Abstract
Aims
Post-traumatic osteoarthritis (PTOA) is a subset of osteoarthritis (OA). The gut microbiome is shown to be involved in OA. However, the effect of exercise on gut microbiome in PTOA remains elusive.
Methods
A total of 18 eight-week Sprague-Dawley rats were assigned into three groups: Sham/sedentary (Sham/Sed), PTOA/sedentary (PTOA/Sed), and PTOA/treadmill-walking (PTOA/TW). PTOA model was induced by transection of the anterior cruciate ligament (ACLT) and the destabilization of the medial meniscus (DMM). Treadmill-walking (15 m/min, 30 min/d, five days/week for eight weeks) was employed in the PTOA/TW group. The response of cartilage, subchondral bone, serology, and gut microbiome and their correlations were assessed.
Results
Eight-week treadmill-walking was effective at maintaining the integrity of cartilage-subchondral bone unit and reducing the elevated systematic inflammation factors and microbiome-derived metabolites. Furthermore, 16S ribosomal ribonucleic acid (rRNA) sequencing showed disease-relevant microbial shifts in PTOA animals, characterized by the decreased abundance of phylum TM7 and the increase of phylum Fusobacteria. At the genus level, the abundance of Lactobacillus, Turicibacter, Adlercreutzia, and Cetobacterium were increased in the PTOA animals, while the increase of Adlercreutzia and Cetobacterium was weakened as a response to exercise. The correlation analysis showed that genus Lactobacillus and Adlercreutzia were correlated to the structural OA phenotypes, while phylum Fusobacteria and genus Cetobacterium may contribute to the effects of exercise on the diminishment of serological inflammatory factors.
Conclusion
Exercise is effective at maintaining the integrity of cartilage-subchondral bone unit, and the exercise-induced modification of disease-relevant microbial shifts is potentially involved in the mechanisms of exercise-induced amelioration of PTOA.
Cite this article: Bone Joint Res 2022;11(4):214–225.
Keywords: Post-traumatic osteoarthritis, Exercise, Microbiome, Cartilage-subchondral bone unit, Serology, mouse model, post-traumatic osteoarthritis, post-traumatic osteoarthritis, subchondral bone, cartilage, inflammation, Osteoarthritis (OA), anterior cruciate ligament, ribonucleic acid, ACLT
Article focus
Gut microbiome is involved in post-traumatic osteoarthritis (PTOA) development. However, the effect of favourable exercise on gut microbiome in PTOA remains elusive. This study aims to investigate the relationships between exercise, gut microbiome, and PTOA.
Key messages
Moderate exercise in the form of treadmill-walking is effective at maintaining the integrity of articular cartilage-subchondral bone unit and eliminating the microbial-relevant low-degree inflammation in PTOA animals, possibly by modifying the PTOA-relevant shifts of the gut microbiome.
The disease-relevant microbial shift in PTOA is characterized by the decreased abundance of phylum TM7 and the increase of phylum Fusobacteria. At the genus level, the abundance of Lactobacillus, Turicibacter, Adlercreutzia, and Cetobacterium were increased in the PTOA animals, while the increase of Adlercreutzia and Cetobacterium was weakened as a response to exercise.
The genera Lactobacillus and Adlercreutzia were correlated to the structural OA phenotypes, while phylum Fusobacteria and genus Cetobacterium may contribute to the effects of exercise on the diminishment of serological inflammatory factors.
Strengths and limitations
This study demonstrates the underlying mechanisms of physical exercise by the modifications of disease-relevant microbial shifts, and sheds light on the exercise-induced modification of gut microbiome in OA management.
These findings support the idea that exercise is potentially associated with PTOA-relevant gut microbial shifts, however the exercise protocol employed in this study has a limited effect on the modification of gut microbiome, which may be attributed to short‐term training, single-ending timepoint, or the limited sample size.
This study only reveals the correlations between gut microbiome, PTOA, and exercise. Experimental validation by microbial manipulation would help to clarify the cause-effect relationships with adequate evidence.
Introduction
Osteoarthritis (OA) is a highly common musculoskeletal disorder involving the whole joint, characterized by the progressive degradation of articular cartilage, osteophyte formation, as well as synovial membrane inflammatory response and subchondral bone sclerosis, causing severe pain, morning stiffness, joint swelling, limited range of motion, physical dysfunction, and restriction of social activity. 1,2 As a subtype of OA, post-traumatic osteoarthritis (PTOA) is characterized by a distinct initial joint-associated structural injury followed by secondary inflammation, immune response, instability, and attenuated neuromuscular feedback. Persistent inflammation in PTOA, locally or systematically, is detectable. 3,4
The gut microbiome is the collection of trillions of microbial organisms that populate the gastrointestinal tract, and play a key role in the development and regulation of host metabolism, immune system, and inflammatory responses. 5 Microbiome alterations have an established role in chronic clinical conditions, including osteoporosis, 6 and two diseases associated with OA, namely obesity and diabetes, 7 by activating the innate immune system, resulting in an increased production of proinflammatory cytokines and bacterial metabolites. Given that low-grade inflammation is well characterized as a key mediator of the pathogenesis of OA, 8 it is reasonable to conclude that the microbiome is involved in OA, via lipopolysaccharide (LPS)-induced low-grade inflammation, metabolic endotoxemia, macrophage activation, and joint damage.
Recent evidence suggests the involvement of the gut microbiome in OA progression. 9,10 When comparing specific pathogen-free (SPF) mice and germ-free (GF) mice, a connection was established in a mouse model between the severity of OA and gut microbiome induced by destabilized medial meniscus (DMM) surgery, which suggests a decline in the severity of PTOA under germ-free circumstances. 11 In this context, the modulation of the gut microbiome is being investigated as a potentially novel strategy in the treatment of OA, which can be reached by maintaining a healthy lifestyle, including a healthy diet supplemented with vitamins, minerals, prebiotics, and probiotics, as well as regular sleeping habits. 12 Moreover, emerging research has shown that exercise, which is effective at alleviating OA symptoms and improving joint function and quality of life, 13-15 can independently alter the composition, abundance, and functional capacity of the gut microbiome. 16 However, the effect of exercise-training on the gut microbiome in experimental animals or OA patients remains elusive.
In our previous study, 17 we found that exercise is effective at maintaining PTOA structural integrity. In this study, we speculated that exercise-training may modify OA progression by modulating the relevant changes of the gut microbiome. We used moderate exercise-training on a PTOA rat model, and assessed the response of the cartilage, subchondral bone, serology, and gut microbiome, and their correlations, to investigate: 1) how the gut microbiome changes during PTOA development; 2) what effects exercise has on gut microbiome in PTOA; and 3) whether exercise ameliorates PTOA by modifiying gut microbiome.
