Abstract
Strategies to improve the acceptance of scaffolds by the body is crucial in tissue engineering (TE) which requires tailoring of the pore structure, mechanical properties and surface characteristics of the scaffolds. In the current study we used a 3-dimensional (3D) printing technique to tailor the pore structure and mechanical properties of (i) nanocellulose based hydrogel scaffolds for soft tissue engineering and (ii) poly lactic acid (PLA) based scaffolds for hard tissue engineering in combination with surface treatment by protein conjugation for tuning the scaffold bioactivity. Dopamine coating of the scaffolds enhanced the hydrophilicity and their capability to bind bioactive molecules such as fibroblast growth factor (FGF-18) for soft TE scaffolds and arginyl glycyl aspartic acid (RGD) peptide for hard TE scaffolds, which was confirmed using MALDI-TOFs. This functionalization approach enhanced the performance of the scaffolds and provided antimicrobial activity indicating that these scaffolds can be used for cartilage or bone regeneration applications. Blood compatibility studies revealed that both the materials were compatible with human red blood cells. Significant enhancement of cell attachment and proliferation confirmed the bioactivity of growth factor functionalized 3D printed soft and hard tissues. This approach of combining 3D printing with biological tuning of the interface is expected to significantly advance the development of biomedical materials related to soft and hard tissue engineering.
3D printed scaffolds with tailored bioactivity using protein conjugation.
Introduction
Tissue engineering (TE) is an interdisciplinary field that applies the principles of engineering and life sciences toward the development of biological substitutes or scaffolds that restore, maintain, or improve tissue function.1 A scaffold should have comparable biological, physical and mechanical properties to the targeted tissue, such as suitable surface properties (such as surface roughness for the attachment of cells and other growth factors) and optimal internal architecture (which includes pore size, pore-size distribution, pore morphology, orientation, pore interconnectivity and surface area to volume ratio).2 As natural tissues are originally developed in a 3-dimensional (3D) environment of extracellular matrix (ECM) having different pore sizes for different functionalities, the success of the scaffold relies in its resemblance to this 3D environment.3 It is believed that different pore sizes influence different cell processes: 150–250 μm promotes cartilage regeneration via production of collagen,4 while 250–500 μm promotes ECM production and cell proliferation.5 In other words, micropores (>300 μm) increase the tendency for better mechanical properties, cell attachment and proliferation, while macropores (<500 μm) facilitate the formation of ECM, tissue growth, nutrient supply, waste removal and bone regeneration.6–8 These contradictory results may be due to the complexity of 3D structures which may affect cell penetration, distribution and nutrient diffusion.9 A smart solution could be the introduction of gradient porosity within the scaffold. In fact, gradient porosity is present in soft and hard natural tissues, such as cartilage, skin and bones. In skin the pore size increase with distance away from the surface and for long and flat bone structural gradient is found in radial and axial direction, respectively, introducing variation in bone density.10
3D printing is a versatile technique to fabricate a scaffold in a layer-by-layer fashion with controlled micro and macro dimensions having high reproducibility. Additionally, images from Computed Tomography (CT), Magnetic Resonance Imaging (MRI) or X-rays can be used to design patient specific scaffolds.11 Nanocellulose is a popular choice of material for soft TE application due to its bio-based origin, cytocompatibility and excellent mechanical properties12 and specific rheological properties such as shear thinning, suitable for 3D printing.13–15 We have shown in our recent studies that 3D printing provides possibilities to tailor the pore structure and also tune the mechanical properties to match the natural tissue.14,15 Likewise, poly lactic acid (PLA) is a popular 3D printable material and PLA based scaffolds are widely used in hard tissue engineering (orthopedic implants) as they have high mechanical strength, biocompatibility, biodegradability and corrosion resistance16–18 as well as allows resorption in the human body after several months. Some of these scaffolds are hydrophobic in nature and nonporous in design which leads to reduced tissue regeneration19,20 primarily due to the lack of hydrophilic moieties that enhances cell proliferation.21
Functional modification of 3D printed scaffolds is clinically important for long-term applications in the biomedical field.22–24 Cellular responses such as adhesion, migration, proliferation and differentiation are highly dependent on surface characteristics of the scaffold. There is a growing interest towards the 3D printing of biopolymers for TE but there are only a few reports on the functional modifications of such scaffolds.25–28 Dopamine surface modification is a facile and simple method introduced by Messersmith's group in 2007 based on mussel inspired polydopamine27 that helps with the robust adhesion of mussels to other substrates. Some studies on the application of polydopamine to convert hydrophobic surfaces into hydrophilic ones are reported.28 Dopamine hydrochloride in the alkaline environment can form polydopamines that can adlayer to various organic and inorganic substances due to the oxidation of the catechol groups of dopamine.29,30 Even though there are reports on the dopamine coating on 3D printed materials, in this study we further improved the bioactivity by the conjugation of growth factors. It can further enhance the cell to cell interaction, proliferation and thus better tissue regeneration. The various functional groups present on dopamine coatings can further provides a platform for the attachment of growth factors, hormones, peptides and chemicals that can be surface immobilized via chemical or physical strategy.31,32 In view of this, we aimed to functionally modify the 3D printed scaffolds with dopamine coating followed by surface functionalization of protein molecules as adhesive layer. PLA has been selected for bone and cellulose nanocrystals (CNC) based hydrogel has been selected for cartilage TE applications. The scaffolds coated with dopamine were examined by FTIR, XRD, SEM and EDS and the presence of proteins was confirmed with MALDI-TOF. Antimicrobial activity and the efficiency of these protein coated surface to enhance the attachment and proliferation of osteoblast and cartilage cells were also studied.
Experimental section
Materials
The supplied raw material used for the processing of cellulose nanocrystals (CNCs) was 17 wt% cellulose suspension which was extracted from unbarked Norway spruce wood chips (Picea abies) using the bioethanol processing plant at SEKAB (Örnsköldsvik, Sweden).33 Sodium alginate (alginic acid sodium salt from brown algae, Mw = 120 000–190 000 g mol−1), gelatin (Bloom 225, Type B, Mw = 40 000–50 000 g mol−1), calcium chloride (CaCl2, Mw = 110.98 g mol−1), glutaraldehyde (50 wt% in H2O, Mw = 100.12 g mol−1), dopamine hydrochloride (Mw = 189.64 g mol−1), arginyl glycyl aspartic acid (RGD) (Mw = 346.34 g mol−1) and Fibroblast Growth Factor-18 (FGF-18) (Mw = 21.2 kDa) were purchased from Sigma-Aldrich (Germany). Poly lactic acid (Ultimaker metallic silver PLA) with a dimeter of 2.85 mm was supplied by Structur3D Printing. The mechanical, electrical and thermal properties of PLA filament are listed in the technical data sheet from Ultimaker. α-Cyano-4-hydroxycinnamic acid and trifluoroacetic acid were purchased from Sigma Aldrich (St.Louis, MO, USA). Analytical grade acetonitrile was purchased from Rathburn Chemicals Ltd (Walkerburn, UK).
