Abstract
Inhibition of α-glucosidase alleviates postprandial high glycemic levels in diabetic or prediabetic population. In Chinese bayberry fruit, myricetin, quercetin and kaempferol are main flavonols, which differ only in their hydroxylation on the B-ring. Kaempferol (4′-OH) showed high IC50 (65.36 ± 0.27 μmol L−1) against α-glucosidase, while quercetin (3′,4′-OH) exhibited stronger inhibition (46.91 ± 0.54 μmol L−1) and myricetin (3′,4′,5′-OH) possessed the strongest inhibitory activity (33.20 ± 0.43 μmol L−1). Molecular docking analysis illustrated that these flavonols could insert to the active cavity of α-glucosidase. Adjacent hydroxyl groups at B-ring of myricetin and quercetin positively contributed to form hydrogen bonds that were important to the stability of flavonol–enzyme complex, while kaempferol had no adjacent hydroxyl groups. Such observation was further validated by molecular dynamics simulations, and in good consistency with in vitro kinetic analysis and fluorescence spectroscopy analysis. Among three flavonols tested, myricetin possessed the strongest inhibition effects on α-glucosidase with the lowest dissociation constant (Ki = 15.56 μmol L−1) of myricetin-α-glucosidase, largest fluorescence quenching constant (Ksv) of (14.26 ± 0.03) × 104 L mol−1 and highest binding constant (Ka) of (1.38 ± 0.03) × 105 L mol−1 at 298 K with the enzyme. Bio-Layer Interferometry (BLI) and circular dichroism (CD) analysis further confirmed that myricetin had high affinity to α-glucosidase and induced conformational changes of enzyme. Therefore, myricetin, quercetin and kaempferol are all excellent dietary α-glucosidase inhibitors and their inhibitory activities are enhanced by increasing number of hydroxyl groups on B-ring.
Inhibition of α-glucosidase alleviates postprandial high glycemic levels in diabetic or prediabetic population.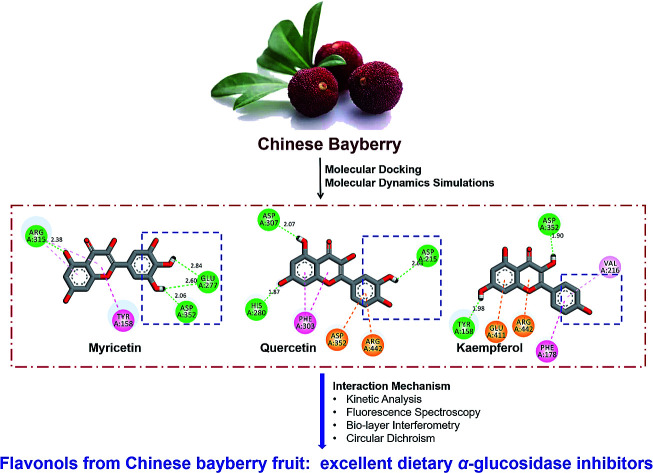
1. Introduction
Diabetes Mellitus (DM) is a chronic metabolic disease characterized by hyperglycemia. With the increase of living conditions and changes in lifestyle, incidence of DM and its complications have increased significantly in the past few decades. In 2017, more than 451 million adults were diagnosed with diabetes worldwide and this number was estimated to reach 693 million by 2045.1 Type 2 Diabetes (T2D), known as non-insulin-dependent DM, is the most prevalent diabetes accounting for more than 90% of all DM and it is commonly associated with overweight and obesity.2 Prevention and control programs are urgently needed to inhibit the dramatic rising incidence of T2D.
Postprandial hyperglycemia is one predominated leading factor for the onset and development of T2D. Carbohydrates are enzymatically digested into monosaccharides such as glucose by intestinal α-glucosidase (EC 3.2.1.20), resulting in rise of glycemic level with the uptake of glucose.3 For T2D patients, rapid postprandial glycemic rise occurs due to impaired blood glucose regulation caused by insulin dysfunction.4
Inhibition of α-glucosidase is an effective way for treatment of T2D by delaying the release of glucose and alleviating postprandial hyperglycemia.5 Drugs such as acarbose, miglitol, and voglibose are clinically used as α-glucosidase inhibitors for treatment of T2D. However, these drugs cause side effects such as gastrointestinal adverse reaction including diarrhea, flatulence, abdominal pain and so on.6,7 Hence, new α-glucosidase inhibitors with better effects and lower side effects are in great demand to alleviate postprandial high glycemic level effectively.
In recent decades, numerous studies have shown that berries or berry extracts possessed significant hypoglycemic effects.8,9 Chinese bayberry (Morella rubra Sieb. et Zucc.), a dark-red colored berry native to China, is rich in bioactive nutrients including anthocyanins and flavonols.10,11 Series investigations conducted by our group have shown that the anthocyanin extracts from Chinese bayberry possesses significant anti-diabetic effects in vivo and in vitro.12–15 However, the flavonol fraction of Chinese bayberry fruit showed much higher inhibitory activity against α-glucosidase than that of the anthocyanin fraction,16 indicating the possible important role that flavonols might also play in antidiabetic effects of the fruit.
The flavonol composition in pulp of 17 Chinese bayberry cultivars was previously investigated by liquid chromatography combined with electrospray ionization mass spectrometry (LC-ESI-MS/MS) by our group.10 Seven flavonols compounds, i.e., myricetin-3-O-rhamnoside, myricetin deoxyhexoside–gallate, quercetin-3-O-galactoside, quercetin-3-O-glucoside, quercetin-3-O-rhamnoside, kaempferol-3-O-galactoside and kaempferol-3-O-glucoside were identified and characterized among the cultivars.10 Therefore, myricetin, quercetin and kaempferol were the major flavonol aglycones in bayberry fruit. Our preliminary study on α-glucosidase inhibition activity of different flavonol aglycones and flavonol glycosides showed that flavonol aglycones exhibited at least ten times higher α-glucosidase inhibition activity than their corresponding flavonol glycosides. Three flavonol aglycones, i.e., myricetin, quercetin and kaempferol, differ only in their hydroxylation on B-ring, and they also showed different enzymatic inhibition activities against α-glucosidase, the mechanism of which is not well studied with the structure-activity point of view.
Molecular docking is one of the most frequently used methods in structure-based drug discovery. It is able to visualized the interaction of small molecule with macromolecules with affinity and binding site prediction.17,18 Such prediction can be validated by molecular dynamics simulation, which is capable of monitoring and evaluating the conformational behaviors of the atoms and molecules.19 Ligand–receptor interaction can be further verified by multiple experimental studies, such as kinetic analysis, fluorescence spectroscopy and circular dichroism (CD) spectra. These approaches have been widely used to illustrate molecular interaction mechanisms such as inhibition type, fluorescence quenching, binding constant, interaction force and conformation change.20–24 Additionally, as a newly label-free technology, bio-layer interferometry (BLI) has been developed rapidly in recent years for molecular interaction studies.25 It can monitor whole process of intermolecular binding in real time and calculate the affinity between molecules.26
Therefore, the present study focused on the investigation of α-glucosidase inhibitory activities of three flavonol aglycones, i.e., myricetin, quercetin and kaempferol, in Chinese bayberry fruit. Their interaction mechanisms with α-glucosidase were systematically explored by molecular docking and molecular dynamics simulations in silico and experimental studies including kinetic analysis, fluorescence spectroscopy, BLI coupled with CD spectra analysis. Such study may provide deeper understanding of inhibitory mechanism and structure-activity difference of dietary flavonols against α-glucosidase and show further evidence about antidiabetic effect of Chinese bayberry fruit.
2. Materials and methods
2.1. Chemicals
Saccharomyces cerevisiae α-glucosidase (23 U mg−1), acarbose, dimethyl sulfoxide (DMSO) and p-nitrophenyl-α-d-glucopyranoside (pNPG) were obtained from Sigma-Aldrich (St. Louis, MO, USA). α-Glucosidase was dissolved in 0.1 mol L−1 potassium phosphate buffer (PPB, pH 6.8). The stock solutions of acarbose and pNPG were freshly prepared just before every experiment. Methanol and acetonitrile for high performance liquid chromatography (HPLC) were of chromatographic grade, and all the other solvents used in the experiment were of analytical grade.
2.2. Flavonols extraction from Chinese bayberry
Three representative cultivars of Chinese bayberry (Morella rubra Sieb. et Zucc.), ‘Biqi’, ‘Dongkui’ and ‘Shuijing’, were harvest at commercial season in June 2018 from Xianju County, Zhejiang Province, China. Fruits free of disease and mechanical damage were selected and frozen in liquid nitrogen. Pulp was freeze-dried (FM 25EL-85, VirTis, Gardiner, NY, USA) and ground into powder. Flavonols was extracted according to our previous report with minor modification.27 The lyophilized powder of Chinese bayberry pulp (1 g) was dissolved in 10 mL of 80% aqueous methanol with sonication for 30 min, centrifuged at room temperature (12 000 rpm, 15 min) and the residue was extracted again as above. Supernatants were then combined and evaporated under reduced pressure at 37 °C by a rotary evaporator. Crude extract powder was totally redissolved in 80% aqueous methanol and 140 μL of solution was transferred to a new centrifuge tube. HCl (3 N, 60 μL) was added for acid hydrolysis and incubated for 1 h at 70 °C. After centrifugation at 12 000 rpm for 15 min, supernatant was used for HPLC analysis.
