
Step 4: With the formulation in Eq. (1), a new synthetic observation is obtained.
| (1) |
Step 5: To generate the desired number of synthetic observations steps 1–4 are repeated.
2.3. Machine learning methods
In this study, the epitope prediction success of different machine learning methods was examined and compared. The methods used in the study are briefly described below.
Decision Tree (DT) decides which class the new data belongs to based on past data. The method creates a tree-like hierarchical structure during the training phase. Thanks to this hierarchical structure, the results are easily understandable and interpretable, and it is one of the most widely used methods because it can be easily adapted to real-life problems (Roiger, 2017). Trees begin with the root node and then propagate the information through internal nodes until it reaches the final leaf nodes. Each node is divided into sub-nodes with basic Yes/No or True/False questions. Deciding which feature will be root, internal nodes or leaf is important to obtain a strong decision tree. It subsets the dataset according to the most important attribute in the dataset. The feature with the highest information gain is determined as the root node. Splitting is performed to create child nodes called decision nodes. The Gini index is calculated for the newly formed nodes until the model reaches the leaves. If the Gini score of the current node is better than the new nodes to be generated from this node, iteration is interrupted for the new node, and in this way, it is decided whether the node is a leaf or an internal node. The Gini index and entropy measures are the most commonly used methods for calculating the impurity of a node (Coppersmith et al., 1999).
Support Vector Machine (SVM) is based on statistical learning theory. The basic operation in SVM is to estimate the most appropriate decision function that can separate the two classes from each other or obtain the hyper-plane that can best distinguish the two classes from each other (Vapnik, 2013). The method was originally built forward to distinguish between two classes that can only be separated linearly. However, in some cases, since it is not possible to separate the data linearly, the model has been adapted and started to be used to separate nonlinear data. In cases where the data is not linearly separate, data is mapped to a high-dimensional feature space with the kernel function and it is tried to be separated linearly. Common kernel functions are of three types: sigmoid, polynomial, and radial-based functions (Goodfellow et al., 2016).
Logistic Regression (logistic) may also be called a linear regression model, but logistic regression uses a more complex cost function. This cost function is called the sigmoid function or the logistic function. The logistic regression hypothesis tends to limit the cost function between 0 and 1. Since linear functions can have a value greater than 1 or less than 0, they cannot be represented by linear functions (Hosmer et al., 2013). The value π(x) = E(Y/x) is known as the conditional mean. For the conditional mean to become linear with the parameters in the model (βo+β1), it needs to be transformed. This transformation is called Logit Transformation. The transformation variable g(x) is linear with the parameters in the model, is continuous, and takes values in the range of -∞ , + ∞. As π(x) increases so does g(x), and if π(x)> 0.5 then g(x) takes positive values (Hosmer et al., 2013).
K-Nearest Neighbor (kNN) method is an algorithm that classifies based on distance. The kNN is frequently preferred in solving classification problems because it is a simple, fast applicable, and successful method. This method calculates the distance measure of the samples in the training set from this sample to give the class label to the sample whose class is unknown. The closest samples (the samples with the smallest distance measure) are selected and the class information of this sample is given to the new sample. The k value here indicates how many nearest neighbors will be looked at, that is, the number of neighbors. Whichever class the majority of these selected k samples belong to is labeled with that class in the new sample (Guo et al., 2003). For this reason, the k value is usually an odd number. Although the distance between neighbors is usually found by the Euclidean distance, distance measures such as Mahalanobis, Hamming, and Manhattan can be used.
Random Forest (RF) is an ensemble method composed of combining many decision trees. In ensemble learning methods, the results of multiple classifiers are brought together and a single decision is made on behalf of the ensemble. Each decision tree in the forest is created by selecting different samples from the original dataset by bootstrap technique and trained with a feature set selected by the random bagging mechanism (Breiman, 2001). Decisions made by a large number of distinct individual trees are then voted on and the class with the most votes as a result of the voting is assigned as the class prediction.
Bagging method also known as Bootstrap Aggregation is one of the ensemble techniques like the random forest method (Breiman, 1996). The method collects predictions of multiple classification algorithms. In estimating numerical values, the estimation of each individual classifier is averaged. In the categorical value estimation, the estimation result of each classifier is evaluated by majority voting and the estimation class with the most votes is determined (Breiman, 2001). In this study, a decision tree was used as a learning model. The steps of the bagging method can be listed as follows:
-
•
T learning dataset (D1, D2,…., DT) is created with bootstrap for learning (Bootstrap operation).
-
•
Learning of the created dataset is started.
-
•
Learning is provided using a learning algorithm.
-
•
In the first step, classification training is performed for each dataset created with bootstrap.
-
•
Estimation is made by combining the results obtained from T learning models.
2.4. Performance evaluation
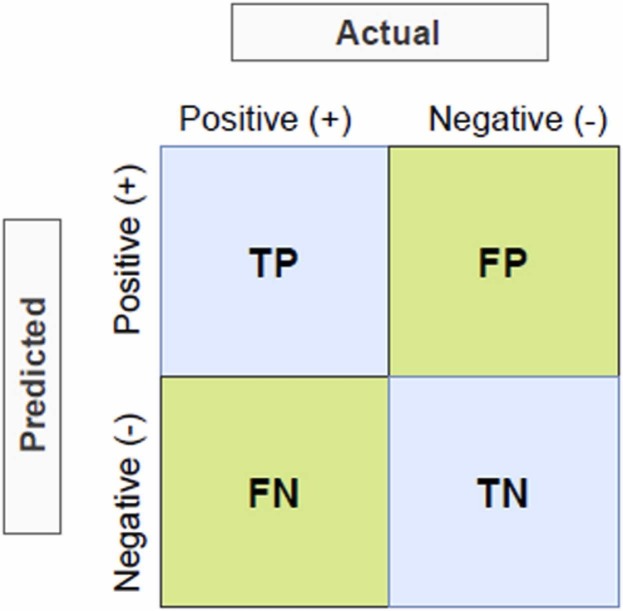
In this study, accuracy (Baldi et al., 2000), precision (Lewis, 1990), f-measure (Powers, 2020), Area Under the ROC Curve (AUC) (Bradley, 1997; Hanley and McNeil, 1982), and precision-recall curve (PRC) (Fawcett, 2006) statistical metrics were used to evaluate the prediction performance of machine learning methods. Overall accuracy is the ratio of correct predictions to all predictions. Precision gives the proportion of samples positively assigned by the model to the correct class. AUC is obtained by placing the selectivity and sensitivity values found according to the different threshold values determined for the positive or negative class of the ROC curve, into the x and y coordinates, respectively, and the relationship of these values is shown graphically with the ROC curve. ROC analysis has a wide range of applications, especially in medicine, veterinary medicine, radiology, psychology, machine learning techniques, and data mining. The AUC gives an average performance value summarizing the ROC curve. The AUC determines the accuracy of the assay in distinguishing epitope and nonepitope peptides. The closer the area under the curve size, which takes a value between 0 and 1, gets closer to 1, the higher the performance of the classifier model (Fawcett, 2006). PRC gives the relationship between precision and recall. The precision-recall curve is an effective evaluation criterion for unbalanced binary classification models due to its minority class focus. These metrics are calculated in Eqs. (2), (3), (4) using the confusion matrix (Deng et al., 2016) given in Fig. 10.
| (2) |
| (3) |
| (4) |
Where, TP is True Positive, TN is True Negative, FP is False Positive and FN is False Negative.
Fig. 10.
Confusion matrix for two-class classification problem.
3. Experimental results
3.1. Prediction of SARS-CoV and B-cell epitopes
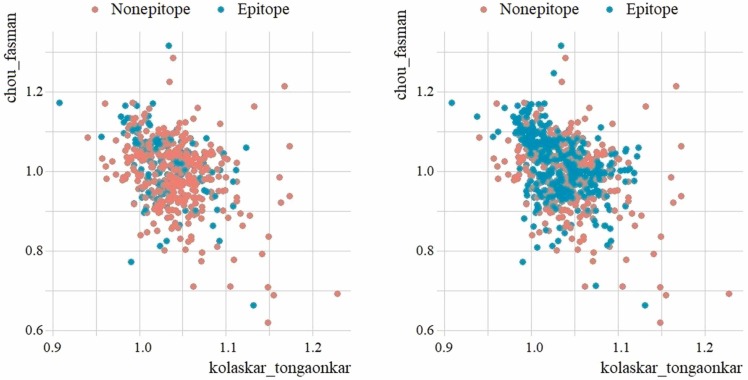
There are a total of 520 samples in the SARS-CoV dataset, of which 380 are in the majority group belonging to the negative (nonepitope) class, and 140 are in the minority group belonging to the positive (epitope) class. Considering the number of class distributions, it is seen that the data belonging to the positive class are in the minority and the dataset has an unbalanced distribution. To increase the performance of the machine learning methods, the samples belonging to the minority group were artificially amplified by the SMOTE method and the SARS-CoV dataset was balanced. There are a total of 758 samples in the dataset balanced with SMOTE, of which 380 are in the negative class and 378 are in the positive class. To visualize the class distribution of the original and SMOTE dataset, the scatter plots are shown in Fig. 11 based on the variables chou_fasman (y axis) and kolaskar_tongaonkar (x axis).
