Abstract
The role of the androgen receptor (AR) in the progression of prostate cancer (PCa) is well established and competitive inhibition of AR ligand binding domain (LBD) has been the mainstay of antiandrogen therapies for advanced and metastatic disease. However, the efficacy of such drugs is often limited by the emergence of resistance, mediated through point mutations and receptor splice variants lacking the AR-LBD. As a result, the prognosis for patients with malignant, castrate-resistant disease remains poor. The amino terminal domain (NTD) of the AR has been shown to be critical for AR function. Its modular activation function (AF-1) is important for both gene regulation and participation in protein–protein interactions. However, due to the intrinsically disordered structure of the domain, its potential as a candidate for therapeutic intervention has been generally overlooked. In this article, we describe the design and development of a functional cell-based assay aimed at identifying small-molecule inhibitors of the AR-NTD. We demonstrate the suitability of the assay for high-throughput screening platforms and validate two initial hits emerging from a small, targeted, library screen in PCa cells.
Keywords: androgen receptor, prostate cancer, castrate-resistant prostate cancer, AR-vs, intrinsically disordered structure, high-throughput screen
Introduction
Worldwide, prostate cancer (PCa) is the second most common cancer of men with an estimated 1.4 million new cases and 375,000 deaths in 2020.1 Blocking androgen receptor (AR) signaling is the mainstay treatment in advanced and metastatic disease and half of men diagnosed with advanced disease are likely to receive hormone therapy. Currently approved drugs block synthesis (e.g., abiraterone) of natural hormones or work as competitive antagonists (e.g., enzalutamide, apalutamide, and darolutamide), binding to the receptor and shutting down the normal signaling cascade.2 The effectiveness of such treatments is eventually compromised by the emergence of resistance leading to lethal castrate-resistant prostate cancer (CRPC), for which there are currently no targeted therapies.3–6
Resistance to current antiandrogens can be multifactorial in origin4,7,8 and includes point mutations that decrease the efficacy of antiandrogen (see Ref. 9) gene rearrangements causing overexpression of the receptor gene10–12 and splice variants lacking the ligand binding domain (LBD).13–16 Darolutamide, and apalutamide, new antiandrogens, recently approved for nonmetastatic CRPC, while effective against known receptor point mutations,17,18 remain limited to targeting the hormone-binding function of the receptor. The amino-terminal domain (NTD) of the AR is essential for receptor function and targeting this region of the protein would be an alternative, but equally effective and selective means of blocking AR activity.19,20
The AR-NTD is structurally intrinsically disordered, is functionally important, underpins potential allosteric regulation, and supports multiple protein–protein interactions involved in target gene regulation (reviewed in Refs. 19 and 21). However, it is a challenging drug target, unsuitable for structure-based drug discovery.19,22 In a ground-breaking study, the group of Marianne Sadar identified a novel small molecule, EPI-001, which bound covalently to and effectively inhibited AR activity.23,24 EPI-001, uniquely, bound the structurally flexible AR-NTD22,23,25 and selectively inhibited receptor–protein interactions.23,24
Moreover, this molecule reduced tumor growth and volume in preclinical models of PCa in vivo.23–25 EPI-506, an analog of EPI-001, was studied in Phase I/II clinical trials (NCT02606123), which ended in 2018, with modest responses observed. Second-generation EPI compounds have been described and are currently in clinical trial (NCT04421222) (see Ref. 26). However, recent work has proposed alternative, AR-independent, mechanisms of action for EPI-001 involving the peroxisome proliferator-activated receptor.27 Although this effect was seen at higher concentrations compared to the reported AR inhibition, it has raised questions of selectivity.
As a consequence of androgen ablation therapy, splice variants in the AR arise through incorporation of cryptic exons located between exon 3 and 4.28 Research has focused primarily on AR splice variant (AR-v)7 and AR-v12 (ARV567es), the former incorporates a cryptic exon adding 16 unique amino acid C-terminal of the AR-DNA binding domain (DBD), while AR-v12 splices exon 8 next to exon 4 (hinge region) resulting in the addition of 10 amino acids.28 While initial reports on receptor splice variants focused on their ability to interact and modulate AR-FL activity, there is now compelling evidence for DNA binding by heterodimers (AR-FL/AR-v), homodimers (AR-v/AR-v), or AR-v monomers.29,30
Crucially, there is growing evidence for AR-vs acting independent of AR-FL and/or hormone to regulate a unique gene signature.28,31–33 Recent studies have indicated that 75% of patients with castrate-resistant disease express constitutively active AR variants lacking the LBD,14 which evade all forms of currently available AR-targeting treatment. CRPC is still driven by AR overexpression and/or splice variants that have lost their ligand-binding regulation and there is priority to identify inhibitors of the AR-NTD to maximize the opportunity of developing new drugs to meet the major clinical challenge of treating CRPC. In this study, we describe the development and validation of a cell-based high-throughput assay for screening and identifying inhibitors of the AR-NTD.
Materials and Methods
Cloning
The pCDNA4/TO/myc-HisB vector was used as the backbone for the newly developed pcDNA4/TO/myc-His AR-v and glucocorticoid receptor N-terminal and DNA binding domain (GRNTD-DBD) expression plasmids. The pcDNA 4/TO/myc-HisB vector (Invitrogen) was sequentially digested with BamHI and HindIII to produce a linear plasmid. AR-v or GRNTD-DBD was amplified using the template plasmids pSVARo and phGR, respectively, using the primers outlined in Supplementary Table S1. Briefly, PCR reactions were set up using the Expand High Fidelity Kit (Roche) and amplification checked on a 1% agarose gel. The remaining amplified DNA was treated with Dpnl to digest methylated parental template DNA and purified (Qiagen quickspin PCR clean-up). An In Fusion™ (Takara Clontech) cloning reaction was then set up for each amplified fragment followed by transformation of Stellar competent cells.
In the case of the pGL4.26-GRE2 luciferase reporter vector, oligonucleotides were designed based on the sequence of the pGRE2-TATA-luciferase plasmid (Supplementary Table S1) (Eurofins MWG Operon). The oligonucleotides contained 15 bps of homology with the target pGL4.26-Luciferase vector, as required for directional In-Fusion cloning. The pGL4.26-Luciferase vector was sequentially digested with BglII and XhoI to give a linear vector, incubated with the annealed glucocorticoid response element (GRE)2 oligonucleotides, and cloned using In-Fusion reaction.
Positive clones for both the expression and reporter constructs were identified by colony PCR screening, analyzed by restriction enzyme digestion, and sequenced to confirm cloning and absence of mutations. Sequencing primers are listed in Supplementary Table S2.
Cell Culture
TREx-293 and VCaP cell lines were obtained from Invitrogen and ATCC, respectively. Cells were routinely grown in T-75 cm2 flasks in Dulbecco's Modified Eagle Medium (DMEM) high glucose (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Sigma).
Transient Transfection of Mammalian Cells
Transient transfection was carried out using the JetPEI® DNA transfection reagent (Polypus-transfection SA). The JetPEI mixture was added to the DNA mix, vortexed briefly, and incubated at room temperature for 30 min. Fresh media and 100 μL of the JetPEI/DNA reaction mix were added to cells in a 24-well plate and incubated for 24 h at 37°C and 5% CO2.
