Abstract
A new combination strategy of an active loading and active targeting approach was applied in this work. The liposomes actively loaded with Curcumin (CRM) (LipCRM) were decorated with cholesterol tagged-anti-nucleolin AS1411 aptamer (NCL) via a new post-insertion approach, utilizing the cholesterol as a wedge to incorporate aptamer into the surface of the liposome bilayer. A successful NCL post-insertion was verified by agarose gel electrophoresis and dynamic light scattering (DLS). The cellular uptake of AptNCL-Lip was investigated using flow cytometry and Confocal Laser Scanning Microscopy (CLSM) on two different human breast cancer cell lines (MCF-7 and MDA-MB-231). The uptake and cytotoxicity of loaded CRM were investigated using flow cytometry and MTT assay. Our results showed successful post insertion of NCL aptamer to the surface of Lip. Also, higher cellular uptake was noted for AptNCL-Alexa-LipRhod compared to blank LipRhod in both cell lines. Moreover, CLSM showed prominent endocytosis and uptake of AptNCL-Alexa–LipRhod into the cytoplasm of breast cancer cells. Furthermore, the results showed a significant increase in the uptake and cytotoxicity of AptNCL-LipCRM compared to LipCRM in both cell lines. Overall, our results demonstrate a successful post-insertion of cholesterol-tagged aptamer into liposomes and the possible combination between active loading and active targeting.
A new combination strategy of an active loading and active targeting approach was applied in this work.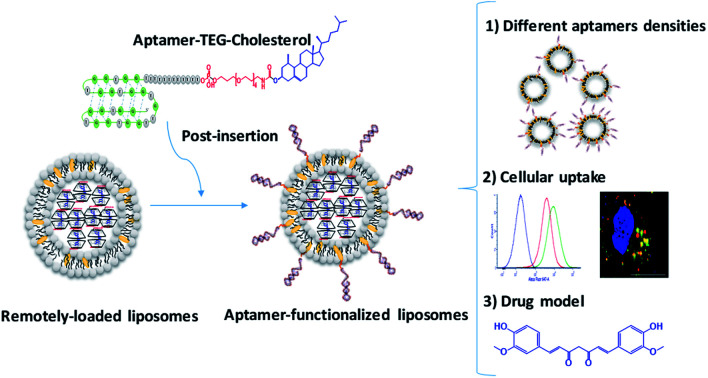
1. Introduction
In nanomedicine, drug delivery systems (DDS) aim at enhancing therapeutic index through specific delivery and release of drugs into targeted tissues and cells. In cancer, DDS can be designed to deliver their therapeutic payloads to cancer cells through improving blood circulation and extravasation of nanomedicine treatments to the abnormal tumor tissues via the enhanced permeability and retention (EPR) effect.1,2 In the last decades, cancer nanomedicine has witnessed success from bench-to-bedside. Despite the fast progress in this field, most nanoparticle-based drug delivery systems lack specificity against their target cells and only a few antitumor nanomedicines have reached the clinical stage.3 Therefore, several strategies have been developed to enhance the targeting capacity of nanomedicines, such as functionalizing nanocarriers with selective targeting ligands and triggering drug release in response to external or intrinsic stimuli.
Liposomes, spherical lipid vesicles in aqueous solution, are one of the most successful drug delivery systems because of their high stability, biocompatibility, easy to synthesize and high drug loading efficiency.4,5 The successful engineering of liposomes functionalized by recognition ligands that target unique or overexpressed tumor biomarkers disclosed promising advances in the drug delivery systems.6 Moreover, intraliposomal active loading has been developed to ensure high intraliposomal drug concentrations and minimal loss of precious chemotherapeutic agents that result from passive loading during liposomes preparation.7
Several types of targeting ligands can be used to guide nanocarriers to tumor cells including receptor binding ligands such as small molecules, peptides, polysaccharides, antibodies, and aptamers.8 Aptamers are synthetic single-stranded oligonucleotides (DNA or RNA) that can be selected by a process called Systematic Evolution of Ligands by EXponential Enrichment (SELEX).9 Aptamers, in their unique three-dimensional (3-D) structure, exhibit high affinity and specificity against a broad range of targets, ranging from small molecules to whole cells or tissues.9–12 In addition to the FDA approved aptamer (Macugen®), many aptamers targeting different diseases are today in clinical trials.13
Liposomes surface can be functionalized with targeting aptamer using different chemical or physical conjugation strategies either during their formation or via conjugation to the surface of preformed liposomes.14 Functionalization of liposomes with aptamers during liposomes formation ends up with aptamers on the surface and in the core of the liposomes. However, this method allows aptamers to occupy the available spaces for drugs in the core of liposomes without any benefit to these aptamers. On the other hand, the conjugation of aptamers to preformed liposomes can be achieved through different methods including covalent binding, such as carbodiimide conjugation and thiol-maleimide crosslinking, affinity coupling such as avidin–biotin interaction and electrostatic adsorption, and post-insertion methods like using aptamer modified with a lipid moiety via polyethylene glycol (PEG) linker, or via complementary DNA strand (cDNA) conjugated on the surface of liposome that is a complementary fit to the intended aptamer.6,15–25
Despite the success in conjugating aptamers to liposomes using previously described methods, these methods suffer from some disadvantages such as complexity, using chemicals for conjugations that may not fit with the functionality of the drug delivery system, and the lower conjugation efficiency. Since conventional liposomes formulation usually contain cholesterol in their membrane to enhance stability, a new aptamer ‘post-insertion’ approach mediated by a cholesterol-conjugated aptamer can be achieved with promising conjugation and targeting efficiency. We have previously published an intraliposomal active loading approach of lipophilic potential drug model “Curcumin” complexed with modified β-cyclodextrins using a transmembrane pH gradient strategy.14 The current work is reporting, for the first time, the functionalization of curcumin-remotely loaded liposomes with cholesterol-tagged anti-nucleolin aptamer using the post-insertion method. The functionality of the aptamer-functionalized liposomes was explored by investigating the cellular uptake and the anti-proliferative effect against nucleolin-expressing breast cancer cell lines (MCF-7 and MDA-MB-231). The current approach in this work can be easily handled and provides an efficient targeted drug delivery system.
2. Materials and methods
2.1. Chemicals
Curcumin, bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione (CRM), was purchased from Sigma Aldrich (St. Louis, MO, USA). 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (PE-Rhod-B), and Cholesterol (CHOL) were obtained from Avanti Polar Lipids, Inc. (Alabaster, Alabama, USA). NCL aptamer sequences: (5′-GGT GGT GGT GGT GGT TGT GGT GGT GGT GGT TTT TTT TTT TT/3CholTEG/-3′) and Alexa flour 647-tagged NCL (NCLAlexa) aptamer (5′-/5Alexa647 N-GGT GGT GGT GGT GGT TGT GGT GGT GGT GGT TTT TTT TTT TT/3CholTEG/-3′) were purchased from Integrated DNA Technologies Inc. (Coralville, IA, USA). Phosphate buffer saline (PBS), Tris-base EDTA buffer (TBE) and [4-(2-hydroxyethyl)-piperazino]-ethanesulfonic acid (HEPES) were obtained from LONZA® (USA). 6-Diamidino-2-phenylindole dihydrochloride (DAPI) from Merck Millipore (USA) and ethidium bromide from Fisher Scientific Ltd (England). Accutase purchased from Capricorn Scientific GmbH (Germany). Cell culture plates from TPP (Switzerland) and Agarose gel from Promega (USA). Acetone, methanol, ethanol, acetonitrile, formaldehyde and 1-propanol were obtained from Carbon group (England). All other chemicals and solvents were of analytical grade and obtained from different sources.