Methods
Animals
All animal experimental protocols were approved by the Experimental Animal Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). A total of 18 eight-week-old male Sprague-Dawley (SD) rats were bred in the SPF condition. Given that animals kept in the same cage tend to have similar microbiomes, the SD rats were individually allocated to ventilated cages filled with irradiated sawdust bedding to avoid this “cage effect”, 18 and maintained on a 12-hour light/dark cycle under a consistent temperature (22°C (± 1°C)). The animals were able to move freely in the cages and had free access to autoclaved food and water. An ARRIVE checklist has been included to demonstrate compliance with the ARRIVE guidelines.
Study design and exercise protocols
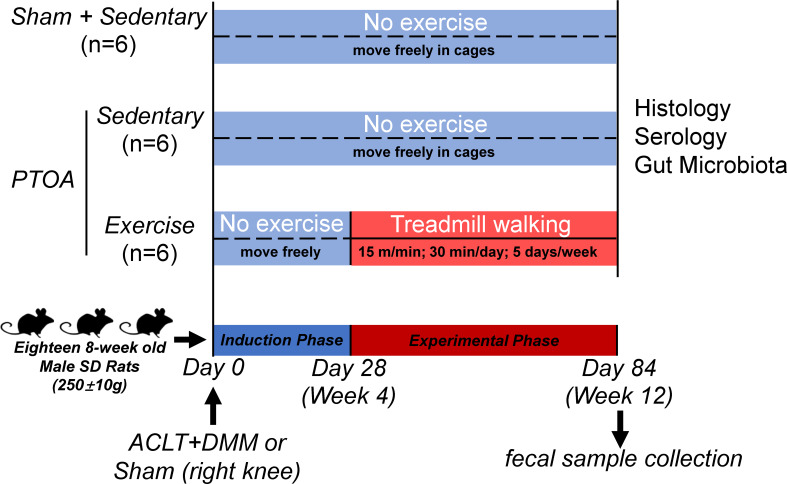
The protocols of this study are presented in Figure 1. The animals were randomly assigned into either the sham group (n = 6) or the PTOA group (n = 12). A minimum sample size of five rats per group is based on the ability to detect a minimal meaningful difference in histological score of the knee joint to provide an α = 0.05 and a power of 80%. Calculation of sample size was performed using G*Power Software (version 3.0.10, Germany). 19 Data for sample size calculations were obtained from a previous study. 17 All surgeries were performed under anaesthesia with Pentobarbital. As described previously, 17 the PTOA group was subjected to the transection of the anterior cruciate ligament (ACLT) and the DMM on the right knee joint, while the incision of skin and medial capsule in sham group was also performed on the right knee joint as an internal control. PTOA animals were further randomly assigned to either the PTOA/sedentary (PTOA/Sed) group (n = 6) or the PTOA/treadmill-walking (PTOA/TW) group (n = 6), while all the sham animals were assigned to the Sham/sedentary (Sham/Sed) group (n = 6). The PTOA/TW group was subjected to treadmill exercise at four weeks after surgery on a flatbed treadmill (Zhenghua Biological Instrument Equipment, China) at a rate of 15 m/min for 30 min/day, five days/week, for eight weeks. The animals in the Sham/Sed and the PTOA/Sed groups were allowed to move freely in cages without any treadmill exercise for eight weeks. The animals were euthanized at the age of 20 weeks, and knee joints were harvested for analysis. Prior to euthanasia, fresh fecal samples were individually collected from each rat within one minute of excretion using sterile 10 ml centrifuge tubes (the tubes having been frozen on dry ice), transported immediately to the laboratory, and then stored at -80°C before total genomic DNA extraction (within 12 hours). Additionally, blood samples were collected through cardiac puncture.
Fig. 1.
Experimental protocols. A total of 18 eight-week-old Sprague Dawley (SD) rats were assigned into three groups: sham/sedentary (Sham/Sed) (n = 6), post-traumatic osteoarthritis (PTOA)/sedentary (PTOA/Sed) (n = 6), and PTOA/treadmill-walking (PTOA/TW) (n = 6). PTOA was induced by the transection of the anterior cruciate ligament (ACLT) and the destabilization of the medial meniscus (DMM). Exercise-training was performed at four weeks after surgery on a flatbed treadmill at a rate of 15 m/min for 30 min/day, five days/week, for eight weeks. After eight weeks of treadmill-walking, the rats were euthanized and assessed with the analysis of micro-CT, histology, serology, and gut microbiome.
Micro-CT analysis of subchondral bone
After the animals were euthanized, all rat knee joints were scanned using a micro-CT system (micro-CT 50 Scanco Medical, Switzerland) with the following parameters: voxel size of 10.5 µm resolution, voltage of 100 kV, and current of 98 µA. The reconstructed images and data were obtained from 3D built-in software. To measure the subchondral bone of tibia, the following parameters were calculated: bone volume/tissue volume fraction (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular separation (Tb.Sp).
Histological analysis of articular cartilage
After micro-CT scanning, knee joints were fixed in 4% paraformaldehyde solution for 48 hours at room temperature, decalcified for four weeks by 10% ethylenediaminetetraacetic acid (EDTA) solution at 4°C, dehydrated in graded ethanol solutions, and embedded in paraffin. The joints were embedded using paraffin in the sagittal plane, and then ten to 20 slides were prepared, each with 4 µm sections collected for histological examination. From each sample, slides from a specific location in the mid-sagittal region were used for safranin O (SafO)/fast green staining to observe the change of chondrocytes, articular cartilage structure, and extracellular matrix. The Osteoarthritis Research Society International (OARSI) scoring system 20 and modified Mankin scoring system 21 were used to describe cartilage lesions. SafO reflecting cartilage proteoglycan content was scored according to a published scoring system. 22 Two areas were evaluated: medial tibial plateau (MTP) and the medial femoral condyle (MFC). These two sites were separately assigned a score based on the standard OARSI score (0 to 6), modified Mankin score (0 to 16), and SafO score (0 to 12). Scoring was performed by three different blinded scorers (XXH, RMC, XRS). The data were double-checked by the organizers (JMZ, TX) and the mean value of three individual scores was calculated for one slide. The mean score of randomly chosen ten slides was calculated to represent one sample. Scores from the tibial and femoral articular cartilage of the medial compartments were summed and reported. A higher score suggests a more advanced histopathological grade of OA.
Serum biochemical analysis
Blood was collected prior to euthanasia through cardiac puncture. Serum was obtained and stored at -80°C until analysis. Serum concentrations of endotoxin, tumour necrosis factor α (TNF-α), lipopolysaccharides (LPS), and interleukin-1β (IL-1β) were measured using enzyme-linked immunosorbent assay kits (ET, RA20354; TNF-α, RA20035-S; LPS, RA20650; IL-1β, RA20020-S; Bio-Swamp, China) according to the manufacturer’s instructions.
