Abstract
The giant magnetoresistance (GMR) effect has seen flourishing development from theory to application in the last three decades since its discovery in 1988. Nowadays, commercial devices based on the GMR effect, such as hard-disk drives, biosensors, magnetic field sensors, microelectromechanical systems (MEMS), etc., are available in the market, by virtue of the advances in state-of-the-art thin-film deposition and micro- and nanofabrication techniques. Different types of GMR biosensor arrays with superior sensitivity and robustness are available at a lower cost for a wide variety of biomedical applications. In this paper, we review the recent advances in GMR-based biomedical applications including disease diagnosis, genotyping, food and drug regulation, brain and cardiac mapping, etc. The GMR magnetic multilayer structure, spin valve, and magnetic granular structure, as well as fundamental theories of the GMR effect, are introduced at first. The emerging topic of flexible GMR for wearable biosensing is also included. Different GMR pattern designs, sensor surface functionalization, bioassay strategies, and on-chip accessories for improved GMR performances are reviewed. It is foreseen that combined with the state-of-the-art complementary metal-oxide-semiconductor (CMOS) electronics, GMR biosensors hold great promise in biomedicine, particularly for point-of-care (POC) disease diagnosis and wearable devices for real-time health monitoring.
Keywords: GMR, flexible sensor, disease diagnosis, genotyping, food safety, brain mapping, cardiac mapping
Graphical Abstract
1. INTRODUCTION
The GMR effect was discovered by Fert and Grünberg for the first time in 1988 from multilayer structures composed of alternating ferromagnetic (FM) and nonmagnetic (NM) conductive layers (Figure 1(A1))1,2 This effect is observed as a “giant” change in the electrical resistance depending on the relative alignments of the magnetizations of adjacent FM layers. The magnetizations of FM layers can be remotely controlled by an external magnetic field, and the resistance of a GMR system is a function of the field magnitude. In this sense, GMR sensors become the preferred option for measuring ultralow magnetic fields, especially in small spaces. Since then, GMR sensors have become extremely widespread throughout the main industries and strategic applications, such as MRAM (magnetoresistive random-access memory), hard disk drives, nonvolatile memory, etc.3-5 The practical significance of the GMR effect has been recognized worldwide and was conferred the Nobel Prize in Physics in 2007.
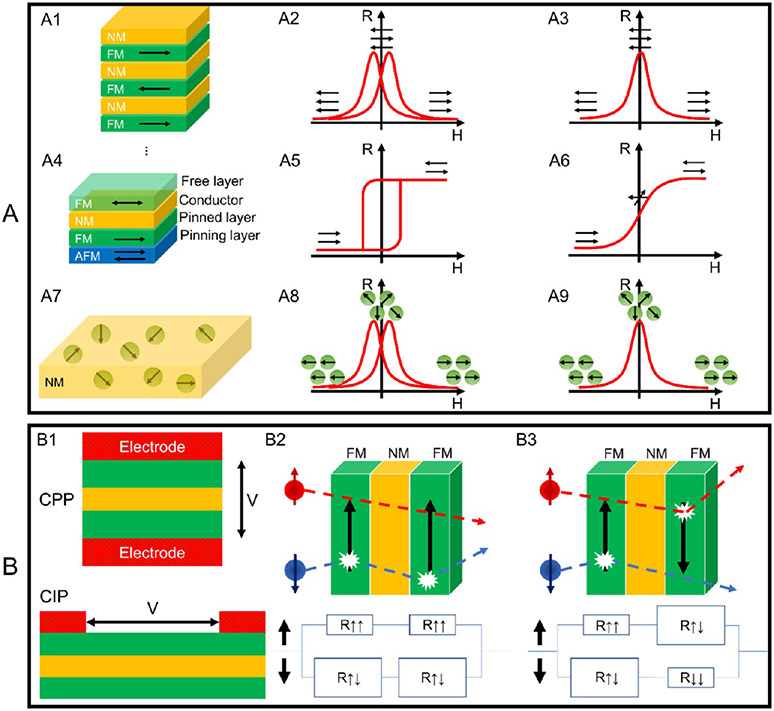
Figure 1.
(A) Schematic views of (A1) GMR multilayer structure, (A4) spin valve structure, and (A7) GMR granular solids, and the R─H curves of the GMR structures in (A2, A3) A1, (A5, A6) A4, and (A8, A9) A7. Schematic views of (B1) the CIP and CPP configurations and (B2, B3) the two-channel models.
GMR systems display superior sensitivities over anisotropic magnetoresistance (AMR) and Hall sensors. Benefiting from the advances in thin-film deposition platforms and micro- and nanofabrication techniques, GMR sensors can be patterned into different shapes and configurations to match different application purposes. In addition, the flexibility of designs allows single GMR, Wheatstone bridge GMR, and GMR array configurations based on needs. Combined with the standard complementary metal-oxide-semiconductor (CMOS) technologies, GMR sensors have been successfully integrated within chips, including in microelectronic circuits, providing compact sensing with better performance.6,7 In the field of biosensors, the enzyme-linked immunosorbent assay (ELISA) and electro-chemical immunoassay (ECIA) platforms are widely used immunoassay tools for disease diagnosis; however, they require not only sophisticated laboratory equipment with frequent equipment maintenance but also professional operation handled by technicians.8-10 These intrinsic characteristics make them unsuitable for point-of-care use.11,12 On the other hand, the optical labels in optical biosensors such as fluorescent dyes, quantum dots, and surface-enhanced Raman spectroscopy (SERS), although quite powerful, are vulnerable to photobleaching and unstable at room temperature, which has greatly restricted their applications for on-field tests. In recent years, significant progress has been made in the field of plasmonic biosensors, with many new plasmonic building blocks such as semiconductors, metals, magnetic materials, and meta-materials being reported to control the light properties in classical and quantum regimes. However, the main challenges in plasmonic biosensors are detection of small molecules at ultralow concentrations and production of compact devices for point-of-care (POC) analysis.13,14 The field-effect transistor (FET)-based biosensors, on the other hand, are versatile tools that directly convert the target molecules and receptor interactions into electrical signals. Although FET-based biosensors have shown high detection sensitivity and fast response, there are still great challenges in building FET-biosensors with miniaturization, low cost, simple operation, reproducibility, and reliability for nonspecialists.15,16 A lateral flow immunoassay (LFIA) is one of the most cost-effective sensors for the detection of multiple diseases including SARS-CoV-2, and the sensor response is read with the naked eye. It is by far the cheapest, fastest, and easiest option to fight the COVID-19 pandemic.17 However, applying the wrong sample size to the LIFA can lead to reliability problems in the test results. Additionally, sample pretreatment may be required for samples that may alter the capillary action (diffusion) of target molecules on the test strip.18,19 Gold nanoparticle-based colorimetry is another biosensing technique that significantly simplifies operating procedures and shortens detection time.20 The entire assay is performed with one solution mixture of the target and the probe without any washing steps, and the color change can be read directly with the naked eye. However, nanoparticle-based colorimetric assays have some disadvantages, such as low detection sensitivity and inability to perform multiplexed detection. Furthermore, it is still in the proof-of-concept stage and the background color of biological fluid samples such as human serum may interfere with the colored signal, affecting the accuracy of the results.21
As they use magnetic nanoparticles (MNPs) as labels, GMR biosensors have a robust labeling system. Furthermore, because biological tissues and fluids are nonmagnetic or diamagnetic, there is negligible background noise from the complex samples, and thus, a higher signal-to-noise ratio is greatly favored for magnetic biosensors such as GMR. Considering these intrinsic advantages of GMR biosensors, researchers are exploiting the potential of developing GMR biosensors into a user-friendly, cost-effective, and rapid testing tool for a growing list of biomedical applications in the field of magnetic field sensors, with an emphasis on biomedical applications such as magnetocardiography (MCG) and magnetoencephalography (MEG). Highly sensitive superconducting quantum interference devices (SQUIDs) are widely used for imaging the biomagnetic fields produced because of electric currents flowing in the active nerve cells in the brain and cardiac muscle. Because this kind of biomagnetic field is very weak, typically several picoteslas (1 pT = 1 × 10−12 T) for MCG and femtoteslas (1 fT = 1 × 10−15 T) for MEG.22 Very limited types of magnetic field sensors meet this requirement. SQUIDs, although highly sensitive, require cryogenic refrigeration to operate, which significantly increases their size, power consumption, and cost. GMR sensors, along with other femtotesla-level detection magnetic field sensors such as spin-exchange relaxation-free (SERF) atomic magnetometers (AMs) that do not require cryogenic operation, are at the forefront of research for MCG and MEG applications.23-27
In this paper, we focus on the recent advances of GMR sensors in biomedical applications. We first introduce different GMR systems including magnetic multilayer structure (Figure 1A1), spin valve trilayer structure (Figure 1A2), and magnetic granular structure (Figure 1A3). Among these, GMR spin valves are more widely used in biomedical areas because of the linear R─H (resistance-field) responses and simple trilayered thin-film structure. Unless otherwise noted, the GMR biosensors mentioned in this paper are GMR spin valves that consist of two FM layers separated by a thin NM spacer. The current in plane (CIP) and current perpendicular to plane (CPP) geometries, spin-dependent scattering, and two-channel resistor model are also explained in Section 2. Flexible GMR sensor is an emerging topic and holds great potential in the field of wearable sensors for in vivo health condition monitoring. Herein, we review some representative configurations of flexible GMR sensors. In Section 3, we review different GMR biosensor designs including biosensor patterns, GMR-based bioassay strategies, and accessories for improved GMR biosensing performances such as microfluidic channels, filters, and magnetic flux concentrator (MFC). Different research groups have independently reported point-of-care (POC) devices based on GMR biosensors, making it possible to carry out all-in-one, fully automatic, and GMR chip-based bioassays at the user end. In Section 4, we review the biomedical applications based on GMR biosensors such as for disease diagnosis, genotyping, food and drug regulation, brain and cardiac mapping, etc. The challenges and opportunities for GMR technology to become a superior healthcare solution are discussed in Section 5.
2. OVERVIEW OF DIFFERENT TYPES OF GMR SENSORS
2.1. GMR Effect in Magnetic Multilayer Systems.
The GMR can be described as the change in electrical resistance of magnetic multilayers in response to an external magnetic field. As shown in Figure 1A3, the FM layers are antiferromagnetically coupled in a zero field. Although the magnetizations of the FM layers are antiparallel, they can be aligned by an external magnetic field. The electrical resistance of a GMR FM/NM multilayer structure is relatively low when the magnetizations of FM layers are in parallel alignment (this resistance is denoted as RP), whereas the resistance is relatively high when magnetizations of FM layers are in antiparallel configurations, resistance denoted as RAP. The GMR ratio is characterized by (RAP – RP)/RP. This effect (i.e., the GMR ratio) is much larger than the anisotropic magnetoresistance (AMR) and therefore is called “giant” magnetoresistance. However, the presence of antiferromagnetic interlayer coupling is not a necessary condition for the GMR effect to occur. The GMR effect can also be achieved by arranging neighboring FM layers with different coercivities. As shown in Figure 1A2, the magnetizations of the soft and hard FM layers switch at different values of the applied magnetic field.
The second example of the GMR system is the spin valve trilayer configuration as shown in Figure 1A4, where the magnetization of one FM layer is pinned by an adjacent antiferromagnetic (AFM) layer, whereas the magnetization of the other FM layer is free to rotate with the external magnetic field. A typical R─H curve of the GMR spin-valve is schematically drawn in Figure 1A5. Although the GMR spin valves show a lower GMR ratio compared to the multilayer GMR systems, they are more popular for sub-picotesla to millitesla-range weak magnetic field detections. For the applications in GMR-based bioassays and magnetic field sensors, spin valves with a linear R─H curve are designed in the way that in zero fields, the magnetizations of FM layers are in a 90° configuration, as depicted in Figure 1A6. This 90° configuration in zero fields can be achieved by introducing magnetic shape anisotropy. Thus, for most spin valves in biomedical applications that require linear R─H responses, the sensors are designed with very high aspect ratios.
The magnetic granular system is another example that shows the GMR effect, denoted as g-GMR. In this g-GMR system, magnetic nanoparticles (MNPs) are either dispersed in conductive gel matrices or embedded in metallic matrices, as shown schematically in Figure 1A7. The R─H curves of g-GMR systems in Figure 1A8, A9 correspond to ferromagnetic and superparamagnetic nanoparticle conditions, respectively. This g-GMR system will be discussed in detail in Section 2.2.
So far, we have introduced three types of GMR structures. For the GMR multilayer structure and GMR granular solids, although they show very high MR ratios, they are not suitable for biomedical applications, because in most biomedical applications, a linear signal output (e.g., sensor voltage) as a function of the external magnetic field is preferred for quantitative bioassays and biomagnetic field recordings. To date, linear response curves are achieved in GMR spin-valve structures (Figure 1A6) by the perpendicular configuration of magnetizations through the manipulation of shape anisotropy or a bias field. It should be noted that there is a trade-off between the sensitivity (MR ratio) and linearity for GMR sensors. Although GMR spin valves show linear response curves and are able to detect weak magnetic fields in many reported works, they usually possess lower MR ratios compared to GMR multilayer structures, because only two FM layers are contributing to the GMR effect.
GMR sensors are generally built following two types of configurations: current in-plane (CIP) and current perpendicular to the plane (CPP), as represented in Figure 1B1. The CIP structure is preferred in industrial applications because it is easier to fabricate and more easily detectable than CPP because of a thicker film layer.28 In the CPP configuration, spin-dependent scattering is produced at the interfaces, consequently generating an interface resistance.29
It is observed that electron spin is conserved if the travel distance is below the mean free path,30 for this specific reason that the NM layer requires a thickness that does not exceed this limiting interval. Because of spin conservation, it is possible to meticulously describe the spin state separately; therefore, electric current flows in two separate channels: one for a spin down and the other for a spin up, as shown in Figure 1B2, B3. Taking the CPP configuration as an example, the resistivity can be self-averaged and modeled by resistors in series. The quantum mechanical tunneling effect of electrons in thin FM layers is primarily due to spin-dependent scattering from 4s to 3d orbitals. Strong scattering happened to spin aligned antiparallel to the direction of magnetization and weak scattering for parallel ones. The conductivity for each spin state is modeled with two resistors, one for each layer. Therefore, two pairs of resistors in parallel are obtained because the scattering of electrons is highly spin dependent in ferromagnetic materials. Spin up states are slightly scattered, and hence, low resistance; and spin down states are scattered in the ferromagnetic configuration, and hence, high resistance. Thus, the total resistance is approximately equal to the low resistance. The second configuration happens when a strong scattering for each spin occurs at least once in a layer.
2.2. GMR Effect in Granular Systems.
Although the GMR effect is prevalently reported in magnetic multilayer systems, any materials and microstructures that have interfaces between FM and NM materials are theoretically able to show the GMR effect.31,32 GMR structures such as granular MNPs embedded in an NM conductive matrix also show a spin-dependent scattering of electrons at the particle–matrix interfaces, as shown in Figure 1A7. In this section, we will discuss several granular GMR systems and their electron transport properties.
The first granular system reported showing the GMR effect was Co─Cu alloy thin film. It was reported by Berkowitz et al. and Xiao et al. independently in 1992.32,33 A microscopic photo of the granular Co─Cu thin film is shown in Figure 2A.32 Since then, other granular thin-film systems such as Co─Ag, Fe─Ag, and Fe─Cr have been demonstrated to show the GMR effect.34-36 To date, most of the granular material systems that exhibit the GMR effect are granular alloys containing two or more immiscible metal elements. The metals are typically deposited on the substrates via magnetron sputtering, molecular beam epitaxy (MBE), or other deposition methods. The magnetic elements then precipitate to form small granules due to low solubility.37
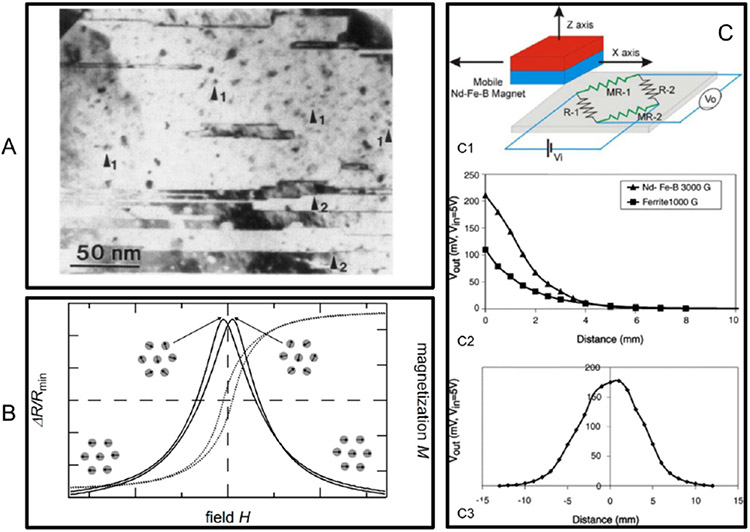
Figure 2.
Granular GMR films and sensors. (A) Micrograph of Co granules embedded in Cu matrix. (B) Schematic illustration of the GMR effect in granular thin films. (C) Granular GMR sensors. (C1) Sensor structure. (C2, C3) Response of the vertical and horizontal movement of the permanent magnet. (A) Reprinted with permission from ref 32. Copyright 1992 American Physical Society. (B) Reprinted from ref 29. Licensed under CC BY 4.0. (C) Reprinted with permission from ref 38. Copyright 2005 Elsevier B.V.