2.2. Cells
MCF-7 (ATCC number: HTB-22) and MDA-MB-231(ATCC number: HTB-26) cells were cultured in RPMI-1640 growth medium (Capricorn Scientific GmbH, Germany) and Eagle's Minimum Essential Medium (EMEM) (Euroclone SpA, Italy), respectively. Both RPMI-1640 and EMEM media were supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) 200 mM l-glutamine, and antibiotics; Penicillin–Streptomycin (100 IU mL−1-100 μg mL−1). Cells were passaged one time every three days. Accutase was used for cell detachment. Hemocytometer counting with trypan blue staining was used to count viable cells. All experiments were performed for passages range 3 to 10. Cells were maintained in humidified 5% CO2 incubator at 37 °C.
2.3. Liposomes preparation
Functionalizing liposomes with NCL aptamer was performed using the new post-insertion protocol to the preformed liposomes. Freshly prepared pH gradient liposomes were prepared using a similar protocol described in our previous work.14 In brief, liposomes (Lip) composed of DPPC and cholesterol in a 65 : 35 molar ratio or fluorescent labeled liposomes (LipRhod) composed of DPPC, cholesterol, and PE-Rhod-B (Ex/Em 560/583 nm) in a 64.5 : 35 : 0.5 molar ratio were mixed and dissolved in 5 mL of chloroform. Then, the thin film obtained through evaporation of chloroform by rotary evaporator. Further, the thin film hydrated with ammonium citrate buffer (pH 4) and extruded 13 times through 100 nm polycarbonate membrane. The resulting liposomes were stored at 4 °C for further active CRM loading, aptamers post-insertion, and cellular assays.
2.4. NCL aptamer post-insertion
Post-insertion of NCL aptamer (AptNCL) to Lip or LipRhod was performed by incubation of fixed amount total lipids (1 mg) of preformed liposomes under stirring with increased concentrations of 3′-cholestrol tagged AptNCL (2.3, 3.3, 5.0 and 6.7 nmol) for 1 h at 60 °C to enable anchoring the hydrophobic cholesterol moieties of the aptamer within the liposomes bilayer. After aptamer post-insertion, the unconjugated free aptamers (MW of 13.7 kD) were removed by ultrafiltration at 13 500 rpm using 100 KD Amicon filters, Merck Millipore Ltd (Ireland). For fluorescent detection purposes and AptNCL quantification, 3′-cholestrol tagged AptNCL composed of 10% (w/w) Alexa 647-labeled (Ex/Em 637/755 nm) NCL aptamers (AptNCL-Alexa) were post inserted into Rhod-labeled liposomes (LipRhod) to prepare dual fluorescent labeled aptamer functionalized liposomes (AptNCL-Alexa–LipRhod). NCL aptamer folding to the G-quadruplex structure was finally enabled using the binding buffer 5 mM PBS buffer with 2.5 mM MgCl2 and 140 mM KCl.26,27 To investigate the impact of different AptNCL density/Lip on the cellular uptake, variable amounts of AptNCL (0.5, 1.6, 3.3, 5, and 6.7 nmol) were post inserted into fixed amount of Lip (1 mg). The ratio of 3.3 nmol of AptNCL per 1 mg of lipids was used for further experiments (corresponds to 1 : 600 molar ratio of AptNCL to total lipids).
2.5. Characterization of aptamer post-insertion to liposomes
2.5.1. Size and zeta potential
The average size, polydisperdity index (PDI), and zeta potential of AptNCL–Lip and free Lip formulations were measured at 25 °C by Dynamic Light Scattering (DLS) using nano ZS (Malvern instruments, UK). The AptNCL–Lip and free Lip dissolved in 1× PBS were diluted in free nuclease water before measurement. The changes in size and PDI for the AptNCL–Lip and blank Lip formulations were investigated at 4 °C in PBS for 5 days.
2.5.2. Agarose gel electrophoresis
Agarose gel electrophoresis was used to confirm the successful aptamer post-insertion into the liposomes bilayer. Samples of free NCL aptamers before and after incubation, blank Lip, and AptNCL–Lip were mixed with loading dye then loaded into 3% agarose gel containing 10 μL of 10 mg mL−1 ethidium bromide followed by running electrophoresis in 1× TBE buffer (pH 8.2) at 70 V for 15 min. Images for analysis were obtained using Chemi Doc™ MP Bio-Rad Imaging Systems (USA).
2.5.3. Transmission electron microscopy (TEM)
Electron micrographs (TEM) for Lip and AptNCL–Lip were obtained after negative staining with uranyl acetate.28 A 200 mesh formvar copper grids (SPI supplies, USA) were double coated with carbon and 1.5% vinyl K solution under low vacuum glow discharge coating system (Leica, Austria). The liposomes samples were diluted (1 : 100 dilution factor) and a drop of diluted liposomes was loaded into double-coated grides followed by air-drying. The loaded grides were then stained with 3% (v/v) of uranyl acetate solution for 20 min at room temperature. After staining, the loaded grides were washed with distilled water and dried at room temperature. TEM micrographs were obtained using Morgagni (FEI, Netherlands) microscope operated at 60 kV.29
2.6. Cellular assays
2.6.1. Flow cytometry
A total of 2 × 105 MCF-7 and MDA-MB-231 cells were seeded into 12-well culture plates for 24 h in tissue culture incubator (37 °C, 95% humidity and 5% CO2) to reach ∼80% confluency. After incubation, the medium was removed and the cells were treated with either AptNCL–Alexa, LipRhod, or AptNCL-Alexa–LipRhod to obtain 200 nM of total aptamer concentration in 500 μL complete medium for 3 h in tissue culture incubator (37 °C, 95% humidity and 5% CO2). After incubation, the cells were then washed with PBS (3 × 0.5 mL), and detached gently with 200 μL of accutase for 5 min and transferred into 5 mL flow tubes. 10 000 events were counted by FACS Canto II and analyzed using BD FACSDiva™ software version 8.0 (BD, USA). A similar cell culturing protocol was used to investigate the cellular uptake of NCLAlexa (200 nM), LipRhod (120 μM), AptNCL-Alexa–LipRhod (200 nM AptNCL/120 μM LipRhod). To investigate the density effect of AptNCL/Lip on the cellular uptake, the cells were treated with AptNCL-Alexa–LipRhod formulations containing increased density of AptNCL (0.5, 1.6, 3.3, 5, and 6.7 nmol) post-inserted into fixed amount of Lip (1 mg). Untreated cells were used as controls for comparison.