2.3. HPLC analysis of flavonols
Flavonol analysis was performed according to Han et al.28 with some modifications. HPLC system (e2695 pump, 2998 PDA detector, Waters, USA), coupled to an Sunfire® C18 analytical column (4.6 × 250 mm, 5 μm) was operated at room temperature, with a flow rate of 1 mL min−1 and injection volume of 10 μL. The mobile phase consisted of 0.1% (v/v) formic acid in water (A) and acetonitrile: 0.1% formic acid (1 : 1, v/v) (B). The elution gradient was as follows: 0–7 min, 10–50% of B, 7–10 min, 50% of B, 10–15 min, 50–100% of B, 15–15.1 min, 100–10% of B; 15.1–20 min, 10% B. Flavonols were detected at 370 nm and quantified by comparison of the peak area with those of the standards.
2.4. Enzyme activity assay
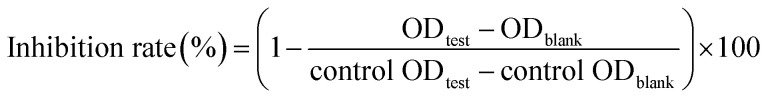
α-Glucosidase inhibitory effects of flavonols were evaluated as previous report with slightly modification.16 In the reaction, 20 μL of α-glucosidase (0.2 U mL−1 in 0.1 mol L−1 PPB) and 112 μL of 0.1 mol L−1 PPB (pH 6.8) were pre-incubated with 8 μL of test sample at 37 °C for 15 min. Reaction started with adding 20 μL of pNPG (2.5 × 10−3 mol L−1) and terminated with 80 μL of Na2CO3 (0.1 mol L−1) after incubation at 37 °C for 15 min. Absorption at 405 nm was measured by a Microplate Reader (Thermo Electron Co., Vantaa, Finland). Acarbose was used as positive control and DMSO was used as negative control. Solution without substrate was used as blank control. The inhibition rate of flavonols against α-glucosidase was calculated as follows:
 |
1 |
2.5. Molecular docking
Molecular docking analysis was performed in silico to predict the interaction between flavonols and α-glucosidase. Docking homology modeling of enzyme was downloaded from the Protein Data Bank (PDB ID: 3A4A).29 It was used as a proper receptor model in docking simulation with flavonols and acarbose by AutoDock Vina, an upgraded approach with significantly improved average accuracy of the binding mode predictions compared to AutoDock 4.30 Receptor was prepared by removing its small molecular ligand and water, add hydrogens and output pdbqt file by AutoDockTools.31 Grid box was set as 60 Å × 60 Å × 60 Å to cover the whole receptor for blind docking, and centered at X = 22.255, Y = −8.621, Z = 23.472. Flexible docking was used in the present study and it allows molecule conformation to change freely during docking process. Defaulted exhaustiveness parameter of 8 was changed as 100 to cover sufficient number of conformation changes. 3D structures of flavonols and acarbose were constructed in Chem 3D 17.1. Intermolecular interactions were analyzed by Discovery Studio and PyMOL (The PyMOL Molecular Graphics System, Version 2.0.6 Schrödinger, LLC).
2.6. Molecular dynamics
Molecular dynamics simulations were conducted with Gromacs (version 5.1.4). Force field was selected as CHARMM36.32 Structure of α-glucosidase was downloaded from the Protein Data Bank (PDB ID: 3A4A). Structures of myricetin, quercetin, kaempferol and acarbose were the same as those used for molecular docking analyses. System equilibration consisted of two steps: 100 ps NVT (constant number of particles, volume and temperature) and 100 ps NPT (constant number of particles, pressure and temperature). Molecular dynamic simulation was conducted for 10 ns, and time step was set as 2 fs. Molecular dynamic simulation results were analyzed with Xmgrace and PyMOL.
2.7. Kinetic analysis of inhibitory type
Inhibition kinetic analysis of flavonols against α-glucosidase was determined by the same method as enzyme assay. Analysis of inhibition mode was based on the inhibitory effect of four different concentrations of flavonol with five different concentrations of substrate (pNPG) on kinetic parameters including the maximal rate (Vmax) and Michaelis–Menten constant (Km) of enzyme. These parameters can be ascertained by Lineweaver–Burk plots, a double-reciprocal plot of enzyme reaction rate (ν) against the substrate concentration (pNPG), i.e., 1/ν vs. 1/[pNPG]. The mixed-type inhibition was expressed as eqn (2):
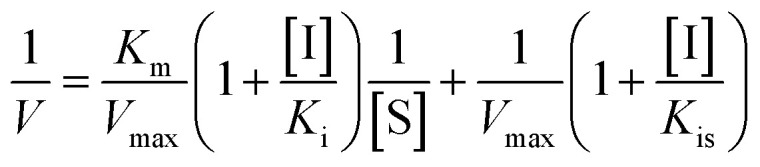 |
2 |
The secondary plots slope/Y-intercept against [flavonol] was plotted from:
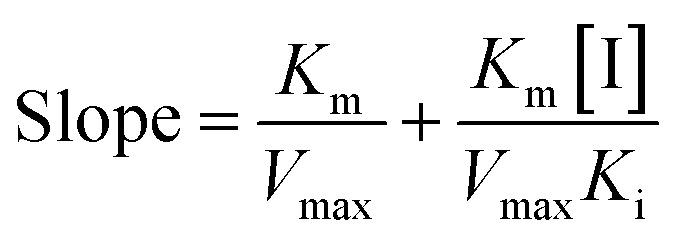 |
3 |
and
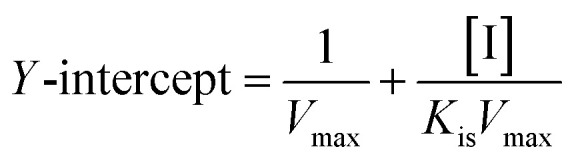 |
4 |
2.8. Fluorescence spectra measurements
Fluorescence spectra analysis was conducted on a spectrofluorometer (Jasco FP-8500, Tokyo, Japan). α-Glucosidase (0.8 μmol L−1) was titrated successively with varying concentrations of myricetin, quercetin and kaempferol from 0 to 19.96 μmol L−1 at three different temperatures (298, 304 and 310 K). The mixtures were equilibrated for 5 min before measurement. Fluorescence emission spectra were then obtained from 290 nm to 500 nm with an excitation wavelength of 280 nm. The excitation bandwidth was set at 1 nm and the emission bandwidth was 2.5 nm. Fluorescence of PPB was subtracted in order to correct the background fluorescence.
2.9. BLI measurements
Binding kinetic analysis of myricetin to α-glucosidase was determined by ForteBio Octet K2 system according to Xu et al.33 with minor modifications. First, α-glucosidase was incubated with biotin. Myricetin were diluted into different concentrations (double dilution from a top concentration of 500 μmol L−1). Then, the affinity constant (KD) and association rate constant (Kon) values were calculated by the super streptavidin (SSA) sensor (Fortebio).
2.10. CD spectroscopy
CD spectra in far-UV region (190–250 nm) were scanned by using a CD spectrometer (J-1500-15ST, Jasco Corp, Tokyo, Japan). α-Glucosidase concentration was fixed on 2 μmol L−1, and molar ratios of myricetin to α-glucosidase were set as 0 : 1, 40 : 1, and 80 : 1, respectively. CD spectra were averaged for three scans under constant nitrogen flush and buffer signal was subtracted.
2.11. Statistical analysis
All data were expressed as means ± standard deviation (SD) with experiments performed in triplicate. Value of half maximal inhibitory concentration (IC50) was calculated using SPSS 19.0 (IBM, Armonk, NY, USA).
3. Results and discussion
3.1. Flavonols in Chinese bayberry pulp
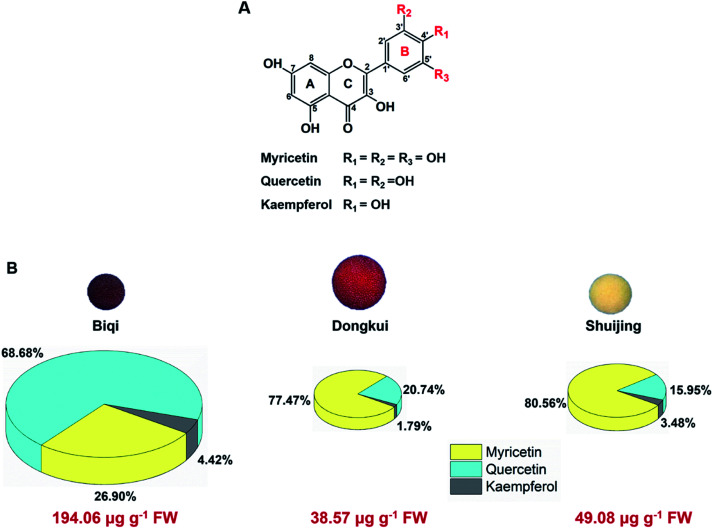
All Chinese bayberry cultivars tested, i.e., dark-red-pulp cultivar ‘Biqi’, pink-pulp cultivar ‘Dongkui’, and white-pulp cultivar ‘Shuijing’, were rich in flavonol glycosides. Their aglycones were myricetin, quercetin and kaempferol, respectively. These three flavonols shared similar chemical structures with only difference in the number of hydroxyl groups on their B-ring (Fig. 1A). Myricetin had three hydroxyl groups at the positions of 3′, 4′, and 5′ of B-ring, while quercetin had two hydroxyl groups (3′,4′-OH) and kaempferol had only one hydroxyl (4′-OH) on the B-ring.