Processing methods
Hydrogel ink preparation
Cellulose nanocrystal based hydrogel ink was formulated according to the procedure described earlier.15 Briefly, 11 wt% CNCs, 6 wt% sodium alginate (SA) and 12 wt% gelatin (Gel) were mixed to have a wet (wt%) composition of CNC/SA/Gel/water: 6.87/1.50/1.50/90.12.
3D printing of CNC hydrogel scaffolds
The 3D printer used was an Ultimaker2+ equipped with Discov3ry Complete for paste printing system from Structur3D Printing (http://www.structur3d.io). Cura was used as the slicing software and the files were saved as gcode to be read by the printer. Prior to printing, syringes filled with the hydrogel ink were gently centrifuged to remove trapped air bubbles. For all prints, a nozzle diameter of 400 μm, a flow rate of 100% and printing speed of 30 mm s−1 were used. Two types of cubic scaffolds (20 mm3) were 3D printed depending on pore structure (1) uniform pores of 400 μm and (2) gradient pores ranging from 110–600 μm. After 3D printing, 3 wt% CaCl2 solution was added dropwise onto the scaffolds until completely wet. After 5 minutes, the scaffolds were gently transferred into a bath of 3 wt% CaCl2 solution for overnight crosslinking of alginate. Then the scaffolds were rinsed with distilled water and left overnight into the bath of 3 wt% glutaraldehyde solution to crosslink gelatin. After double crosslinking, the 3D printed scaffolds were washed thoroughly and stored in distilled water.
3D printing of PLA scaffolds
PLA was printed with nozzle diameter of 250 μm, nozzle temperature of 210 °C, print bed temperature of 90 °C, flow rate 100% and printing speed of 100 mm s−1. Two types of porous cubic scaffolds (20 mm3) were 3D printed depending on the pore structure, (1) uniform pores of 400 μm and (2) gradient pores in the range of 100–500 μm. Non-porous PLA was also 3D printed in the form of a screw with a length of 18 mm and head diameter of 5 mm.
Dopamine coating
Prior to dopamine coating, both hydrogel and PLA 3D printed scaffolds were washed with ethanol, acetone and distilled water followed by room temperature drying. Dopamine solution was prepared by dissolving 20 mg mL−1 of dopamine hydrochloride in 10 mM Tris buffer of pH 8.5 and coating was achieved via direct immersion technique at room temperature for 16 hours. Afterwards, the scaffolds were washed with ethanol and distilled water.
FGF-18 immobilization
Fibroblast growth factor-18 solution (FGF-18) was prepared in 5 mM Tris pH 8.0 to a concentration of 50 ng mL−1, in which 3D printed hydrogel scaffolds were immersed for 5 hours at 40 °C with gentle stirring. Afterwards, the scaffolds were washed with distilled water.
RGD conjugation
RGD solution was prepared in 0.1 N acetic acid (50 ng mL−1). Dopamine coated 3D printed PLA scaffolds were immersed in the solution for 5 hours at 40 °C with gentle stirring. After which the scaffolds were washed with distilled water.
Characterization
Fourier transform infrared spectroscopy was recorded on a Varian 670-IR FTIR spectrometer equipped with ATR accessory (Specac, UK). The measurements were done at room temperature with a resolution of 4 cm−1 and an accumulation of 50 scans in the spectral range of 390–4000 cm−1. X-ray diffraction spectra were collected on a PANalytical X'Pert PRO MPD (PANalytical, Netherlands) with copper radiation (λ = 1.54056 Å) with a scan range of 5°–50°. Scanning electron microscopy of the scaffolds was performed using a JEOL JSM-7401F (JEOL, Japan). The hydrogel samples were freeze dried. All samples were sputter coated with gold for 30 seconds prior to analysis. Energy Dispersive X-ray spectroscopy was performed on a Hitachi TM3000 (Hitachi, Japan). Matrix-assisted laser desorption/ionization (MALDI) analysis was performed on Voyager-DE™ STR mass spectrometer (Perspective Biosystem, Framingham, MA, USA), equipped with a nitrogen laser (337 nm), in reflector mode and positive ionization. The acquisition was performed with an accelerating potential of 20 kV, grid voltage set at 94%, and a delay time of 100 ns. Spectra were acquired in a mass range of 50 to 500 or 1000 m/z for RGD were processed and calculated by the Data Explorer V4 software (Applied Biosystems Inc., Foster City, CA, USA). In case of FGF-18, mass acquisition range was from 20 000 to 80 000 m/z. prior to analysis, 1 μL of matrix solution was dropped on the scaffold surface and air dried. The matrix solution for RGD was composed of saturated alpha-cyano-4-hydroxy-cinnamic acid (CHCA) in 50% acetonitrile (CAN) and trifluoroacetic acid (TFA) 0.05% v/v and for FGF-18 the matrix solution was the same. Water was purified using a Millipore water purification system to a resistance >18 MΩ cm−1.
In vitro antimicrobial activity and blood compatibility study
Antimicrobial activity was performed by disc diffusion method to measure the zone of inhibition method.34 Two strains were selected, Gram positive S. aureus and Gram negative E. coli to study the antimicrobial activity of dopamine coated protein immobilized scaffolds. 10 mL of overnight grown cultures were prepared (1 × 106 cfu mL−1) and from this 10 μL were uniformly spread on Luria broth agar plates. After these scaffolds were placed on the plates in triplicates followed by incubation for 24 hours at 37 °C and the zone size were measured.
RBC aggregation study was carried out to understand the blood compatibility of the dopamine coated protein immobilized scaffolds. For this blood samples were collected from healthy volunteers and transferred to a tube containing 3.8% sodium citrate at a ratio of 9 : 1 (blood : anticoagulant). Red Blood Cells (RBCs) were collected after a centrifugation at 750 rpm for 15 minutes. 3D printed scaffolds with dopamine coating and protein conjugation were used for this study. Briefly CNC, Dop-CNC, Dop-CNC-FGF18, PLA, Dop-PLA and Dop-PLA-RGD were cut in to small pieces of 1 × 1 cm2 and sterilized for 20 minutes under UV irradiation. Further the sterilized samples were incubated with diluted RBC at 37 °C. After the incubation the samples were observed under a phase contract microscope (Leica DMIRB, Germany).