2.6.2. Confocal Laser Scanning Microscopy (CLSM)
MCF-7 and MDA-MB-231 cells were seeded into 24-well culture plates containing glass coverslips and incubated with their compatible culture medium, for 24 h in tissue culture incubator (37 °C, 95% humidity and 5% CO2). After incubation, the culture medium was removed and the cells were treated with 300 μL of AptNCL-Alexa–LipRhod mixture (200 nM AptNCL/120 μM LipRhod prepared in complete culture medium) on ice for 7 min. Unbound AptNCL-Alexa–LipRhod was removed by washing the cells two times in cold PBS and then the internalization of AptNCL-Alexa–LipRhod was performed in complete culture medium at 37 °C for the indicated time-points. The cells were then washed twice with PBS and fixed with 4% formaldehyde for 15 min at room temperature in dark. The fixed cells were then quenched in 50 mM ammonium chloride (NH4Cl) for 5 min to decrease autofluorescence. DAPI was used to stain the nucleus in cells. The confocal images were acquired using LSM 780 (Zeiss, Germany). The objective lens used for acquiring the images was Plan-Apochromat 63×/1.4 Oil DIC M27. Lasers 405 nm, 560 nm, and 633 nm were activated for excitation of the nuclear stain DAPI, Rhodamine B, and Alexa-Fluor 647, respectively. The detection range for fluorescence emission signals were: 420–520 nm for DAPI, 535–624 nm for Rhodamine B, and 637–755 nm for Alexa-Fluor 647.
2.6.3. CRM actively-loaded liposomes cellular uptake
CRM was complexed with E-βCD and actively loaded into liposomes using pH gradient method described in our previous work.14 In brief, CRM-in-E-βCD (1.25 mM) was actively loaded into 1.25 mM freshly prepared pH-gradient preformed liposomes core and the CRM was quantified using the absorbance at maximum wavelength (λmax) = 420 nm and showed 41% encapsulation efficiency. The actively loaded liposomes were further functionalized with cholesterol tagged NCL aptamer via post-insertion protocol described before. Following similar cell culture and flow cytometry protocol, the seeded cells were treated with 500 μL of complete medium containing either; empty Lip (120 μM), CRM actively loaded liposomes (LipCRM) (120 μM), or aptamer functionalized-actively loaded liposomes (NCLAlexa–LipCRM) (200 nM AptNCL/50 μM LipCRM). Untreated cells were used as control for comparison. The cells were then washed three times with PBS, detached gently with 200 μL of accutase for 10 min, and transferred into flow tubes for flow cytometry analysis. All experiments were performed in triplicate.
2.6.4. Cytotoxicity assay
MDA-MB-231 and MCF-7 (5 × 103 cells per well) were seeded in 96-well plates (TPP, Switzerland). After 24 h, the cells were treated with different concentrations of CRM in LipCRM and AptNCL–LipCRM that range from 0 to 40 μM. After 72 h of incubation at 37 °C, treatment was removed from the wells, followed by adding 15 μL of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) solution (Bioworld, USA) and 100 μL of medium. After 3 h of incubation, the medium was removed, and the cells were mixed with 50 μL of dimethyl sulphoxide (DMSO). The absorbance was measured at a wavelength of 570 nm using Glomax microplate reader (Promega, USA).
2.7. Statistical analysis
The results were presented as the mean ± standard deviation of three independent experiments. Statistical significance was determined using unpaired two-tailed student's t-test with GraphPad Prism 6 (GraphPad Software Inc., USA). A value of P < 0.05 was considered to assign a statistically significant difference.
3. Results and discussion
3.1. NCL aptamer post-insertion into liposomes surface
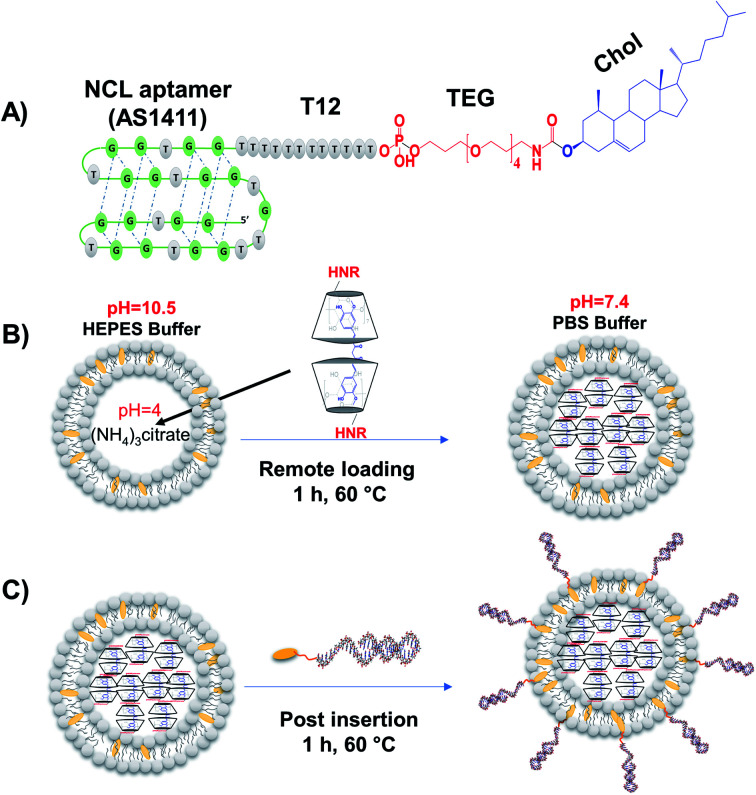
Several aptamers have been successfully functionalized to different nanoparticles for selective targeting and drug delivery into targeted tissues.6,30,31 In this study, we developed a new strategy to functionalize liposomes with aptamers through post-insert cholesterol tagged-aptamer into actively-loaded preformed liposomes bilayer utilizing the fluidity of DPPC-based liposomes at 60 °C (Fig. 1A–C).14,32 The sequence of the 3′-cholesterol-tagged anti-NCL aptamer (Fig. 1A) was designed with TEG and 12 thymine unit linker region to act as a spacer to separate the aptamer active recognition sequence from the hydrophobic cholesterol moiety.33 The 5′-Alexa flour 647-labeled NCL aptamer (NCLAlexa) was used for qualitative and qualitative measurements.
Fig. 1. AptNCL aptamer design. (A) Chemical representation of the quadruplex structure of cholesterol-conjugated NCL aptamer via poly thymine (T12) and tetraethyleneglycol (TEG) linker (AptNCL). (B) Intraliposomal active loading of CRM-modified β-cyclodextrins complexes into liposomes core via pH gradient method. (C) AptNCL post-insertion on the surface of preformed actively loaded liposomes.