DNA extraction, 16S rRNA amplification, and Illumina Miseq sequencing
Bacterial genomic DNA was extracted from 200 mg of each fecal sample using Fast DNA SPIN extraction kits (MP Biomedicals, USA) according to the manufacturer’s instructions. The quantity and quality of extracted DNA were confirmed by agarose gel electrophoresis and NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA), respectively. The V3–V4 region of the bacterial 16S ribosomal ribonucleic acid (rRNA) genes was amplified by polymerase chain reaction (PCR) with the forward primer 338 F (5’-ACTCCTACGGGAGGCAGCA-3’) and the reverse primer 806 R (5’-GGACTACHVGGGTWTCTAAT-3’). Each PCR reaction contained 5 μl of Q5 reaction buffer (5×), 5 μl of Q5 High-Fidelity GC buffer (5×), 0.25 μl of Q5 High-Fidelity DNA Polymerase (5 U/μl), 2 μl (2.5 mM) of dNTPs, 1 μl (10 uM) of forward and reverse primer, 2 μl of DNA Template, and 8.75 μl of double-distilled water (ddH2O). The following PCR thermocycler protocol was used: initial denaturation at 98°C for two minutes, 25 cycles consisting of denaturation at 98°C for 15 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension of five minutes at 72°C. PCR products were purified with Agencourt AMPure Beads (Beckman Coulter, USA) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, USA). After the individual quantification step, products were sequenced using paired-end method and Illumina MiSeq platform with MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology (China).
Bioinformatics analysis of sequencing data
Sequences with a mean Phred score lower than 20, ambiguous bases, mononucleotide repeats exceeding 8 bp, and sequence lengths shorter than 150 bp were removed. Only sequences with overlaps longer than 10 bp and without any mismatch were assembled according to their overlap sequences. Barcode and sequencing primers were trimmed from the assembled sequence. The remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence similarity by USAEARCH. 16 Taxonomic classification was performed by Basic Local Alignment Search Tool (BLAST) searching of the representative sequences, set against the Greengenes Database 23 using the best hit method. 24 Sequence data analyses were mainly performed using QIIME and R packages (v3.2.0, R Foundation for Statistical Computing, Austria). An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. The abundance of bacteria in each taxon depends on the number of reads in the sequence. A Venn diagram was generated for comparisons among the OTUs of the groups. The α diversity values of each sample were assessed on the basis of the observed OTUs, bias-corrected Chao richness estimator (Chao1), Shannon index, and Simpson index. β diversity measures depending on Bray-Curtis distance were calculated using mothur software. Linear discriminant analysis Effect Size (LEfSe) was performed to identify abundant taxa differentially across groups. 25
Statistical analysis
Comparisons between two groups were performed by two-tailed independent-samples t-test and one-way analysis of variance (ANOVA) for comparison among multiple groups. All data were displayed as means and standard deviations, and analyzed using GraphPad Prism software version 7.0 (GraphPad, USA). A p-value < 0.05 was considered statistically significant. Correlations were analyzed by using Spearman’s correlation in R (version 3.5.3). A p-value < 0.05 was considered statistically significant.
Results
Exercise attenuates PTOA-relevant phenotypes of cartilage-subchondral bone unit and decreases the serological inflammatory factors and microbiome-derived metabolites
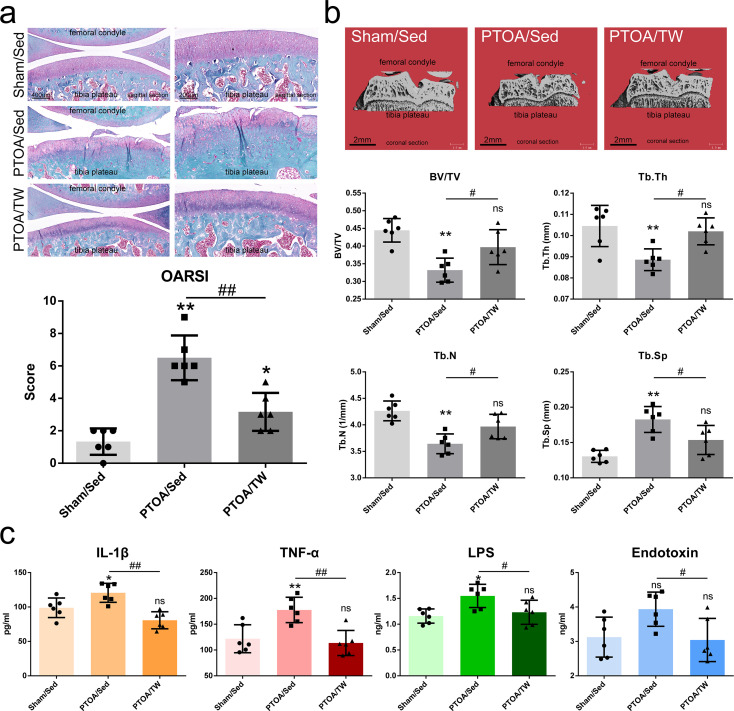
Since obesity and overloading are risk factors for OA, body weight was strictly controlled. As shown in Supplementary Figure a, there was no significant difference among three groups at the beginning and the ending timepoint. The PTOA-relevant histological phenotypes of cartilage-subchondral bone unit were notably induced in the PTOA/Sed group at the 12th week after surgery, characterized by articular cartilage degeneration and subchondral bone lesion. As shown in Figure 2a, the cartilage degeneration observed in the PTOA/Sed group was characterized by cartilage fissures, disorganized chondrocyte sequence, and cartilage matrix loss, which were consistent with OARSI score, modified Mankin score, and SafO score (Supplementary Figure b). The lesion of the subchondral bone in the PTOA/Sed group was characterized by bone loss (Figure 2b). These changes suggest the attenuated integrity of cartilage-subchondral bone unit in the PTOA animal model. However, it is striking that eight weeks of treadmill-walking efficiently attenuated these trauma-induced OA phenotypes, consequently maintaining the integrity of cartilage-subchondral bone unit in PTOA. Compared with the PTOA/Sed group, the cartilage degeneration was alleviated as a response to eight-week treadmill-walking in the PTOA/TW group after surgery (Figure 2a, Supplementary Figure b). The response of subchondral bone characterized by the reduction of bone loss was also consistently observed in the exercised animals (Figure 2b). The serum concentrations of pro-inflammatory factors IL-1β and TNF-α were elevated as a response to PTOA as expected (Figure 2c), while the levels of IL-1β and TNF-α were decreased significantly in PTOA/TW group compared with PTOA/Sed group as a response to eight-week treadmill-walking. Considering microbiome-derived metabolite LPS can activate inflammatory responses via intestinal barrier into the circulation, 26 we then assessed the levels of serum LPS, which were shown to be decreased in PTOA/TW group compared with PTOA/Sed group. Interestingly, an increased concentration of LPS in the PTOA/Sed group may provide association between gut microbiome, inflammation, and joint damage. As a biomarker related to intestinal permeability, serum endotoxin was not significantly increased in PTOA/Sed group (p = 0.065, one-way ANOVA) and decreased significantly in PTOA/TW group compared with PTOA/Sed group. It is striking that exercise was also able to reduce the increased LPS and endotoxin, suggesting that the modification of microbiome induced by exercise may involve systematic inflammation and/or OA. Together these findings support the hypothesis that treadmill-walking prevents the progression of PTOA-relevant phenotypes, maintains the integrity of articular cartilage-subchondral bone unit, and reduces pro-inflammatory mediators and microbiome-derived metabolites in serum.
