Abstract
COVID-19 is a form of disease triggered by a new strain of coronavirus. Automatic COVID-19 recognition using computer-aided methods is beneficial for speeding up diagnosis efficiency. Current researches usually focus on a deeper or wider neural network for COVID-19 recognition. And the implicit contrastive relationship between different samples has not been fully explored. To address these problems, we propose a novel model, called deep contrastive mutual learning (DCML), to diagnose COVID-19 more effectively. A multi-way data augmentation strategy based on Fast AutoAugment (FAA) was employed to enrich the original training dataset, which helps reduce the risk of overfitting. Then, we incorporated the popular contrastive learning idea into the conventional deep mutual learning (DML) framework to mine the relationship between diverse samples and created more discriminative image features through a new adaptive model fusion method. Experimental results on three public datasets demonstrate that the DCML model outperforms other state-of-the-art baselines. More importantly, DCML is easier to reproduce and relatively efficient, strengthening its high practicality.
Keywords: COVID-19 recognition, Deep mutual learning, Contrastive learning, Fast AutoAugment, Adaptive model fusion
1. Introduction
In 2020, the World Health Organization (WHO) officially declared the outbreak of COVID-19 [1], the disease caused by SARS-CoV-2, a pandemic. COVID-19 is highly infectious and can potentially evolve to fatal acute respiratory distress syndrome (ARDS). Early detection and effective diagnosis are very helpful to control the spreading of COVID-19 [2]. As we know, the most common screening method to detect COVID-19 is reverse-transcription polymerase chain reaction (RT-PCR) testing. However, it is very laborious and some studies have reported its low sensitivity in the early stages. Additionally, the availability of RT-PCR is still limited in many parts of the world. Therefore, many medical images, including medical computed tomography (CT) and X-ray, may be the next options for effectively detecting this virus because most hospitals usually have the corresponding equipment to generate these medical images. Moreover, CT or X-ray images are easily obtained without RT-PCR. Contrarily, health workers need to receive proper training to collect PCR samples, while the processing and generation of CT or X-ray images are relatively easy. Therefore, the computer-aided COVID-19 recognition method has significant practical value, which helps reduce the burden of RT-PCR and speed up diagnosis efficiency[3], [4].
From 2012, deep learning methods, such as convolutional neural network (CNN), have greatly promoted the development of computer vision (CV) [5] field. Recently, deep learning has become an emerging field that can play an important role in the detection of COVID-19 [6], [7], [8]. Some deep learning methods used X-rays or CT images to complete COVID-19 recognition. Their researches have demonstrated great progress in COVID-19 recognition.
However, current researches usually focus on designing deeper or wider neural networks for COVID-19 recognition [9], [10], [11]. Owing to deeper or wider network, the corresponding training procedure of neural network will be faced with several problems, such as gradient vanishing or gradient exploding, which may affect the final performance and practicality of recognition model. Moreover, owing to annotation costs or other ethical reasons, high-quality image samples are usually scarce [12], [13]. This will increase the risk of overfitting. Lastly, the implicit contrastive relationship between different samples has not been fully explored, which can strengthen the discriminative ability of the extracted features. To address the above issues, we propose a novel model, called deep contrastive mutual learning (DCML), for automatic COVID-19 recognition. First, we use a multi-way data augmentation method to enrich the original training dataset. Second, we employ the well-known mutual learning idea to train a relatively lightweight recognition model, which can fully mine the pathological knowledge between the heterogeneous networks and reduce the dependence on computing resources. Third, we incorporate the popular contrastive learning strategy into the mutual learning model. We intend to learn more discriminative features for effective and robust COVID-19 recognition. Finally, we propose a novel adaptive model fusion method to train a more powerful COVID-19 classifier. Conceptually and empirically, the main contributions of this paper can be summarized as follows:
-
1)
We propose a novel DCML model which seamlessly combines many mainstream technologies, including data augmentation, mutual learning, and contrastive learning, to complete effective and robust COVID-19 recognition. DCML tries to imitate practical diagnosis scenarios as much as possible to focus on decision-making. And it is also a good prototype combining contrastive learning and mutual learning.
-
2)
We propose a new adaptive feature fusion method, which focuses on adaptively fusing the image features generated from the sub-networks in the DCML framework.
-
3)
Extensive experiments were conducted on three benchmark datasets. The corresponding results demonstrate the superior classification of our model over other state-of-the-art baseline methods. The code of our method is available at https://github.com/ME-liang/DCML.
The remainder of this paper is organized as follows: Section 2 presents related works and our research motivations. The DCML model is described in Section 3. Experiments on two well-known datasets and the corresponding results are illustrated in Section 4. Finally, Section 5 provides the conclusions and future scope of the work.
2. Related works
2.1. COVID-19 recognition
A world-class infectious disease has outbroken in 2020, it was later found that the source of infection was a novel coronavirus called COVID-19. Owing to the low efficiency of manual examination of the medical images including CT or X-ray images, more and more researchers in the fields of CV or machine learning tried to develop a computer-aided intelligent system, hoping that these medical images can be used for automatic intelligent detection of COVID-19 and improving the corresponding recognition efficiency.
Recently, deep learning technologies, including CNN [14], [15], [16], [17], [18] and Transformer [19], have achieved great success in most CV tasks, which also plays an important role in the detection of COVID-19. For example, Jaiswal et al. [20] fine-tuned the DenseNet 201 model for COVID-19 recognition. Sen et al. [21] used a bi-stage deep feature selection approach for the COVID-19 detection problem. Butt et al. [22] aimed to establish a screening model for distinguishing COVID-19 pneumonia from that Influenza-A viral pneumonia and healthy cases using a ResNet18 with a location-attention mechanism. Some following-up methods based on transfer learning have been proposed, and most of them used those existing networks, such as VGG [23], ResNet [24], [25], [26], and DenseNet [27]. Apostolopoulos et al. [28] relied on the MobileNet [29] with better interpretability for helping radiologists to understand how the predictions were produced. Angelov et al. [30] used the GoogLeNet architecture (a non-pretrained model) for extracting features. Then they used the extracted features to train a multi-layer perception classifier for the recognition of COVID-19 CT images. Panwar et al. [31] used the pre-trained VGG-19 model with five fully-connected layers to complete COVID-19 recognition. Panwar et al. [31] also employed a Grad-CAM [32] -based color visualization approach to better interpret the corresponding diagnosis results.
Recently, several new network architectures for COVID-19 recognition emerged, such as the COVID-Net [33], a representative work which achieved a promising accuracy for image-level diagnosis on the chest X-ray (CXR). Based on the COVID-Net, Javaheri et al. [34] later proposed the CovidCTNet to differentiate positive COVID-19 infections from community-acquired pneumonia and other lung diseases. Soltanian et al. [35] proposed a lightweight model for cough recognition. S. Ghosh et al. [36] proposed a modified residual network-based enhancement (ENResNet) scheme for the visual clarification of COVID-19 pneumonia impairment from CXR images and classification of COVID-19 under deep learning framework. P. Gaur et al. [37] proposed a new method for preprocessing and classifying COVID-19 positive and negative from CT scan images. This method which is being proposed uses the concept of empirical wavelet transformation for preprocessing, selecting the best components of the red, green, and blue channels of the image are trained on the proposed network. Series Adapter [38] and Parallel-Adapter [39] use series domain adapter for joint learning from multiple image datasets. MS-Net [40] constructs a multi-site recognition model. They can also be used for COVID-19 recognition. Zhao et al. [41] proposed the latest modified version of COVID-Net for the recognition of COVID-19 CT images. It absorbed a contrastive learning objective into its model and explicitly regularized the class-sensitive and domain-invariant latent semantic feature space. Moreover, a group of state-of-the-art models, including Fast ConvNets [42], neural architecture search net (NasNet) [43], gray-level co-occurrence matrix (GLCM) [44], and EfficientNet [45] have been used to complete COVID-19 recognition on CXR images.
Recently, Huang et al. [46] proposed a new Evidential COVID-Net for COVID-19 recognition, which is composed of CNN-based feature extraction and belief function-based classification module. An alternative redesigned framework based on Capsule Network [47] aims to handle small-scale datasets more effectively, which is of valuable significance given the emergency of COVID-19 initial outbreak. Unlike the above researches, Gozes et al. [48] presented a system that can utilize both 2D and 3D deep learning models, relying on modifying and adapting out-of-the-box artificial intelligence models and combining them with domain-wise clinical understanding. Similarly, Li et al. [49] also proposed a 3D deep learning architecture for COVID-19 recognition. Moreover, some researchers focused on the feature fusion method. Tang et al. [50] tackled automated severity assessment (i.e., differentiating non-severe and severe) for COVID-19 recognition using CT images through the exploration of those identified severity-related features. Rahimzadeh et al. [51] developed a neural network that concatenate the corresponding features extracted from Xception and ResNet50V2 networks, which can boost the final recognition performance.
All the above-mentioned works have achieved great progress on the task of COVID-19 recognition. However, on the one hand, the corresponding COVID-19 images are very scarce, which may lead to overfitting. On the other hand, the above networks are very complex and hard to reproduce. Notably, they have not explored the potential ability of heterogeneous networks.
2.2. Deep mutual learning
Deep neural networks have achieved remarkable results in the fields of CV, speech recognition, and natural language processing. To complete more complex tasks, the corresponding network must use a deeper or wider structure. Although these neural networks have achieved satisfactory performance on specific tasks, lots of computing requirements make them difficult to be deployed on those resource-constrained environments, which may limit their practicalities. To address this issue, Hinton [52] proposed the famous model distillation method. It regards a pre-trained large network as a teacher that provides additional knowledge to a small network (student). The student network imitates the category probability estimated by the teacher network. And the student network can even obtain better performance. The basic principle behind model distillation [52] is using the additional supervision from the teacher model to train the student model, which surpasses the traditional supervised learning objective. Since then, most offline distillation methods have followed this principle. In [52], [53], [54], [55], the classification probability distribution of the corresponding teacher model was used as the additional supervision. However, the above traditional model distillation methods need a pre-trained large network. And they only employ a one-way knowledge transfer, which cannot take full advantage of the teacher and student networks.