Different methods have been successfully used to functionalize liposomes with different aptamers including NCL aptamer.6,15–25,34 However, these methods suffer some disadvantages including complexity, using chemicals for conjugations that may not fit with the functionality of the drug delivery system, and the lower conjugation efficiency. The current study provided a direct, simple, controlled, and efficient method for functionalizing of preformed liposomes with NCL aptamer that can be applied for different types of liposomes and aptamers. Regarding the mechanism of post-insertion from a chemical point of view. Our liposomes composed of DPPC and cholesterol and the phase transition temperature (Tc) of DPPC is known to be ∼41 °C where the transition of the liposome membrane structure from titled-gel into the fluid-crystalline phase can occur.35 In the current work, the AptNCL–cholesterol as amphiphilic molecules were post inserted into the surface of the liposomes at 60 °C which is above the Tc of DPPC and thereby the liposomes will be in the fluid-crystalline phase at this temperature. Therefore, the fluid-crystalline phase facilitates the anchoring of AptNCL–cholesterol into the surface of pre-formed liposomes via the hydrophobic–hydrophobic (co-assembly) interactions of cholesterol with the bilayer of liposomes membranes. Moreover, the hydrophilic nature and the large size of aptamers force the aptamers to the outer surface of the liposomes.
3.2. Characterization of AptNCL–Lip post-insertion
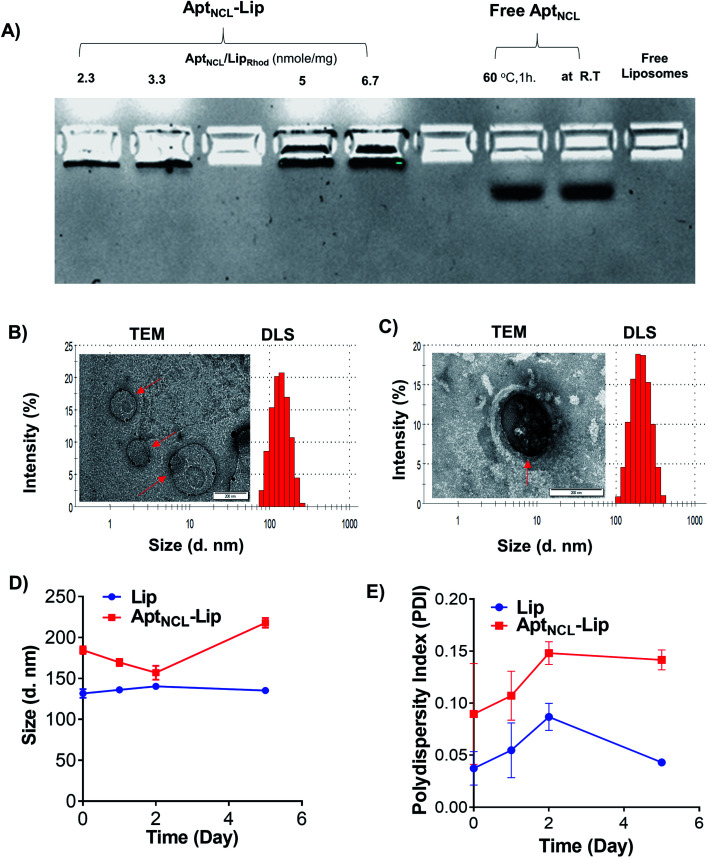
To investigate the successfulness of the post-insertion of NCL aptamer to the surface of liposomes, fixed amounts of Lip were then incubated with increasing concentrations of AptNCL for 1 h at 60 °C. The AptNCL–Lip conjugates then analyzed using agarose gel-electrophoresis. Free AptNCL and blank Lip were used as controls for comparison. Fig. 2A shows the trapping of AptNCL–Lip in the wells compared to the normal migration of free AptNCL. Moreover, the intensity of bands was associated with increasing concentrations of AptNCL–Lip conjugate, and no detectable free AptNCL was observed after post-insertion (∼2%), which indicate the high insertion efficiency of AptNCL into Lip. In addition, gel electrophoresis showed the stability of AptNCL aptamer during the post-insertion conditions when the migration of free AptNCL from stock compared to the free AptNCL running in the same post-insertion conditions. Also, there was no observed signal for the blank liposomes. The effect of AptNCL post-insertion on the morphology, size, polydesbersity, and zeta potential of preformed Lip was also investigated and the results shown in Fig. 2B and C and Table 1. An observable 32.7 nm increasing in the average Lip size for the NCL-Lip (174.8 ± 4.1) compared to blank Lip (142.1 ± 1.6 nm) was defined. In addition, the decrease in the Lip surface charge to around −16.6 mV for AptNCL–Lip (−19.6 ± 0.9 mV) compared to blank Lip (−2.9 ± 0.4 mV) is another evidence that confirmed the successful of AptNCL post-insertion. Such an increase in the size and decrease in the surface charge of AptNCL functionalized Lip can be explained by the functionalization of Lip with negative charges of AptNCL molecules. The size results of TEM (Fig. 2B and C) are in agreement with the results of DLS and showed round liposomes morphology for both Lip and AptNCL–Lip. Noteworthy, grafting of AptNCL into the surface of the Lip converted the Lip from neutral into anionic Lip. The impact of liposomes charge on the cellular interactions is well reported in the literature. In a recent study, Vu and coworkers investigated the cellular interactions of three neutral, cationic, and anionic liposomes in human blood using a platform of dynamic flow of human blood. Interestingly, all liposomes showed strong interactions with B cells and low interactions with other types of blood cells. Moreover, the interactions of neutral liposomes with B cells were the lowest compared to cationic and anionic liposomes.36 In the current study, AptNCL is an anionic macromolecule used for selective targeting of specific cells through affinity binding. Consequently, a further well-designed investigation for the cellular interactions of aptamers-functionalized liposomes with human blood is highly valued.
Fig. 2. Characterization of liposomes before and after AptNCL post-insertion using: (A) agarose gel electrophoresis with increasing aptamer density from 2.3, 3.3, 5 and 6.7 nmol. compared to free aptamer, undergo post insertion condition or directly from the stock solution and free liposomes. (B) Representative DLS histogram and TEM image for Lip and (C) representative DLS histogram and TEM image for AptNCL–Lip. (D) The change hydrodynamic diameter and (E) the polydispersity index of Lip and AptNCL–Lip over 5 days incubation at 4 °C in PBS (mean ± SD, n = 3).