Stable Transfection
TREx-293 or TREx-GRE2 cells were grown in DMEM high glucose (Life Technologies) +10% FBS +5 μg/mL blasticidin (+ 50 μg/mL hygromycin for TREx-GRE2 cells). Cells were transfected with 4 μg of pGL4.26-GRE2 or pcDNA4-TO-AR-v or pcDNA4-TO-GRNTD-DBD using JetPEI reagent according to the manufacturer's protocol. Cells were incubated for 48 h at 37°C and 5% CO2 before medium was replaced with DMEM +10% FBS +5 μg/mL blasticidin plus selection antibiotics as appropriate (−100 μg/mL hygromycin or 200 μg/mL zeocin). Medium containing selection antibiotic was replenished every 2 days until colony formation was observed. Cells were then split 1:3 and grown to 60% confluence in T-75cm2 flasks containing selection antibiotic(s). This process was repeated twice to generate stocks of polyclonal cells.
After the third passage, antibiotic concentration was reduced to 50 μg/mL hygromycin or 100 μg/mL zeocin, as appropriate for maintenance.
Monoclonal cultures of TREx-GRE2 -AR-v or GRNTD-DBD cell lines were generated using limiting dilution in 96-well plates. Cells were split and seeded at a density of 10 cells/well in DMEM +10% FBS +5 μg/mL blasticidin +50 μg/mL hygromycin +100 μg/mL zeocin. Once cells reached 40% confluence, they were split again and seeded at a density of 1 cell/well in 96-well plates. This process was repeated twice. Wells with a single cell were identified using light microscopy. Once cells in these wells reached 60% confluence, they were trypsinized and transferred to six-well plates. They were grown to 80% confluence and transferred to T-75 cm2 flasks for propagation. At each passage in T-75 cm2 flasks, a portion of cells were frozen for long-term storage in liquid nitrogen.
Luciferase reporter gene assay I
Cells were seeded in 24-well plates at a density of 5 × 104 cells per well in maintenance medium with 10% charcoal stripped FBS (Sigma). After 24 h, the medium was replaced, and cells were transfected with reporter and receptor expression plasmids as required. For transient transfection, the medium was changed after a further 24 h, and cells were treated with drugs, lead compounds, or hormone at concentrations indicated. Cells stably expressing TREx-GRE2-AR-v/-GRNTD-DBD AR or GR polypeptides were induced with 1 μg/mL tetracycline, and were treated with drug or test compounds as described. After drug treatments for 24 h, cells were washed with phosphate-buffered saline, lysed using 1 × passive lysis buffer (Promega) with agitation for 15 min at room temperature, and cell debris removed by centrifugation.
The supernatants were transferred to new Eppendorf tubes on ice and 17 μL of each lysate was placed, in triplicate, in white-walled 96-well plates. Luciferase activity was measured using a Glomax 96 microplate luminometer (Promega) with an injection of 100 μL/well luciferase assay buffer (13 mM MgSO4.7H2O, 30 mM GlyGly pH 7.8, 1.7 mM Na2ATP, and 11 μM luciferin). Relative light units (RLUs), which signified luciferase activity, were normalized for protein levels by dividing the raw RLUs by the sample protein concentration, which was determined by DC™ protein assay (BIO-RAD).
Luciferase reporter gene assay II (high-throughput screening)
An Echo 550 acoustic liquid handler (Labcyte, Inc.) was used to transfer test compounds from the BioAscent compound cloud,34,35 the NIH Clinical Collection (NCC), or Selleckchem FDA-approved drug library compounds (6–10 mM stock concentration in dimethyl-sulfoxide [DMSO]) from 384-well low dead volume source plates (Labcyte, #LP-0200) into white-walled 384-well assay plates (Greiner, #781080). All wells were treated with the same final concentration of DMSO. Twenty-five microliters of TREx-GRE2 or TREx-GRE2-AR-v/-GRNTD-DBD cells was seeded at a density of 50,000 cells/mL in DMEM (without phenol red) plus 10% charcoal stripped FBS +1 μg/mL tetracycline using a WellMate automated liquid handler (Thermo Scientific Matrix).
Maximum inhibition control cells not treated with tetracycline. plates were incubated at room temperature for 1 h, before incubation for 16 h at 37°C, 5% CO2 and 95% O2. After 24 h, OneGlo EX reagent was reconstituted and 15 μL was added to each well with a WellMate. Plates were incubated for 3 min at room temperature in the dark, before RLUs were measured using an EnVision (PerkinElmer) or PHERAstar (BMG Labtech) microplate reader. Data were exported into Excel and analyzed using ActivityBase (ID Business Solutions) and Vortex Software (Dotmatics). An high-throughput screen (HTS) assay protocol is included as Supplementary Table S3.
Resazurin (Alamar blue) cell viability assay
The reduction of resazurin (7-hydroxy-3H-phenoxazin-3-one 10-oxide) to red fluorescent resorufin in the mitochondria of viable cells by NADPH dehydrogenase results in fluorescence proportional to the number of viable cells in a population. Test compounds were transferred as described previously into black-walled, clear-bottom 384-well assay plates, creating 7- or 10-point dose–response curves with a maximum final assay concentration of 40 μM and final DMSO concentration of 0.67%. Positive control cells were treated with 40 μM doxorubicin and negative control cells were treated with DMSO only at a final concentration of 0.67% (Supplementary Fig. S5). TREx-GRE2 or TREx-GRE2/AR-NTD cells were seeded at 50,000 cells/mL (25 μL/well) in DMEM (without phenol red) plus 10% C/S FBS (+1 μg/mL tetracycline where appropriate).
Plates were incubated at room temperature for 1 h before incubation at 37°C and 5% CO2. After 24 h, resazurin reagent (Sigma) was defrosted and diluted 1 in 92.5 in DMEM +10% C/S FBS to give a working concentration of 270 μM. Resazurin reagent (5 μL) was added to each well of the assay plate using an automatic multichannel pipette to give a final assay concentration of 45 μM. Cells were incubated for 60–90 min at 37°C, before fluorescence was measured using an EnSpire (PerkinElmer) microplate reader at a single time point, excitation 560 nm and emission 587 nm. Data were exported for analysis in ActivityBase Software.
CellTox™ Green cytotoxicity assay
The CellTox™ Green cytotoxicity assay (Promega) uses an asymmetric cyanine dye to bind DNA of cells with compromised membrane integrity. Fluorescence is proportional to the number of dead cells. Test compounds were transferred as described previously into a black-walled, clear-bottom 384-well assay plate. All wells were treated with DMSO at a final concentration of either 0.1% or 0.25% and positive control cells were treated with 0.6 μL cell lysis reagent/well. No cell control was included.
TREx-GRE2/AR-NTD cells were seeded at 50,000 cells/mL (25 μL/well) using a WellMate automatic liquid handler in DMEM (without phenol red) +10% C/S FBS +1 μg/mL tetracycline and incubated at room temperature for 1 h before incubation overnight at 37°C and 5% CO2. The following day, 5 μL 5 × CellTox Green reagent was added to each well using an automatic multichannel pipette. Plates were incubated at room temperature in the dark for 90 min before fluorescence intensity, relative fluorescence units, was measured using an EnSpire plate reader: excitation 490 nm and emission 525 nm. Data were exported for analysis in GraphPad Prism or ActivityBase software.
Enzyme Inhibition Assay: Purified Recombinant Luciferase
Test compounds were transferred as described previously into white-walled opaque 384-well assay plates. Purified recombinant firefly luciferase (QuantiLum, Promega) was diluted 106-fold in cell lysis buffer (Promega) plus 1 mg/mL bovine serum albumin (BSA) and 1 fmole/L was preincubated with compound for 30 min before addition of 10 μL SteadyGlo reagent. Negative control samples contained BSA only. After 3 min of incubation at room temperature in the dark, luminescence was measured using the EnVision microplate reader. Data were exported to Excel and analyzed in ActivityBase.