To address this problem, Zhang [56] proposed a deep mutual learning (DML) strategy in which a group of student networks learns and guides each other throughout the whole training process. Instead of the statically one-way knowledge conversion between the teacher and student networks, DML uses multiple networks to train at the same time. Each network not only accepts the supervision from the ground-truth but also refers to the learning experience from the peer network. All these can further improve the generalization ability of the whole framework. Finally, the two networks share learning experiences (dark knowledge) to achieve mutual learning and common great progress. More importantly, online knowledge distillation (KD) between heterogeneous networks can be realized easily. Anil et al. [57] and Gao et al. [58] further extended the DML idea to accelerate the training procedure of a large-scale distributed neural network. Although the above researches have promoted the progress of KD, none of them absorbed the contrastive learning idea to the distillation procedure. Moreover, to the best of our knowledge, few works have used the DML idea to complete COVID-19 recognition.
2.3. Research motivations
Most of the above-mentioned methods usually used a deeper or wider network structure of a single model to improve the final recognition performance. Objectively speaking, this is an effective method for improving COVID-19 recognition. However, as we know, owing to complex network structure, the corresponding training procedure of neural network will be faced with several problems, such as gradient vanishing or gradient exploding, which may affect the final performance and practicality of recognition model. Meanwhile, owing to the lack of sufficient training samples [12], [13], the final recognition model is prone to overfitting, which may also reduce its practicality to a certain degree. Lastly, the implicit contrastive relationship between different samples has not been explored, which helps strengthen the discriminative ability of the extracted features.
Naturally, our research motivations are three-folds. First, we used a multi-way data augmentation method, which can automatically search the most suitable augmentation strategy for the COVID-19 image datasets, and obtain high-quality augmented image samples. This lays a firm data foundation for the subsequent training. Second, we employed the well-known DML framework to train a relatively lightweight recognition model, which can make full use of the pathological knowledge between heterogeneous networks and reduce the dependence on computing resources. More importantly, we incorporated the popular contrastive learning strategy into our DML-based recognition model. Hence, we obtained the DCML model. It is also a good prototype combining contrastive learning and mutual learning. We aim to learn more discriminative features through contrastive learning for more effective COVID-19 recognition. Finally, based on DCML, we proposed a novel adaptive model fusion method to generate more discriminative features and train a more powerful COVID-19 image classifier.
3. The proposed DCML model
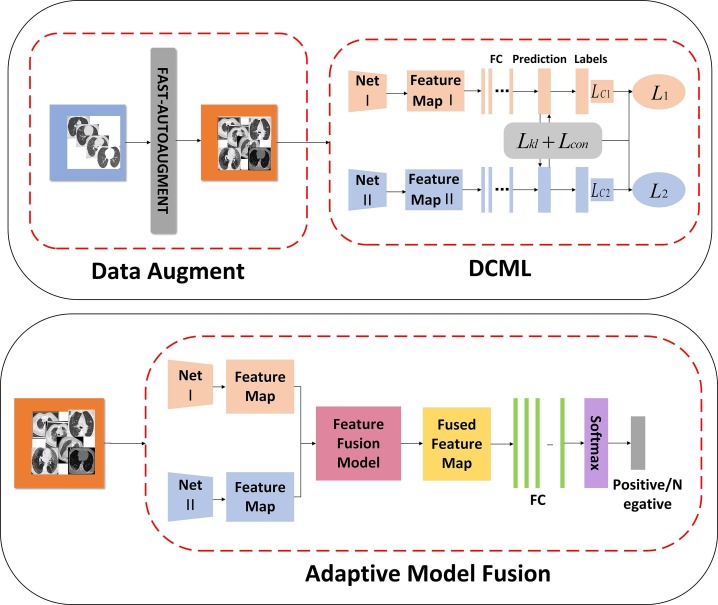
The core idea of the DCML model is to imitate practical diagnosis scenarios as much as possible to focus on decision-making. Our model consists of four parts. First, the proposed DCML model uses the AutoAugment-based method to obtain more high-quality image samples. Secondly, our model employs the DML framework to break the isolation between different networks and establishes a strong foundation for the fusion of these complementary networks. This can imitate the mutual communication and learn from multiple experienced pathologists (or radiologists) to a certain extent. Notably, unlike the traditional DML model, DCML absorbs the contrastive learning idea into the knowledge transfer procedure, which enhances the model’s ability to distinguish different classes. Finally, we propose the adaptive model fusion strategy to further utilize the implicit complementary correlations among heterogeneous networks and train a more powerful image classifier with better generalization capabilities, which can mimic the centralized decision-making process of these pathologists (or radiologists) as much as possible. The corresponding technological pipeline of our method is illustrated in Fig. 1 .
Fig. 1.
The technological pipeline of the DCML model.
3.1. Model framework
Fig. 1 shows the technological pipeline of the DCML model.
At the first stage, we employed the Fast AutoAugment (FAA) [59] method to enrich the original training dataset. Compared with other traditional data augmentation methods, FAA can automatically search effective data augmentation strategies for the target datasets. In this way, high-quality image samples can be obtained. Additionally, unlike other AutoAugment-based method [60], FAA can avoid the GPU-consuming of repeatedly training the network by adjusting different data augmentation parameters, thereby achieving a speed increase of 100 ∼ 1000 times.
At the second stage, we used any two networks (Net I and Net II) to build the DCML model, which ensures that the proposed DCML model is a general and versatile framework. Each of the two networks form a parallel queue of the DCML model. Additionally, we modified the DML framework by adding a contrastive loss to the original mutual learning procedure, which can strengthen each network’s ability to distinguish different classes. In this knowledge transfer procedure, the two networks can mine out sufficient pathological knowledge for independent training and form complementary advantages for the whole framework.
At the last stage, we proposed a new adaptive model fusion strategy. It further strengthens the implicit complementarity by fusing the corresponding heterogeneous layers from the two networks. To fulfill this goal, each network is frozen. The two networks are only used to extract the corresponding deep-level features of CT images, including “Feature Map I” and “Feature Map II”. And the corresponding feature maps from heterogeneous layers are adaptively fused to create more discriminative features for training the final COVID-19 image classifier.
3.2. Model formulation
Stage 1: FAA Strategy.
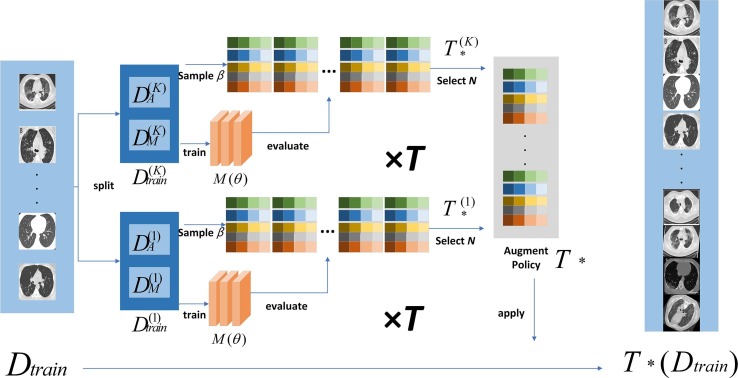
Fig. 2 shows the procedure of FAA.
Fig. 2.
The technological procedure of FAA.
Data augmentation is a useful strategy that can simultaneously increase the amount and diversity of the training data through some random techniques. As we know, popular augmentation methods include shifting, flipping, mirroring, and other spatial operations. However, fixed data augmentation methods are not suitable for all datasets. Contrarily, AutoAugment-based methods [59], [60] can adaptively search effective data augmentation strategies for a target dataset. It first creates a search space for data augmentation strategies and then directly evaluates the quality of a specific strategy on the target dataset. A strategy in the search space contains many sub-strategies. Each sub-strategy is randomly selected for each image sample in each mini-batch. Each sub-strategy consists of two traditional operations such as translation, rotation or shearing, and the probability of applying these operations. As analyzed above, owing to good performance and rapid speed, we employed FAA [59] to implement data augmentation. The FAA method is shown as Algorithm 1.
In Algorithm 1, is the relevant model parameter of FAA. is the training dataset. The dataset is divided into parts. represents the number of Bayesian optimizations on the same sub-dataset. is the number of iterations of the Bayesian optimizer. Each Bayesian optimization selects the best samples as a candidate strategy and divide into and , which are used to learn model parameters and explore enhancement strategy respectively. represents augmentation strategy.
| Algorithm 1: Fast AutoAugment |
|---|
| Input: () |
| Split into K-fold data |
| Fordo |
| Train and |
| Fordo |
| Return |
Through the FAA algorithm, we can find the best augmentation strategy suitable for the COVID-19 datasets and obtain high-quality image samples, which lays a firm data foundation for the subsequent model training.
Stage 2: DCML.
In the DCML model, we first use the mutual distillation characteristic of the DML framework to explore the complementary relationship of different networks. The DML framework can enable multiple networks (we used two heterogeneous networks in DCML) to realize online mutual knowledge transfer during the training process. And this two-way knowledge transfer strategy lays a powerful foundation for the subsequent adaptive model fusion.
In addition to mining the complementary knowledge under the mutual learning mode, another goal is to promote intra-class cohesion and inter-class separation of infected semantic embedding (i.e., COVID-19) and cross-modal non-infected cases. Hence, we adopt the popular contrastive learning idea into our model and merge a contrastive loss with the Kullback-Leibler (KL) loss. The contrastive loss can make full use of the label information than the traditional cross-entropy loss. After adding the contrastive loss, the clusters of samples belonging to the same class are pulled closer in the embedding space whereas the clusters of samples from different classes are pushed far away than before. This characteristic helps improve the final recognition accuracy. Here we formulate the DCML model in a general way.
samples from categories are described as , while the set of the corresponding labels is expressed as with . In each iterative optimization process, the same mini-batch is input into the two networks, respectively, and the same calculation process is performed on each network.
