Abstract
Post-translational modifications of histones play important roles in regulating chromatin structure and gene expression programs, and the modified histones can be passed on to subsequent generations as an epigenetic memory. The fission yeast has been a great model organism for studying histone modifications in heterochromatin assembly and epigenetic inheritance. Here, we review findings in this organism that cemented the idea of chromatin-based inheritance and highlight recent studies that reveal the role of histone turnover in regulating this process.
Keywords: Epigenetic inheritance, histone turnover, heterochromatin, H3K9 methylation, Clr4
Graphical Abstract
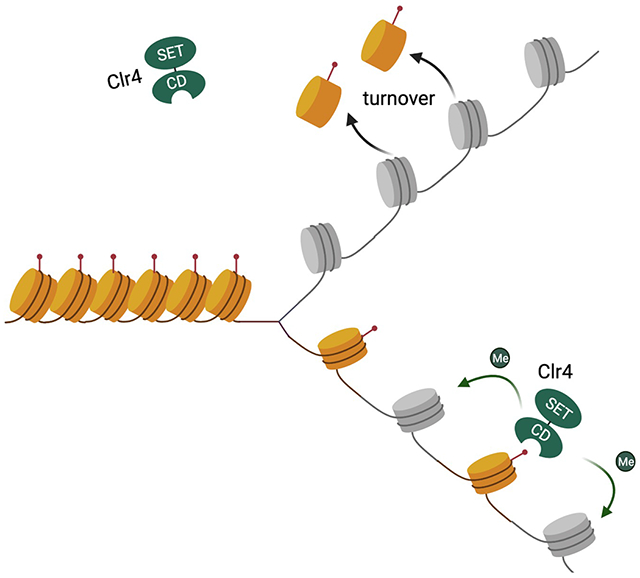
During DNA replication, parental histone (H3-H4)2 tetramers are deposited back at their original locations and segregated equally onto daughter strands to direct the formation of nucleosomes. The H3K9me3 modification on parental histones recruits the H3K9 methyltransferase Clr4 to modify nearby nucleosomes formed by newly synthesized histones, therefore duplicating H3K9me3 at both daughter strands. The turnover of parental histones breaks this “read-write” cycle for chromatin structure inheritance.
Introduction
Eukaryotic genomic DNA is folded with histones and non-histone proteins into chromatin. Covalent modifications of histones, chromatin remodeling, as well as DNA modifications, play important roles in setting up the gene expression programs that determine cell identity. Once these chromatin states have been established, they can form an epigenetic memory of gene activity that will be inherited by subsequent generations of cells [1].
Histones play major roles in transmitting epigenetic information [1–3]. The prevailing view is that during DNA replication, the passage of the replication fork disrupts parental nucleosomes. Parental (H3-H4)2 tetramers, which contain original histone modifications, are deposited back to the original location and equally segregated onto both daughter strands to direct the formation of nucleosomes [4–8]. The remaining gaps in DNA are filled by nucleosomes formed by newly synthesized (H3-H4)2, resulting in the intermingling of nucleosomes containing either parental or new (H3-H4)2. For certain histone modifications involved in gene silencing, such as trimethylation of histone H3 at lysine 9 (H3K9me3) and lysine 27 (H3K27me3), the existing histone modifications on parental histones recruit the corresponding modifying enzymes, which lead to modifications of nearby nucleosomes, and therefore restore the original histone modification profiles on both replicated chromatids (Fig. 1) [1, 9].
Fig. 1.
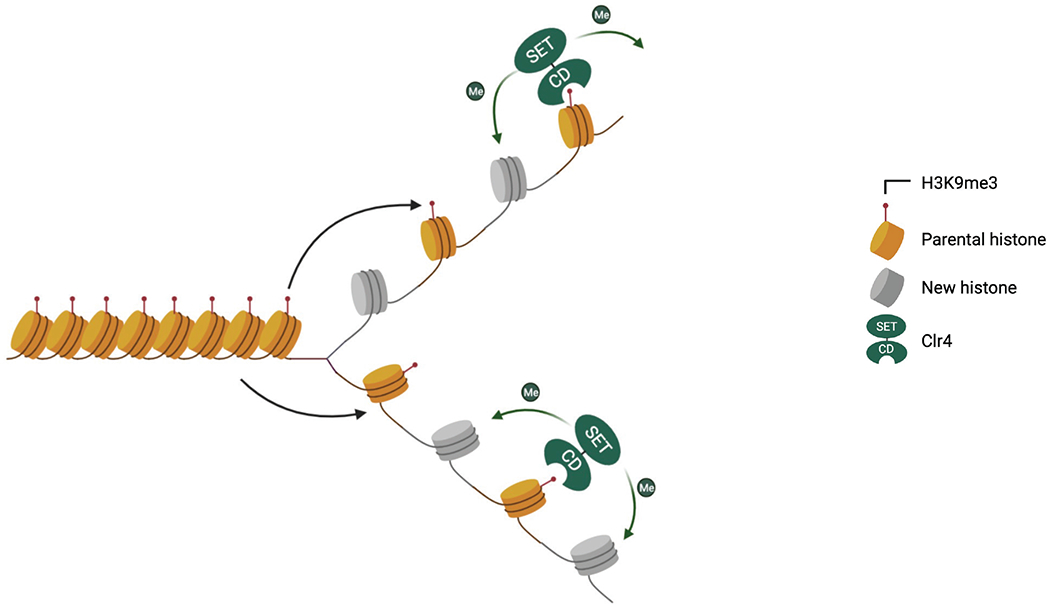
A model for histone-based epigenetic inheritance. At heterochromatin regions, the histones are marked with H3K9me3. During DNA replication, parental (H3-H4)2 tetramers are deposited back to the original location and segregated equally onto both daughter strands. The histone H3K9 methyltransferase Clr4 contains the catalytic SET domain and a chromodomain (CD) that recognizes H3K9me3. Parental histones containing H3K9me3 recruit Clr4 to modify nearby nucleosomes formed by newly synthesized histones, therefore duplicating the histone modification on both replicated DNA strands.