Western Blotting
Cells were lysed using passive lysis buffer (Promega) and proteins resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Hybond-P PVDF membrane (Amersham). Membranes were incubated with primary antibody overnight at 4°C and after wash steps, incubated for 1 h at room temperature with secondary antibody conjugated to horseradish peroxidase. Antibody–protein complexes were detected by enhanced chemiluminescence using a Li COR Odyssey Imaging System.
Statistical Analysis
Statistical analysis was carried out using parametric or nonparametric (Kruskal–Wallis) analysis of variance (ANOVA) followed by Dunnett's post-test comparing treated groups with vehicle control. All results are expressed as mean ± standard error of the mean, unless stated otherwise, values were considered significant when p < 0.05. The analysis was performed using GraphPad Prism 5.0. Robust statistics were utilized for HTS applications and are outlined in detail in main text.
Results and Discussion
Construction of Reporter Cell Line
The AR-NTD is essential for receptor activity and there is a growing recognition that targeting this region with small molecule inhibitors would be clinically beneficial (Fig. 1A). However, the intrinsically disordered structure of the AR-NTD renders the use of structure-based drug design approaches limited.
Fig. 1.
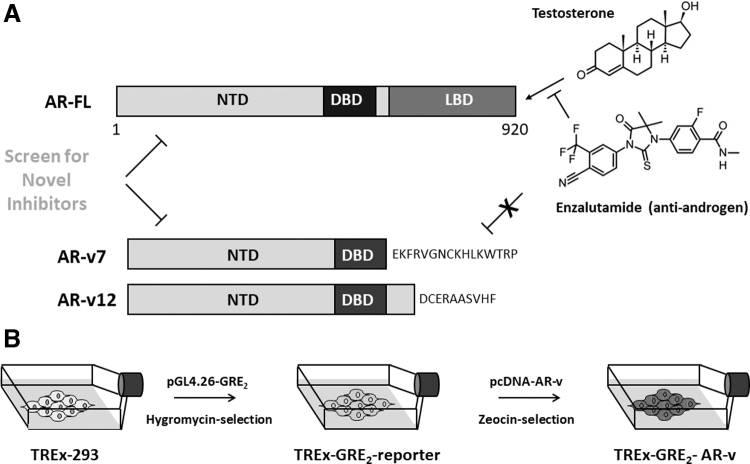
AR splice variant screening assay. (A) Schematic representation of the AR-FL and splice variants AR-v7 and AR-v12 lacking the LBD. The figure illustrates binding of testosterone, and the antiandrogen enzalutamide to the AR-FL, but not AR-vs. (B) Two-step design for the generation of double-stable cell lines expressing a receptor-dependent reporter gene and an inducible AR-v polypeptide. AR-FL, full-length androgen receptor; AR-v, androgen receptor splice variant; DBD, DNA binding domain; LBD, ligand binding domain; NTD, amino terminal domain.
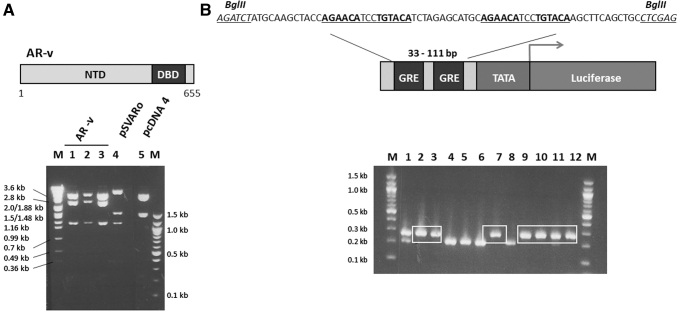
To overcome this difficulty, we have developed a cell-based functional assay to allow high-throughput screening of large compound libraries for candidate inhibitors of the AR-NTD (Fig. 1B). The AR NTD-DBD (amino acids 1 to 655) representing a generic AR-v, lacking the LBD, but retaining the amino-terminal and DBDs (NTD and DBD respectively), was cloned as a PCR fragment (1,997 bp) into the pCDNA4/TO/myc-His vector (Supplementary Fig. S1), creating a tetracycline-inducible expression construct for AR-v. Positive clones were confirmed by diagnostic restriction enzyme digests (Fig. 2A) and DNA sequencing.
Fig. 2.
Design and construction of expression and reporter gene plasmids. (A) Schematic representation of AR-v, amino acids 1–655, representing a generic splice variant containing the nuclear localization signal and part of the hinge domain. Agarose gel illustrating the expected digestion pattern of three fragments of 1,110, 2,335, and 3,443 bp for pcDNA-AR-v after cleavage with NcoI restriction enzyme (lanes 1 to 3). For comparison, the parental pcDNA4/TO/myc-HisB plasmid and pSVARo plasmids were also digested with NcoI. The base-pair size DNA ladder (M) is shown. (B) The upstream region of the GRE2-TATA-Luciferase vector containing two palindromic hormone response elements (highlighted in bold and underlined). Oligonucleotides containing this sequence were designed with flanking asymmetric BglII and XhoI restriction sites (italics and underlined), and cloned into the pGL4.26-Luciferase vector. Following cloning and transformation of pGL4.26-GRE2-luc, single colonies were selected from the plate and subjected to a colony PCR reaction and products resolved on a 2% agarose gel. A 78bp increase can be detected in positive clones of the pGL4.26-GRE2-Luciferase vector where the hormone response elements have been introduced compared to the parental pGL4.26-Luciferase plasmid, seen in lanes 2, 3, 7, 9, to 12. Selected sizes for the DNA ladder (M) are shown.
The pCDNA4/TO/myc-His plasmid was chosen as it has two tetracycline operator 2 (TetO2) sites within the human cytomegalo-virus promoter, allowing tetracycline-regulated expression of integrated genes.36 These TetO2 sequences interact with the stably expressed tetracycline repressor protein preventing expression of the transfected protein in the absence of tetracycline. This was deemed important as steroid receptors lacking the LBD are constitutively active and have been shown to cause inhibition of gene transcription and cell growth due to overexpression, a phenomenon called “squelching” (Ref. 37 and references therein). In parallel, a luciferase reporter gene driven by two GREs from the tyrosine amino transferase gene were cloned upstream of the TATA box in plasmid pGL4.26, to create pGL-GRE2-luciferase (Fig. 2B) and positive clones confirmed by DNA sequencing.
To create a double stable reporter cell assay suitable for high-throughput screening, the human embryonic kidney cell line HEK (TREx) 293 was chosen (Fig. 1B) as these cells are AR negative and easily transfected. These cells have the additional advantage of stably expressing the tetracycline repressor protein and thus give an inducible expression system. HEK (TREx) 293 cells were transfected with pGL-GRE2-luciferase. Stable transfectants were selected after treatment with the antibiotic, hygromycin (100 μg/mL), creating the stable reporter cell line TREx-GRE2 (Fig. 1B).
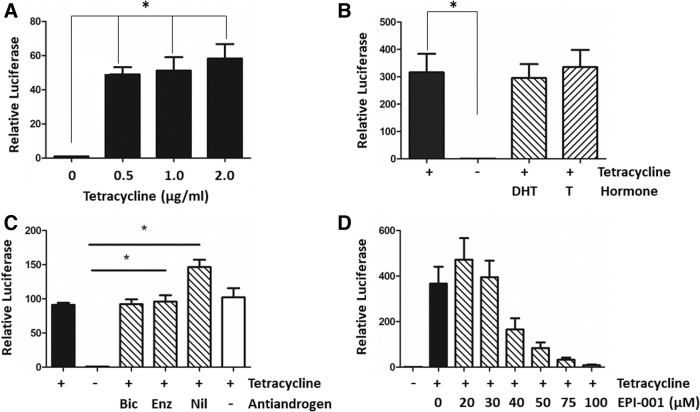
To confirm inducible expression of the AR-v polypeptide and a robust functional readout from the integrated luciferase reporter gene, TREx-293 or TREx-GRE2 cells were transiently transfected with pcDNA-AR-v. Increasing concentrations of tetracycline resulted in strong induction of luciferase activity and AR-v polypeptide expression (Fig. 3A and Supplementary Fig. S2).