The probability of class for sample given by the neural network Net I is computed as:
| (1) |
where the logit is the output of the softmax layer.
For a multi-classification task, we first need a supervised loss. And the cross-entropy error between the predicted values and the correct labels is defined as the supervised loss of the neural network Net I:
| (2) |
where is an indicator function. If the label is equal to the predicted value, is set to 1. Otherwise, it is set to 0.
| (3) |
Similarly, we can obtain the supervised loss, called , of the neural network Net II, which is shown as follow:
| (4) |
Besides the supervised loss introduced above, we also need a loss to complete mutual learning in our DCML framework. As shown in Fig. 1, the peer network Net II is introduced to improve the prediction accuracy and generalization ability of the network Net I. The network Net II uses its posterior probability to provide its training experience (or dark knowledge) to the network Net I. Meanwhile, the network Net I uses its posterior probability to offer its training experience to the network Net II. Hence, this builds a two-way knowledge transfer mode. To ensure that and can play a positive role in the whole training procedure, we employ the KL divergence as shown in Equation (5) to quantify the matching degree of the predictions of the two networks.
| (5) |
Similarly, we can obtain the mutual learning loss, called , which is shown as follow:
| (6) |
As described above, the contrastive loss focuses on learning a kind of mapping relationship, which contributes to making the following contrastive learning: the samples belonging to the same class are pulled closer while the samples from different categories are pushed far away than before. Based on this idea, we employ the contrastive loss [61] shown as follows to complete contrastive learning:
| (7) |
Where represents the direct Euclidean distance between and . w is the network weight. is the label of whether the two samples match, means that the two samples are similar or matched. On the contrary, means not matching, represent the set threshold, and is the number of samples.
Summarily, based on the above-mentioned mutual learning loss, contrastive loss, and supervised loss, we define the final loss, namely and , of the two networks Net I and Net II, respectively:
| (8) |
| (9) |
Therefore, in the proposed DCML model, any network no longer learns in isolation. The two networks can learn from each other and encourage each other to mine sufficient pathological knowledge for better depicting COVID-19 images. Meanwhile, the contrastive loss can promote intra-class cohesion and inter-class separation of all the samples. Moreover, the corresponding feature layer of each network also builds a firm foundation for the subsequent adaptive model fusion. The pathological knowledge and diagnosis experience from different “pathologists or radiologists” (Net I or Net II) can complement each other, which is a necessary basis for centralized decision-making.
Stage 3: Adaptive Model Fusion.
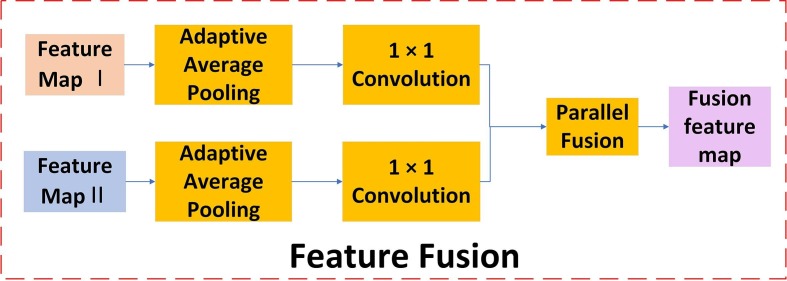
As shown in Fig. 1, we fuse two heterogeneous features after implementing DCML, which can mimic the centralized decision-making process of these pathologists (or radiologists) as much as possible. To fuse two heterogeneous features, we should match their sizes. First, the pooling-sampling operations are used to transform the length and width of each feature map, and the 1 × 1 convolution is employed in turn to transform the number of channels of each feature map. Particularly, we used an adaptive average pooling strategy. In this adaptive average pooling procedure, it is necessary to set the size of the output tensor according to the one desired without considering the size of the input feature maps. The fused features are input into the final softmax layer to train a classifier. Therefore, the length and width of each feature map are set to 1. Moreover, the 1 × 1 convolution changes the number of the corresponding channels, which enhances the abstract expression ability of the local module. Hence, the proposed model fusion method can fuse any two feature maps from heterogeneous networks, adaptively. This can cleverly avoid the traditional operators and does not need to set different pooling and convolution parameters for different feature maps. From another perspective, this helps strengthen the practicality of the proposed COVID-19 recognition model. Our adaptive fusion procedure is shown in Fig. 3 .
Fig. 3.
The adaptive model fusion process.
As mentioned above, the adaptive average pooling and 1 × 1 convolution can match any two feature maps with arbitrary sizes. Then, we used the concatenation mode to complete the final model fusion. Given feature maps, namely , the principle of the concatenation mode is shown as follows:
| (10) |
Here, is a collection of all the feature maps. This means that makes the new feature more diverse, which is beneficial to the final COVID-19 image classification.
After the fused feature maps have been obtained, the result of the adaptive model fusion method is implemented through the dense connect function shown as follows:
| (11) |
where and refer to the corresponding weight and bias, respectively. A fusion classifier is built in turn based on . And it is fine-tuned to achieve the best classification performance on the test dataset. In this equation, for the concatenation operation, the shapes of and will change as the fusion changes. This requires the same length and width of any feature map. The concatenation mode makes the number of channels of the fused feature map equal to the sum of the number of channels of each single feature map. Therefore, when we use the concatenation mode to fuse the extracted feature maps, the corresponding length and width of each feature map should be converted to the same size. As described above, the adaptive average pooling and 1 × 1 convolution guarantee that the model fusion method can adaptively match any two feature maps with arbitrary sizes. Hence, we can obtain a more powerful fusion classifier for COVID-19 image recognition.
In summary, the proposed data augmentation method lays a firm data foundation for model training. The DCML framework establishes the complementary correlation among different networks, which enables our recognition model to fully mine the complementary dark knowledge and implement effective contrastive learning. Finally, the adaptive model fusion method is proposed to create the fusion classifier for COVID-19 image recognition.
4. Dataset preparation
4.1. Datasets
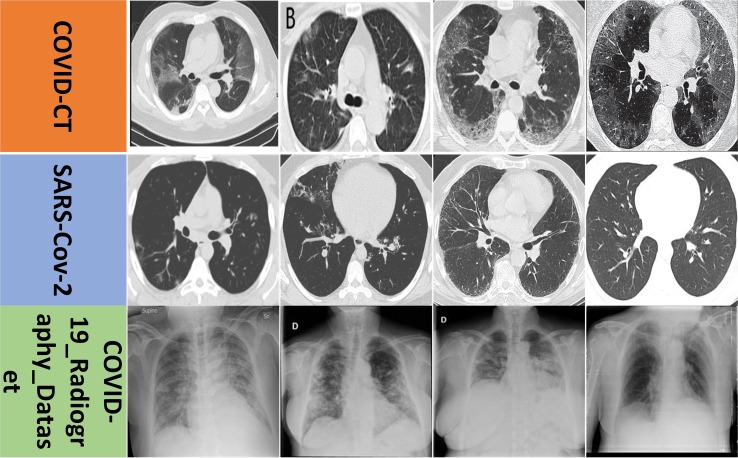
According to Refs. [62], [63], [64], [65], we adopted three public COVID-19 CT image datasets, including COVID-CT [62], SARS-CoV-2 [63], and COVID-19_Radiography_Dataset [64], [65], to evaluate our model. The corresponding URL link for the COVID-19 CT image is shown as follows: https://github.com/UCSD-AI4H/COVID-CT. The corresponding URL link for the SARS-Cov-2 image is shown as follows: https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset. The corresponding URL link for the COVID-19_Radiography_Dataset image is shown as follows: https://www.kaggle.com/tawsifurrahman/COVID19-radiography-database?select=COVID-19_Radiography_Dataset. The SARS-CoV-2 dataset consists of 2482 CT images from 120 patients, in which 1252 images are positive with COVID-19 and other 1230 images are non-COVID but with other types of lung disease manifestations. The spatial sizes of these images range from 119 × 104 to 416 × 512. The COVID-CT dataset includes 349 CT images from 216 patients containing clinical findings of COVID-19 and 397 CT images from 171 patients without COVID-19. The resolutions of these images range from 102 × 137 to 1853 × 1485. COVID-19_Radiography_Dataset is a popular database of CXR images for COVID-19 positive cases along with normal and infection images. In its first version, 219 COVID-19 and 1341 normal CXR images were release. In its first update, the corresponding COVID-19 samples were increased to 1200 CXR images. In its current 2nd update, the authors increased the database to 3616 COVID-19 positive cases along with 10,192 normal images on January 06th, 2021. The resolution of each image in the new dataset is uniform 299 × 299. We used the 2nd update of COVID-19_Radiography_Dataset in our experiments. Compared with CT image, CXR image has the following three evident advantages: 1) lower acquisition cost; 2) lower radiation dosage; 3) faster imaging speed. Hence, CXR images are widely used as lung disease screening, such as COVID-19 recognition.
Summarily, the three datasets have some apparent differences in imaging mode, acquisition time, and sample source. These help firmly and comprehensively validate the effectiveness and robustness of the proposed DCML model. In our experiments, all images are resized to 224 × 224 in an axial plane. Fig. 4 shows some representative images from the three datasets.
Fig. 4.
Datasets exhibition.