Fig. 2.
Exercise attenuates post-traumatic osteoarthritis (PTOA)-relevant phenotypes of cartilage-subchondral bone unit and dismisses the serological inflammatory factors and microbiome-derived metabolites. a) The representative images of Safranin O-Fast staining (scale bar = 400 μm, black) in a sagittal plane and Osteoarthritis Research Society International (OARSI) score of the Sham/sedentary (Sed), PTOA/Sed, and PTOA/treadmill-walking (TW) groups. b) Micro‐CT representative images of the tibial subchondral bone of each group (scale bar = 2 mm, black) in a coronal plane, and histograms representing the parameters of tibial trabecular bone: trabecular number (Tb.N), volume/tissue volume (BV/TV), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp). c) Quantitative analysis of the serological inflammatory factors interleukin beta (IL-1β) and tumour necrosis factor alpha (TNF-α) and the microbiome-derived metabolites liposaccharide (LPS) and endotoxin. Data are presented as the mean and standard deviation, n = 6. *p < 0.05, *p < 0.01, PTOA/Sed or PTOA/TW versus Sham/Sed, one-way analysis of variance (ANOVA); #p < 0.05, ##p < 0.01, PTOA/Sed versus PTOA/TW, one-way ANOVA. ns, not significant.
Exercise modifies the PTOA-relevant changes of gut microbiome
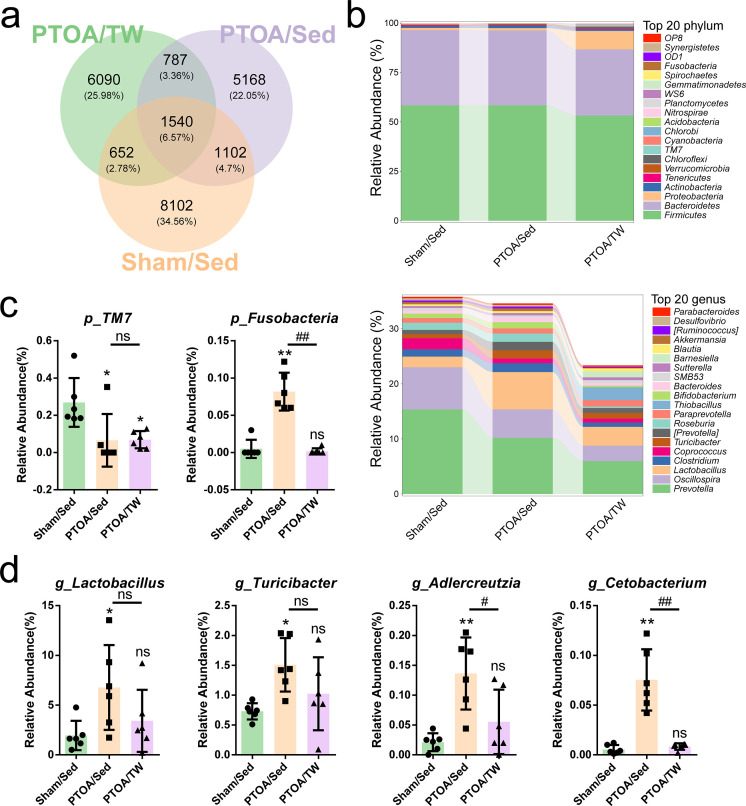
Due to the finding that treadmill-walking suppressed LPS and endotoxin, we speculated that the gut microbiome may be involved in the mechanisms of exercise-induced favourable outcomes. To address this question, the microbiome composition and diversity of 18 fecal samples were assessed by deep sequencing of the V3–V4 region of the 16S rRNA genes. On the basis of 97% sequence similarity, we obtained 23,441 OTUs, 1,540 of which existed in all groups and were thus defined as core OTUs. The core OTUs comprised 6.57% of the total OTUs, whereas 8,102, 5,168, and 6,090 OTUs were uniquely identified at the Sham/Sed, PTOA/Sed, and PTOA/TW groups, respectively (Figure 3a). The OTUs were classified into 36 bacterial phyla and 432 genera. According to the number of reads in the sequence, the top 20 phyla and genera in relative abundance of the fecal microbiome are displayed in Figure 3b and Supplementary Figure c. Firmicutes and Bacteroidetes were the most dominant phyla in these three groups, followed by Proteobacteria and Actinobacteria. These four phyla accounted for 98.43%, 98.88%, and 96.99% of the reads in Sham/Sed, PTOA/Sed, and PTOA/TW. Other phyla (Tenericutes, Verrucomicrobia, Chloroflexi, TM7, Cyanobacteria, Chlorobi, Acidobacteria, Nitrospirae, Planctomycetes, WS6, Gemmatimonadetes, Spirochaetes, Fusobacteria, OD1, Synergistetes, and OP8) were present at very low relative abundances. At the genus level, Prevotella was the most dominant in three groups. Other major genera included Oscillospira, Lactobacillus, Clostridium, Coprococcus, and Turicibacter. These six genera accounted for 29.03%, 26.09%, and 14.71% of the sequences of the Sham/Sed, PTOA/Sed, and PTOA/TW groups.
Fig. 3.
The altered abundance of gut microbiome in the exercised post-traumatic osteoarthritic (PTOA) animals. a) A Venn diagram was generated to compare operational taxonomic units (OTUs) among three groups and to depict OTUs that were unique to the three groups. b) Taxonomic profiles of the fecal bacteria from 16S ribosomal ribonucleic acid (rRNA) gene sequencing show the relative abundance of the top 20 phyla and the top 20 genera of fecal bacteria presented in the Sham/sedentary (Sed), PTOA/Sed, and PTOA/treadmill-walking (TW) groups. Changes in the c) two bacterial phyla and d) five genera in the gut microbiome composition of three groups. p < 0.05, p < 0.01, PTOA/Sed or PTOA/TW versus Sham/Sed, one-way analysis of variance (ANOVA); #p < 0.05, ##p < 0.01, PTOA/Sed versus PTOA/TW, one-way ANOVA; ns, not significant. Data are presented as the mean and standard deviation, n = 6.
