Abstract
Objectives
In this study, we aimed to differentiate normal cervical graphs and graphs of diseases that cause mechanical neck pain by using deep convolutional neural networks (DCNN) technology.
Materials and methods
In this retrospective study, the convolutional neural networks were used and transfer learning method was applied with the pre-trained VGG-16, VGG-19, Resnet-101, and DenseNet-201 networks. Our data set consisted of 161 normal lateral cervical radiographs and 170 lateral cervical radiographs with osteoarthritis and cervical degenerative disc disease.
Results
We compared the performances of the classification models in terms of performance metrics such as accuracy, sensitivity, specificity, and precision metrics. Pre-trained VGG-16 network outperformed other models in terms of accuracy (93.9%), sensitivity (95.8%), specificity (92.0%), and precision (92.0%) results.
Conclusion
The results of this study suggest that the deep learning methods are promising support tool in automated control of cervical graphs using the DCNN and the exclusion of normal graphs. Such a supportive tool may reduce the diagnosis time and provide radiologists or clinicians to have more time to interpret abnormal graphs.
Keywords: Cervical radiography, convolutional neural network, deep learning, disc space narrowing, osteoarthritic changes, transfer learning.
Introduction
Although the cervical spine supports the head, it has quite functional features such as flexion, extension, lateral flexion, and rotation. Due to this wide range of motion, there is a susceptibility to trauma and degenerative changes. Neck pain due to cervical spine problems is a very common medical problem in the adult population. Cervical strain, cervical spondylosis, and discopathies are common causes of mechanical neck pain. Apart from these, traumas, tumors and reflected pain are also important causes of neck pain.[1] Patients with non-traumatic neck pain can be examined in various polyclinics such as family medicine, internal medicine, orthopedics, physical medicine and rehabilitation, neurology, neurosurgery, and rheumatology outpatient clinics. Almost every patient is evaluated with plain radiography in terms of obtaining easy and fast results at the diagnosis stage.[2] However, it is not always possible to evaluate radiographs by a radiologist, and cervical radiographs may be incorrectly evaluated. According to statistics, the misdiagnosis rate by human in the interpretation of plain radiographies can reach 10 to 30%.[3]
With the prominence of the digital systems in medical imaging methods, the X-rays can be evaluated instantaneously, as well as remotely with computer and internet support. It can be also evaluated retrospectively by storing the images. With the development of computer technology, automatic processing of large amount of data becomes possible for decision support through machine learning techniques.
Deep convolutional neural networks (DCNN) and similar machine learning methodologies have started to take an important place in the medical field such as deep learning, drug development, genetic, audio and visual objects recognition. Deep learning has attracted an increasing amount of attention from medical sciences, as it has proved to result in better performance than traditional machine learning algorithms in processing massive quantities of data. These methods have been applied successfully in various medical domains to support and enhance diagnosis processes. To illustrate, deep learning is applied to classify patients with brain stroke based on data gathered from patients and tomography scans and magnetic resonance images and predictive features that are risky for the mortality rate are identified and their effect on mortality is predicted with reasonable accuracy.[4] Similarly, deep learning technique is used in the diagnosis of three common dermatopathology diseases based on the whole-slide images (WSI) where fully convolutional neural network (CNN) is created that improved accuracy in classifying the dermatopathology diseases.[5] In the prediction of colorectal cancer, deep learning method was fed with digitized hemotoxylin and eosin (H-E) stained histology slides which resulted in high accuracy rates for normal and cancer slides.[6]
The CNN is an artificial neural network (ANN) algorithm that applies multilayer neural network structure. It is a powerful algorithm for image processing and pattern recognition, as it is capable of catching spatial and temporal information in an image by the use of appropriate filters. The CNN requires a large amount of data to create the learning model. In case of inadequate data, transfer learning can be utilized that provides effective classification solution. Transfer learning is a recent trend in deep learning that uses existing knowledge to solve different problems.[7] Transfer learning is proved to be quite effective, particularly in domains where data sources are limited in nature.[8]
The ImageNet is a project that aims to categorize images into almost 22,000 separate object categories using around 1.2 million training images with the goal of training learning models to classify an input image into 1,000 separate object categories. The CNN networks are pre-trained on ImageNet data and have the capacity to be used for images other than ImageNet data set via transfer learning. Among many pre-trained models, VGG-16 and VGG-19 architectures have 16 and 19 layers respectively with 3x3 convolutional layers.[9] ResNet is introduced as a network-in-network architecture that is built on micro-architecture modules.[10] DenseNet[11] is a more recent architecture that is characterized by its simplicity that expects smaller number of parameters with respect to ResNet network providing similar accuracy.