4.2. Data augmentation and image preprocessing
In this study, the well-known data augmentation technology named FAA was first used to enrich the three public datasets. This can not only improve the final classification performance but also alleviate the overfitting problem and enhance the robustness of the proposed recognition model. For fair performance comparison, we only enrich the training dataset. Suggested data augmentation techniques include rotation, horizontal flip, mirror flip, and random cropping. Therefore, the training set is doubled after data augmentation. In addition to data augmentation, we also performed normalized preprocessing on CT or CXR images. Each image was normalized into zero mean and unit variance for intensity values along channel dimension. The statistical data of the three datasets are exhibited in Table 1 . As shown in Table 1, COVID-19_Radiography_Dataset has more image samples compared to the COVID-CT and SARS-Cov-2 datasets. Notably, the sample imbalance phenomenon is more evident on this dataset. Hence, each dataset has its own characteristic, which puts forward higher requirements for the proposed recognition model.
Table 1.
The statistical data of the three public COVID image datasets. Unit: image.
| Dataset | Train |
Test |
|||
|---|---|---|---|---|---|
| Pos | Neg | Pos | Neg | ||
| COVID-CT | Original | 251 | 292 | 98 | 105 |
| After FAA | 502 | 584 | 98 | 105 | |
| SARS-Cov-2 | Original | 939 | 923 | 313 | 307 |
| After FAA | 1878 | 1846 | 313 | 307 | |
| COVID-19_Radiography_Dataset | Original | 2712 | 7644 | 904 | 2548 |
| After FAA | 5424 | 15,288 | 904 | 2548 | |
5. Experimental results and analysis
5.1. Evaluation metrics and baselines
5.1.1. Evaluation metrics
To evaluate the proposed COVID-19 recognition model comprehensively and objectively, several evaluation metrics, including accuracy, the area under the receiver operating characteristic (ROC) curve (AUC), precision, recall (sensitivity), and F1, are employed in this study.
Accuracy is a popular metric for image classification. For our prediction results, it represents the ratio of correct predictions to the total number of samples in the prediction results, which is shown as follow:
| (12) |
where is the total number of the COVID-19 images, and is the number of the images that have been correctly classified by the model.
Precision is another mainstream metric for image classification. It is the ratio of images that are correctly classified as positive (COVID) and all images that are classified as positive. It indicates how many of the image samples predicted to be positive are true positive samples. Hence, the Precision metric is shown as follows:
| (13) |
where means true positive and means false positive.
Recall (Sensitivity) is another important metric for image classification. It is the ratio of correct predictions (positive) to the total number of samples in the prediction results. The Sensitivity metric is shown as follows:
| (14) |
where is false negative.
F1 is used to evaluate the overall classification performance of a model. It is the harmonic mean of the Precision and Sensitivity values, which is shown as follows:
| (15) |
AUC represents the area under the ROC curve. It is another important metric that is usually used to evaluate the overall classification performance of a classification model. A large AUC indicates satisfactory performance, which means that the corresponding ROC curve is very close to the (0, 1) point and far from the 45◦ diagonal of the coordinate axis.
5.1.2. Implementation details
We used the PyTorch backend on our server with three NVIDIA GeForce GTX 2080Ti GPUs, and our memory is 94 GB. Each dataset is split randomly into 70% for training and 30% for testing. We used Adam with weight decay and set the initial learning rate to 0.001. The momentum value is 0.9 the weight decay for regularization is 1e-4, and the decay for learning rate is 0.1. We set the batch size to 40 and trained our model for 50 epochs. According to our experimental results, we chose SeResNet50 (Net I) and SeResNetxt101 (Net II) as the two basic networks to create the DCML model.
5.1.3. Baselines
We compare the proposed DCML model with the following three types of baselines:
-
1)
The fine-tuned deep learning networks include ResNet50, ResNet101, DenseNet201, VGG-16, VGG-19, SqueezeNet, GoogleNet, AlexNet, InceptionResNetV2, and InceptionV3.
-
2)
Two sub-nets we used in DCML: SeResNet50 (SeR50), and SeResNetxt101 (SeR101).
-
3)
Mainstream COVID-19 image recognition models include Evidential Covid-Net [46], Angelov et al.’s model [30], Panwar et al.’s model [31], Sen et al.’s model [21], Jaiswal et al.’s model [20], COVID-Net [33], Series Adapter [38], Parallel Adapter [39], MS-Net [40], Zhao et al.’s model [41], Fast-CovNet [42], NasNet [43], GLCM [44], and EfficientNet [45].
5.2. Quantitative results
First, we compare the DCML model with all the baselines introduced above, and the corresponding experimental results are shown in Table 2 . As shown in Table 2, the proposed DCML model performs well on both datasets. We made a deep-level analysis from the following three perspectives.
Table 2.
Performance comparisons with baselines. The best value of each column is shown as 0.8818. “w” means with FAA while “w/o” means without FAA.
| Dataset | Model | Accuracy↑ | F1↑ | Sensitivity↑ | Precision↑ | AUC↑ |
|---|---|---|---|---|---|---|
| COVID-CT | ResNet50 | 0.7600 | 0.7300 | 0.6700 | 0.8000 | 0.7600 |
| ResNet101 | 0.7900 | 0.7800 | 0.8000 | 0.7600 | 0.7900 | |
| DenseNet201 | 0.7900 | 0.7700 | 0.7300 | 0.8100 | 0.7900 | |
| VGG-19 | 0.8000 | 0.7900 | 0.7900 | 0.8100 | 0.8000 | |
| SqueezeNet | 0.7300 | 0.7000 | 0.6500 | 0.8000 | 0.7300 | |
| SeR50 | 0.7488 | 0.7302 | 0.7041 | 0.7582 | 0.7588 | |
| SeR101 | 0.7537 | 0.7573 | 0.7959 | 0.7222 | 0.7301 | |
| Evidential Covid-Net | 0.7310 | 0.7020 | / | / | 0.8700 | |
| COVID-Net | 0.6312 | 0.6109 | 0.5773 | 0.6403 | 0.7109 | |
| Series Adapter | 0.7001 | 0.6708 | 0.7491 | 0.6304 | 0.7392 | |
| Parallel Adapter | 0.7493 | 0.7346 | 0.7181 | 0.7984 | 0.8029 | |
| MS-Net | 0.7623 | 0.7654 | 0.7407 | 0.7929 | 0.8219 | |
| Zhao et al. | 0.7869 | 0.7883 | 0.7971 | 0.7802 | 0.8532 | |
| SeR50 (DML) | 0.8079 | 0.7983 | 0.8102 | 0.8087 | 0.8083 | |
| SeR101 (DML) | 0.8621 | 0.8516 | 0.8029 | 0.9190 | 0.8559 | |
| DCML (w/o FAA) | 0.8768 | 0.8737 | 0.8807 | 0.8829 | 0.8807 | |
| DCML (w FAA) | 0.8818 | 0.8779 | 0.8811 | 0.8800 | 0.8811 | |
| SARS-Cov-2 | ResNet101 | 0.9496 | 0.9503 | 0.9715 | 0.9300 | 0.9498 |
| GoogleNet | 0.9173 | 0.9182 | 0.9350 | 0.9020 | 0.9179 | |
| VGG-16 | 0.9496 | 0.9497 | 0.9543 | 0.9402 | 0.9496 | |
| AlexNet | 0.9375 | 0.9361 | 0.9228 | 0.9498 | 0.9368 | |
| SeR50 | 0.8824 | 0.8828 | 0.8786 | 0.8871 | 0.8821 | |
| SeR101 | 0.9098 | 0.9088 | 0.8914 | 0.9269 | 0.9311 | |
| Angelov et al. | 0.8860 | 0.8915 | 0.8860 | 0.8970 | / | |
| Panwar et al. | 0.9404 | 0.9450 | 0.9400 | 0.9500 | / | |
| Sen et al. | 0.9532 | 0.9530 | 0.9530 | 0.9530 | / | |
| Jaiswal et al. | 0.9625 | 0.9629 | 0.9629 | 0.9629 | / | |
| COVID-Net | 0.7712 | 0.7603 | 0.7097 | 0.8004 | 0.8408 | |
| Series Adapter | 0.8573 | 0.8619 | 0.8191 | 0.9098 | 0.9293 | |
| Parallel Adapter | 0.8213 | 0.8239 | 0.8002 | 0.8351 | 0.8999 | |
| MS-Net | 0.8798 | 0.8873 | 0.8491 | 0.9378 | 0.9437 | |
| Zhao et al. | 0.9083 | 0.9087 | 0.8589 | 0.9575 | 0.9624 | |
| SeR50 (DML) | 0.9274 | 0.9251 | 0.9423 | 0.9103 | 0.9229 | |
| SeR101 (DML) | 0.9452 | 0.9400 | 0.9350 | 0.9494 | 0.9419 | |
| DCML (w/o FAA) | 0.9565 | 0.9553 | 0.9596 | 0.9561 | 0.9596 | |
| DCML (w FAA) | 0.9742 | 0.9733 | 0.9755 | 0.9731 | 0.9731 | |
| COVID-19_Radiography_Dataset | VGG19 | 0.9000 | 0.8800 | 0.9400 | 0.8300 | 0.8700 |
| ResNet101 | 0.8751 | 0.9220 | 0.8701 | 0.8923 | 0.8910 | |
| ResNet50 | 0.8866 | 0.8744 | 0.8996 | 0.8612 | 0.9566 | |
| InceptionResNetV2 | 0.9485 | 0.9617 | 0.9401 | 0.9447 | 0.9201 | |
| Fast-CovNet | 0.8211 | 0.8006 | 0.9805 | 0.6765 | 0.7664 | |
| Inceptionv3 | 0.7668 | 0.8163 | 0.7009 | 0.9771 | 0.8021 | |
| NasNet | 0.9583 | 0.9594 | 0.9337 | 0.9866 | 0.9858 | |
| GLCM | 0.9222 | 0.9030 | 0.8895 | 0.7911 | 0.8156 | |
| EfficientNet | 0.8084 | 0.7707 | 0.7244 | 0.8602 | 0.8200 | |
| SeR50 | 0.8674 | 0.8719 | 0.9271 | 0.8065 | 0.8696 | |
| SeR101 | 0.8786 | 0.8878 | 0.9392 | 0.8148 | 0.8873 | |
| SeR50 (DML) | 0.9545 | 0.9442 | 0.9532 | 0.8834 | 0.9412 | |
| SeR101 (DML) | 0.9539 | 0.9576 | 0.9591 | 0.9470 | 0.9667 | |
| DCML (w/o FAA) | 0.9826 | 0.9762 | 0.9770 | 0.9778 | 0.9771 | |
| DCML (w FAA) | 0.9870 | 0.9825 | 0.9770 | 0.9837 | 0.9865 |
Compared with the fine-tuned deep learning networks, the corresponding performance of the DCML model was greatly improved on both datasets. For example, on the COVID-CT dataset, compared with the most competitive VGG-19 model, the corresponding improvements of accuracy, F1-score, Sensitivity, Precision, and AUC are 8.18%, 8.79%, 9.11%, 7.00%, and 8.11%, respectively. On the SARS-Cov-2 dataset, compared with the most competitive ResNet101 model, the corresponding improvements are 2.46%, 2.30%, 0.40%, 4.31%, and 2.33%, respectively. On COVID-19_Radiography_Dataset, compared with the most competitive VGG-19 model, the corresponding improvements of accuracy, F1-score, Sensitivity, Precision, and AUC are 8.70%, 10.25%, 3.70%, 15.37%, and 11.65%, respectively. As we know, deep learning networks are very complex (i.e., ResNet101 and DenseNet201) and require sufficient high-quality training samples, which makes them prone to overfitting. Especially for the medical image recognition tasks, owing to the lack of sufficient image samples, the overfitting problem becomes more serious or evident than before. Contrarily, the proposed DCML model can better deal with this problem. On the one hand, it can obtain high-quality image samples through data augmentation, which helps alleviate the problem of data scarcity to a certain degree. On the other hand, the corresponding network trained by the proposed DCML framework is relatively lightweight with fewer parameters and lower complexity. Hence, our recognition model is not prone to overfitting. Importantly, it has a powerful generalization ability. Compared with the sub-nets we used in DCML, the proposed DCML model also performs better on both datasets. For example, on the COVID-CT dataset, compared with the SeR101, the corresponding improvements of accuracy, F1-score, Recall, Precision, and AUC are 12.81%, 12.06%, 8.52%, 15.78%, and 15.1%, respectively. In the DCML model, we make the two sub-nets learn from each other, which enhances the heterogeneous information transfer between the networks. In addition, the contrastive loss between different samples is always added into the process of mutual learning, which strengthens the classification ability of the model, and finally, the two heterogeneous networks are adaptively merged. All these contribute to the improvement of the final performance of the DCML model.