Fig. 3.
Validation of stable TREx-GRE2-reporter cell lines. (A) TREx-293 cells were transiently transfected with pcDNA-AR-v and pGL4.26-GRE2-luc. Chart showing luciferase reporter gene activity (relative luciferase) after induction of AR-v with increasing concentrations of tetracycline. (B, C) Effects of hormone agonists and antagonists on AR-v activity in stable cells with integrated luciferase reporter and AR-v constructs. Luciferase activity in the absence (−) or presence (+) of tetracycline (1 μg/mL). Addition of 10 nM testosterone (T) or DHT did not significantly change AR-v-dependent transcription (B). (C) AR-v-dependent transcription activity is resistant to the antiandrogens bicalutamide (Bic, 10 μM), enzalutamide (Enz, 1 μM), and nilutamide (Nil, 10 μM) that target the AR-LBD (Fig. 1A). (D) Dose-dependent inhibition of AR-v activity was seen with increasing concentrations of EPI-001, an inhibitor of the AR-NTD. Kruskal–Wallis one-way ANOVA with Dunnett's posthoc test (*p < 0.05, n = 3). ANOVA, analysis of variance; DHT, dihydro-testosterone.
The TREx-GRE2 cells (Fig. 1B) were transfected with the expression plasmid pcDNA-AR-v, and after treatment with zeocin (200 μg/mL) stable transfected clones were selected, resulting in the cell line TREx-GRE2-AR-v (Fig. 1B). Treatment of TREx-GRE2-AR-v cells with tetracycline led to almost 100 × fold induction of luciferase activity. As expected, there was no significant effect of either dihydro-testosterone (DHT) or testosterone on transcriptional activation by AR-v (Fig. 3B). Furthermore, AR-v-dependent transcriptional activity was resistant to treatment of cells with a number of nonsteroidal antiandrogens, including bicalutamide, enzalutamide, and nilutamide (Fig. 3C). Interestingly, nilutamide (10 μM) resulted in a further increase in reporter gene activity. In contrast, the recently described AR-NTD inhibitor EPI-00123 demonstrated a dose-dependent inhibition of luciferase activity, with an IC50 value of 37.90 μM (Fig. 3D).
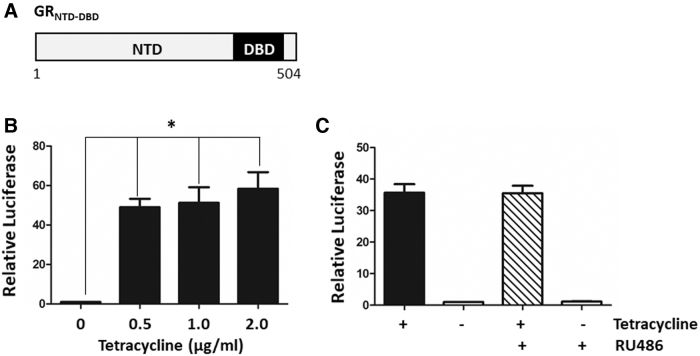
In addition to the AR-v cell line, cells expressing an inducible GR (glucocorticoid receptor) polypeptide (pcDNA-GRNTD-DBD), lacking the LBD (Fig. 4A), were constructed in a similar manner: TREx-GRE2-GRNTD-DBD. Transiently transfected TREx-293 or TREx-GRE2 cells demonstrated robust activation of the reporter gene with increasing concentrations of tetracycline and expression of GRNTD-DBD (Fig. 4B and Supplementary Fig. 2SA), and this activity was unaffected by adding 1 μM of the antagonist mifepristone (RU486) (Fig. 4C).
Fig. 4.
Stable reporter gene cell line expressing the GRNTD-DBD. (A) Schematic representation of the GRNTD-DBD polypeptide, amino acids 1 to 504. (B) Transient transfection of TREx-293 cells with pcDNA-GRNTD-DBD and pGL4.26-GRE2-luc. Expression of GRNTD-DBD with increasing concentrations of tetracycline resulted in a significant increase in luciferase activity. (C) Transient transfection of TREx-GRE2-reporter cells with pcDNA-GRNTD-DBD resulted in increased reporter gene activity (as in B), which was not inhibited with the anti-glucocorticoid RU486 (1 μM mifepristone). Kruskal–Wallis one-way ANOVA with Dunnett's posthoc test (*p < 0.05, n = 3).
Development of a High-Throughput Screening Assay for AR-v Inhibitors
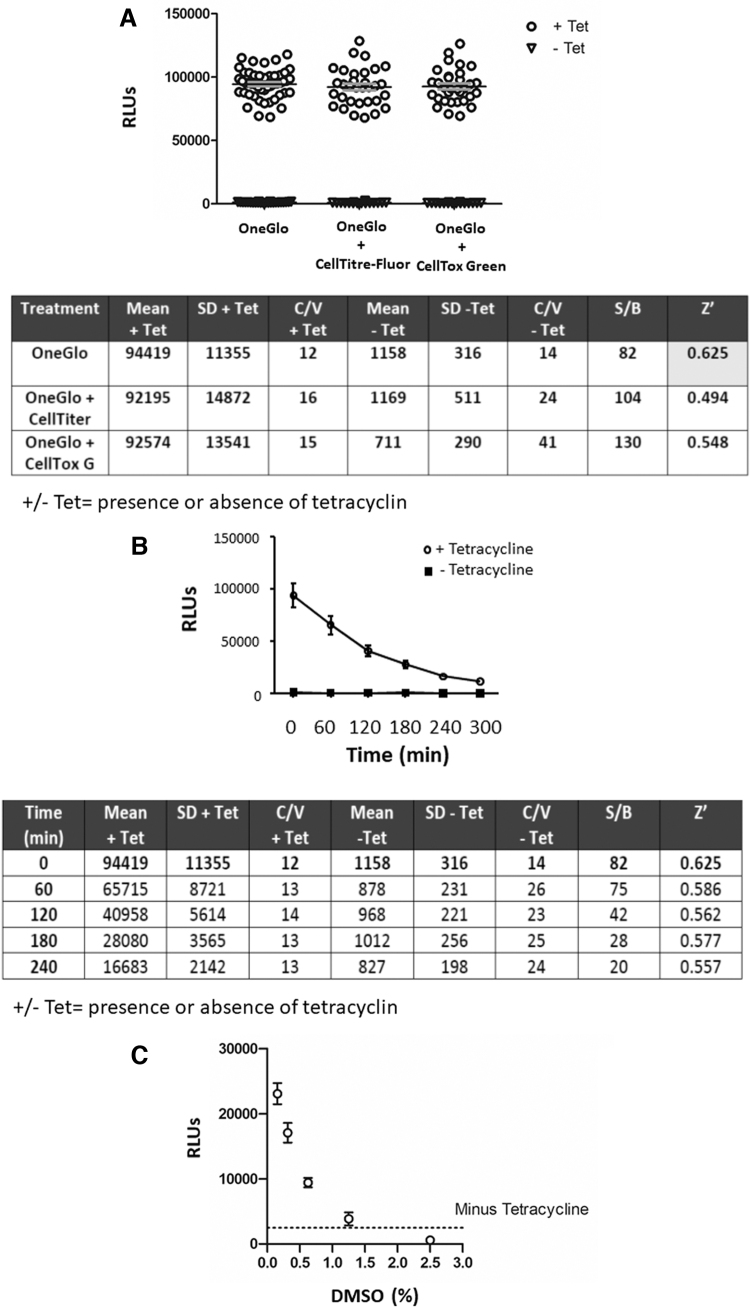
To demonstrate the suitability of TRE-x-GRE2-AR-v cells for drug screening, it was necessary to miniaturize the assay to a 384-well format and optimize a number of parameters, including cell density, induction with tetracycline and best reagents to measure luciferase activity, and cell viability. Assuring the quality of the assay is arguably the most important part of the assay development process. A high signal to background (S/B) ratio with low variability is key for identifying positive, relatively low-efficacy/affinity hit compounds.