Fig. 1. (A) Chemical structures of main flavonols in Chinese bayberry fruit. (B) Pi chart representation of flavonol composition in three cultivars of Chinese bayberry pulp. Flavonol contents were analyzed by high performance liquid chromatography.
Among the three cultivars, ‘Biqi’ showed the highest total flavonol content (194.06 μg g−1 fresh weight, FW), followed by ‘Shuijing’ (49.08 μg g−1 FW) and ‘Dongkui’ (38.57 μg g−1 FW) (Fig. 1B). In ‘Biqi’ pulp, quercetin was the most predominated flavonol with the content of 133.28 μg g−1 FW, while ‘Dongkui’ or ‘Shuijing’ pulp showed the largest proportion of myricetin in its flavonol composition as 29.88 and 39.54 μg g−1 FW, respectively (Fig. 1B). Kaempferol content was relative low in Chinese bayberry pulp (Fig. 1B). In bilberry and blueberry fruits, myricetin content were shown as 17 and 31 μg g−1 FW, respectively.34 In addition, it was reported that the bark or leaf of Chinese bayberry also contained these flavonols.35,36 Hence, Chinese bayberry fruit was a good source for dietary flavonol supplements.
3.2. α-Glucosidase inhibitory effects of flavonols
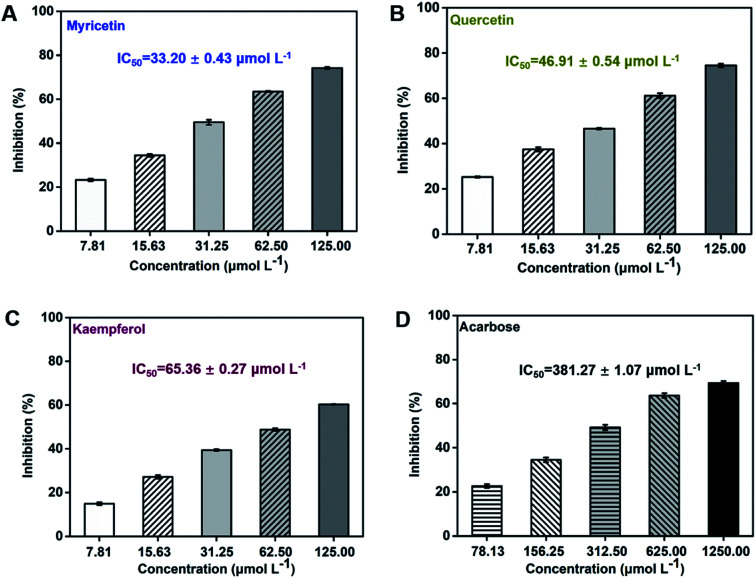
Flavonol glycosides will be hydrolyzed into their respective aglycones in lumen of the small intestine before absorption.37 Hence, the present study focused on investigation of the α-glucosidase inhibitory activity of three flavonol aglycones in Chinese bayberry. Inhibitory activities of myricetin, quercetin and kaempferol from Chinese bayberry fruit on α-glucosidase were shown in Fig. 2. These three flavonols all exhibited significant inhibition effects against α-glucosidase in a dose-dependent manner. At the concentration of 125 μmol L−1, myricetin or quercetin caused approximately 75% inhibition of the activity of α-glucosidase, while kaempferol caused approximately 60% inhibition of α-glucosidase activity. Among them, myricetin possessed the lowest IC50 value (33.20 ± 0.43 μmol L−1), followed by quercetin (46.91 ± 0.54 μmol L−1) and kaempferol (65.36 ± 0.27 μmol L−1) (Fig. 2). Such values were in agreement with that of the previous reports as 11.63 μmol L−1 for myricetin,38 40.89 μmol L−1 for quercetin39 and 56.70 μmol L−1 for kaempferol,40 respectively. Besides, IC50 values of these three flavonols were all much lower than that of the positive control acarbose (381.27 ± 1.07 μmol L−1) (Fig. 2), the value of which was consistent to the previous reports.41,42 Therefore, myricetin (3′,4′,5′-OH on the B-ring) showed the most potent inhibitory activity against α-glucosidase, followed by quercetin (3′,4′-OH on the B-ring) and kaempferol (4′-OH on the B-ring), indicating that the increased number of hydroxyl groups on its B-ring enhanced inhibitory activity on α-glucosidase.
Fig. 2. Inhibitory effects of flavonols from Chinese bayberry on α-glucosidase (acarbose was tested as the positive control).
3.3. Molecular docking analysis
Molecular docking is widely applied to bioactivity screening and structure-activity study in the field of drug discovery.43,44 As a computational analogue technique, it is recommended as an alternative way for experimental pre-screening and has shown significant success rate in drug discovery process.38,45,46 In the present study, docking analysis was performed to predict the interaction of flavonols and acarbose with α-glucosidase. Each docking calculation was executed with a high exhaustiveness and eventually twenty predicted modes were generated for every small molecule. The one with low binding energy and small dist. from the best mode was selected to visually illustrate receptor–ligand binding and analyze interaction mechanism.
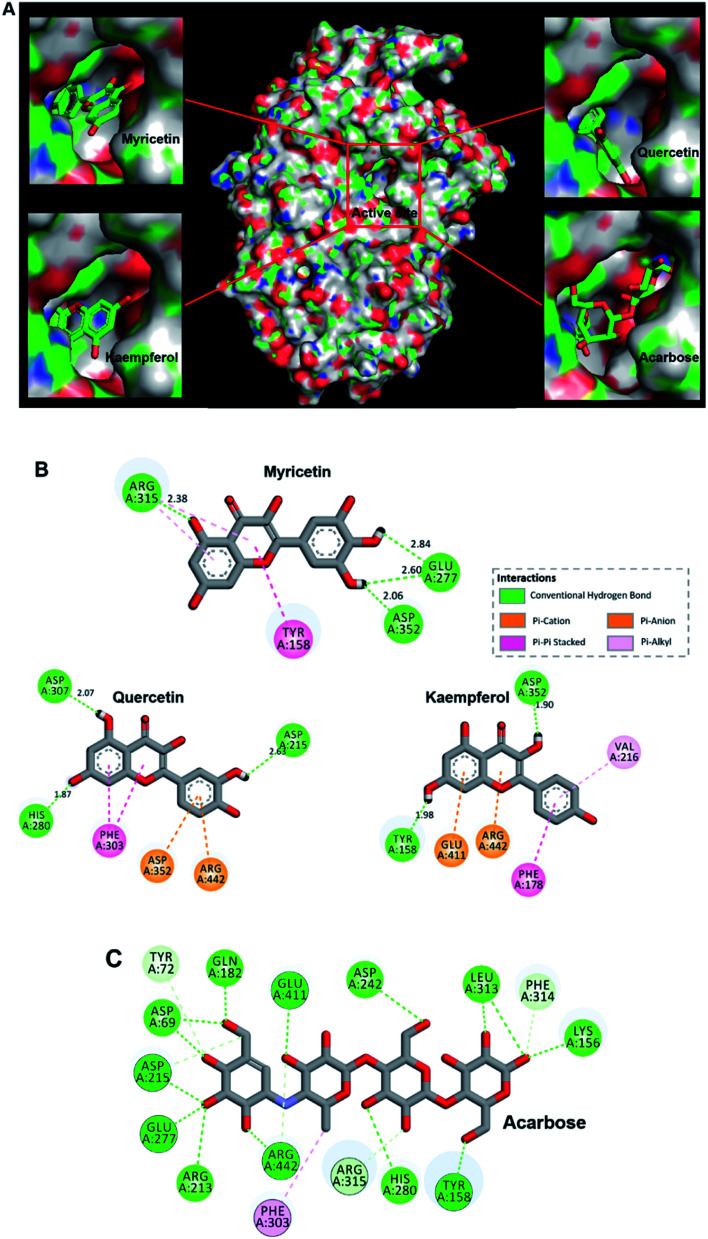
Results showed flavonols or acarbose all deeply inserted to active cavity of α-glucosidase (Fig. 3A). Related interaction residues as well as forces were shown in Fig. 3B and C. Competitive inhibitor acarbose was surrounded by amino acid residues in the catalytically active cavity of α-glucosidase with binding energy of −8.7 kcal mol−1 (Fig. 3C, Table 1). These residues were reported to play vital roles in the catalytic process.47,48 Thus, residues displayed as stick style in Fig. 3C were the ones that also involved in binding of flavonols to α-glucosidase, demonstrating the binding site of flavonols was close to that of acarbose.
Fig. 3. (A) Predicted binding poses of myricetin, quercetin, kaempferol and acarbose docked into α-glucosidase. (B) Schematic 2D diagram of myricetin, quercetin and kaempferol interacted with key residues within the active pocket of α-glucosidase. (C) Acarbose interacted with amino acid residues located in the active cavity of α-glucosidase.
Predicted affinity and kinetic analysis of myricetin, quercetin, kaempferol and acarbose binding to α-glucosidase.
| Inhibitors | Affinitya (kcal mol−1) | K i b (μmol L−1) | K is c (μmol L−1) | Inhibitory type |
|---|---|---|---|---|
| Myricetin | −9.2 | 15.56 | 31.64 | Mixed-type |
| Quercetin | −9.0 | 16.83 | 37.13 | Mixed-type |
| Kaempferol | −8.8 | 18.75 | 33.67 | Mixed-type |
| Acarbose | −8.7 | 109.95 | — | Competitive |
Affinity is predicted by molecular docking in silico.