Compared with those mainstream models for COVID-19 classification, our DCML model also performs better on both datasets. For example, on the COVID-CT dataset, compared with the most competitive baseline (Zhao et al.), the corresponding improvements of accuracy, F1-score, Recall, Precision, and AUC are 9.49%, 8.96%, 8.40%, 9.98%, and 2.79%, respectively. Similar results can also be observed on the SARS-Cov-2 and COVID-19_Radiography_Dataset datasets. DCML can process both CT and CXR images well. And it also solves the data imbalance problem well. Hence, the proposed DCML model is effective and robust for COVID-19 image recognition. Contrarily, Zhao et al. only used the traditional data augmentation method, which cannot effectively alleviate the data scarcity problem. However, the DCML model employs the state-of-the-art FAA method to enrich the original training dataset. This builds a firm data foundation for the subsequent model training. More importantly, unlike Zhao et al. and Jaiswal et al., the DCML model employs several convincing losses, including supervised loss, mutual learning loss, and contrastive loss, to complete COVID-19 image classification. All these losses form a kind of joint force to promote the corresponding performance improvement of the DCML model. Notably, compared with these baselines, our model is lightweight, which can be deployed on the most common devices.
Summarily, the proposed DCML model is superior to all the baselines, and it also has the following important characteristics: simplicity, lightweight, easier to implement, and high efficiency. These help to enhance the practical value of the proposed model. Certainly, owing to few training samples, challenges remain on the current COVID-CT datasets. In the future, we intend to introduce the attention mechanism to better capture the key lesions of COVID CT images.
5.3. Cross-Validation
Cross-validation is an important method for evaluating recognition models objectively. The most commonly-used cross-validation is k-fold method. The specific k-fold cross-validation procedure is: we randomly divide the training set into k parts, and take one of them as the validation set which is used to evaluate the proposed model. The remaining k-1 copies are the training set. We repeat this step k times, and a different subset of each time is chosen for validation. Hence, k scores are obtained. We use five-fold (k = 5) cross-validation on COVID-19_Radiography_Dataset to evaluate the DCML model. Five metrics, including Accuracy, F1, Sensitivity, Precision, and AUC, are used here. The corresponding experimental results are shown in Table 3 .
Table 3.
Five-fold cross-validation. Avg means average value. Std means Standard Deviation.
| 1st fold | 2nd fold | 3rd fold | 4th fold | 5th fold | Avg | Std | |
|---|---|---|---|---|---|---|---|
| Accuracy | 0.9849 | 0.9693 | 0.9880 | 0.9841 | 0.9913 | 0.9835 | 0.0084 |
| F1 | 0.9794 | 0.9386 | 0.9842 | 0.9785 | 0.9879 | 0.9737 | 0.0200 |
| Sensitivity | 0.9822 | 0.9346 | 0.9876 | 0.9764 | 0.9893 | 0.9740 | 0.0226 |
| Precision | 0.9833 | 0.9509 | 0.9879 | 0.9838 | 0.9891 | 0.9790 | 0.0159 |
| AUC | 0.9819 | 0.9588 | 0.9873 | 0.9757 | 0.9893 | 0.9786 | 0.0123 |
According to Table 3, we find that the corresponding results of each fold are close (low standard deviations), and the corresponding average values are satisfactory too. This experimental phenomenon proves that our DCML model has good generalization and stability.
5.4. Real-Time efficiency
In this section, we evaluate the real-time efficiency of the DCML model and make comparisons with several mainstream baselines on the COVID-CT, COVID-19_Radiography_Dataset, and SARS-Cov-2 datasets. For a fair comparison, we calculated the test time of each model. The corresponding experimental results are shown in Table 4 . The DCML model has the best real-time efficiency. This also reflects the lightweight characteristics of our model, demonstrating its high practicality. Hence, our model is efficient as well as effective for COVID-19 recognition.
Table 4.
Real-time efficiency. The best value on each dataset is shown as 5.21.
| Dataset | Model | Real-Time Efficiency (e-4)/s ↓ |
|---|---|---|
| COVID-CT | SeR50 | 6.86 |
| SeR101 | 5.22 | |
| SeR50 (DML) | 5.52 | |
| SeR101 (DML) | 5.22 | |
| DCML | 5.21 | |
| SARS-Cov-2 | SeR50 | 2.55 |
| SeR101 | 3.53 | |
| SeR50 (DML) | 2.03 | |
| SeR101 (DML) | 1.98 | |
| DCML | 1.93 | |
| COVID-19_Radiography_Dataset | SeR50 | 6.37 |
| SeR101 | 6.29 | |
| SeR50 (DML) | 5.79 | |
| SeR101 (DML) | 5.50 | |
| DCML | 4.98 |
5.5. Running procedure of DCML
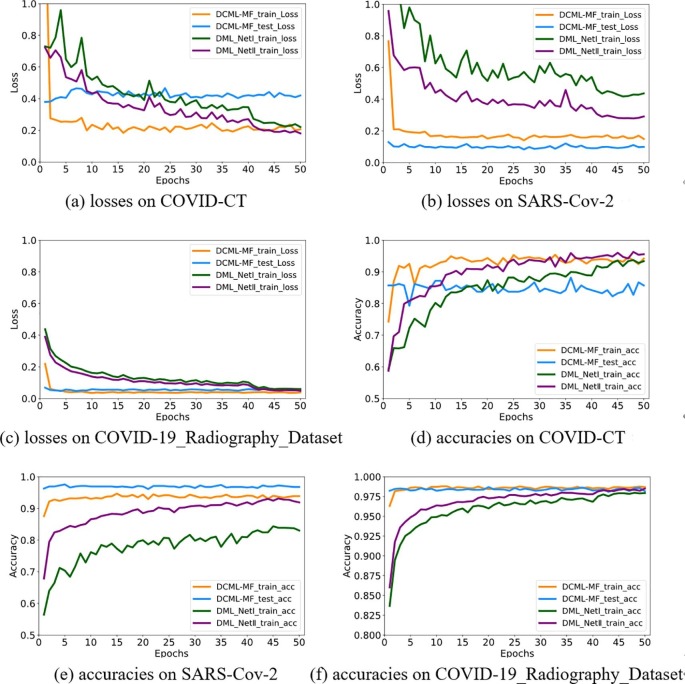
In this section, we illustrate the running procedure of the DCML model on the COVID-CT, COVID-19_Radiography_Dataset, and SARS-Cov-2 datasets. This can help us better understand the implicit running mechanism of our model. All the results are shown in Fig. 5 . Both the results of the DML and DCML models are illustrated and compared objectively in Fig. 5.
Fig. 5.
Real-time running curves.
As shown in Fig. 5 (a) ∼ (c), with the increase of training epoch, the corresponding training loss generally decreases and shows a downward trend. This validates that our training procedure is effective. The DCML model tries to converge to an optimal value. Notably, compared with the traditional DML model, our model not only converges quickly but also converges to a relatively better value on each dataset. Our model is effective for COVID-19 image recognition. Moreover, owing to more high-quality training data generated by FAA, smoother curves of training and test can be observed on the COVID-19_Radiography_Dataset dataset. In addition, the testing loss is relatively stable and very close to the corresponding training loss, especially for COVID-19_Radiography_Dataset. This indicates that on the one hand, our model is not overfitting owing to the application of the FAA data augmentation method and sufficient knowledge mined among two heterogeneous networks. On the other hand, owing to the combination of three convincing losses, robust but effective features are extracted to better characterize CT or CXR images. Moreover, large loss fluctuations are observed on the more challenging COVID-CT dataset because this dataset contains relatively few image samples. This also demonstrates recognition challenge remains on this dataset (you can also refer to Table 2).