In the present study, we aimed to evaluate osteoarthritic changes, loss of cervical lordosis and disc space narrowing, which can be seen in the lateral cervical radiography of patients admitted to the hospital with neck pain, with deep learning technology using transfer learning.
Patients and Methods
Data set
After the approval of the Ankara City Hospital Ethics Committee (Date: 25/06/2020, No: E1-20-392), the lateral cervical radiography images of 416 patients admitted to the Rheumatology Department of Ankara City Hospital between January 2019 and December 2019 were obtained from the hospital's electronic registration system. Patients between the ages of 20 to 65 who suffered from neck pain were included in the study. Patients who had cervical trauma, patients with systemic disease with cervical spine involvement such as ankylosing spondylitis, patients who had cervical spine surgery and patients with malignancy with cervical spine involvement were excluded from the study. Radiographs of a total of 85 patients who did not meet the criteria were not included in the study.
Cervical radiographs were evaluated by dividing into following two groups: Group 1, normal cervical radiographs; normal disc space without osteoarthritic changes; and Group 2, pathological cervical radiographs; loss of cervical lordosis, narrowing of the disc space and/or degenerative changes in the vertebral corpus.
Radiographic images of the patients were evaluated by a radiologist and a rheumatology specialist, both of whom have more than 10 years of experience, without being aware of each other. Radiographs agreed by these two investigators as normal or abnormal were included in the study. The intraclass correlation coefficient for intra-observer and inter-observer reliability was 0.94 and 0.92, respectively, for cervical radiographic evaluation. The images were excluded from the study in case of disputes.
Data preprocessing
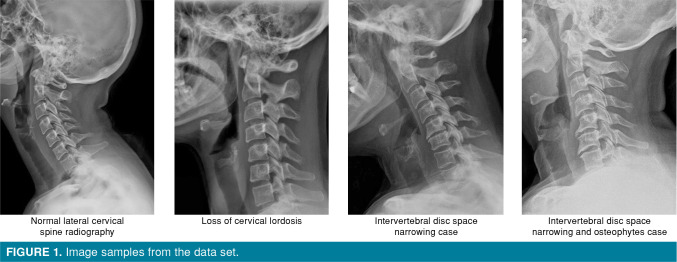
Lateral cervical radiographs used in this study were in JPG format, different width and height pixels. All images, in between C1 and C7, were cropped to include cervical vertebrae. A white color padding was added to the right and left side of each image to prevent data loss and distortion so that the square frame was obtained and the resolution remained unchanged. All of the images were resized to 224x224 pixels. As a result, 224x224x3 (3 means RGB image format) dimension images were obtained. Figure 1 shows some cropped and resized images samples.
Figure 1. Image samples from the data set.
Data set is divided into training (70%), validation (15%) and test set (15%) randomly (Table I). In deep learning methods, the performance of the model improves as the number of data increases. Statistically, the number of samples allocated for training should be sufficient for training, and the number of test data should show the performance of the model and give similar results in repetitions of the test. Since it was observed that the same ratios were used in relevant studies previously, the above partitioning was preferred as a suitable choice to achieve the purpose of this study
Table 1. Number of images used for training, validation and testing stages.
| Training | Validation | Test | Total | |
| Abnormal | 119 | 26 | 25 | 170 |
| Normal | 113 | 24 | 24 | 161 |
| Total | 232 | 50 | 49 | 331 |
Validation data set was used for hyperparameter tuning and test set was used for evaluating the model accuracy. The hyperparameters used to tune each model are given in Table II.