DLS measurements of the hydrodynamic diameter, polydispersity index and zeta potential characterization of liposomes before and after AptNCL post insertion (mean ± SD, n = 3).
| Lip | AptNCL–Lip | |
|---|---|---|
| Hydrodynamic diameter (d nm ± SD) | 142.1 ± 1.6 | 174.8 ± 4.1 |
| Polydispersity index (PDI ± SD) | 0.16 ± 0.01 | 0.27 ± 0.02 |
| ζ-potential (mV ± SD) | −2.9 ± 0.4 | −19.6 ± 0.9 |
3.3. Cellular uptake
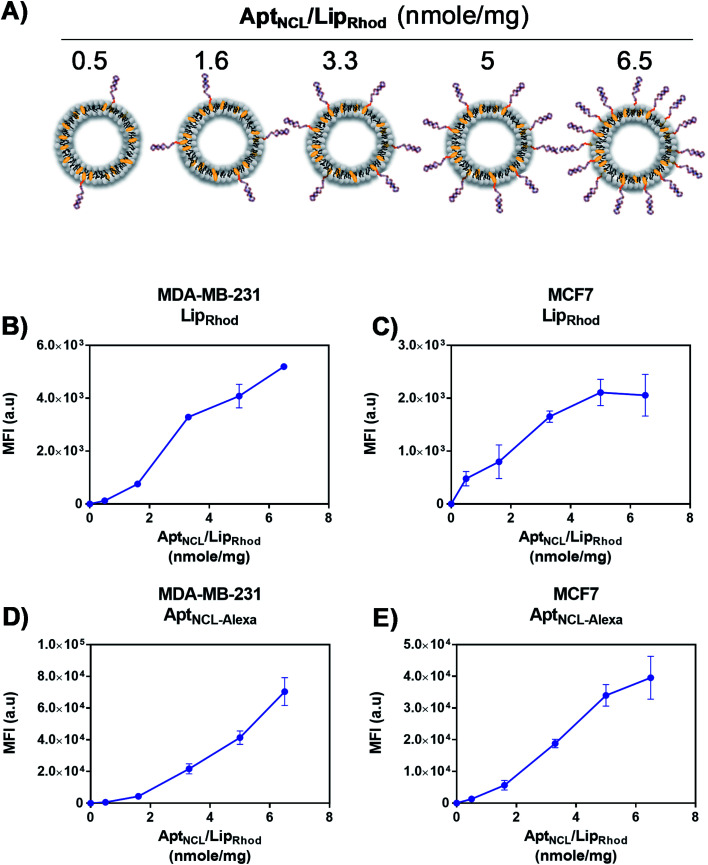
Following our successful functionalization of Lip with AptNCL, we investigated the selective cellular uptake of AptNCL–Lip using two human breast cancer cell lines, MCF-7 and MDA-MB-23 that are known for the expression of nucleolin receptor. The selectivity of binding of AptNCL-Alexa–LipRhod toward the NCL receptors was determined by comparing the binding between AptNCL-Alexa–LipRhod, free NCL-Alexa and LipRhod with cellular NCL receptors using flow cytometry. First, to investigate the correlation between the AptNCL density and the cellular uptake of AptNCL–LipRhod (Fig. 3A), both MCF-7 and MDA-MB-23 were treated with increased densities of AptNCL post-inserted into fixed amount of LipRhod in the AptNCL-Alexa–LipRhod formulation and followed by determination of the fluorescent intensity using flow cytometry (Fig. 3B–E). The results showed that the average density of AptNCL required for reaching LipRhod uptake plateau was range between 3.3 and 5 nmol of AptNCL per 1 mg of Lip in the MCF-7 and MDA-MB-231 cells. This result showed that there is a correlation between the ligand density and the cellular uptake and the need for the optimization of ligand-density for subsequent applications. Therefore, we decided to conduct all the cellular assays using the density of 3.3 nmol of AptNCL per 1 mg of Lip and the concentration of 200 nM of AptNCL either free or conjugated.
Fig. 3. Optimization AptNCL/Lip surface density and cellular uptake. (A) Variable amounts of AptNCL post-inserted into fixed amount of Lip. (B) and (C) Mean Fluorescent Intensity (MFI) of rhodamine (LipRhod), obtained from flow cytometry analysis, vs. different AptNCL densities of AptNCL–Alexa–LipRhod of the formulation in MDA-MB-231 and MCF-7 cell lines. (D) and (E) Mean Fluorescent Intensity (MFI) of Alexa flour 647-tagged NCL (NCLAlexa) aptamer, obtained from flow cytometry analysis, vs. different AptNCL densities of the AptNCL-Alexa–LipRhod formulation in MDA-MB-231 and MCF-7 cell lines (mean ± SD, n = 3).
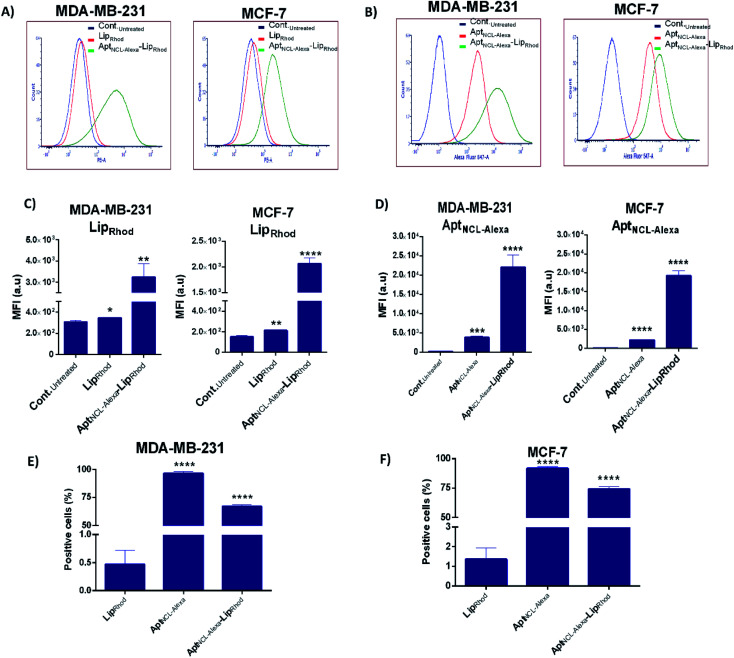
Following optimization of the concentration of AptNCL-Alexa–LipRhodin vitro, the selective binding of AptNCL-Alexa–LipRhod toward the overexpressing NCL receptors was investigated by comparing the uptake of AptNCL-Alexa–LipRhod with the uptake of NCL-Alexa and blank LipRhod. Untreated cells were used for background fluorescence. A higher mean fluorescent intensity was obtained for both MCF-7 and MDA-MB-231 cells when treated with AptNCL-Alexa–LipRhod compared to free NCL-Alexa and blank LipRhod (Fig. 4). In both cell lines, the results showed significant differences in uptake for AptNCL-Alexa–LipRhod compared to free AptNCL-Alexa or blank LipRhod (Fig. 4A and B), where the AptNCL-Alexa–LipRhod showed 9.4 and 9.6 folds higher cellular uptake compared to blank LipRhod in MDA-MB-231 and MCF-7, respectively (Fig. 4C). Moreover, AptNCL-Alexa–LipRhod showed 5.8 and 8.7 folds higher cellular uptake for the conjugated AptNCL-Alexa than free AptNCL-Alexa in MDA-MB-231 and MCF-7, respectively (Fig. 4D). The higher signal obtained from AptNCL-Alexa–LipRhod compared to free AptNCL-Alexa can be explained by the polyvalent interaction between aptamers-conjugated liposomes and the targeted cells.31Fig. 4E and F show a significant increase in the percent of positive cells treated with AptNCL-Alexa-LipRhod compared to LipRhod in both tested cell lines.
