Introduction
Maize rough dwarf disease (MRDD) is a worldwide disease caused by a virus (Bai et al., 2002; Harpaz, 1959). Rice black‐streaked dwarf virus (RBSDV) was identified as the major pathogen causing MRDD in maize (Zea mays) and dwarfing disease in other cereal crops, seriously threatening crop production in Asia (Bai et al., 2002). The recessive allele conferring MRDD resistance has been cloned and characterized (Liu et al., 2020). The natural variant with an alternative exon 10 caused by a helitron transposon insertion, designated ZmGDIα‐hel, weakened the interaction between the RBSDV P7‐1 protein and the encoded ZmGDIα‐hel protein, leading to quantitative resistance in maize plants. True loss‐of‐function alleles in ZmGDIα were expected to confer MRDD resistance at the cost of deleterious effects (Liu et al., 2020), as GDIα regulates small Rab GTPases, which are critical for vesicle membrane trafficking in eukaryotes (Schalk et al., 1996). However, plant Rab GTPases form the largest protein family and have evolved a unique set of 8 RAB sub‐families with divergent profiles in monocot and dicot species (Tripathy et al., 2021). Essential but redundant plant factors, such as Eukaryotic Translation Initiation Factor 4E, have been a major target for engineering viral resistance without affecting plant fitness (Bastet et al., 2017). We therefore aimed to explore the possibility of generating null alleles of ZmGDIα to engineer viral resistance. Our efforts may provide a novel approach to MRDD resistance breeding beyond the natural allele of ZmGDIα‐hel.
To generate a null mutant line, we constructed a CRISPR/Cas9 vector (Figure 1a) targeting exon 1 of ZmGDIα (Figure 1b) about 30 bp downstream of the translation start codon (Figure 1c). Stable transformation of maize inbred line ZC01 was performed as previously described (Li et al., 2017). We obtained 61 independent T0 maize plants that contained both the Bar and Cas9 genes, as determined by PCR amplification. Of those, 25 T0 plants harboured mutations at the intended target site based on PCR and sequencing. We selected the two edited event T0 plants E1 and E2, which were homozygous for a 1‐bp insertion (E1) or a 32‐bp deletion (E2), respectively (Figure 1c). The T1 plants were screened for transgene‐free with neither Cas9 nor Bar. The target sequencing data of both zmgdiα mutants confirmed their stable inheritance across the T0, T1 and T2 generations. The T1 and transgene‐free T2 mutant plants were characterized further.
Figure 1.
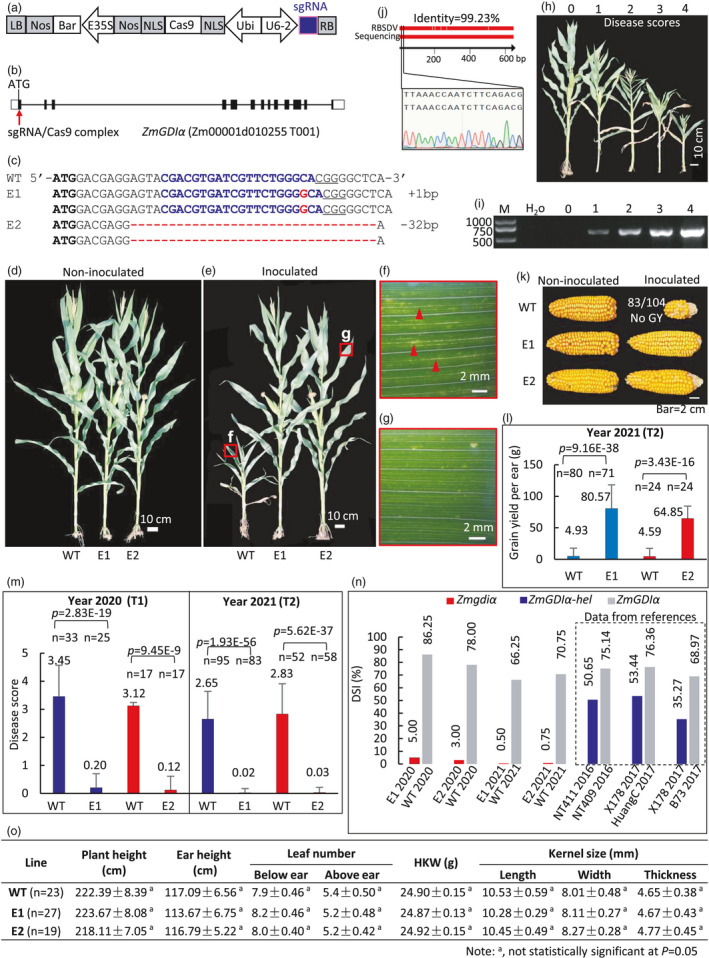
Genome editing ZmGDIα confers resistance against MRDD without agronomic penalty. (a) Constructed vector. (b) The designed target site. (c) Genotype of the selected homozygous mutations. (d, e) Comparison of non‐inoculated (d) and inoculated (e) plants. The leaves in the red rectangles in (e) are magnified to show MRDD symptoms in the WT (f) and E2 (g). (f, g) Typical waxy enation symptom of MRDD is seen on the WT (f) but not in mutant (g). (h) Representative plants with scored MRDD severity from 0 to 4 from resistance to susceptibility. (i) RT‐PCR detection of RBSDV showing that the virus titre is proportional to scored MRDD severity. (j) Verification of RBSDV identity by sequencing RT‐PCR amplicon in (i). (k, l) Comparison of representative