Fig. 11.
Scatter plot of original SARS-CoV dataset (left) vs SMOTE dataset (right).
In Fig. 11, it is seen that the positive classes in the minority group approach the majority group. It is seen that the nearest k-neighbor values are sampled in the region where positive samples are concentrated.
Medical datasets encountered in real life are often unbalanced datasets. Because of the low prevalence of the disease, the small number of samples in the data related to the disease, the diagnosis of the disease, and the diagnostic tests that require cost limit the datasets. Although the samples belonging to the negative (nonepitope) class are in the majority of the datasets used in the study, the samples that are required to be classified belong to the positive (epitope) class. Because the positive label in the dataset indicates that a peptide is an epitope and is the information that will be used in vaccine design. In unbalanced datasets, classes with large sample numbers dominate in the learning phase, and some imbalances can be seen while classifying the observation values belonging to minority classes. In this study, it was examined how increasing the minority group samples in the dataset and making it balanced affects the performance of classification models, especially in predicting the positive (epitope) class. For this purpose, the original SARS-CoV dataset (520 samples) and the SMOTE dataset (758 samples) were divided into 80% training and 20% testing. The classification performances of the models in test sets were compared in Table 4.
Table 4.
Performance comparison of classification methods for epitope prediction in original SARS-CoV and SMOTE SARS-CoV dataset.
| Method | Accuracy |
Precision |
F-Measure |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive (+) |
Negative (-) |
Positive (+) |
Negative (-) |
Positive (+) |
Negative (-) |
|||||||
| Original | SMOTE | Original | SMOTE | Original | SMOTE | Original | SMOTE | Original | SMOTE | Original | SMOTE | |
| RF | 0.565 | 0.855 | 0.889 | 0.831 | 0.591 | 0.808 | 0.878 | 0.873 | 0.578 | 0.831 | 0.883 | 0.852 |
| SVM | 0.130 | 0.754 | 0.975 | 0.542 | 0.600 | 0.578 | 0.798 | 0.726 | 0.258 | 0.484 | 0.870 | 0.779 |
| Logistic | 0.130 | 0.609 | 0.975 | 0.795 | 0.600 | 0.712 | 0.798 | 0.710 | 0.214 | 0.654 | 0.878 | 0.621 |
| Bagging | 0.826 | 0.841 | 0.802 | 0.831 | 0.543 | 0.806 | 0.942 | 0.863 | 0.214 | 0.656 | 0.878 | 0.750 |
| kNN | 0.435 | 0.783 | 0.864 | 0.675 | 0.476 | 0.667 | 0.843 | 0.789 | 0.655 | 0.823 | 0.876 | 0.847 |
| DT | 0.696 | 0.826 | 0.840 | 0.795 | 0.552 | 0.770 | 0.907 | 0.846 | 0.455 | 0.720 | 0.854 | 0.727 |
As previously reported, positive samples, i.e. peptides which are the epitope, were tried to be determined in this study. As can be seen in Table 4, samples belonging to the positive class, which were generally balanced with SMOTE, had better results than samples from the original dataset. The most successful method in classifying positive samples was the RF method with 85.5% accuracy, 80.8% precision, and 85.5% f-measure rate.
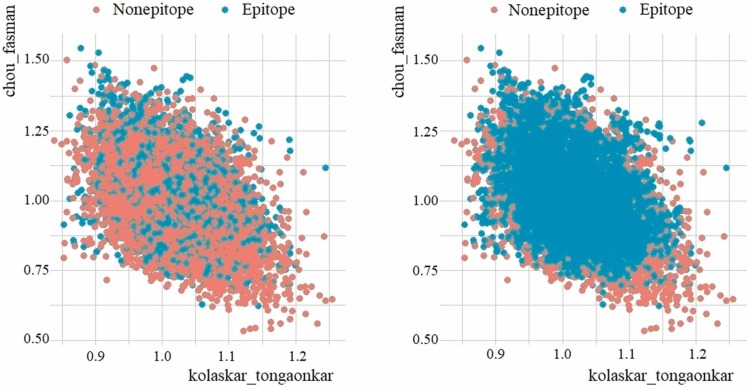
The class distribution of the original B-cell dataset and the dataset balanced with SMOTE is visualized in Fig. 12. In the scatter plot, the variables chou_fasman on the x-axis and kolaskar_tongaonkar on the y-axis were taken as bases. There are 14732 peptides in the B-cell dataset, of which 10,485 are nonepitope (negative) and 3902 are epitope (positive). The SMOTE dataset contains 10,485 nonepitopes (negative), and 10,457 epitopes (positive), a total of 20942 peptides.
Fig. 12.
Scatter plot of original B-cell dataset (left) vs SMOTE dataset (right).
The models were trained with 80% of the samples in the original B-cell and SMOTE B-cell dataset, and the classification success of the models was measured with the remaining 20%. Test performances of the methods used in the study in B-cell epitope prediction are given in Table 5. When the accuracy, precision, and f-measure results of the methods in predicting the positive class were examined, all methods were more successful in the SMOTE dataset compared to the original dataset. The results show that the RF method performs successful classification with 91.4% accuracy, 88.7% precision, and 90.0% f-measure value in the SMOTE dataset.
Table 5.
Performance comparison of classification methods for epitope prediction in original B-cell and SMOTE B-cell dataset.
| Method | Accuracy |
Precision |
F-Measure |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive (+) |
Negative (-) |
Positive (+) |
Negative (-) |
Positive (+) |
Negative (-) |
|||||||
| Original | SMOTE | Original | SMOTE | Original | SMOTE | Original | SMOTE | Original | SMOTE | Original | SMOTE | |
| RF | 0.712 | 0.914 | 0.929 | 0.887 | 0.801 | 0.887 | 0.889 | 0.915 | 0.754 | 0.900 | 0.908 | 0.901 |
| SVM | 0.413 | 0.795 | 0.944 | 0.782 | 0.749 | 0.778 | 0.800 | 0.798 | 0.172 | 0.637 | 0.824 | 0.576 |
| Logistic | 0.050 | 0.660 | 0.987 | 0.589 | 0.603 | 0.607 | 0.721 | 0.642 | 0.532 | 0.787 | 0.866 | 0.790 |
| Bagging | 0.702 | 0.903 | 0.925 | 0.873 | 0.790 | 0.873 | 0.885 | 0.903 | 0.092 | 0.633 | 0.833 | 0.614 |
| kNN | 0.680 | 0.878 | 0.902 | 0.849 | 0.738 | 0.849 | 0.875 | 0.878 | 0.744 | 0.888 | 0.905 | 0.888 |
| DT | 0.552 | 0.714 | 0.863 | 0.748 | 0.619 | 0.739 | 0.827 | 0.724 | 0.708 | 0.863 | 0.889 | 0.864 |
3.2. Determining of epitopes on the SARS-CoV-2 spike protein
The SAR-CoV-2 dataset used in the study consists of 20312 peptides. It is not possible to analyze and physically test so many peptides in vaccine design. Furthermore, since the dataset is unlabeled, it is not known which peptide is the epitope and therefore which peptide can be used in the vaccine design. After determining the machine learning method that successfully predicts epitopes in SARS-CoV and B-cell datasets, it is possible to make successful SARS-CoV-2 epitope prediction with this method.
In this study, SARS-CoV-2 epitope prediction was made using both SARS-CoV and B-cell datasets separately for training. After the proposed SMOTE-RF-SVM method was trained with the SARS-CoV dataset, the epitopes in the SARS-CoV-2 dataset were determined. With the proposed method, 1483 peptides were classified as epitopes (positive), and 18,829 peptides were classified as nonepitopes (negative). Later, the B-cell dataset was used for training and the SARS-CoV-2 dataset was used for testing. Here, 1875 peptides were classified as epitopes (positive), and 18,437 peptides were classified as nonepitopes (negative) were estimated with the proposed method. Peptides determined as epitopes in both classifications were selected and as a result, 252 peptides were identified as epitopes by the SMOTE-RF-SVM method.