Biocompatibility and cell proliferation
The pre-osteoblast and cartilage like cell line MC3T3-E1 were obtained from NCCS (Pune) and used to perform in vitro cell studies. Osteoblast and skeletal cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin in a 5% CO2 incubator at 37 °C and seeded onto 48-well plates at a density of 5 × 104 cells per well. After 24 hours, the cell culture medium was exchanged with fresh medium and the scaffolds after proper sterilization were cultured for a period of 72 h. For the cell proliferation study of the dopamine and protein loaded scaffolds, the cells were drop seeded onto the scaffolds at a density of 5 × 104 cells per well in 1 mL of media. Cell proliferation was determined at different time intervals. The cytotoxicity and cell proliferation were evaluated by using the MTT assay (n = 4) and live dead staining assay respectively. The absorbance of the medium was measured at 450 nm using a microplate reader (ELISA, Bio-Rad, Hercules, CA, USA). After 72 hours of proliferation, cells were stained with PI nuclear stain and observed under the fluorescent microscopy at 405 nm.
To study the chondrogenic differentiation stem cells were seeded on CNC scaffolds at a density of 1 × 104 cells per well and incubated at 37 °C, 5% CO2 for 3 h in DMEM containing 10% FBS and antibiotics. After that 1 mL of medium was added to each well then incubated for 24 h. After the incubation scaffolds were placed in a new culture plate to remove the dead cells. Culture medium changed 3 times a week. On the 5th, 10th 15th, 20th, 25th and 30th day scaffolds were checked for DNA content and GAG content. For DNA analysis samples were digested with papain buffer (Sigma-Aldrich, Germany), 10 mM EDTA, 10 mM phosphate buffer and 10 mM cysteine. DNA content was measured fluorometrically. Dimethylmethylene blue assay were performed to analyse the GAG content using bovine chondroitin sulfate as a slandered.
Stem cell osteogenesis was calculated with the help of alkaline phosphatase (ALP) activity. The cell lines were cultured in a 24 well cell culture plate. The concentration of the cells was 1 × 104 in each well and the scaffolds were incubated for a period of 5, 10, 15 and 20 days. The medium was removed and wells were washed with PBS. Alkaline phosphatase reagent (Sigma-Aldrich, Germany) was added in dark and kept for 30 min and the measurement of absorption were recorded with an ELISA reader at 405 nm wavelength.
Results and discussions
3D printing of scaffolds
We have used nanocellulose hydrogel ink and PLA filaments for 3D printing of scaffolds with uniform and gradient porosity. Nanocellulose based gel printing is found to be more challenging compared to PLA melt printing. In gel printing, the hydrogel ink should be in the form of smooth paste with appropriate rheological properties (shear thinning) to allow the continuous extrusion in the form of a filament. The developed hydrogel ink showed an apparent viscosity of 1.55 × 105 Pa s and storage modulus of 4.42 × 107 Pa. Moreover, the swelling of the hydrogel ink can adversely affect the printing resolution. The pre-crosslinking of the 3D printed scaffold with CaCl2 on the print bed is required in order to stabilize the fragile print so that it can be transferred into the crosslinking bath. Further post treatment with CaCl2 and gluteraldehyde is used to crosslink CNC with gelatin and alginate forming a stable gel.
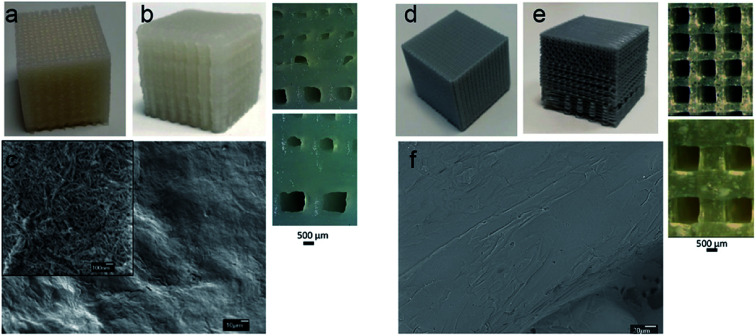
Fig. 1a and b shows the 3D printed scaffolds of CNCs based hydrogel ink with uniform porosity of 400 μm and gradient porosity of 110–600 μm where the pore sizes in the range suitable for cartilage regeneration. Due to the hydrogel nature of the ink a variation in pore size of ±30 μm was observed. The SEM image, Fig. 3c, shows the morphology of the walls of the 3D printed scaffold. It may be noted that CNC scaffold walls show nanoscale porosity and nanoscaled surface roughness hydrogel scaffold which is expected to favour cell interactions.
Fig. 1. CNCs based hydrogel ink scaffolds with (a) uniform pores of 400 μm and (b) gradient pore of 110–600 μm and (c) SEM image of the walls of 3D printed scaffold (inset image at high resolution) (d) PLA scaffolds with uniform pores of 400 μm, (e) gradient pores of 100–500 μm and (f) SEM image of the walls of PLA 3D printed scaffold.
Fig. 3. MALDI-TOF pattern of (a) Dop-CNC, (b) Dop-CNC-FGF18 (c) Dop-PLA and (d) Dop-PLA-RGD. The chemical structures of dopamine FGF18 and RGD are given inset.
PLA melt printing is less challenging compared to nanocellulose hydrogel printing and do not require post treatment or curing of the printed constructs. The heated print bed and the simultaneous cooling of the print during extrusion process enhance the print resolution. PLA scaffolds were 3D printed to target hard tissue with a variety of pore structure (Fig. 1d and e) uniform pores of 400 μm and gradient pores of 100–500 μm. Unlike CNC scaffolds, no nanoscaled porosity was expected on the walls of PLA scaffold (Fig. 1f).
The hydrogel scaffolds have a compression modulus in the range of 0.20–0.45 MPa when tested in simulated in vivo conditions (in water at 37 °C).15 PLA scaffolds showed a significantly higher compression modulus of 7–9 MPa. The modulus data justifies the use of CNC hydrogel scaffolds for soft tissue engineering and the PLA scaffolds for hard tissue engineering. All further surface treatments of the scaffolds in this study is guided by the mechanical property data.
Dopamine coating
Polydopamine coating is a non-toxic and solvent free coating which can be done on any substrate and the amine or thiol group of dopamine can promote cell adhesion by adsorbing serum protein such as fibronectin and vitronectin.35–37 In this study a uniform coating of dopamine on the surface of 3D printed gradient porous scaffolds was achieved by simple immersion in buffered dopamine (pH 8.5). This leads to the spontaneous deposition of polydopamine films on the scaffold surface through strong covalent and non-covalent interactions with the organic surfaces.38 The coatings were further characterized by FTIR, XRD, SEM and EDS.