Some differences identified at the levels of phylum and genus of fecal microbiome among the three groups are displayed in Figure 3c. The relative abundance of the phylum TM7 was consistently lower in both PTOA/Sed and PTOA/TW groups compared to the Sham/Sed group. The phylum Fusobacteria, which was robustly abundant in the gut of the PTOA rats without exercise, was nearly absent in the Sham/Sed and PTOA/TW groups. In this context, phylum TM7 and Fusobacteria were identified as the PTOA-relevant phyla and exercise decreased the abundance of Fusobacteria. At the genus level, the relative abundance of Lactobacillus, Turicibacter, Adlercreutzia, and Cetobacterium was increased in the PTOA/Sed group compared to Sham/Sed group, while the PTOA-induced Adlercreutzia and Cetobacterium were reduced by exercise (Figure 3d).
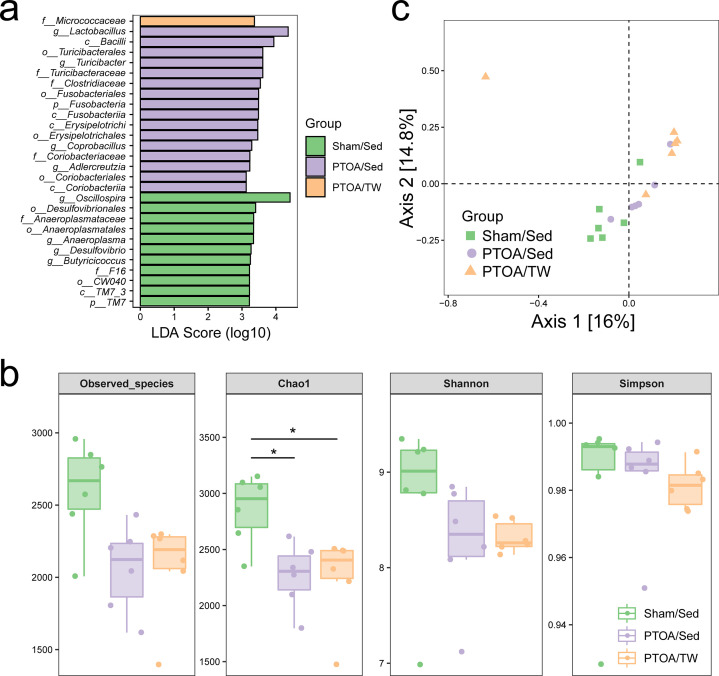
Microbial composition was further analyzed using the LEfSe. At the phylum level, the LEfSe analysis revealed that the phylum TM7 was significantly enriched in fecal samples of the Sham/Sed group while phylum Fusobacteria was significantly enriched in the PTOA/Sed group (Figure 4a). At the genus level, four genera (Oscillospira, Anaeroplasma, Desulfovibrio, and Butyricicoccus) were significantly enriched in the Sham/Sed group and other four genera (Lactobacillus, Turicibacter, Coprobacillus, and Adlercreutzia) were significantly enriched in PTOA groups. We also observed that the class TM7_3 and the three orders (Desulfovibrionales, Anaeroplasmatales, and CW040) were enriched in the Sham/Sed group, whereas the four classes (Bacilli, Fusobacteriia, Erysipelotrichi, and Coriobacteriia) and the four orders (Turicibacterales, Fusobacteriales, Erysipelotrichales, and Coriobacteriales) were enriched in the PTOA/Sed group. Additionally, at the family level, Anaeroplasmataceae and F16 were highly enriched in the Sham/Sed group, while Turicibacteraceae, Clostridiaceae, and Coriobacteriaceae were enriched in PTOA/Sed group and Micrococcaceae was enriched in the PTOA/TW group. Moreover, we also assessed α and β diversity of the fecal microbiome. The Chao 1 index in PTOA groups consistently decreased as a response to experimental injury compared to the Sham animals, while no remarkable difference was noted in the Shannon and Simpson indexes among the three groups (Figure 4b). The principal coordinate analysis (PCoA) based on Bray-Curtis distance showed no connection between β diversity and OA or exercise (Figure 4c). These results indicated that both joint injury and exercise induce the alternations of the intestinal microbiome composition, and that microbiome offers a potential connection between OA and exercise due to the effects of exercise on OA-relevant microbial shifts.
Fig. 4.
The composition and diversity of gut microbiome in the exercised post-traumatic osteoarthritic (PTOA) animals. a) Linear discriminant analysis effect size (LEfSe) analysis of the fecal bacterial community of the sham/sedentary (Sed), PTOA/Sed, and PTOA/treadmill-walking (TW) groups. b) Comparison of the fecal microbial community diversity of the Sham/Sed, PTOA/Sed, and PTOA/TW group, including comparison of the number of observed operational taxonomic units, Chao 1 index, Shannon index, and Simpson index. c) Trajectory of the gut microbiome structure of rats based on Bray-Curtis distance. LDA, linear discriminant analysis.
Exercise-induced modification of microbial shifts is associated with the integrity of cartilage-subchondral bone unit and systematic inflammation
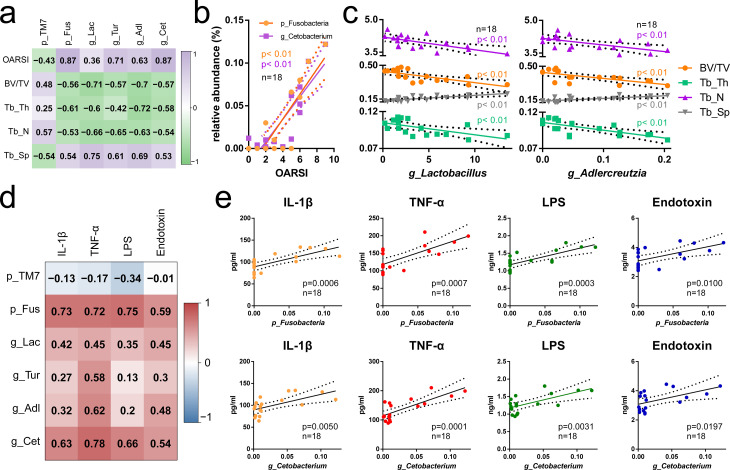
Given that the effects of exercise on cartilage-subchondral bone unit and gut microbiome were defined respectively, we further investigated the relationship of these effects by correlation analysis. As shown in Figure 5a, the correlations of joint structural features, including OARSI score and micro-CT data, and PTOA-relevant microorganisms, including two phyla (phylum TM7 and phylum Fusobacteria) and four genera (genus Lactobacillus, Turicibacter, Adlercreutzia, and Cetobacterium) were defined. Interestingly, phylum Fusobacteria and genus Cetobacterium showed strong positive correlations with OARSI (Figure 5b), indicating their potential involvement in cartilage biology and the PTOA-relevant cartilage changes, while genus Lactobacillus and Adlercreutzia showed stronger negative correlations with subchondral bone loss than the other PTOA-relevant microorganisms (Figure 5c). These results indicated a strong connection between structural phenotypes and PTOA-relevant microorganisms, among which exercise-responsive genus Lactobacillus and genus Adlercreutzia may be at least partially involved in the mechanisms of the exercise-induced maintenance of cartilage-subchondral bone unit integrity. Moreover, we performed correlation analysis between serological factors and the relative abundance of the OA-relevant microbiome. As shown in Figures 5d and 5e, phylum Fusobacteria and genus Cetobacterium were positively correlative to the levels of IL-1β, TNF-α, LPS, and endotoxin. These results suggested the potential mechanism that exercise reduces the PTOA-relevant genera Lactobacillus and Adlercreutzia, resulting in the dismission of systematic inflammation and subsequent PTOA structural amelioration.
