Abstract
Macrophages are notable immune cells that are recruited to the injury sites after peripheral nerve injury. Following peripheral nerve injury, increasing numbers of macrophages engulf debris and promote nerve regeneration. However, changes of pro-inflammatory (M1) and anti-inflammatory (M2) macrophages, two types of macrophages with dissimilar biological functions, have not been discovered. In the current study, the expression profiles of M1 and M2 macrophage marker genes in the sciatic nerve stumps and dorsal root ganglions (DRGs) after rat sciatic nerve injury were determined using RNA sequencing. Robust up-regulation of macrophage marker genes was observed in the injured sciatic nerve stumps as compared with in the DRGs. Measurement of the dynamic expression levels of M1 macrophage specific marker genes CD38 and Gpr18 as well as M2 macrophage specific marker genes Egr2 and Myc suggested that M1 macrophages were highly involved at all tested time points after peripheral nerve injury while M2 macrophage might be more involved in the later phase after nerve injury. Dynamic changes of M1 macrophage-inducing miRNAs showed that miR-18a, miR-19b, miR-21, miR-29a, and miR-29b were elevated in the injured nerve stump. These up-regulated miRNAs might mediate macrophage polarization by targeting multiple genes, such as Pten. Collectively, our study explored the unique temporal patterns of pro-inflammatory and anti-inflammatory macrophages after peripheral nerve injury for genetic aspects and provided a deeper understanding of the cellular and molecular basis of microenvironment reconstruction after peripheral nerve injury.
The temporal patterns of pro-inflammatory and anti-inflammatory macrophages after peripheral nerve injury.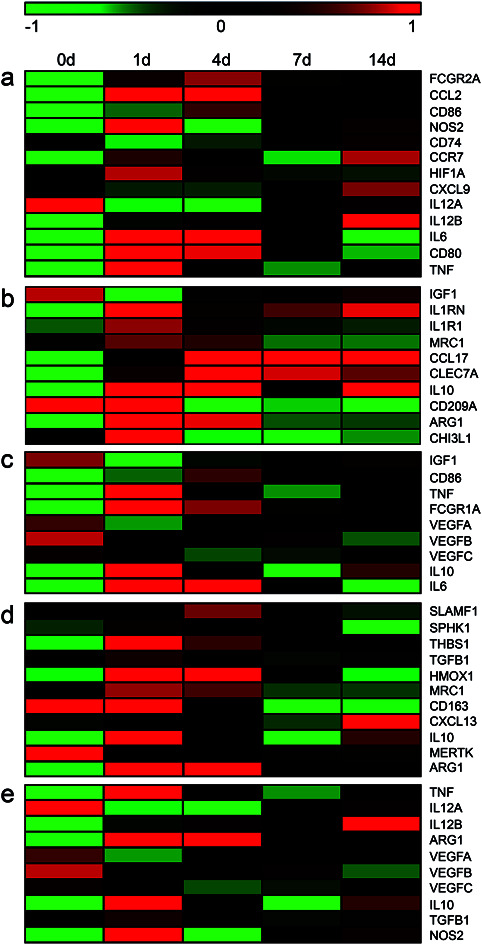
Introduction
Macrophages are large, phagocytic immune cells that can be found essentially in all tissues.1 Macrophages help to maintain tissue homeostatis by engulfing, digesting, and disrupting pathogens, microbes, cancer cells, dead cells, and cellular debris. Moreover, macrophages respond to environment cues and function as key mediators of tissue repair and remodeling.2 Considering the essential roles of macrophages in tissue injury and repair, macrophages are considered as potential therapeutic targets.3,4 Detailed investigations of macrophages find that macrophages can generally be classified into two main groups: pro-inflammatory (M1) macrophages and anti-inflammatory (M2) macrophages according to their specific bioactivities.5 M1 and M2 macrophages contribute to different phases of wound healing.6,7 M1 macrophages stimulate local inflammation and remove debris at the injury site while M2 macrophages promote cell proliferation, supply cell population, and restore tissue homeostatis.8
Emerging studies demonstrate that macrophages play essential roles in peripheral nerve injury and repair.9 Following peripheral nerve injury, Wallerian degeneration occurs at the distal nerve stumps and leads to the breakdown of axons and myelin sheaths.10 Inflammatory cells, especially macrophages, accumulate at the injury sites to engulf axon and myelin debris, clear the regenerative path, and rebuild the local microenvironment to promote nerve regeneration.11 Assessments of endoneurial cell populations show that macrophages account for 2.4%–9.0% of cell populations in the intact peripheral nerve stumps.12,13 The percentage of macrophages of robustly increased to 22.0% at 1 day after peripheral nerve injury and 13.2% at 4 days after peripheral nerve injury.13 These findings, from the aspect of cell numbers, indicated the essential roles of macrophages in peripheral nerve regeneration. However, it is worth noting that M1 and M2 macrophages exhibit diverse biological effects. Pro-inflammatory M1 macrophages mainly secret cytokines and drive inflammatory responses while anti-inflammatory M2 macrophages mainly regulate inflammation and induce tissue repair and remodeling.2 Actually, the fine turning of M1 and M2 macrophages is a key point that need to be considered in the construction of biomaterials in tissue engineering and regenerative medicine.14,15 Therefore, for designing an ideal tissue engineered nerve that fits the physiological conditions after peripheral nerve injury and benefits peripheral nerve regeneration, it is of great importance to identify specific changes of different types of macrophages after peripheral nerve injury first.