Studies of heterochromatin formation in the fission yeast Schizosaccharomyces pombe have been instrumental in revealing the mechanisms of heterochromatin assembly and chromatin-mediated epigenetic inheritance [9]. In this article, we will highlight the discoveries in fission yeast on epigenetic inheritance and also discuss recent work revealing the critical role of controlling histone turnover rate during this process.
Fission yeast heterochromatin assembly
In fission yeast, large domains of heterochromatin are formed at pericentric regions, the silent mating-type region, and subtelomeres [10]. The nucleosomes within these regions are hypoacetylated and contain H3K9me3, which is catalyzed by the histone methyltransferase Clr4 [11, 12]. Clr4 also associates with a E3 ubiquitin ligase (Cul4, Rik1, Raf1, Raf2) to form the Clr4 complex (CLRC) [13–15]. CLRC ubiquitylates H3K14 (H3K14ub), and H3K14ub increases the affinity of Clr4 with the H3 tail to enhance its enzymatic activity [16–18].
In addition to the catalytic SET domain, Clr4 contains a chromodomain that recognizes H3K9me3 [19]. Therefore, Clr4 not only “writes” the H3K9me3 mark, but also “reads” the mark it creates. Existing H3K9me3 recruits Clr4 and enhances its enzymatic activity [20], forming a self-reinforcing loop for H3K9me3, leading to heterochromatin spreading and the formation of large H3K9me3 domains. H3K9me3 creates binding sites for HP1 family proteins Swi6 and Chp2, which in turn recruit diverse proteins to regulate transcription, recombination, and other cellular processes, such as chromosome segregation and mating-type switching [10].
Heterochromatin establishment and maintenance
The formation of heterochromatin is separated into two distinct steps: establishment and maintenance [9] (Fig. 2A, 2B, and Table 1). During the establishment step, Clr4 is recruited to specific genomic loci to initiate H3K9me3. During the maintenance step, Clr4 restores heterochromatin structure after DNA replication based on existing H3K9me3 on parental histones, independent of the recruitment signal. The pathways responsible for each step have been clearly delineated in this organism.
Fig. 2.
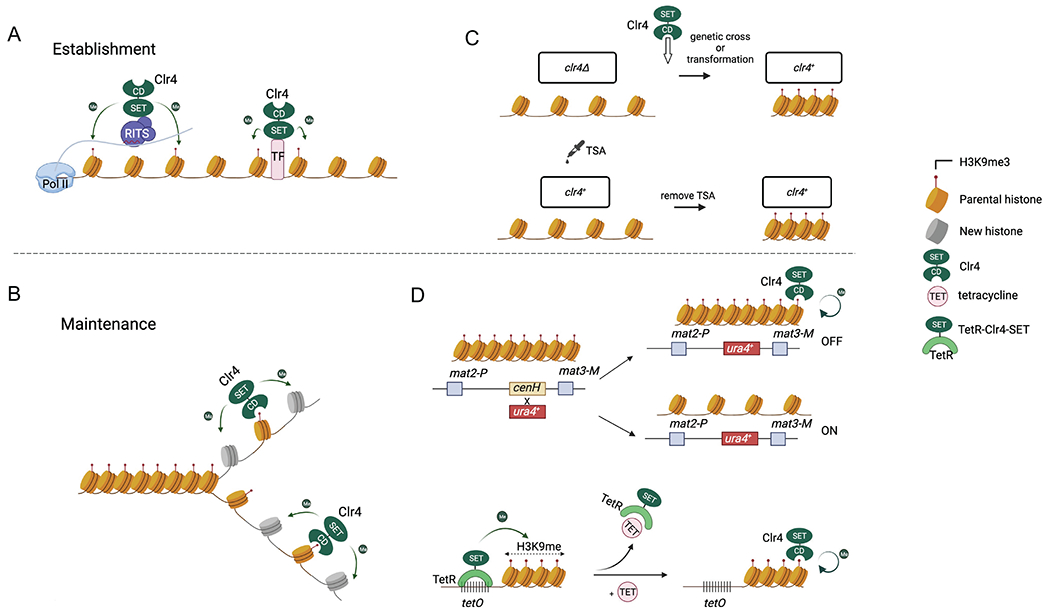
Heterochromatin establishment and maintenance. (A) Heterochromatin establishment refers to the targeting of Clr4 to defined genomic locations by RNAi or transcription factors to initiate H3K9me3. (B) Heterochromatin maintenance refers to the process of replicating H3K9me3 during DNA replication without initiation signals. (C) Methods used to examine heterochromatin establishment. Pre-existing heterochromatin is first erased by clr4Δ or the addition of TSA, followed by the reintroduction of Clr4 or removal of TSA. (D) Methods used to examine heterochromatin maintenance. Top, the replacement of cenH, which mediates heterochromatin establishment through RNAi, with a ura4+ reporter results in two metastable epigenetic states. The ura4-off state can be maintained through mitosis and meiosis. Bottom, TetR-Clr4-SET is recruited to tetO sites to establish heterochromatin. The addition of tetracycline release TetR-Clr4-SET and heterochromatin is maintained by endogenous Clr4.
Table 1.
Factors involved in heterochromatin establishment and maintenance.
| Protein Name | Establishment | Maintenance |
|---|---|---|
| CLRC | ✓ | ✓ |
| Swi6 | ✓ | ✓ |
| Dicer | ✓ | |
| RITS | ✓ | |
| Atf1/Pcr1 | ✓ | |
| shelterin | ✓ |
The RNA interference (RNAi) pathway is critical for the recruitment of Clr4 to establish heterochromatin at repetitive DNA present at all major heterochromatin domains [10]. The DNA repeats are transcribed, producing double-stranded RNAs (dsRNAs) [21]. The ribonuclease Dicer (Dcr1) processes these dsRNAs into small interfering RNAs (siRNAs), which are loaded onto the RNA-induced transcriptional silencing complex (RITS) [22]. The Argonaute protein (Ago1) within RITS binds siRNAs and directs RITS to nascent RNA transcripts from repeat regions [22]. RITS then recruits CLRC to initiate H3K9me3 [19, 23].