A schematic illustration of the origin of the GMR effect in granular systems is presented in Figure 2B. In the absence of an external magnetic field, the magnetization directions of the small magnetic granules are randomly distributed, and the resistance of the system is at its maximum.29 When a magnetic field is applied, the magnetization directions of these granules are aligned along the external field direction, which results in the reduction of resistance. Different theoretical models have been proposed to describe the GMR effect in granular alloys. Among them, the very first and most frequently used was reported by Zhang and Levy.39,40 They stated the GMR effect in granular films is similar to the CPP multilayer GMR systems, and therefore, their previous formalism for a CPP-GMR multilayer structure was used.41 By limiting the discussion to low temperature and assuming that the granules are single-domain MNPs, they found that the GMR ratio of granular thin films could be further increased by (1) reducing impurities in the matrix, (2) increasing the spin-dependent to spin-independent potentials ratios, (3) increasing interfaces’ spin-dependent scattering roughness, (4) decreasing the sizes of granules until these granules can be magnetically saturated by the applied field (for a fixed magnetic field and temperature condition, if the applied field cannot magnetically saturate the granules, then the GMR ratio decreases with the size of the granules), and (5) increasing the concentration of the magnetic granules. They also pointed out that the size distribution of the granules was a key factor that affect the GMR ratio of the granular system.
On the basis of the granular alloy films, several granular GMR sensors have been developed. For example, Arana et al. reported a linear position sensor based on Ag─Co alloy thin films.38 The schematic sensor structure is shown in Figure 2C1. They demonstrated that the granular GMR sensors can be used to measure the position of the permanent magnets, and the results are shown in Figure 2C2, C3.
2.3. Flexible GMR Sensors.
2.3.1. Printable GMR Sensors.
Recent years have witnessed increasing research interests in flexible sensors, in that these devices possess great potentials in the field of wearable appliances. One of the most cost-efficient ways to fabricate such flexible sensors is printing. Although printable electronics have been developed for many years, printable GMR sensors were first demonstrated in 2012 by Karnaushenko et al.42 To get a printable magneto-sensitive ink, they first deposited [Co/Cu]50 GMR multilayer films on a buffer-layer-coated silicon substrate via magnetron sputter deposition. Next, the as-deposited GMR multilayers are released from the substrate, dried, and ball milled into GMR flakes. These [Co/Cu]50 GMR flakes were then dispersed in a conductive polymer-based binder solution, and the resulting magnetic ink was printed on various substrates including flexible ones such as paper, as shown in Figure 3A. These printable sensors were proven to be able to achieve a maximum GMR ratio of 8% at room temperature.
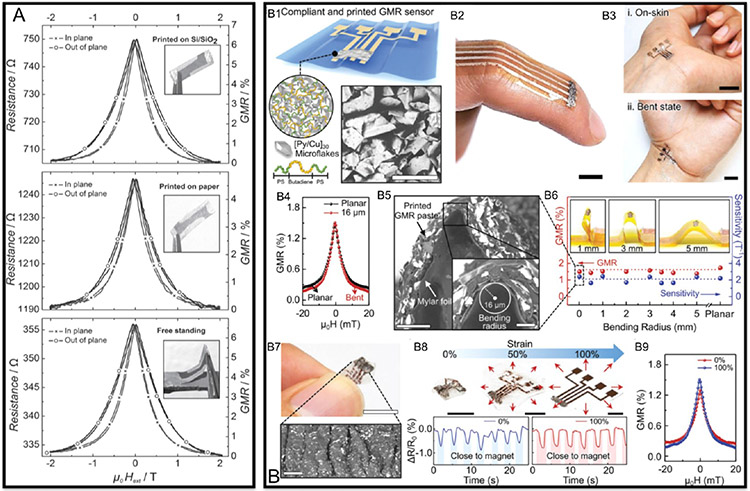
Figure 3.
GMR effect in polymer mixed GMR flakes. (A) GMR ratios for sensors painted on Si wafer (top), paper (middle), and the free-standing sensor (bottom). (B) Printable and stretchable GMR sensors. (B1) Schematic illustration of the printable GMR sensor. (B2, B3) Photographs of flexible GMR sensors on skin. (B4, B5) show the GMR ratio and morphology of the sensor under a bending radius of 16 μm. (B6) GMR ratios and sensitivities under different bending conditions. (B7) Image of the sensor surface after stretching. (B8, B9) Performance and GMR ratio of the sensor under stretching conditions. (A) Reprinted with permission from ref 42. Copyright 2012 Wiley–VCH. (B) Reprinted from ref 44. Licensed under CC BY 4.0.
The metallic multilayer flakes are the cause of the GMR effect, whereas the conductive polymeric solution prevents the flakes from oxidation and serves as a binder to make the flakes printable. Therefore, these are two key factors that impact the performance of such printable GMR sensors. In 2015, Karnaushenko et al. prepared another type of printable GMR sensors with the same kind of GMR flakes but different conductive polymer matrix, and they increased the maximum GMR ratio to 37%.43 Very recently, Ha et al. fabricated a stretchable GMR sensor with high performance by screen printing.44 They used poly(styrene–butadiene-styrene) (SBS) elastomer as the binder solution and mixed it with [Pu/Cu]30 GMR multilayer microflakes, then printed the GMR paste onto a thin polymeric foil to obtain a flexible sensor as shown in Figure 3B1-B3. This screen-printed GMR sensor exhibited a GMR ratio of 1.95% under an external field of 0.88 mT and shows no obvious decrease in GMR ratio under bending (see Figure 3B4-B6) and stretching conditions (see Figure 3B7-B9), which made it a good candidate for wearable electronics.
2.3.2. GMR Sensors on Ultrathin Flexible Substrates.
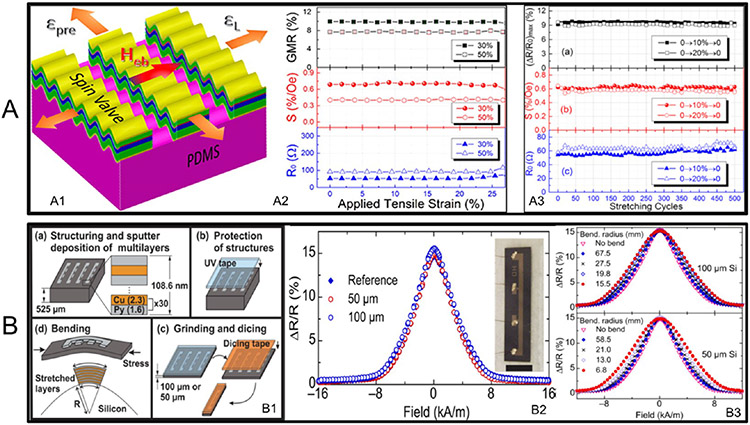
Besides printing, a more traditional way of fabricating flexible GMR sensors is directly depositing the GMR multilayers on flexible substrates. In 1992, Parkin et al. demonstrated that Co/Cu multilayers deposited on thin Kapton substrates still show the GMR effect, and the GMR ratio was comparable to the Co/Cu multilayers on rigid silicon substrates.45 Since then, various organic thin films have been found to be suitable as substrates for flexible GMR sensors, such as polyethylene naphthalate (PEN)46 and polyethylene terephthalate (PET),47 because of their good mechanical properties including light weight and high strength.48 However, using organic thin films as substrates also has severe limitations.49 For one thing, the GMR multilayers are metallic films that can only be stretched or bent to a very small extent. For the other, because of the large elastic property differences between the metallic films and the substrates, the large residual strain could cause low robustness of GMR sensors. To overcome these limitations, Li et al. deposited dual spin-valve GMR structures on polydimethylsiloxane (PDMS) substrates with uniaxial tensile prestrains.50 The functional layers formed periodic wrinkled ribbons patterns as shown in Figure 4A1. This shape enabled a nearly constant GMR ratio and field sensitivity over a tensile strain range of 0–25% (see Figure 4A2). The fatigue tests revealed that these sensors can maintain their high performance after hundreds of stretching and releasing cycles, as shown in Figure 4A3.
Figure 4.
GMR sensors on flexible substrates. (A) GMR sensors on prestressed PDMS substrates. (A1) Schematic illustration of the sensor. (A2) GMR ratio of the sensor under strain from 0 to 25%. (A3) GMR ratio during stretching–releasing cycles. (B) GMR sensors on ultrathin Si membrane. (B1) Schematic illustration of the fabrication process of the GMR sensor. (B2) GMR ratio of sensors on rigid Si wafer, 50 μm thick substrates, and 100 μm thick substrates. (B3) GMR ratio of the sensors under different bending conditions. (A) Reprinted with permission from ref 50. Copyright 2016 American Chemical Society. (B) Reprinted with permission from ref 54. Copyright 2015 AIP Publishing LLC.
Instead of polymer films, rigid substrates such as silicon wafers and glasses can also become flexible by decreasing their thickness to a micrometer range.51-53 For example, Pérez et al. reported the flexible GMR sensors on an ultrathin silicon substrate with thicknesses of 100 and 50 μm.54 As presented in Figure 4(B1), the fabrication process started with the deposition of GMR multilayers on silicon wafers, followed by a grinding process from the backsides of the wafers to reduce the thickness to 100 or 50 μm. The minimum acceptable bending radii were 15.5 mm and 6.8 mm for 100 and 50 μm silicon substrates, respectively. Within these ranges, a GMR ratio of around 15.3% was achieved regardless of membrane thickness and bending radius, as shown in Figure 4B2, B3.
3. GMR BIOSENSOR DESIGNS
3.1. Different GMR Biosensor Patterns.
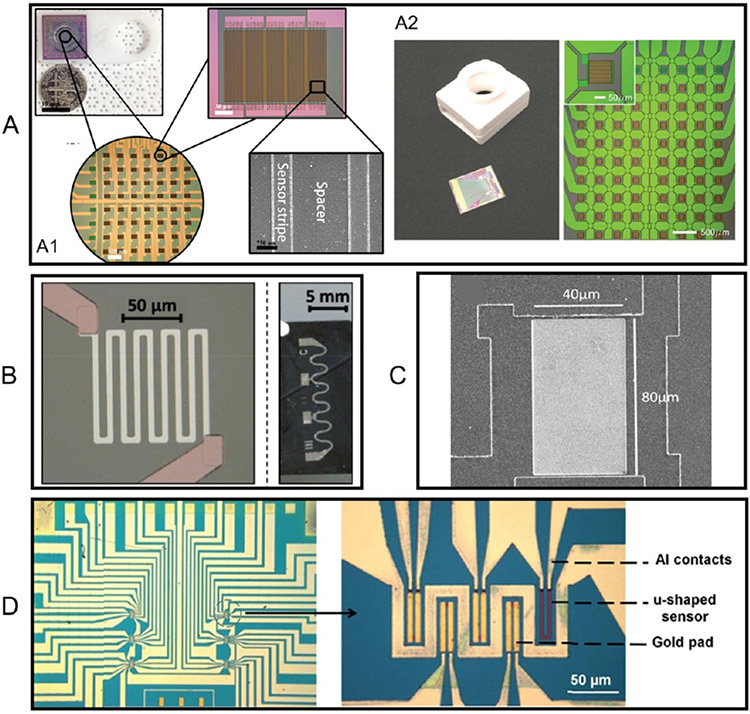
Benefiting from the current micro- and nanofabrication techniques, each GMR biosensor is typically in the size of tens to hundreds of micrometers. As shown in Figure 5, theoretically, GMR biosensors could be scaled to more than 1 million sensors/cm2.55,56 To date, different GMR biosensor patterns have been reported, for example, a stripe shape57,58 (Figure 5A), a spiral shape59,60 (not shown), a meander shape61 (Figure 5B), large-area62 (Figure 5C), and a “U” shape63 (Figure 5D). It is reported that for GMR-based bioassays, the MNP labels landing at the sensor edges cause larger signal differences compared to the same MNP labels landing on top of the sensor.20 This conclusion favors the design of GMR biosensors with higher aspect ratios, such as the stripe and spiral shapes. On the other hand, the large-area GMR biosensors realize the detection of MNP labels through a local-magnetic-reversal-nucleation mechanism.62,64
Figure 5.
Different GMR biosensor patterns. (A) GMR stripe shape biosensors. (A1, A2) Two types of GMR stripe biosensor designs reported independently by two research groups. (B) Spiral shape GMR biosensor pattern. (C) Large-area GMR biosensor pattern. (D) “U” shape GMR biosensor pattern. (A1) Reprinted from ref 57. Licensed under CC BY 4.0. (A2) Reprinted from ref 58. Licensed under CC BY 4.0. (B) Reprinted with permission from ref 61. Copyright 2015 Wiley–VCH. (C) Reprinted with permission from ref 62. Copyright 2017 AIP Publishing LLC. (D) Reprinted with permission from ref 63. Copyright 2016 The Royal Society of Chemistry.
In addition to the sensing mechanism-driven sensor pattern design, some practical issues should be considered when designing the GMR biosensors. Sensors with high aspect ratios (such as the stripe, spiral, and meander shapes in Figure 5A, B) have higher risks of open circuitry. Minor defects in the GMR thin films/substrates and even some photoresist residues (before the sensor pattern etching step) may cause breakage in these GMR sensors. This issue is mitigated when connecting dozens of GMR stripes in series and parallel. For example, in Figure 5A1, one GMR biosensor consists of 5 groups of GMR stripes connected in series and each group consists of 10 GMR stripes connected in parallel. GMR stripes connected in series can avoid the risks of open circuitry, whereas GMR stripes connected in parallel can increase the overall resistance as well as the signal-to-noise ratio (SNR).
Another major issue of stripe-shaped GMR biosensors is the small effective sensing area. Compared to the large area GMR biosensors, the effective sensing area of stripe-shaped GMR biosensors is small because of the spacers between sensor strips (see Figure 5A1). Some MNP labels may land on the spacers and do not contribute to the GMR signal. In addition, bioassay repeatability is a concern when measuring low abundancy target analytes because the captured MNP labels are randomly distributed on the sensor and spacer regions. In view of this, the large-area and “U”-shaped GMR biosensors are favored.
3.2. GMR Biosensor Surface Functionalization.
The sensitivity of the GMR biosensor is strongly dependent on the reactivity of capture probes that are immobilized on the sensor surface. Thus, the GMR sensor surface modification is a crucial step before transforming a GMR sensor into a functional biosensor. To date, different kinds of surface modification methods have been reported to modify the surface of rigid substrates including glass, silicon, polystyrene, etc.65-69 Herein, we introduce two frequently used GMR sensor surface modification methods, the 3-aminopropyltriethoxysilane (APTES)/glutaraldehyde (Glu) [APTES/Glu] and the 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS) [EDC/NHS]70 methods. The steps to carry out each surface modification method are summarized in Table 1.
Table 1.
GMR Sensor Surface Modification Methods
| method | APTES/Glu method | EDC/NHS method |
|---|---|---|
| pretreatment | GMR chips are subjected to oxygen plasma or washed with acetone followed by methanol and finally isopropanol then air-dried with nitrogen to remove organic residues adsorbed onto the surface | |
| modification steps | 1. add 0.5–10% APTES solution (in toluene/acetone) to the GMR sensor surface and incubate at room temperature for 5 min, then rinse with acetone, followed by deionized water, and dry with N2 | 1. add polyamine solution to the GMR sensor surface and incubate at room temperature for 5 min, then rinse with deionized water and dry with N2 |
| 2. add 5% Glu solution to the GMR sensor surface and incubate at room temperature for 5 h; rinse the GMR chip with deionized water and dry with N2 | 2. add polyacid solution to the GMR sensor surface and incubate at room temperature for 5 min, then rinse with deionized water and dry with N2 | |
| 3. GMR sensor is ready for immobilizing capture probes | 3. add EDC/NHS solution to GMR sensor surface and incubate at room temperature for 1 h; rinse the GMR chip with deionized water and dry with N2 | |
| 4. GMR sensor is ready for immobilizing capture probes | ||
It is worth mentioning that GMR sensors are usually coated with an ultrathin passivation layer (typically Al2O3, SiO2, and Si3N4 thin films) to avoid the corrosion caused by interfacing with biological samples. There is a trade-off on the thickness of this passivation layer. It should be thin enough to allow magnetic signals from MNP labels to be detectable by the GMR biosensors, and it should be thick enough to avoid the leakage of liquid through defects/pores in the passivation layer. Typically, this passivation layer is around tens of nanometers.
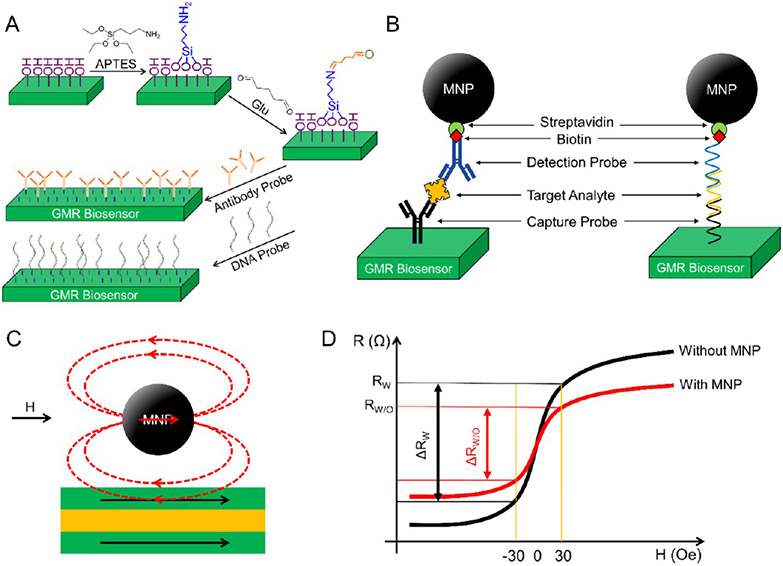
Before the surface modification steps, the substrate surfaces should be pretreated to remove organic residues and increase the surface hydrophilicity for better chemical treatment. For example, in some reported pretreatment steps, substrates are usually subjected to oxygen plasma (or ultraviolet ozone) and then treated with H2O2 solution.57,71,72 Substrate surfaces are more hydrophilic and are easier for further chemical treatment and grafting of proteins after the pretreatment. As shown in Figure 6A, in the APTES/Glu modification method, the GMR sensor surface is then treated with APTES, where the silanol groups will attach to the hydroxyl groups (─OH) from the substrate surface. Once on the surface, these silanes polymerize internally to form Si─O─Si (siloxane) linkages, exposing amine groups (─NH2) on the other end. Next, the Glu solution is added, where one of the two aldehyde groups (─CHO) is covalently bonded with the amine group from APTES. The other reactive aldehyde group can form a covalent bond with the amine groups from capture probes. Other GMR sensor surface modification methods using polyethylenimine (PEI),72 polyvinyl chloride (PVC),73 epoxysilane (3-glycidoxypropyl-trimethoxysilane),74 etc. have also been reported. After the surface modification with functional groups, GMR biosensors/chips can be stored under a dry environment before anchoring capture probes.