M1 and M2 macrophages express common or exclusive marker genes.16 To discover the significant involvement of M1 and M2 macrophages during peripheral nerve injury and repair, in the current study, we performed a bioinformatic analysis of the transcriptional signature of rat sciatic nerve stumps after nerve crush injury and determined changes of macrophage marker genes. It was worth noting that besides the direct effect on nerve stumps, peripheral nerve injury also stimulate changes in dorsal root ganglions (DRGs). In DRGs, peripheral nerve injury induces the recruitment of macrophages, activates recruited macrophages, and stimulates immune responses.17 But the involvement and functions of macrophages in DRGs were not fully investigated. Therefore, in the current study, we also evaluated gene profiles in DRGs and examined the expression patterns of macrophage marker genes in DRGs at different time points after nerve injury.
Experimental
Animal surgery and tissue collection
Adult Sprague-Dawley (SD) male rats (180–220 g) were induced to anesthetic depth with mixed narcotics containing 85 mg kg−1 trichloroacetaldehyde monohydrate (RichJoint, Shanghai, China), 42 mg kg−1 magnesium sulphate (Xilong Scientific, Guangzhou, Guangdong, China), and 17 mg kg−1 sodium pentobarbital (Sigma-Aldrich, St. Louis, MO, USA). Rat sciatic nerves at 10 mm above the bifurcation into the tibial and common fibular nerves were crushed with forceps for 10 seconds for 3 times as previously described.18 Rat sciatic nerve stumps at the crush site and L4–L6 DRGs were harvested at multiple time points after sciatic nerve crush for subsequent sequencing. Rats underwent sham-surgery were designated as 0 day control. SD rats were obtained from the Experimental Animal Center of Nantong University. Animal surgery were conducted in accordance with Institutional Animal Care guidelines of Nantong University, China and approved ethically by the Administration Committee of Experimental Animals, Nantong University, Jiangsu Province, China.
RNA sequencing and bioinformatic analysis
mRNA sequencing of rat sciatic nerve stumps and DRGs after nerve crush injury were conducted in previous studies.18,19 Sequencing data of sciatic nerve stumps were deposited in NCBI database (accession number PRJNA394957; SRP113121).18,20 Sequencing data of DRGs were deposited under accession number PRJNA547681 (SRP200823).19,21 miRNA sequencing of rat sciatic nerve stumps and DRGs after nerve transection injury were performed and differentially expressed miRNAs were reported.22 The expression patterns of mRNAs and miRNAs were displayed in heatmaps using MeV software. Connections between miRNAs and their target genes were built using Cytoscape software.23
RT-PCR
RNA samples were isolated from collected rat sciatic nerve stumps using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and reverse transcribed to cDNA using the Prime-Script Reagent Kit (TaKaRa, Dalian, China). qRT-PCR was performed with SYBR Premix Ex Taq (TaKaRa) on an ABI Stepone system (Applied Biosystems, Foster City, CA). Relative quantifications of target genes CD86, CD38, Gpr18, Egr2, and Myc were conducted using the comparative 2−ΔΔCt method and normalized against reference gene GAPDH. Primers were synthesized by Sangon Biotech (Shanghai, China). Primer sequences were as follows: CD86 (forward) 5′-CAACTAATGAGTATGGCGACAAC-3′ and (reverse) 5′-AAACTGGGGCTGCGAAAA-3′; CD38 (forward) 5′-GTCGCCTTGGTAGTGGTCGT-3′ and (reverse) 5′-CAGTGGTGCGTAGTCTTCATTG-3′; Gpr18 (forward) 5′-TCACTGCCCTATGGGTTTTC-3′ and (reverse) 5′-GTTCTTGAGTTCCTTGGCGTAT-3′; Egr2 (forward) 5′-GGTTGCGACAGGAGGTTCT-3′ and (reverse) 5′-GGATGTGAGTGGTGAGGTGG′-3′; Myc (forward) 5′-TGATGACCGAGCTACTTGGAG-3′ and (reverse) 5′-AGGCTGGTGCTGTCTTTGC-3; and GAPDH (forward) 5′-AACGACCCCTTCATTGAC-3′ and (reverse) 5′-TCCACGACATACTCAGCAC-3′.
Statistical analysis
Ordinary one-way ANOVA and Dunnett's multiple comparisons test were applied for comparison between groups using GraphPad Prism 6.0 software (GraphPad Software, Inc., San Diego, CA, USA). A p-value < 0.05 was considered as statistical significance.