Fig. 4. Flow cytometry analyses for the uptake of AptNCL-Alexa-LipRhod in cultured cells. (A) and (B) Flow cytometry histogram analysis and (C) and (D) the percent of positive cells after treatment with blank LipRhod, AptNCL-Alexa, and AptNCL-Alexa–LipRhod. (E) and (F) The mean fluorescent intensity (MFI) measurements of LipRhod and AptNCL-Alexa signals for the MDA-MB-231 and MCF-7 cell lines. Cells were treated with free AptNCL-Alexa (200 nM), or blank LipRhod (120 μM), or AptNCL-Alexa–LipRhod (200 nM AptNCL/120 μM LipRhod) and compared to untreated cells (mean ± SD, n = 3).
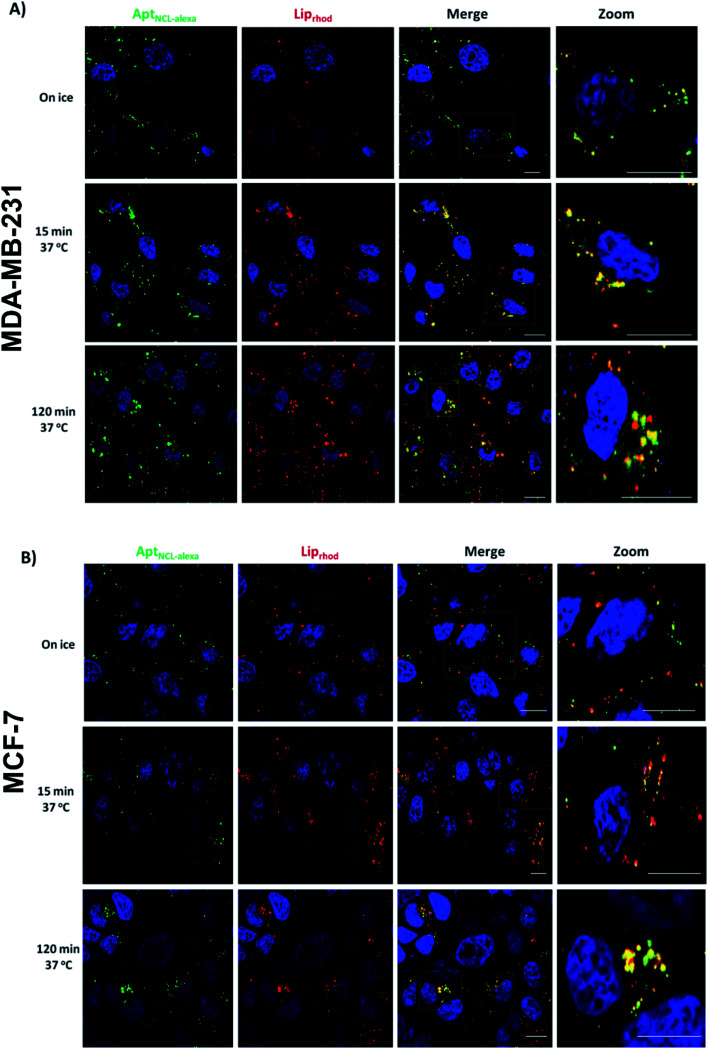
To determine the internalization kinetics of AptNCL-Alexa–LipRhod from the cell surface and the intracellular trafficking into the endocytic pathway, an internalization assay was employed followed by Confocal Laser Scanning Microscopy (CLSM). Breast cancer cell lines (MDA-MB-231 and MCF-7) were treated with AptNCL-Alexa–LipRhod and incubated on ice (Fig. 5A and B, 0 min). Surface bound AptNCL-Alexa–LipRhod complexes were then internalized at 37 °C over a 120 min period. The AptNCL-Alexa–LipRhod complexes were efficiently internalized; by 15 min chase the surface AptNCL-Alexa–LipRhod had been internalized and was located to endosomal like structures distributed in the cell periphery (Fig. 5A and B, 15 min). At the 120 min chase time point the AptNCL-Alexa–LipRhod were still located in endosomal like structures with considerable amount of the AptNCL-Alexa–LipRhod can be seen concentrated near the perinuclear region of the cell (Fig. 5A and B, 120 min). Notably, there was a marked overlap of AptNCL-Alexa signal with LipRhod signal at all-time points. The observed colocalization between fluorescent signals of both AptNCL-Alexa and LipRhod further validates successful post-insertion of AptNCL-Alexa in Lip and indicates appropriate cellular uptake and trafficking of intact AptNCL-Alexa–LipRhod particles.
Fig. 5. Confocal laser scanning microscopy (CLSM) analyses for the uptake of AptNCL-Alexa–LipRhod in cultured cells. (A) MDA-MB-231 or (B) MCF7 cells were incubated with AptNCL-Alexa–LipRhod for 7 min on ice. Cells for the 0 min time-point were fixed immediately in ice chilled 4% paraformaldehyde. For internalization, cells were washed in PBS and chased at 37 °C for the indicated time points. All cells were fixed in 4% paraformaldehyde and nuclei stained with DAPI (blue color). Alexa Fluor 647 conjugated NCL aptamer signal (AptNCL-Alexa) is shown in green color, Rhod-labeled liposomes (LipRhod) signal is shown in Red color. Bars represent 10 μm.
Important to mention here that there are some evidences from previous published studies that showed NCL aptamers could hyperstimulate macropinocytosis upon binding to the nucleolin receptor on cancer cells, which suggest that nucleolin receptor does not just act as a classic cell surface receptor but is also essential for the stimulation of macropinocytosis, which leads to further uptake of the NCL aptamers and induces cell death.37,38 However, further experiments are needed to dissect endocytosis pathways responsible for uptake of AptNCL-Alexa-LipRhod in breast cancer cells.
3.4. CRM as a drug model
Previously, we have successfully modified β-cyclodextrin (βCD) with secondary amine (E-βCD) that can work as a carrier and solubilize for CRM through the formation of CRM–E-βCD inclusion complexes.14 Further, the CRM–E-βCD inclusion complexes were successfully loaded into liposomes using a transmembrane pH gradient method. The encapsulation efficiency showed CRM shoed ∼5 folds higher compared to the conventional passive loading into the same liposomes. Moreover, the apparent formation constant (Kf) for CRM–E-βCD was 100.9 mM2 compared to 51.6 mM2 resulted in CRM-βCD. The complex stoichiometric ratio showed that the formation of the inclusion of complexes with 1 : 2 and 1 : 1 (guest : host) stoichiometry is possible. In the current work, the encapsulation efficiency was ∼41% and post-insertion of AptNCL into preformed Lip showed no effect on the encapsulation efficiency compared to the unfunctionalized and remotely loaded Lip.