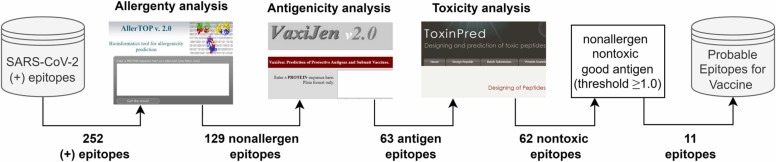
After identifying possible epitope peptides with the proposed hybrid method, allergenicity, antigenicity and toxicity analysis of epitopes were performed with bioinformatics tools. For an epitope to be used in vaccine design, it must be nonallergen, antigen, and nontoxic. In this study, allergenicity, antigenicity, and toxicity analyzes were performed with AllerTop 2.0 (AllerTop, 2021), Vaxijen 2.0 (VaxiJen, 2021), and ToxiPred (ToxinPred, 2021) bioinformatics tools, respectively, this process and the results obtained are summarized in Fig. 13.
Fig. 13.
Flowchart of bioinformatics analysis stage in this study.
AllerTop (2021) is a bioinformatics tool that estimates allergenicity. This tool has a database of 2427 allergens and 2427 nonallergens and classifies the test sample according to the kNN (k = 1) method. A peptide to be used as a vaccine should not be allergic, that is, it should not be allergenic to the host system (Yashvardhini et al., 2021). As seen in Fig. 13, allergenicity analysis was performed for 252 epitopes using the AllerTop 2.0 tool. According to the allergenicity analysis, it was determined that 129 peptides were nonallergen and 123 peptides were allergen. Obtained allergenicity analysis results are given in Table 6.
Table 6.
Determined peptides and peptides' allergenicity results.
| Peptide | Allergenicity | Peptide | Allergenicity | Peptide | Allergenicity |
|---|---|---|---|---|---|
| QTNSPS | Nonallergen | EAEVQIDRLITGR | Allergen | SSGWTAGAAAYYVG | Nonallergen |
| VGGNYNY | Nonallergen | LIRAAEIRASANL | Allergen | SGWTAGAAAYYVGY | Allergen |
| GPKKSTN | Nonallergen | IRAAEIRASANLA | Allergen | GWTAGAAAYYVGYL | Nonallergen |
| LPDPSKPS | Nonallergen | RAAEIRASANLAA | Allergen | WTAGAAAYYVGYLQ | Nonallergen |
| PGDSSSGWT | Nonallergen | AAEIRASANLAAT | Allergen | TAGAAAYYVGYLQP | Nonallergen |
| GDSSSGWTA | Nonallergen | AEIRASANLAATK | Allergen | AGAAAYYVGYLQPR | Nonallergen |
| DSSSGWTAG | Nonallergen | EIRASANLAATKM | Allergen | AAYYVGYLQPRTFL | Nonallergen |
| NLYFQGGGG | Allergen | DFCGKGYHLMSFP | Nonallergen | VGYLQPRTFLLKYN | Nonallergen |
| LYFQGGGGS | Nonallergen | HGVVFLHVTYVPA | Nonallergen | GYLQPRTFLLKYNE | Nonallergen |
| YFQGGGGSG | Nonallergen | GVVFLHVTYVPAQ | Nonallergen | LLKYNENGTITDAV | Allergen |
| FQGGGGSGY | Nonallergen | EKNFTTAPAICHD | Allergen | YNENGTITDAVDCA | Allergen |
| QLPPAYTNSF | Allergen | KNFTTAPAICHDG | Allergen | PLSETKCTLKSFTV | Allergen |
| IAWNSNNLDS | Allergen | NFTTAPAICHDGK | Allergen | VQPTESIVRFPNIT | Allergen |
| AWNSNNLDSK | Allergen | FTTAPAICHDGKA | Allergen | QPTESIVRFPNITN | Nonallergen |
| WNSNNLDSKV | Allergen | QIITTDNTFVSGN | Nonallergen | ESIVRFPNITNLCP | Nonallergen |
| NSNNLDSKVG | Allergen | IITTDNTFVSGNC | Allergen | PFGEVFNATRFASV | Nonallergen |
| SNNLDSKVGG | Allergen | ITTDNTFVSGNCD | Nonallergen | EVFNATRFASVYAW | Allergen |
| QAGSTPCNGV | Nonallergen | TTDNTFVSGNCDV | Nonallergen | VFNATRFASVYAWN | Allergen |
| VNFNFNGLTG | Nonallergen | TDNTFVSGNCDVV | Nonallergen | FNATRFASVYAWNR | Allergen |
| NFNFNGLTGT | Allergen | ELDKYFKNHTSPD | Allergen | SVYAWNRKRISNCV | Allergen |
| QIYKTPPIKD | Allergen | LDKYFKNHTSPDV | Nonallergen | VYAWNRKRISNCVA | Allergen |
| GFNFSQILPD | Nonallergen | KYFKNHTSPDVDL | Allergen | YAWNRKRISNCVAD | Nonallergen |
| NTVYDPLQPE | Nonallergen | YFKNHTSPDVDLG | Allergen | AWNRKRISNCVADY | Allergen |
| ENLYFQGGGG | Allergen | FKNHTSPDVDLGD | Allergen | WNRKRISNCVADYS | Nonallergen |
| NLYFQGGGGS | Nonallergen | KNHTSPDVDLGDI | Nonallergen | NRKRISNCVADYSV | Allergen |
| LYFQGGGGSG | Nonallergen | NHTSPDVDLGDIS | Nonallergen | RKRISNCVADYSVL | Allergen |
| RQIAPGQTGKI | Nonallergen | SPDVDLGDISGIN | Allergen | KRISNCVADYSVLY | Allergen |
| QIAPGQTGKIA | Nonallergen | DVDLGDISGINAS | Allergen | RISNCVADYSVLYN | Allergen |
| YNYLYRLFRKS | Nonallergen | DLGDISGINASVV | Allergen | ISNCVADYSVLYNS | Allergen |
| AGSTPCNGVEG | Nonallergen | LGDISGINASVVN | Allergen | SNCVADYSVLYNSA | Allergen |
| APAICHDGKAH | Allergen | SGINASVVNIQKE | Allergen | NCVADYSVLYNSAS | Allergen |
| LDKYFKNHTSP | Nonallergen | NASVVNIQKEIDR | Allergen | CVADYSVLYNSASF | Allergen |
| DKYFKNHTSPD | Allergen | ASVVNIQKEIDRL | Allergen | LYRLFRKSNLKPFE | Allergen |
| FKNHTSPDVDL | Allergen | SVVNIQKEIDRLN | Nonallergen | YRLFRKSNLKPFER | Allergen |
| LGKYEQYIKGS | Allergen | VVNIQKEIDRLNE | Nonallergen | RLFRKSNLKPFERD | Allergen |
| GKYEQYIKGSG | Allergen | VNIQKEIDRLNEV | Allergen | LFRKSNLKPFERDI | Allergen |
| QYIKGSGRENL | Allergen | NIQKEIDRLNEVA | Nonallergen | FRKSNLKPFERDIS | Nonallergen |
| YIKGSGRENLY | Allergen | IQKEIDRLNEVAK | Nonallergen | RKSNLKPFERDIST | Nonallergen |
| AIHVSGTNGTKR | Nonallergen | ESLIDLQELGKYE | Nonallergen | KSNLKPFERDISTE | Nonallergen |
