Abstract
Macrolide susceptibility was investigated in clinical group B streptococci obtained from neonates or pregnant women in 2000 in France. Of 490 consecutive isolates, 18% were resistant to erythromycin. The erm(B), erm(A) subclass erm(TR), and mef(A) genes were harbored by 47, 45, and 6% of these strains, respectively. Two isolates did not harbor erm or mef genes.
Group B streptococci (GBS) are a leading cause of neonatal infections. Intrapartum antibiotic prophylaxis is now recommended for colonized women to prevent neonatal GBS disease, with penicillin G being the drug of choice (1). Women allergic to β-lactam antibiotics can receive intravenous clindamycin or erythromycin (1). Although penicillin resistance in GBS has not yet been reported, isolates resistant to erythromycin and related antibiotics have been previously described (2, 10, 18, 19, 23, 24, 32).
The known mechanisms of macrolide resistance in streptococci are targets of modification by a ribosomal methylase associated with erm genes (17, 26, 33), a macrolide-specific efflux mechanism encoded by the mef(A) gene (7), and mutations in the 23S rRNA and ribosomal L4 and L22 proteins (9, 30, 31; A. Canu, B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1927, p. 118, 2000). The prevalences and mechanisms of macrolide resistance have been widely reported worldwide for group A streptococci (GAS) and Streptococcus pneumoniae (4, 5, 8, 11, 12, 14, 16, 22, 28); relevant data on GBS are rare (3). The aims of this study were to assess the macrolide sensitivity of clinical GBS strains recently isolated in France and determine the genetic mechanisms of resistance.
In 2000, 88 erythromycin-resistant GBS isolates were identified among 490 consecutive isolates in the Paris (France) area. The isolates were recovered from genital specimens of pregnant women (n = 67) or from gastric fluid or ear specimens of colonized or infected newborns (n = 21). β-hemolytic colonies and suspected nonhemolytic colonies were identified as GBS by using a commercial agglutination technique (Murex Diagnostics, Dartford, United Kingdom). The GBS serotypes were as follows: serotype Ia, n = 2; serotype Ib, n = 9; serotype II, n = 6; serotype III, n = 28; serotype IV, n = 10; serotype V, n = 26; and nontypeable, n = 7.
The detection of erythromycin-resistant GBS isolates and determination of resistance phenotypes were performed as previously described (11, 27). The MICs of erythromycin azithromycin, josamycin, spiramycin, clindamycin, and streptogramin B were determined for all isolates with erythromycin inhibition zone diameters of less than 21 mm (20, 21). MICs were determined by the agar dilution method in Mueller-Hinton medium supplemented with 5% defibrinated sheep blood. The plates were incubated overnight at 35°C in air.
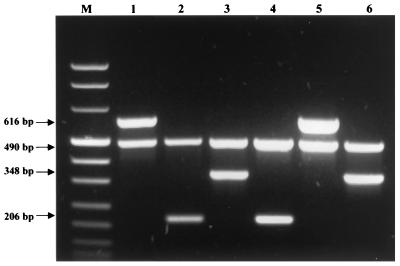
All erythromycin-resistant isolates were screened for erythromycin resistance genes. The mef and erm genes were detected by multiplex PCR amplification with previously described primers (5, 15, 26, 29). The internal PCR control was the mreA gene. The primers used to detect the mreA gene were 5′-AGA CAC CTC GTC TAA CCT TC-3′ and 5′-TCT GCA GGT AAG TAA GTG CG-3′ (6). Streptococcus agalactiae BM 132, S. agalactiae SBI, and Streptococcus pyogenes 02 C1110 were used as positive PCR controls for the erm(B), mef(A), and erm(A) subclass erm(TR) genes, respectively (3, 5, 7). Five erythromycin-susceptible GBS isolates were used as negative controls. Amplification of DNA from the positive controls with the corresponding primers yielded PCR products of the expected sizes [616, 490, 348, and 206 bp for erm(B), mreA, erm(A) subclass erm(TR), and mef(A), respectively] (Fig. 1). These PCR products were used for direct sequencing in an Applied Biosystems model 373 DNA sequencer by a modification of Sanger's method (25). The amplimers were found to be identical to the erm(B), erm(A) subclass erm(TR), and mef(A) genes (7, 26, 33).
FIG. 1.
PCR analysis of erythromycin-resistant control strains. Primers specific for the detection of erm(B) (lane 1), mef(A) (lane 2), and erm(A) subclass erm(TR) (lane 3) were used, followed by three representative clinical isolates (lanes 4, 5, and 6). Lanes 1 to 6, internal PCR control, the mreA gene, Lane M, DNA molecular size marker VIII (Boehringer Mannheim).