While RNAi is the major pathway recruiting CLRC to pericentric repeats, DNA-based mechanisms are also involved in heterochromatin establishment at other locations. For example, at the silent mating-type region, the cenH repeat, which is homologous to pericentric repeats, recruits Clr4 through the RNAi pathway [24]. Adjacent to cenH are binding sites for transcription factor Atf1/Pcr1, which also independently recruits Clr4 [25–29]. At subtelomeres, a similar repeat sequence present in the tlh genes promotes RNAi-mediated recruitment of Clr4 [30]. In addition, the telomere-protecting complex shelterin recruits Clr4 to telomere repeats to establish heterochromatin [30, 31].
Heterochromatin maintenance is the process of restoring histone modification profiles after DNA replication. As the replication fork moves forward, parental histones are deposited back to the original location and intermixed with newly synthesized histones. Parental histones containing H3K9me3 serve as signals for the recruitment of Clr4 to modify newly synthesized histones, restoring the H3K9me3 domain [9, 19]. This self-template mechanism is expected to maintain heterochromatin structure even after the signals for heterochromatin establishment are removed.
In normal conditions, both establishment and maintenance pathways are required for heterochromatin integrity, but genetic manipulations allow clear determination whether a factor is involved specifically in heterochromatin establishment or maintenance. For example, to examine the role of RNAi in heterochromatin establishment, pre-existing heterochromatin is first completely erased by clr4Δ, followed by the re-introduction of clr4+ through transformation or genetic crosses [23, 24, 32–34]. Alternatively, the histone deacetylase inhibitor tricostatin A (TSA) is used to erase existing heterochromatin, followed by TSA removal to examine heterochromatin integrity. RNAi mutants failed to establish H3K9me3 in these assays, suggesting that it is required for heterochromatin establishment [24, 25] (Fig. 2C).
Examining heterochromatin maintenance requires the removal of heterochromatin establishment pathways. At the silent mating-type locus, cenH mediates Clr4 recruitment through RNAi [24]. Replacing cenH with a ura4+ reporter gene leads to cells with one of two stably maintained states: “ura4-on” (the reporter is expressed) and “ura4-off” (the reporter is repressed) [35] (Fig. 2D). The cells are identical in DNA sequence, but differ in their chromatin environment at the silent mating-type locus [24, 36]. Because the conversion from ura4-on to ura4-off is at a very low frequency, it is presumed that the heterochromatin in ura4-off cells is maintained independent of the initiation signal. Genetic analyses demonstrate that these epigenetic states are inherited through both mitosis and meiosis similar to gene alleles [35], thus firmly establishing the role of chromatin structure in epigenetic inheritance.
However, such a simplified explanation is complicated by later findings of an additional, albeit less robust mechanism of heterochromatin establishment involving DNA binding proteins Atf1/Pcr1, which cooperate with RNAi to recruit Clr4 to the silent mating-type region [25–29]. To avoid complications from possible unknown mechanisms at endogenous heterochromatin loci, ectopic heterochromatin is established by the recruitment of Clr4 to tetO binding sites through a TetR and Clr4-SET domain (TetR-Clr4-SET) fusion protein, leading to the silencing of adjacent report genes [33, 37] (Fig. 2D). As the addition of tetracycline quickly releases TetR-Clr4-SET from tetO binding sites, heterochromatin is expected to be maintained through the self-templated restoration of H3K9me3 by the endogenous Clr4. However, silencing and H3K9me3 are quickly lost upon the release of TetR-Clr4-SET, suggesting that heterochromatin is not stably inherited in wild-type cells.
Interestingly, removal of an anti-silencing protein Epe1 allows the inheritance of this artificial heterochromatin after TetR-Clr4-SET release [33, 37]. Moreover, the inheritance of such chromatin structure is critically dependent on the ability of the chromodomain of Clr4 to recognize H3K9me3. These results clearly demonstrate that cells can indeed mediate epigenetic inheritance through chromatins structure by coupling the “reading” and “writing” of H3K9me3. However, they also indicate that there are mechanisms that normally counteract histone-based heterochromatin maintenance in wild-type cells.
Epe1 contains a JmjC domain, which is the catalytic domain of histone demethylases [38]. It localizes to heterochromatin regions through its interaction with Swi6 [39–41]. It is proposed that Epe1 is a H3K9 demethylase and that H3K9me3 removal by Epe1 opposes epigenetic inheritance [33, 37]. However, no enzymatic activity of Epe1 has been demonstrated in vitro, and the commonly used mutations that are expected to only abolish Epe1 demethylase activity also influence its interaction with Swi6 [42]. Moreover, Epe1 also exerts its function on heterochromatin independent of its JmjC domain [43–45]. For example, it recruits the SAGA histone acetyltransferase complex and the BET family bromodomain protein Bdf2 [43, 44]. Therefore, the mechanisms that counteract histone-based heterochromatin maintenance remain unclear.
Mutations that bypass RNAi reveal the role of histone turnover in regulating epigenetic inheritance
Further clues about the mechanisms that counteract heterochromatin inheritance come from many mutations that bypass RNAi for heterochromatin integrity. At pericentric repeats, RNAi is the major pathway to establish heterochromatin [21]. However, heterochromatin is not properly maintained in RNAi mutants, even though RNAi is not expected to be required for heterochromatin inheritance [24, 33]. This result is consistent with the idea that there are pathways actively preventing heterochromatin maintenance. Interestingly, inactivating a number of pathways, such as the Mst2 histone acetyltransferase complex, the INO80 chromatin remodeling complex, the JmjC domain protein Epe1, and the transcription associated Paf1 complex, allows heterochromatin formation at pericentric repeats in the absence of RNAi [32, 33, 40, 46–49]. Analyses of these factors demonstrate a critical role of suppressing histone turnover in regulating epigenetic inheritance (Fig. 3).