Table 2. Hyperparameters used in the training.
| Optimizer | SGDM |
| Mini-batch size | 16 |
| Initial learning rate | 3e-4 |
| Learning rate drop factor | 0.2 |
| Learning rate drop period | 8 |
| L2 regularization factor | 0.004 |
| Validation frequency | 16 |
| Momentum | 0.9 |
| Maximum # of epochs | 20 |
| SGDM: Stochastic gradient descent with momentum. | |
Data processing environment
This study was performed on a LENOVO Intel® Core™ i7-9750H / Y540 / 16G / 512 GeForce RTX2060 computer and in MATLAB® R2018b environment. A source code, including also MATLAB® commands provided by Deep Learning Toolbox™, was prepared by using MATLAB macro environment.
Transfer learning, data augmentation
The CNN algorithm requires a large number of images to train the network. When the number of images is not sufficient to train the network, transfer learning can be used where an already trained CNN model with large number of images is reused as a starting point for pattern recognition. Several such networks are available such as AlexNet, VGG, ResNet, MobileNet, DenseNet, and SqueezeNet. In fairly recent studies, these networks have been applied effectively for classifying medical images.[12-15] In this study, to classify 331 images, we have applied VGG-16,[9] VGG-19,[9,16] Resnet-101,[17] DenseNet-201[11] pre-trained CNN networks.
Statistical analysis
Statistical analysis was performed using the MATLAB R2018b (MathWorks, Natick, MA, USA). As a result of the classification, performances of the networks were evaluated with performance parameters such as accuracy, sensitivity, specificity, and precision. These performance metrics were calculated from the confusion matrix obtained during the testing of the models (TP= True Positive; FP= False Positive; TN= True Negative; FN= False Negative).