Among the 88 GBS erythromycin-resistant isolates, 71, 23 and 6% expressed the inducible macrolide-lincosamide-streptogramin B (MLSB), constitutive MLSB, and M resistance phenotypes, respectively. Table 1 shows MICs for the isolates according to erythromycin resistance genotype. PCR amplification showed that all the resistant isolates with the constitutive MLSB, inducible MLSB, and M phenotypes harbored the erm(B) or erm(A) subclass erm(TR) and mef(A) genes, respectively. All strains carried the mreA gene, but two erythromycin-resistant strains did not yield amplified products with the erm and mef primers tested; the mechanisms of resistance are under investigation. The MICs of various drugs for these two isolates were as follows: ≥128 μg/ml for all macrolides and clindamycin and 16 μg/ml for streptogramin B for the first isolate and 32 μg/ml for macrolides, ≥128 μg/ml for clindamycin, and 8 μg/ml for streptogramin B for the second isolate. The distributions of the erythromycin resistance genes are shown in Table 2 according to serotype.
TABLE 1.
MICs of macrolides and related agents for 86 erythromycin-resistant GBS isolates according to known mechanisms of resistance
| Antimicrobial agent | MIC (μg/ml)a
|
||
|---|---|---|---|
| 50% | 90% | Range | |
| erm(B) (n = 41) | |||
| Erythromycin | ≥128 | ≥128 | 2–≥128 |
| Azithromycin | ≥128 | ≥128 | 1–≥128 |
| Josamycin | ≥128 | ≥128 | 2–≥128 |
| Spiramycin | ≥128 | ≥128 | 2–≥128 |
| Clindamycin | ≥128 | ≥128 | 0.06–≥128 |
| Streptogramin B | 32 | 128 | 1–≥128 |
| erm(A) subclass erm(TR) | |||
| (n = 40) | |||
| Erythromycin | 4 | 32 | 1–≥128 |
| Azithromycin | 8 | 64 | 1–≥128 |
| Josamycin | 2 | 32 | 0.5–≥128 |
| Spiramycin | 2 | 32 | 0.5–64 |
| Clindamycin | 0.064 | ≥128 | 0.064–≥128 |
| Streptogramin B | 4 | 8 | 2–16 |
| mef(A) (n = 5) | |||
| Erythromycin | 2 | 2 | 2 |
| Azithromycin | 2 | 4 | 1–4 |
| Josamycin | 0.5 | 1 | 0.5–1 |
| Spiramycin | 0.5 | 1 | 0.5–1 |
| Clindamycin | ≤0.032 | 0.064 | ≤0.032–≤0.064 |
| Streptogramin B | 2 | 4 | 2–4 |
50 and 90%, MICs at which 50 and 90% of isolates are inhibited, respectively.
TABLE 2.
Serotype distribution according to genetic mechanism of macrolide resistance in 88 erythromycin-resistant GBS isolates
| Genetic mechanism of resistance | No. of isolates belonging to serotype:
|
||||||
|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | NTa | |
| erm(B) (n = 41) | 4 | 3 | 15 | 3 | 13 | 3 | |
| erm(A) subclass erm(TR) (n = 40) | 4 | 3 | 10 | 6 | 13 | 4 | |
| mef(A) (n = 5) | 2 | 3 | |||||
| Unknown (n = 2) | 1 | 1 | |||||
| Total (%) | 2 (2) | 9 (10) | 6 (7) | 28 (32) | 10 (11) | 26 (30) | 7 (8) |
NT, nontypeable.
Erythromycin resistance in GBS has mainly been investigated in North America. In the most recent studies, the rates of resistance ranged from 4 to 25% (2, 10, 18, 19, 23, 24, 32). In our study of GBS isolates of similar origins collected in the Paris area in 2000, the prevalence of erythromycin resistance was 18%. A previous North American study has shown an increase in GBS erythromycin resistance from 1995 to 1998, which could be related to the implementation of American guidelines recommending intrapartum antibiotic prophylaxis for GBS infection (1). In our institutions, the level of GBS erythromycin resistance varied only from 16% in 1997 to 18% in 2000, with no significant change in the consumption of macrolides during the last 5 years (E. Bingen, unpublished data).