Figure 6.
(A) Schematic view of the APTES/Glu method for GMR sensor surface modification and functionalization of capture probes. (B) Schematic view of the sandwich assay. (C) Upon the application of an external magnetic field, the stray fields generated by MNPs can disturb the local magnetizations in the GMR biosensor’s free layer. (D) Schematic view of the R─H curves of the GMR sensor with and without the MNPs captured on the sensor surface. (A) Reprinted from ref 57. Licensed under CC BY 4.0.
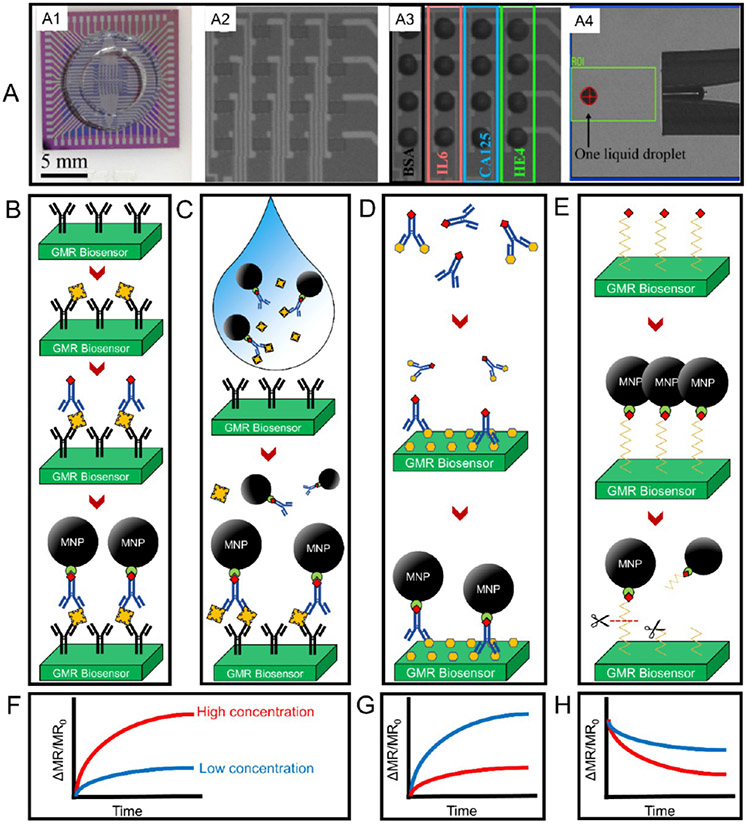
As is mentioned in Section 3.1, the modern micro- and nanofabrication techniques allow the integration of dozens to thousands of GMR biosensors in a fingernail-sized chip. Combined with the external CMOS electronics, each GMR biosensor works independently, which allows the multiplexed detection of a panel of target analytes in real time. For example, Figure 7A1 is a 2 cm × 2 cm GMR chip consisting of an 8 × 8 GMR biosensor array. Theoretically, these independently working biosensors allow the multiplexed detection of up to 64 analytes. With a nanoplotter facility (such as the SCIENION sciFLEXARRAYER product lines and the GeSiM microarray printers), a fixed volume of capture probe liquid droplet (see the photograph of one 400 pL droplet from Figure 7A4) can be precisely dispensed on each GMR biosensor with submicrometer and subnanoliter resolutions. Figure 7A2, A3 are the photographs of a 4 × 4 GMR biosensor array before and after dispensing capture probes (i.e., IL6, CA125, and HE4, BSA (bovine serum albumin) serves as negative control), respectively. The nanoliter volume droplets containing capture probes are incubated on the GMR biosensors overnight at 4 °C under a relative humidity of ~90% to allow immobilization. After anchoring the capture probes to GMR biosensors, the residues (unbound capture probes) are washed out and the biosensors are further blocked with BSA to avoid the nonspecific binding of ligands from biological samples.
Figure 7.
(A) GMR biosensor arrays designed for multiplexed bioassays. (A1) One example of a 2 cm × 2 cm GMR biosensor chip consists of 64 GMR biosensors working independently. (A2, A3) 4 × 4 GMR biosensor array before and after dispensing nanoliter volume capture probes and BSA. (A4) Photograph of one 400 pL liquid droplet dispensed by a nanoplotter facility. Steps of carrying out (B) a traditional sandwich bioassay, (C) a wash-free sandwich bioassay, and (D) a competitive bioassay on GMR biosensors. (E) Steps of carrying out enzyme detection on GMR biosensors. (F–H) Schematic drawings of GMR signals in the scenarios of high and low concentrations of target analytes for B–E, respectively. (A) Reprinted with permission from ref 77. Copyright 2018 Elsevier B.V.
It should be noted that the surface modification methods of GMR sensors described above are to increase the adsorption capacity of capture probes (such as capture antibodies, capture nucleic acids, etc.), to obtain a larger number (higher area density) of capture probes. After the functionalization step, it is crucial to apply a blocking layer (e.g., BSA) to cover the remaining binding sites on the sensors. The blocking layer can effectively reduce the nonspecific binding of reagents from the test sample, thus reducing the biosensor noise (note: this noise is caused by nonspecific binding, which is different from the inherent noise of the GMR sensor itself). In addition, the selection of suitable capture probes with the highest binding affinity and specificity for the target analyte is also of great significance for the overall implementation of GMR biosensors. The binding affinity and specificity information on capture probes for the target analyte can be either obtained from the manufacturer (if purchased from the manufacturer) or confirmed with the help of ELISA and Western blot. GMR biosensors can also be used for profiling the affinity and specificity of capture probes.55 Choosing the right type of capture probe with high binding affinity and specificity for the target analyte can further increase the detection limit of the GMR biosensor (in a biological manner) and reduce the time required for binding to reach equilibrium.75
3.3. Different GMR-Based Bioassay Strategies.
The GMR-based bioassay is similar to the standard immunoassay platforms such as ELISA (enzyme-linked immunosorbent assay). In Figure 6B, take the sandwich bioassay scheme as an example, where the capture probes (i.e., capture antibody, capture DNA/RNA, etc.) are first immobilized on the GMR biosensor surface, then the target analytes (i.e., protein biomarkers, peptides, ssDNA or RNA) of interest are introduced and specifically bind to the capture probes. Next, the MNP-labeled detection probes (i.e., detection antibody, detection DNA/RNA, etc.) are introduced and specifically bind to the target analytes and form a “sandwich” structure. The greater number of target analytes from the biological sample, the more MNPs are coupled on the GMR biosensor surface. As shown in Figure 6C, upon the application of an AC magnetic field, the stray fields generated by captured MNP labels can disturb the local magnetizations in the GMR biosensor’s free layer, resulting in a resistance change of the GMR biosensor (see Figure 6D). The change in resistance is usually monitored as the GMR biosensor signal (in the form of voltage or ΔMR, or ΔMR/MR0), which is then mapped into the domain of target analyte amount/concentration. The sandwich bioassay scheme itself is intrinsically highly sensitive and robust as the capture/detection probes bind to two sites from the target analyte. Thus, it has been widely used in the GMR-based bioassays for disease diagnosis, genotyping, toxin and pathogen detection, etc.71,76-80
However, the traditional GMR-based sandwich bioassay described above requires several wash steps to remove the unbound reagents, and thus, it is limited in lab-based bioassays handled by professionals, which as a result, limits its application for bioassays in nonclinical settings (i.e., school, home, clinic, in he field) by a layperson. Figure 7B shows the steps to carry out one sandwich assay on GMR biosensors. It requires the operator to wash out the unbound analytes after the incubation with a biological sample and then wash out the unbound detection probes before adding the MNP labels. In view of this disadvantage, a wash-free sandwich bioassay was reported in recent years to free the users’ hands.81,82 As shown in Figure 7C, the MNP labels, detection probes, and the biological sample containing target analytes are first incubated to allow the specific binding reach to equilibrium. This mixture is then added to the capture probe-functionalized GMR biosensors for a one-step assay. In this scenario, only the MNP label captured by the GMR biosensor surface through the capture probe–target analyte–detection probe structure causes the GMR signal change, whereas the unbound (or free) MNP labels are floating in the solution and their stray fields are too weak to be picked up by the GMR biosensors because magnetic fields decay with distance cubed. The GMR signal in the form of ΔMR/MR0 monotonically increases with the increase in target analyte concentrations as schematically shown in Figure 7F.
Other bioassay schemes such as the competitive bioassay, although less sensitive to experimental errors as it only requires one binding site on the target analyte, have also been reported for detecting small molecules that usually lack two binding sites/epitopes required for the sandwich assay.83-85 As shown in Figure 7D, the detection probes are at first incubated with a biological sample containing target analytes to allow the specific binding to saturate. The mixture is then added to GMR biosensors that are densely covered with target analytes, where the unoccupied detection probes can bind to GMR biosensors. After washing out the free reagents, MNP labels are added to bind to the immobilized detection probes through biotin–streptavidin interaction. For the GMR-based competitive bioassays, the GMR signal monotonically decreases with the increase in target analyte concentrations as schematically shown in Figure 7G.
As an honorable mention, the GMR biosensor has also been reported for the detection of enzyme/protease.86 As shown in Figure 7E, in an enzyme/protease assay, MNP labels are first bound to the GMR biosensor surface through a biotinylated peptide; with the addition of target enzyme/protease, peptides are cleaved, and MNP labels are released from the surface, causing a GMR signal drop (Figure 7H). With a higher concentration of target enzyme/protease, a higher GMR signal drop is expected. For example, Adem et al. reported the quantitative detection of the cysteine protease, papain. They immobilized MNPs to the surface of a GMR biosensor through the linkage of papain peptide.86 The GMR sensor array containing immobilized MNPs was then incubated with papain for 160 min, and the MNPs were released from the surface in a time-dependent manner. They detected a significant release of MNPs after 3.5 min of incubation using 4 nM of the cysteine protease, papain. They have also confirmed the detection of papain from urine samples.
On a separate note, all the GMR-based biosensors rely on the detection of MNP labels that are captured to or released from (i.e., in enzyme/protease assay shown in Figure 7E, H) the sensor surface. When an external magnetic field is applied, the stray field generated by these captured MNPs will interfere with the local magnetization in the free layer of the GMR biosensor, which will cause the sensor’s MR (and resistance) to change and achieve quantitative detection of the target analyte. There are several technical requirements for choosing MNP labels. First, MNPs should be superparamagnetic, and thus, they show zero magnetic moments in the absence of an external magnetic field. This superparamagnetic property of MNPs can effectively avoid the aggregation of nanoparticles, which may lead to a nonlinear increase in the magnetic signal with the concentration/amount of the target analyte. The requirement of superparamagnetism limits the magnetic core sizes of MNPs to tens of nanometers, as well as limiting the maximum magnetic moment per MNP. In pursuit of higher sensitivity for detecting an ultralow number of target analytes, MNP labels with larger magnetic moments are preferred. Thus, researchers can use multicore magnetic beads (usually several hundred nanometers to several micrometers) that are composed of smaller be superparamagnetic MNPs embedded in a matrix to maintain superparamagnetism and meanwhile high magnetic moment per MNP/bead. Second, MNPs should exhibit high colloidal stability (can be measured by the zeta potential). MNPs with low colloidal stability may precipitate during the bioassay process and cause false positive/negative results. The MNP surface chemical modifications and the pH of the solution can both affect the colloidal stability, and thus, special attention should be paid when designing MNPs. Third, MNPs should show uniform size distribution to improve the repeatability of results. For detecting samples with very low concentrations/number of target analytes, only very low amounts of MNPs are captured onto or released from the GMR biosensors. Thus, the variation in magnetic signal/moment generated by each MNP may cause repeatability issues. Although it is impossible to synthesize MNPs with identical sizes and shapes, higher size uniformity is preferred for GMR bioassays. More detailed discussions on how to choose and design MNPs for GMR-based bioassays are available in our previous work.87,88
3.4. GMR Point-of-Care Devices and Implementation Details.
3.4.1. GMR Point-of-Care (POC) Devices.
Point-of-care (POC) devices are intended for collecting clinical information on patients in resource-limited regions (mostly), clinics, and laboratories.89 The global POC diagnostics market is rapidly growing with acute and chronic diseases worldwide and overwhelming pandemics such as COVID-19.90-95 POC devices allow users with minimum training requirements to run tests at nonclinical settings; if combined with the Internet of Medical Things (IoMT), it could help patients receive proper healthcare at home and establish a comprehensive disease management database for government and healthcare organizations, benefiting the fast disease surveillance and control.94 The usefulness of the POC device is largely dependent on the cost (including cost per device and cost per bioassay), portability, accuracy, accessibility of assay reagents, and the average assay waiting time.
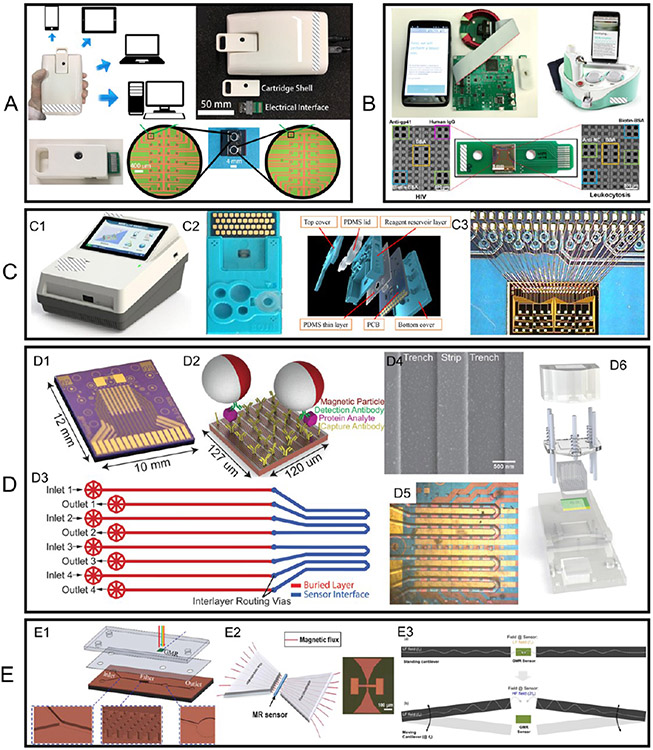
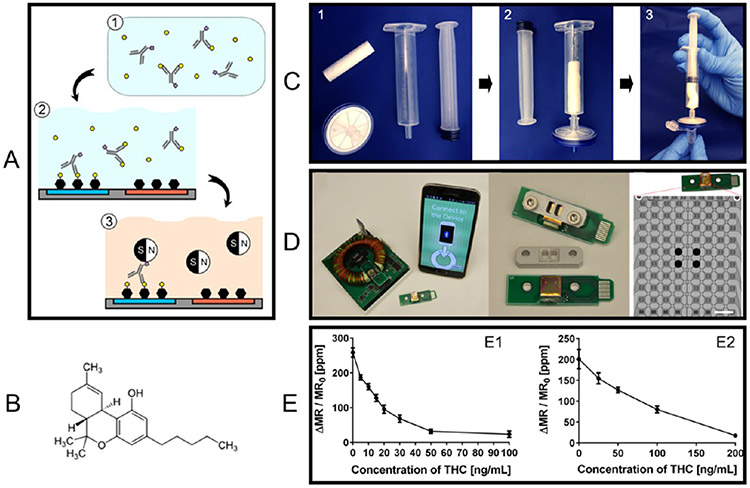
GMR biosensing platforms, after decades of development, have also been developed into POC devices by different research groups. As shown in Figure 8A, a University of Minnesota research group has developed a GMR POC device, Z-Lab.96 Z-Lab realizes real-time GMR signal collection and transmission wirelessly to a smartphone or through a USB connection to a desktop. This Z-Lab POC device consists of three parts: a plastic cartridge, an electrical interface connecting the electrodes from GMR biosensors to the PCB, and a hand-held device. The disposable, flash drive-sized cartridge contains one GMR chip that hosts 29 individual GMR biosensors. A Stanford University research group reported a GMR POC device called the Eigen Diagnosis Platform, as shown in Figure 8B.97 Sharing similar designs, the cartridge contains one GMR chip consisting of 80 individual sensors. Another group from the Chinese Academy of Sciences reported a GMR POC platform that added a microfluidic channel, as shown in Figure 8C, which allows for fully automatic assays.73 Instead of a cartridge with an electrical interface connecting the sensor electrodes, the GMR chip consisting of 40 individual sensors is wire-bonded to the PCB. The electrical parts of the GMR POC device are around $100 and each assay costs around $5 (including one GMR chip embedded in a disposable cartridge, MNP labels, and reagents) if mass produced.82
Figure 8.