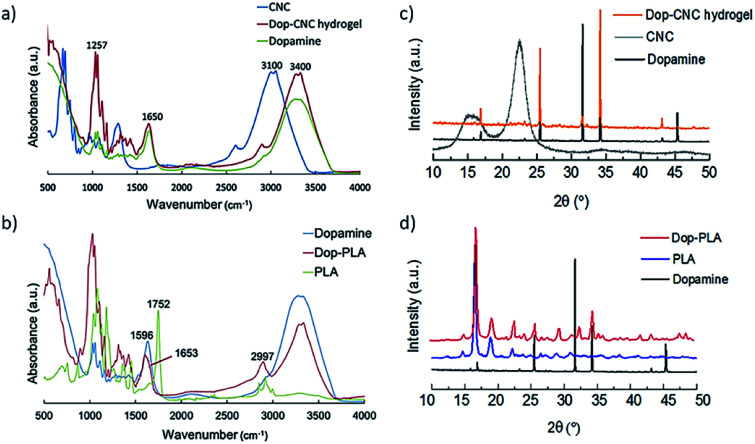
Fig. 2a and b shows FTIR spectra of dopamine coated scaffolds which confirms the presence of its components and conjugations. In Fig. 2a, the absorption band of cellulose at 3100 cm−1 corresponds to the O–H stretching vibrations and the band at 2900 cm−1 is assigned to C–H stretching vibrations of aliphatic chains of sodium alginate.39,40 The presence of dopamine is indicated by the peaks at 1091 and 1257 cm−1that are assigned to C–O stretching of catechol groups. Another band at 1285 cm−1 is from the phenolate C–O stretching which is an important indicator of catechol coordination to the dopamine surface.41–43 Characteristic primary amine bending and stretching of dopamine can be seen at 1650 cm−1. Fig. 2b shows the PLA characteristic stretching frequencies for C O, –CH3 asymmetric, and C–O, at 1746, 2995 and 1080 cm−1, respectively.44,45
Fig. 2. FTIR spectra of (a) Dop-CNC hydrogel scaffolds and (b) Dop-PLA scaffolds, XRD patterns of (c) Dop-CNC hydrogel scaffolds and (d) Dop-PLA scaffolds.
Bending frequencies for –CH3 asymmetric and –CH3 symmetric have been identified at 1452 and 1361 cm−1, respectively. In the dopamine-coated sample, a shift in the absorption bands of 1752 to 1653 cm−1 is due to the CO stretching vibrations indicating the polymerization of polydopamine on the surface of PLA. In addition, OH stretching modes and aliphatic CH stretching modes at 2997 cm−1 of pure PLA are absent in dopamine coated PLA. The NH stretching in the dopamine molecules was also shifted to 2928 cm−1 and 1598 cm−1 in case of dopamine coated PLA. It is mainly due to the molecular interactions between the amine groups of dopamine and PLA surface.46,47
In Fig. 2c CNC hydrogel showed peaks around 7.5°, 10.5° and 16° and it is a typical peak of cellulose with d-values of 5.42, 3.85 and 2.54 Å corresponding to 110, 200 and 004 planes.14 In Fig. 2d, the well-defined peaks between 9° to 35° are the contributions from crystalline PLA phase. The absence of crystalline peaks further than 35° indicates the presence of amorphous PLA. After coating in both types of scaffolds, dopamine is the prominent phase. In case of Dop-CNC hydrogel scaffolds, dopamine solution can be absorbed deep into the surface due to its hydrogel nature and therefore cellulose I crystal structure is not evident in the XRD pattern. However, in case of Dop-PLA, the coating stays at the top and cannot penetrate inside and therefore PLA structure is still evident in the XRD pattern.
Surface modification with growth factor (FGF-18) and peptide (RGD)
Limited acceptance of scaffolds in tissue engineering is usually attributed to its improper contact between the scaffold and the neighboring tissue. In view of this, for cartilage tissue regeneration, we immobilized the growth factor (FGF-18) on the polydopamine coated surface of 3D printed cartilage scaffold. FGF-18 was first reported in 1998 as a novel growth factor and current evidence suggests that FGF-18 can have a positive influence in chondrogenesis and osteogenesis during skeletal development and are actively involved in the cell growth, morphogenesis, tissue repair, inflammation and also in developmental process.48,49 See the chemical structure shown in Fig. 3b (inset).
In the case of bone tissue regeneration also there is a critical need to proliferate osteoblasts on the mechanical surface, so the coating of these implants with cell adhesive molecules provides a strong affinity for the cells to proliferate. Pierschbacher and Ruoslahti (1984) first reported RGD as the cell adhesion motif displayed on many extracellular matrix (ECM) and plasma proteins.50 RGD as a bio mimetic peptide can increase the cell adhesion and attachment on the matrix surface by preventing cell apoptosis.51 To improve the osteogenesis of the hard tissue implants we conjugated RGD on dopamine surface. RGD chemical structure shown in Fig. 3d (inset).
The SEM images (Fig. S1, ESI†) show the topography of the 3D printed hydrogel and PLA scaffolds after the above mentioned modifications. The CNC hydrogel scaffolds showed smooth surfaces after coating with dopamine and increased surface roughness after coating with growth factor FGF-18. In the case of PLA scaffolds the coating of dopamine did not show any significant impact on the scaffold surface morphology except for some insoluble particles observed on the surface due to polymerization to polydopamine. The coating with peptide RGD increased the surface roughness of PLA scaffold. In the case of both soft and hard scaffolds the increased surface roughness was expected to be of advantage.
To understand the surface chemistry changes of the scaffolds after dopamine coating and protein immobilization EDS was performed (Fig. S2, ESI†). Successful coating of dopamine on both soft and hard scaffolds was confirmed by the presences of unique amine group as there was an increase in the nitrogen content of the scaffolds52,53 as compared to uncoated scaffold surface. Further protein immobilization was confirmed by an enhancement in nitrogen content as compared dopamine coating. In case of PLA scaffolds, nitrogen content of 10.23% after dopamine coating was increased to 11.19% after RGD immobilization. Similar trend was found for hydrogel scaffolds where nitrogen content of 11.56% after dopamine coating was increased to 12.53% after the FGF-18 conjugation which confirm the presence of dopamine and proteins on the surface of hydrogel and PLA scaffolds. These findings agree with the previous reports of an increase in nitrogen content due to dopamine coating.48
The main goal of this study was effective transfer of RGD and FGF-18 on to the scaffold surface through dopamine chemistry which provides a suitable environment for cellular proliferation and differentiation. As discussed earlier dopamine have catechol and amine group which allow the polymer to form on any substrate under alkaline condition which can conjugate to biomolecules via imine formation.54 In our study we used very low concentration of growth factors for the conjugation and therefore the final immobilization was confirmed by the molecular weight analysis using MALDI-TOF mass spectrometry.