accuracy=(TP+TN)/(TP+TN+FP+FN)
sensitivity (recall)=TP/(TP+FN)
specificity=TN/(TN+FP)
precision=TP/(TP+FP)
Results
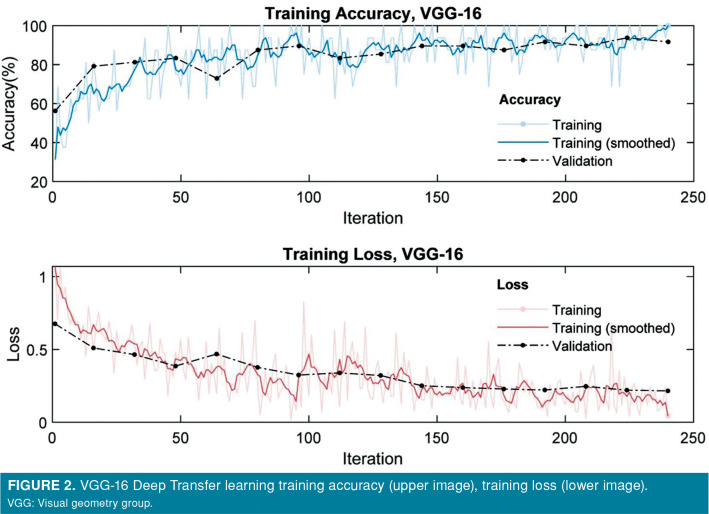
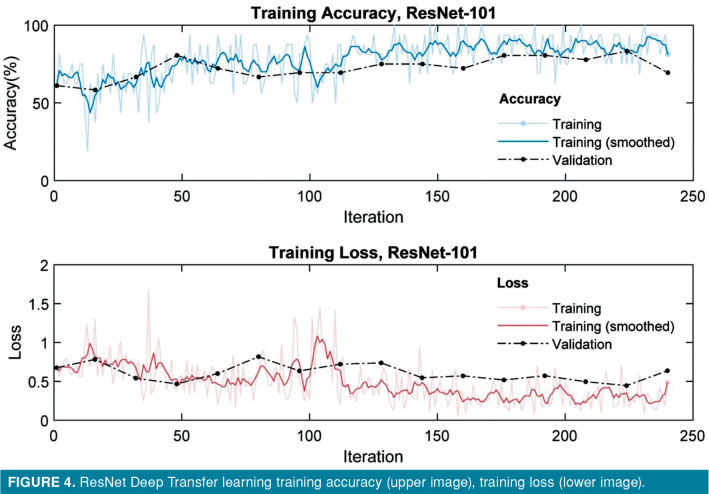
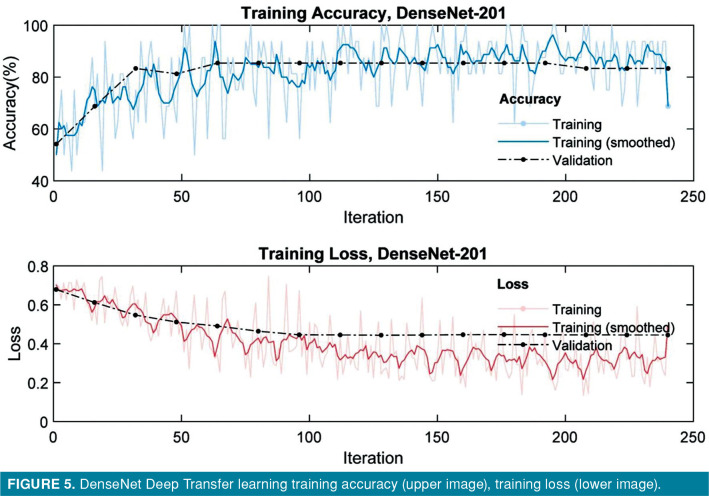
The accuracy and loss of VGG-16, VGG-19, Resnet-101, DenseNet-201 model for training and validation sets are presented in plots shown in Figures 2-5. Both training and validation losses decreased and converge as the iteration increased, whereas the accuracy increased for those data sets as the iteration increased. All models were run for 20 epochs; training time was 5 min on VGG-16 network, 6 min on VGG-19 network, 6 min on ResNet network, and 33 min on DenseNet network.
Figure 2. VGG-16 Deep Transfer learning training accuracy (upper image), training loss (lower image). VGG: Visual geometry group.
Figure 3. VGG-19 Deep Transfer learning training accuracy (upper image), training loss (lower image). VGG: Visual geometry group.
Figure 4. ResNet Deep Transfer learning training accuracy (upper image), training loss (lower image).
Figure 5. DenseNet Deep Transfer learning training accuracy (upper image), training loss (lower image).
The model performance was evaluated in terms of performance parameters in confusion matrices (Figure 6). The deep transfer learning models were tested with the images, which were separate from neither training nor validating data sets. The overall accuracy achieved by the VGG-16, VGG-19, Resnet-101, DenseNet-201 models were 93.9%, 91.8%, 89.8% and 85.7% respectively. Table III shows the results obtained by testing the models.
Figure 6. Confusion matrices for deep transfer learning using VGG-16, VGG-19, ResNet-101, DenseNet-201. VGG: Visual geometry group.