While the prevalence and mechanisms of erythromycin resistance in S. pneumoniae and GAS have been widely investigated (4, 5, 8, 12, 14, 22, 28), to our knowledge such data are not available for GBS. In our study, erythromycin resistance in GBS was mainly associated with the erm(B) and erm(A) subclass erm(TR) genes (47 and 45% of isolates, respectively), with only 6% of isolates harboring the mef(A) gene. None of the strains carried both erm(A) and erm(B) or both mef and erm, as previously observed with GAS isolates (13). The mreA gene, initially considered a novel macrolide efflux gene, was detected for all our strains (6). Indeed, the mreA gene is now considered a housekeeping gene for the GBS species (G. Clarebout, and R. Leclercq, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 840, p. 115, 1999). Erythromycin resistance in two of our strains was not associated with either the mef or the erm genes. Similar results have recently been reported with GAS isolates (22). Such resistance in beta-hemolytic streptococci may be related to mutations in ribosomal proteins, as previously reported for S. pneumoniae (9, 30, 31).
Interestingly, the mechanisms of macrolide resistance in our GBS isolates differed from those previously described for pneumococcal and GAS isolates in France (5, 11). While erythromycin resistance in pneumococci is mainly associated with erm(B), erythromycin-resistant GAS strains bore the erm(B) or mef(A) gene and, sporadically, the erm(A) subclass erm(TR) gene. Insufficient data are available to compare the genetic mechanisms underlying erythromycin resistance in GBS in France and elsewhere. However, several recent North American studies showed a rate of erythromycin- and clindamycin-resistant GBS of 4 to 16% (2, 10, 18, 19), pointing to the involvement of the erm(B) and/or erm(A) subclass erm(TR) genes.
Our study shows that erythromycin resistance is not equally distributed among the different GBS serotypes, with higher rates being associated with serotypes III and V. This is a matter of concern, as these serotypes are usually associated with invasive strains. Thus, antibiotic intrapartum prophylaxis for patients allergic to penicillin must be guided by macrolide susceptibility testing of each GBS isolate.
Surveillance of macrolides and patterns of resistance in GBS, associated with a survey of macrolide consumption, should continue.
Acknowledgments
We thank Joyce Sutcliffe for providing reference strains S. pyogenes 02C1110, Corinne Arpin for providing S. agalactiae SB1 and BM 132, and R. Leclercq for helpful discussion.
REFERENCES
- 1.American Academy of Pediatrics, Committee on Infectious Diseases and Committee on Fetus and Newborn. Revised guidelines for prevention of early-onset group B streptococcal (GBS) infection. Pediatrics. 1997;99:489–496. doi: 10.1542/peds.99.3.489. [DOI] [PubMed] [Google Scholar]
- 2.Andrews J I, Diekema D J, Hunter S K, Rhomberg P R, Pfaller M A, Jones R N, Doern G V. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the western hemisphere. Am J Obstet Gynecol. 2000;183:859–862. doi: 10.1067/mob.2000.108839. [DOI] [PubMed] [Google Scholar]
- 3.Arpin C, Daube H, Tessier F, Quentin C. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob Agents Chemother. 1999;43:944–946. doi: 10.1128/aac.43.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baquero F, García-Rodríguez J A, García de Lomas J, Aguilar L The Spanish Surveillance Group for Respiratory Pathogens. Antimicrobial resistance of 1,113 Streptococcus pneumoniae isolates from patients with respiratory tract infections in Spain: results of a 1-year (1996–1997) multicenter surveillance study. Antimicrob Agents Chemother. 1999;43:357–359. doi: 10.1128/aac.43.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingen E, Fitoussi F, Doit C, Cohen R, Tanna A, George R, Loukil C, Brahimi N, Le Thomas I, Deforche D. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob Agents Chemother. 2000;44:1453–1457. doi: 10.1128/aac.44.6.1453-1457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clancy J, Dib-Hajj F, Petitpas J W, Yuan W. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae Antimicrob. Agents Chemother. 1997;41:2719–2723. doi: 10.1128/aac.41.12.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 8.Cornaglia G. Macrolide resistance and Streptococcus pyogenes: molecular basis, epidemiology and clinical significance. Rev Med Microbiol. 1999;10:245–258. [Google Scholar]