K i represents the dissociation constant for inhibitors binding to free enzyme.
K is represents the dissociation constant for inhibitors binding to enzyme–substrate complex.
Flavonols occupied the entrance to active center of α-glucosidase. Their B-ring were more deeply penetrated to interact with catalytic residues (Fig. 3A). Myricetin formed four hydrogen bonds with three amino acids residues in the active pocket of α-glucosidase, including Glu 277, Arg 315 and Asp 352 (Fig. 3B). Its binding energy to α-glucosidase was predicted as −9.2 kcal mol−1 (Table 1), which is lower than that of acarbose, suggesting the greater stability of myricetin-α-glucosidase complex. In addition, hydrophobic was generated in the interaction as Pi–Pi stacked with Tyr 158, and Pi-alkyl with Arg 315. Main residues involved in the interaction between quercetin and α-glucosidase were Asp215, His 280, Phe 303, Asp 307, Asp 352 and Arg 442. Kaempferol was surrounded by Tyr 158, Phe 178, Val 216, Asp 352, Glu 411 and Arg 442. Predicted affinity of both compounds binding to α-glucosidase were calculated as −9.0 kcal mol−1 and −8.8 kcal mol−1, respectively (Table 1). Such values were similar as previous reports with the energy of −10.0 kcal mol−1 for quercetin or −9.3 kcal mol−1 for kaempferol binding to α-glucosidase.49 Hydrogen bonds and Pi interactions were also formed between their interactions with the enzyme (Fig. 3B). Pi-effects have an important contribution to protein–ligand recognition since they provide a significant amount of binding enthalpy.50 Compared with the interaction forces between acarbose and α-glucosidase, stronger Pi-interactions were shown in flavonols possessing more aromatic rings (Fig. 3B, C). It might be one reason that flavonols showed stronger inhibitory effects on α-glucosidase compared with acarbose. To summarize, hydrogen bonding and hydrophobic forces might be predominant interaction forces between flavonols and α-glucosidase.
Interestingly, myricetin (3′,4′,5′-OH) and quercetin (3′,4′-OH) had adjacent hydroxyl groups on B-ring where powerful hydrogen bonds were formed with distances up to 2.60 Å (Fig. 3B). For kaempferol (4′-OH), it lacks of adjacent hydroxyl groups, and hydrogen bonds were formed between single hydroxyl group on its A or C ring and α-glucosidase, with a smaller distance about 1.90 Å (Fig. 3B). These might be the main reason why myricetin, quercetin and kaempferol showed quite similar structures but different inhibitory effects on α-glucosidase. Adjacent hydroxyl groups on B ring of flavonols might increase its electron cloud density, which was beneficial for hydrogen atoms donation to form hydrogen bonds easier with active-center residues of α-glucosidase. It was reported that hydrogen bonds formation could help to increase the stability of complex,51,52 so the inhibitor bound more tightly to enzyme and enhanced its inhibition activity. Therefore, it was proposed that α-glucosidase inhibitory effects of these flavonols were enhanced with increase number of hydroxyl groups at their B-ring, which was consistent with the results of in vitro enzyme activity test.
3.4. Molecular dynamics simulations
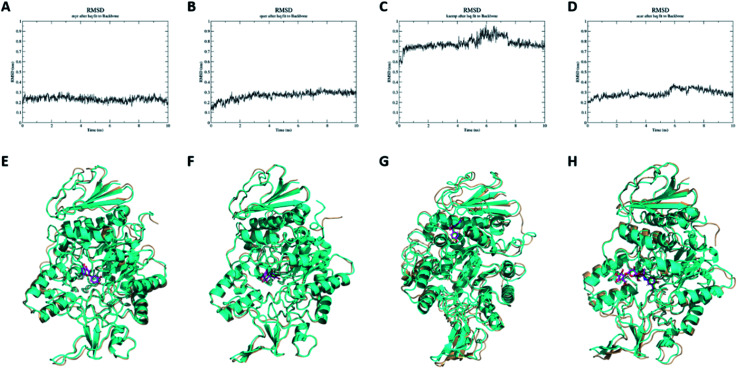
With purpose of validating the reliability of molecular docking results, molecular dynamics simulations were conducted on myricetin, quercetin, kaempferol and acarbose interaction with α-glucosidase. As shown in Fig. 4A–D, RMSD values of molecular dynamics simulation results of myricetin, quercetin, kaempferol and acarbose reached 0.16 nm, 0.30 nm, 0.75 nm and 0.26 nm after 10 ns of simulation, which indicated that the molecular docking results were reliable. As shown in Fig. 4E–H and ESI Movie 1–4,† binding patterns of myricetin, quercetin, kaempferol and acarbose remained unchanged, but their binding conformations were changed slightly. For α-glucosidase, conformations of α-helixes and β-sheets showed minor changes, while coils were changed in relatively large extents. These results indicated that conformations of α-glucosidase were changed when these four ligands bound to the enzyme. To confirm the structure-activity difference, interaction mechanism of these flavonols and α-glucosidase were further explored by kinetic analysis and fluorescence spectrum study.
Fig. 4. RMSD and conformation analyses of molecular dynamics simulation results. Conformation of α-glucosidase before molecular dynamics simulation is colored in cyan, conformation of α-glucosidase after molecular dynamics simulation is colored in wheat, conformation of ligand before molecular dynamics simulation is colored in purple, conformation of ligand after molecular dynamics simulation is colored in green. (A) RMSD of myricetin. (B) RMSD of quercetin. (C) RMSD of kaempferol. (D) RMSD of acarbose. (E) Conformation analyses of myricetin. (F) Conformation analyses of quercetin. (G) Conformation analyses of kaempferol. (H) Conformation analyses of acarbose.
3.5. Kinetic type of inhibition on α-glucosidase
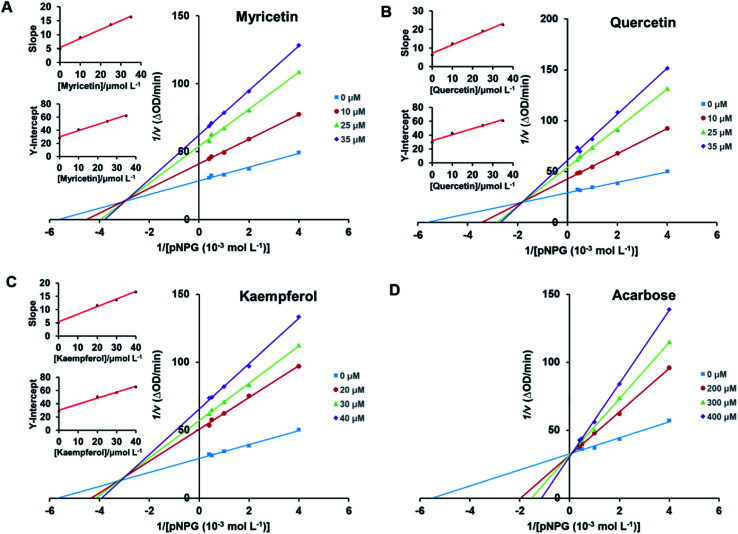
Lineweaver–Burk plots were conducted to analyze inhibitory type of myricetin, quercetin, and kaempferol on α-glucosidase. According to double-reciprocal plot of v against different concentrations of substrate [pNPG], all lines were linear and intersected in the second quadrant (Fig. 5A–C). Furthermore, vertical intercept (1/Vmax) and horizontal intercept (1/Km) changed simultaneously, as the Vmax value decreased with Km increased. These results suggested that myricetin, quercetin and kaempferol were all mixed-type inhibitors, and they could bind to free enzyme as well as enzyme–substrate complex. It was in good agreement with those reported by Phan et al.53 Acarbose induced a competitive type of inhibition on α-glucosidase since its Vmax value did not change but its Km value gradually increased with the rising concentration of acarbose (Fig. 5D), which was consistent with the previous reports.54,55
Fig. 5. Lineweaver–Burk plots for kinetic analysis of α-glucosidase inhibition by myricetin (A), quercetin (B), kaempferol (C) and the positive control acarbose (D). c(α-glucosidase) = 0.83 × 10−7 mol L−1. The secondary plots of slope and Y-intercept vs. [flavonols] were in the inset.
In addition, the secondary plots (inset of Fig. 5A–C) of slope and Y-intercept against flavonols concentrations were linearly fitted, indicating that they bound in a single inhibition site or a single class of inhibition sites on α-glucosidase.56 According to eqn (3) and (4), values of Ki and Kis in the presence of myricetin were calculated as 15.56 μmol L−1 and 31.64 μmol L−1, respectively, where Ki value was obviously much lower than Kis (Table 1). The two values represented the dissociation constants for inhibitor binding to free enzyme and enzyme–substrate complex, respectively.57 The smaller value of the constant suggested the stronger binding and more potent inhibition of flavonol on enzyme. Therefore, such results indicated that myricetin was more firmly bound to free α-glucosidase than to α-glucosidase–substrate complex, and so did quercetin and kaempferol. Furthermore, dissociation constant for myricetin-α-glucosidase Ki was the minimum, followed by quercetin, meaning myricetin (3′,4′,5′-OH) had stronger affinity to free enzyme than that of quercetin (3′,4′-OH) or kaempferol (4′-OH).