We measured the molecular weight of dopamine on the surface of both scaffolds; the mass spectra showed a sharp peak in the position of 153 and/or 186 kDa indicates the presence of polydopamine (Fig. 3a and c). A mass peak around at 21 kDa proves the identity and immobilization of our target compound in the case of FGF-18 (Fig. 3d). Likewise, the molecular ion peak 137, 217, 347 and 422 m/z could be assigned to the conjugation of RGD on the surface (Fig. 3c). Due to proper conjugation of RGD to Dopamine there was a shift in the peak position from 130 to 422 m/z. As suggested by the reviewer we also measured the molecular weight of RGD alone. We observed a sharp peak at 346 kDa (Fig. 3b). The results showed the well-coated dopamine and further conjugation of growth factors on the scaffold surface.
Water absorption by PLA is significantly lower (5 + 0.14 wt%) compared to 3D printed cellulose hydrogel scaffolds with 90 ± 0.1% water. It was noted that the pH decreased in the case of PLA scaffolds from 7.4 to 6.7 by the 20th day (see Fig. S3, ESI†), attributable to the slow degradation of PLA in aqueous medium.55 On the other hand, the pH remained stable around 7.4 for the CNC scaffolds in aqueous medium.
In vitro antimicrobial activity and blood compatibility
In this study antibacterial activity of the dopamine coatings can be directly visualized from the agar plates and the zone of inhibition after polydopamine coatings reveals moderate bactericidal effect. The antibacterial activities of coatings are mainly associated with the damage of cell wall surfaces.
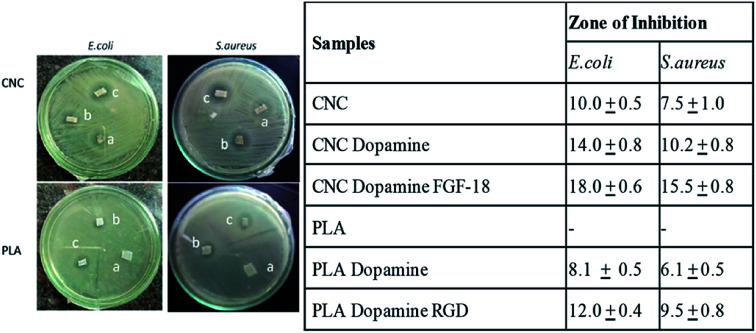
On comparing uncoated samples (a) in Fig. 4, it is clear that 3D printed PLA scaffolds do not show any antimicrobial activity whereas CNC scaffolds show a zone of inhibition dopamine coated cellulose as well as PLA (samples b) showed clear zone of inhibition compared to the uncoated ones. Fig. 4 also shows that antimicrobial activity of dopamine clogged the multiplication of both Gram positive and negative bacteria. Bacterial infections are the major challenge associated with the implantation of medical devices and developing antimicrobial surfaces is of paramount importance in scaffolds. Recent studies on the effect of antibacterial coatings with polydopamine concluded that dopamine coatings exhibit moderate antibacterial effect56 and the antibacterial effect of polydopamine coatings on E. coli and S. aureus,57 attributable to the functional groups which present on the surface of dopamine, especially benzene group.56Fig. 4 further confirms that the immobilization of growth factors RGD and FGF-18 enhanced the antibacterial activity (see table). It has been shown that polymers functionalized with RGD peptides possess anti-adhesive properties against certain bacterial species. The ability of RGD and FGF-18 peptides coatings to kill the bacteria on contact was proven with the zone of inhibition study either by inhibiting the growth or directly killing the bacteria. Mechanism of this peptide towards the bacteria can be attributed to the electrostatic attraction with the negatively charged bacterial cell wall. Since the tissue engineered scaffolds are in direct contact with blood it needs to be tested for blood compatibility. In view of this we tested the blood compatibility of dopamine coated protein immobilized scaffolds. For this RBCs were collected from blood samples followed by incubation with scaffolds and observed under microscope. If the material is having any kind of toxicity it will causes aggregation of RBC which can't be used for any kind of biomedical applications.
Fig. 4. Antimicrobial activity of (a) CNC and PLA scaffolds and their corresponding (b) Dop coated and (c) growth factor conjugated systems. Table (right) summarises the quantitative information on the inhibition zone towards E. coli and S. aureus.
Observations from our study revealed that both cellulose and PLA based scaffolds were blood compatible before and after coating (Fig. S4, ESI†). The microscopic images showed healthy cells without any damage to the RBCs. From these observations we can conclude that these scaffolds are blood compatible and useful for various tissue engineering applications.
Cell viability evaluations
Here we evaluated the toxicity of biomaterials and peptides which we used for the coating of 3D printed scaffolds and also the scaffolds itself for respective target applications.
Cartilage and osteoblast cells adhesion and proliferation
Surface characteristics of biomaterials play a major role in the cell adhesion, proliferation and differentiation. Cell adhesion is the primary event that occurs when cells come in contact with a biomaterial and poor cell adhesion to orthopedic and cartilage implants result in failure.58
We evaluated the biocompatibility of CNC and PLA scaffolds and the corresponding surface modified materials using cartilage and osteoblast cell lines and the results are summarized in Fig. 5. Cartilage cells were seeded on UV light sterilized CNC, dopamine-CNC, dopamine-CNC-FGF-18 scaffolds and osteoblast cells were seeded on PLA, dopamine-PLA, dopamine-PLA RGD scaffolds Dop-CNC-FGF18 (Fig. 5a) and Dop-PLA-RGD (Fig. 5b) clearly showed biocompatibility and indicate that these peptides enhanced the cell proliferation compared to negative control as well as the unmodified and dopamine coated scaffolds. The observations confirm our hypothesis that growth factors targeted for specific interactions, induce fast proliferation of respective cells on the materials for better success after implantation.
Fig. 5. The cytotoxicity of (a) cartilage and (b) osteoblast cells cultured on the CNC and PLA scaffolds respectively (c) cell attachment on CNC and PLA scaffolds showing the effect of dopamine coating and growth factors. CNC scaffolds were seeded with cartilage cells and PLA scaffold were seeded with osteoblast cells.