Fig. 3.
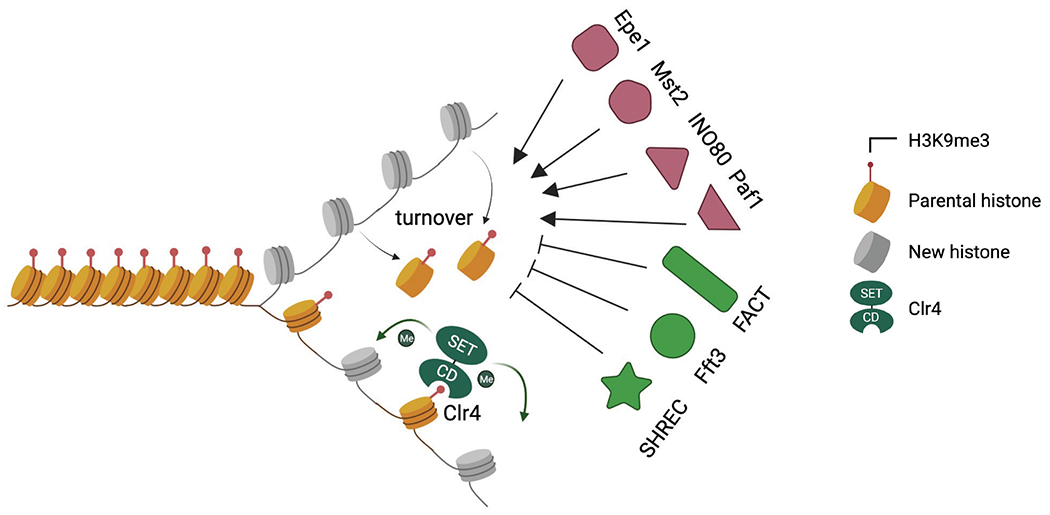
Low histone turnover rate is critical for preserving parental histones for epigenetic inheritance. The turnover of parental histones breaks the read-write mechanism of Clr4 to duplicate parental histone modifications. Epe1, the Mst2 complex, the INO80 complex, and the Paf1 complex promote histone turnover, while SHREC, Fft3, and FACT ensure low histone turnover rate.
Mutations of the Mst2 histone acetyltransferase complex bypass RNAi for heterochromatin maintenance [32]. The Mst2 complex includes the MYST family histone acetyltransferase Mst2 as the catalytic subunit and other proteins such as Nto1, Pdp3, Tfg3, Ptf1 and Ptf2 [50, 51]. The subunit composition is similar to the NuA3 complex in budding yeast and the MOZ/MORF complex in mammalian systems [52]. The Mst2 complex is normally excluded from heterochromatin regions because the PWWP domain of the Pdp3 subunit recognizes H3K36me3, which is normally enriched at euchromatin regions [53]. However, in the absence of RNAi, the Mst2 complex gains access to heterochromatin and acetylates histones within heterochromatin regions [32]. The Mst2 complex is a highly specific H3K14 acetyltransferase, and abolishing its enzymatic activity is critical for bypassing RNAi [32, 51]. It is proposed that H3K14 acetylation promotes transcription at pericentric repeats, which leads to a higher histone turnover rate and the loss of the parental histones containing H3K9me3, therefore breaking the “read-write” cycle for chromatin-based epigenetic inheritance. As a result, RNAi is needed to reestablish heterochromatin during every cell cycle [32]. The Mst2 complex was later demonstrated to stimulate histone turnover at heterochromatin [54].
Mutations of the INO80 chromatin remodeling complex also bypass RNAi for heterochromatin maintenance [49]. The INO80 complex regulates diverse processes such as nucleosome positioning, histone turnover, and the incorporation of histone variant H2A.Z [55]. Fission yeast INO80 complex is composed of a conserved catalytic core containing Ino80, Rvb1, Rvb2, Ies2, Ies4, Ies6, Act1, Arp4, Arp5, Arp8, and Taf14 [49, 56, 57]. It also contains accessory subunits, such as Iec1, Hap2, Nht1, Iec3, and Iec5, although how they regulate INO80 function is not clear. Interestingly, mutations bypassing RNAi are all accessory subunits of INO80, and this function is largely dependent on the Iec5 subunit. Further analyses show that Iec5 regulates histone turnover, but has no impact on nucleosome positioning or histone H2A.Z exchange at heterochromatin regions [49]. INO80 localizes to heterochromatin and interacts with heterochromatin protein Swi6, suggesting that it plays a direct role at heterochromatin regions [49, 58]. As the repeat regions are transcribed during the S phase of the cell cycle [59, 60], RNA polymerase II (Pol II) inevitably collides with the advancing replication fork [61]. In budding yeast, INO80 is critical for resolving transcription-replication conflicts by removing Pol II [62, 63]. It is possible that such a function is conserved in fission yeast, and the presence of INO80 at heterochromatin ensures the proper replication of heterochromatin. However, an undesired effect of INO80 action is an increase in histone turnover rate at heterochromatin regions. Histone turnover reduces the amounts of parental histones deposited onto daughter DNA strands, therefore affecting the propagation of H3K9me3 required for heterochromatin inheritance.
Mutations of Epe1 bypass RNAi [40], and later it was shown that epe1Δ bypasses heterochromatin maintenance in RNAi mutants [33]. As discussed earlier, the biochemical activity of Epe1 remains elusive. Interestingly, Epe1 also regulates transcription and promotes histone turnover at heterochromatin regions [64], further corroborating the idea that low histone turnover rate is critical for maintaining parental histone modification for heterochromatin inheritance. Although the mechanism of how Epe1 affects histone turnover is unclear, the association of Epe1 with SAGA [44] suggests that Epe1 might stimulate histone turnover through histone acetylation.