In clinical, commonly used antidiabetic drugs, such as acarbose, miglitol and voglibose all belong to competitive inhibitors.3 They compete with substrate for the active site of α-glucosidase, prevent ligand–substrate binding and catalysis of substrate, and thus inhibit the activity of enzyme. While they cannot bind to α-glucosidase–substrate complex. Plenty of natural α-glucosidase inhibitors, e.g., luteolin, genistein, morin and galangin, were all reported to work in a mixed-type manner.5,41,58 Such dietary flavonoids could form enzyme–inhibitor complexes as well as enzyme–substrate–inhibitor complexes to interrupt enzyme–substrate intermediate.58
3.6. Fluorescence quenching studies and binding parameters
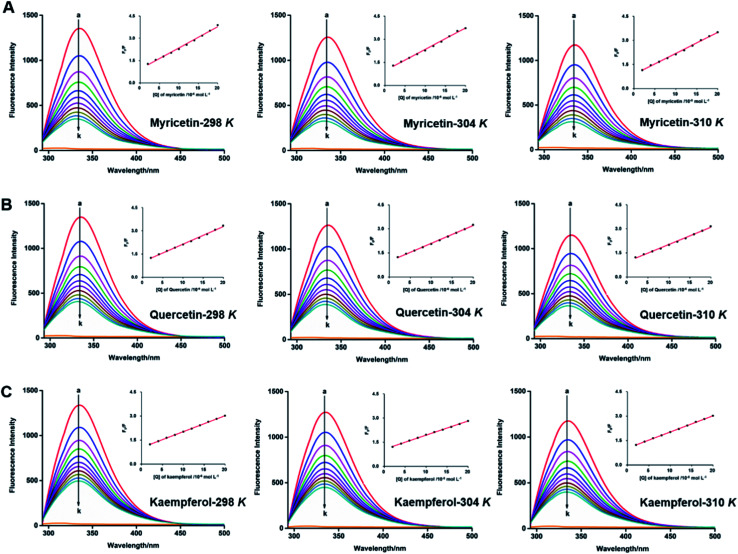
Interaction between flavonols and α-glucosidase were investigated by fluorescence spectroscopy. Fluorescence intensity of α-glucosidase, along with treatment of gradually increasing concentrations of myricetin (Fig. 6A), quercetin (Fig. 6B) or kaempferol (Fig. 6C) were scanned from 290 nm to 500 nm. Excitation wavelength was set as 280 nm and detections were at three different temperatures of 298 K, 304 K and 310 K. Results showed that α-glucosidase had a strong fluorescence intensity at around 336 nm, while flavonols did not display fluorescence under the same conditions. It meant that flavonol itself had no intrinsic fluorescence and such spectra was a proper way to explore effects of flavonols on endogenous fluorescence of α-glucosidase. As shown in Fig. 6, with increase concentrations of flavonols, intrinsic fluorescence intensity of α-glucosidase successively decreased but the maximum peak wavelength did not shift. It indicated that flavonols could interact with α-glucosidase and quench its intrinsic fluorescence. Related fluorescence quenching mechanisms of α-glucosidase by flavonols were investigated by the following Stern–Volmer equation:59
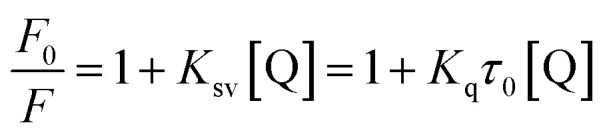 |
5 |
where F0 and F represented fluorescence intensities of α-glucosidase before and after addition of flavonols, respectively. [Q] was the concentration of flavonols. τ0 meant fluorophore lifetime without flavonol and was about 10−8 s for α-glucosidase. Ksv and Kq displayed Stern–Volmer quenching constant and biomolecule quenching rate constant, respectively.
Fig. 6. Synchronous fluorescence spectra of α-glucosidase in the absence and presence of myricetin (A), quercetin (B), kaempferol (C) at different temperatures 298 K, 304 K and 310 K. c(α-glucosidase) = 0.8 × 10−6 mol L−1. c(flavonols) = (0, 2, 4, 6, 7.99, 9.99, 11.99, 13.98, 15.97, 17.97, 19.96) × 10−6 mol L−1 for curves a→k, respectively. Plots of F0/F vs. [Q] were in the inset.
According to eqn (5), plots of F0/F vs. [Q] were plotted to calculate values of Ksv and Kq at different temperatures of 298, 304 and 310 K. All lines were linearly fitted and the slope equal to the Ksv value (inset of Fig. 6). As shown in Table 2, the Ksv values for myricetin were (14.26 ± 0.03) × 104, (13.81 ± 0.04) × 104, and (13.05 ± 0.03) × 104 L mol−1 at 298, 304 and 310 K, respectively. Such values gradually decreased along with increasing temperatures. Moreover, Ksv values for myricetin at 298 K was much larger than that of quercetin [(11.42 ± 0.06) × 104 L mol−1] and kaempferol [(9.91 ± 0.08) × 104 L mol−1], indicating myricetin (3′,4′,5′-OH) showed the strongest ability to quench intrinsic fluorescence of α-glucosidase, followed by quercetin (3′,4′-OH) and kaempferol (4′-OH). In addition, as Kq = Ksv/τ0 (τ0 = 10−8 s), corresponding Kq values were about 1012 L mol−1 s−1, which were much greater than the maximum quenching constant of diffusion collision (2 × 1010 L mol−1 s−1). These results demonstrated that static quenching is the main quenching process of flavonols on the intrinsic fluorescence of α-glucosidase.
Quenching constants Ksv, binding constants Ka and relative thermodynamic parameters of the interaction of myricetin, quercetin, kaempferol with α-glucosidase at different temperatures of 298 K, 301 K and 310 Ka.
| T (K) | K sv (×104 L mol−1) | R a b | K a (×105 L mol−1) | n | R b c | ΔH° (kJ mol−1) | ΔS° (J mol−1K−1) | ΔG° (kJ mol−1) |
|---|---|---|---|---|---|---|---|---|
| Myricetin | ||||||||
| 298 | 14.26 ± 0.03 | 0.9963 | 1.38 ± 0.03 | 0.99 ± 0.01 | 0.9970 | −16.16 ± 0.57 | 44.22 ± 1.17 | −29.34 ± 0.05 |
| 304 | 13.81 ± 0.04 | 0.9953 | 1.23 ± 0.03 | 1.04 ± 0.01 | 0.9949 | −29.60 ± 0.04 | ||
| 310 | 13.05 ± 0.03 | 0.9967 | 1.07 ± 0.01 | 1.06 ± 0.01 | 0.9983 | −29.87 ± 0.04 | ||
| Quercetin | ||||||||
| 298 | 11.42 ± 0.06 | 0.9978 | 1.25 ± 0.01 | 0.97 ± 0.01 | 0.9988 | −12.23 ± 0.98 | 56.51 ± 3.27 | −29.07 ± 0.03 |
| 304 | 11.20 ± 0.06 | 0.9990 | 1.13 ± 0.03 | 0.99 ± 0.01 | 0.9991 | −29.40 ± 0.03 | ||
| 310 | 10.76 ± 0.09 | 0.9986 | 1.03 ± 0.03 | 1.01 ± 0.01 | 0.9977 | −29.74 ± 0.04 | ||
| Kaempferol | ||||||||
| 298 | 9.91 ± 0.08 | 0.9995 | 1.23 ± 0.01 | 0.87 ± 0.01 | 0.9995 | −9.71 ± 0.99 | 64.84 ± 3.25 | −29.03 ± 0.03 |
| 304 | 8.81 ± 0.05 | 0.9993 | 1.12 ± 0.03 | 0.93 ± 0.01 | 0.9998 | −29.42 ± 0.01 | ||
| 310 | 8.04 ± 0.02 | 0.9998 | 1.05 ± 0.01 | 0.98 ± 0.01 | 0.9997 | −29.80 ± 0.02 | ||
Values are presented means ± standard deviation (SD).
R a is the correlation coefficient for the Ksv values.
R b is the correlation coefficient for the Ka values.
As for static quenching process, it was assumed that flavonols had an independent binding site n on α-glucosidase and related binding parameters could be calculated by the equation below:60
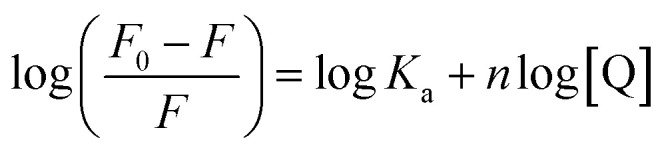 |
6 |
where F0, F and [Q] denoted the same as eqn (5). Ka represented binding constant of flavonol and α-glucosidase. n was the number of binding sites per protein molecule.