We imaged the attachment and spreading of osteoblast and cartilage cells on the PLA and CNC surface, respectively (Fig. 5c). The images show that cells do not stay alive on PLA, but shows better viability on cellulose surface, attributable to its hydrophilicity. The moisture binding capability of cellulose hydrogel scaffolds is considered an added advantage for cell attachment and proliferation and supports earlier studies where wet surface facilitated on the proliferation of mammalian cells.59
Dopamine coating enhanced the attachment and spreading of both the cell lines and on both type of scaffolds. Even though PLA is a polymer of preference in medical implants its poor hydrophilic nature prevents the cells from attachment and proliferation. In this study we used dopamine chemistry to provide hydrophilicity to PLA scaffold and also to augment the cell proliferation.32 As can be observed from Fig. 5c cartilage and osteoblast cells were spread uniformly over dopamine-cellulose FGF-18 and dopamine-PLA RGD indicating cell attachment and proliferation. Cells cultured on growth factor functionalized surface have a well extended morphology with a larger cell area than that of cells cultured on bare 3D printed surfaces.
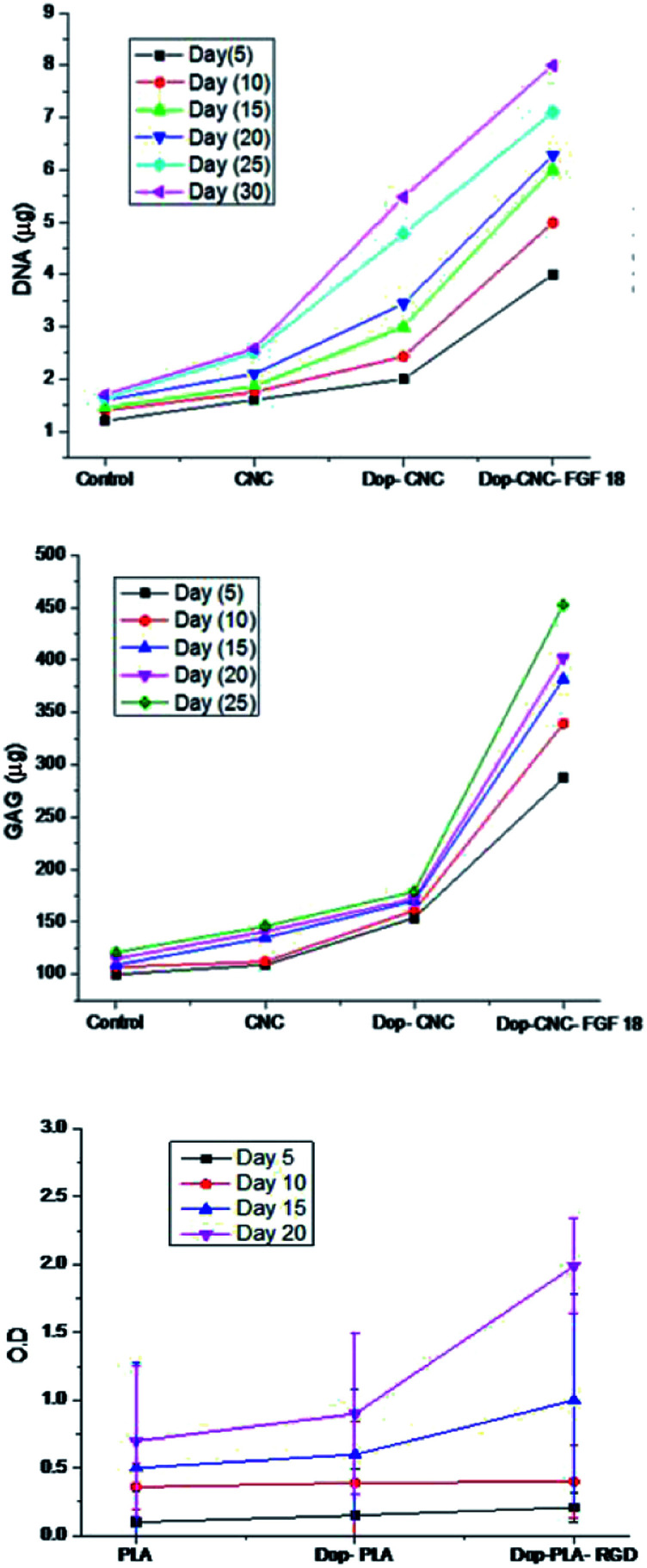
We further investigated the effect of growth factor (FGF-18) on cartilage tissue regeneration through chondrogenic differentiation assay on CNC, Dop-CNC and Dop-CNC-FGF18 scaffolds. As can be seen from the Fig. 6 the DNA content and GAG amount increased drastically from bare CNC scaffold to FGF-18 immobilized scaffold during the incubation period of 30 days. This increase in GAG content can directly correlate with chondrogenic differentiation since GAG synthesis is considered as a biomarker of chondrogenesis. Alkaline phosphatase (ALP) level is considered to be a marker for the analysis of osteogenic differentiation and the production of ALP indicated the presence of osteoblast cells and the formation of new bone cells. Results from ALP assay indicated that RGD conjugated PLA supported the growth of osteoblast cells and significantly enhanced the production of ALP, whereas the ALP production was less in the other two scaffolds such as PLA and Dop-PLA. After 15 days there was a significant increase in the APL production by RGD conjugated PLA scaffolds.
Fig. 6. Chondrogenic differentiation of CNC scaffolds and osteogenic differentiation of PLA scaffolds at different time intervals. DNA and GAG content was tracked for CNC scaffold and ALP production was tracked for PLA scaffolds.
In this study we tailored the surface chemistry and surface roughness of 3D printed scaffolds with dopamine and further immobilized with FGF-18 and RGD to attain a specific biological response. Our findings highlight that FGF-18 enhanced the cartilage cells growth and RGD promoted osteoblast growth in a significant way. This study provides a new insight in to the post treatment with amino functionalized molecules as surface modification of 3D printed biomaterials and its possible application in the biomedical field.