| IHVSGTNGTKRF | Allergen | SLIDLQELGKYEQ | Nonallergen | SNLKPFERDISTEI | Nonallergen |
| HVSGTNGTKRFD | Allergen | LIDLQELGKYEQY | Nonallergen | NLKPFERDISTEIY | Allergen |
| EFQFCNDPFLGV | Allergen | FQGGGGSGYIPEA | Nonallergen | LKPFERDISTEIYQ | Nonallergen |
| LKSFTVEKGIYQ | Allergen | RKDGEWVLLSTFL | Nonallergen | KPFERDISTEIYQA | Nonallergen |
| KSFTVEKGIYQT | Allergen | KDGEWVLLSTFLG | Nonallergen | PFERDISTEIYQAG | Nonallergen |
| NSNNLDSKVGGN | Nonallergen | GILPSPGMPALLSL | Nonallergen | YRVVVLSFELLHAP | Nonallergen |
| SNNLDSKVGGNY | Allergen | TNSFTRGVYYPDKV | Nonallergen | RVVVLSFELLHAPA | Nonallergen |
| KKFLPFQQFGRD | Allergen | STEKSNIIRGWIFG | Nonallergen | VVVLSFELLHAPAT | Nonallergen |
| NSYECDIPIGAG | Allergen | SNIIRGWIFGTTLD | Nonallergen | PKKSTNLVKNKCVN | Nonallergen |
| SYECDIPIGAGI | Nonallergen | SKTQSLLIVNNATN | Allergen | KKSTNLVKNKCVNF | Nonallergen |
| YECDIPIGAGIC | Nonallergen | TQSLLIVNNATNVV | Allergen | KCVNFNFNGLTGTG | Allergen |
| VASQSIIAYTMS | Allergen | QSLLIVNNATNVVI | Nonallergen | CVNFNFNGLTGTGV | Allergen |
| IAYTMSLGAENS | Nonallergen | SLLIVNNATNVVIK | Nonallergen | NFNFNGLTGTGVLT | Allergen |
| DEMIAQYTSALL | Allergen | LLIVNNATNVVIKV | Allergen | NGLTGTGVLTESNK | Allergen |
| DVVIGIVNNTVY | Allergen | IVNNATNVVIKVCE | Allergen | LTGTGVLTESNKKF | Allergen |
| VIGIVNNTVYDP | Allergen | NNATNVVIKVCEFQ | Nonallergen | TGTGVLTESNKKFL | Allergen |
| IGIVNNTVYDPL | Allergen | NATNVVIKVCEFQF | Nonallergen | ADQLTPTWRVYSTG | Nonallergen |
| GIVNNTVYDPLQ | Allergen | ATNVVIKVCEFQFC | Nonallergen | DQLTPTWRVYSTGS | Nonallergen |
| NTVYDPLQPELD | Nonallergen | KQGNFKNLREFVFK | Nonallergen | QLTPTWRVYSTGSN | Nonallergen |
| TVYDPLQPELDS | Allergen | QGNFKNLREFVFKN | Nonallergen | LTPTWRVYSTGSNV | Nonallergen |
| VYDPLQPELDSF | Allergen | GNFKNLREFVFKNI | Nonallergen | TPTWRVYSTGSNVF | Nonallergen |
| YDPLQPELDSFK | Nonallergen | NFKNLREFVFKNID | Allergen | TWRVYSTGSNVFQT | Nonallergen |
| DPLQPELDSFKE | Nonallergen | FKNLREFVFKNIDG | Allergen | WRVYSTGSNVFQTR | Nonallergen |
| PELDSFKEELDK | Allergen | KNLREFVFKNIDGY | Allergen | RVYSTGSNVFQTRA | Nonallergen |
| YVRKDGEWVLLS | Allergen | NLREFVFKNIDGYF | Allergen | VYSTGSNVFQTRAG | Nonallergen |
| VRKDGEWVLLST | Allergen | LREFVFKNIDGYFK | Allergen | YSTGSNVFQTRAGC | Nonallergen |
| DGVYFASTEKSNI | Nonallergen | REFVFKNIDGYFKI | Nonallergen | ASYQTQTNSPSGAG | Nonallergen |
| GVYFASTEKSNII | Allergen | EFVFKNIDGYFKIY | Allergen | SYQTQTNSPSGAGS | Nonallergen |
| VYFASTEKSNIIR | Allergen | FVFKNIDGYFKIYS | Allergen | SPSGAGSVASQSII | Nonallergen |
| YFASTEKSNIIRG | Allergen | VFKNIDGYFKIYSK | Allergen | LTGIAVEQDKNTQE | Nonallergen |
| YVGYLQPRTFLLK | Nonallergen | FKNIDGYFKIYSKH | Allergen | GIAVEQDKNTQEVF | Allergen |
| VGYLQPRTFLLKY | Nonallergen | VRDLPQGFSALEPL | Allergen | IAVEQDKNTQEVFA | Nonallergen |
| SFSTFKCYGVSPT | Allergen | DLPQGFSALEPLVD | Nonallergen | AVEQDKNTQEVFAQ | Nonallergen |
| FSTFKCYGVSPTK | Nonallergen | LPQGFSALEPLVDL | Allergen | FGGFNFSQILPDPS | Nonallergen |
| STFKCYGVSPTKL | Nonallergen | PQGFSALEPLVDLP | Allergen | FNFSQILPDPSKPS | Nonallergen |
| LNDLCFTNVYADS | Nonallergen | QGFSALEPLVDLPI | Nonallergen | LICAQKFNGLTVLP | Nonallergen |
| NDLCFTNVYADSF | Allergen | NITRFQTLLALHRS | Nonallergen | ICAQKFNGLTVLPP | Nonallergen |
| LCFTNVYADSFVI | Allergen | ITRFQTLLALHRSY | Nonallergen | CAQKFNGLTVLPPL | Nonallergen |
| GGVSVITPGTNTS | Nonallergen | FQTLLALHRSYLTP | Nonallergen | AQKFNGLTVLPPLL | Allergen |
| GVSVITPGTNTSN | Nonallergen | YLTPGDSSSGWTAG | Nonallergen | QKFNGLTVLPPLLT | Allergen |
| NTSNEVAVLYQDV | Allergen | TPGDSSSGWTAGAA | Nonallergen | KFNGLTVLPPLLTD | Allergen |
| FNSAIGKIQDSLS | Nonallergen | PGDSSSGWTAGAAA | Nonallergen | FNGLTVLPPLLTDE | Allergen |
| AIGKIQDSLSSTA | Allergen | GDSSSGWTAGAAAY | Nonallergen | NGLTVLPPLLTDEM | Allergen |
| IGKIQDSLSSTAS | Allergen | DSSSGWTAGAAAYY | Nonallergen | GLTVLPPLLTDEMI | Nonallergen |
| PEAEVQIDRLITG | Allergen | SSSGWTAGAAAYYV | Nonallergen | LTVLPPLLTDEMIA | Allergen |
According to the allergenicity analysis, 129 nonallergen peptides were selected (Table 6) and their antigenicity score was calculated. For this, the VaxiJen 2.0(VaxiJen, 2021) bioinformatics tool was used. VaxiJen makes an alignment-independent prediction of protective antigens using the physicochemical properties of proteins. Antigenicity is based on the vaccine's ability to bind to B-cell receptors and increase the immune response in the host (Yashvardhini et al., 2021). The default threshold value in the VaxiJen tool is 0.4, and epitopes with antigenicity higher than this value are called antigens. Antigenicity analysis results of 129 nonallergen epitopes are presented in Table 7.
Table 7.
Results of antigenicity analysis on probable non-allergens.