Results
Transcriptional characteristics of macrophage markers in sciatic nerve stumps after peripheral nerve injury
Besides the classification of macrophages to classically activated M1 macrophages and alternatively activated M2 macrophages, M2 macrophages can be further subdivided to M2a, M2b, M2c, and M2d macrophages. M2a macrophages promote the removal of apoptosis cells and the proliferation and migration of cells and contribute to wound healing. M2b macrophages encourage cell maturation and stabilization and modulate the synthesis of extracellular matrix. M2c macrophages also influence extracellular matrix. Besides, M2c macrophages benefit growth factor production, immune-suppression, and tissue remodeling. M2d macrophages stimulate angiogenesis and would healing.24,25
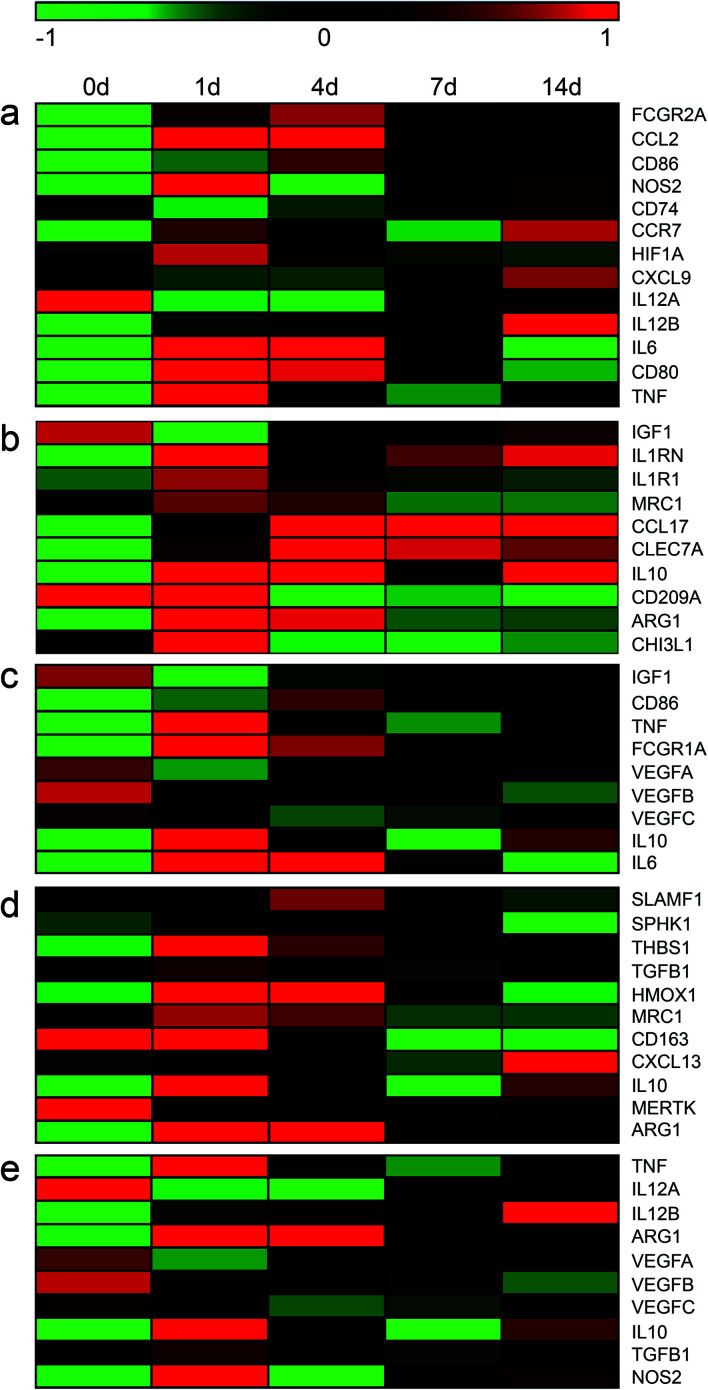
M1 macrophages and sub-types of M2 macrophages possess common and distinct markers.24,25 To obtain a cue of changes of M1 and M2 macrophages in the sciatic nerve stumps following peripheral nerve injury, the expression levels of M1 and M2 macrophage markers were screened from sequencing data and visualized in heatmaps (Table S1† and Fig. 1). The expression levels of these macrophage markers at multiple time points after nerve injury were compared with their expression levels at 0 day (uninjured control). Red color indicated relatively highly expression levels while green color indicated relatively lower expression levels. It was observed that most M1 macrophage markers, except for IL12A, exhibited elevated expressions in the injured sciatic nerve stumps as compared with the 0 day control (Fig. 1a). Many M2a macrophage marker genes, except for CD209A, were also up-regulated in the sciatic nerve stumps after nerve injury as compared with the 0 day control. And numerous M2a macrophage markers, such as CCL17, CLEC7A, and IL10, were kept expressed at relatively higher level post injury (Fig. 1b). Similar as M2a macrophage marker genes, the majority of M2c macrophage marker genes, except for MERTK, were significantly up-regulated (Fig. 1d). On the other hand, M2b and M2d macrophage marker genes were not dramatically increased as compared with M1, M2a, and M2c macrophage marker genes (Fig. 1c and e). This might because that some common marker genes of M2b and M2d macrophages, VEGFA, VEGFB, and VEGFC, were not significantly altered after nerve injury.
Fig. 1. The temporal expression patterns of markers of (a) M1, (b) M2a, (c) M2b, (d) M2c, and (e) M2d macrophages in sciatic nerve stumps at 0 day, 1 day, 4 days, 7 days, and 14 days after nerve injury. Red color indicated up-regulation and green color indicated down-regulation.