As part of our optimization and development of a high-throughput screening assay, the minimal criteria required for accessing the European Lead Factory38,39 were used as guideline: 384-well format; no wash steps (homogenous); preferably greater than threefold signal to background ratio; Z prime (Z′) > 0.6; maximum assay volume ≤30 μL; minimum DMSO tolerance 0.5%; and stable reporter in a mycoplasma free cell line. Mean (μ), SD (σ), S/B, Z′, and coefficient of variance (%CV) were calculated for each time point, reagent type and volume, and microplate reader. The Z′ statistic is a measure of statistical effect size and the variability and is used as an indicator of assay quality The formulae for calculating Z′ and S/B are outlined below:
Optimization of TREx GRE2-AR-v Cell Assay for HTS
Seeding density of cells was optimized based on both assay signal window (S/B) and variance of controls (%CV). At the minimum density tested of 312 cells per well, an assay signal window >20-fold was achieved, up to a maximum of 50-fold change at the highest density tested. As a result, variance within the high and low controls was the driving factor in defining an appropriate seeding density. As no suitable positive control inhibitor was available at the time of the screen (EPI-001 showed considerable variance in efficacy), minimizing variance of the controls was critical to minimizing the false discovery rate during hit selection, resulting in the selection of 1,250 cells/well as the condition with the least variance (Supplementary Fig. S3A).
Screening plates were also quality controlled, with a robust Z′> 0.6 required between minimum and maximum controls on all plates (Supplementary Fig. S3). Pilot experiments also demonstrated that addition of tetracycline at the same time as the cells were seeded into assay plates gave the most consistent readout of reporter gene activity (Supplementary Fig. S3B). TREx-GRE2-AR-v cells were incubated overnight, and the following morning treated with OneGlo™ reagent alone, OneGlo and CellTiter-Fluor™ reagents, or OneGlo and CellTox Green reagents. All three data sets showed the same Max (+ Tetracycline) and Min (− Tetracycline) signals, and there no significant difference between them (Fig. 5A).
Fig. 5.
Optimization of luciferase assays using the TREx-GRE2-AR-v cell line. (A) TREx-GRE2-AR-v cells were seeded at 1,250 cells/well using the Matrix WellMate in the presence or absence of 1 μg/mL tetracycline and incubated for 60 min at room temperature before incubation at 37°C, 95% O2 and 5% CO2. Twenty-four hours later, cells were treated with OneGlo luciferase reagent and RLUs were read using the PHERAstar microplate reader. All data plotted with mean (bar). The Z′ for OneGlo alone exceeded the 0.6 threshold (highlighted), with a signal to background ratio of 82. (B) The TREx-GRE2-AR-v luciferase assay was stable up to 4 h with a Z′ exceeding 0.5. AR expression induced with tetracycline (as described in A) resulted in significant luciferase activity at all time points (mean ± SD; two-way ANOVA with Bonferonni posthoc test (p < 0.05). (C) DMSO tolerance of the TREx-GRE2-AR-v cell line in the optimized 384-well format. TREx-GRE2-AR-v cells were seeded in 384-well plates at a density of 1,250 cells/well in the presence or absence of tetracycline as indicated. DMSO was added to give final concentrations indicated using the Biomek® laboratory automation workstation. Cells were incubated overnight as previously described and luciferase assays were conducted the following day using OneGlo reagent. DMSO significantly inhibited luciferase activity at all concentrations, but the levels of inhibition were acceptable up to 1.25%. Data are expressed with mean ± SEM and were analyzed using Kruskal–Wallis one-way ANOVA and Dunnett's posthoc test (p < 0.05). SEM, standard error of the mean.
The signal to background (Max/Min) was intrinsically very variable as the Max effect value is so much higher than the Min and was judged not the best statistic for cross assay QC. As such, variance of the +/− tetracycline surrogate conditions was prioritized over the absolute S/B (which remained large) to minimize false negatives in the final screen. We therefore used Z′ as the primary driver of assay quality and it was the highest for OneGlo only (Fig. 5A). Multiplexing with cytotoxicity using either the CellTiter-Fluor or CellTox Green assays appears to increase the variability of the experiment and thus reduces the Z′ to below the 0.6 threshold (Fig. 5A). As a result, screening was conducted as a single read luciferase assay only, with cytotoxicity assessed as a secondary assay.
Importantly, we also demonstrated that the TREx-GRE2-AR-v assay was stable over time with a Z′ of greater than 0.5 for 4 h (OneGlo reagent alone), and a very high, although decreasing, S/B ratio (Fig. 5B).
DMSO Tolerance Testing
TREx-GRE2-AR-v cells were seeded in 384-well plates at a density of 1,250 cells/well in the presence or absence of tetracycline. A Biomek® laboratory automation workstation (Beckman Coulter) was used to conduct a two-fold dilution series of DMSO in media, from a starting concentration of 100%. Five microliters of DMSO was then transferred from compound plate to assay plate to give a final maximum concentration of 20%. Following incubation overnight, cells were lysed using OneGlo reagent and luciferase activity was measured. DMSO significantly inhibited the signal at all concentrations tested, but the degree of inhibition was acceptable up to 1.25% (Fig. 5C) and was within the concentrations used in the HTS of compound libraries, which did not exceed 0.25%.
High-Throughput Screening of Chemical Libraries
As proof of principal, three chemical libraries (Supplementary Fig. S4A) were selected for a pilot screen of the TREx-GRE2-AR-v cell line for AR-NTD inhibitors. Compounds from the NCC Library and the Selleckchem FDA-approved drug library were assessed at 10 μM with 0.1% DMSO final concentration (n = 1). A selection of compounds from the BioAscent compound cloud of diverse lead-like structures was also assessed at 10 μM, but with 0.25% DMSO final concentration (n = 1). All compounds were screened in 384-well plates in the conditions previously optimized for the TREx-GRE2-AR-v cell line: 6,759 compounds were screened in total, over 2 days.
Following measurement of luciferase activity, compound effect in RLU was normalized to percentage effect relative to internal plate controls (Supplementary Fig. S4B). RLUs were consistently higher in outer wells and edge effects are commonly observed in plate-based cellular assays and are due to differences in the rate of gas exchange and temperature fluctuation affecting cell growth in these positions (Supplementary Fig. S4B). As a result, the data were adjusted for well/column/row effect by applying median polishing as described by Gribbon et al.40 (Supplementary Fig. S4C).
After combining results of the BioAscent screen (Day 1) and NCC/Selleckchem screen (Day 2), the robustness of the assay was such that a selection cutoff of >54% effect was calculated to be statistically significant, defined as those compounds that produced a percentage effect greater than three times the robust standard deviation (scaled median absolute deviation) from the median percentage effect for all compounds. One hundred eighty-two compounds were identified as hits using this criterion (2.69% primary hit rate) (Fig. 6).