Fig. 5.
Exercise-induced modification of microbial shifts is associated with the integrity of cartilage-subchondral bone unit and systematic inflammation. a) Correlation analysis of Osteoarthritis Research Society International (OARSI) score and subchondral bone micro-CT parameters and the abundance of the post-traumatic osteoarthritis (PTOA)-relevant gut microbiome. b) Scatter plot of the correlation of phylum Fusobacteria and genus Cetobacterium and OARSI. c) Scatter plot of the correlation of genus Lactobacillus and genus Adlercreutzia and micro-CT parameters. d) Correlation analysis of serological inflammatory factors and microbiome-derived metabolites and PTOA-relevant gut microbiome. e) Scatter plots of the correlation of the phylum Fusobacteria and the genus Cetobacterium with serological inflammatory factors and microbiome-derived metabolites. BV/TV, volume/tissue volume; g_Adl, genus Adlercreutzia; g_Get, genus Cetobacterium; g_Lac, genus Lactobacillus; g_Pha, genus Phascolarctobacterium; g_Tur, genus Turicibacter; p_Fus, phylum Fusobacteria; p_TM7, phylum TM7; Tb.N, trabecular number; Tb.Sp, trabecular separation; Tb.Th, trabecular thickness.
Discussion
Low-grade inflammation is well characterized as a key mediator of the pathogenesis of OA. 8,27 It is widely accepted that the gut microbiome produces a wide range of molecules, including enzymes and pro-inflammatory metabolites, making their way from the ‘leaky gut’ to the systemic circulation and inducing systematic inflammation. 28,29 In this context, it is feasible to hypothesize that the modulation of gut microbiome by external approaches may modify OA progression.
Exercise therapy not only reduces pain and improves functioning for OA individuals, 30 but also may modulate the composition, functions, and metabolites of gut microbiome to exert possible benefits for the host. 31 In our previous study, treadmill training is a favourable intervention to slow down the development of PTOA by remodelling the cartilage-subchondral unit. 17 The aim of this study was to identify potential cross-sectional associations between gut microbiome, OA histological severity, and inflammatory biomarkers in the context of injury-induced OA. Our results support the idea that treadmill-training attenuates PTOA progress in a possible mechanism of modifying the PTOA-relevant microbial shifts and reducing the systematic pro-inflammatory cytokines and metabolites to maintain the integrity of the cartilage-subchondral unit.
Before exploring mechanisms potentially responsible for the effects of gut microbiome on OA severity, we investigated a specific bacterial product, LPS, reported to be increased in serum and synovial samples of osteoarthritic individuals. 32 Studies have shown that gut bacterial products, including LPS, can enter the systemic circulation and affect both organs and joints by causing low-grade inflammation. 33 Similar to LPS, endotoxin was shown to be recognized by Toll-like receptors (TLRs) and associated with inflammatory response. 34 In particular, endotoxins trigger macrophage-mediated inflammation, which is thought to have a causal role in OA-related pain and severity. 35 Although some lines of evidence are conflicted, 36,37 systematic inflammatory response and serum LPS are responsive to antibiotic perturbations on gut microbiome. Exercise not only downregulates genes associated with the inflammatory process, 38 but also improves the clearance of endotoxins by Kupffer cells, 39 which indicates that the elimination of systematic inflammatory factors and microbiome-derived metabolites may be the mechanisms of exercise-induced cartilage-subchondral bone unit integrity maintenance. Indeed, our study showed a substantial elimination of LPS and endotoxin in the PTOA exercised animals, whereas PTOA/Sed rats might develop increased gut permeability with endotoxemia in association with upregulation of circulating proinflammatory molecules consistent with an activated innate immune system. The cytokines elevated in the PTOA/Sed rats, IL-1β, and TNF-α are well characterized as the key mediators in the pathological joint destruction in PTOA. 40 Taking these lines of evidence together, it is not surprising to infer the favourable effects of exercise on the integrity of gut permeability and the elimination of systematic inflammation in PTOA animals.
Since these results suggest a connection between gut microbiome and OA, we examined the differences in OTU abundance between the Sham/Sed, PTOA/Sed, and PTOA/TW rats. The abundance of Fusobacteria was increased in the PTOA/Sed group and consistently positively correlated with OARSI score, serum concentrations of IL-1β, TNF-α, LPS, and endotoxin, which were suppressed by exercise in the PTOA/TW group. Our results are supported by Huang et al 41 who demonstrated a positive correlation of abundance of Fusobacterium with OA histological severity and inflammatory biomarker concentrations. The links between Fusobacterium species and host immune response are supported by some lines of evidence. Okahashi et al 42 showed that Fusobacterium species can produce LPS and enhance IL-1 production by murine peritoneal macrophages. Ye et al 43 showed that Fusobacterium induced CCL20 protein expression in monocytes and stimulated the monocyte/macrophage activation and migration in an in vitro co-culture assay. Most recently, Martin-Gallausiaux et al 44 showed that Fusobacterium-derived extracellular vesicles trigger innate immunity of gut epithelial cells by promoting nuclear factor kappa B (NF-κB) activation via dynamin-mediated endocytosis. Together with these findings, we speculated that exercise might reduce IL-1β, TNF-α, LPS, and endotoxin by reducing the abundance of Fusobacteria and/or increasing the integrity of the intestinal barrier to block its effect, which suppresses systematic low-grade inflammation and, in doing so, benefits the osteoarthritic joint. Besides Fusobacteria, we also found that Cetobacterium positively correlated with OARSI score and concentrations of serological inflammatory factors and microbiome-derived metabolites, while there is no evidence to directly support the links between Cetobacterium and OA. Moreover, we did not identify similar changes in the previously reported bacterial families/species associated with OA, 45 such as Lactobacillus, which could be attributed to the differences in sample size and the type of OA models (injury-induced vs obesity-associated OA). Still, this study suggests that gut microbiome may be involved in OA by modifying low-grade inflammation, and that exercise has a favourable effect on osteoarthritic joints, possibly by manipulating gut microbiome.