Transcriptional characteristics of macrophage markers in DRGs after peripheral nerve injury
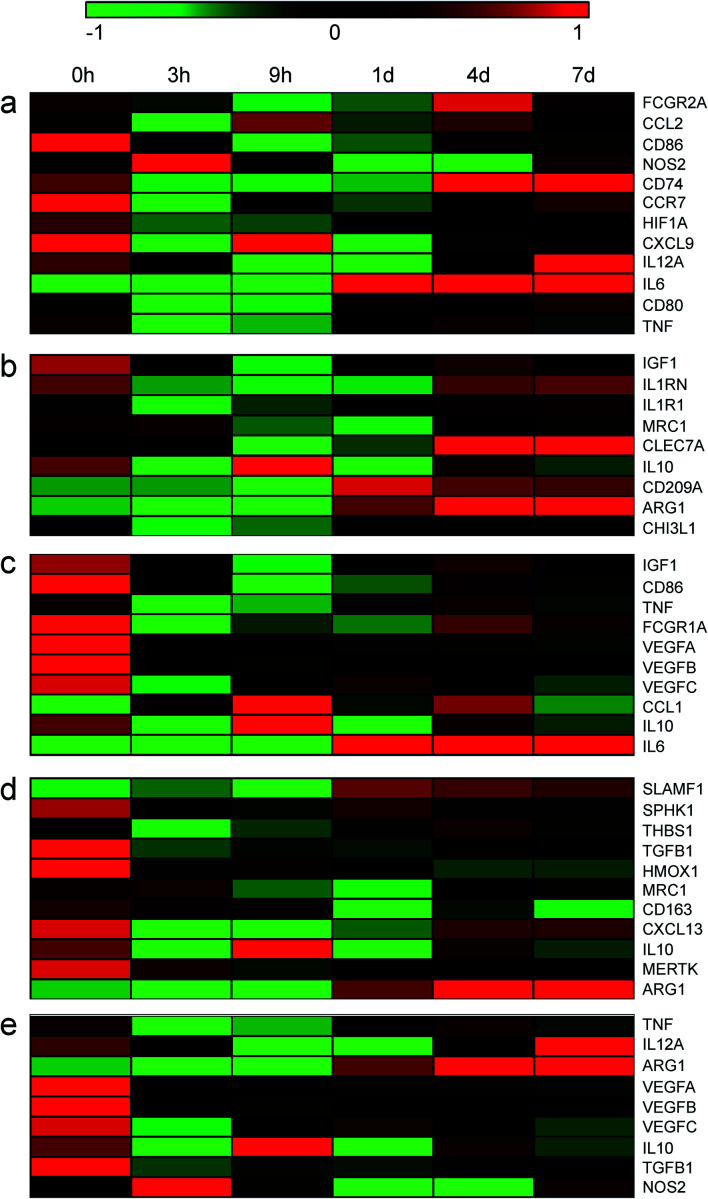
Changes of these marker genes in DRGs after nerve injury were investigated as well (Fig. 2 and Table S2†). The expression levels of these marker genes were not robustly altered in DRGs as compared with in sciatic nerve stumps. Among M1 macrophage marker genes, only a total of 5 genes, i.e., FCGR2A, CCL2, NOS2, CD74, and IL6, were significantly up-regulated after nerve injury. And their relative expression levels in DRGs were much lower as compared with values in sciatic nerve stumps. In addition, multiple genes, i.e., CD86, CCR7, CXCL9, IL12A, were found to be down-regulated. CD80 was found to be first down-regulated and then up-regulated (Fig. 2a). Consistent with the expression profiles of M1 macrophage marker genes, in DRGs, M2a, M2b, M2c, and M2d macrophage marker genes were not altered as robust as in sciatic nerve stumps (Fig. 2b–e). Heatmaps showed that different from the overall increasing trend of marker genes in sciatic nerve stumps, marker genes in DRGs generally exhibited a decreasing trend (Fig. 2).
Fig. 2. The temporal expression patterns of markers of (a) M1, (b) M2a, (c) M2b, (d) M2c, and (e) M2d macrophages in DRGs at 0 hour, 3 hours, 9 hours, 1 day, 4 days, and 7 days after nerve injury. Red color indicated up-regulation and green color indicated down-regulation.
Expressions of M1 and M2 macrophage markers in sciatic nerve stumps after peripheral nerve injury
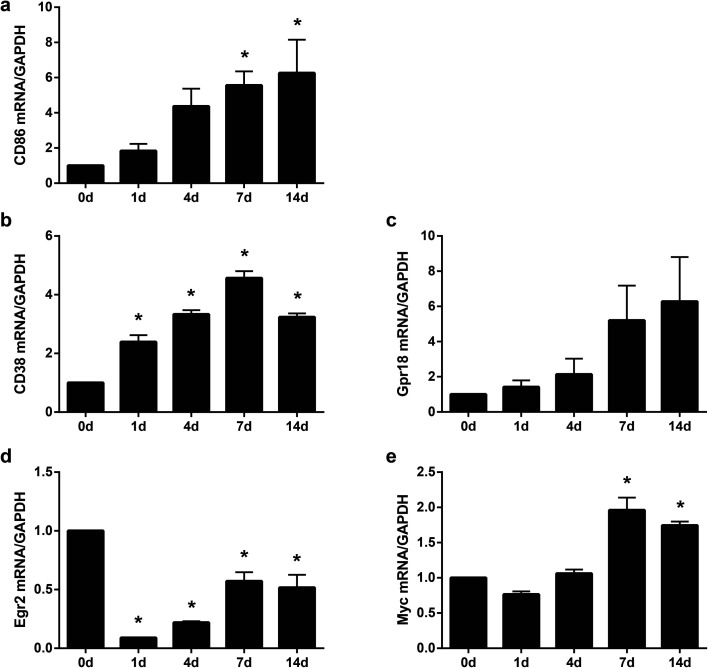
Sequencing data indicated that compared with DRGs, in sciatic nerve stumps, the up-regulation of M1 and M2 macrophage markers were vigorous. To validate the accuracy of sequencing outcomes, the temporal expression patterns of critical macrophage markers were further determined by RT-PCR. The expression levels of CD86, a canonical macrophage marker that was demonstrated to be up-regulated by sequencing, were first examined. Sequencing data showed that as compared with the 0 day control, the expression levels of CD86 were elevated to 2.81 folds (log2 ratio = 1.49), 5.59 folds (log2 ratio = 2.48), 3.97 folds (log2 ratio = 1.99), and 3.90 folds (log2 ratio = 1.93) at 1, 4, 7, and 14 days after peripheral nerve injury (Table S1†). RT-PCR results showed that consistent with sequencing data, the relative abundances of CD86 largely increased in the sciatic nerve stumps after nerve injury (Fig. 3a), indicating that our sequencing data were accurate.