| Peptide | Vaxijen score | Antigenicity | Peptide | Vaxijen score | Antigenicity |
|---|---|---|---|---|---|
| QTNSPS | 0.0301 | Nonantigen | NATNVVIKVCEFQF | 0.3036 | Nonantigen |
| VGGNYNY | 1.3327 | Antigen | ATNVVIKVCEFQFC | -0.3036 | Nonantigen |
| GPKKSTN | 0.3011 | Nonantigen | KQGNFKNLREFVFK | 0.1686 | Nonantigen |
| LPDPSKPS | -0.2699 | Nonantigen | QGNFKNLREFVFKN | 0.0923 | Nonantigen |
| PGDSSSGWT | 0.1337 | Nonantigen | GNFKNLREFVFKNI | 0.0817 | Nonantigen |
| GDSSSGWTA | 0.3077 | Nonantigen | REFVFKNIDGYFKI | -0.0602 | Nonantigen |
| DSSSGWTAG | 0.2444 | Nonantigen | DLPQGFSALEPLVD | 0.3503 | Nonantigen |
| LYFQGGGGS | 0.6074 | Antigen | QGFSALEPLVDLPI | 0.2838 | Nonantigen |
| YFQGGGGSG | 0.3571 | Nonantigen | NITRFQTLLALHRS | 0.1039 | Nonantigen |
| FQGGGGSGY | 0.3826 | Nonantigen | ITRFQTLLALHRSY | 0.1883 | Nonantigen |
| QAGSTPCNGV | 0.1004 | Nonantigen | FQTLLALHRSYLTP | 0.4991 | Antigen |
| VNFNFNGLTG | 1.5867 | Antigen | YLTPGDSSSGWTAG | 0.4578 | Antigen |
| GFNFSQILPD | 0.6074 | Antigen | TPGDSSSGWTAGAA | 0.1487 | Nonantigen |
| NTVYDPLQPE | 0.5004 | Antigen | PGDSSSGWTAGAAA | 0.1889 | Nonantigen |
| NLYFQGGGGS | 0.7834 | Antigen | GDSSSGWTAGAAAY | 0.2846 | Nonantigen |
| LYFQGGGGSG | 0.4798 | Antigen | DSSSGWTAGAAAYY | 0.4142 | Antigen |
| RQIAPGQTGKI | 1.4465 | Antigen | SSSGWTAGAAAYYV | 0.3218 | Nonantigen |
| QIAPGQTGKIA | 1.4618 | Antigen | SSGWTAGAAAYYVG | 0.3269 | Nonantigen |
| YNYLYRLFRKS | -0.4485 | Nonantigen | GWTAGAAAYYVGYL | 0.5673 | Antigen |
| AGSTPCNGVEG | 0.0073 | Nonantigen | WTAGAAAYYVGYLQ | 0.5999 | Antigen |
| LDKYFKNHTSP | -0.2323 | Nonantigen | TAGAAAYYVGYLQP | 0.7174 | Antigen |
| AIHVSGTNGTKR | 0.736 | Antigen | AGAAAYYVGYLQPR | 1.0663 | Antigen |
| NSNNLDSKVGGN | 0.6962 | Antigen | AAYYVGYLQPRTFL | 0.5125 | Antigen |
| SYECDIPIGAGI | 1.0008 | Antigen | VGYLQPRTFLLKYN | 0.5523 | Antigen |
| YECDIPIGAGIC | 0.668 | Antigen | GYLQPRTFLLKYNE | 0.3921 | Nonantigen |
| IAYTMSLGAENS | 0.9403 | Antigen | QPTESIVRFPNITN | 0.055 | Nonantigen |
| NTVYDPLQPELD | 0.3363 | Nonantigen | ESIVRFPNITNLCP | 0.6583 | Antigen |
| YDPLQPELDSFK | 0.1219 | Nonantigen | PFGEVFNATRFASV | 0.1918 | Nonantigen |
| DPLQPELDSFKE | -0.0625 | Nonantigen | YAWNRKRISNCVAD | 0.2786 | Nonantigen |
| DGVYFASTEKSNI | 0.524 | Antigen | WNRKRISNCVADYS | 0.2138 | Nonantigen |
| YVGYLQPRTFLLK | 0.472 | Antigen | FRKSNLKPFERDIS | 0.6091 | Antigen |
| VGYLQPRTFLLKY | 0.4736 | Antigen | RKSNLKPFERDIST | 0.4607 | Antigen |
| FSTFKCYGVSPTK | 0.9029 | Antigen | KSNLKPFERDISTE | 0.4643 | Antigen |
| STFKCYGVSPTKL | 1.153 | Antigen | SNLKPFERDISTEI | 0.2788 | Nonantigen |
| LNDLCFTNVYADS | 0.9334 | Antigen | LKPFERDISTEIYQ | -0.1738 | Nonantigen |
| GGVSVITPGTNTS | 0.3461 | Nonantigen | KPFERDISTEIYQA | -0.3184 | Nonantigen |
| GVSVITPGTNTSN | 0.4725 | Antigen | PFERDISTEIYQAG | -0.2817 | Nonantigen |
| FNSAIGKIQDSLS | 0.1406 | Nonantigen | YRVVVLSFELLHAP | 0.8065 | Antigen |
| DFCGKGYHLMSFP | 0.3697 | Nonantigen | RVVVLSFELLHAPA | 0.7038 | Antigen |
| HGVVFLHVTYVPA | 0.8662 | Antigen | VVVLSFELLHAPAT | 0.7845 | Antigen |
| GVVFLHVTYVPAQ | 1.1232 | Antigen | PKKSTNLVKNKCVN | 0.5391 | Antigen |
| QIITTDNTFVSGN | 0.244 | Nonantigen | KKSTNLVKNKCVNF | 1.0894 | Antigen |
| ITTDNTFVSGNCD | 0.1017 | Nonantigen | ADQLTPTWRVYSTG | 0.6906 | Antigen |
| TTDNTFVSGNCDV | 0.0517 | Nonantigen | DQLTPTWRVYSTGS | 0.7635 | Antigen |
| TDNTFVSGNCDVV | 0.0787 | Nonantigen | QLTPTWRVYSTGSN | 0.9924 | Antigen |
| LDKYFKNHTSPDV | -0.0794 | Nonantigen | LTPTWRVYSTGSNV | 0.8582 | Antigen |
| KNHTSPDVDLGDI | 1.4147 | Antigen | TPTWRVYSTGSNVF | 0.1616 | Nonantigen |
| NHTSPDVDLGDIS | 1.5909 | Antigen | TWRVYSTGSNVFQT | 0.1548 | Nonantigen |
| SVVNIQKEIDRLN | 0.3254 | Nonantigen | WRVYSTGSNVFQTR | 0.4314 | Antigen |
| VVNIQKEIDRLNE | 0.1308 | Nonantigen | RVYSTGSNVFQTRA | 0.3248 | Nonantigen |
| NIQKEIDRLNEVA | 0.0144 | Nonantigen | VYSTGSNVFQTRAG | 0.4252 | Antigen |
| IQKEIDRLNEVAK | -0.1773 | Nonantigen | YSTGSNVFQTRAGC | 0.6965 | Antigen |
| ESLIDLQELGKYE | 0.6804 | Antigen | ASYQTQTNSPSGAG | 0.5246 | Antigen |
| SLIDLQELGKYEQ | 0.9235 | Antigen | SYQTQTNSPSGAGS | 0.4818 | Antigen |
| LIDLQELGKYEQY | 0.8932 | Antigen | SPSGAGSVASQSII | 0.4354 | Antigen |
| FQGGGGSGYIPEA | 0.0156 | Nonantigen | LTGIAVEQDKNTQE | 0.6711 | Antigen |
| RKDGEWVLLSTFL | 0.727 | Antigen | IAVEQDKNTQEVFA | 0.3395 | Nonantigen |
| KDGEWVLLSTFLG | 0.9298 | Antigen | AVEQDKNTQEVFAQ | 0.1637 | Nonantigen |
| GILPSPGMPALLSL | 0.3727 | Nonantigen | FGGFNFSQILPDPS | 0.5927 | Antigen |
| TNSFTRGVYYPDKV | 0.1154 | Nonantigen | FNFSQILPDPSKPS | 0.3471 | Nonantigen |
| STEKSNIIRGWIFG | -0.5204 | Nonantigen | LICAQKFNGLTVLP | 0.3627 | Nonantigen |
| SNIIRGWIFGTTLD | -0.3339 | Nonantigen | ICAQKFNGLTVLPP | 0.1843 | Nonantigen |
| QSLLIVNNATNVVI | 0.4427 | Antigen | CAQKFNGLTVLPPL | 0.1016 | Nonantigen |
| SLLIVNNATNVVIK | 0.4772 | Antigen | GLTVLPPLLTDEMI | 0.4082 | Antigen |
| NNATNVVIKVCEFQ | 0.0357 | Nonantigen | |||
As a result of allergenicity and antigenicity analysis, 63 peptides were determined as nonallergen and antigen. ToxinPred (2021) tool was used to measure the toxicity of these peptides. Toxicity represents amount or degree of poisonous and measures the damaging capacity of a substance. In drug and vaccine design, the active substance is expected to be nontoxic. The ToxinPred web server estimates the toxicity of peptides based on their physicochemical properties using the SVM method. The results obtained by toxicity analyzing 63 nonallergen and antigen peptides from a biochemical perspective are given in Table 8. SVM score of < 0.0 indicates that the peptide is nontoxic. In order for the vaccine to initiate an immune response in the host cell, the epitope must have a hydrophilic nature (Solanki et al., 2019, Gupta et al., 2013). Low molecular weight indicates that the peptide is nontoxic and less allergenic (Pooja et al., 2017). Nontoxic 62 peptides were determined and these are given in Table 8.
Table 8.