Although these findings suggest that exercise is potentially associated with PTOA-relevant gut microbial shifts, the exercise protocol employed in this study has a limited effect on the modification of gut microbiome. In this way, we admit several limitations in the present study. First, the sample size of the rat study was relatively small. Second, the difference in gut microbiome between species should be considered, and validation in human samples would be valuable in the future. Furthermore, this study did not reveal the effects of exercise on gut microbiome in the sham animals, because we did not include a sham group with treadmill training. Moreover, this study only reveals the correlations between gut microbiome, PTOA, and exercise. Experimental validation by microbial manipulation, such as fecal microbiome transplantation, 41 co-housing, 46 antibiotic intervention, 37 and the use of germ-free animals, 41 would help to speculate the cause-effect relationships with adequate evidence. Finally, limited effects of exercise on the modification of gut microbiome may be attributed to short‐term training and a single-ending timepoint. As a result, the modified protocols should be expected in different stages of OA in animals.
In conclusion, exercise is effective at maintaining the integrity of cartilage-subchondral bone unit, and exercise-induced modification of PTOA-relevant gut microbial shifts may be the key mechanism of the favourable outcomes of exercised PTOA animals.
Author contributions
X. X. Hao: Investigation, Resources, Data curation, Writing – original draft.
J. M. Zhang: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Visualization.
X. R. Shang: Resources.
K. Sun: Resources.
J. Zhou: Data curation, Writing – review & editing.
J. Liu: Resources.
R. M. Chi: Investigation.
T. Xu: Project administration, Funding acquisition.
Funding statement
The authors disclose receipt of the following financial or material support for the research, authorship, and/or publication of this article: support from the National Natural Science Foundation of China (Grant No. 82072556).
ICMJE COI statement
The authors declared no conflict of interests about the publication of this paper.
Ethical review statement
This study was approved by the Experimental Animal Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China).
Open access funding
The open access funding for this study was provided by the National Natural Science Foundation of China (Grant No. 82072556).
Supplementary material
Additional information of body weight, histopathological evaluations, and the relative abundance of fecal bacteria in the individual rats, as well as the ARRIVE checklist.
© 2022 Author(s) et al. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial No Derivatives (CC BY-NC-ND 4.0) licence, which permits the copying and redistribution of the work only, and provided the original author and source are credited. See https://creativecommons.org/licenses/by-nc-nd/4.0/.
Contributor Information
Xiaoxia Hao, Email: 1561174394@qq.com.
Jiaming Zhang, Email: jiaming_zhangtjmc@icloud.com.
Xingru Shang, Email: 13783836979@163.com.
Kai Sun, Email: 1085844308@qq.com.
Jun Zhou, Email: jzhou30@bwh.harvard.edu.
Jiawei Liu, Email: Xiaoliu578578@163.com.
Ruimin Chi, Email: Crm1020@163.com.
Tao Xu, Email: xutdoc@163.com.
References
- 1. Sanghi D, Avasthi S, Mishra A, Singh A, Agarwal S, Srivastava RN. Is radiology a determinant of pain, stiffness, and functional disability in knee osteoarthritis? A cross-sectional study. J Orthop Sci. 2011;16(6):719–725. 10.1007/s00776-011-0147-y [DOI] [PubMed] [Google Scholar]
- 2. Glyn-Jones S, Palmer AJR, Agricola R, et al. . Osteoarthritis. Lancet. 2015;386(9991):376–387. 10.1016/S0140-6736(14)60802-3 [DOI] [PubMed] [Google Scholar]
- 3. Wang LJ, Zeng N, Yan ZP, Li JT, Ni GX. Post-traumatic osteoarthritis following ACL injury. Arthritis Res Ther. 2020;22(1):57. 10.1186/s13075-020-02156-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lieberthal J, Sambamurthy N, Scanzello CR. Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthritis Cartilage. 2015;23(11):1825–1834. 10.1016/j.joca.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31(1):69–75. 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li S, Mao Y, Zhou F, Yang H, Shi Q, Meng B. Gut microbiome and osteoporosis: a review. Bone Joint Res. 2020;9(8):524–530. 10.1302/2046-3758.98.BJR-2020-0089.R1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care. 2010;33(10):2277–2284. 10.2337/dc10-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson WH, Lepus CM, Wang Q, et al. . Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12(10):580–592. 10.1038/nrrheum.2016.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hernandez CJ. The microbiome and bone and joint disease. Curr Rheumatol Rep. 2017;19(12):77. 10.1007/s11926-017-0705-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Favazzo LJ, Hendesi H, Villani DA, et al. . The gut microbiome-joint connection: implications in osteoarthritis. Curr Opin Rheumatol. 2020;32(1):92–101. 10.1097/BOR.0000000000000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ulici V, Kelley KL, Azcarate-Peril MA, et al. . Osteoarthritis induced by destabilization of the medial meniscus is reduced in germ-free mice. Osteoarthritis Cartilage. 2018;26(8):1098–1109. 10.1016/j.joca.2018.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szychlinska MA, Di Rosa M, Castorina A, Mobasheri A, Musumeci G. A correlation between intestinal microbiota dysbiosis and osteoarthritis. Heliyon. 2019;5(1):e01134. 10.1016/j.heliyon.2019.e01134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nelson AE, Allen KD, Golightly YM, Goode AP, Jordan JM. A systematic review of recommendations and guidelines for the management of osteoarthritis: the chronic osteoarthritis management initiative of the U.S. bone and joint initiative. Semin Arthritis Rheum. 2014;43(6):701–712. 10.1016/j.semarthrit.2013.11.012 [DOI] [PubMed] [Google Scholar]
- 14. McAlindon TE, Bannuru RR, Sullivan MC, et al. . OARSI guidelines for the non-surgical management of knee osteoarthritis. Osteoarthritis Cartilage. 2014;22(3):363–388. 10.1016/j.joca.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 15. He Z, Nie P, Lu J, et al. . Less mechanical loading attenuates osteoarthritis by reducing cartilage degeneration, subchondral bone remodelling, secondary inflammation, and activation of NLRP3 inflammasome. Bone Joint Res. 2020;9(10):731–741. 10.1302/2046-3758.910.BJR-2019-0368.R2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 17. Hao X, Wang S, Zhang J, Xu T. Effects of body weight-supported treadmill training on cartilage-subchondral bone unit in the rat model of posttraumatic osteoarthritis. J Orthop Res. 2021;39(6):1227–1235. 10.1002/jor.24791 [DOI] [PubMed] [Google Scholar]