Fig. 3. Expression levels of (a) CD86, (b) CD38, (c) Gpr18, (d) Egr2, and (e) Myc in sciatic nerve stumps at 0 day, 1 day, 4 days, 7 days, and 14 days after nerve injury. The relative expression levels of target genes CD86, CD38, Gpr18, Egr2, and Myc were normalized with reference gene GAPDH. Data were presented as means ± SEM. Asterisks indicated statistically significant differences compared with 0 day control (*, p-value < 0.05; ordinary one-way anova; n = 3).
Many macrophage markers lack macrophage phenotype specificity. For instance, CD86 are marks of both M1 macrophages and M2 macrophages. To further clarify changes of M1 and M2 macrophages after peripheral nerve injury, the expression profiles of some recently discovered specific macrophage phenotype markers were further examined. CD38 and Grp18 were identified as M1 macrophage-exclusive genes whereas Egr2 and Myc were identified as M2 macrophage-exclusive genes.16 RT-PCR experiments were performed to evaluate the expression trends of these novel M1 and M2 macrophage markers in the injured sciatic nerve stumps. CD38 and Gpr18 were found to be up-regulated after nerve injury (Fig. 3b and c). Egr2 were identified to be down-regulated after nerve injury and the expressions of Egr2 slightly recovered to its basal level at later time points after nerve injury (Fig. 3d). Another novel M2 macrophage marker Myc was found to be up-regulated at 7 and 14 days after nerve injury (Fig. 3e). These findings implied that M1 macrophages played great roles at all test time points after peripheral nerve injury, especially at early time points. The large amount of pro-inflammatory M1 macrophages might switch to anti-inflammatory M2 macrophages at later time points after peripheral nerve injury to enhance nerve regeneration.
Identification of the expression profiles of macrophage polarization-regulating miRNAs
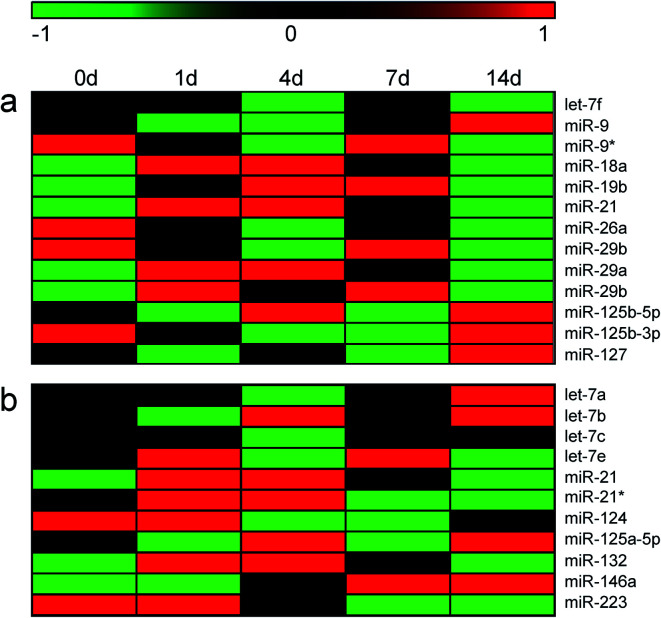
Sequencing data and RT-PCR results, from the genetic aspect, suggested that activation of macrophages in sciatic nerve stumps after peripheral nerve injury. Emerging studies showed that macrophages may be activated and polarized by microRNAs (miRNAs).26 Moreover, miRNAs may specifically promote the polarization of M1 or M2 macrophages. It have been demonstrated that let-7f, miR-9, miR-18a, miR-19a, miR-19b, miR-21, miR-26a, miR-26b, miR-29a, miR-29b, miR-125b, miR-127, and miR-155 induce M1 macrophage polarization while let-7a, let-7b, let-7c, let-7e, miR-21, miR-34, miR-124, miR-125a-5p, miR-132, miR-146a, miR-223, and miR-511 induce M2 macrophage polarization.27 Previously obtained miRNA sequencing data22 was applied to screen differentially expressed modulating miRNAs. miR-21, a miRNA that modulates both M1 and M2 macrophages, were found to be up-regulated in the injured peripheral nerve stumps (Fig. 4). Some other M1 macrophage-inducing miRNAs, such as miR-18a, miR-19b, miR-29a, and miR-29b, were also up-regulated after nerve injury (Fig. 4a). Up-regulated M2 macrophage-induced miRNAs, including miR-132 and miR-146a, were also discovered (Fig. 4b). However, the association of these miRNAs with specific sub-group of macrophages (i.e., M2a, M2b, M2c, and M2b) have not been revealed and the expressions of these miRNAs could not be categorized to M2 sub-groups.