By plotting log[(F0 − F)/F] vs. log[Q], binding constant (Ka) and the number of binding sites (n) at 298, 304 and 310 K were calculated from intercept (log Ka) and slope (n) of corresponding linearly fitted plots, respectively. Such data were all listed in Table 2. Since correlation coefficients (Rb) were all above 0.99, assumption underlying the derivation of eqn (6) was reasonable.61
Since values of n were all approximately equal to 1 (Table 2), there was just one binding site on α-glucosidase in the presence of each flavonol. It was consistent with the results obtained from Lineweaver–Burk plot analysis. Ka values of each flavonol at different temperatures were in the order of 105 L mol−1 (Table 2), indicating a high affinity existing between the three flavonols and α-glucosidase. Thus, Ka values tended to decrease with increase of temperature, which was accord with the variations of Ksv values, suggesting that stability of myricetin-α-glucosidase complex would decrease at higher temperature, as well as quercetin-α-glucosidase and kaempferol-α-glucosidase complexes. At the temperature of 298 K, Ka value of myricetin-α-glucosidase system was much larger than that of quercetin/kaempferol-α-glucosidase system [(1.25 ± 0.01) × 105 L mol−1, (1.23 ± 0.01) × 105 L mol−1, respectively]. It indicated that myricetin (3′,4′,5′-OH) showed stronger affinity on α-glucosidase, which was in consistent with the results that myricetin showed the strongest α-glucosidase inhibitory effects and next is quercetin (3′,4′-OH) and kaempferol (4′-OH).
3.7. Thermodynamic analysis
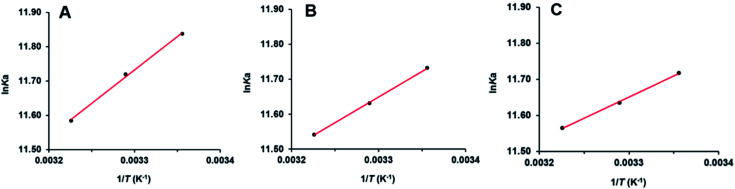
Interaction forces between flavonols and α-glucosidase were evaluated by the van't Hoff equation:
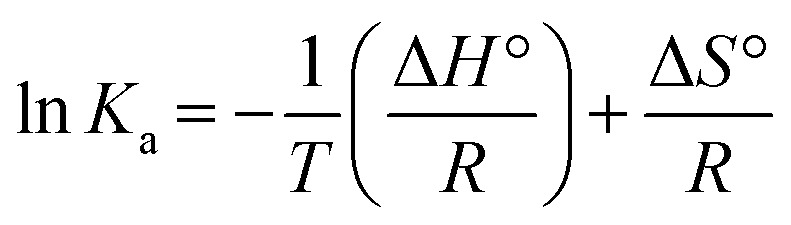 |
7 |
where Ka (L mol−1) was the binding constant at the temperature of T (K). R (8.314 J mol−1 K−1) was the gas constant. ΔH° and ΔS° represented enthalpy change (kJ mol−1) and entropy change (J mol−1 K−1), respectively. Values of ΔH° and ΔS° were determined by the slope and intercept of the linear fitted line of ln Kavs. 1/T (Fig. 7) and summarized in Table 2. Free energy change ΔG° (kJ mol−1) was further calculated by the following equation:62
| ΔG° = ΔH° − TΔS° = −RT ln Ka | 8 |
Fig. 7. van't Hoff’ plots of myricetin (A), quercetin (B), kaempferol (C) and α-glucosidase.
Negative values of ΔG° meant that binding process of flavonols and α-glucosidase was spontaneous. As shown in Table 2, ΔH° were calculated as negative values. It indicated that interaction between each flavonol and α-glucosidase was an exothermic process and decrease of temperature would be beneficial for the binding. According to the theory of Ross and Subramanian,63 ΔH° < 0, ΔS° > 0 implied that hydrophobic forces and hydrogen bonding might be the predominant interaction forces between flavonols and α-glucosidase, which validated the results from molecular docking.
3.8. Further investigation on the strongest α-glucosidase inhibitor myricetin
As the results shown above, myricetin had the strongest α-glucosidase inhibitory activity in our assay. It worked as a mixed-type inhibitor, and showed high affinity to α-glucosidase. Their binding induced conformational changes of enzyme by forming myricetin-α-glucosidase complex and thus resulted in the inhibition activity on enzyme. As the most potent α-glucosidase inhibitor, myricetin was selected as a representative flavonol to further investigate the above results by reversibility, CD spectra and BLI analysis.
3.8.1. Reversibility
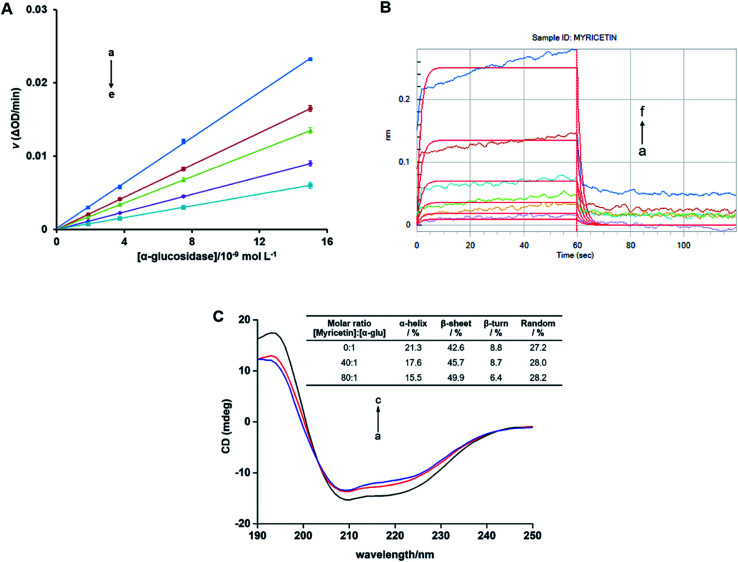
According to plots of v vs. [α-glucosidase] at various concentrations (Fig. 8A), all lines passed through the origin and showed high linearity, and slopes of lines reduced with the increase of myricetin concentrations. It indicated that myricetin just reduced catalytic rate rather than inactive the enzyme completely. For irreversible inhibitor, it may inactive the enzyme by covalent binding to form a stable complex.64 Therefore, such result confirmed that myricetin was a reversible inhibitor. It helped to understand that myricetin showed α-glucosidase inhibitory effect in a mixed-type manner, which belongs to reversible inhibition.
Fig. 8. (A) Plots of v vs. [α-glucosidase]. c(pNPG) = 3.13 × 10−4 mol L−1. Myricetin concentrations were 0, 0.78, 1.56, 3.13, 6.25 × 10−5 mol L−1 for curves a→e, respectively. (B) Binding kinetic analysis for the interaction of myricetin with α-glucosidase by bio-layer interferometry (BLI). c(myricetin) = (0.16, 0.31, 0.63, 1.25, 2.50, 5.00) × 10−4 mol L−1 for curves a→f. The red vertical line at 60 s marks the start of dissociation of the compound from the sensor surface. (C) Circular Dichroism (CD) spectra of α-glucosidase in the presence of increasing amounts of myricetin. c(α-glucosidase) = 2.01 × 10−6 mol L−1, the molar ratios of myricetin to α-glucosidase were 0 : 1 (a), 40 : 1 (b), 80 : 1 (c), respectively.
3.8.2. BLI analysis
Kinetic analysis of myricetin binding to α-glucosidase was carried out by BLI technology. As shown in Fig. 8B, the association responses improved with increase of myricetin concentrations. Affinity constant of KD was 6.19 × 10−4 mol L−1 and association rate constant of Kon was 2.59 × 103 mol−1 L−1 S−1. Such results confirmed that myricetin binds to α-glucosidase in a high affinity manner.
3.8.3. CD spectra analysis
CD spectra was used to study the secondary structure changes of α-glucosidase induced by myricetin. As shown in Fig. 8C, far-UV (190–250 nm) CD spectra of α-glucosidase were characterized by two negative bands at 210 nm and 222 nm. These negative peaks were reported as characteristics of α-helical structure and generated from electron transfer of n→π* and π→π* for peptide bonds of α-helix.65 After addition of 80 μmol L−1 or 160 μmol L−1 of myricetin to α-glucosidase, CD intensities at both bands were regularly decreased (Fig. 8C), suggesting conformational changes of α-glucosidase due to its interaction with myricetin.
For free α-glucosidase, percentages of α-helix, β-sheet, β-turn and random coil was 21.3%, 42.6%, 8.8% and 27.2%, respectively. With the increase molar ratios of myricetin to α-glucosidase (from 40 : 1 to 80 : 1), contents of α-helix and β-turn tended to decrease from 17.6% to 15.5% and from 8.7% to 6.4%, respectively. While proportions of β-sheet increased from 45.7% to 49.9%, random coil percentages were slightly enhanced from 28.0% to 28.2% (Fig. 8C). Such secondary structure changes demonstrated that binding of myricetin to α-glucosidase might induce an unfolding of protein structure, and thus result in enzyme activity inhibition. It helped explain the results deduced from molecular docking, kinetic analysis and fluorescence spectrum that myricetin induced conformational changes of α-glucosidase by inhibitor–enzyme complex formation. Whereby myricetin prevented substrate from binding to enzyme and inhibited α-glucosidase activity.