(A–C) Photographs (and drawings) of three GMR POC devices reported by different groups. (D) (D3, D5) Microfluidic channels D3 and D5 (D1) aligned on the GMR sensor arrays with the sensor array compartmentalized into four complementary independent compartments. (D2) Sandwich assay. (D4) SEM image of MNP labels on GMR sensor stripes. (D6) Schematic view of cartridge integrating microfluidic chip and sensor array. (E1) Microfabricated micropillar arrays for on-chip filtering that is compatible with a microfluidic chip. (E2) (Left) Schematic view of MFC to amplify the magnetic field in the sensor region (left) and (right) a photograph of a pair of microfabricated MFCs. (E3) Schematic view of cantilever mechanical movement induced AC magnetic field in the GMR sensor region. (A) Reprinted with permission from ref 96. Copyright 2017 American Chemical Society. (B) Reprinted with permission from ref 97. Copyright 2021 Elsevier B.V. (C) Reprinted with permission from ref 73. Copyright 2018 Elsevier B.V. (D) Reprinted with permission from ref 100. Copyright 2015 The Royal Society of Chemistry. (E1) Reprinted with permission from ref 74. Copyright 2018 The Royal Society of Chemistry. (E2) Reprinted with permission from ref 101. Copyright 2020 Elsevier B.V. (E3) Reprinted from ref 102. Licensed under CC BY 4.0.
Although differing in the functional circuit implementations, all three of these devices constitute the same conceptual blocks for (1) signal generation, (2) signal decoupling, and (3) user accessibility. The signal generation methodology constitutes sinusoid generation for magnetic field and modulating control signal for the GMR array to facilitate implementation of the frequency-multiplexed scheme. These sinusoids are generated using high-resolution DAC devices. The core functionality of the signal decoupling stage is the removal of carrier tones from the GMR signal, which is usually ~30 dB larger than the higher harmonics of interest.82 This can be achieved either by the implementation of a Wheatstone bridge96 or through subtraction of a compensation signal.82,97 Post decoupling, the GMR signal is processed by filter implementations and the data are digitized using high-resolution ADC for frequency-domain processing. Finally, user accessibility is enabled through a customized mobile application guiding the user through individual process steps and providing the final bioassay results. The surface-functionalized GMR biosensors operate on the principle of localized proximity sensing where the binding of MNP labels to sensor surfaces causes a change to the sensor’s electrical resistance. This electrical resistance change is monitored in real-time in the form of magnetoresistance change (ΔMR/MR0, sometimes use ΔMR) in parts per million (ppm).
To clarify, we would like to bring to readers’ attention that although many groups reported GMR-based bioassays and biomagnetic field recordings (reviewed in Section 4), most of them are broadly defined as biosensors that may not have been integrated into POC devices yet. To prevent confusion, we use the broad term “biosensors” to refer to both POC devices and non-POC devices (such as benchtop systems) in the following context.
3.4.2. On-Chip Microfluidic Channels, Filters, and Magnetic Flux Concentrator (MFC).
Polymer-based microfluidic channels hold an important role in biological applications, especially for potential point-of-care measurements. By adding a microfluidic channel to a bioassay platform, it can effectively reduce the reagent cost (as a smaller volume of reagents is needed), enhance the bioassay speed, and meanwhile allow the integration of several processing steps into a single system. It can also make high-throughput biological screening possible by parallel processing multiple samples achieved with multiple fluidic paths and minimizing cross-contamination.98,99 For example, Bechstein et al. compartmentalized a GMR biosensor array using microfluidic channels to address individual sensor compartments with reagents.100 As shown in Figure 8D1, D3, D5), four microfluidic channels split the 64 GMR biosensors into four individual groups. The cartridge in Figure 8D6 integrates microfluidic channels and a GMR biosensor array chip and provides a pressure seal between each component. Thus, this compartmentalization approach allows the separation and cross-reactivity-free measurement of different kinds of analytes on a single GMR biosensor array chip. They used this design to observe pairwise cross-reactivities from multiplexed detection of cancer biomarkers epidermal growth factor receptor (EGFR), epithelial cell adhesion molecule (EpCAM), and transmembrane glycoprotein encoded by the Tacstd2 gene (Trop-2). For the control sample (channel 1), all three biomarkers (EGFR, EpCAM, and Trop-2) are mixed and multiplexed by GMR biosensors functionalized with these three corresponding capture antibodies, whereas for active samples (channels 2, 3, 4), only one kind of biomarker is added. Figure 8D) shows the sandwich assay strategy used for this experiment and the SEM image in Figure 8D4 confirms the captured MNPs on the GMR biosensor surface.
On top of the microfluidic channel, Tsai et al. fabricated an array of micropillars for filtering blood cells and passing only plasma to the guided-mode resonance biosensors area at the other terminal, as shown in Figure 8E1.74 Although this work is not designed for GMR biosensors, the idea of using micropillar arrays for on-chip filtering is worth learning. The filtering step is of great significance when dealing with raw biological samples such as whole blood, saliva, etc., where the large cells, viscous mucus, and debris may interfere with the biosensor signals. On the other hand, magnetic separation or ultra-centrifugation or a washing step is used to remove these signal interference factors in lab-based settings. This micropillar array as an on-chip filter makes it possible to carry all-in-one, fully automatic, and chip-based bioassays on site.
The detection of biomagnetic fields in the picotesla (pT) range is favored for many biomedical applications including brain and cardiac mapping (discussed in Sections 4.5 and 4.5). Magnetic flux concentrator (MFC) has been proposed as a possible method to improve the detection limit of many kinds of magnetic sensors including GMR, MTJ (magnetic tunnel junction), AMR, MEMS, etc.101,103-106 As shown in Figure 8E2, an MFC is usually made of high-permeability materials to improve the sensor sensitivity by concentrating the magnetic flux to the sensor area. It is reported that a good MFC can increase the magnetic flux by a factor of several hundred times.107 There are several factors affecting the flux concentration factor including the relative permeability of the material and the geometry of MFC structure such as the area ratio between the end and start sections of the concentrator. Guedes et al. reported that with the addition of a Co93Zr3Nb4 soft MFC, the detection limit of the GMR sensor in the thermal noise regime is improved from 1.3 to 0.064 nT/Hz1/2,103 In addition to MFC, MEMS piezoelectric cantilevers are also reported to assist the MFC in lowering the sensor noise floor as well as improving the detection limit.102,108,109 To be specific, as shown in Figure 8E3, a pair of MEMS piezoelectric cantilevers modulate the low-frequency magnetic signals into a high-frequency regime where the 1/f noise in the GMR sensor can be neglected. The results confirmed that this hybrid GMR-MEMS device shows an improved detection limit from 1 nT/Hz1/2 (low-frequency regime–1/f noise limited) to 1 pT/Hz1/2 (high-frequency regime–thermal noise limited).102
4. GMR BIOSENSOR-BASED BIOMEDICAL APPLICATIONS
4.1. GMR Biosensors for Disease Diagnosis.
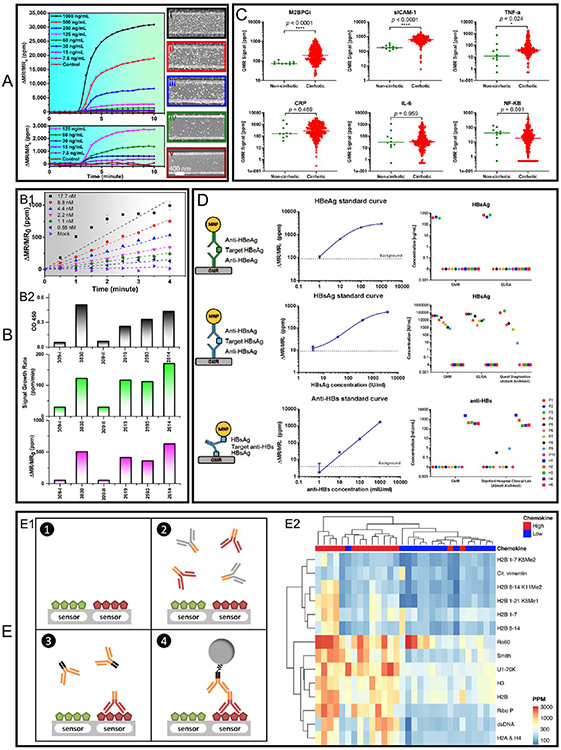
The past decade has seen a spike in applying GMR biosensors for the detection of a wide range of diseases and pathogens including SARS-CoV-2,110 influenza A virus (IAV),57,81,96 cirrhosis,71 hepatitis B virus (HBV),78,111,112 ovarian cancer,77 etc. In this section, we will cover some representative works in applying GMR biosensors for disease diagnosis with a focus on protein biomarkers detection. In 2017, Wu et al. reported a GMR POC device for the detection of IAV nucleoprotein (NP).96 They used the traditional sandwich assay strategy where the active GMR biosensors are surface-functionalized with IAV monoclonal capture antibodies (catalog #BE0159, clone H16-L10–4R5 (HB-65), Bio X Cell, West Lebanon, NH), and the control GMR biosensors are covered by BSA (negative control) or biotinylated BSA (positive control). Fifty microliter samples of recombinant influenza H1N1 NP (Sino Biological Inc. Beijing, China) of varying concentrations are tested. Followed by the addition of monoclonal detection antibody (catalog #MAB8257B, EMD Millipore Corporation, Temecula, CA,) and MNP labels (catalog #130-048-101, Miltenyi Biotec, Inc. Auburn, CA). The real-time GMR signal in the form of ΔMR/MR0 is recorded in a 10 min time window, as shown in Figure 9A. The GMR signal is averaged from 24 active biosensors and five negative control sensors. MNP labels are added at 3 min and, because of the high binding affinity of streptavidin (from MNP label) and biotin (from detection antibody), the binding saturates at 10 min. The GMR signal monotonically increases with the concentration of H1N1 NP. SEM images are taken on the biosensors after the assay to further confirm the binding of MNP labels on the GMR biosensor surfaces. As shown in Figure 9A(i-v), the density of captured MNP labels increases from control sensor (Figure 9A(v), to active sensors (Figure 9A(iv): 60 ng/mL, Figure 9A(iii): 250 ng/mL, Figure 9A(ii): 500 ng/mL, and Figure 9A(i): 1000 ng/mL). This confirms that the GMR signal is proportional to the concentration of target analytes (H1N1 NP in this scenario) as well as the MNP labels captured through the sandwich structure.
Figure 9.
(A, B1) Real-time GMR signal recorded from samples with varying concentrations of IV NP. A used a traditional sandwich assay scheme and B used a wash-free sandwich assay scheme. SEM images i–v of GMR biosensors after the bioassay show a varying number of immobilized MNP labeled on the sensor surface. The real-time GMR signal in A is the average of 24 active GMR sensors; error bars are not shown in this selected figure. The real-time GMR signal in B) is the average of 24 active GMR sensors; error bars are marked at 2 and 4 min and represent the standard deviation. (B2) Comparison of results from six nasal swab samples tested by ELISA (top) and GMR platform, where the GMR signal is represented by ΔMR/MR0 (bottom) and ΔMR/MR0/Δt (middle). Signal growth rate is the average of 24 GMR sensors, error bars are not shown. (C) Scatter plots of individual cirrhosis biomarkers from noncirrhotic and cirrhotic patients. Lines on the plots mark median biomarker signal levels. Respective p-values were calculated and shown. (D) HBeAg, HBsAg, and anti-HBs sandwich magnetic immunoassay structures (left column), the standard curves obtained from GMR biosensors (middle column), the analyte concentrations from serum samples calculated from GMR signals compared with other diagnostic methods (ELISA and/or Abbott Architect). The error bars in standard curves represent standard deviations of 80 identical sensor signals. (E) GMR multiplexed assay to identify interferon-associated autoantibodies in systemic lupus erythematosus (SLE). (E1) Another version of the sandwich assay involves antigen–primary antibody–secondary antibody–MNP label. (E2) An unsupervised hierarchically clustered heatmap shows the candidate autoantigens identified by SAM (significance analysis of microarrays). (A) Reprinted with permission from ref 96. Copyright 2017 American Chemical Society. (B) Reprinted from ref 81. Licensed under CC BY 4.0. (C) Reprinted with permission from ref 71. Copyright 2020 American Chemical Society. (D) Reprinted from ref 78. Licensed under CC BY 4.0. (E) Reprinted from ref 58. Licensed under CC BY 4.0.
In 2019, Su et al. reported the detection of IAV NP with a wash-free sandwich assay strategy.81 The preincubated MNP label-detection antibody-H1N1 NP complexes are added to GMR biosensors and GMR signals are recorded in real-time (Figure 9B1 plots the averaged GMR signal from 24 active sensors). Because the binding affinity between H1N1 NP and capture antibodies is weaker than streptavidin–biotin conjugations. It takes a longer time for the dynamic binding process to reach equilibrium (as well as a long time for the GMR signal to saturate). Thus, they only record the GMR signal for 4 min (before saturates) and use both GMR signal (ΔMR/MR0) and GMR signal growth rate (expressed as ΔMR/MR0/Δt) as indicators of H1N1 NP concentration. Because of the relative abundance of H1N1 NP from the biological sample and the capture antibodies from the GMR biosensor surface, the real-time binding curves show different slopes (growth rates) as seen in Figure 9B1. These two indicators are later used to calibrate the relative abundancies of IAV from swine nasal swab samples. The GMR signal and signal growth rate are compared with the ELISA results in Figure 9B2. All four positive and two negative results from GMR biosensors are consistent with those from ELISA, which proves the wash-free sandwich assay is a reliable method for the “yes or no” IAV level test in the raw samples taken from the field.
GMR-based sandwich assays have also been reported for the multiplexed detection of six cirrhosis biomarkers: mac-2-binding protein glycan isomer (M2BPGi), soluble intercellular adhesion molecule-1 (sICAM-1), c-reactive protein (CRP), nuclear factor kappa-light chain-enhancer of activated B cells (NF-κB), tumor necrosis factor-alpha (TNF-α), and interleukin-6 (IL-6). As shown in Figure 9C, the GMR signals (ΔMR/MR0) are scatter plotted for each biomarker from noncirrhotic and cirrhotic patients. Demonstrating the powerful abilities of GMR biosensor arrays in protein biomarker screening, allowing for faster, less expensive, and more accurate disease early stage detection. In addition, quantitative, multiplexed GMR bioassays are possible by plotting the standard GMR signal–analyte concentration curve and using it as a look-up table for calculating the analyte concentrations of unknown samples. For example, Gani et al. reported the multiplexed detection of HBV “e” antigen (HBeAg), HBV surface antigen (HBsAg), and the antibody against HBsAg (anti-HBs) from serum samples.78 They first obtained the assay standard curve for each analyte from 80 identical GMR sensors (Figure 9D middle column) then used these curves as look-up tables to calculate the analyte concentrations from 15 serum samples based on the GMR signal. As shown in the right column in Figure 9D, the quantitative results from GMR biosensors are compared with other diagnostic methods (ELISA and/or Abbott Architect). It shows that the GMR biosensor holds great promise as a more accessible alternative for HBV screening, diagnosis, and treatment monitoring.
Lee et al. reported the multiplexed detection of multiple autoantibodies associated with interferon (IFN) dysregulation in systemic lupus erythematosus (SLE).58 In this work, a different version of the sandwich bioassay strategy is used. As shown in Figure 9(E1), the GMR biosensors are functionalized with autoantigens to identify autoantibodies associated with dysregulation of the IFN pathway in SLE. Serum samples from SLE patients with high (n = 15) and low (n = 15) chemokine scores are tested on GMR biosensors. Once the autoantibodies specifically bind to the GMR biosensor surface, the residuals are washed out and antihuman IgG biotinylated antibodies are added as the bridge between autoantibodies and MNP labels added in the next step. An unsupervised hierarchically clustered heatmap is plotted and significance analysis of microarrays (SAM) is used to identify autoantigens with significantly different antibody reactivity between the groups, as shown in Figure 9E2. In this work, they discovered that 12 autoantibodies identified by SAM have not previously been associated with chemokine scores. They included histones (H2A and H4, H2B, and H3), methylated (K5Me1, K5Me2, and K11Me2), and unmodified peptides from the N-terminal tail of H2B, Ribo P, Sm, Ro60, and U1–70K. This work shows that GMR biosensor arrays can identify autoantibodies associated with relevant clinical manifestations of SLE, with potential for use as biomarkers in clinical practice.
4.2. GMR Biosensors for Genotyping.
GMR bioassay platforms combined with PCR (polymerase chain reaction) or LAMP (loop-mediated isothermal amplification) technologies have also been reported for the detection of viral nucleic acid (DNA or RNA) including human papillomavirus (HPV),113 hepatitis B virus (HBV),111,112 and cDNA (complementary DNA),114 and DNA mutations for cancer prognostics,79 etc. The first report of GMR multilayer (Si/(Ni80Fe20)1.6 nm/[Cu 1.9 nm/(Ni80Fe20) 1.6 nm]10/Ta 3 nm) biosensor for nucleic acid detection was by Schotter et al. in 2004.115 They used a spiral-shaped GMR multilayer biosensor that covers a circular area of 70 μm diameter. It achieved a dynamic detection range of 16 ng/mL to 10 μg/mL for DNA detection. Although the first report of applying GMR spin-valve biosensors for nucleic acid detection was in 2008 by Xu et al.113 They are able to detect as low as 10 pM HPV DNA (after PCR amplification) and achieved an accuracy of ~90% from 12 assays of different HPV genotypes (i.e., HPV16, 18, and 45).