Fig. 4. The temporal expression patterns of miRNAs that induce (a) M1 and (b) M2 macrophages in sciatic nerve stumps at 0 day, 1 day, 4 days, 7 days, and 14 days after nerve injury. Red color indicated up-regulation and green color indicated down-regulation.
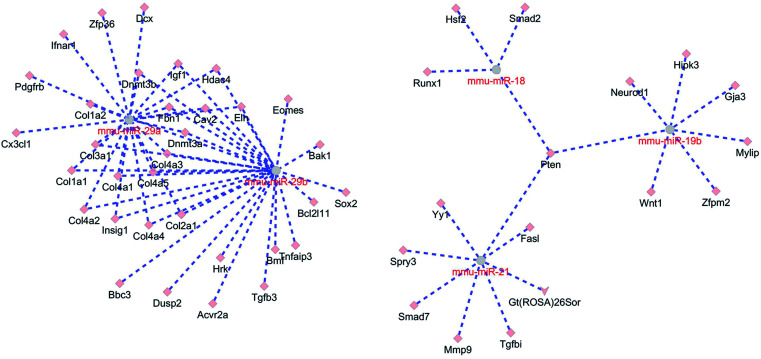
Macrophage polarization-regulating miRNAs execute their biological functions by binding to and affecting various target genes. Considering that the polarization of M1 macrophages obviously detected after peripheral nerve injury, up-regulated M1 macrophage-inducing miRNAs, i.e., miR-18a, miR-19b, miR-21, miR-29a, and miR-29b were subjected to bioinformatic analysis to construct a miRNA-mRNA network. miR-18a, miR-19b, miR-21, miR-29a, and miR-29b were screening in the mice (Mus musculus) miRNA database using GluoGO and GluePedia tools in Cytoscape software since mice (Mus musculus) miRNA database are more complete and miRNAs are highly conserved among species. miTarBase database was applied to identify target genes of these mmu-miRNAs and Cytoscape software was used to build the interactions between miRNAs and target genes. miR-18, miR-19b, and miR-21 were associated due to the same target gene Pten. miR-29a and miR-29b were closely related with common target genes Dnmt3b, Igf1, Hdac4, Fbn1, Cav2, Elm, Dnmt3a, Insig1, Col1a1, Col2a1, Col3a1, Col4a1, Col4a3, Col4a4, and Col4a5 (Fig. 5). These miRNAs might bind to their target genes, inhibited the expressions of target genes, and conduct critical biological functions.
Fig. 5. The miRNA–mRNA interaction network of miR-18, miR-19b, miR-21, miR-29a, and miR-29b. Interactions between miRNAs and target genes were labeled in dotted lines.
Discussion
Inflammation and immune responses occur immediately after peripheral nerve injury and are vital for the normal regeneration of injured peripheral nerves.11,28 As key immune cells, the polarization of macrophages after peripheral nerve injury are essential for the degeneration of distal nerve stumps, the clearance of axon growth path, and the rebuilding of a permissible microenvironment. Preventing the infiltration of circulating macrophages to the injured site leads to the failure of myelin clearance and nerve regeneration.29 Macrophages are thus considered as a key promoting factor for the repair and regeneration of injured peripheral nerves.
However, studies of the repair and regeneration of the injured peripheral nerves in aged animals revealed the complexity of macrophages. Stratton et al. found that monocyte migration and macrophage accumulation were delayed in aged injured nerves. Diminished macrophage infiltration and impaired macrophage-associated secreted protein signaling lead to regenerative deficits in aged animals.30 On the other hand, Buttner et al. found that there existed chronic inflammation and elevated macrophage infiltration in the intact peripheral nerves in aged animals and concluded that increased macrophage infiltration would reduce the regenerative ability of aged injured nerves.31 These controversial observations might be due to alternations of different types of macrophages and implied that success regeneration might require the co-action of M1 and M2 macrophages. Actually, a specific investigation of the biological roles of polarized pro-inflammatory (M1) and pro-healing (M2a and M2c) macrophages on peripheral nerve regeneration demonstrated that M1 and M2 macrophages exhibited different effects on the in vitro migration of Schwann cells and the in vivo infiltration of Schwann cells. The in vivo implantation of M2 macrophages, encouraged Schwann cell migration, stimulated Schwann cell infiltration, and promoted axon growth while the in vivo implantation of M2 macrophages did not display similar benefiting effects.32 Therefore, for the potential application of macrophages in tissue remodeling and regenerative medicine, it is of great importance to decipher alternations and fine tuning of M1 and M2 macrophages.
Here, using previously obtained sequencing data, we found that many marker genes of macrophages, especially marker genes of M1 macrophages, were strongly up-regulated in the injured nerve stumps immediately after nerve injury. Gene expressions were not that significantly altered in DRGs although peripheral nerve injury would also polarize macrophages and mediate immune responses in DRGs. RT-PCR validation of the expression levels of macrophage marker genes indicated the accuracy of sequencing data. It would be helpful to determine the temporal expression patterns of protein products in future studies.