Mutations of the Paf1 complex (Paf1C) bypass RNAi and increase the spreading of heterochromatin to neighboring regions, and Paf1C counteracts both heterochromatin establishment and maintenance [48, 65, 66]. The Paf1 complex is associated with transcriptional elongation and it augments other histone modifications such as H2B ubiquitination and H3K4 methylation [67]. However, the impact of Paf1C on heterochromatin is independent of these histone modifications [48]. Interestingly, Paf1C also regulates histone turnover at heterochromatin regions, further supporting a role of low histone turnover rate in heterochromatin inheritance [48].
Finally, mutations of telomere shelterin bypass RNAi for heterochromatin maintenance [47]. Shelterin is required for chromosome end protection and telomere length homeostasis [68]. It also directly associates with Clr4 to promote heterochromatin assembly at the telomere region [31]. Separation-of-function mutations of shelterin allow the clear demonstration that telomere heterochromatin assembly, but not chromosome end protection or telomere length control, is involved in bypassing RNAi [47]. Mutations of shelterin disrupt telomere heterochromatin, leading to an increase in the availability of Swi6 to facilitate heterochromatin maintenance at centromeres in RNAi mutants. Consistent with this idea, overexpression Swi6 also bypasses RNAi [47]. As HP1 proteins can undergo liquid-liquid phase separation [69–71], it is possible that higher levels of Swi6 create a favorable environment for retaining parental histones containing H3K9me3, which interact with Swi6.
Regulating histone turnover at heterochromatin in wild-type cells
The histone turnover rate at heterochromatin regions is normally low in wild-type cells [64, 72]. This could be the consequence of low transcriptional activity within heterochromatin regions. However, there are also mechanisms that actively maintain low histone turnover rate at heterochromatin, involving proteins such as the SHREC complex, chromatin remodeling protein Fft3, and the histone chaperone FACT.
SHREC is a complex that includes the histone deacetylase Clr3, the chromatin remodeling protein Mit1, and additional proteins [73, 74]. It is recruited to heterochromatin regions through its interaction with HP1 proteins Swi6 and Chp2, as well as other DNA-based mechanisms [41, 73–75]. The resulting histone deacetylation and chromatin remodeling lead to regularly spaced nucleosomes, the exclusion of the transcription machinery, and low histone turnover rate [64, 73, 74]. Since both SHREC and Epe1 are recruited by HP1 proteins to heterochromatin, they compete with each other to regulate histone turnover [64].
Fft3 is a chromatin remodeling protein homologous to SMARCAD1 in humans and FUN30 in budding yeast, and is required for heterochromatin integrity [76]. It regulates heterochromatin maintenance, but not establishment [77]. Fft3 is recruited to heterochromatin through Swi6 and associates with the replication machinery. It is proposed that Fft3 reduces histone turnover during DNA replication to prevent the accumulation of RNA-DNA hybrids [77].
FACT is a histone chaperone that maintains nucleosome structure during transcription and replication [78]. FACT associates with Swi6 and this association requires the nuclear rim protein Amo1 and the RNA processing RIXC complex. FACT, Amo1, and RIXC are all required for heterochromatin maintenance, but dispensable for heterochromatin establishment [79–82]. The tethering of heterochromatin to Amo1 at the nuclear periphery may create a specialize domain with a higher concentration of silencing proteins, which aids the recruitment of FACT to suppress histone turnover [80].
While SHREC regulates histone turnover throughout the cell cycle, Fft3 and Amo1-RIXC-FACT might have more dedicated roles in suppressing histone turnover during DNA replication. As DNA replication machinery requires the disruption of nucleosomes to advance the replication fork, such forces may consequently increase histone turnover rate. The combined actions of these factors counteract disruptions caused by DNA replication and preserves parental histones for epigenetic inheritance. For example, sequences adjacent to cenH at the mating-type region recruit diverse factors to maintain low histone turnover rate, therefore allowing heterochromatin inheritance even in the presence of Epe1 [25–29].
The studies of heterochromatin in wild-type and RNAi mutant cells all demonstrate that low turnover rate of parental histones is important for epigenetic inheritance. The process that generates higher degree of H3K9me3 is very slow in mammalian cells [83, 84], which might also be the case in fission yeast given that Clr4 is inefficient in catalyzing H3K9me3 compare to H3K9me2 [20]. Therefore, the parental histones need to remain at their original locations for longer periods to serve as the template for H3K9me3 inheritance.
Histone turnover and epigenetic stability
Low histone turnover rate not only preserves existing heterochromatin, but also stabilizes ectopic heterochromatin. In certain cases, such ectopic heterochromatin creates epigenetic alleles that allow cells to adapt to environmental challenges.
In addition to large constitutive heterochromatin domains, the fission yeast genome also contains smaller regions of H3K9me2 scattered within the genome, termed heterochromatin islands [85]. These islands are dynamic and can frequently change in response to environmental signals [85–87]. Mutations of the Mst2 complex, INO80 complex, Epe1, and Paf1C all lead to increased H3K9me levels at existing heterochromatin islands, the expansion of H3K9me2 into neighboring regions, and the formation of new ectopic heterochromatin islands [17, 45, 48, 54, 85]. Such expansion is especially dramatic when cells lose both Mst2 and Epe1, leading to the inactivation of essential genes nearby and growth arrest [54]. However, these cells quickly recover by the formation of an ectopic heterochromatin island at the clr4+ locus, which in turn down-regulates the levels of Clr4 to constrain heterochromatin domains in the genome. When the formation of heterochromatin at the clr4+ locus is blocked, cells accumulate heterochromatin at the rik1+ locus, which encodes the Rik1 protein of CLRC required for H3K9 methylation. Given that independent colonies form ectopic heterochromatin at the same locus, it is conceivable that there are weak heterochromatin initiation signals at these loci. In wild-type cells, such heterochromatin islands are quickly erased due to high histone turnover rate by the combined actions of Mst2 and Epe1. However, when both proteins are absent, these ectopic heterochromatin loci are stabilized and selected as they are advantageous for cell growth. Such epigenetic changes can be inherited through mitosis and meiosis, conferring future generations greater resistance to heterochromatic stress [54].