Results of toxicity analysis on probable antigens.
| Peptide/Probable antigen | SVM score | Hydrophilicity | Molecular weight | Toxicity |
|---|---|---|---|---|
| VGGNYNY | -0.79 | -0.81 | 785.91 | Nontoxic |
| LYFQGGGGS | -0.59 | -0.68 | 885.08 | Nontoxic |
| VNFNFNGLTG | -1.27 | -0.81 | 1082.33 | Nontoxic |
| GFNFSQILPD | -1.40 | -0.49 | 1137.40 | Nontoxic |
| NTVYDPLQPE | -0.90 | 0.04 | 1175.40 | Nontoxic |
| NLYFQGGGGS | -0.57 | -0.59 | 999.20 | Nontoxic |
| LYFQGGGGSG | -0.61 | -0.61 | 942.15 | Nontoxic |
| RQIAPGQTGKI | -0.91 | 0.17 | 1168.53 | Nontoxic |
| QIAPGQTGKIA | -0.98 | -0.15 | 1083.42 | Nontoxic |
| AIHVSGTNGTKR | -1.17 | 0.12 | 1240.56 | Nontoxic |
| NSNNLDSKVGGN | -1.19 | 0.34 | 1218.42 | Nontoxic |
| SYECDIPIGAGI | -0.31 | -0.24 | 1237.56 | Nontoxic |
| YECDIPIGAGIC | -0.23 | -0.35 | 1235.62 | Nontoxic |
| IAYTMSLGAENS | -0.41 | -0.40 | 1256.56 | Nontoxic |
| DGVYFASTEKSNI | -1.57 | 0.06 | 1430.71 | Nontoxic |
| YVGYLQPRTFLLK | -1.73 | -0.63 | 1598.12 | Nontoxic |
| VGYLQPRTFLLKY | -1.53 | -0.63 | 1598.12 | Nontoxic |
| FSTFKCYGVSPTK | -0.91 | -0.31 | 1464.87 | Nontoxic |
| STFKCYGVSPTKL | -0.77 | -0.25 | 1430.86 | Nontoxic |
| LNDLCFTNVYADS | -1.19 | -0.39 | 1474.78 | Nontoxic |
| GVSVITPGTNTSN | -1.42 | -0.38 | 1246.53 | Nontoxic |
| HGVVFLHVTYVPA | -1.57 | -1.12 | 1438.89 | Nontoxic |
| GVVFLHVTYVPAQ | -1.35 | -1.06 | 1429.88 | Nontoxic |
| KNHTSPDVDLGDI | -0.44 | 0.50 | 1410.69 | Nontoxic |
| NHTSPDVDLGDIS | -0.57 | 0.29 | 1369.59 | Nontoxic |
| ESLIDLQELGKYE | -0.96 | 0.46 | 1536.90 | Nontoxic |
| SLIDLQELGKYEQ | -1.06 | 0.25 | 1535.92 | Nontoxic |
| LIDLQELGKYEQY | -1.07 | 0.05 | 1612.02 | Nontoxic |
| RKDGEWVLLSTFL | -1.27 | -0.07 | 1564.01 | Nontoxic |
| KDGEWVLLSTFLG | -1.46 | -0.30 | 1464.88 | Nontoxic |
| QSLLIVNNATNVVI | -0.83 | -0.82 | 1497.98 | Nontoxic |
| SLLIVNNATNVVIK | -0.94 | -0.62 | 1498.02 | Nontoxic |
| FQTLLALHRSYLTP | -1.23 | -0.74 | 1660.16 | Nontoxic |
| YLTPGDSSSGWTAG | -0.92 | -0.35 | 1398.64 | Nontoxic |
| DSSSGWTAGAAAYY | -0.83 | -0.46 | 1406.60 | Nontoxic |
| GWTAGAAAYYVGYL | -1.26 | -1.14 | 1462.82 | Nontoxic |
| WTAGAAAYYVGYLQ | -1.14 | -1.13 | 1533.90 | Nontoxic |
| TAGAAAYYVGYLQP | -1.30 | -0.89 | 1444.80 | Nontoxic |
| AGAAAYYVGYLQPR | -1.45 | -0.64 | 1499.88 | Nontoxic |
| AAYYVGYLQPRTFL | -1.46 | -0.91 | 1662.12 | Nontoxic |
| VGYLQPRTFLLKYN | -1.58 | -0.57 | 1712.24 | Nontoxic |
| ESIVRFPNITNLCP | -0.88 | -0.29 | 1603.07 | Nontoxic |
| FRKSNLKPFERDIS | -1.78 | 0.73 | 1737.18 | Nontoxic |
| RKSNLKPFERDIST | -1.56 | 0.88 | 1691.11 | Nontoxic |
| KSNLKPFERDISTE | -1.70 | 0.88 | 1664.04 | Nontoxic |
| YRVVVLSFELLHAP | -1.57 | -0.67 | 1643.17 | Nontoxic |
| RVVVLSFELLHAPA | -1.58 | -0.54 | 1551.07 | Nontoxic |
| VVVLSFELLHAPAT | -1.47 | -0.79 | 1495.99 | Nontoxic |
| PKKSTNLVKNKCVN | 0.10 | 0.48 | 1573.09 | Toxic |
| KKSTNLVKNKCVNF | -0.15 | 0.30 | 1623.15 | Nontoxic |
| ADQLTPTWRVYSTG | -1.34 | -0.30 | 1594.94 | Nontoxic |
| DQLTPTWRVYSTGS | -1.49 | -0.24 | 1610.94 | Nontoxic |
| QLTPTWRVYSTGSN | -1.64 | -0.44 | 1609.96 | Nontoxic |
| LTPTWRVYSTGSNV | -1.40 | -0.56 | 1580.96 | Nontoxic |
| WRVYSTGSNVFQTR | -1.09 | -0.36 | 1701.07 | Nontoxic |
| VYSTGSNVFQTRAG | -0.97 | -0.36 | 1486.80 | Nontoxic |
| YSTGSNVFQTRAGC | -0.66 | -0.33 | 1490.80 | Nontoxic |
| ASYQTQTNSPSGAG | -0.91 | -0.19 | 1368.57 | Nontoxic |
| SYQTQTNSPSGAGS | -0.95 | -0.13 | 1384.57 | Nontoxic |
| SPSGAGSVASQSII | -0.87 | -0.31 | 1260.56 | Nontoxic |
| LTGIAVEQDKNTQE | -1.55 | 0.44 | 1545.88 | Nontoxic |
| FGGFNFSQILPDPS | -1.55 | -0.51 | 1525.88 | Nontoxic |
| GLTVLPPLLTDEMI | -0.91 | -0.47 | 1512.06 | Nontoxic |
4. Discussion
The SARS-CoV-2 virus continues to spread rapidly all over the world, naturally mutating. It has been determined that some mutations seen recently are more resistant to vaccines (Thomson et al., 2021). The rise of these mutant viruses could force the development of second-generation vaccines. It is important to determine the epitopes that can be used in vaccine design by in silico methods so that the production of a new generation vaccine can be fast, effective, and low cost. This study, aimed to identify candidate epitopes for epitope-based SARS-CoV-2 vaccine design with artificial intelligence/machine learning and bioinformatics tools.
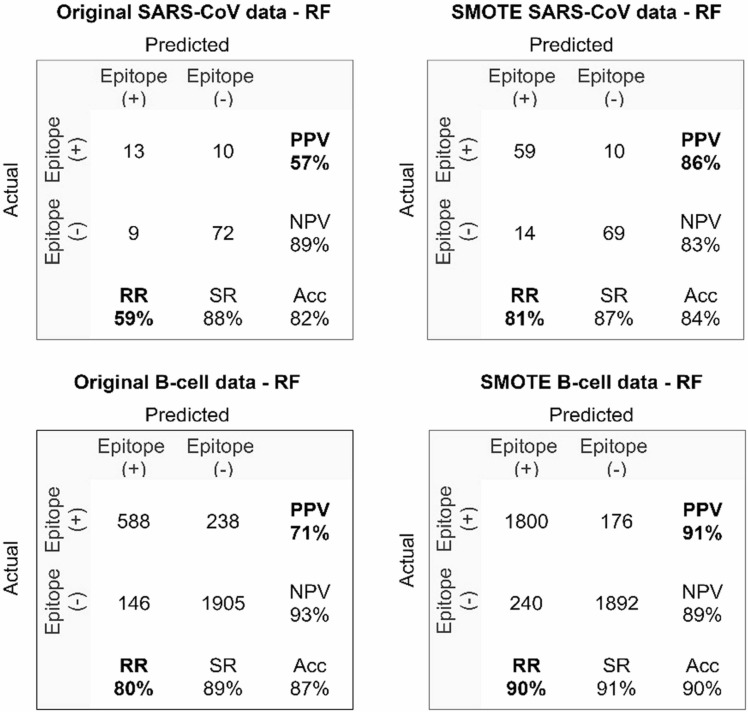
SARS-CoV, B-cell and SARS-CoV-2 datasets were used in the study. Since the labeled SARS-CoV and B-cell datasets have an unbalanced distribution, the datasets were balanced with the SMOTE method. After increasing the positive class in the minority group with SMOTE, the classification performance of machine learning methods was compared with the original dataset. In the datasets balanced with SMOTE, the prediction success of epitopes was higher than that of original dataset, and the most successful results were obtained with the RF method. In Fig. 14, the prediction results of the RF method in the original and SMOTE datasets are presented with confusion matrices for comparison.
Fig. 14.
Confusion matrices of the most successful method (RF) for original vs SMOTE datasets.