4. Conclusions
The present study demonstrated potent inhibitory effects and substrate–enzyme interaction mechanism of myricetin, quercetin and kaempferol from Chinese bayberry fruit on α-glucosidase. Three flavonols tested all exhibited strong inhibitory effects on α-glucosidase, and such activities were enhanced with increase of hydroxyl groups attached to ring B. Molecular docking combined with molecular dynamics simulation illustrated that adjacent hydroxyl groups at B-ring of flavonols could positively contribute to form hydrogen bonds that are more powerful. It helped to increase the stability of flavonol–enzyme complexes, and different B-ring hydroxylations might be the main reason for their activity difference on α-glucosidase. Such in silico results were further validated by kinetic analysis, fluorescence spectroscopy, CD spectra and BLI analysis. These findings indicated that flavonols in Chinese bayberry fruit were excellent α-glucosidase inhibitors and fruit rich in flavonols might be promising nutritional supplements for prevention and treatment of T2D.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This research was funded by the National Natural Science Foundation of China (31872067), the National Key R&D Program of China (No. 2017YFE0102200), the 111 project (B17039), and the Fundamental Research Funds for the Central Universities.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05015f
References
- Cho N. H. Shaw J. E. Karuranga S. Huang Y. da Rocha F. J. D. Ohlrogge A. W. Malanda B. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- World Health Organization, Global report on diabetes, Geneva, 2016 [Google Scholar]
- Bischoff H. Pharmacology of α-glucosidase inhibition. Eur. J. Clin. Invest. 1994;24:3–10. doi: 10.1111/j.1365-2362.1994.tb02418.x. [DOI] [PubMed] [Google Scholar]
- Kheirandish M. Mahboobi H. Yazdanparast M. Kamal M. A. Challenges related to glycemic control in type 2 diabetes mellitus patients. Curr. Drug Metab. 2017;18:157–162. doi: 10.2174/1389200218666170116105023. [DOI] [PubMed] [Google Scholar]
- Zeng L. Ding H. Hua X. Zhang G. Gong D. Galangin inhibits α-glucosidase activity and formation of non-enzymatic glycation products. Food Chem. 2019;271:70–79. doi: 10.1016/j.foodchem.2018.07.148. [DOI] [PubMed] [Google Scholar]
- Cheng A. Y. Fantus I. G. Oral antihyperglycemic therapy for type 2 diabetes mellitus. Can. Med. Assoc. J. 2005;172:213–226. doi: 10.1503/cmaj.1031414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujusawa T. Ikegami H. Inoue K. Kawabata Y. Ogihara T. Effect of two α-glucosidase inhibitors, voglibose and acarbose, on postprandial hyperglycemia correlates with subjective abdominal symptoms. Metabolism. 2005;54:387–390. doi: 10.1016/j.metabol.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Castro-Acosta M. L. Lenihan-Geels G. N. Corpe C. P. Hall W. L. Berries and anthocyanins: promising functional food ingredients with postprandial glycaemia-lowering effects. Proc. Nutr. Soc. 2016;75:342–355. doi: 10.1017/S0029665116000240. [DOI] [PubMed] [Google Scholar]
- Vendrame S. Del Bo' C. Ciappellano S. Riso P. Klimis-Zacas D. Berry fruit consumption and metabolic syndrome. Antioxidants. 2016;5:E34. doi: 10.3390/antiox5040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Huang H. Zhang Q. Fan F. Xu C. Sun C. Li X. Chen K. Phytochemical characterization of Chinese bayberry (Myrica rubra Sieb. et Zucc.) of 17 cultivars and their antioxidant properties. Int. J. Mol. Sci. 2015;16:12467–12481. doi: 10.3390/ijms160612467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Zhang X. Xie W. Zheng Y. Cao J. Cao P. Chen Q. Li X. Sun C. The growth of SGC-7901 tumor xenografts was suppressed by Chinese bayberry anthocyanin extract through upregulating KLF6 gene expression. Nutrients. 2016;8:599. doi: 10.3390/nu8100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C. Zhang B. Zhang J. Xu C. Wu Y. Li X. Chen K. Cyanidin-3-glucoside-rich extract from Chinese bayberry fruit protects pancreatic β cells and ameliorates hyperglycemia in streptozotocin-induced diabetic mice. J. Med. Food. 2012;15:288–298. doi: 10.1089/jmf.2011.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. Buya M. Qin W. Sun C. Cai H. Xie Q. Xu B. Wu Y. Anthocyanins from Chinese bayberry extract activate transcription factor Nrf2 in β cells and negatively regulate oxidative stress-induced autophagy. J. Agric. Food Chem. 2013;61:8765–8772. doi: 10.1021/jf4012399. [DOI] [PubMed] [Google Scholar]
- Zhang X. Huang H. Zhao X. Lv Q. Sun C. Li X. Chen K. Effects of flavonoids-rich Chinese bayberry (Myrica rubra Sieb. et Zucc.) pulp extracts on glucose consumption in human HepG2 cells. J. Funct. Foods. 2015;14:144–153. doi: 10.1016/j.jff.2015.01.030. [DOI] [Google Scholar]
- Zhang X. Lv Q. Jia S. Chen Y. Sun C. Li X. Chen K. Effects of flavonoid-rich Chinese bayberry (Morella rubra Sieb. et Zucc.) fruit extract on regulating glucose and lipid metabolism in diabetic KK-Ay mice. Food Funct. 2016;7:3130–3140. doi: 10.1039/C6FO00397D. [DOI] [PubMed] [Google Scholar]
- Yan S. Zhang X. Wen X. Lv Q. Xu C. Sun C. Li X. Purification of flavonoids from Chinese bayberry (Morella rubra Sieb. et Zucc.) fruit extracts and α-glucosidase inhibitory activities of different fractionations. Molecules. 2016;21:1148. doi: 10.3390/molecules21091148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. G. Dos Santos R. N. Oliva G. Andricopulo A. D. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S. Bordoloi M. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr. Drug Targets. 2019;20:501–521. doi: 10.2174/1389450119666181022153016. [DOI] [PubMed] [Google Scholar]
- Chinnasamy S. Selvaraj G. Kaushik A. C. Kaliamurthi S. Chandrabose S. Singh S. K. Thirugnanasambandam R. Gu K. Wei D. Q. Molecular docking and molecular dynamics simulation studies to identify potent AURKA inhibitors: assessing the performance of density functional theory, MM-GBSA and mass action kinetics calculations. J. Biomol. Struct. Dyn. 2019:1–11. doi: 10.1080/07391102.2019.1674695. [DOI] [PubMed] [Google Scholar]
- Tayyab S. Izzudin M. M. Kabir M. Z. Feroz S. R. Tee W. V. Mohamad S. B. Alias Z. Binding of an anticancer drug, axitinib to human serum albumin: fluorescence quenching and molecular docking study. J. Photochem. Photobiol., B. 2016;162:386–394. doi: 10.1016/j.jphotobiol.2016.06.049. [DOI] [PubMed] [Google Scholar]
- Chen Y. C. Wang H. M. Niu Q. X. Ye D. Y. Liang G. W. Binding between saikosaponin C and human serum albumin by fluorescence spectroscopy and molecular docking. Molecules. 2016;21:153. doi: 10.3390/molecules21020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya N. Maurya J. K. Singh U. K. Dohare R. Zafaryab M. Rizvi M. M. A. Kumari M. Patel R. In vitro cytotoxicity and interaction of noscapine with human serum albumin: effect on structure and esterase activity of HAS. Mol. Pharm. 2019;16:952–966. doi: 10.1021/acs.molpharmaceut.8b00864. [DOI] [PubMed] [Google Scholar]
- Zhang Z. C. Wang H. B. Zhou Q. Hu B. Wen J. H. Zhang J. L. Screening of effective xanthine oxidase inhibitors in dietary anthocyanins from purple sweet potato (Ipomoea batatas L. Cultivar Eshu No. 8) and deciphering of the underlying mechanisms in vitro. J. Funct. Foods. 2017;36:102–111. doi: 10.1016/j.jff.2017.06.048. [DOI] [Google Scholar]
- Wang Y. Zhang G. Yan J. Gong D. Inhibitory effect of morin on tyrosinase: insights from spectroscopic and molecular docking studies. Food Chem. 2014;163:226–233. doi: 10.1016/j.foodchem.2014.04.106. [DOI] [PubMed] [Google Scholar]
- Cooper M. A. Current biosensor technologies in drug discovery. Drug Discovery World. 2006;7:68–82. [Google Scholar]
- Rich R. L. Myszka D. G. Higher-throughput, label-free, real-time molecular interaction analysis. Anal. Biochem. 2007;361:1–6. doi: 10.1016/j.ab.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Cao Y. Xie L. Ma Y. Ren C. Xing M. Fu Z. Wu X. Yin X. Xu C. Li X. PpMYB15 and PpMYBF1 transcription factors are involved in regulating flavonol biosynthesis in peach fruit. J. Agric. Food Chem. 2019;67:644–652. doi: 10.1021/acs.jafc.8b04810. [DOI] [PubMed] [Google Scholar]
- Han Y. Vimolmangkang S. Soria-Guerra R. E. Rosales-Mendoza S. Zheng D. Lygin A. V. Korban S. S. Ectopic expression of apple F3'H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010;153:806–820. doi: 10.1104/pp.109.152801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M. Pan J. Hu X. Gong D. Zhang G. Inhibitory effect of corosolic acid on α-glucosidase: kinetics, interaction mechanism and molecular simulation. J. Sci. Food Agric. 2019;99:5881–5889. doi: 10.1002/jsfa.9862. [DOI] [PubMed] [Google Scholar]