Interestingly, when fission yeast cells are treated with caffeine, the levels of Epe1 are reduced and a shortened form of Mst2 is produced [88]. These changes lead to the formation of an ectopic heterochromatin at hba1+, leading to caffeine resistance. Therefore, there are mechanisms that maintain epigenetic stability through higher histone turnover rate. When subjected to environmental challenges, cells modulate these activities to stabilize heterochromatin, leading to a pool of epigenetic alleles for the selection of new traits and adaption to the environment.
Concluding remarks
The fission yeast has been instrumental in elucidating the mechanism of heterochromatin assembly. The advanced genetic tools allow clear distinction of establishment and maintenance steps to further dissect the mechanism of epigenetic inheritance. The discoveries in fission yeast and other systems firmly established chromatin-based model of epigenetic inheritance: the parental histones are equally deposited into both daughter strands at the original location after DNA replication, and the existing modifications on parental histones recruit the enzymes to modify newly incorporated histones. Recent studies in fission yeast also demonstrate the role of low histone turnover rate in preserving the parental histones during DNA replication for epigenetic inheritance. Interestingly, in Neurospora crassa, heterochromatin regions also show reduced histone turnover, which is dependent on the function of H3K9 methyltransferase, HP1 protein, and histone deacetylase [89], suggesting that the role of low histone turnover in epigenetic inheritance is conserved. Factors that regulate histone turnover not only are critical for maintaining the epigenetic stability of the genome, but also provide ample opportunities for regulatory inputs to change the epigenetic landscapes, allowing cells to adapt to changing environmental conditions. It would be interesting to examine how the activities of these factors are regulated and whether mechanisms of epigenetic inheritance are conserved in higher organisms.
Acknowledgements
We thank Rachel Ding and Qiulin Zhu for editing help. The work in the Jia lab was supported by National Institutes of Health grant R35-GM126910 to SJ.
Abbreviations
- dsRNA
double-strand RNA
- CLRC
Clr4 methyltransferase complex
- FACT
facilitates chromatin transcription
- H3K9me3
trimethylation of histone H3 lysine 9
- RIXC
Rix1 complex
- RITS
RNA-induced transcriptional silencing complex
- RNAi
RNA interference
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- SET
Suv39, Enhancer of Zeste, and Trithorax
- SHREC
Snf2/Hdac-containing Repressor Complex
- TetO
tetracycline operator
- TetR
tetracycline repressor
- TSA
tricostatin A
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Escobar TM, Loyola A & Reinberg D (2021) Parental nucleosome segregation and the inheritance of cellular identity, Nat Rev Genet. 22, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serra-Cardona A & Zhang Z (2018) Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity, Trends Biochem Sci. 43, 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart-Morgan KR, Petryk N & Groth A (2020) Chromatin replication and epigenetic cell memory, Nat Cell Biol. 22, 361–371. [DOI] [PubMed] [Google Scholar]
- 4.Gan H, Serra-Cardona A, Hua X, Zhou H, Labib K, Yu C & Zhang Z (2018) The Mcm2-Ctf4-Polalpha Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands, Mol Cell. 72, 140–151 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R & Groth A (2018) MCM2 promotes symmetric inheritance of modified histones during DNA replication, Science. 361, 1389–1392. [DOI] [PubMed] [Google Scholar]
- 6.Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu RM & Zhang Z (2018) A mechanism for preventing asymmetric histone segregation onto replicating DNA strands, Science. 361, 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escobar TM, Oksuz O, Saldana-Meyer R, Descostes N, Bonasio R & Reinberg D (2019) Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication, Cell. 179, 953–963 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlissel G & Rine J (2019) The nucleosome core particle remembers its position through DNA replication and RNA transcription, Proc Natl Acad Sci U S A. 116, 20605–20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allshire RC & Madhani HD (2018) Ten principles of heterochromatin formation and function, Nat Rev Mol Cell Biol. 19, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grewal SI & Jia S (2007) Heterochromatin revisited, Nat Rev Genet. 8, 35–46. [DOI] [PubMed] [Google Scholar]
- 11.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD & Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases, Nature. 406, 593–9. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama J, Rice JC, Strahl BD, Allis CD & Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly, Science. 292, 110–3. [DOI] [PubMed] [Google Scholar]
- 13.Hong EJ, Villen J, Gerace EL, Gygi SP & Moazed D (2005) A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation, RNA Biol. 2, 106–11. [DOI] [PubMed] [Google Scholar]
- 14.Horn PJ, Bastie JN & Peterson CL (2005) A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation, Genes Dev. 19, 1705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia S, Kobayashi R & Grewal SI (2005) Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin, Nat Cell Biol. 7, 1007–13. [DOI] [PubMed] [Google Scholar]
- 16.Oya E, Nakagawa R, Yoshimura Y, Tanaka M, Nishibuchi G, Machida S, Shirai A, Ekwall K, Kurumizaka H, Tagami H & Nakayama JI (2019) H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly, EMBO Rep. 20, e48111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan CM, Kim JK, Wang J, Bao K, Sun Y, Chen H, Yue JX, Stirpe A, Zhang Z, Lu C, Schalch T, Liti G, Nagy PL, Tong L, Qiao F & Jia S (2021) The histone H3K9M mutation synergizes with H3K14 ubiquitylation to selectively sequester histone H3K9 methyltransferase Clr4 at heterochromatin, Cell Rep. 35, 109137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stirpe A, Guidotti N, Northall SJ, Kilic S, Hainard A, Vadas O, Fierz B & Schalch T (2021) SUV39 SET domains mediate crosstalk of heterochromatic histone marks, eLife. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Mosch K, Fischle W & Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin, Nat Struct Mol Biol. 15, 381–8. [DOI] [PubMed] [Google Scholar]
- 20.Al-Sady B, Madhani HD & Narlikar GJ (2013) Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread, Mol Cell. 51, 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI & Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi, Science. 297, 1833–7. [DOI] [PubMed] [Google Scholar]