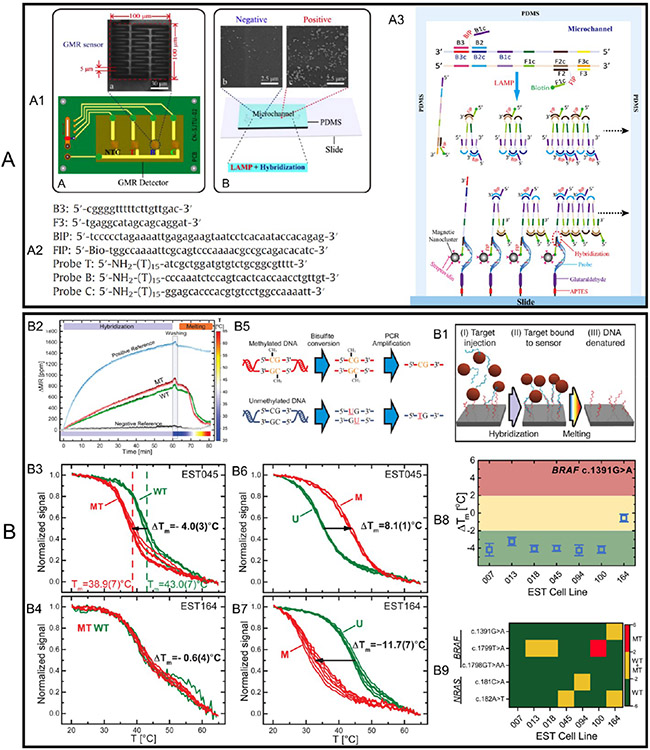
Following these pioneering works, another group reported the detection of HBV genotypes by integrating the GMR multilayer biosensors (NiFeCo 6 nm/[Cu 2.1 nm/(NiFeCo) 1.5 nm]10/Ta 100 nm.), LAMP technology, and a microfluidic chip.112 As shown in Figure 10A1, the microfluidic chip is used to shorten the assay time. The GMR multilayer biosensor is in a zigzag pattern with a line width of 5 μm and overall sensor area of 100 μm × 100 μm. Four GMR multilayer biosensors (3 active and 1 control) are welded on a PCB with a digital multimeter (not shown) for signal collection. A microfluidic channel with 300 μm (length) × 300 μm (width) × 100 μm (depth) is made from biocompatible PDMS. The primers B3, F3, BIP (B1c-B2), and biotinylated primer FIP (F1c-F2) are used to amplify the conserved domain of the HBV genome. Probe T hybridizes with all genotypes of HBV DNA and probes B and C uniquely hybridize with HBV genotype B and C, respectively. The sequences of primers and probes are shown in Figure 10A2. The LAMP amplification and DNA hybridization steps are schematically drawn in Figure 10A3. In short, primers were incubated with an HBV DNA template at 63 °C for 1 h. Meanwhile, the DNA probes from the GMR multilayer biosensor surface were hybridized with target HBV DNA fragments produced by LAMP. Finally, streptavidin-conjugated MNP labels were added and captured by the hybridized DNA pairs through the biotin at the terminal end of target HBV DNA, followed by a wash step to remove unbound reagents from the microfluidic channel. The GMR signal is collected and mapped to the HBV DNA concentration (before LAMP amplification). In this work, we achieved a detection limit of 10 copies/mL target HBV DNA through a 1 h LAMP amplification, 15 min MNP labeling, and 3 min wash step. We want to bring it to readers’ attention that some works report the limit of detection (LOD) by using the concentrations of nucleic acids before amplification, whereas other works report the LOD by using the concentrations after amplification.
Figure 10.
(A) GMR multilayer biosensors combined with LAMP and microfluidic channel for genotyping HBV DNA. (A1) Photographs of one GMR multilayer biosensor and four biosensors assembled on a PCB. (A2) Schematic view of the microfluidic channel made from PDMS and MNP labels bound on GMR multilayer biosensors. (A2) Sequences of primers and probes. (A3) LAMP amplification and DNA hybridization steps. (B) Simultaneous profiling of DNA mutation and methylation by melting analysis using GMR (SV) biosensors. (B1) Schematic protocol for the profiling DNA mutation and methylation using GMR biosensors. (B2) Real-time GMR signal (ΔMR) recorded in a complete cycle of hybridization, wash, and melting curve measurement. (B3, B4) Normalized melting curves from WT (wild type) and MT (mutant type) probes targeting BRAF c.1391 G > A mutation, where the EST045 (B3) and EST164 (B4) cell lines are wild type and heterozygous mutant, respectively. (B5) Schematic of the bisulphite conversion process. (B6, B7) are the curves from U (unmethylated) and M (methylated) probes (i.e., GMR biosensors functionalized with each type of probe). The melting curves are measured for (B6) the hypermethylated cell line EST045 and (B7) the unmethylated cell line EST164. (B8) Mutation profiling of melanoma cell lines. ΔTm measured for BRAF c.1391G > A mutation for the seven investigated EST cell lines. Error bars in B8 are one standard deviation (n = 4–6). (B9) Heat map of measured ΔTm with an applied threshold to genotype mutations: WT in green, heterozygous MT in yellow, homozygous MT in red. (A) Reprinted with permission from ref 112. Copyright 2013 Elsevier B.V. (B) Reprinted with permission from ref 79. Copyright 2017 American Chemical Society.
Rizzi et al. reported the GMR spin-valve biosensors for simultaneous profiling of DNA mutation and methylation by means of measuring the DNA melting point.79 DNA methylation is gaining increasing interest as complementary information to DNA mutations for cancer diagnostics and prognostics. The basic idea of this work is to first PCR amplify the genomic (mutation) or bisulphite-treated (methylation) DNA. The amplicons are then magnetically labeled with MNPs. After magnetic column separation and denaturation, the ssDNA-conjugated MNPs (i.e., MNP-ssDNA complexes) are introduced to the GMR biosensor array where multiple DNA probes are functionalized on each sensor. With the hybridization of MNP-ssDNA to probes, the GMR signal (in the form of ΔMR) is recorded in real time followed by a wash step, a melting curve measurement is carried out where the temperature is swept from 20 to 65 °C, as shown in Figure 10B1. For example, in Figure 10B2, in a DNA mutation analysis, two probes complementary to the wild type (WT) and mutant type (MT) are functionalized on different GMR biosensors. During the hybridization step, the GMR signal (ΔMR) increases with the binding of MNP labels. To obtain single-base specificity, at 60 min, we used stringent washing after hybridization. The hybrids are then challenged by increasing the temperature and continuously measuring DNA melting simultaneously for all probes (MT, WT, negative, and positive control probes) on the GMR biosensor array. A biotinylated DNA probe is used as a positive reference and a DNA probe with an unspecific sequence is used as a negative reference. Panels B3 and B4 in Figure 10 show the melting curves of WT BRAF amplicons hybridized to WT and MT probes for the c.1391G > A mutation. The GMR signal (ΔMR) is normalized by the initial signal at T = 20 °C and the melting temperature Tm is defined as the temperature at which the signal dropped 50%. ΔTm (= Tm(MT) − Tm(WT)) indicates a higher complementarity of the target to the WT probes than the MT probe, and hence that the target is WT. The EST045 cell lines (Figure 10B3) are wild type and a single base mismatch between the WT targe and MT probe causes great differences in the melting curves, whereas the EST164 cell lines (Figure 10B4) are heterozygous samples containing both MT and WT targets that can hybridize to both WT and MT probes. The resulting melting curves are overlapping.
Similarly, DNA methylation can also be detected by employing bisulphite treatment on the genomic DNA to convert a methylation event into a single base substitution, as shown in Figure 10B5. After the bisulphite conversion, the gene promoter region of interest is amplified by PCR. The amplicons are then hybridized to probes complementary to unmethylated (U) or methylated (M).Panels B6 and B7 in Figure 10 show melting curves to analyze the methylation status. For the hypermethylated cell line EST045 in Figure 10B6, the GMR biosensors functionalized with M probes show higher melting temperature and thus higher complementarity of the target ssDNA, proving the hypermethylated target DNA. On the other hand, for unmethylated cell line EST164 in Figure 10B7, the GMR biosensors functionalized with U probes show higher melting temperature, thus proving that target DNA strands are unmethylated. The multiplexed DNA profiling of melanoma cell lines is then carried out on a GMR biosensor array consisting of 64 individual sensors. Using the mutation and methylation detection techniques described above, they have simultaneously probed three mutation sites in BRAF, two mutation sites in NRAS, two methylation sites in the KIT promoter, and two methylation sites in the RARB promoter in triplicate. Figure 10B8 shows the ΔTm values measured for the BRAF c.1391 G > A mutation for all cell lines. Error bars are standard deviations (n = 4–6). ΔTm = −4 °C indicates a homozygous WT sequence, whereas a ΔTm value close to zero indicates a heterozygous mutation (i.e., EST 164). Furthermore, Figure 10B9 is the heatmap of ΔTm for all investigated mutations for each cell line. Classifying WT (ΔTm < −2 °C), heterozygous MT (−2 °C < ΔTm < 2 °C), and homozygous MT (ΔTm > 2 °C). They also applied it to detect the DNA methylation density. In its present format, the GMR biosensor platform can be used for the simultaneous triplicate investigation of about 20 mutation and methylation sites. Table 2 provides a summary of various disease/pathogen detection based on GMR biosensors.
Table 2.
Summary of GMR Biosensors for Detection of Biological Analytes
| disease/pathogen | target analyte | matrices | limit of detection (LOD) |
assay time | ref |
|---|---|---|---|---|---|
| ovarian cancer | cancer antigen 125 (CA125) | PBS buffer | 3.7 U/mL | 20 min | 77 |
| human epididymis protein 4 (HE4) | 7.4 pg/mL | ||||
| interleukin 6 (IL6) | 7.4 pg/mL | ||||
| cirrhosis | Mac-2 binding protein glycan isomer (M2BPGi) | serum | NA | 15 min | 71 |
| soluble intercellular adhesion molecule-1 (sICAM-1) | 96.9 pg/mL | ||||
| C-reactive protein (CRP) | 14.2 pg/mL | ||||
| nuclear factor kappa-light chain-enhancer of activated B cells (NF-κB) | NA | ||||
| tumor necrosis factor-alpha (TNF-α) | 12.6 pg/mL | ||||
| Interleukin 6 (IL6) | 4.9 pg/mL | ||||
| cardiovascular disease | pregnancy-associated plasma protein-A (PAPP-A) | PBS buffer, serum | 1 ng/mL | 40 min | 76 |
| proprotein convertase subtilisin/kexintype9 (PCSK9) | 433.4 pg/mL | ||||
| suppression of tumorigenicity 2 (ST2) | 40 pg/mL | ||||
| C-reactive protein (CRP) | blood | 1 ng/mL | 15 min | 85 | |
| tumor | alpha-fetoprotein (AFP) | serum | 0.52 ng/mL | 15 min | 73 |
| carcinoembryonic antigen (CEA) | 0.27 ng/mL | ||||
| cytokeratin 19 fragment (CYFRA21–1) | 0.25 ng/mL | ||||
| neuron specific enolase (NSE) | 0.5 ng/mL | ||||
| free β-subunit of human chorionic gonadotropin (free-β-hCG) | 0.3 ng/mL | ||||
| squamous cell carcinoma (SCC) | 0.3 ng/mL | ||||
| pepsinogen I (PG I) | 1 ng/mL | ||||
| pepsinogen II (PG II) | 0.5 ng/mL | ||||
| total prostate-specific antigen (tPSA) | 0.02 ng/mL | ||||
| free prostate-specific antigen (fPSA) | 0.07 ng/mL | ||||
| thyroglobulin (Tg) | 1 ng/mL | ||||
| carbohydrate antigen 19–9 (CA19–9) | 2 ng/mL | ||||
| hepatitis B virus (HBV) | HBV e antigen (HBeAg) | serum | 1 IU/mL | NA | 78 |
| HBV surface antigen (HBsAg) | 3 IU/mL | ||||
| antibody against HBsAg (Anti-HBs) | 1 mIU/mL | ||||
| HBV DNA | NA | 10 copies/mL | 1 h | 112 | |
| HBV DNA | NA | 200 IU/mL | 15 min | 111 | |
| human papillomavirus (HPV) | HPV DNA | NA | 10 pM | 10 min | 113 |
| influenza A virus (IAV) | IAV nucleoprotein (IAV NP) | PBS buffer | 15 ng/mL | 10 min | 96 |
| H3N2 virus | 125 TCID50/mL | ||||
| IAV nucleoprotein (IAV NP) | PBS buffer | 0.3 nM | 4 min | 81 | |
| H3N2 virus | nasal swab | 250 TCID50/mL | |||
| heart failure and other cardiac diseases | N-terminal pro-B type natriuretic peptide (NT-proBNP) | PBS buffer, human plasma | 0.01 ng/mL | <10 min | 116 |
| SARS-CoV-2 | anti-SARS-CoV-2 IgG | NA | 5 ng/mL | 10 min | 110 |
| anti-SARS-CoV-2 IgM | 10 ng/mL | ||||
| E. coli O157H:H7 | E. coli O157H:H7 bacteria particle | PBS buffer | 100 CFU/mL | NA | 80 |
| NA | NA | 117 | |||
| systemic lupus erythematosus (SLE) | histone H2A & H4 | serum | NA | 15 min | 58 |
| histone H2B 1–7 K5Me2 | |||||
| histone H2B 8–14 | |||||
| histone H2B 8–14 K11Me2 | |||||
| Histone H2B 1–21 K5Me1 | |||||
| histone H2B 1–7 | |||||
| histone H2B | |||||
| histone H3 | |||||
| plasmid double-stranded DNA (dsDNA) | |||||
| ribonucleoprotein 60 kDa (Ro60) | |||||
| ribosomal phosphoprotein P0 (Ribo P) | |||||
| citrullinated vimentin peptide (Cit. vimentin) | |||||
| small nuclear ribonuclearprotein 68 (U1–70K) | |||||
| Smith | |||||
| cancer | Jurkat leukemia cells | PBS buffer | NA | NA | 99 |
| HIV | HIV-1 glycoprotein 41 (gp41) | saliva | NA | 16 min | 97 |
| leukocytosis | neutrophil elastase (NE) | plasma | NA |
4.3. GMR Biosensors for Food and Drug Regulation.
Foodborne pathogens are a growing global public health concern that not only affects human health but also affects productivity and international trade.118-120 To prevent, control, and mitigate the impact of potential foodborne disease outbreaks. A growing number of biosensors, such as surface plasmon resonance (SPR), electrochemical, impedimetric, etc., have been reported for the rapid detection of foodborne pathogens.121-127 Among them, although GMR biosensors are still developing, they play an important role in the early and accurate detection of foodborne pathogens,80,117 allergens,128 and toxins.70
For example, most Escherichia coli (E. coli) are harmless and are part of a healthy intestinal tract. However, if it is transmitted to humans through consumption of contaminated or raw/undercooked food. It may cause illnesses including diarrhea, urinary tract infections, respiratory illness, and bloodstream infections. Many foodborne illness outbreaks related to E. coli have been reported in recent years.129,130 With faster detection time and high sensitivity, it will be a valuable tool for the real-time diagnosis of foodborne pathogens. Sun et al. reported that their GMR sandwich assay protocol can detect E. coli O157H:H7 as low as 100 CFU/mL.80 On the other hand, Kokkinis et al. reported an innovative method to detect E. coli bacteria particles dynamically, using GMR sensors as an MNP label speedometer.117 They labeled the E. coli bacteria by MNPs and passed the MNP-E. coli complexes through a microfluidic channel directed by an external magnetic field. A GMR sensor is placed at each end of the channel to detect the MNPs when pass through. For the control group, MNPs without conjugation of E. coli are passed through the channel. The velocity of MNPs in the microfluidic channel is calculated and used as an indicator of the presence of a pathogen in the sample fluid.
Drug abuse can affect several aspects of a person’s physical and mental health. Herein, we introduce a GMR competitive assay for detecting marijuana directly from saliva.84 Tetrahy-drocannabinol (THC), as shown in Figure 11B, the main psychoactive component of marijuana, is one kind of small molecule with a molecular weight of 314.5 Da. The traditional THC test is chromatography combined with mass spectroscopy using blood samples; the bulky facility limits its application in field and road tests. The urine tests, which are also noninvasive compared to blood tests, can only detect THC metabolites and are subjected to a time delay (urine at least several hours after ingesting cannabis).131 Thus, for on-site and road testing, oral fluid (i.e., saliva) samples provide a viable way for noninvasive and rapid measurement.132 Lee et al. reported a miniaturized platform with a disposable chip cartridge that has Bluetooth communication with a smartphone, as shown in Figure 11D, each GMR chip consists of 80 individual biosensors for multiplexed detection.84 A saliva sample collection kit consists of a cotton swab, syringe, and filter unit, as shown in Figure 11C. The saliva sample is at first collected with a cotton swab, and then this swab is inserted into the syringe to squeeze and release the fluid (as a simple replacement of traditional centrifuge step). The filter attached to the end of the syringe removes viscous mucus, food particles, debris, etc.
Figure 11.
(A) GMR-based competitive assay for the detection of small molecule THC. (B) Chemical structure of THC. (C) Saliva collection scheme and step. (D) GMR biosensor platform with a smartphone interface (left), GMR signal reader with a dimension of 105 mm × 90 mm (middle), and GMR biosensor array consisting of 80 sensors. (E) THC assay concentration curves. GMR signal collected from filtered saliva samples with (E1) 15 min preincubation and 15 min chip incubation and (E2) 0 min preincubation and 2 min chip incubation. The error bars represent standard deviations of eight identical sensor signals. Figure reprinted with permission from ref 84. Copyright 2016 American Chemical Society.