- Trott O. Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M. Huey R. Lindstrom W. Sanner M. F. Belew R. K. Goodsell D. S. Olson A. J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. MacKerell A. D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. Bao Q. Jia J. Liu F. Guo X. Zhang M. Wei J. Lu M. Xu L. Zhang X. You Q. Sun H. CPUY201112, a novel synthetic small-molecule compound and inhibitor of heat shock protein Hsp90, induces p53-mediated apoptosis in MCF-7 cells. Sci. Rep. 2016;6:19004. doi: 10.1038/srep19004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttä-Riihinen K. R. Kamal-Eldin A. Mattila P. H. González-Paramás A. M. Törrönen A. R. Distribution and contents of phenolic compounds in eighteen Scandinavian berry species. J. Agric. Food Chem. 2004;52:4477–4486. doi: 10.1021/jf049595y. [DOI] [PubMed] [Google Scholar]
- Akazawa H. Fujita Y. Banno N. Watanabe K. Kimura Y. Manosroi A. Manosroi J. Akihisa T. Three new cyclic diarylheptanoids and other phenolic compounds from the bark of myrica rubra and their melanogenesis inhibitory and radical scavenging activities. J. Oleo Sci. 2010;59:213–221. doi: 10.5650/jos.59.213. [DOI] [PubMed] [Google Scholar]
- Yang H. H. Ge Y. Q. Sun Y. J. Liu D. D. Ye X. Q. Wu D. Identification and characterisation of low-molecular-weight phenolic compounds in bayberry (Myrica rubra Sieb. et Zucc.) leaves by HPLC-DAD and HPLC-UV-ESIMS. Food Chem. 2011;128:1128–1135. doi: 10.1016/j.foodchem.2011.03.118. [DOI] [Google Scholar]
- Cassidy A. Minihane A.-M. The role of metabolism (and the microbiome) in defining the clinical efficacy of dietary flavonoids. Am. J. Clin. Nutr. 2017;105:10–22. doi: 10.3945/ajcn.116.136051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y. Ma Y. Cheng G. Zhang Y. Cai S. Comparative study of dietary flavonoids with different structures as α-glucosidase inhibitors and insulin sensitizers. J. Agric. Food Chem. 2019;67:10521–10533. doi: 10.1021/acs.jafc.9b04943. [DOI] [PubMed] [Google Scholar]
- Huang G. J. Hsieh W. T. Chang H. Y. Huang S. S. Lin Y. C. Kuo Y. H. α-Glucosidase and aldose reductase inhibitory activities from the fruiting body of Phellinus merrilli. J. Agric. Food Chem. 2011;59:5702–5706. doi: 10.1021/jf2003943. [DOI] [PubMed] [Google Scholar]
- Varghese G. K. Bose L. V. Habtemariam S. Antidiabetic components of Cassia alata leaves: identification through α-glucosidase inhibition studies. Pharm. Biol. 2013;51:345–349. doi: 10.3109/13880209.2012.729066. [DOI] [PubMed] [Google Scholar]
- Zeng L. Zhang G. Liao Y. Gong D. Inhibitory mechanism of morin on α-glucosidase and its anti-glycation properties. Food Funct. 2016;7:3953–3963. doi: 10.1039/C6FO00680A. [DOI] [PubMed] [Google Scholar]
- Xu Y. Xie L. Xie J. Liu Y. Chen W. Pelargonidin-3-O-rutinoside as a novel α-glucosidase inhibitor for improving postprandial hyperglycemia. Chem. Commun. 2019;55:39. doi: 10.1039/C8CC07985D. [DOI] [PubMed] [Google Scholar]
- Turkson J., in The Molecular Basis of Human Cancer, Springer, 2017, pp. 695–707 [Google Scholar]
- Ziedan N. I. Hamdy R. Cavaliere A. Kourti M. Prencipe F. Brancale A. Jones A. T. Westwell A. D. Virtual screening, SAR and discovery of 5-(indole-3-yl)-2-[(2-nitrophenyl) amino][1,3,4]-oxadiazole as a novel Bcl-2 inhibitor. Chem. Biol. Drug Des. 2017;90:147–155. doi: 10.1111/cbdd.12936. [DOI] [PubMed] [Google Scholar]
- Rasouli H. Hosseini-Ghazvini S. M. Adibi H. Khodarahmi R. Differential α-amylase/α-glucosidase inhibitory activities of plant-derived phenolic compounds: a virtual screening perspective for the treatment of obesity and diabetes. Food Chem. 2017;8:1942–1954. doi: 10.1039/c7fo00220c. [DOI] [PubMed] [Google Scholar]
- Sarkar A. Sen S. A comparative analysis of the molecular interaction techniques for in silico drug design. Int. J. Pept. Res. Ther. 2020;26:209–223. doi: 10.1007/s10989-019-09830-6. [DOI] [Google Scholar]
- Yamamoto K. Miyake H. Kusunoki M. Osaki S. Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose. FEBS J. 2010;277:4205–4214. doi: 10.1111/j.1742-4658.2010.07810.x. [DOI] [PubMed] [Google Scholar]
- Peng X. Zhang G. Liao Y. Gong D. Inhibitory kinetics and mechanism of kaempferol on α-glucosidase. Food Chem. 2016;190:207–215. doi: 10.1016/j.foodchem.2015.05.088. [DOI] [PubMed] [Google Scholar]
- Ning Z. Zhai L. Huang T. Peng J. Hu D. Xiao H. Wen B. Lin C. Zhao L. Bian Z. Identification of α-glucosidase inhibitors from Cyclocarya paliurus tea leaves using UF-UPLC-Q/TOF-MS/MS and molecular docking. Food Funct. 2019;10:1893. doi: 10.1039/C8FO01845F. [DOI] [PubMed] [Google Scholar]
- Meyer E. A. Castellano R. K. Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem., Int. Ed. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- Roy S. Nandi R. K. Ganai S. Majumdar K. C. Das T. K. Binding interaction of phosphorus heterocycles with bovine serum albumin: a biochemical study. J. Pharm. Anal. 2017;7:19–26. doi: 10.1016/j.jpha.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire J. B. Boyken S. E. Baker D. Kuhlman B. Rapid sampling of hydrogen bond networks for computational protein design. J. Chem. Theory Comput. 2018;14:2751–2760. doi: 10.1021/acs.jctc.8b00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan M. A. T. Wang J. Tang J. Lee Y. Z. Ng K. Evaluation of α-glucosidase inhibition potential of some flavonoids from Epimedium brevicornum. LWT--Food Sci. Technol. 2013;53:492–498. doi: 10.1016/j.lwt.2013.04.002. [DOI] [Google Scholar]
- Yee H. S. Fong N. T. A review of the safety and efficacy of acarbose in diabetes mellitus. Pharmacotherapy. 1996;16:792–805. [PubMed] [Google Scholar]
- Liu Z. Yang Y. Dong W. Liu Q. Wang R. Pang J. Xia X. Zhu X. Liu S. Shen Z. Xiao Z. Liu Y. Investigation on the enzymatic profile of mulberry alkaloids by enzymatic study and molecular docking. Molecules. 2019;24:1776. doi: 10.3390/molecules24091776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. P. Chen Q. X. Song K. K. Xie J. J. Inhibitory effects of salicylic acid family compounds on the diphenolase activity of mushroom tyrosinase. Food Chem. 2006;95:579–584. doi: 10.1016/j.foodchem.2005.01.042. [DOI] [Google Scholar]
- Xiong Z. Liu W. Zhou L. Zou L. Chen J. Mushroom (Agaricus bisporus) polyphenoloxidase inhibited by apigenin: multi-spectroscopic analyses and computational docking simulation. Food Chem. 2016;203:430–439. doi: 10.1016/j.foodchem.2016.02.045. [DOI] [PubMed] [Google Scholar]
- Tadera K. Minami Y. Takamatsu K. Matsuoka T. Inhibition of α-glucosidase and α-amylase by flavonoids. J. Nutr. Sci. Vitaminol. 2006;52:149–153. doi: 10.3177/jnsv.52.149. [DOI] [PubMed] [Google Scholar]
- Boguta P. Sokolowska Z. Zinc binding to fulvic acids: assessing the impact of pH, metal concentrations and chemical properties of fulvic acids on the mechanism and stability of formed soluble complexes. Molecules. 2020;25:1297. doi: 10.3390/molecules25061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G. W. Chen Y. C. Wang Y. Wang H. M. Pan X. Y. Chen P. H. Niu Q. X. Interaction between saikosaponin D, paeoniflorin, and human serum albumin. Molecules. 2018;23:249. doi: 10.3390/molecules23020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. Zhao H. Xu Q. Liu R. Investigation of the interaction between indigotin and two serum albumins by spectroscopic approaches. J. Pharm. Anal. 2013;3:257–269. doi: 10.1016/j.jpha.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C. Wang H. M. Niu Q. X. Ye D. Y. Liang G. W. Binding between saikosaponin C and human serum albumin by fluorescence spectroscopy and molecular docking. Molecules. 2016;21:153. doi: 10.3390/molecules21020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. D. Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- Xu J. C. Nie X. L. Hong Y. P. Jiang Y. Wu G. Q. Yin X. L. Wang C. R. Wang X. Q. Synthesis of water soluble glycosides of pentacyclic dihydroxytriterpene carboxylic acids as inhibitors of α-glucosidase. Carbohydr. Res. 2016;424:42–53. doi: 10.1016/j.carres.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Liu B. S. Yan X. N. Cao S. N. Chong B. H. Lü Y. K. Studies on the interaction of palmatine hydrochloride with bovine hemoglobin. Luminescence. 2014;29:211–218. doi: 10.1002/bio.2529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.