As shown in Fig. 5 (d) ∼ (f), with the epoch growing, the corresponding training and test accuracies show an upward trend. It is worth noting that our model converges quickly than a single model. We guess our DCML model takes full advantage of every single network. This further validates the effectiveness of the proposed DCML model from another perspective. Our model gets sufficient training and is not overfitting. More importantly, On the COVID-CT dataset, we obtained the best test accuracy at the 35th epoch. However, The DML model needs more training steps to get the best optimal value (the 44th epoch). On the SARS-Cov-2 dataset, we obtained the best test accuracy at the 5th epoch (DML needs 44 epochs). And the performance gap between the test and training curves is more evident on the COVID-CT dataset. On the COVID-19_Radiography_Dataset dataset, we got the best test accuracy at the 4th epoch (DML requires 45 epochs). Hence, as analyzed above, the corresponding training of the COVID-CT dataset is still a large challenge. In summary, the DCML model gets sufficient training. And it is robust and effective for COVID-19 image recognition.
5.6. Comparisons with baselines using FAA
In order to evaluate the effect of the data augmentation method on baselines, we chose four models, including ResNet18, COVIDNet-Small [33], COVIDNet-Large [33], and WildCat [66], to complete recognition experiments on the COVID-19_Radiography_Dataset dataset. All the accuracies are listed in Table 5 .
Table 5.
Comparisons with baselines. The best value of each column is shown as 0.9870. Here, “w” means with FAA whereas “w/o” means without FAA.
| Model | Accuracy |
|---|---|
| ResNet18 (w/o FAA) | 0.9000 |
| ResNet18 (w FAA) | 0.9300 |
| COVIDNet-Small (w/o FAA) | 0.9000 |
| COVIDNet-Small (w FAA) | 0.9200 |
| COVIDNet-Large (w/o FAA) | 0.9400 |
| COVIDNet-Large (w FAA) | 0.9600 |
| WildCat (w/o FAA) | 0.9032 |
| WildCat (w FAA) | 0.9174 |
| DCML (w/o FAA) | 0.9826 |
| DCML (w FAA) | 0.9870 |
It can be seen from Table 5 that each baseline has been improved by using FAA. This also validates that FAA is actually an effective data augmentation method. However, the DCML model outperforms other baselines with or without FAA.
5.7. Comparison with DML
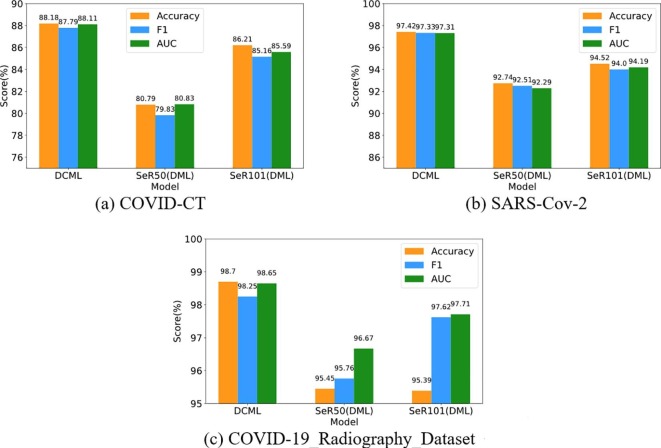
In this section, we compare the DCML model (Net I is SeR50 and Net II is SeR101) with the traditional DML model (each network including Net I and Net II was trained under the DML framework) using more metrics on the COVID-CT, COVID-19_Radiography_Dataset, and SARS-Cov-2 dataset. Three key evaluation metrics, including Accuracy, F1, and AUC, are chosen to complete the overall performance comparisons more comprehensively. All the results are illustrated in Fig. 6 .
Fig. 6.
Comparisons with the conventional DML model.
As shown in Fig. 6, the DCML model outperforms the traditional DML model in any evaluation metric. Although the DML model employs lightweight networks to complete training, three key factors were not considered in this model. Unlike the traditional DML model, we absorbed both adaptive model fusion strategy and a contrastive loss into DCML, which plays a very important role in COVID-19 recognition (Please refer to the ablation analysis section for the details of their contributions). The first factor is the lack of high-quality images. Contrarily, our DCML model uses the FAA augmentation method to alleviate this issue. The second factor is the ignorance of the implicit relationship between positive and negative samples. In contrast, the DCML model introduces the well-known contrastive learning idea, which can distinguish positive and negative samples from the perspective of contrastive learning. This can decrease the distance of the samples from the same category and increase the distance of the samples from different categories, making the corresponding classification margin more robust and effective than before. The last factor is the lack of adaptive model fusion. The two single networks in the DCML framework contain sufficient but complementary dark knowledge for effective COVID-19 recognition. We proposed the adaptive model fusion strategy to fully mine these complementary correlations among heterogeneous layers, which mimics the centralized decision-making process of these pathologists (or radiologists) as much as possible. The adaptive model fusion method further uses the complementary correlations among the heterogeneous layers to train an effective and robust COVID-19 image classifier. More importantly, this can further boost the final classification performance.
Summarily, the DCML model derives from the DML framework. However, owing to absorbing several new characteristics, DCML beats DML on each COVID-19 image dataset. This validates that each modification of the DML model is effective and robust.
5.8. Ablation analysis
The DCML model consists of the single network (Net I is SeR50 and Net II is SeR101), basic DML framework, FAA method, proposed adaptive model fusion strategy, and the proposed contrastive loss. To evaluate the real contribution of each component of the DCML model more objectively, a detailed ablation analysis experiment should be performed carefully. We used three key metrics, including Accuracy, F1, and AUC, to complete our ablation analysis experiments on the COVID-CT, COVID-19_Radiography_Dataset, and SARS-Cov-2 datasets. Here, “Evaluate DML (Net I)” intends to evaluate the actual contribution of the DML framework from the perspective of Net I (SeR50). “Evaluate DML (Net II)” intends to evaluate the actual contribution of the DML framework from the perspective of Net II (SeR101). “Evaluate MF” tries to evaluate the real contribution of the proposed adaptive model fusion module. “Evaluate CL” intends to evaluate the actual contribution of the proposed contrastive loss. “Evaluate FAA” tries to evaluate the actual contribution of the FAA data augmentation method. All the ablation analysis results are shown in Fig. 7 .
Fig. 7.
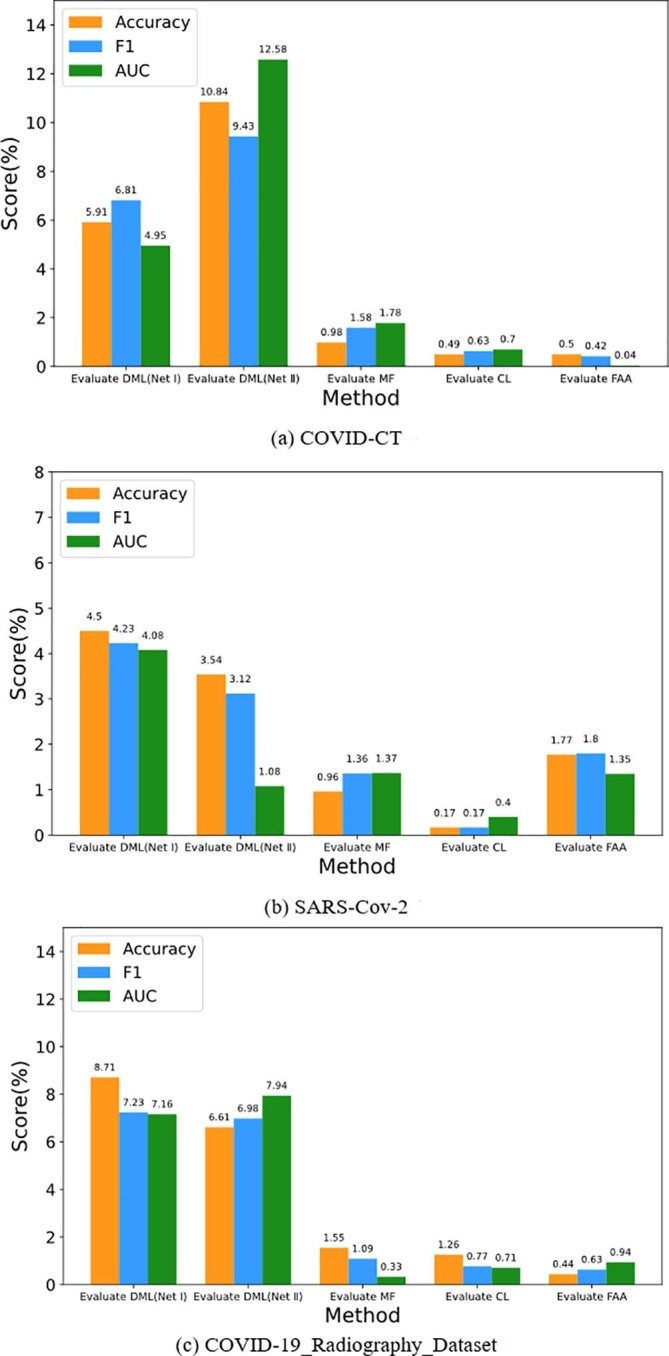
Ablation analysis on each dataset.