The traditional sandwich assay scheme is not applicable because (1) small molecules may lack the two binding sites (epitopes) required for sandwich assay, and (2) the sandwich structure cannot be formed because of steric hindrance. A competitive assay scheme is applied for detecting THC. As shown in Figure 11A, in the preincubation step, the anti-THC biotinylated antibody is mixed with the biological sample and incubated for 15 min to bind to THC. The mixture is then added to the GMR biosensors that functionalized with BSA (negative control) or THC-conjugated BSA (THC-BSA) and incubated for another 15 min. After a final wash step to remove unbound reagents, streptavidin-coated MNPs are added to label the residual antibodies and the GMR signal is recorded. It should be noted that in this competitive assay, the GMR signal is proportional to the abundance of the MNP labels and inversely proportional to the concentration of THC in the sample. As shown in Figure 11E1, after 15 min of preincubation (using 1 μg/mL antibody) and 15 min of on-chip incubation, the concentration–response curve shows a dynamic detection range of 0–50 ng/mL of THC. Meanwhile, by using a higher concentration of antibody in the preincubation step, faster binding between THC and antibody can reduce the waiting time for detection. Thus, without any preincubation, the anti-THC biotinylated antibodies (5 μg/mL) and filtered saliva samples are immediately added to the chip and incubated for 2 min. The GMR biosensors show a wider dynamic range of 0–200 ng/mL of THC, see Figure 11E2. In Figure 11E, the error bars marked on concentration curves represent standard deviations of eight identical sensor signals. The reduced assay time on the user side is expected to enable a rapid roadside drug detection. In addition, GMR biosensors have also been reported for the detection of heavy metal (Hg2+) in water133 and the characterization of alkali mineral water.134
4.4. GMR Biosensors for Brain Mapping.
GMR biosensors are micrometer to nanometer sized and can be fabricated on flexible substrates. Hence, they provide a better fit into the gyri and sulci of the brain during in vivo imaging. To date, GMR biosensors with a detection range of nT/Hz1/2 to pT/Hz1/2 incorporated into an implantable probe have been examined for neuronal activity mapping by several research groups.26,135,136 Some special mentions include the works of Chopin et al., where they have reported that GMRs with a sensitivity of around 1–2%/mT can facilitate brain mapping.25 Another prominent example is the report of “magnetrodes” to record neuronal activity.25,26
As shown in Figure 12A1, GMR spin valves are microfabricated on a 200 μm thick silicon substrate, and the whole device is etched to form a needle shape for ease of tissue penetration.26 The equivalent-noise spectrum in Figure 12A2 shows that the sensitivities of GMR sensors are on the order of 1–20 nT/Hz1/2 for frequencies within 1–100 Hz. This GMR “magnetrode” is electrically insulated and inserted 1 mm into the brain of an anesthetized cat using micromanipulators. The design of the GMR sensors are such that it is sensitive to magnetic fields, which appears to be orthogonal to the tip, i.e., it is parallel to the cortical surface. A tungsten electrode is implanted 1 mm away from the magnetrode to obtain a standard reference electric recording. As shown in Figure 12A3, a flash of light is directed to the eye of the cat to physiologically activate the recorded brain area. The visual cortex of a cat brain undergoes a visual stimulation inducing brain signals in response to the light, which is recorded by GMR sensors (i.e., magnetic recording) and tungsten electrodes (i.e., electric recording). The output signals from the tungsten electrode and magnetrode are averaged over 1000 times of recording using externally applied visual stimulus. The resulting averaged signals are noted as the event-related potential (ERP) and event-related field (ERF), respectively. Panels A4-A and A4-B in Figure 12 are the simultaneously recorded ERP and ERF from cat 1 upon a 100 ms long visual stimulus (time 0–0.1 s within the dashed vertical lines). The direct comparison of averaged ERP and ERF signals within the stimulus window is given in Figure 12A4-C, where the ERF showed a strong negative component at 40 ms and a positive peak around 60 ms with a peak-to-peak amplitude of 2.5 nT. On the other hand, ERP shows a comparable signal trend with a slightly shorter time lag to the stimulus. The Pearson correlation coefficient (a value in the interval from −1 to +1 to determine the linear correlation between two generic variables, where the extremes −1 and +1 are characterized by perfect linear correlation for the same and opposite site, respectively, and 0 with no correlation between the two variables) is used to determine the linear correlation between ERF and ERP as a function of time lag as shown in Figure 12A4-D: positive values indicate that the ERF lagged the ERP. The correlation function peaked at a value of 0.55, for a lag of approximately 2 ms. The side peaks and troughs are due to the partially rhythmic nature of the ERP and ERF. Similar results are also obtained from cat 2A in Figure 12A5.
Figure 12.
(A1) SEM picture of the GMR magnetrode. The element is deposited onto a silicon substrate 200 μm thick and 150 μm wide. (A2) Equivalent-noise spectral density for different DC input voltages equivalent to AC inputs of 0.5 V (blue) and 1 V (red). (A3) Experimental setup of the in vivo neuronal activity recording. The visual cortex of the cat undergoes visual stimulation while the GMR magnetrode records the signal. (A4-A, A5-E) Event-related field (ERF) of one GMR sensor output averaged over 1000 times for a visual stimulus 100 ms long, error bars not shown. (A4-B, A5-F) Event-related potential (ERP) by averaging the signal from an independent tungsten electrode over 1000 stimulus repetitions, error bars not shown. (A4-C, A5-G) Comparison between ERF (red) and ERP (green). (A4-D, A5-H) Pearson correlation coefficient between ERF and ERP as a function of time lag. (B1) Brain slice of the hippocampus, where the region of insertion of the GMR sensor probe is visible along with the field direction. (B3-a) Sequence of pulses measured in one of the GMR sensors from the array located in the pyramidal cell bodies region, after hippocampus excitation in the CA3 region. (B3-b, B3-c) Signal readouts after the application of CNQX and TTX, respectively. (A) Reprinted with permission from ref 26. Copyright 2017 Elsevier. (B) Reprinted with permission from ref 136. Copyright 2011 AIP Publishing LLC.
In addition, GMR sensors have also been reported for in vitro synaptic potential measurements on mice hippocampus brain slices.136 The authors designed 15 GMR spin valves on a chip and integrated them in a recording chamber underneath the brain slice, as shown in Figure 12B1, B2. The hippocampus excitation takes place in the CA3 region, which delivers a pulse to the pyramidal neurons in region CA1. Because the hippocampus acts like a closed loop, the synaptic and action potential generates along a particular route. Figure 12B3-a shows a sequence of pulses measured by GMR biosensors underneath the CA1 region. The GMR sensor readout consists of pulses with 20 μV amplitude and 20–30 ms length. On the basis of the R─H response curves of GMR biosensors, the authors calculated the source magnetic field to be 2.5 μT assuming the neuron and biosensors are separated by 10 μm. Furthermore, postsynaptic receptor blocker CNQX (Figure 12B3-b) and sodium channel blocker TTX (Figure 12B3-c) are applied to prove that the magnetic signal arises from the biological action potential.
4.5. GMR Biosensors for Cardiac Mapping.
Super-conducting quantum interference devices (SQUIDs) are frequently used for cardiac mapping. However, SQUIDs are very bulky and expensive and require an external cooling system and magnetic shielding, not to mention a relevant maintenance cost. GMR biosensors for cardiac imaging are more feasible compared to their SQUID counterparts. The integration of GMR sensors with MFCs, MEMS cantilevers, and Wheatstone bridges for detecting sub-nT down to fT biomagnetic fields is practically achievable.22,102,137 The low power consumption, ultrasensitive magnetic field detection, and micrometer-scale sized GMR biosensors are extremely advantageous in comparison to other technologies for cardiac mapping.138 Together with other standard monitoring purposes, GMR sensor technology for cardiac mapping has a major advantage in comparison to SQUIDs especially because of the operation in an electromagnetic environment at room temperature without any shielding, as well as the considerable reduction of the overall noise associated with the measurement.27,102
In 2011, Pannetier-Lecoeur et al. reported GMR sensors for measuring the magnetic signatures of the electric activity from the human heart.22 They mounted four GMR sensors on a large Niobium loop and wired them to form a Wheatstone bridge. The GMR sensors are electrically isolated by a 300 nm silicon nitride layer and immersed in liquid helium (4 K). At low frequencies, the sensitivity is limited by the 1/f noise and thus gives a field detection range equivalent to 3 pT/Hz1/2 at 1 Hz. For recording MCG signals, GMR sensors are placed at a distance of 25–30 mm from the chest. The MCG recordings are taken horizontally across a 6 × 6-point grid separated by 3 cm in the left–right direction and 5 cm in the foot–head direction. At each spot, the MCG signal is recorded for 1 min and then averaged from 46–54 readings. The characteristic components of normal ECG, the P-wave, QRS complex, and T-wave are all present in MCG signals. A magnetic field map is generated based on the MCG signal. The polarity inversion of the QR-segment is observed along the diagonal of the measurement plane, whereas this pattern is a little different for the T-wave. It should be noted that in this work, the GMR biosensors are working under a condition of 4 K (in liquid helium) and within a magnetically shielded room.
On the other hand, Fujiwara et al. reported MCG recording on a healthy human chest in a similar manner, but with magnetic tunnel junctions (MTJs).139 The MTJ sensors are also configured in a Wheatstone bridge but without magnetic shielding or low-temperature conditions (i.e., liquid helium is not required). The sensing system noise decreases with a higher number of averaging on the MCG signals. Note that MCG signal averaging is achieved by synchronizing the R peak of the ECG waveform. This work demonstrated that MTJ sensors can perform room-temperature detection of the QRS complex within 1 min, which is very useful for the diagnosis of heart diseases.
5. CONCLUSIVE REMARKS
GMR systems have been extensively used as magnetic field sensors for hard disk drives, biosensors, microelectromechanical systems (MEMS), etc. Compared to its industrial applications, the biomedical applications based on the GMR effect have received less attention. In this paper, we emphasized the great potential of GMR biosensors for a wide variety of biomedical applications including point-of-care (POC) disease diagnosis, genotyping, and food and drug regulation, as well as a more recent applications in brain and cardiac imaging.
An emerging research topic lies in the flexible GMR biosensors for wearable health condition monitoring in real time. Herein, we have reviewed flexible GMR sensors synthesized by different strategies. A traditional way of fabricating flexible GMR sensors is to directly deposit the magnetic multilayer stacks onto flexible substrates. In view of this, several types of polymer films, as well as silicon and glass, are reported to serve as flexible substrates provided that their thicknesses are in the micrometer range. Because of the large elastic property differences between GMR thin films and substrates, large residual strain is expected, which could cause unstable GMR sensitivity (low robustness and low experiment repeatability). On the other hand, printable GMR sensors that are based on ball-milled GMR flakes dispersed in a conductive polymer are an alternative solution. The magnetic ink can be screen printed onto various flexible substrates including paper. The high robustness and elasticity of screen-printed GMR sensors make it a good candidate for on-skin electronics. It can be foreseen that flexible GMR biosensors in this form will be a good fit for real-time brain and cardiac mapping.
On the other hand, GMR biosensors based on a solid silicon substrate have developed into a mature platform. Several research groups and companies have reported POC devices based on GMR sensor arrays on silicon substrates. Although these platforms are powerful and capable of achieving rapid and multiplexed detections. The integration with on-chip filtration (to remove blood cells, viscous mucus, food particles, debris, etc. from biofluidic samples) and microfluidic channels are still in the development stage, which is the main challenge before transforming GMR POC devices into on-field test kits. We can see the gap from current GMR POC devices to “press one button and get the result within a few minutes”. The on-chip microfluidic channels with a filtration function can push this GMR-based bioassay platform into a more automatic, user-friendly, easy-to-use, on-site, and bedside healthcare solution.
To date, the technical and biological/clinical challenges related to GMR-based biosensing for various applications still exist. In the past two decades, attempts have been made to address some of these challenges. For example, the growing quest for high-throughput disease screening (i.e., multiplexed biomarkers detection) and genotyping has facilitated the design of GMR biosensor arrays that are capable of multiplexing different analytes in parallel. Each GMR biosensor works independently and is functionalized with different capture probes (e.g., capture antibodies or nucleic acids, etc.) to specifically detect target analytes.71,73,76,77 Furthermore, to transform GMR bioassays from lab settings to on-site tests (which applies for on-site disease diagnosis, genotyping, and food and drug regulation), researchers have reported a one-step, wash-free assay strategy to simplify the testing protocol with minimal sacrifice of the performance. For example, a strategy is reported to mix the detection probe, the MNP functionalized with the capture probe, and the biological fluid sample containing the target analyte, without the need for repeated washing procedures in the traditional bioassay process.81 Another GMR wash-free bioassay protocol is proposed by integrating a fully automatic microfluidic channel.100,140 Using a syringe pump system, the reagents are sequentially injected into the microfluidic chip and flowed over the GMR sensors. Another technical challenge to which not much attention has been paid is the sample filtration step before the GMR bioassay, especially when dealing with unprocessed clinical samples. Although we commented that GMR sensors are intrinsically more sensitive over optical biosensors because negligible magnetic background noise is expected from a biological matrix. The impurities from raw biological samples such as large cells, viscous mucus, and food debris may still interfere with the GMR biosensors. These impurities may cause accidental precipitation or nonspecific binding of MNP, thereby increasing the false positive/negative rate. A filter unit attached to the syringe has been reported to remove viscous mucus and food particles before the GMR bioassay step.84 However, this filter unit is independent of the GMR bioassay platform and it is relatively rudimentary, which makes the filtration performance vary from sample to sample, potentially reducing the reproducibility of the bioassay. An on-chip filtration system that is compatible with the GMR bioassay chip is highly recommended. Several works have reported this type of on-chip filtration design for working with unprocessed clinical samples.141-143
The application of GMR biosensors in the brain and cardiac mapping has continued to emerge in recent years. Although there is no doubt on the capability of GMR biosensors for detecting nT/Hz1/2 to pT/Hz1/2 range biomagnetic fields. Special attention should be paid to the tissue–device interfaces. The potential risks of implanting or attaching GMR biosensors in the brain and heart tissues include (1) electric leakage of GMR biosensors and electrodes, (2) corrosion of GMR biosensors after long-term exposure to the complex biochemical fluid environment, (3) biological contamination and tissue inflammation problem. To solve these challenges, we should surface encapsulate GMR biosensors with a thin layer of a biocompatible material such as polyimide, silicones, epoxy resins, and parylene C. Another technical challenge is introduced with the encapsulation of GMR biosensors by biocompatible materials, the increased distance from biomagnetic field sources (i.e., neuron cells, heart tissues) to the sensors, because magnetic field drops with distance cubed. The increased distance from biological tissue to GMR biosensors increases the difficulty of magnetic mapping/recording and requires the GMR sensor to have better detection capabilities.
On top of these challenges in applying GMR biosensors to clinical settings and on-field tests, compared to the optical biosensors, magnetic biosensors such as GMR are intrinsically more sensitive given that biological samples are nonmagnetic (or diamagnetic). Therefore, negligible magnetic background noise is expected from a biological matrix. In addition, the state-of-the-art micro- and nanofabrication techniques make it possible to assemble 0.1–1 million GMR biosensors per square centimeter. By integrating with CMOS electronics, GMR biosensor arrays can make high-sensitivity multiplexed bioassays and high spatial resolution brain and cardiac imaging feasible and affordable. All these unique characteristics of the GMR biosensors have pushed researchers to further exploit its potential for a growing list of real-life biomedical applications.
ACKNOWLEDGMENTS
This study was financially supported by the U.S. Department of Agriculture–National Institute of Food and Agriculture (NIFA) under Award 2020-67021-31956. Research reported in this publication was supported by the National Institute Of Dental & Craniofacial Research of the National Institutes of Health under Award R42DE030832. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Kai Wu, Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Denis Tonini, Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Shuang Liang, Department of Chemical Engineering and Materials Science, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Renata Saha, Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Vinit Kumar Chugh, Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Jian-Ping Wang, Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, Minnesota 55455, United States; Department of Chemical Engineering and Materials Science, University of Minnesota, Minneapolis, Minnesota 55455, United States.