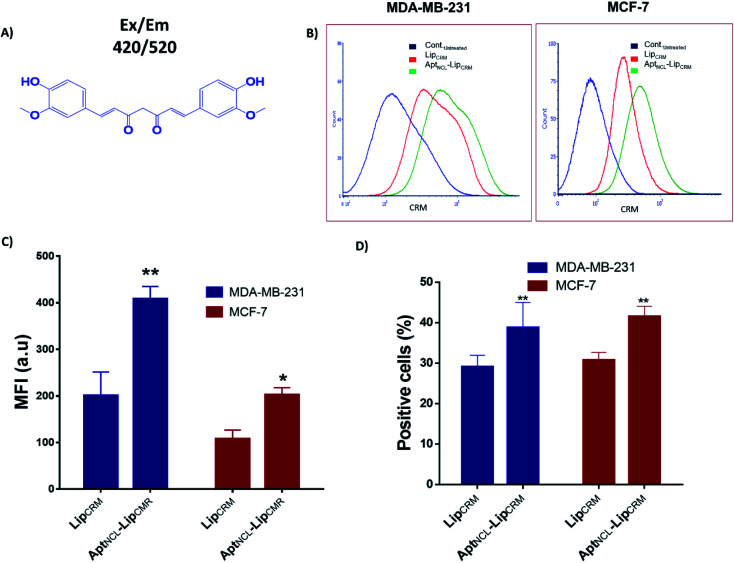
To investigate the functionality of the new active loading and active targeting combination strategy, we examined the cellular uptake and the cytotoxic effect of our final formulation of the aptamer-functionalized and actively CRM-loaded liposomes (AptNCL–LipCRM) on the MDA-MB-231 and MCF-7 cell lines. The fluorescence properties have been characterized and can be detected using fluorescent based detection methods. Therefore, we have investigated the uptake of CRM using flow cytometry at Ex/Em 420/520 (Fig. 6A). CRM has been remotely loaded into Lip using pH gradient method followed by post-insertion of AptNCL. The flow cytometry analysis showed that CRM signal was observed in both LipCRM and AptNCL–LipCRM (Fig. 6B–D). The results showed ∼2.0 folds apparent increase of CRM cellular uptake for AptNCL–LipCRM compared to LipCRM in both cell lines which confirm our previous cellular uptake results. Also, the percentages of positive cells treated with AptNCL–LipCRM were significantly higher compared to the percentages of cells treated with LipCRM. Besides, the results demonstrate that aptamer post insertion conditions do not influence the targeting capacity of the actively loaded liposomes.
Fig. 6. Uptake and cytotoxicity of CRM loaded into Lip (LipCRM) and AptNCL–Lip (AptNCL–LipCRM). (A) The chemical structure of curcumin with its excitation/emission wavelength. (B) Flow cytometry histogram analysis of MDA-MB-231 and MCF-7 cell lines for LipCRM signal after incubation with treated with AptNCL–LipCRM compared to LipCRM. (C) Mean fluorescent intensity measurements of CRM fluorescence signals obtained from LipCRM and AptNCL–LipCRM for the MDA-MB-231 and MCF-7 cell lines. (D) The percent of positive cells for CRM signal after treatment with LipCRM and AptNCL–LipCRM (mean ± SD, n = 3).
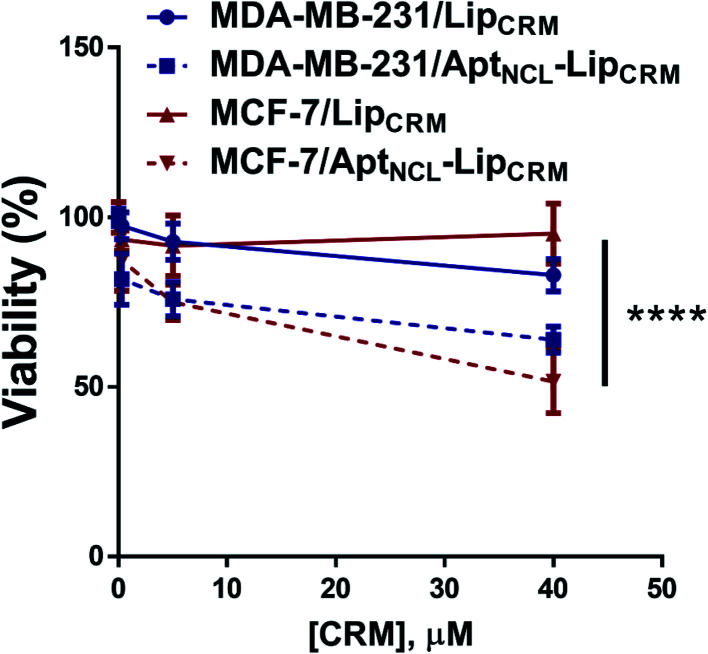
Furthermore, we have investigated the cytotoxicity of LipCRM and AptNCL–LipCRM against the MDA-MB-231 and MCF-7 cell lines. The results showed a significant reduction in the viability of both MDA-MB-231 and MCF-7 cells when treated with AptNCL–LipCRM compared to LipCRM treatment (Fig. 7). These results are in agreement with the cellular uptake results and demonstrate the preservation of CRM cytotoxic efficacy in our model.
Fig. 7. Viability assay of both MDA-MB-231 and MCF-7 cells when treated with AptNCL–LipCRM compared to LipCRM. Cells were treated with different concentrations of CRM in LipCRM and AptNCL–LipCRM range from 0 to 40 μM for 72 h (mean ± SD, n = 3).
4. Conclusions
In this work, we combined active loading and active targeting DDS and developed a new aptamer-liposomes conjugation strategy based on post-insertion approach. Cholesterol attached NCL aptamer was successfully post inserted in the bilayer of actively loaded preformed liposomes that were reported in our previous work. Cellular uptake studies, either by flow cytometry or CLSM, showed the key role of aptamer conjugation for liposomes active targeting and entry into the cancer cells. The NCL aptamer functionalized liposomes actively loaded with curcumin payload enhanced cellular internalization toxicity to MDA-MB-231 and MCF-7 breast cancer cells as compared to non-functionalized liposomes.