Table 3. Performance metrics obtained by testing pre-trained VGG-16, VGG-19, ResNet-101, DenseNet-201 models.
| VGG-16 | VGG-19 | Resnet-101 | DenseNet-201 | |
| Accuracy (%) | 93.9 | 91.8 | 89.8 | 85.7 |
| Sensitivity (%) | 95.8 | 92.0 | 95.5 | 84.6 |
| Specificity (%) | 92.0 | 91.7 | 85.2 | 87.0 |
| Precision (%) | 92.0 | 92.0 | 84.0 | 88.0 |
| VGG: Visual geometry group. | ||||
Discussion
In the current study, we attempted to classify normal and pathological cervical radiographs using VGG-16, VGG-19, ResNet-101, DenseNet-201 networks where current methods of data augmentation and transfer learning were applied. Among the pre-trained networks used, pre-trained VGG-16 network outperformed other models in terms of accuracy (93.9%), sensitivity (95.8%), specificity (92.0%), and precision (92.0%). In this study, VGG networks performed a bit better than others. We observed similar performance values in our previous works, as well.[18] This may be due to differences in the kernel size or the architecture of the networks. The kernel size (receptive-field filter size) of VGG networks is 3x3, the ResNet kernel size is 7x7, and the DenseNet kernel size is 7x7. Kernel is the filter size applied on the input image, and data loss is high with a large kernel size.
Cervical radiography is the first step imaging method for evaluating patients with neck pain. This method is widely used for its applicability.[1] However, radiologists are not always available and, sometimes, it is not possible to reach them under daily workload. The labeling and separating normal radiographic images through deep learning methods enable radiologist to use their time more efficiently by sparing more time for analyzing pathological radiographs, which increases the effectiveness of the diagnosis activities. The objective of this study is to distinguish between normal and pathological cervical radiographs by artificial intelligence and deep learning methods.
The need for large amounts of training data poses a challenge for deep learning within radiology, as large data sets with accurate annotations are rarely directly available. Therefore, many of the deep learning-based projects used in radiology would require a significant manual effort to collect the data needed before starting training. As a result, access to relevant training data limits the development and spread of applications based on deep learning in radiology.[19]
There are successful clinical studies evaluating changes in the skeletal system due to trauma or osteoarthritis with DCNN technology[2,20-23] which has been used in computer plain imaging, particularly in the field of medical radiology.[24-31] Accordingly, this method has become increasingly popular in musculoskeletal radiology. Recently, Al Arif et al.[32] defined a fully automatic cervical vertebra segmentation framework for radiographic images in cervical trauma patients.
For the osteoarthritis diagnosis, transfer learning has successfully applied resulting in high classification performance.[18,23] There are magnetic resonance and computerized tomography imaging studies on intervertebral disc lesions that use CNN as an artificial intelligence method. Several studies related to the use of this technique in the spine have been reported.[33-37] Galbusero et al.[33] showed that, in biplanar spine radiographs, kyphosis, lordosis, Cobb angle of scoliosis, pelvic incidence, sacral slope, and pelvic tilt evaluations were successful using deep learning approach. Jamaludin et al.[34] showed that the usefulness of automation of reading of radiological features from magnetic resonance images in patients with lumbar pain. Computer-aided diagnosis of degenerative intervertebral disc diseases from lumbar magnetic resonance images has been reported previously.[35,36] Galbusera et al.[37] demonstrated the successful recognition of radiological anatomic points on lumbar spine lateral radiographs using neural network-based methods.
One of the limitations of our study is that it evaluates whether there is only cervical degenerative change or disc space narrowing in patients presenting with neck pain. The prevalence of osteoarthritic changes in the cervical spine with advancing age decreases, the expected benefit of using this method in this age group. In addition, it cannot make the differential diagnosis of conditions that can cause neck pain such as trauma, tumor, rheumatic diseases. However, our study may be a guide for studies that can make more detailed radiological analysis.
In conclusion, our study results suggest that deep transfer learning methods may be a beneficial assistant for evaluation of cervical arthrosis. To the best of our knowledge, this is the first study that evaluates cervical osteoarthritis. We suggest further large-scale, prospective studies to draw more reliable conclusions.
Footnotes
Conflict of Interest: The authors declared no conflicts of interest with respect to the authorship and/or publication of this article.
Financial Disclosure: The authors received no financial support for the research and/or authorship of this article.