REFERENCES
- (1).Binasch G; Grünberg P; Saurenbach F; Zinn W Enhanced Magnetoresistance in Layered Magnetic Structures with Antiferromagnetic Interlayer Exchange. Phys. Rev. B 1989, 39 (7), 4828–4830. [DOI] [PubMed] [Google Scholar]
- (2).Baibich MN; Broto JM; Fert A; Van Dau FN; Petroff F; Etienne P; Creuzet G; Friederich A; Chazelas J Giant Magnetoresistance of (001) Fe/(001) Cr Magnetic Superlattices. Phys. Rev. Lett 1988, 61 (21), 2472. [DOI] [PubMed] [Google Scholar]
- (3).Tehrani S; Slaughter JM; Chen E; Durlam M; Shi J; DeHerren M Progress and Outlook for MRAM Technology. IEEE Trans. Magn 1999, 35 (5), 2814–2819. [Google Scholar]
- (4).Su D; Wu K; Saha R; Peng C; Wang J-P Advances in Magnetoresistive Biosensors. Micromachines 2020, 11 (1), 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kanai H; Noma K; Hong J Advanced Spin-Valve GMR Head. Fujitsu Sci. Technol. J 2001, 37 (2), 174–182. [Google Scholar]
- (6).Cubells-Beltrán M-D; Reig C; Madrenas J; De Marcellis A; Santos J; Cardoso S; Freitas PP Integration of GMR Sensors with Different Technologies. Sensors 2016, 16 (6), 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ren C; Bayin Q; Feng S; Fu Y; Ma X; Guo J Biomarkers Detection with Magnetoresistance-Based Sensors. Biosens. Bioelectron 2020, 165, 112340. [DOI] [PubMed] [Google Scholar]
- (8).Beavis KG; Matushek SM; Abeleda APF; Bethel C; Hunt C; Gillen S; Moran A; Tesic V Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for Detection of IgA and IgG Antibodies. J. Clin. Virol 2020, 129, 104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pastucha M; Farka Z; Lacina K; Mikušová Z; Skládal P Magnetic Nanoparticles for Smart Electrochemical Immunoassays: A Review on Recent Developments. Microchim. Acta 2019, 186 (5), 312. [DOI] [PubMed] [Google Scholar]
- (10).Khanmohammadi A; Aghaie A; Vahedi E; Qazvini A; Ghanei M; Afkhami A; Hajian A; Bagheri H Electrochemical Biosensors for the Detection of Lung Cancer Biomarkers: A Review. Talanta 2020, 206, 120251. [DOI] [PubMed] [Google Scholar]
- (11).Michel M; Bouam A; Edouard S; Fenollar F; Di Pinto F; Mège J-L; Drancourt M; Vitte J Evaluating ELISA, Immunofluorescence, and Lateral Flow Assay for SARS-CoV-2 Serologic Assays. Front. Microbiol 2020, 11, DOI: 10.3389/fmicb.2020.597529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Beitollahi H; Ivari SG; Torkzadeh-Mahani M Application of Antibody–Nanogold–Ionic Liquid–Carbon Paste Electrode for Sensitive Electrochemical Immunoassay of Thyroid-Stimulating Hormone. Biosens. Bioelectron 2018, 110, 97–102. [DOI] [PubMed] [Google Scholar]
- (13).Mejía-Salazar JR; Oliveira ON Jr, Plasmonic Biosensing: Focus Review. Chem. Rev 2018, 118 (20), 10617–10625. [DOI] [PubMed] [Google Scholar]
- (14).Liu J; Jalali M; Mahshid S; Wachsmann-Hogiu S Are Plasmonic Optical Biosensors Ready for Use in Point-of-Need Applications? Analyst 2020, 145 (2), 364–384. [DOI] [PubMed] [Google Scholar]
- (15).Chao L; Liang Y; Hu X; Shi H; Xia T; Zhang H; Xia H Recent Advances in Field Effect Transistor Biosensor Technology for Cancer Detection: A Mini Review. J. Phys. Appl. Phys 2022, 55, 153001. [Google Scholar]
- (16).Wadhera T; Kakkar D; Wadhwa G; Raj B Recent Advances and Progress in Development of the Field Effect Transistor Biosensor: A Review. J. Electron. Mater 2019, 48 (12), 7635–7646. [Google Scholar]
- (17).Pérez-López B; Mir M Commercialized Diagnostic Technologies to Combat SARS-CoV2: Advantages and Disadvantages. Talanta 2021, 225, 121898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sajid M; Kawde A-N; Daud M Designs, Formats and Applications of Lateral Flow Assay: A Literature Review. J. Saudi Chem. Soc 2015, 19 (6), 689–705. [Google Scholar]
- (19).Bishop JD; Hsieh HV; Gasperino DJ; Weigl BH Sensitivity Enhancement in Lateral Flow Assays: A Systems Perspective. Lab. Chip 2019, 19 (15), 2486–2499. [DOI] [PubMed] [Google Scholar]
- (20).Aldewachi H; Chalati T; Woodroofe MN; Bricklebank N; Sharrack B; Gardiner P Gold Nanoparticle-Based Colorimetric Biosensors. Nanoscale 2018, 10 (1), 18–33. [DOI] [PubMed] [Google Scholar]
- (21).Chang C-C; Chen C-P; Wu T-H; Yang C-H; Lin C-W; Chen C-Y Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications. Nanomaterials 2019, 9 (6), 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pannetier-Lecoeur M; Parkkonen L; Sergeeva-Chollet N; Polovy H; Fermon C; Fowley C Magnetocardiography with Sensors Based on Giant Magnetoresistance. Appl. Phys. Lett 2011, 98 (15), 153705. [Google Scholar]
- (23).Savukov IM Spin Exchange Relaxation Free (SERF) Magnetometers. In High Sensitivity Magnetometers; Springer, 2017; pp 451–491. [Google Scholar]
- (24).Wyllie R; Kauer M; Smetana GS; Wakai RT; Walker TG Magnetocardiography with a Modular Spin-Exchange Relaxation-Free Atomic Magnetometer Array. Phys. Med. Biol 2012, 57 (9), 2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chopin C; Torrejon J; Solignac A; Fermon C; Jendritza P; Fries P; Pannetier-Lecoeur M Magnetoresistive Sensor in Two-Dimension on a 25 Mm Thick Silicon Substrate for in Vivo Neuronal Measurements. ACS Sens. 2020, 5 (11), 3493–3500. [DOI] [PubMed] [Google Scholar]
- (26).Caruso L; Wunderle T; Lewis CM; Valadeiro J; Trauchessec V; Trejo Rosillo J; Amaral JP; Ni J; Jendritza P; Fermon C; et al. In Vivo Magnetic Recording of Neuronal Activity. Neuron 2017, 95 (6), 1283–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pannetier-Lecoeur M; Polovy H; Sergeeva-Chollet N; Cannies G; Fermon C; Parkkonen L Magnetocardiography with GMR-Based Sensors. Journal of Physics: Conference Series 2011, 303, 012054. [Google Scholar]
- (28).Jogschies L; Klaas D; Kruppe R; Rittinger J; Taptimthong P; Wienecke A; Rissing L; Wurz MC Recent Developments of Magnetoresistive Sensors for Industrial Applications. Sensors 2015, 15 (11), 28665–28689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ennen I; Kappe D; Rempel T; Glenske C; Hütten A Giant Magnetoresistance: Basic Concepts, Microstructure, Magnetic Interactions and Applications. Sensors 2016, 16 (6), 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ramli R; Haryanto F; Khairurrijal K; Djamal M GMR Biosensors for Clinical Diagnostics. In Biosensors for Health, Environment and Biosecurity; IntechOpen: Rijeka, Croatia, 2011. [Google Scholar]
- (31).Tsymbal EY; Pettifor DG Perspectives of Giant Magnetoresistance. Solid State Phys. 2001, 56, 113–237. [Google Scholar]
- (32).Berkowitz AE; Mitchell JR; Carey MJ; Young AP; Zhang S; Spada FE; Parker FT; Hutten A; Thomas G Giant Magnetoresistance in Heterogeneous Cu-Co Alloys. Phys. Rev. Lett 1992, 68 (25), 3745. [DOI] [PubMed] [Google Scholar]
- (33).Xiao JQ; Jiang JS; Chien CL Giant Magnetoresistance in Nonmultilayer Magnetic Systems. Phys. Rev. Lett 1992, 68 (25), 3749. [DOI] [PubMed] [Google Scholar]
- (34).Xiong P; Xiao G; Wang JQ; Xiao JQ; Jiang JS; Chien CL Extraordinary Hall Effect and Giant Magnetoresistance in the Granular Co-Ag System. Phys. Rev. Lett 1992, 69 (22), 3220. [DOI] [PubMed] [Google Scholar]
- (35).Spizzo F; Angeli E; Bisero D; Ronconi F; Vavassori P; Allia P; Selvaggini V; Coisson M; Tiberto P; Vinai F GMR as a Function of Temperature in FeAg Granular Samples: The Effect of Magnetic Interactions. J. Magn. Magn. Mater 2003, 262 (1), 88–91. [Google Scholar]
- (36).Brüx U; Schneider T; Acet M; Wassermann EF Giant Magnetoresistance in Cr 100– x Fe x Bulk Granular Alloys. Phys. Rev. B 1995, 52 (5), 3042. [DOI] [PubMed] [Google Scholar]
- (37).Peng D; Wang J; Wang L; Liu X; Wang Z; Chen Y Electron Transport Properties of Magnetic Granular Films. Sci. China Phys. Mech. Astron 2013, 56 (1), 15–28. [Google Scholar]
- (38).Arana S; Arana N; Gracia FJ; Castano E High Sensitivity Linear Position Sensor Developed Using Granular Ag–Co Giant Magnetoresistances. Sens. Actuators Phys 2005, 123, 116–121. [Google Scholar]
- (39).Zhang S Theory of Giant Magnetoresistance in Magnetic Granular Films. Appl. Phys. Lett 1992, 61 (15), 1855–1857. [Google Scholar]
- (40).Zhang S; Levy PM Conductivity and Magnetoresistance in Magnetic Granular Films. J. Appl. Phys 1993, 73 (10), 5315–5319. [Google Scholar]
- (41).Zhang S; Levy PM Conductivity Perpendicular to the Plane of Multilayered Structures. J. Appl. Phys 1991, 69 (8), 4786–4788. [Google Scholar]
- (42).Karnaushenko D; Makarov D; Yan C; Streubel R; Schmidt OG Printable Giant Magnetoresistive Devices. Adv. Mater 2012, 24 (33), 4518–4522. [DOI] [PubMed] [Google Scholar]
- (43).Karnaushenko D; Makarov D; Stöber M; Karnaushenko DD; Baunack S; Schmidt OG High-Performance Magnetic Sensorics for Printable and Flexible Electronics. Adv. Mater 2015, 27 (5), 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ha M; Cañón Bermúdez GS; Kosub T; Mönch I; Zabila Y; Oliveros Mata ES; Illing R; Wang Y; Fassbender J; Makarov D Printable and Stretchable Giant Magnetoresistive Sensors for Highly Compliant and Skin-Conformal Electronics. Adv. Mater 2021, 33 (12), 2005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Parkin SSP; Roche KP; Suzuki T Giant Magnetoresistance in Antiferromagnetic Co/Cu Multilayers Grown on Kapton. Jpn. J. Appl. Phys 1992, 31 (9A), L1246. [Google Scholar]
- (46).Ota S; Ando A; Chiba D A Flexible Giant Magnetoresistive Device for Sensing Strain Direction. Nat. Electron 2018, 1 (2), 124–129. [Google Scholar]
- (47).Melzer M; Kaltenbrunner M; Makarov D; Karnaushenko D; Karnaushenko D; Sekitani T; Someya T; Schmidt OG Imperceptible Magnetoelectronics. Nat. Commun 2015, 6 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Makarov D; Melzer M; Karnaushenko D; Schmidt OG Shapeable Magnetoelectronics. Appl. Phys. Rev 2016, 3 (1), 011101. [Google Scholar]
- (49).Sheng P; Wang B; Li R Flexible Magnetic Thin Films and Devices. J. Semicond 2018, 39 (1), 011006. [Google Scholar]
- (50).Li H; Zhan Q; Liu Y; Liu L; Yang H; Zuo Z; Shang T; Wang B; Li R-W Stretchable Spin Valve with Stable Magnetic Field Sensitivity by Ribbon-Patterned Periodic Wrinkles. ACS Nano 2016, 10 (4), 4403–4409. [DOI] [PubMed] [Google Scholar]
- (51).Rance WL; Burst JM; Meysing DM; Wolden CA; Reese MO; Gessert TA; Metzger WK; Garner S; Cimo P; Barnes TM 14%-Efficient Flexible CdTe Solar Cells on Ultra-Thin Glass Substrates. Appl. Phys. Lett 2014, 104 (14), 143903. [Google Scholar]
- (52).Dutta P; Rathi M; Zheng N; Gao Y; Yao Y; Martinez J; Ahrenkiel P; Selvamanickam V High Mobility Single-Crystalline-like GaAs Thin Films on Inexpensive Flexible Metal Substrates by Metal-Organic Chemical Vapor Deposition. Appl. Phys. Lett 2014, 105 (9), 092104. [Google Scholar]
- (53).Chen X; Mi W Mechanically Tunable Magnetic and Electronic Transport Properties of Flexible Magnetic Films and Their Heterostructures for Spintronics. J. Mater. Chem. C 2021, 9 (30), 9400–9430. [Google Scholar]
- (54).Pérez N; Melzer M; Makarov D; Ueberschär O; Ecke R; Schulz SE; Schmidt OG High-Performance Giant Magnetoresistive Sensorics on Flexible Si Membranes. Appl. Phys. Lett 2015, 106 (15), 153501. [Google Scholar]
- (55).Gaster RS; Xu L; Han S-J; Wilson RJ; Hall DA; Osterfeld SJ; Yu H; Wang SX Quantification of Protein Interactions and Solution Transport Using High-Density GMR Sensor Arrays. Nat. Nanotechnol 2011, 6 (5), 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Wang SX; Guanxiong Li. Advances in Giant Magnetoresistance Biosensors with Magnetic Nanoparticle Tags: Review and Outlook. IEEE Trans. Magn 2008, 44 (7), 1687–1702. [Google Scholar]
- (57).Krishna VD; Wu K; Perez AM; Wang JP Giant Magnetoresistance-Based Biosensor for Detection of Influenza A Virus. Front. Microbiol 2016, 7, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Lee J-R; Haddon DJ; Wand HE; Price JV; Diep VK; Hall DA; Petri M; Baechler EC; Balboni IM; Utz PJ; Wang SX Multiplex Giant Magnetoresistive Biosensor Microarrays Identify Interferon-Associated Autoantibodies in Systemic Lupus Erythematosus. Sci. Rep 2016, 6, 27623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Glathe S; Hübner U; Mattheis R; Seidel P Influence of Transverse Fields on Domain Wall Pinning in Ferromagnetic Nanostripes. J. Appl. Phys 2012, 112 (2), 023911. [Google Scholar]
- (60).Reiss G; Brueckl H; Huetten A; Schotter J; Brzeska M; Panhorst M; Sudfeld D; Becker A; Kamp PB; Puehler A; Wojczykowski K; Jutzi P Magnetoresistive Sensors and Magnetic Nanoparticles for Biotechnology. J. Mater. Res 2005, 20 (12), 3294–3302. [Google Scholar]
- (61).Melzer M; Karnaushenko D; Lin G; Baunack S; Makarov D; Schmidt OG Direct Transfer of Magnetic Sensor Devices to Elastomeric Supports for Stretchable Electronics. Adv. Mater 2015, 27 (8), 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Feng Y; Liu J; Klein T; Wu K; Wang J-P Localized Detection of Reversal Nucleation Generated by High Moment Magnetic Nanoparticles Using a Large-Area Magnetic Sensor. J. Appl. Phys 2017, 122 (12), 123901. [Google Scholar]
- (63).Dias TM; Cardoso FA; Martins SAM; Martins VC; Cardoso S; Gaspar JF; Monteiro G; Freitas PP Implementing a Strategy for On-Chip Detection of Cell-Free DNA Fragments Using GMR Sensors: A Translational Application in Cancer Diagnostics Using ALU Elements. Anal. Methods 2016, 8 (1), 119–128. [Google Scholar]
- (64).Su D; Wu K; Wang J-P Large-Area GMR Bio-Sensors Based on Reverse Nucleation Switching Mechanism. J. Magn. Magn. Mater 2019, 473, 484–489. [Google Scholar]
- (65).Kim J; Park H; Jung D; Kim S Protein Immobilization on Plasma-Polymerized Ethylenediamine-Coated Glass Slides. Anal. Biochem 2003, 313 (1), 41–45. [DOI] [PubMed] [Google Scholar]
- (66).Yin HB; Brown T; Wilkinson JS; Eason RW; Melvin T Submicron Patterning of DNA Oligonucleotides on Silicon. Nucleic Acids Res. 2004, 32 (14), e118–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Shang X; Zhu Y; Li Z Surface Modification of Silicon Carbide with Silane Coupling Agent and Hexadecyl Iodiele. Appl. Surf. Sci 2017, 394, 169–177. [Google Scholar]
- (68).Saengdee P; Chaisriratanakul W; Bunjongpru W; Sripumkhai W; Srisuwan A; Jeamsaksiri W; Hruanun C; Poyai A; Promptmas C Surface Modification of Silicon Dioxide, Silicon Nitride and Titanium Oxynitride for Lactate Dehydrogenase Immobilization. Biosens. Bioelectron 2015, 67, 134–138. [DOI] [PubMed] [Google Scholar]
- (69).Wang W; Wang Y; Tu L; Klein T; Feng YL; Wang JP Surface Modification for Protein and DNA Immobilization onto GMR Biosensor. Ieee Trans. Magn 2013, 49 (1), 296–299. [Google Scholar]
- (70).Mak AC; Osterfeld SJ; Yu H; Wang SX; Davis RW; Jejelowo OA; Pourmand N Sensitive Giant Magnetoresistive-Based Immunoassay for Multiplex Mycotoxin Detection. Biosens. Bioelectron 2010, 25 (7), 1635–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ng E; Le AK; Nguyen MH; Wang SX Early Multiplexed Detection of Cirrhosis Using Giant Magnetoresistive Biosensors with Protein Biomarkers. ACS Sens. 2020, 5 (10), 3049–3057. [DOI] [PubMed] [Google Scholar]
- (72).Osterfeld SJ; Yu H; Gaster RS; Caramuta S; Xu L; Han S-J; Hall DA; Wilson RJ; Sun S; White RL; et al. Multiplex Protein Assays Based on Real-Time Magnetic Nanotag Sensing. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (52), 20637–20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Gao Y; Huo W; Zhang L; Lian J; Tao W; Song C; Tang J; Shi S; Gao Y Multiplex Measurement of Twelve Tumor Markers Using a GMR Multi-Biomarker Immunoassay Biosensor. Biosens. Bioelectron 2019, 123, 204–210. [DOI] [PubMed] [Google Scholar]