As shown in Fig. 7, adding the DML framework leads to the largest performance improvement on each dataset. It is the most basic module for the DCML model. Second, adding the proposed adaptive model fusion module leads to a significant performance improvement on each dataset. This module adaptively fuses two heterogeneous feature maps extracted from the two basic networks, which mimics the centralized decision-making process of these pathologists (or radiologists) as much as possible. Certainly, mutual learning and contrastive learning all build very firm foundation for the proposed model fusion strategy. Different modules excite each other. This is an interesting issue. Third, adding the FAA strategy brings a significant improvement to the SARS-Cov-2 dataset. Contrarily, owing to too few original samples (please refer to Table 1), the corresponding performance improvement on the COVID-CT dataset is not evident. Hence, robust but effective feature learning (or feature fusion strategy) is more important for this dataset. Another sample refinement method [67] may address this issue well. Fourth, using the contrastive loss only brings evident performance improvements on the COVID-19_Radiography_Dataset datasets. This indicates that the more image samples, the more contrastive learning strategy is needed. In the future, we plan to apply independent contrastive learning [68] to complete COVID-19 recognition. This loss can vary the semantic distance between different samples.
As an important conclusion of the paper, the descend order of real contribution of the COVID-CT dataset is “DML > MF > CL > FAA” while the corresponding contribution order of the SARS-Cov-2 dataset is “DML > FAA > MF > CL”. The corresponding contribution order of COVID-19_Radiography_Dataset is “DML > MF > CL > FAA”. Hence, different COVID image datasets need diverse improvement strategies. In general, the basic DML framework and adaptive feature fusion module play a relatively important role in the DCML model. But other necessary modifications, such as data augmentation and contrastive learning, can also boost the final recognition performance.
In a word, the DCML model makes full use of each component to train a powerful COVID-19 image classifier with better generalization ability. Certainly, some modules need to be further improved for more effective recognition.
6. Conclusion
In this article, we propose a novel but effective and efficient DCML model for COVID-19 image recognition. We first used the well-known FAA method to enrich the original datasets. High-quality image samples are obtained in turn, which lays a firm data foundation for the subsequent model training. Then, we absorbed the popular contrastive learning idea into the conventional DML framework. We want to break the isolation of heterogeneous neural networks and provide sufficient complementary correlations (or dark knowledge) for adaptive model fusion. Meanwhile, we intend to mine certain contrastive knowledge to learn more discriminative image features for effective and robust COVID-19 recognition. Finally, we proposed the adaptive model fusion module, which uses the complementary correlations between multiple heterogeneous networks to train a more powerful classifier. Experimental results on three public datasets firmly demonstrate that DCML is a general and effective model which outperforms other state-of-the-art baseline methods. And it is also a robust model. Moreover, each module of the DCML model contributes to improving the corresponding COVID-19 recognition performance. Different modules excite each other and all the modules form a kind of joint force to promote the final recognition performance. Certainly, different contribution ranks were obtained on different datasets. Moreover, DCML tries to imitate practical diagnosis scenarios as much as possible, which helps narrow the gap between theoretical research and clinical application.
Although DCML has achieved satisfactory performance on three publicly available datasets, the limitations of the DCML model should be explained objectively. The scale of these three datasets is relatively small, so the trained recognition model may not generalize to biopsy images which have very large scale. Additionally, the COVID-19 dataset is still scarce, and different datasets are heterogeneous. Therefore, how to simultaneously reduce the impact of heterogeneity among them has become an important issue.
In the future, we plan to combine the state-of-the-art image generation method to generate more realistic images of lungs infected by COVID-19. We hope this can further alleviate the issue of the lack of high-quality CT images. We also intend to use state-of-the-art image segmentation methods, such as UNet [69], to locate the key lesions in COVID-19 CT images, which can bring more interpretable results. Finally, a recent popular transformer model like ViT [70] is another feasible and interesting research route.
7. Data availability
The data used to support the findings of this study are included within the article.
8. Funding statement
This research was funded by the National Natural Science Foundation of China, grant numbers 62161011 and 61861016, the Natural Science Foundation of Jiangxi Provincial Department of Science and Technology, grant numbers 20202BABL202044, 20202BABL212006, and 20212BAB202006, the Key Research and Development Plan of Jiangxi Provincial Science and Technology Department, grant number 20192BBE50071, the Humanity and Social Science Fund of Ministry of Education of China, grant numbers 20YJAZH142, the Science and Technology Projects of Jiangxi Provincial Department of Education, grant number GJJ190323, and the Humanity and Social Science Foundation of Jiangxi University, grant number TQ20108, and TQ21203.
Credit authorship contribution statement
Hongbin Zhang: Conceptualization, Validation, Investigation, Data curation, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition. Weinan Liang: Software, Validation, Investigation, Resources, Visualization, Writing – original draft. Chuanxiu Li: Investigation, Visualization, Software, Validation. Qipeng Xiong: Methodology, Software, Writing – original draft. Haowei Shi: Software, Validation. Lang Hu: Formal analysis. Guangli Li: Investigation, Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We should give sincere thanks to Xingyi Yang, Xuehai He, Jinyu Zhao, Yichen Zhang, Shanghang Zhang, and Pengtao Xie who provided the COVID-CT dataset. We should give sincere thanks to Eduardo Soares, Plamen Angelov, Sarah Biaso, Michele Higa Froes, and Daniel Kanda Abe who provided the SARS-Cov-2 dataset. We should give sincere thanks to a team of researchers from Qatar University, Doha, Qatar, and the University of Dhaka, Bangladesh along with their collaborators from Pakistan and Malaysia in collaboration with medical doctors who provided the COVID-19_Radiography_Dataset. We also give our thanks to Jingyi Hou, Xiang Zhong, Guangxin Xu, and Guangting Wu who gave many valuable pieces of advice about the DCML model. Finally, we would like to thank the editor and the reviewers for their helpful suggestions.
References
- 1.Shah F.M., Joy S.K.S., Ahmed F., Hossain T., Humaira M., Ami A.S., Paul S., Jim M.A.R.K., Ahmed S. A Comprehensive Survey of COVID-19 Detection Using Medical Images. SN Comput. Sci. 2021;2(6) doi: 10.1007/s42979-021-00823-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Islam M.d., Milon F.K., Alhajj R., Zeng J. A Review On Deep Learning Techniques For The Diagnosis Of Novel Coronavirus (COVID-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahman, Sejuti, Sujan Sarker, Abdullah Al Miraj, Ragib Amin Nihal, Anamul Haque and Abdullah Al Noman. “Deep Learning Driven Automated Detection Of COVID-19 From Radiography Images: A Comparative Analysis.” Cognit Comput (2020): n. pag. [DOI] [PMC free article] [PubMed]
- 4.Serena Low W.C., Chuah J.H., Tee C.A.T.H., Anis S., Shoaib M.A., Faisal A., Khalil A., Lai K.W., Jolfaei A. An Overview Of Deep Learning Techniques On Chest X-Ray And CT Scan Identification Of COVID-19. Comput. Math. Methods Med. 2021;2021:1–17. doi: 10.1155/2021/5528144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017;60(6):84–90. [Google Scholar]
- 6.Chikontwe P., Luna M., Kang M., Hong K.S., Ahn J.H., Park S.H. Dual attention multiple instance learning with unsupervised complementary loss for COVID-19 screening. Med. Image Anal. 2021;72:102105. doi: 10.1016/j.media.2021.102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., Nayak D.R., Guttery D.S., Zhang X., Zhang Y.-D. “COVID-19 Classification By CCSHNet with deep fusion using transfer learning and discriminant correlation analysis. Int. J. Inf. Fusion. 2021;68:131–148. doi: 10.1016/j.inffus.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva P., Luz E., Silva G., Moreira G., Silva R., Lucio D., Menotti D. COVID-19 Detection In CT images with deep learning: a voting-based scheme and cross-datasets analysis. Inf. Med. Unlocked. 2020;20:100427. doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zulkifley M.A., Abdani S.R., Zulkifley N.H. COVID-19 screening using a lightweight convolutional neural network with generative adversarial network data augmentation. Symmetry. 2020;12(9):1530. [Google Scholar]
- 10.Wang Shuihua, Satapathy Suresh Chandra, Anderson Donovan, Chen Shi-Xin, Zhang Yu‐Dong. “Deep Fractional Max Pooling Neural Network for COVID-19 Recognition. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.726144. n. pag. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Liang S., Liu H., Gu Y.u., Guo X., Li H., Li L.i., Wu Z., Liu M., Tao L. Fast Automated Detection Of COVID-19 From Medical Images Using Convolutional Neural Networks. Commun. Biol. 2021;4(1) doi: 10.1038/s42003-020-01535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bemportato P., Casalino G., Castellano G., Vessio G. Automatic Clustering Of CT Scans Of COVID-19 Patients Based On Deep Learning. MDAI. 2021 [Google Scholar]
- 13.Zhou Z., Shin J.Y., Zhang L., Gurudu S.R., Gotway M.B., Liang J. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Fine-tuning Convolutional Neural Networks For Biomedical Image Analysis: Actively And Incrementally; pp. 4761–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeCun, Yann, Léon Bottou, Yoshua Bengio and Patrick Haffner. “Gradient-based Learning Applied To Document Recognition.” (1998).
- 15.Simonyan, Karen and Andrew Zisserman. “Very Deep Convolutional Networks For Large-Scale Image Recognition.” CoRR abs/1409.1556 (2015): n. pag.
- 16.Szegedy, Christian, Sergey Ioffe, Vincent Vanhoucke and Alexander Amir Alemi. “Inception-v4, Inception-ResNet And The Impact Of Residual Connections On Learning.” AAAI (2017).
- 17.Hu J., Shen L.i., Albanie S., Sun G., Wu E. Squeeze-and-excitation networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020;42(8):2011–2023. doi: 10.1109/TPAMI.2019.2913372. [DOI] [PubMed] [Google Scholar]
- 18.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference On Computer Vision And Pattern Recognition (CVPR) 2016. Deep Residual Learning for Image Recognition; pp. 770–778. [Google Scholar]
- 19.Vaswani, Ashish, Noam M. Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N. Gomez, Lukasz Kaiser and Illia Polosukhin. “Attention Is All You Need.” ArXiv abs/1706.03762 (2017): n. pag.