Fig. 6.
High-throughput screening using the TREx-HRE2-AR-v cell line. Greiner opaque white-walled 384-well plates were preloaded with compounds from the BioAscent (green), NCC (red), and Selleckchem (yellow) chemical libraries using the ECHO liquid handler to give a final concentration of 10 μM/well. Cells were seeded in the presence of tetracycline and incubated for 60 min at room temperature, before incubation for 16 h at 37°C, 5% CO2, as described above. The following day, cells were lysed using OneGlo reagent and luciferase activity was measured using the PHERAstar plate reader. Data were uploaded to Activity base and hits were identified as those compounds with significantly decreased luciferase activity compared to the maximum. Normalized effect was plotted against object ID in Activity base, and the 182 hits were highlighted using enlarged dots. Data were analyzed using robust statistics based on the median and robust SD (n = 1) and a total of 6,759 compounds tested. SD, standard deviation. Color images are available online.
Hit Confirmation and Counter screening
A 2.69% hit rate is high, but not unusual for a luciferase reporter, cell-based screening assay.41 Off-target effects and spurious hits may often account for a large proportion of hits in the initial screen. For this reason, in addition to hit confirmation in the primary screening assay, it is essential to profile hits for off-target activity, which can be achieved by employing counter screens relevant to the cell line and reporter technology used. All 182 compounds were cherry picked and screened in duplicate against the TREx-GRE2-AR-v cell line using the same conditions as the pilot screen to give n = 3.
Counter screen I: Cytotoxic compounds
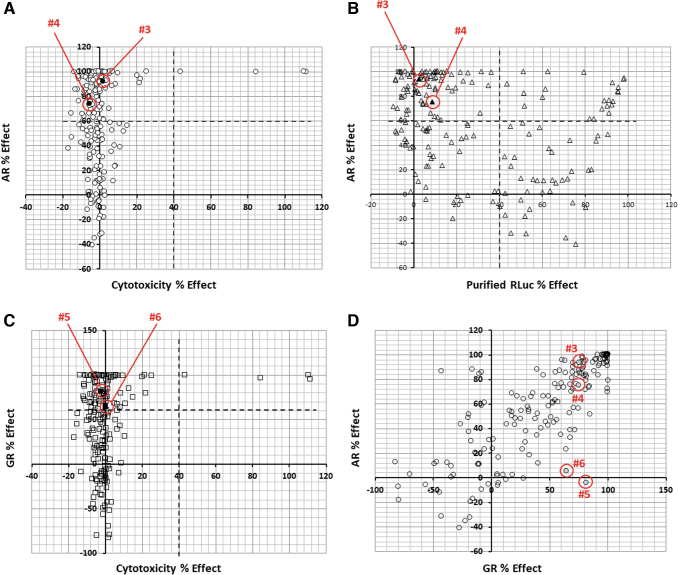
One possible explanation for the effect of test compounds on luciferase expression is that they may cause cell toxicity. To this end, the 182 hits were tested in a CellTox Green assay at 10 μM in duplicate in the TREx-GRE2 cells. Very few compounds showed significant levels of cytotoxicity when compared against efficacy for AR-v inhibition (Fig. 7A).
Fig. 7.
Screening of libraries of drug-like molecules for AR-v inhibitors. (A) Inhibition of AR-v activity is plotted as “% Effect” against “Cytotoxicity % Effect.” Upper left quadrant indicates small molecules with significant inhibitory activity (>60%) of AR-v transactivation with little/no general cytotoxicity (<30%). (B) AR % Effect plotted against inhibition of luciferase alone activity. Upper left quadrant indicates small molecules with significant inhibitory activity (>60%) of AR-v transactivation with little nonspecific activity against the reporter gene (<40%). (C) Inhibition of GRNTD-DBD transcriptional activity plotted as “GR Effect %” against general cytotoxicity. Upper left quadrant indicates small molecules with significant inhibitory activity (>60%) of GR transactivation with little/no general cytotoxicity (<30%). (D) Inhibition of AR-v activity plotted against inhibition of GRNTD-DBD. The position of hits #3, #4, #5, and #6 is indicated with red circles (A–D). Color images are available online.
Counter screen II: Inhibitors of luciferase enzyme activity
Inhibitors of firefly luciferase are prevalent in screening libraries41 and can complicate hit to lead and lead optimization in drug discovery.42,43 To eliminate these compounds from the hit list, 1 fM purified recombinant firefly luciferase enzyme was diluted in buffer and preincubated with 10 μM test compound (n = 2) for 1 h before monitoring luciferase activity SteadyGlo reagent.
A large number of compounds were identified as inhibitors of luciferase, although there was no correlation with the TREx-GRE2-AR-v reporter activity. Compounds that showed less than 40% activity against luciferase and greater than 60% activity in the TREx-GRE2-AR-v cell line were considered of interest (Fig. 7B).
Counter screen III: compounds with activity at the GRNTD-DBD
The TREx-GRE2-GR-NTD-DBD cell line was judged to be suitable for small-scale counter screening and provided a quick way to determine if a compound was selective for the AR. Plates were preloaded with 10 μM of the 182 initial hit compounds, in duplicate, and TREx-GRE2-GR-NTD-DBD cells were screened as previously described.
Luciferase assays were conducted with OneGlo reagent and RLUs were converted to GR % effect relative to internal plate controls (compared to maximum effect of cells incubated in absence of compound and tetracycline). Figure 7C compares inhibition of GR activity against cytotoxicity measurements (CellTox Green assay).
Comparison of AR-v and GR-NTD-DBD shows a correlation for compound activity in both cell assays with “hits” having over 60% efficacy at the AR significantly overlapping with those that have greater than 60% efficacy at the GR (Fig. 7D). These compounds were noted, but were not dismissed from the screen at this stage due to the emerging therapeutic possibility for dual AR/GR inhibitors in CRPC (reviewed in Refs. 44–46). There are several possible reasons that could account for compounds with activity in both the TREx-GRE2-AR-v and TREx-GRE2-GR-NTD-DBD cell lines. They may target a common structural property of the intrinsically disordered NTD, show activity at the conserved DBDs, inhibit nuclear import through a common mechanism, or have a more pleiotropic effect on gene transcription or protein degradation.
Interestingly, seven compounds emerged, which appeared to be specific for the AR-v, with greater than 60 AR% effect and less than 30 GR% effect. Similarly, a small number of compounds were observed with selectivity for GR over AR, for example, compounds #5 and #6 (Fig. 7D).
Overall, the compound re-test and three counter screens combined to produce a hit list of 70 compounds (38% of the initial 182 hits) from the three chemical libraries, an overall hit rate of 1.03%, with seven compounds appearing specific for the AR-v.
Initial Hit List
Assessment of the compound structures for chemical tractability allowed them to be divided into four groups: chemically tractable (suitable for hit optimization and manipulation); not chemically tractable; concerns about chemical tractability and off-target effects; or synthetically complex structures, which would require parent and analog compounds for hit optimization (“complex”). The BioAscent compounds were all deemed chemically tractable, as expected from a library designed as “lead-like” structures. Chemical tractability, as defined above, varied for the NCC/Selleckchem libraries. However, it is possible that these compounds (if effective) could be used in their current state as they are all previously licensed therapies. It was also discovered that there was some overlap between drugs identified in the NCC and Selleckchem libraries.