- (74).Tsai M-Z; Hsiung C-T; Chen Y; Huang C-S; Hsu H-Y; Hsieh P-Y Real-Time CRP Detection from Whole Blood Using Micropost-Embedded Microfluidic Chip Incorporated with Label-Free Biosensor. Analyst 2018, 143 (2), 503–510. [DOI] [PubMed] [Google Scholar]
- (75).Bostrom J; Lee CV; Haber L; Fuh G Improving Antibody Binding Affinity and Specificity for Therapeutic Development. In Therapeutic Antibodies; Springer, 2009; pp 353–376. [DOI] [PubMed] [Google Scholar]
- (76).Wang Y; Wang W; Yu LN; Tu L; Feng YL; Klein T; Wang JP Giant Magnetoresistive-Based Biosensing Probe Station System for Multiplex Protein Assays. Biosens. Bioelectron 2015, 70, 61–68. [DOI] [PubMed] [Google Scholar]
- (77).Klein T; Wang W; Yu L; Wu K; Boylan KL; Vogel RI; Skubitz AP; Wang J-P Development of a Multiplexed Giant Magnetoresistive Biosensor Array Prototype to Quantify Ovarian Cancer Biomarkers. Biosens. Bioelectron 2019, 126, 301–307. [DOI] [PubMed] [Google Scholar]
- (78).Gani AW; Wei W; Shi R-Z; Ng E; Nguyen M; Chua M-S; So S; Wang SX An Automated, Quantitative, and Multiplexed Assay Suitable for Point-of-Care Hepatitis B Virus Diagnostics. Sci. Rep 2019, 9 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Rizzi G; Lee JR; Dahl C; Guldberg P; Dufva M; Wang SX; Hansen MF Simultaneous Profiling of DNA Mutation and Methylation by Melting Analysis Using Magnetoresistive Biosensor Array. ACS Nano 2017, 11 (9), 8864–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Sun X; Lei C; Guo L; Zhou Y Separable Detecting of Escherichia Coli O157H: H7 by a Giant Magneto-Resistance-Based Bio-Sensing System. Sens. Actuators B Chem 2016, 234, 485–492. [Google Scholar]
- (81).Su D; Wu K; Krishna V; Klein T; Liu J; Feng Y; Perez AM; Cheeran MC; Wang J-P Detection of Influenza a Virus in Swine Nasal Swab Samples With a Wash-Free Magnetic Bioassay and a Handheld Giant Magnetoresistance Sensing System. Front. Microbiol 2019, 10, 1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Choi J; Gani AW; Bechstein DJ; Lee J-R; Utz PJ; Wang SX Portable, One-Step, and Rapid GMR Biosensor Platform with Smartphone Interface. Biosens. Bioelectron 2016, 85, 1–7. [DOI] [PubMed] [Google Scholar]
- (83).Srinivasan B; Li Y; Jing Y; Xing C; Slaton J; Wang J-P A Three-Layer Competition-Based Giant Magnetoresistive Assay for Direct Quantification of Dndoglin from Human Urine. Anal. Chem 2011, 83 (8), 2996–3002. [DOI] [PubMed] [Google Scholar]
- (84).Lee J-R; Choi J; Shultz TO; Wang SX Small Molecule Detection in Saliva Facilitates Portable Tests of Marijuana Abuse. Anal. Chem 2016, 88 (15), 7457–7461. [DOI] [PubMed] [Google Scholar]
- (85).Meng F; Zhang L; Huo W; Lian J; Jesorka A; Shi X; Gao Y Dynamic Range Expansion of the C-Reactive Protein Quantification with a Tandem Giant Magnetoresistance Biosensor. ACS Omega 2021, 6, 12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Adem S; Jain S; Sveiven M; Zhou X; o’Donoghue AJ; Hall DA Giant Magnetoresistive Biosensors for Real-Time Quantitative Detection of Protease Activity. Sci. Rep 2020, 10 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Wu K; Su D; Liu J; Saha R; Wang J-P Magnetic Nanoparticles in Nanomedicine: A Review of Recent Advances. Nanotechnology 2019, 30 (50), 502003. [DOI] [PubMed] [Google Scholar]
- (88).Liu J; Su D; Wu K; Wang J-P High-Moment Magnetic Nanoparticles. J. Nanoparticle Res 2020, 22 (3), 1–16. [Google Scholar]
- (89).Konwar AN; Borse V Current Status of Point-of-Care Diagnostic Devices in the Indian Healthcare System with an Update on COVID-19 Pandemic. Sens. Int 2020, 1, 100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Rajan A; Glorikian H Point-of-Care Diagnostics: Market Trends and Growth Drivers. Expert Opin. Med. Diagn 2009, 3 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- (91).Heidt B; Siqueira WF; Eersels K; Diliën H; van Grinsven B; Fujiwara RT; Cleij TJ Point of Care Diagnostics in Resource-Limited Settings: A Review of the Present and Future of PoC in Its Most Needed Environment. Biosensors 2020, 10 (10), 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Huckle D Point-of-Care Diagnostics: An Advancing Sector with Nontechnical Issues. Expert Rev. Mol. Diagn 2008, 8 (6), 679–688. [DOI] [PubMed] [Google Scholar]
- (93).Rezaei M; Razavi Bazaz S; Zhand S; Sayyadi N; Jin D; Stewart MP; Ebrahimi Warkiani M Point of Care Diagnostics in the Age of COVID-19. Diagnostics 2021, 11 (1), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Yang T; Gentile M; Shen C-F; Cheng C-M Combining Point-of-Care Diagnostics and Internet of Medical Things (IoMT) to Combat the COVID-19 Pandemic; Multidisciplinary Digital Publishing Institute: Basel, Switzerland, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Vandenberg O; Martiny D; Rochas O; van Belkum A; Kozlakidis Z Considerations for Diagnostic COVID-19 Tests. Nat. Rev. Microbiol 2021, 19 (3), 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Wu K; Klein T; Krishna VD; Su D; Perez AM; Wang J-P Portable GMR Handheld Platform for the Detection of Influenza A Virus. ACS Sens 2017, 2, 1594–1601. [DOI] [PubMed] [Google Scholar]
- (97).Ng E; Yao C; Shultz TO; Ross-Howe S; Wang SX Magneto-Nanosensor Smartphone Platform for the Detection of HIV and Leukocytosis at Point-of-Care. Nanomedicine Nanotechnol Biol. Med 2019, 16, 10–19. [DOI] [PubMed] [Google Scholar]
- (98).Situma C; Hashimoto M; Soper SA Merging Microfluidics with Microarray-Based Bioassays. Biomol Eng. 2006, 23 (5), 213–231. [DOI] [PubMed] [Google Scholar]
- (99).Kokkinis G; Cardoso S; Keplinger F; Giouroudi I Microfluidic Platform with Integrated GMR Sensors for Quantification of Cancer Cells. Sens. Actuators B Chem 2017, 241, 438–445. [Google Scholar]
- (100).Daniel JB Microfluidic Multiplexed Partitioning Enables Flexible and Effective Utilization of Magnetic Sensor Arrays. Lab Chip 2015, 15 (22), 4273–4276. [DOI] [PubMed] [Google Scholar]
- (101).Feng Y; Chen J-Y; Wu K; Wang J-P Design and Fabrication of Integrated Magnetic Field Sensing System with Enhanced Sensitivity. J. Magn. Magn. Mater 2020, 511, 166728. [Google Scholar]
- (102).Guedes A; Macedo R; Jaramillo G; Cardoso S; Freitas PP; Horsley DA Hybrid GMR Sensor Detecting 950 PT/Sqrt (Hz) at 1 Hz and Room Temperature. Sensors 2018, 18 (3), 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Guedes A; Almeida JM; Cardoso S; Ferreira R; Freitas PP Improving Magnetic Field Detection Limits of Spin Valve Sensors Using Magnetic Flux Guide Concentrators. IEEE Trans. Magn 2007, 43 (6), 2376–2378. [Google Scholar]
- (104).Edelstein AS; Fischer GA; Pedersen M; Nowak ER; Cheng SF; Nordman CA Progress toward a Thousandfold Reduction in 1/ f Noise in Magnetic Sensors Using an Ac Microelectromechanical System Flux Concentrator. J. Appl Phys 2006, 99 (8), 08B317. [Google Scholar]
- (105).Hu J; Ji M; Qiu W; Pan L; Li P; Peng J; Hu Y; Liu H; Pan M Double-Gap Magnetic Flux Concentrator Design for High-Sensitivity Magnetic Tunnel Junction Sensors. Sensors 2019, 19 (20), 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Chen J; Wurz MC; Belski A; Rissing L Designs and Characterizations of Soft Magnetic Flux Guides in a 3-D Magnetic Field Sensor. IEEE Trans. Magn 2012, 48 (4), 1481–1484. [Google Scholar]
- (107).Fescenko I; Jarmola A; Savukov I; Kehayias P; Smits J; Damron J; Ristoff N; Mosavian N; Acosta VM Diamond Magnetometer Enhanced by Ferrite Flux Concentrators. Phys. Rev. Res 2020, 2 (2), 023394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Guedes A; Patil SB; Cardoso S; Chu V; Conde JP; Freitas PP Hybrid Magnetoresistive/ Microelectromechanical Devices for Static Field Modulation and Sensor 1/ f Noise Cancellation. J. Appl. Phys 2008, 103 (7), 07E924. [Google Scholar]
- (109).Guedes A; Jaramillo G; Buffa C; Vigevani G; Cardoso S; Leitao DC; Freitas PP; Horsley DA Towards PicoTesla Magnetic Field Detection Using a GMR-MEMS Hybrid Device. IEEE Trans. Magn 2012, 48 (11), 4115–4118. [Google Scholar]
- (110).Bayin Q; Huang L; Ren C; Fu Y; Ma X; Guo J Anti-SARS-CoV-2 IgG and IgM Detection with a GMR Based LFIA System. Talanta 2021, 227, 122207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Zhi X; Liu QS; Zhang X; Zhang YX; Feng J; Cui DX Quick Genotyping Detection of HBV by Giant Magnetoresistive Biochip Combined with PCR and Line Probe Assay. Lab. Chip 2012, 12 (4), 741–745. [DOI] [PubMed] [Google Scholar]
- (112).Zhi X; Deng M; Yang H; Gao G; Wang K; Fu H; Zhang Y; Chen D; Cui D A Novel HBV Genotypes Detecting System Combined with Microfluidic Chip, Loop-Mediated Isothermal Amplification and GMR Sensors. Biosens. Bioelectron 2014, 54, 372–377. [DOI] [PubMed] [Google Scholar]
- (113).Xu L; Yu H; Akhras MS; Han S-J; Osterfeld S; White RL; Pourmand N; Wang SX Giant Magnetoresistive Biochip for DNA Detection and HPV Genotyping. Biosens. Bioelectron 2008, 24 (1), 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Ravi N; Rizzi G; Chang SE; Cheung P; Utz PJ; Wang SX Quantification of CDNA on GMR Biosensor Array towards Point-of-Care Gene Expression Analysis. Biosens. Bioelectron 2019, 130, 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Schotter J; Kamp PB; Becker A; Puhler A; Reiss G; Bruckl H Comparison of a Prototype Magnetoresistive Biosensor to Standard Fluorescent DNA Detection. Biosens. Bioelectron 2004, 19 (10), 1149–1156. [DOI] [PubMed] [Google Scholar]
- (116).Wang W; Klein T; Collins J Giant Magnetoresistive Based Handheld System for Rapid Detection of Human NT-ProBNP. In Frontiers in Biomedical Devices; American Society of Mechanical Engineers: New York, 2019; Vol. 41037, p V001T04A003. [Google Scholar]
- (117).Kokkinis G; Cardoso SF; Cardoso FA; Giouroudi I Microfluidics for the Rapid Detection of Pathogens Using Giant Magnetoresistance Sensors. IEEE Trans. Magn 2014, 50 (11), 1–4. [Google Scholar]
- (118).Cliver DO; Riemann HP Foodborne Diseases; Gulf Professional Publishing: Houston, TX, 2002. [Google Scholar]
- (119).Ananchaipattana C; Hosotani Y; Kawasaki S; Pongsawat S. Md.; Latiful B; Isobe S; Inatsu Y Prevalence of Foodborne Pathogens in Retailed Foods in Thailand. Foodborne Pathog. Dis 2012, 9 (9), 835–840. [DOI] [PubMed] [Google Scholar]
- (120).Dhama K; Rajagunala S; Chakrabort S; Verma AK; Kumar A; Tiwari R; Kapoor S Food-Borne Pathogens of Animal Origin-Diagnosis, Prevention, Control and Their Zoonotic Significance: A Review. Pak. J. Biol. Sci PJBS 2013, 16 (20), 1076–1085. [DOI] [PubMed] [Google Scholar]
- (121).Arora P; Sindhu A; Dilbaghi N; Chaudhury A Biosensors as Innovative Tools for the Detection of Food Borne Pathogens. Biosens. Bioelectron 2011, 28 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- (122).Sharma H; Mutharasan R Review of Biosensors for Foodborne Pathogens and Toxins. Sens. Actuators B Chem 2013, 183, 535–549. [Google Scholar]
- (123).Koubova V; Brynda E; Karasova L; Škvor J; Homola J; Dostalek J; Tobiška P; Rošický J Detection of Foodborne Pathogens Using Surface Plasmon Resonance Biosensors. Sens. Actuators B Chem 2001, 74 (1–3), 100–105. [Google Scholar]
- (124).Zhang Z; Zhou J; Du X Electrochemical Biosensors for Detection of Foodborne Pathogens. Micromachines 2019, 10 (4), 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Saravanan A; Kumar PS; Hemavathy RV; Jeevanantham S; Kamalesh R; Sneha S; Yaashikaa PR Methods of Detection of Food-Borne Pathogens: A Review. Environ. Chem. Lett 2021, 19 (1), 189–207. [Google Scholar]
- (126).Zhao X; Lin C-W; Wang J; Oh DH Advances in Rapid Detection Methods for Foodborne Pathogens. J. Microbiol. Biotechnol 2014, 24 (3), 297–312. [DOI] [PubMed] [Google Scholar]
- (127).Kim G; Om AS; Mun JH Nano-Particle Enhanced Impedimetric Biosensor for Detedtion of Foodborne Pathogens. Journal of Physics: Conference Series 2007, 61, 112. [Google Scholar]
- (128).Ng E; Nadeau KC; Wang SX Giant Magnetoresistive Sensor Array for Sensitive and Specific Multiplexed Food Allergen Detection. Biosens. Bioelectron 2016, 80, 359–365. [DOI] [PubMed] [Google Scholar]
- (129).Arnade C; Calvin L; Kuchler F Consumer Response to a Food Safety Shock: The 2006 Food-borne Illness Outbreak of E. Coli O157: H7 Linked to Spinach. Appl Econ. Perspect. Policy 2009, 31 (4), 734–750. [Google Scholar]
- (130).Osman KM; Kappell AD; Elhadidy M; ElMougy F; El-Ghany WAA; Orabi A; Mubarak AS; Dawoud TM; Hemeg HA; Moussa IMI; et al. Poultry Hatcheries as Potential Reservoirs for Antimicrobial-Resistant Escherichia Coli: A Risk to Public Health and Food Safety. Sci. Rep 2018, 8 (1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Allen KR Screening for Drugs of Abuse: Which Matrix, Oral Fluid or Urine? Ann. Clin. Biochem 2011, 48 (6), 531–541. [DOI] [PubMed] [Google Scholar]
- (132).Molnar A; Lewis J; Doble P; Hansen G; Prolov T; Fu S A Rapid and Sensitive Method for the Identification of Delta-9-Tetrahydrocannabinol in Oral Fluid by Liquid Chromatography–Tandem Mass Spectrometry. Forensic Sci. Int 2012, 215 (1–3), 92–96. [DOI] [PubMed] [Google Scholar]
- (133).Wang W; Wang Y; Tu L; Klein T; Feng Y; Li Q; Wang J-P Magnetic Detection of Mercuric Ion Using Giant Magnetoresistance-Based Biosensing System. Anal. Chem 2014, 86 (8), 3712–3716. [DOI] [PubMed] [Google Scholar]
- (134).Lee J-H; Kim D-W; Kim M-J; Kang J-H; Lee S-S Fabrication and Characteristics of Alkali Mineral Water Sensor Using GMR-SV Biosensor. In Proceedings of the Korean Magnestics Society Conference; The Korean Magnetics Society: Seoul, Korea, 2008; p 249.1–249.1. [Google Scholar]
- (135).Amaral J; Gaspar J; Pinto V; Costa T; Sousa N; Cardoso S; Freitas P Measuring Brain Activity with Magnetoresistive Sensors Integrated in Micromachined Probe Needles. Appl. Phys. A: Mater. Sci. Process 2013, 111 (2), 407–412. [Google Scholar]
- (136).Amaral J; Cardoso S; Freitas P; Sebastião A Toward a System to Measure Action Potential on Mice Brain Slices with Local Magnetoresistive Probes. J. Appl. Phys 2011, 109 (7), 07B308. [Google Scholar]
- (137).Li G; Sun S; Wilson RJ; White RL; Pourmand N; Wang SX Spin Valve Sensors for Ultrasensitive Detection of Superparamagnetic Nanoparticles for Biological Applications. Sens. Actuators Phys 2006, 126 (1), 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Shen H-M; Hu L; Fu X Integrated Giant Magnetoresistance Technology for Approachable Weak Biomagnetic Signal Detections. Sensors 2018, 18 (1), 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Fujiwara K; Oogane M; Kanno A; Imada M; Jono J; Terauchi T; Okuno T; Aritomi Y; Morikawa M; Tsuchida M; et al. Magnetocardiography and Magnetoencephalography Measurements at Room Temperature Using Tunnel Magneto-Resistance Sensors. Appl. Phys. Express 2018, 11 (2), 023001. [Google Scholar]
- (140).Bechstein DJ; Lee J-R; Ooi CC; Gani AW; Kim K; Wilson RJ; Wang SX High Performance Wash-Free Magnetic Bioassays through Microfluidically Enhanced Particle Specificity. Sci. Rep 2015, 5, 11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Geng Z; Ju Y; Wang W; Li Z Continuous Blood Separation Utilizing Spiral Filtration Microchannel with Gradually Varied Width and Micro-Pillar Array. Sens. Actuators B Chem 2013, 180, 122–129. [Google Scholar]
- (142).Geng Z; Ju Y; Wang Q; Wang W; Li Z Multi-Component Continuous Separation Chip Composed of Micropillar Arrays in a Split-Level Spiral Channel. RSC Adv. 2013, 3 (34), 14798–14806. [Google Scholar]
- (143).Chen X; Cui D; Chen J Design, Fabrication and Characterization of Nano-filters in Silicon Microfluidic Channels Based on MEMS Technology. Electrophoresis 2009, 30 (18), 3168–3173. [DOI] [PubMed] [Google Scholar]