Conclusions
3D printed implants are explored today to overcome the limitations of conventional implants by providing customizable shape, sizes, porosity and we demonstrate that nanocellulose and PLA can be used to 3D print scaffolds via gel and melt printing respectively to achieve application specific (soft and hard tissue regeneration) mechanical properties. The functional modification of the surfaces of 3D printed biopolymer scaffolds with proteins after dopamine coating was found to provide an efficient methodology to enhance the scaffold bioactivity and biocompatibility in both soft and hard tissue regeneration scaffolds. MALDI analysis revealed the proper immobilization of growth factors such as Arginine–Glycine–Aspartic (RGD) and FGF-18 on the dopamine surface. While the hydrophilicity and moisture in cellulose hydrogel scaffolds resulted in inherent interaction with cells whereas in the case of PLA, the dopamine coating and protein immobilization was crucial in enhancing the cell interactions and viability. The peptide (RGD) functionalized PLA enhanced the osteoblast proliferation and osteogenic differentiation whereas FGF-18 immobilized soft scaffold enhanced the cartilage cells proliferation and chondrogenic differentiation. The study demonstrates that combining 3D printing with tailoring the biological interface provides a viable and universal process to develop tissue specific scaffolds with significantly enhanced the rate of acceptance by the human body.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors acknowledge the financial support from Swedish Research Council, (VR) under projects DNR 2016–05709 (Bioheal), and DNR 2017–04254.
Electronic supplementary information (ESI) available: SEM_EDS_blood compatibility. See DOI: 10.1039/d0ra08295c
Notes and references
- Langer R. Vacanti J. P. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- Vasireddi R. Basu B. Rapid Prototyp. Rep. 2015;21:716–724. doi: 10.1108/RPJ-12-2013-0123. [DOI] [Google Scholar]
- Fernandus H. M. Morones L. Clemens B. Jan D. B. J. Mater. Chem. 2009;19:5474–5484. doi: 10.1039/B822177D. [DOI] [Google Scholar]
- Zhang Q. Lu H. Kawazoe N. Chen G. Acta Biomater. 2014;10:2005–2013. doi: 10.1016/j.actbio.2013.12.042. [DOI] [PubMed] [Google Scholar]
- Lien S. M. Ko L. Y. Huang T. J. Acta Biomater. 2009;5:670–679. doi: 10.1016/j.actbio.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Loh Q. L. Choong C. Composites, Part B. 2013;19:485–502. doi: 10.1089/ten.teb.2012.0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V. C. Dunn C. K. Zhang Z. Deng Y. Qi H. J. Sci. Rep. 2017;7:8018–8026. doi: 10.1038/s41598-017-07771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava M. M. Draghi L. Giordano C. Pietrabissa R. J. Appl. Biomater. Funct. Mater. 2016;14:223–229. doi: 10.5301/jabfm.5000302. [DOI] [PubMed] [Google Scholar]
- Luca A. Ostrowska B. Moldero B. I. L. Lepedda A. Swieszkowski W. Blitterswijk C. V. Moroni C. V. L. Sci. Rep. 2016;6:22898. doi: 10.1038/srep22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltom A. Zhong G. Muhammed A. Adv. Mater. Sci. Eng. 2019:3429527–3429540. [Google Scholar]
- Bücking T. M. Hill E. R. Robertson J. L. Maneas E. Plumb A. A. Nikitichev D. I. PLoS One. 2017;12:0178540. doi: 10.1371/journal.pone.0178540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guise C. and Fangueiro R., in Natural Fibres: Advances in Science and Technology Towards Industrial Applications. Rilem bookseries, ed. Fangueiro R. and Rana S., Springer, Dordrecht, 2016, vol. 12 [Google Scholar]
- Chinga-Carrasco G. Biomacromolecules. 2018;19:701–711. doi: 10.1021/acs.biomac.8b00053. [DOI] [PubMed] [Google Scholar]
- Sultan S. Mathew A. P. Nanoscale. 2018;10:4421–4431. doi: 10.1039/C7NR08966J. [DOI] [PubMed] [Google Scholar]
- Sultan S. Mathew A. P. J. Visualized Exp. 2019;146:59401. doi: 10.3791/59401. [DOI] [PubMed] [Google Scholar]
- Giordano R. A. Wu B. M. Borland S. W. Cima L. G. Sachs E. M. Cima M. J. J. Biomater. Sci. Polym. Ed. 1997;8:63–75. doi: 10.1163/156856297X00588. [DOI] [PubMed] [Google Scholar]
- Kao C. T. Chi-Chang L. Yi-Wen C. Yeh C. H. Yuan Fang H. YouShie M. Mater. Sci. Eng. C. 2015;56:165–173. doi: 10.1016/j.msec.2015.06.028. [DOI] [PubMed] [Google Scholar]
- Song Y. Li W. Song K. Yee Y. Lee V. L. Mater. Des. 2017;123:154–164. doi: 10.1016/j.matdes.2017.03.051. [DOI] [Google Scholar]
- Zhu W. Ma X. Gou M. Mei D. Zhang K. Chen S. Curr. Opin. Biotechnol. 2016;40:103–112. doi: 10.1016/j.copbio.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Cai Y. Li J. Khoon Poh C. Chuan Tan C. H. Thian E. S. Fu Y. H. J. Sun J. J. Mater. Chem. B. 2013;1:5971–5976. doi: 10.1039/C3TB20680G. [DOI] [PubMed] [Google Scholar]
- Liu X. Yuk H. Lin S. Parada G. A. Tang T. C. Tham E. Fuente C. D. Lu T. K. Zhao X. Adv. Mater. 2018;30:1704821. doi: 10.1002/adma.201704821. [DOI] [PubMed] [Google Scholar]
- Tsai W. B. Chen W. T. Chien H. W. Kuo W. H. Wang M. J. Acta Biomater. 2011;7:4187–4194. doi: 10.1016/j.actbio.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Jaidev L. R. Chatterjee K. Mater. Des. 2019;161:44–54. doi: 10.1016/j.matdes.2018.11.018. [DOI] [Google Scholar]
- Uday M. G. Subha K. R. Rath N. J. Clean. Prod. 2019;230:412–419. doi: 10.1016/j.jclepro.2019.05.082. [DOI] [Google Scholar]
- Li B. Agarwal V. Ho D. Vede J. P. Iyer K. S. New J. Chem. 2018;42(9):7237–7240. doi: 10.1039/C7NJ04914E. [DOI] [Google Scholar]