- 22.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI & Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex, Science. 303, 672–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J & Allshire RC (2010) Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity, Cell. 140, 666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A & Grewal SI (2002) Establishment and maintenance of a heterochromatin domain, Science. 297, 2232–7. [DOI] [PubMed] [Google Scholar]
- 25.Jia S, Noma K & Grewal SI (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins, Science. 304, 1971–6. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, Choi ES, Shin JA, Jang YK & Park SD (2004) Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6, The Journal of biological chemistry. 279, 42850–9. [DOI] [PubMed] [Google Scholar]
- 27.Wang X & Moazed D (2017) DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation, Science. 356, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Paulo JA, Li X, Zhou H, Yu J, Gygi SP & Moazed D (2021) A composite DNA element that functions as a maintainer required for epigenetic inheritance of heterochromatin, Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenstein RA, Jones SK, Spivey EC, Rybarski JR, Finkelstein IJ & Al-Sady B (2018) Noncoding RNA-nucleated heterochromatin spreading is intrinsically labile and requires accessory elements for epigenetic stability, eLife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanoh J, Sadaie M, Urano T & Ishikawa F (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres, Curr Biol. 15, 1808–19. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Cohen AL, Letian A, Tadeo X, Moresco JJ, Liu J, Yates JR 3rd, Qiao F & Jia S (2016) The proper connection between shelterin components is required for telomeric heterochromatin assembly, Genes Dev. 30, 827–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bahler J, Smith GR & Jia S (2011) Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions, Genes Dev. 25, 214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragunathan K, Jih G & Moazed D (2015) Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment, Science. 348, 1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadaie M, Iida T, Urano T & Nakayama J (2004) A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast, Embo J. 23, 3825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grewal SI & Klar AJ (1996) Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis, Cell. 86, 95–101. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama J, Klar AJ & Grewal SI (2000) A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis, Cell. 101, 307–17. [DOI] [PubMed] [Google Scholar]
- 37.Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL & Allshire RC (2015) Epigenetics. Restricted epigenetic inheritance of H3K9 methylation, Science. 348, 132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P & Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins, Nature. 439, 811–6. [DOI] [PubMed] [Google Scholar]
- 39.Zofall M & Grewal SI (2006) Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats, Mol Cell. 22, 681–92. [DOI] [PubMed] [Google Scholar]
- 40.Trewick SC, Minc E, Antonelli R, Urano T & Allshire RC (2007) The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin, Embo J. 26, 4670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K & Nakayama J (2008) Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast, Mol Cell Biol. 28, 6973–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raiymbek G, An S, Khurana N, Gopinath S, Larkin A, Biswas S, Trievel RC, Cho US & Ragunathan K (2020) An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase, eLife. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Tadeo X, Hou H, Tu PG, Thompson J, Yates JR 3rd & Jia S (2013) Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries, Genes Dev. 27, 1886–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao K, Shan CM, Moresco J, Yates J 3rd & Jia S (2019) Anti-silencing factor Epe1 associates with SAGA to regulate transcription within heterochromatin, Genes Dev. 33, 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorida M, Hirauchi T, Ishizaki H, Kaito W, Shimada A, Mori C, Chikashige Y, Hiraoka Y, Suzuki Y, Ohkawa Y, Kato H, Takahata S & Murakami Y (2019) Regulation of ectopic heterochromatin-mediated epigenetic diversification by the JmjC family protein Epe1, PLoS Genet. 15, e1008129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reyes-Turcu FE, Zhang K, Zofall M, Chen E & Grewal SI (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin, Nat Struct Mol Biol. 18, 1132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F & Jia S (2013) Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly, Genes Dev. 27, 2489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadeghi L, Prasad P, Ekwall K, Cohen A & Svensson JP (2015) The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast, EMBO Rep. 16, 1673–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shan CM, Bao K, Diedrich J, Chen X, Lu C, Yates JR 3rd & Jia S (2020) The INO80 Complex Regulates Epigenetic Inheritance of Heterochromatin, Cell Rep. 33, 108561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomez EB, Espinosa JM & Forsburg SL (2005) Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing, Mol Cell Biol. 25, 8887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez-Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, Yates JR 3rd, Bahler J, O’Connell MJ & Jia S (2012) Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast, The Journal of biological chemistry. 287, 4386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee KK & Workman JL (2007) Histone acetyltransferase complexes: one size doesn’t fit all, Nat Rev Mol Cell Biol. 8, 284–95. [DOI] [PubMed] [Google Scholar]
- 53.Flury V, Georgescu PR, Iesmantavicius V, Shimada Y, Kuzdere T, Braun S & Buhler M (2017) The Histone Acetyltransferase Mst2 Protects Active Chromatin from Epigenetic Silencing by Acetylating the Ubiquitin Ligase Brl1, Mol Cell. 67, 294–307 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Reddy BD & Jia S (2015) Rapid epigenetic adaptation to uncontrolled heterochromatin spreading, eLife. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrison AJ & Shen X (2009) Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes, Nat Rev Mol Cell Biol. 10, 373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh PP, Shukla M, White SA, Lafos M, Tong P, Auchynnikava T, Spanos C, Rappsilber J, Pidoux AL & Allshire RC (2020) Hap2-Ino80-facilitated transcription promotes de novo establishment of CENP-A chromatin, Genes Dev. 34, 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hogan CJ, Aligianni S, Durand-Dubief M, Persson J, Will WR, Webster J, Wheeler L, Mathews CK, Elderkin S, Oxley D, Ekwall K & Varga-Weisz PD (2010) Fission yeast Iec1-ino80-mediated nucleosome eviction regulates nucleotide and phosphate metabolism, Mol Cell Biol. 30, 657–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iglesias N, Paulo JA, Tatarakis A, Wang X, Edwards AL, Bhanu NV, Garcia BA, Haas W, Gygi SP & Moazed D (2020) Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance, Mol Cell. 77, 51–66 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M & Grewal SI (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly, Nature. 451, 734–7. [DOI] [PubMed] [Google Scholar]