- 18. Hildebrand F, Nguyen TLA, Brinkman B, et al. . Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14(1):R4. 10.1186/gb-2013-14-1-r4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. 10.3758/bf03193146 [DOI] [PubMed] [Google Scholar]
- 20. Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage. 2010;18 Suppl 3:S17-23. 10.1016/j.joca.2010.05.025 [DOI] [PubMed] [Google Scholar]
- 21. Mankin HJ, Dorfman H, Lippiello L, Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am. 1971;53-A(3):523–537. [PubMed] [Google Scholar]
- 22. McNulty MA, Loeser RF, Davey C, Callahan MF, Ferguson CM, Carlson CS. A comprehensive histological assessment of osteoarthritis lesions in mice. Cartilage. 2011;2(4):354–363. 10.1177/1947603511402665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeSantis TZ, Hugenholtz P, Larsen N, et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altschul SF, Madden TL, Schäffer AA, et al. . Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Segata N, Izard J, Waldron L, et al. . Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moreira APB, Texeira TFS, Ferreira AB, Peluzio M do CG, Alfenas R de CG. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br J Nutr. 2012;108(5):801–809. 10.1017/S0007114512001213 [DOI] [PubMed] [Google Scholar]
- 27. Huang Z, Kraus VB. Does lipopolysaccharide-mediated inflammation have a role in OA? Nat Rev Rheumatol. 2016;12(2):123–129. 10.1038/nrrheum.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biver E, Berenbaum F, Valdes AM, et al. . Gut microbiota and osteoarthritis management: An expert consensus of the European society for clinical and economic aspects of osteoporosis, osteoarthritis and musculoskeletal diseases (ESCEO). Ageing Res Rev. 2019;55:100946. 10.1016/j.arr.2019.100946 [DOI] [PubMed] [Google Scholar]
- 29. Hao X, Shang X, Liu J, Chi R, Zhang J, Xu T. The gut microbiota in osteoarthritis: where do we stand and what can we do? Arthritis Res Ther. 2021;23(1):42. 10.1186/s13075-021-02427-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang W, Moskowitz RW, Nuki G, et al. . OARSI recommendations for the management of hip and knee osteoarthritis, Part II: OARSI evidence-based, expert consensus guidelines. Osteoarthritis Cartilage. 2008;16(2):137–162. 10.1016/j.joca.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 31. Monda V, Villano I, Messina A, et al. . Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev. 2017;2017:3831972. 10.1155/2017/3831972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang ZY, Stabler T, Pei FX, Kraus VB. Both systemic and local lipopolysaccharide (LPS) burden are associated with knee OA severity and inflammation. Osteoarthritis Cartilage. 2016;24(10):1769–1775. 10.1016/j.joca.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Creely SJ, McTernan PG, Kusminski CM, et al. . Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292(3):E740-7. 10.1152/ajpendo.00302.2006 [DOI] [PubMed] [Google Scholar]
- 34. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 35. Boer CG, Radjabzadeh D, Medina-Gomez C, et al. . Intestinal microbiome composition and its relation to joint pain and inflammation. Nat Commun. 2019;10(1):4881. 10.1038/s41467-019-12873-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Y, Zhao X, Zhao J, et al. . A mixture of Lactobacillus species isolated from traditional fermented foods promote recovery from antibiotic-induced intestinal disruption in mice. J Appl Microbiol. 2018;124(3):842–854. 10.1111/jam.13687 [DOI] [PubMed] [Google Scholar]
- 37. Guan Z, Jia J, Zhang C, et al. . Gut microbiome dysbiosis alleviates the progression of osteoarthritis in mice. Clin Sci (Lond). 2020;134(23):3159–3174. 10.1042/CS20201224 [DOI] [PubMed] [Google Scholar]
- 38. González-Chávez SA, Pacheco-Tena C, Quiñonez-Flores CM, Espino-Solis GP, Burrola-De Anda JI, Muñoz-Morales PM. Positive transcriptional response on inflammation and joint remodelling influenced by physical exercise in proteoglycan-induced arthritis: an animal study. Bone Joint Res. 2020;9(1):36–48. 10.1302/2046-3758.91.BJR-2019-0055.R2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Komine S, Akiyama K, Warabi E, et al. . Exercise training enhances in vivo clearance of endotoxin and attenuates inflammatory responses by potentiating Kupffer cell phagocytosis. Sci Rep. 2017;7(1):11977. 10.1038/s41598-017-12358-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han PF, Wei L, Duan ZQ, et al. . Contribution of IL-1β, 6 and TNF-α to the form of post-traumatic osteoarthritis induced by “idealized” anterior cruciate ligament reconstruction in a porcine model. Int Immunopharmacol. 2018;65:212–220. 10.1016/j.intimp.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 41. Huang Z, Chen J, Li B, et al. . Faecal microbiota transplantation from metabolically compromised human donors accelerates osteoarthritis in mice. Ann Rheum Dis. 2020;79(5):646–656. 10.1136/annrheumdis-2019-216471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okahashi N, Koga T, Nishihara T, Fujiwara T, Hamada S. Immunobiological properties of lipopolysaccharides isolated from Fusobacterium nucleatum and F. necrophorum. J Gen Microbiol. 1988;134(6):1707–1715. 10.1099/00221287-134-6-1707 [DOI] [PubMed] [Google Scholar]
- 43. Ye X, Wang R, Bhattacharya R, et al. . Fusobacterium Nucleatum subspecies Animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res (Phila). 2017;10(7):398–409. 10.1158/1940-6207.CAPR-16-0178 [DOI] [PubMed] [Google Scholar]
- 44. Martin-Gallausiaux C, Malabirade A, Habier J, Wilmes P. Fusobacterium nucleatum extracellular vesicles modulate gut epithelial cell innate immunity via FomA and TLR2. Front Immunol. 2020;11:583644. 10.3389/fimmu.2020.583644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Collins KH, Paul HA, Reimer RA, Seerattan RA, Hart DA, Herzog W. Relationship between inflammation, the gut microbiota, and metabolic osteoarthritis development: studies in a rat model. Osteoarthritis Cartilage. 2015;23(11):1989–1998. 10.1016/j.joca.2015.03.014 [DOI] [PubMed] [Google Scholar]
- 46. Liu X, Li X, Xia B, et al. . High-fiber diet mitigates maternal obesity-induced cognitive and social dysfunction in the offspring via gut-brain axis. Cell Metab. 2021;33(5):923–938. 10.1016/j.cmet.2021.02.002 [DOI] [PubMed] [Google Scholar]