A literature review was conducted for the compounds identified in the NCC and Selleckchem libraries to determine their approved use, their likelihood of mediating their effects through cell death or senescence, and any previous suggestion of activity at the AR or in PCa. From this analysis the number of compounds in the NCC/Selleckchem libraries with the potential to mediate their effects through the AR-v was reduced from to 51 to 21, based on their likelihood of causing cell death unrelated to the AR. However, several compounds were also identified, which had been identified in earlier screens for receptor inhibitors (e.g., Teniposide47) or previously shown to disrupt AR signaling or expression (e.g., Camptothecin,48,49 pitavastatin calcium,50,51 and Temosirolimus52), providing confirmation that our screening assay was capable of identifying compounds with activity at the target AR-vs.
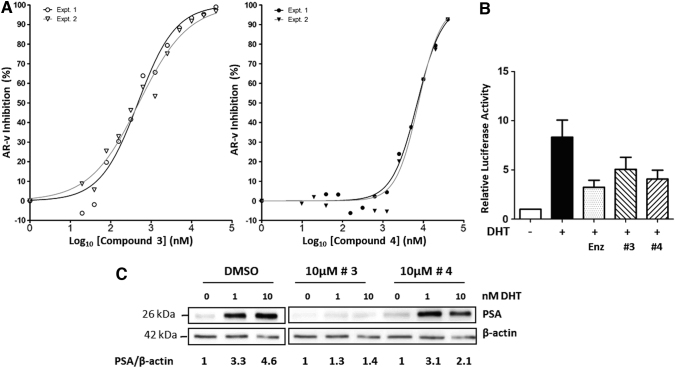
Following counter screens, two compounds (compounds #3 and #4 in Fig. 7) from the BioAccent library were chosen for further validation experiments using repurchased solids. IC50 values of 0.48 μM and 7.3 μM were determined for compounds #3 and #4, respectively, against AR-v transcriptional activity in TREx-GRE2-AR-v cells (Fig. 8A). To determine if compounds #3 and #4 could inhibit the full-length AR in a prostate cell model, we used VCaP cells. VCaP cells were transfected with pGL4.26-GRE2-luciferase and treated with 10 nM DHT, resulting in an eightfold hormone-dependent activation of transcription.
Fig. 8.
Inhibition of AR-FL in PCa cells. (A) IC50 measurements for inhibition of AR-v in TREx-GRE2-AR-v cells: dose–response of AR-v activity in presence of increasing concentrations of compound #3 gave IC50 values of 470 and 497 nM in experiments 1 and 2, respectively. Dose–response of AR-v activity in presence of compound #4 just approached a plateau at the highest concentration tested and IC50 values of 7.0 and 7.6 μM were calculated for experiments 1 and 2, respectively. All data were plotted, and analysis was conducted using GraphPad Prism nonlinear regression with four parameters (variable slope) (n = 2). Percentage inhibition was calculated with + tetracycline control as 0% and −tetracycline control as 100% in each independent experiment. (B) In the prostate cell line, VCaP, endogenous AR-FL induced reporter gene activity in response to DHT (10 nM). Enzalutamide significantly inhibited reporter gene activity at a concentration of 10 μM. Compounds #3 and #4 inhibited reporter gene activity at a similar level to enzalutamide, at concentrations of 5 and 10 μM, respectively. All results are expressed as mean ± SEM. (C) Inhibition of endogenous expression of the well-validated androgen-responsive protein PSA in VCaP PCa cells. Hormone (DHT) treatment resulted in threefold-to-fivefold increase in PSA expression (first three lanes). Compound #3 reduced expression by 60%–70%, while compound #4 inhibited AR activity maximally by ca.50%.
Compound #3 and #4 inhibited receptor-dependent transcription by 40%–45%, which was comparable to the reduction seen with the antiandrogen enzalutamide (50%) (Fig. 8B). Importantly, to test the effectiveness of our novel inhibitors, we also examined expression of prostate-specific antigen (PSA) a well-validated AR target gene and a biomarker used in PCa diagnosis. Robust inhibition of PSA protein expression by compound #3 was also observed in the VCaP cell model, with a more modest inhibition seen with compound #4 (Fig. 8C).
Conclusions
The emergence of AR-vs, lacking the LBD, poses a significant clinical challenge for the treatment of advanced and metastatic PCa. We have developed a cell-based reporter assay for AR-v activity and demonstrated this is suitable for high-throughput screening of chemical libraries. In a pilot screen, we identified several promising candidate molecules. Two of these have been further validated, demonstrating submicro- to micro-Molar IC50 values against AR-v and inhibition of full-length AR in a PCa cell model in both a reporter gene assay and against endogenous expression of PSA, an androgen-regulated protein upregulated in PCa. In conclusion, we have produced and validated a novel screening assay for inhibitors of AR-vs.
Supplementary Material
Acknowledgments
A.E.M. was supported by a BBSRC-CASE studentship together with support from IOmet Pharma (Nine Edinburgh Bioquarter, Edinburgh, Scotland, United Kingdom), NHS Grampian Endowments, and the charity Friends of Anchor. The high-throughput assay and screen were developed with support in kind from the Scottish Life Science Alliance (SULSA) assay development program. We are also grateful to support from Dr Alan Wise (IOmet Pharma) for advice and critical input.
Abbreviations Used
- AR
androgen receptor
- AR-v
AR splice variant
- CRPC
castrate-resistant prostate cancer
- DBD
DNA binding domain
- DMEM
Dulbecco's Modified Eagle Medium
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- GRNTD-DBD
glucocorticoid receptor N-terminal and DNA binding domains
- LBD
ligand binding domain
- NCC
NIH Clinical Collection
- NTD
amino terminal domain
- PCa
prostate cancer
- PSA
prostate-specific antigen
- RLU
relative light unit
- S/B
signal to background
- TetO2
tetracycline operator 2
- Z′
Z prime
Disclosure Statement
No competing financial interests exist.
Funding Information
Biotechnology and Biological Sciences Research Council (Grant code - RGD1371), National Health Service (NHS) (Grant 16/11/040) and Friends of Anchor (Grant RS2015003).
Supplementary Material
References
- 1. Globocan (2020). https://gco.iarc.fr/today/data/factsheets/cancers/27-Prostate-fact-sheet.pdf (accessed February 28, 2022).