Besides the dynamic changes of macrophage marker genes, we also screened a group of macrophage-associated miRNAs that were differentially expressed after nerve injury as miRNAs play regulatory roles in the expression signatures of macrophages and inflammatory responses during wound healing.33 Moreover, considering that M1 macrophages were robustly involved in peripheral nerve regeneration, we used up-regulated M1 macrophage-inducing miRNAs to construct a miRNA-mRNA interaction network and identified several essential target genes. The enrichment of these essential target genes to gene ontology (GO) terms reveled that macrophage-associated miRNAs and mRNAs were associated with cellular responses, cellular behavior, and tissue development and organization (Table S3†).
Pten, a gene coding for tumor suppressor phosphatase and tensin homologue,34 was found to be the target gene of miR-18, miR-19b, and miR-21. Pten has been recognized as a star gene in the neuroscience field since He's lab found that deletion of Pten would activate mTOR pathway and promote axon regeneration and functional repair after central nerve injury.35–37 In the peripheral nervous system, Pten was also identified to be altered expressed after nerve injury.38 Inhibition of Pten could stimulate neurite outgrowth from DRGs.39 However, the effect of Pten on inflammation/anti-inflammation remain largely undetermined. Here, our results showed that Pten was a center gene in the pro-inflammatory macrophage-associated miRNA–mRNA interaction network. Elevated expressions of miR-18, miR-19b, and miR-21 might induce reduced expression of their target gene Pten. Down-regulated Pten might further affect M1 macrophage-associated inflammatory responses and induce the reconstruction of microenvironment after nerve injury.
Notably, multiple genes coding for collagens, including Col1a1, Col2a1, Col3a1, Col4a1, Col4a3, Col4a4, and Col4a5, were found to be targeted by miR-29a and miR-29b. Col6a1 (collagen VI) was demonstrated to a promoting factor of macrophage recruitment and peripheral nerve regeneration.40,41 The biological functions of other collagen genes were not well discovered. Our bioinformatic analysis indicated that increased amount of miR-29a and miR-29b might decrease the expression levels of Col1a1, Col2a1, Col3a1, Col4a1, Col4a3, Col4a4, and Col4a5. These genes might execute different functions as Col6a1 in macrophage recruiting, implying the complexity of collagens and extracellular matrix in microenvironment remodeling and wound healing.
Conclusions
In the current study, we explored changes of pro-inflammatory and anti-inflammatory macrophages in sciatic nerve stumps and DRGs after peripheral nerve injury. These findings might increase the understanding of molecular mechanisms of peripheral nerve injury and provide a cue for the dual-roles of macrophages in wound healing.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China, No. 31971276, and Natural Science Foundation of Jiangsu Higher Education Institutions of China (Major Program), No. 19KJA320005.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06607a
References
- Ovchinnikov D. A. Genesis. 2008;46:447–462. doi: 10.1002/dvg.20417. [DOI] [PubMed] [Google Scholar]
- Mosser D. M. Edwards J. P. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. J. Lantis J. Kirsner R. S. Shah V. Molyneaux M. Carter M. J. Wound Repair Regen. 2016;24:613–629. doi: 10.1111/wrr.12444. [DOI] [PubMed] [Google Scholar]
- Suzuki T. Arumugam P. Sakagami T. Lachmann N. Chalk C. Sallese A. Abe S. Trapnell C. Carey B. Moritz T. Malik P. Lutzko C. Wood R. E. Trapnell B. C. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C. D. Crit. Rev. Immunol. 2012;32:463–488. doi: 10.1615/CritRevImmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- Minutti C. M. Knipper J. A. Allen J. E. Zaiss D. M. Semin. Cell Dev. Biol. 2017;61:3–11. doi: 10.1016/j.semcdb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Vannella K. M. Wynn T. A. Annu. Rev. Physiol. 2017;79:593–617. doi: 10.1146/annurev-physiol-022516-034356. [DOI] [PubMed] [Google Scholar]
- Caputa G. Flachsmann L. J. Cameron A. M. Immunol. Cell Biol. 2019;97:268–278. doi: 10.1111/imcb.12237. [DOI] [PubMed] [Google Scholar]
- Chen P. Piao X. Bonaldo P. Acta Neuropathol. 2015;130:605–618. doi: 10.1007/s00401-015-1482-4. [DOI] [PubMed] [Google Scholar]
- Chen Z. L. Yu W. M. Strickland S. Annu. Rev. Neurosci. 2007;30:209–233. doi: 10.1146/annurev.neuro.30.051606.094337. [DOI] [PubMed] [Google Scholar]
- Benowitz L. I. Popovich P. G. Curr. Opin. Neurol. 2011;24:577–583. doi: 10.1097/WCO.0b013e32834c208d. [DOI] [PubMed] [Google Scholar]