References
- 1.Cohen SP. Epidemiology, diagnosis, and treatment of neck pain. Mayo Clin Proc. 2015;90:284–299. doi: 10.1016/j.mayocp.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Lim J, Kim J, Cheon S. A deep neural network-based method for early detection of osteoarthritis using statistical data. Int J Environ Res Public Health. 2019;16:1281–1281. doi: 10.3390/ijerph16071281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerlikowske K, Carney PA, Geller B, Mandelson MT, Taplin SH, Malvin K, et al. Performance of screening mammography among women with and without a firstdegree relative with breast cancer. Ann Intern Med. 2000;133:855–863. doi: 10.7326/0003-4819-133-11-200012050-00009. [DOI] [PubMed] [Google Scholar]
- 4.Someeh N, Asghari Jafarabadi M, Shamshirgaran SM, Farzipoor F. The outcome in patients with brain stroke: A deep learning neural network modeling. J Res Med Sci. 2020;25:78–78. doi: 10.4103/jrms.JRMS_268_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen TG, Jackson BH, Feeser TA, Kent MN, Moad JC, Krishnamurthy S, et al. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform. 2018;9:32–32. doi: 10.4103/jpi.jpi_31_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L, Walker B, Liang PI, Tong Y, Xu C, Su YC, et al. Colorectal cancer detection based on deep learning. J Pathol Inform. 2020;11:28–28. doi: 10.4103/jpi.jpi_68_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samala RK, Chan HP, Hadjiiski LM, Helvie MA, Cha KH, Richter CD. Multi-task transfer learning deep convolutional neural network: Application to computeraided diagnosis of breast cancer on mammograms. Phys Med Biol. 2017;62:8894–8908. doi: 10.1088/1361-6560/aa93d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks. Advances in Neural Information Processing Systems. 2014;27:3320–3328. [Google Scholar]
- 9.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition; 3rd International Conference on Learning Representations (ICLR); May 7-9, 2015; San Diego. 2015.. arXiv: 1409.1556. ICLR. [Google Scholar]
- 10.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016:770–778. [Google Scholar]
- 11.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017:2261–2269. [Google Scholar]
- 12.Polat H, Danaei Mehr H. Classification of pulmonary CT images by using hybrid 3D-deep convolutional neural network architecture. Applied Sciences. 2019;9:940–940. [Google Scholar]
- 13.Yahalomi E, Chernofsky M, Werman M, editors. Detection of distal radius fractures trained by a small set of X-ray images and faster R-CNN. Cham: Springer; 2019. [Google Scholar]
- 14.Kim DH, MacKinnon T. Artificial intelligence in fracture detection: Transfer learning from deep convolutional neural networks. Clin Radiol. 2018;73:439–445. doi: 10.1016/j.crad.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, et al. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. 2017;88:581–586. doi: 10.1080/17453674.2017.1344459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. [Google Scholar]
- 17.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015:1–9. [Google Scholar]
- 18.Üreten K, Erbay H, Maraş HH. Detection of hand osteoarthritis from hand radiographs using convolutional neural networks with transfer learning. Turkish Journal of Electrical Engineering And Computer Sciences. 2020;28:2968–2978. [Google Scholar]
- 19.Greenspan H, van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Transactions on Medical Imaging. 2016;35:1153–1159. [Google Scholar]
- 20.Cheng CT, Ho TY, Lee TY, Chang CC, Chou CC, Chen CC, et al. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur Radiol. 2019;29:5469–5477. doi: 10.1007/s00330-019-06167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyaz S, Açıcı K, Sümer E. Femoral neck fracture detection in X-ray images using deep learning and genetic algorithm approaches. Jt Dis Relat Surg. 2020;31:175–183. doi: 10.5606/ehc.2020.72163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badgeley MA, Zech JR, Oakden-Rayner L, Glicksberg BS, Liu M, Gale W, et al. Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit Med. 2019;2:31–31. doi: 10.1038/s41746-019-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Üreten K, Arslan T, Gültekin KE, Demir AND, Özer HF, Bilgili Y. Detection of hip osteoarthritis by using plain pelvic radiographs with deep learning methods. Skeletal Radiol. 2020;49:1369–1374. doi: 10.1007/s00256-020-03433-9. [DOI] [PubMed] [Google Scholar]