- 2. Estébanez-Perpiñá E, Bevan CL, McEwan IJ: Eighty years of targeting androgen receptor activity in prostate cancer: the fight goes on. Cancers (Basel) 2021;13:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meng MV, Grossfeld GD, Sadetsky N, Mehta SS, Lubeck DP, Carroll PR: Contemporary patterns of androgen deprivation therapy use for newly diagnosed prostate cancer. Urology 2002;60(3 Suppl 1):7–11. [DOI] [PubMed] [Google Scholar]
- 4. Scher HI, Sawyers CL: Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol 2005;23:8253–8261. [DOI] [PubMed] [Google Scholar]
- 5. Bourke L, Kirkbride P, Hooper R, Rosario AJ, Chico TJ, Rosario DJ: Endocrine therapy in prostate cancer: time for reappraisal of risks, benefits and cost-effectiveness? Br J Cancer 2013;108:9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ged Y, Horgan AM: Management of castrate-resistant prostate cancer in older men. J Geriatr Oncol 2016;7:57–63. [DOI] [PubMed] [Google Scholar]
- 7. Chen CD, Welsbie DS, Tran C, et al. : Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004;10:33–39. [DOI] [PubMed] [Google Scholar]
- 8. Yap TA, Smith AD, Ferraldeschi R, Al-Lazikani B, Workman P, de Bono JS: Drug discovery in advanced prostate cancer: translating biology into therapy. Nat Rev Drug Discov 2016;15:699–718. [DOI] [PubMed] [Google Scholar]
- 9. Hay CW, McEwan IJ: The impact of point mutations in the human androgen receptor: classification of mutations on the basis of transcriptional activity. PLoS ONE 2012;7:e32514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linja MJ, Savinainen KJ, Saramäki OR, et al. : Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res 2001;61:3550–3555. [PubMed] [Google Scholar]
- 11. Quigley DA, Dang HX, Zhao SG, et al. : Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y, Yang R, Henzler CM, et al. : Diverse AR gene rearrangements mediate resistance to androgen receptor inhibitors in metastatic prostate cancer. Clin Cancer Res 2020;26:1965–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dehm SM, Tindall DJ: Alternatively spliced androgen receptor variants. Endocr Relat Cancer 2011;18:R183–R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Antonarakis ES, Lu C, Wang H, et al. : AR-v7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Laere B, van Dam PJ, Whitington T, et al. : Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. J Eur Urol 2017;72:192–200. [DOI] [PubMed] [Google Scholar]
- 16. Sharp A, Coleman I, Yuan W, et al. : Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest 2019;129:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Killock D: 2016 ARAMIS - is darolutamide set to become the ‘third musketeer’ of nmCRPC? Nat Rev Clin Oncol 2019;16:273. [DOI] [PubMed] [Google Scholar]
- 18. Sugawara T, Baumgart SJ, Nevedomskaya E, et al.: Darolutamide is a potent androgen receptor antagonist with strong efficacy in prostate cancer models. Int J Cancer 2019;145:1382–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monaghan AE, McEwan IJ: A sting in the tail: the N-terminal domain of the androgen receptor as a drug target. Asian J Androl 2016;18:687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sadar MD: Discovery of drugs that directly target the intrinsically disordered region of the androgen receptor. Expert Opin Drug Discov 2020;5:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McEwan IJ: Intrinsic disorder in the androgen receptor: identification, characterisation and drugability. Mol Biosyst 2012;8:82–90. [DOI] [PubMed] [Google Scholar]
- 22. De Mol E, Fenwick RB, Phang CT, et al. : EPI-001, a compound active against castration-resistant prostate cancer, targets transactivation unit 5 of the androgen receptor. ACS Chem Biol 2016;11:2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andersen RJ, Mawji NR, Wang J, et al. : Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010;17:535–546. [DOI] [PubMed] [Google Scholar]
- 24. Yang YC, Banuelos CA, Mawji NR, et al. : Targeting androgen receptor activation function-1 with EPI to overcome resistance mechanisms in castration-resistant prostate cancer. Clin Cancer Res 2016;22:4466–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Myung JK, Banuelos CA, Fernandez JG, et al. : An androgen receptor Nterminal domain antagonist for treating prostate cancer. J Clin Invest 2013;123:2948–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirayama Y, Tam T, Jian K, Andersen RJ, Sadar MD: Combination therapy with androgen receptor N-terminal domain antagonist EPI-7170 and enzalutamide yields synergistic activity in AR-V7-positive prostate cancer. Mol Oncol 2020;14:2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brand LJ, Olson ME, Ravindranathan P, et al. : EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget 2015;6:3811–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu J, Van der Steen T, Tindall DJ: Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol 2015;12:137–144. [DOI] [PubMed] [Google Scholar]
- 29. Antonarakis ES, Luo J: Prostate cancer: AR splice variant dimerization-clinical implications. Nat Rev Urol 2015;12:431–433. [DOI] [PubMed] [Google Scholar]
- 30. Xu D, Zhan Y, Qi Y, et al. : Androgen receptor splice variants dimerize to transactivate target. Genes Cancer Res 2015;75:3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krause WC, Shafi AA, Nakka M, Weigel NL: Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol 2014;54:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones D, Wade M, Nakjang S, et al. : FOXA1 regulates androgen receptor variant activity in models of castrate-resistant prostate cancer. Oncotarget 2015;6:29782–29794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang J, Wang L, Poluben L, et al. : Androgen receptor splice variant 7 functions independently of the full length receptor in prostate cancer cells. Cancer Lett 2021;519:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aitken L, Baillie G, Pannifer A, et al. : In vitro assay development and HTS of small-molecule human ABAD/17β-HSD10 inhibitors as therapeutics in Alzheimer's disease. SLAS Discov 2017;22:676–685. [DOI] [PubMed] [Google Scholar]
- 35. Gatta V, Ilina P, Porter A, McElroy S, Tammela P: Targeting quorum sensing: high-throughput screening to identify novel LsrK inhibitors. Int J Mol Sci 2019;20. pii: E3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao F, Svensjö T, Winkler T, Lu M, Eriksson C, Eriksson E: Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum Gene Ther 1998;9:1939–1950. [DOI] [PubMed] [Google Scholar]
- 37. McEwan IJ, Gustafsson J-Å: Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci U S A 1997;94:8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kingwell K: European Lead Factory hits its stride. Nat Rev Drug Discov 2016;15:221–222. [DOI] [PubMed] [Google Scholar]
- 39. McElroy SP, Jones PS, Barrault DV: The SULSA Assay Development Fund: accelerating translation of new biology from academia to pharma. Drug Discov Today 2017;22:199–203 [DOI] [PubMed] [Google Scholar]
- 40. Gribbon P, Lyons R, Laflin P, et al. : Evaluating real-life high-throughput screening data. J. Biomol Screen 2005;10:99–107. [DOI] [PubMed] [Google Scholar]
- 41. Thorne N, Shen M, Lea WA, et al. : Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem Biol 2012;19:1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Auld DS, Lovell S, Thorne N, et al. : Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc Natl Acad Sci U S A 2010;107:4878–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McElroy SP, Nomura T, Torrie LS, et al. : A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol 2013;2013;11:e1001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pungsrinont T, Baniahmad A: Targeting the glucocorticoid receptor for a combinatory treatment strategy for castration resistant prostate cancer. AME Med J 2018;3:41. [Google Scholar]
- 45. Hirayama Y, Sadar MD: Does increased expression of glucocorticoid receptor support application of antagonists to this receptor for the treatment of castration resistant prostate cancer? AME Med J 2018;3:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar R: Emerging role of glucocorticoid receptor in castration resistant prostate cancer: a potential therapeutic target. J Cancer 2020;11:696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones JO, Diamond MI: A cellular conformation-based screen for androgen receptor inhibitors. ACS Chem Biol 2008;3:412–418. [DOI] [PubMed] [Google Scholar]
- 48. Liu S, Yuan Y, Okumura Y, Shinkai N, Yamauchi H: Camptothecin disrupts androgen receptor signaling and suppresses prostate cancer cell growth. Biochem Biophys Res Commun 2010;394:297–302. [DOI] [PubMed] [Google Scholar]
- 49. Minelli R, Cavalli R, Ellis L, et al. : Nanosponge-encapsulated camptothecin exerts anti-tumor activity in human prostate cancer cells. Eur J Pharm Sci 2012;47:686–694. [DOI] [PubMed] [Google Scholar]
- 50. Murtola TJ, Visakorpi T, Lahtela J, Syvälä H, Tammela TL: Statins and prostate cancer prevention: where we are now, and future directions. Nat Clin Pract Urol 2008;5:376–387. [DOI] [PubMed] [Google Scholar]
- 51. Yokomizo A, Shiota M, Kashiwagi E, et al. : Statins reduce the androgen sensitivity and cell proliferation by decreasing the androgen receptor protein in prostate cancer cells. Prostate 2011;71:298–304. [DOI] [PubMed] [Google Scholar]
- 52. Wang Y, Mikhailova M, Bose S, Pan CX, deVere White RW, Ghosh PM: Regulation of androgen receptor transcriptional activity by rapamycin in prostate cancer cell proliferation and survival. Oncogene 2008;27:7106–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.