- Entezar A. Iman R. Li G. Colin R. D. Rognon P. Li Q. Jiang X. Zreiquat H. Adv. Healthcare Mater. 2018:1801353. doi: 10.1002/adhm.201801353. [DOI] [Google Scholar]
- Lee H. Dellatore S. M. Miller W. M. Messersmith P. B. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Zhu L. Zhu L. Zhu B. Xu Y. Langmuir. 2011;27:14180–14187. doi: 10.1021/la202877k. [DOI] [PubMed] [Google Scholar]
- Bernsmann F. Richert L. Senger B. Lavalle P. Voegel J. C. Schaaf P. Vincent B. Soft Matter. 2008;4:1621–1624. doi: 10.1039/B806649C. [DOI] [PubMed] [Google Scholar]
- Wei Q. Zhang F. Li J. Li B. Zhao C. Polym. Chem. 2010;1:1430–1433. doi: 10.1039/C0PY00215A. [DOI] [Google Scholar]
- Ozhukil Kollath V. Mullens S. Luyten J. Traina K. Cloots R. Mater. Technol. 2016;31:241–245. doi: 10.1179/1753555715Y.0000000048. [DOI] [Google Scholar]
- Lee S. J. Lee D. Yoon T. R. Kim H. K. Jo H. H. Park S. J. Lee J. H. Kim W. D. Kwon K. Park S. A. Acta Biomater. 2016;40:182–191. doi: 10.1016/j.actbio.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Mathew A. P. Oksman K. Karim Z. Liu P. Khan S. A. Naseri N. Ind. Crops Prod. 2014;58:212–219. doi: 10.1016/j.indcrop.2014.04.035. [DOI] [Google Scholar]
- Bauer A. W. Kirby W. M. M. Sherris J. C. Turck M. Am. J. Clin. Pathol. 1966;36:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- Huang J. Grater S. V. Corbellini F. Rinck S. Bock E. Kemkemer R. Kessler H. Ding J. Spatz J. P. Nano Lett. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X. Z. Ghosh K. Liu Y. Palumbo F. S. Luo Y. Clark R. A. Prestwich G. D. J. Biomed. Mater. Res., Part A. 2004;68:365–375. doi: 10.1002/jbm.a.20002. [DOI] [PubMed] [Google Scholar]
- Tsai W. B. Chen W. T. Chien H. W. Kuo W. H. Wang M. J. J. Biomater. Appl. 2014;28:837–848. doi: 10.1177/0885328213483842. [DOI] [PubMed] [Google Scholar]
- Ryu J. H. Messersmith P. B. Lee H. ACS Appl. Mater. Interfaces. 2018;10:7523–7540. doi: 10.1021/acsami.7b19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W. Fan H. Wang L. Jin Z. Langmuir. 2015;31:11671–11677. doi: 10.1021/acs.langmuir.5b02757. [DOI] [PubMed] [Google Scholar]
- Oh S. Y. Yoo D. Shin Y. Kim H. C. Kim H. Y. Chung Y. S. Park W. H. Youk J. H. Carbohydr. Res. 2005;340:2376–2391. doi: 10.1016/j.carres.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Bella C. D. Duchi S. O'Connell C. D. Blanchard R. Augustine C. Yue Z. Thompson F. Richards C. Beirne S. OnofrilloC C. Bauquier S. H. Ryan S. D. Pivonka P. Wallace G. G. Choong P. F. J. Tissue Eng. Regener. Med. 2018;12:611–621. doi: 10.1002/term.2476. [DOI] [PubMed] [Google Scholar]
- Zangmeister R. A. Morris T. A. Tarlov M. J. Langmuir. 2013;29:8619–8628. doi: 10.1021/la400587j. [DOI] [PubMed] [Google Scholar]
- Zhou W. H. Lu C. H. Guo X. C. Chen F. R. Yang H. H. Wang X. R. Huang-Hao Y. J. Mater. Chem. 2010;20:880–883. doi: 10.1039/B916619J. [DOI] [Google Scholar]
- Garlotta D. J. Polym. Environ. 2001;9:63–84. doi: 10.1023/A:1020200822435. [DOI] [Google Scholar]
- Yang S. L. Wu Z. H. Yang W. Yang M. B. Polym. Test. 2008;27:957–963. doi: 10.1016/j.polymertesting.2008.08.009. [DOI] [Google Scholar]
- Gartner H. Li Y. Almenar E. Appl. Surf. Sci. 2015;332:488–493. doi: 10.1016/j.apsusc.2015.01.157. [DOI] [Google Scholar]
- Gupta K. K. Mishra P. K. Srivastava P. Gangwar M. Nath G. Maiti P. Appl. Surf. Sci. 2013;264:375–382. doi: 10.1016/j.apsusc.2012.10.029. [DOI] [Google Scholar]
- Jiang J. H. Zhu L. P. Li X. L. Xu Y. Y. Zhu B. K. J. Membr. Sci. 2010;2:194–202. doi: 10.1016/j.memsci.2010.08.017. [DOI] [Google Scholar]
- Teven C. M. Farina E. M. Rivas J. Reid R. R. Genes Dis. 2014;1(2):199–213. doi: 10.1016/j.gendis.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierschbacher M. D. Ruoslahti E. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- Leal M. S. Briones X. Villalobos V. Queneau Y. Leiva A. Rios H. E. Pavez J. Silva C. P. Carassco C. Neirra-Carrillo A. Roth A. D. Tamayo L. Urzua M.D. ACS Appl. Mater. Interfaces. 2019;11:19751–19762. doi: 10.1021/acsami.9b02503. [DOI] [PubMed] [Google Scholar]
- Liao Y. Wang Y. Feng X. Wang W. Xu F. Zhang L. Mater. Chem. Phys. 2010;121:534–540. doi: 10.1016/j.matchemphys.2010.02.019. [DOI] [Google Scholar]
- Zhu B. Edmondson S. Polymer. 2011;52:2141–2149. doi: 10.1016/j.polymer.2011.03.027. [DOI] [Google Scholar]
- Lee H. Rho J. Messersmith P. Adv. Mater. 2009;21:431–434. doi: 10.1002/adma.200801222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-H. Chen Y.-W. Lee A. K. Yao C.-H. Shie M. Y. J. Mater. Sci.: Mater. Med. 2019;30:1–12. doi: 10.1007/s10856-018-6202-x. [DOI] [PubMed] [Google Scholar]
- Chalkiadaki G. Nikitovic D. Berdiaki A. Sifaki M. Krasagakis K. Tzanakakis G. N. Int. J. Biochem. Cell Biol. 2009;41(6):1323–1331. doi: 10.1016/j.biocel.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Lei S. Yang Y. Yanshuang Z. Feng L. Xueji Z. Sci. Rep. 2016;6:24420. doi: 10.1038/srep24420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L. Wang M. Li P. Gan D. Yan L. Xu J. Wang K. Fang L. Chan C. W. Zhang H. Yuan H. Lu X. ACS Appl. Mater. Interfaces. 2018;22:28015–28026. doi: 10.1021/acsami.8b05314. [DOI] [PubMed] [Google Scholar]
- Lampin M. Warocquier-Clérout R. Legris C. Degrange M. Sigot-Luizard M. F. J. Biomed. Mater. Res. 1997;36:99–108. doi: 10.1002/(SICI)1097-4636(199707)36:1<99::AID-JBM12>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.