Contributions
H. N., I. M., D. A., H. A., R. Z., M. Q., and W. A. Carried out all experiments, data curation, software, and writing-original draft. H. N., I. M., F. O., and W. A. Contributed to data investigation and writing-original draft. F. O., A. A., S. I., W. A., and A. Aw. Contributed to supervision, data validation, and review-editing. H. N., I. M., F. O., S. I., and W. A. contributed to conceptualization and project administration.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Prabhakar U. Maeda H. Jain R. K. Sevick-Muraca E. M. Zamboni W. Farokhzad O. C. Barry S. T. Gabizon A. Grodzinski P. Blakey D. C. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo L. Y. Theek B. Storm G. Kiessling F. Lammers T. Curr. Opin. Biotechnol. 2013;24:1159–1166. doi: 10.1016/j.copbio.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D. Karp J. M. Hong S. Farokhzad O. C. Margalit R. Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- Noble G. T. Stefanick J. F. Ashley J. D. Kiziltepe T. Bilgicer B. Trends Biotechnol. 2014;32:32–45. doi: 10.1016/j.tibtech.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Hafner A. Lovric J. Lakos G. P. Pepic I. Int. J. Nanomed. 2014;9:1005–1023. doi: 10.2147/IJN.S55359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshaer W. Hillaireau H. Fattal E. Adv. Drug Deliv. Rev. 2018;134:122–137. doi: 10.1016/j.addr.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Fenske D. B. Cullis P. R. Expet Opin. Drug Deliv. 2008;5:25–44. doi: 10.1517/17425247.5.1.25. [DOI] [PubMed] [Google Scholar]
- Bertrand N. Wu J. Xu X. Kamaly N. Farokhzad O. C. Adv. Drug Deliv. Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandola C. Kalme S. Casteleijn M. G. Urtti A. Neerathilingam M. J. Biosci. 2016;41:535–561. doi: 10.1007/s12038-016-9632-y. [DOI] [PubMed] [Google Scholar]
- Maier K. E. Levy M. Mol. Ther.–Methods Clin. Dev. 2016;3:16014. doi: 10.1038/mtm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K.-i. Kimoto M. Hanson C. Sanford M. Young H. A. Hirao I. Sci. Rep. 2015;5:18478. doi: 10.1038/srep18478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff K. Brown J. Clippinger A. J. Biotechnol. Adv. 2015;33:1787–1798. doi: 10.1016/j.biotechadv.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Ismail S. I. Alshaer W. Adv. Drug Delivery Rev. 2018;134:51–64. doi: 10.1016/j.addr.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Odeh F. Nsairat H. Alshaer W. Alsotari S. Buqaien R. Ismail S. Awidi A. Al Bawab A. RSC Adv. 2019;9:37148–37161. doi: 10.1039/C9RA07560G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S. E. Lee K. H. Park Y. S. Oh D. K. Oh S. Kim K. S. Kim D. E. J. Contr. Release. 2014;196:234–242. doi: 10.1016/j.jconrel.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Mann A. P. Bhavane R. C. Somasunderam A. Liz Montalvo-Ortiz B. Ghaghada K. B. Volk D. Nieves-Alicea R. Suh K. S. Ferrari M. Annapragada A. Gorenstein D. G. Tanaka T. Oncotarget. 2011;2:298–304. doi: 10.18632/oncotarget.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Liu M. Tong X. Sun N. Zhou L. Cao Y. Wang J. Zhang H. Pei R. Biomacromolecules. 2015;16:2618–2623. doi: 10.1021/acs.biomac.5b00250. [DOI] [PubMed] [Google Scholar]
- Ara M. N. Matsuda T. Hyodo M. Sakurai Y. Hatakeyama H. Ohga N. Hida K. Harashima H. Biomaterials. 2014;35:7110–7120. doi: 10.1016/j.biomaterials.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kang H. O'Donoghue M. B. Liu H. Tan W. Chem. Commun. 2010;46:249–251. doi: 10.1039/B916911C. [DOI] [PubMed] [Google Scholar]
- Ninomiya K. Yamashita T. Kawabata S. Shimizu N. Ultrason. Sonochem. 2014;21:1482–1488. doi: 10.1016/j.ultsonch.2013.12.023. [DOI] [PubMed] [Google Scholar]
- Wu Y. Sefah K. Liu H. Wang R. Tan W. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yari H. Nkepang G. Awasthi V. Materials. 2019;12:756. doi: 10.3390/ma12050756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z. Tong R. Mishra A. Xu W. Wong G. C. Cheng J. Lu Y. Angew. Chem. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- Willis M. C. Collins B. D. Zhang T. Green L. S. Sebesta D. P. Bell C. Kellogg E. Gill S. C. Magallanez A. Knauer S. Bendele R. A. Gill P. S. Janjic N. Bioconjugate Chem. 1998;9:573–582. doi: 10.1021/bc980002x. [DOI] [PubMed] [Google Scholar]
- Allen T. M. Cullis P. R. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Li Z. Liu Z. Yin M. Yang X. Yuan Q. Ren J. Qu X. Biomacromolecules. 2012;13:4257–4263. doi: 10.1021/bm301563q. [DOI] [PubMed] [Google Scholar]
- Deng K. Hou Z. Li X. Li C. Zhang Y. Deng X. Cheng Z. Lin J. Sci. Rep. 2015;5:7851. doi: 10.1038/srep07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płaczek M. Kosela M. Acta Pharm. 2016;66:1–22. doi: 10.1515/acph-2016-0003. [DOI] [PubMed] [Google Scholar]
- Alshaer W. Zraikat M. Amer A. Nsairat H. Lafi Z. Alqudah D. A. Al Qadi E. Alsheleh T. Odeh F. Alkaraki A. Zihlif M. Bustanji Y. Fattal E. Awidi A. RSC Adv. 2019;9:30976–30988. doi: 10.1039/C9RA05636J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Nissenbaum E. Radovic-Moreno A. F. Wang A. Z. Langer R. Farokhzad O. C. Trends Biotechnol. 2008;26:442–449. doi: 10.1016/j.tibtech.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Alshaer W. Hillaireau H. Vergnaud J. Ismail S. Fattal E. Bioconjugate Chem. 2015;26:1307–1313. doi: 10.1021/bc5004313. [DOI] [PubMed] [Google Scholar]
- Alshaer W. Hillaireau H. Vergnaud J. Mura S. Delomenie C. Sauvage F. Ismail S. Fattal E. J. Controlled Release. 2018;271:98–106. doi: 10.1016/j.jconrel.2017.12.022. [DOI] [PubMed] [Google Scholar]
- Xing H. Tang L. Yang X. Hwang K. Wang W. Yin Q. Wong N. Y. Dobrucki L. W. Yasui N. Katzenellenbogen J. A. Helferich W. G. Cheng J. Lu Y. J. Mater. Chem. B. 2013;1:5288–5297. doi: 10.1039/C3TB20412J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odeh F. Nsairat H. Alshaer W. Ismail M. A. Esawi E. Qaqish B. Bawab A. A. Ismail S. I. Molecules. 2020;25:3. doi: 10.3390/molecules25010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sułkowski W. W. Pentak D. Nowak K. Sułkowska A. J. Mol. Struct. 2005;744–747:737–747. doi: 10.1016/j.molstruc.2004.11.075. [DOI] [Google Scholar]
- Vu M. N. Kelly H. G. Wheatley A. K. Peng S. Pilkington E. H. Veldhuis N. A. Davis T. P. Kent S. J. Truong N. P. Small. 2020:e2002861. doi: 10.1002/smll.202002861. doi: 10.1002/smll.202002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Reyes E. M. Teng Y. Bates P. J. Cancer Res. 2010;70:8617. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Reyes E. M. Salipur F. R. Shams M. Forsthoefel M. K. Bates P. J. Mol. Oncol. 2015;9:1392–1405. doi: 10.1016/j.molonc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]