maize ears (k) and grain yield (l) in the field. Eighty‐three out of 104 inoculated WT plants produced no grain. (m) Disease scores across years 2020 and 2021. (n) Comparison of DSI observed in the null mutants and the previously reported ZmGDI‐hel. NT411/NT409 and X178/Huang C data were reproduced from Liu C. et al. (2020). X178/B73 data were reproduced from Liu Q. et al. (2016). DSI (%) = ∑(disease score × number of plants with this disease score) × 100 / (maximum disease score × total number of plants). (o) The edited lines show no agronomic penalty without MRDD infection in the field. The differences were not statistically significant at P = 0.05.
We inoculated maize seedlings with RBSDV and then transplanted them to the field (Liu et al., 2016). The non‐inoculated edited plants showed no obvious differences compared to WT plants (Figure 1d). In sharp contrast, inoculated WT plants had much shorter overall height and internodes compared to E1 and E2 plants (Figure 1e). In addition, we only observed another typical MRDD symptom consisting of waxy enations on the abaxial surface of WT upper leaves (Figure 1f) but not in the mutants (Figure 1g), thus validating their resistant phenotype.
We mixed seeds from the WT and each null mutant line separately in a 1:1 ratio for blind artificial inoculation, transplanting and field phenotyping to exclude possible bias. We assigned a five‐grade disease score (Liu et al., 2016) to each plant (Figure 1h). We ascertained that the virus titre is proportional to MRDD severity (Figure 1i) by RT‐PCR using the primer pair that is specific to a 652‐bp region in the S4 segment of RBSDV. Sanger sequencing of the amplicon yielded the sequences that were 99.2% identical to the deposited RBSDV sequence at NCBI (#KY662121.1), confirming infection by this virus (Figure 1j).
We sequenced ZmGDIα in all blind‐mixed individuals for genotyping and scoring agronomic traits. We then calculated and compared the agronomic performance (Figure 1k, l) and disease score (Figure 1m) of the WT and mutant groups. Most inoculated WT plants produced few to no kernels. By contrast, inoculated edited mutant lines bore ears comparable to those seen on non‐inoculated plants (Figure 1k). In 2020, we identified each WT:mutant ratios were a 1:1 ratio from blind‐mixed population, as determined by a chi‐square test at P = 0.05 (Figure 1m, left). The average disease score of the E1 line was significantly different from that of the corresponding WT plants, as was the average disease score of the E2 line relative to its corresponding WT. In 2021, we further repeated this analysis using about three times as many plants as in 2020. Again, we obtained WT:mutant ratios consistent with a 1:1 ratio. The average disease scores for E1 and E2 were significantly different from those of their corresponding WT, respectively (Figure 1m, right). The resistance conferred by zmgdiα was higher than that of previously reported resistant materials carrying ZmGDIα‐hel, as evidenced by their much lower disease severity index (DSI) (Liu C. et al., 2016; Liu Q. et al 2020) (Figure 1n). These data indicated that the ZmGDIα locus identified by Liu et al. (2020) was a valuable target for engineering MRDD resistance and the generated null mutants might confer higher resistance.