- Mueller M. Wacker K. Ringelstein E. B. Hickey W. F. Imai Y. Kiefer R. Am. J. Pathol. 2001;159:2187–2197. doi: 10.1016/S0002-9440(10)63070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian T. Wang P. Chen Q. Yi S. Liu Q. Wang H. Wang S. Geng W. Liu Z. Li S. RSC Adv. 2018;8:41181–41191. doi: 10.1039/C8RA08298G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodriguez P. Chen H. Erndt-Marino J. D. Liu F. Totsingan F. Gross R. A. Hahn M. S. ACS Appl. Bio Mater. 2019;2:601–612. doi: 10.1021/acsabm.8b00799. [DOI] [PubMed] [Google Scholar]
- Chen H. Erndt-Marino J. Diaz-Rodriguez P. Kulwatno J. Jimenez-Vergara A. C. Thibeault S. L. Hahn M. S. J. Biomed. Mater. Res., Part B. 2019;107:1056–1067. doi: 10.1002/jbm.b.34198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski K. A. Amici S. A. Webb L. M. Ruiz-Rosado Jde D. Popovich P. G. Partida-Sanchez S. Guerau-de-Arellano M. PLoS One. 2015;10:e0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J. Woolf C. J. Nat. Neurosci. 2007;10:1361–1368. doi: 10.1038/nn1992. [DOI] [PubMed] [Google Scholar]
- Yi S. Zhang H. Gong L. Wu J. Zha G. Zhou S. Gu X. Yu B. PLoS One. 2015;10:e0143491. doi: 10.1371/journal.pone.0143491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L. Wu J. Zhou S. Wang Y. Qin J. Yu B. Gu X. Yao C. Biochem. Biophys. Res. Commun. 2016;478:206–212. doi: 10.1016/j.bbrc.2016.07.067. [DOI] [PubMed] [Google Scholar]
- Zhao L. Yi S. J. Cell. Physiol. 2019;234:6876–6885. doi: 10.1002/jcp.27446. [DOI] [PubMed] [Google Scholar]
- Shen Y. Y. Gu X. K. Zhang R. R. Qian T. M. Li S. Y. Yi S. Neural Regener. Res. 2020;15:1502–1509. doi: 10.4103/1673-5374.274343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B. Zhou S. Wang Y. Ding G. Ding F. Gu X. PLoS One. 2011;6:e24612. doi: 10.1371/journal.pone.0024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. Markiel A. Ozier O. Baliga N. S. Wang J. T. Ramage D. Amin N. Schwikowski B. Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. X. Zhang S. X. Wu H. J. Rong X. L. Guo J. J. Leukocyte Biol. 2019;106:345–358. doi: 10.1002/JLB.3RU1018-378RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. Peng J. Han G. H. Ding X. Wei S. Gao G. Huang K. Chang F. Wang Y. Neural Regener. Res. 2019;14:1335–1342. doi: 10.4103/1673-5374.253510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essandoh K. Li Y. Huo J. Fan G. C. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feketea G. Bocsan C. I. Popescu C. Gaman M. Stanciu L. A. Zdrenghea M. T. Cells. 2019;8:420. doi: 10.3390/cells8050420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Gu X. Yi S. Front. Cell. Neurosci. 2016;10:274. doi: 10.3389/fncel.2016.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuche W. Friede R. L. J. Neurocytol. 1984;13:767–796. doi: 10.1007/BF01148493. [DOI] [PubMed] [Google Scholar]
- Stratton J. A. Eaton S. Rosin N. L. Jawad S. Holmes A. Yoon G. Midha R. Biernaskie J. Front. Aging Neurosci. 2020;12:174. doi: 10.3389/fnagi.2020.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner R. Schulz A. Reuter M. Akula A. K. Mindos T. Carlstedt A. Riecken L. B. Baader S. L. Bauer R. Morrison H. Aging Cell. 2018;17:e12833. doi: 10.1111/acel.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokarram N. Merchant A. Mukhatyar V. Patel G. Bellamkonda R. V. Biomaterials. 2012;33:8793–8801. doi: 10.1016/j.biomaterials.2012.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos Jimenez V. Bradley E. J. Willemsen A. M. van Kampen A. H. Baas F. Kootstra N. A. Physiol. Genomics. 2014;46:91–103. doi: 10.1152/physiolgenomics.00140.2013. [DOI] [PubMed] [Google Scholar]
- Song M. S. Salmena L. Pandolfi P. P. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Park K. K. Liu K. Hu Y. Smith P. D. Wang C. Cai B. Xu B. Connolly L. Kramvis I. Sahin M. He Z. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. Lu Y. Lee J. K. Samara R. Willenberg R. Sears-Kraxberger I. Tedeschi A. Park K. K. Jin D. Cai B. Xu B. Connolly L. Steward O. Zheng B. He Z. Nat. Neurosci. 2010;13:1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X. Qiao M. Bei F. Kim I. J. He Z. Sanes J. R. Neuron. 2015;85:1244–1256. doi: 10.1016/j.neuron.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Xiang J. Wu J. He B. Lin T. Zhu Q. Liu X. Zheng C. Neurosci. Lett. 2018;676:78–84. doi: 10.1016/j.neulet.2018.04.016. [DOI] [PubMed] [Google Scholar]
- Zhou S. Shen D. Wang Y. Gong L. Tang X. Yu B. Gu X. Ding F. PLoS One. 2012;7:e44768. doi: 10.1371/journal.pone.0044768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. Cescon M. Zuccolotto G. Nobbio L. Colombelli C. Filaferro M. Vitale G. Feltri M. L. Bonaldo P. Acta Neuropathol. 2015;129:97–113. doi: 10.1007/s00401-014-1369-9. [DOI] [PubMed] [Google Scholar]
- Lv D. Zhou L. Zheng X. Hu Y. Eur. J. Neurosci. 2017;45:1258–1267. doi: 10.1111/ejn.13558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.