As seen in the confusion matrix, while the RF method correctly classifies 13 positive samples in the original SARS-CoV dataset, SMOTE correctly classifies 59 positive samples in the SARS-CoV dataset. While the positive predicted value (PPV) rate was 57% in the original dataset, it increased to 86% in the SMOTE SARS-CoV dataset. The recall rate increased from 59% in the original dataset to 81% in the SMOTE dataset. When the prediction performance of the RF method is compared with the original B-cell dataset and the dataset balanced with the SMOTE method, it is seen that the performance of the balanced dataset is considerably higher than of the original dataset. While PPV was 71% and RR was 80% in the original B-cell dataset, PPV increased to 91% and RR to 90% in the SMOTE dataset. The results obtained from this study showed that the dataset balanced with SMOTE improved the performance of machine learning methods in epitope prediction. The performance of the machine learning methods to predict epitopes in the SMOTE SARS-CoV and SMOTE B-cell datasets are given in Table 9.
Table 9.
AUC and PRC results of methods for SMOTE SARS-CoV and SMOTE B-cell dataset.
| Method | SMOTE SARS-CoV |
SMOTE B-cell |
||
|---|---|---|---|---|
| AUC | PRC | AUC | PRC | |
| RF | 0.940 | 0.944 | 0.956 | 0.953 |
| SVM | 0.725 | 0.709 | 0.816 | 0.839 |
| Logistic | 0.719 | 0.721 | 0.656 | 0.635 |
| Bagging | 0.883 | 0.856 | 0.947 | 0.953 |
| kNN | 0.802 | 0.762 | 0.864 | 0.814 |
| DT | 0.839 | 0.798 | 0.757 | 0.733 |
The AUC is a criterion often used to measure the quality of a classification algorithm. The PRC relates the positive predictive value of a classifier to its true positive rate and is used to evaluate classification performance. When the AUC and PRC results of the methods are compared in epitope prediction, the success of the RF method in estimating epitopes in the SMOTE SARS-CoV dataset is 94.0% and 94.4%, respectively. In SMOTE B-cell dataset epitope prediction, the RF method achieved successful results compared to other methods with 95.6% AUC and 95.3% PRC values. Jain et al. (2021) performed epitope prediction using SARS-CoV and B-cell datasets. In this study, epitopes in the SARS-CoV dataset were predicted with 91.9% AUC. The dataset defined as SARS-CoV-2 was obtained by combining the SARS-CoV and B-cell datasets. Epitopes in this dataset were estimated with 92.3% AUC. Ghoshal et al. (2021) used Bayesian neural networks (BNNs) with the dropweights method for B-cell epitope estimation. Epitope prediction was made with 85% accuracy in the study using SARS-CoV and B-cell datasets. Noumi et al. (2021) used the long short-term memory network (LSTM) method for epitope prediction. The highest accuracy value obtained in the study was 79%. When epitope prediction studies were examined using machine learning methods, the SARS-CoV-2 dataset was not used, and SARS-CoV-2 epitopes were not predicted. Experimental results obtained from the study show that epitopes in SARS-CoV and B-cell datasets in this study were predicted more successfully than other studies (94.0% AUC for SARS-CoV, 95.6% for B-cell). Furthermore, allergenicity, antigenicity and toxicity analyses of the determined SARS-CoV-2 epitopes were performed in our study.
It is important to identify peptides with high epitope potential from the SARS-CoV-2 proteins that can be used in the vaccine to reduce the experiments to be performed physically in the laboratory environment. With the proposed SMOTE-RF-SVM method, 252 of 20312 peptides were determined to be probable epitopes (positive). For a peptide to be used in a vaccine, it must be nonallergenic, antigen, and nontoxic, in addition to being an epitope. Allergenicity, antigenicity and toxicity analyses of 252 peptides were performed using AllerTop, VaxiJen and ToxinPred bioinformatics tools. It was determined that 62 of these epitopes are nonallergenic, antigenic and nontoxic. The threshold level for a peptide to be an antigen in the VaxiJen tool was selected by default as > 0.4. However, in this study, a threshold level of ≥ 1.0 was chosen to identify good (high) antigen epitopes. As a result, 11 probable nonallergen, highly antigenic and nontoxic epitopes were selected from 20312 SARS-CoV-2 peptides that can be used for vaccine design. Analyses of the determined candidate epitopes are given in Table 10.
Table 10.
Probable nonallergen, high antigen and nontoxic epitopes.
| Peptide | Allergenicity analysis |
Antigenicity analysis |
Toxicity analysis |
||||
|---|---|---|---|---|---|---|---|
| Allergenicity | Antigen score | Antigenicity | SVM score | Hydro- philicity | Molecular weight | Toxicity | |
| VGGNYNY | Nonallergen | 1.3327 | Antigen | -0.79 | -0.81 | 785.91 | Nontoxic |
| VNFNFNGLTG | Nonallergen | 1.5867 | Antigen | -1.27 | -0.81 | 1082.33 | Nontoxic |
| RQIAPGQTGKI | Nonallergen | 1.4465 | Antigen | -0.91 | 0.17 | 1168.53 | Nontoxic |
| QIAPGQTGKIA | Nonallergen | 1.4618 | Antigen | -0.98 | -0.15 | 1083.42 | Nontoxic |
| SYECDIPIGAGI | Nonallergen | 1.0008 | Antigen | -0.31 | -0.24 | 1237.56 | Nontoxic |
| STFKCYGVSPTKL | Nonallergen | 1.1530 | Antigen | -0.77 | -0.25 | 1430.86 | Nontoxic |
| GVVFLHVTYVPAQ | Nonallergen | 1.1232 | Antigen | -1.35 | -1.06 | 1429.88 | Nontoxic |
| KNHTSPDVDLGDI | Nonallergen | 1.4147 | Antigen | -0.44 | 0.50 | 1410.69 | Nontoxic |
| NHTSPDVDLGDIS | Nonallergen | 1.5909 | Antigen | -0.57 | 0.29 | 1369.59 | Nontoxic |
| AGAAAYYVGYLQPR | Nonallergen | 1.0663 | Antigen | -1.45 | -0.64 | 1499.88 | Nontoxic |
| KKSTNLVKNKCVNF | Nonallergen | 1.0894 | Antigen | -0.15 | 0.30 | 1623.15 | Nontoxic |
5. Conclusion
The COVID-19 outbreak showed that the world is not prepared for an epidemic and showed how important it is to design a rapid vaccine. It takes more than 15 years to develop a vaccine using conventional methods (Krammer, 2020). After COVID-19 was declared a state of emergency, large companies worked collaboratively to produce a vaccine quickly. Despite this, the first vaccination took more than a year, and millions of people died from the epidemic (Cihan, 2021). It seems possible to quickly design a vaccine for future epidemics by utilizing machine learning methods. The novelty of this study is to propose a successful method for epitope prediction and to show researchers the usability of machine learning and bioinformatics tools in vaccine design.
In the study, it was observed that the epitope prediction success of the models increased in general after the SARS-CoV and B-cell datasets used for model training were balanced. When the epitope prediction performances of ML methods were compared for the datasets balanced with the SMOTE method, it was seen that the RF method made more successful predictions than other methods. Epitopes determined by the developed hybrid approach (SMOTE-RF-SVM) were analyzed with the bioinformatics tools AllerTop, VaxiJen, ToxinPred, and allergen, antigen, and toxic epitopes not suitable for use in vaccine design were eliminated.
With the proposed SMOTE-RF-SVM hybrid approach, 252 positive epitope candidates that can be used in vaccine design were determined from 20312 peptides. Then, the AllerTop tool was used to determine nonallergen peptides, and it was determined that 123 of 252 candidate epitopes were allergen and 129 were nonallergen. Antigenicity and toxicity analyses were performed on nonallergen epitope candidates using the VaxiJen and ToxinPred tools, respectively. As a result of the analyses, 11 possible nonallergen, high antigen and nontoxic peptides were determined that can be used in the design of vaccines against SARS-CoV-2 (“VGGNYNY”, “VNFNFNGLTG”, “RQIAPGQTGKI”, “QIAPGQTGKIA”, “SYECDIPIGAGI”, “STFKCYGVSPTKL”, “GVVFLHVTYVPAQ”, “KNHTSPDVDLGDI”, “NHTSPDVDLGDIS”, “AGAAAYYVGYLQPR”, “KKSTNLVKNKCVNF”). It is anticipated that the findings from this study will help medical biotechnologists design fast, useful, and effective vaccines.
CRediT authorship contribution statement
Pınar Cihan: Conceptualization, Writing – original draft preparation, Methodology, Validation, Software; Visualization, Writing – review and editing. Zeynep Banu Ozger: Writing – original draft preparation, Methodology, Validation, Software; Visualization, Writing – review and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by Turkish Scientific and Technical Research Council, Turkey-TÜBİTAK (Project Number: 121E326).
References
- AllerTop, 2021. Bioinformatics tool for allergenicity prediction.