- 60.Kloc A, Zaratiegui M, Nora E & Martienssen R (2008) RNA interference guides histone modification during the S phase of chromosomal replication, Curr Biol. 18, 490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, Cande WZ, Antequera F, Arcangioli B & Martienssen RA (2011) RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II, Nature. 479, 135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B & Papamichos-Chronakis M (2015) INO80 Chromatin Remodeler Facilitates Release of RNA Polymerase II from Chromatin for Ubiquitin-Mediated Proteasomal Degradation, Mol Cell. 60, 784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poli J, Gerhold CB, Tosi A, Hustedt N, Seeber A, Sack R, Herzog F, Pasero P, Shimada K, Hopfner KP & Gasser SM (2016) Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress, Genes Dev. 30, 337–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aygun O, Mehta S & Grewal SI (2013) HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin, Nat Struct Mol Biol. 20, 547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verrier L, Taglini F, Barrales RR, Webb S, Urano T, Braun S & Bayne EH (2015) Global regulation of heterochromatin spreading by Leo1, Open Biol. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalik KM, Shimada Y, Flury V, Stadler MB, Batki J & Buhler M (2015) The Paf1 complex represses small-RNA-mediated epigenetic gene silencing, Nature. 520, 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Francette AM, Tripplehorn SA & Arndt KM (2021) The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin, J Mol Biol. 433, 166979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jain D & Cooper JP (2010) Telomeric strategies: means to an end, Annu Rev Genet. 44, 243–69. [DOI] [PubMed] [Google Scholar]
- 69.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S & Narlikar GJ (2017) Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin, Nature. 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X & Karpen GH (2017) Phase separation drives heterochromatin domain formation, Nature. 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD & Narlikar GJ (2019) HP1 reshapes nucleosome core to promote phase separation of heterochromatin, Nature. 575, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svensson JP, Shukla M, Menendez-Benito V, Norman-Axelsson U, Audergon P, Sinha I, Tanny JC, Allshire RC & Ekwall K (2015) A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin, Genome Res. 25, 872–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R & Grewal SI (2007) SHREC, an effector complex for heterochromatic transcriptional silencing, Cell. 128, 491–504. [DOI] [PubMed] [Google Scholar]
- 74.Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S & Moazed D (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms, Mol Cell. 32, 778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamada T, Fischle W, Sugiyama T, Allis CD & Grewal SI (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast, Mol Cell. 20, 173–85. [DOI] [PubMed] [Google Scholar]
- 76.Stralfors A, Walfridsson J, Bhuiyan H & Ekwall K (2011) The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation, PLoS Genet. 7, e1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taneja N, Zofall M, Balachandran V, Thillainadesan G, Sugiyama T, Wheeler D, Zhou M & Grewal SI (2017) SNF2 Family Protein Fft3 Suppresses Nucleosome Turnover to Promote Epigenetic Inheritance and Proper Replication, Mol Cell. 66, 50–62 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Formosa T & Winston F (2020) The role of FACT in managing chromatin: disruption, assembly, or repair?, Nucleic Acids Res. 48, 11929–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC & Ladurner AG (2007) The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi, Curr Biol. 17, 1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, Sun LL, Wheeler D, Zofall M & Grewal SIS (2020) Positioning Heterochromatin at the Nuclear Periphery Suppresses Histone Turnover to Promote Epigenetic Inheritance, Cell. 180, 150–164 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shipkovenska G, Durango A, Kalocsay M, Gygi SP & Moazed D (2020) A conserved RNA degradation complex required for spreading and epigenetic inheritance of heterochromatin, eLife. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahata S, Chida S, Ohnuma A, Ando M, Asanuma T & Murakami Y (2021) Two secured FACT recruitment mechanisms are essential for heterochromatin maintenance, Cell Rep. 36, 109540. [DOI] [PubMed] [Google Scholar]
- 83.Xu M, Wang W, Chen S & Zhu B (2011) A model for mitotic inheritance of histone lysine methylation, EMBO Rep. 13, 60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen ON, Imhof A & Groth A (2015) Two distinct modes for propagation of histone PTMs across the cell cycle, Genes Dev. 29, 585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C & Grewal SI (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation, Science. 335, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gallagher PS, Larkin M, Thillainadesan G, Dhakshnamoorthy J, Balachandran V, Xiao H, Wellman C, Chatterjee R, Wheeler D & Grewal SIS (2018) Iron homeostasis regulates facultative heterochromatin assembly in adaptive genome control, Nat Struct Mol Biol. 25, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei Y, Lee NN, Pan L, Dhakshnamoorthy J, Sun LL, Zofall M, Wheeler D & Grewal SIS (2021) TOR targets an RNA processing network to regulate facultative heterochromatin, developmental gene expression and cell proliferation, Nat Cell Biol. 23, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Torres-Garcia S, Yaseen I, Shukla M, Audergon P, White SA, Pidoux AL & Allshire RC (2020) Epigenetic gene silencing by heterochromatin primes fungal resistance, Nature. 585, 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Storck WK, Abdulla SZ, Rountree MR, Bicocca VT & Selker EU (2020) A Light-Inducible Strain for Genome-Wide Histone Turnover Profiling in Neurospora crassa, Genetics. 215, 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]