- 24.Arif M, Schoots IG, Castillo Tovar J, Bangma CH, Krestin GP, Roobol MJ, et al. Clinically significant prostate cancer detection and segmentation in low-risk patients using a convolutional neural network on multi-parametric MRI. Eur Radiol. 2020;30:6582–6592. doi: 10.1007/s00330-020-07008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu M, Yan H, Xia J, Zhu L, Zhang T, Zhu Z, et al. Deep convolutional neural networks for tongue squamous cell carcinoma classification using Raman spectroscopy. Photodiagnosis Photodyn Ther. 2019;26:430–435. doi: 10.1016/j.pdpdt.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Kim TK, Yi PH, Wei J, Shin JW, Hager G, Hui FK, et al. Deep learning method for automated classification of anteroposterior and posteroanterior chest radiographs. J Digit Imaging. 2019;32:925–930. doi: 10.1007/s10278-019-00208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakhani P, Sundaram B. Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 28.Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S, et al. A new 2. 5D representation for lymph node detection using random sets of deep convolutional neural network observations. Med Image Comput Comput Assist Interv. 2014;17:520–527. doi: 10.1007/978-3-319-10404-1_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trebeschi S, van Griethuysen JJM, Lambregts DMJ, Lahaye MJ, Parmar C, Bakers FCH, et al. Deep learning for fullyautomated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep. 2017;7:5301–5301. doi: 10.1038/s41598-017-05728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghafoorian M, Karssemeijer N, Heskes T, van Uden IWM, Sanchez CI, Litjens G, et al. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci Rep. 2017;7:5110–5110. doi: 10.1038/s41598-017-05300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Açıcı , K , Sümer E, Beyaz S. Comparison of different machine learning approaches to detect femoral neck fractures in X-ray images. Health Technol. 2021;11:643–653. [Google Scholar]
- 32.Al Arif SMMR, Knapp K, Slabaugh G. Fully automatic cervical vertebrae segmentation framework for X-ray images. Comput Methods Programs Biomed. 2018;157:95–111. doi: 10.1016/j.cmpb.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Galbusera F, Niemeyer F, Wilke HJ, Bassani T, Casaroli G, Anania C, et al. Fully automated radiological analysis of spinal disorders and deformities: A deep learning approach. Eur Spine J. 2019;28:951–960. doi: 10.1007/s00586-019-05944-z. [DOI] [PubMed] [Google Scholar]
- 34.Jamaludin A, Lootus M, Kadir T, Zisserman A, Urban J, Battié MC, et al. ISSLS PRIZE IN BIOENGINEERING SCIENCE 2017: Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J. 2017;26:1374–1383. doi: 10.1007/s00586-017-4956-3. [DOI] [PubMed] [Google Scholar]
- 35.Koh J, Chaudhary V, Dhillon G. Disc herniation diagnosis in MRI using a CAD framework and a two-level classifier. Int J Comput Assist Radiol Surg. 2012;7:861–869. doi: 10.1007/s11548-012-0674-9. [DOI] [PubMed] [Google Scholar]
- 36.Oktay AB, Albayrak NB, Akgul YS. Computer aided diagnosis of degenerative intervertebral disc diseases from lumbar MR images. Comput Med Imaging Graph. 2014;38:613–619. doi: 10.1016/j.compmedimag.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Galbusera F, Bassani T, Costa F, Brayda-Bruno M, Zerbi A, Wilke HJ. Artificial neural networks for the recognition of vertebral landmarks in the lumbar spine. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization. 2018;6:447–452. [Google Scholar]