To evaluate whether there are agronomic penalties due to the E1 or E2 mutations, we compared the agronomic performance under conditions free from RBSDV inoculation in the field, using a random‐block design with two repeats each consisting of about 60 plants. We observed no obvious phenotypic differences for plant growth or development between the WT and the null zmgdiα mutants (Figure 1d). In addition, all other measured parameters were comparable between WT, E1, and E2 plants (Figure 1o).
In summary, our data indicate that both null mutants generated through CRISPR/Cas9 editing exhibit stronger resistance against MRDD than the natural ZmGDIα‐hel allele. Our study also alleviates concerns about possible agronomic penalties associated with ZmGDIα loss‐of‐function mutants. Targeted editing of Rab GDIα might be extended in other monocot crops to engineer RBSDV resistance.
Conflict of interest
A related patent had been submitted to the State Intellectual Property Office of China.
Authors contribution
CL, MK, FY, JZ, XQ, JW, DD, and CX performed the experiments. CX and CL wrote the manuscript.
Acknowledgements
This research was supported by the National Science Foundation of China (32001551), the National Key Research and Development Program of China (2020YFE0202300), and the Agricultural Science and Technology Innovation Program of the CAAS (S2021ZD03).
Liu, C. , Kong, M. , Yang, F. , Zhu, J. , Qi, X. , Weng, J. , Di, D. and Xie, C. (2022) Targeted generation of Null Mutants in ZmGDIα confers resistance against maize rough dwarf disease without agronomic penalty. Plant Biotechnol. J., 10.1111/pbi.13793
References
- Bai, F.W. , Yan, J. , Qu, Z.C. , Zhang, H.W. , Xu, J. , Ye, M.M. and Shen, D.L. (2002) Phylogenetic analysis reveals that a dwarfing disease on different cereal crops in China is due to rice black streaked dwarf virus (RBSDV). Virus Genes, 25, 201–206. [DOI] [PubMed] [Google Scholar]
- Bastet, A. , Robaglia, C. and Gallois, J.L. (2017) eIF4E resistance: natural variation should guide gene editing. Trends Plant Sci. 22, 411–419. [DOI] [PubMed] [Google Scholar]
- Harpaz, I. (1959) Needle transmission of a new maize virus. Nature, 184, BA77–BA78. [Google Scholar]
- Li, C. , Liu, C. , Qi, X. , Wu, Y. , Fei, X. , Mao, L. et al. (2017) RNA‐guided Cas9 as an in vivo desired‐target mutator in maize. Plant Biotechnol. J. 15, 1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.L. , Hua, J. , Liu, C. , Zhang, D. , Hao, Z. , Yong, H. et al. (2016) Fine mapping of a quantitative trait locus conferring resistance to maize rough dwarf disease. Theor. Appl. Genet, 129, 2333–2342. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Deng, S. , Liu, B. , Tao, Y. , Ai, H. , Liu, J. et al. (2020) A helitron‐induced RabGDIα variant causes quantitative recessive resistance to maize rough dwarf disease. Nat. Commun. 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk, I. , Zeng, K.E. , Wu, S.‐K. , Stura, E.A. , Matteson, J. , Huang, M. , Tandon, A. et al. (1996) Structure and mutational analysis of Rab GDP‐dissociation inhibitor. Nature, 381, 42–48. [DOI] [PubMed] [Google Scholar]
- Tripathy, M.K. , Deswal, R. and Sopory, S.K. (2021) Plant RABs: role in development and in abiotic and biotic stress responses. Curr. Genomics, 22, 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]