- 9.Depardieu F, Courvalin P. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2001;45:319–323. doi: 10.1128/AAC.45.1.319-323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez M, Hickman M E, Baker C J. Antimicrobial susceptibilities of group B streptococci isolated between 1992 and 1996 from patients with bacteremia or meningitis. Antimicrob Agents Chemother. 1998;42:1517–1519. doi: 10.1128/aac.42.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitoussi F, Doit C, Geslin P, Brahimi N, Bingen E. Mechanisms of macrolide resistance in clinical pneumococcal isolates in France. Antimicrob Agents Chemother. 2001;45:636–638. doi: 10.1128/AAC.45.2.636-638.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geslin P. Rapport d'activité année 1997. Creteil, France: Centre National de Référence des Pneumocoques; 1998. [Google Scholar]
- 13.Giovanetti E, Montanari M P, Mingoia M, Varaldo P E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother. 1999;43:1935–1940. doi: 10.1128/aac.43.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston N J, de Azavedo J C, Kellner J D, Low D E. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2425–2426. doi: 10.1128/aac.42.9.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klugman K P, Capper T, Widdowson C A, Koornhof H J, Moser W. Increased activity of 16-membered lactone ring macrolides against erythromycin-resistant Streptococcus pyogenes and Streptococcus pneumoniae: characterization of South African isolates. J Antimicrob Chemother. 1998;42:729–734. doi: 10.1093/jac/42.6.729. [DOI] [PubMed] [Google Scholar]
- 16.Lagrou K, Peetermans W E, Verhaegen J, Van Lierde S, Verbist L, Van Eldere J. Macrolide resistance in Belgian Streptococcus pneumoniae. J Antimicrob Chemother. 2000;45:119–121. doi: 10.1093/jac/45.1.119. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin F-Y, Azimi C P H, Weisman L E, Philips III J B, Regan J, Clark P, Rhoads G G, Clemens J, Troendle J, Pratt E, Brenner R A, Gill V. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995–1998. Clin Infect Dis. 2000;31:76–79. doi: 10.1086/313936. [DOI] [PubMed] [Google Scholar]
- 19.Morales W J, Dickey S S, Bornick P, Lim D V. Change in antibiotic resistance of group B Streptococcus: impact on intrapartum management. Am J Obstet Gynecol. 1999;181:310–314. doi: 10.1016/s0002-9378(99)70553-3. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests; approved standard M2–A7. 7th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7–A5. 5th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 22.Palavecino E L, Riedel I, Berrios X, Bajaksouzian S, Johnson D, Kaplan E, Jacobs M R. Prevalence and mechanisms of macrolide resistance in Streptococcus pyogenes in Santiago, Chile. Antimicrob Agents Chemother. 2001;45:339–341. doi: 10.1128/AAC.45.1.339-341.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearlman M D, Pierson C L, Faix R G. Frequent resistance of clinical group B streptococci isolates to clindamycin and erythromycin. Obstet Gynecol. 1998;92:258–261. doi: 10.1016/s0029-7844(98)00155-0. [DOI] [PubMed] [Google Scholar]
- 24.Rouse D J, Andrews W W, Lin F Y C, Mott C W, Ware J C, Philips J B., III Antibiotic susceptibility profile of group B Streptococcus acquired vertically. Obstet Gynecol. 1998;92:931–934. doi: 10.1016/s0029-7844(98)00263-4. [DOI] [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seppälä H, Skurnik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seppäla H, Nissinen A, Yu Q, Huovinen P. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J Antimicrob Chemother. 1993;32:885–891. doi: 10.1093/jac/32.6.885. [DOI] [PubMed] [Google Scholar]
- 28.Shortridge V D, Doern G V, Brueggemann A B, Beyer J M, Flamm R K. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic surveillance study conducted in the United States in 1994–1995. Clin Infect Dis. 1999;29:1186–1188. doi: 10.1086/313452. [DOI] [PubMed] [Google Scholar]
- 29.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tait-Kamradt A, Davies T, Appelbaum P C, Depardieu F, Courvalin P, Petitpas J, Wondrack L, Walker A, Jacobs M R, Sutcliffe J. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob Agents Chemother. 2000;44:3395–3401. doi: 10.1128/aac.44.12.3395-3401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tait-Kamradt A, Davies T, Cronan M, Jacobs M R, Appelbaum P C, Sutcliffe J. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–2125. doi: 10.1128/aac.44.8.2118-2125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyrrell G J, Senzilet L D, Spika J S, Kertesz D A, Alagaratnam M, Lovgren M, Talbot J A the Sentinel Health Unit Surveillance System Site Coordinators. Invasive disease due to group B streptococcal infection in adults: results from a Canadian, population-based, active-laboratory surveillance study—1996. J Infect Dis. 2000;182:168–173. doi: 10.1086/315699. [DOI] [PubMed] [Google Scholar]
- 33.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]