- Ansari H.R., Raghava G.P. Identification of conformational B-cell Epitopes in an antigen from its primary sequence. Immunome Res. 2010;6:1–9. doi: 10.1186/1745-7580-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- Batista G.E., Prati R.C., Monard M.C. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor. Newsl. 2004;6:20–29. [Google Scholar]
- Bradley A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30:1145–1159. [Google Scholar]
- Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. [Google Scholar]
- Breiman L. Using iterated bagging to debias regressions. Mach. Learn. 2001;45:261–277. [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Cao K., Wei C., Gaidon A., Arechiga N., Ma T. Learning imbalanced datasets with label-distribution-aware margin loss. Adv. Neural Inf. Process. Syst. 2019;32 [Google Scholar]
- Ceylan Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002;16:321–357. [Google Scholar]
- Chen H.-Z., Tang L.-L., Yu X.-L., Zhou J., Chang Y.-F., Wu X. Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2. Infect. Dis. Poverty. 2020;9:1–10. doi: 10.1186/s40249-020-00713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cihan P. Forecasting fully vaccinated people against COVID-19 and examining future vaccination rate for herd immunity in the US, Asia, Europe, Africa, South America, and the World. Appl. Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cihan P. The machine learning approach for predicting the number of intensive care, intubated patients and death: The COVID-19 pandemic in Turkey. Sigma J. Eng. Nat. Sci. 2022;40:85–94. [Google Scholar]
- Cooper M.D., Alder M.N. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Coppersmith D., Hong S.J., Hosking J.R. Partitioning nominal attributes in decision trees. Data Min. Knowl. Discov. 1999;3:197–217. [Google Scholar]
- Delves P.J., Roitt I.M. The immune system. N. Engl. J. Med. 2000;343:37–49. doi: 10.1056/NEJM200007063430107. [DOI] [PubMed] [Google Scholar]
- Deng X., Liu Q., Deng Y., Mahadevan S. An improved method to construct basic probability assignment based on the confusion matrix for classification problem. Inf. Sci. 2016;340:250–261. [Google Scholar]
- Douzas G., Bacao F., Last F. Improving imbalanced learning through a heuristic oversampling method based on k-means and SMOTE. Inf. Sci. 2018;465:1–20. [Google Scholar]
- Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006;27:861–874. [Google Scholar]
- Ghoshal B., Ghoshal B., Swift S. and Tucker A.. Uncertainty estimation in SARS-CoV-2 B-cell epitope prediction for vaccine development. In: Proceedings of the International Conference on Artificial Intelligence in Medicine. Springer, 2021, p. 361–6.
- Goodfellow I., Bengio Y., Courville A. MIT Press; 2016. Deep Learning. [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R., et al. Severe acute respiratory syndrome-related coronavirus: the species and its viruses – a statement of the Coronavirus Study Group. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(671–80) doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Wang H., Bell D., Bi Y., Greer K. OTM Confederated International Conferences" On the Move to Meaningful Internet Systems". Springer; 2003. KNN model-based approach in classification; pp. 986–996. [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Kapoor P., Chaudhary K., et al. In silico approach for predicting toxicity of peptides and proteins. PLOS One. 2013;8 doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(271–80) doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmer D.W., Jr, Lemeshow S., Sturdivant R.X. John Wiley & Sons; 2013. Applied Logistic Regression. [Google Scholar]
- Hosseini A., Hashemi V., Shomali N., et al. Innate and adaptive immune responses against coronavirus. Biomed. Pharmacother. 2020 doi: 10.1016/j.biopha.2020.110859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundi P., Shahsavari R. Comparative studies among machine learning models for performance estimation and health monitoring of thermal power plants. Appl. Energy. 2020;265 [Google Scholar]
- Jain N., Jhunthra S., Garg H., et al. Prediction modelling of COVID using machine learning methods from B-cell dataset. Results Phys. 2021;21 doi: 10.1016/j.rinp.2021.103813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaggle. COVID-19/SARS B-cell epitope prediction. 2021.
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLOS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DD. Representation quality in text classification: An introduction and experiment. Speech and Natural Language: Proceedings of a Workshop Held at Hidden Valley, Pennsylvania, June 24–27, 1990. 1990.
- Medzhitov R., Janeway C., Jr Innate immune recognition: mechanisms and pathways. Immunol. Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- Melo R., Lemos A., Preto A.J., et al. Computational approaches in antibody-drug conjugate optimization for targeted cancer therapy. Curr. Top. Med. Chem. 2018;18:1091–1109. doi: 10.2174/1568026618666180731165222. [DOI] [PubMed] [Google Scholar]
- Misbah S., Ahmad A., Butt M.H., Khan Y.H., Alotaibi N.H., Mallhi T.H. A systematic analysis of studies on corona virus disease 19 (COVID-19) from viral emergence to treatment. J. Coll. Physicians Surg. Pak. 2020;30:9–18. doi: 10.29271/jcpsp.2020.Supp1.S9. [DOI] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noumi T., Inoue S., Fujita H., et al. Epitope prediction of antigen protein using attention-based LSTM network. J. Inf. Process. 2021;29:321–327. [Google Scholar]
- Pathak S., Palan U. Science Publishers; 2005. Immunology: Essential and Fundamental. [Google Scholar]
- Pooja K., Rani S., Kanwate B., Pal G.K. Physico-chemical, sensory and toxicity characteristics of dipeptidyl peptidase-IV inhibitory peptides from rice bran-derived globulin using computational approaches. Int. J. Pept. Res. Ther. 2017;23:519–529. [Google Scholar]
- Powers D.M. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv Prepr. 2020 [Google Scholar]
- Quadeer A.A., Ahmed S.F., McKay M.R. Landscape of epitopes targeted by T cells in 852 individuals recovered from COVID-19: Meta-analysis, immunoprevalence, and web platform. Cell Reports Medicine. 2021;2(6):37–63. doi: 10.1016/j.xcrm.2021.100312. 100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9:231. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiger R.J. Chapman Hall/CRC; 2017. Data Mining: A Tutorial-Based Primer. [Google Scholar]
- Saha S., Raghava G.P.S. Prediction of continuous B‐cell epitopes in an antigen using recurrent neural network. Protein.: Struct. Funct. Bioinform. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T-and B-cell epitope prediction. J. Immunol. Res. 2017:2017. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygılı A. A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl. Soft Comput. 2021;105 doi: 10.1016/j.asoc.2021.107323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukat M.S., Foers A.D., Woodmansey S., Evans S.C., Fowler A., Soilleux E.J. Use of machine learning to identify a T cell response to SARS-CoV-2. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M.S., Ahmed S.F., Quadeer A.A., McKay M.R. In silico T cell epitope identification for SARS-CoV-2: progress and perspectives. Adv. Drug Deliv. Rev. 2021;171:29–47. doi: 10.1016/j.addr.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki V., Tiwari M., Tiwari V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci. Rep. 2019;9:1–19. doi: 10.1038/s41598-019-41496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir ul Qamar M., Saleem S., Ashfaq U.A., Bari A., Anwar F., Alqahtani S. Epitope‐based peptide vaccine design and target site depiction against Middle East Respiratory Syndrome Coronavirus: an immune-informatics study. J. Transl. Med. 2019;17:1–14. doi: 10.1186/s12967-019-2116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(1171–87) doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ToxinPred, 2021. Designing and prediction of toxic peptides.
- Turlapati V.P.K., Prusty M.R. Outlier-SMOTE: a refined oversampling technique for improved detection of COVID-19. Intell. -Based Med. 2020;3 doi: 10.1016/j.ibmed.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V. Springer Science & Business Media; 2013. The Nature of Statistical Learning Theory. [Google Scholar]
- VaxiJen, 2021. VaxiJen: Prediction of Protective Antigens and Subunit Vaccines. [DOI] [PMC free article] [PubMed]
- Vita R., Mahajan S., Overton J.A., et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashvardhini N., Kumar A., Jha D.K. Immunoinformatics Identification of B-and T-Cell Epitopes in the RNA-Dependent RNA Polymerase of SARS-CoV-2. Can. J. Infect. Dis. Med. Microbiol. 2021:1–8. doi: 10.1155/2021/6627141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani Z., Rafiei A., Yazdani M., Valadan R. Design an efficient multi-epitope peptide vaccine candidate against SARS-CoV-2: an in silico analysis. Infect. Drug Resist. 2020;13:3007. doi: 10.2147/IDR.S264573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li Y. Forecasting the COVID-19 vaccine uptake rate: an infodemiological study in the US. Hum. Vaccin. Immunother. 2022:1–8. doi: 10.1080/21645515.2021.2017216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]