- 20.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification Of The Covid-19 Infected Patients Using Densenet201 Based Deep Transfer Learning. J. Biomol. Struct. Dyn. 2021;39(15):5682–5689. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 21.Sen S., Saha S., Chatterjee S., Mirjalili S., Sarkar R. A bi-stage feature selection approach for covid-19 prediction using chest CT images. Appl. Intell. 2021;51(12):1–16. doi: 10.1007/s10489-021-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Butt, J. Gill, D. Chun, and B. A. Babu, “Deep Learning System To Screen Coronavirus Disease 2019 Pneumonia,” Appl. Intell., Apr. 2020. [Online]. Available: doi:10.1007/S10489-020-01714-3. [DOI] [PMC free article] [PubMed]
- 23.L. Hall, D. Goldgof, R. Paul, and G. M. Goldgof, “Finding Covid-19 From chest x-rays Using Deep Learning On A Small Dataset,” May 2020. [Online]. Available: doi:10.36227/techrxiv.12083964.
- 24.A. Narin, C. Kaya, and Z. Pamuk, “Automatic Detection Of Coronavirus Disease (covid-19) Using x-ray Images And Deep Convolutional Neural Networks,” 2020, arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- 25.A. Abbas, M. Abdelsamea, and M. Gaber, “Classification Of covid19 In chest x-ray Images Using Detrac Deep Convolutional Neural Network,” Apr. 2020. [Online]. Available: doi:10.1101/2020.03. 30.20047456. [DOI] [PMC free article] [PubMed]
- 26.M. Farooq and A. Hafeez, “Covid-resnet: A Deep Learning Framework For Screening Of Covid19 From Radiographs,” 2020, arXiv:2003.14395.
- 27.X. Li, C. Li, and D. Zhu, “Covid-mobilexpert: On-device Covid-19 Patient Triage And Follow-up Using Chest x-rays,” 2020, arXiv:2004.03042.
- 28.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting Possibly Representative COVID-19 Biomarkers from X-ray Images with Deep Learning Approach and Image Data Related to Pulmonary Diseases. J. Med. Biol. Eng. 2020;40(3):462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard, Andrew G., Menglong Zhu, Bo Chen, Dmitry Kalenichenko, Weijun Wang, Tobias Weyand, Marco Andreetto and Hartwig Adam. “MobileNets: Efficient Convolutional Neural Networks For Mobile Vision Applications.” ArXiv abs/1704.04861 (2017): n. pag.
- 30.P. Angelov, E. Almeida Soares, Explainable-by-design Approach For Covid-19 Classification via Ct-Scan, medRxiv, 2020.
- 31.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-cam based color visualization approach for fast detection of Covid-19 cases using chest x-ray and ct-scan Images. Chaos, Solit Fractals. 2020 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vision. 2020;128(2):336–359. [Google Scholar]
- 33.Z. Q. L. Linda Wang and A. Wong, “Covid-net: A Tailored Deep Convolutional Neural Network Design For Detection Of Covid-19 Cases From Chest Radiography Images,” 2020, arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- 34.T. Javaheri et al., “Covidctnet: An Open-source Deep Learning Approach To Identify Covid-19 Using CT Image,” 2020, arXiv:2005.03059.
- 35.Soltanian, Mohammad and Keivan Borna. “Covid-19 Recognition From Cough Sounds Using Lightweight Separable-quadratic Convolutional Network.” Biomedical Signal Processing and Control 72 (2021): 103333 – 103333. [DOI] [PMC free article] [PubMed]
- 36.Ghosh, Swarup Kr and Anupam Ghosh. “ENResNet: A Novel Residual Neural Network For Chest X-ray Enhancement Based COVID-19 Detection.” Biomedical Signal Processing and Control 7 2 (2021): 103286 - 103286. [DOI] [PMC free article] [PubMed]
- 37.Gaur, Pramod, V S Malaviya, Abhay Gupta, Gautam Bhatia, Ram Bilas Pachori and Divyesh Sharma. “COVID-19 Disease Identification From Chest CT Images Using Empirical Wavelet Transformation And Transfer Learning.” Biomedical Signal Processing and Control 71 (2021): 103076 - 103076. [DOI] [PMC free article] [PubMed]
- 38.S.-A. Rebuffi, H. Bilen, and A. Vedaldi, “Learning Multiple Visual Domains With Residual Adapters,” in Proc. Adv. Neural Inf. Process. Syst., 2017, pp. 506–516.
- 39.S.-A. Rebuffi, H. Bilen, and A. Vedaldi, “Efficient Parametrization Of Multi-Domain Deep Neural Networks,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2018, pp. 8119–8127.
- 40.Liu Q., Dou Q., Yu L., Heng P.A. MS-NET: multi-site network for improving prostate segmentation with heterogeneous MRI Data. IEEE Trans. Med. Imag. 2020;39(9):2713–2724. doi: 10.1109/TMI.2020.2974574. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Wang, Quande Liu, and Qi Dou, Contrastive Cross-Site Learning With Redesigned Net For COVID-19 CT Classification, doi:10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed]
- 42.Lebedev V., Lempitsky V.S. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Fast ConvNets Using Group-Wise Brain Damage; pp. 2554–2564. [Google Scholar]
- 43.Zoph B., Vasudevan V., Shlens J., Quoc V.L. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018. Learning Transferable Architectures For Scalable Image Recognition; pp. 8697–8710. [Google Scholar]
- 44.Garg M., Dhiman G. A novel content-based image retrieval approach for classification using GLCM features and texture fused LBP variants. Neural Comput. Appl. 2021;33(4):1311–1328. [Google Scholar]
- 45.Tan, Mingxing and Quoc V. Le. “EfficientNet: Rethinking Model Scaling For Convolutional Neural Networks.” ArXiv abs/1905.11946 (2019): n. pag.
- 46.Huang, Ling, Su Ruan and Thierry Denoeux. “Covid-19 Classification With Deep Neural Network And Belief Functions.” The Fifth International Conference on Biological Information and Biomedical Engineering (2021): n. pag.
- 47.P. Afshar, S. Heidarian, F. Naderkhani, A. Oikonomou, K. N. Plataniotis, and A. Mohammadi, “Covid-caps: A Capsule Network-based Framework For Identification Of Covid-19 Cases From x-ray Images,” Pattern Recognit. Letters, Sep. 2020. [Online]. Available: doi:10.1016/j.patrec. 2020.09.010. [DOI] [PMC free article] [PubMed]
- 48.O. Gozes et al., “Rapid ai Development Cycle For The Coronavirus (covid-19) Pandemic: Initial Results For Automated Detection & Patient Monitoring Using Deep Learning CT Image Analysis,” 2020, arXiv:2003.05037.
- 49.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Artificial intelligence distinguishes Covid-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Z. Tang et al., “Severity Assessment Of Coronavirus Disease 2019 (Covid-19) Using Quantitative Features From Chest CT Images,” 2020, arXiv:2003.11988.
- 51.Rahimzadeh M., Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inf. Med. Unlocked. 2020;19:100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinton, G., Vinyals, O., Dean, J. (2015). Distilling The Knowledge In A Neural Network. arXiv preprint arXiv:1503.02531.
- 53.Ba, L. J., Caruana, R. (2013). Do Deep Nets Really Need To Be Deep?. arXiv preprint arXiv:1312.6184.
- 54.Wen T., Lai S., Qian X. Preparing lessons: improve knowledge distillation with better supervision. Neurocomputing. 2021;454:25–33. [Google Scholar]
- 55.Cho J.H., Hariharan B. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. On the efficacy of knowledge distillation; pp. 4794–4802. [Google Scholar]
- 56.Ying Z., Xiang T., Hospedales T.M., Huchuan L.u. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018. Deep Mutual Learning; pp. 4320–4328. [Google Scholar]
- 57.Anil, R., Pereyra, G., Passos, A., Ormandi, R., Dahl, G. E., Hinton, G. E. (2018). Large Scale Distributed Neural Network Training Through Online Distillation. arXiv preprint arXiv:1804.03235.
- 58.Gao L., Lan X., Mi H., Feng D., Xu K., Peng Y. Multistructure-based collaborative online distillation. Entropy. 2019;21(4):357. doi: 10.3390/e21040357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim, Sungbin, Ildoo Kim, Taesup Kim, Chiheon Kim and Sungwoon Kim. “Fast AutoAugment.” NeurIPS (2019).
- 60.Cubuk, Ekin Dogus, Barret Zoph, Dandelion Mané, Vijay Vasudevan and Quoc V. Le. “AutoAugment: Learning Augmentation Policies From Data.” ArXiv abs/1805.09501 (2018): n. pag.
- 61.Hadsell, Raia, Sumit Chopra and Yann LeCun. “Dimensionality Reduction By Learning An Invariant Mapping.” 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'06) 2 (2006): 1735-1742.
- 62.Yang, Xingyi, Jinyu Zhao, Yichen Zhang, Xuehai He and Pengtao Xie. “COVID-CT-Dataset: A CT Scan Dataset About COVID-19.” ArXiv abs/2003.13865 (2020): n. pag.
- 63.Soares, Eduardo A., Plamen P. Angelov, Sarah Biaso, Michele Higa Froes and Daniel Kanda Abe. “SARS-CoV-2 CT-scan Dataset:A Large Dataset Of Real Patients CT Scans For SARS-CoV-2 Identification.” medRxiv (2020): n. pag.
- 64.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N., Reaz M.B.I., Islam M.T. Can AI Help In screening Viral And COVID-19 Pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 65.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., Islam M.T., Al Maadeed S., Zughaier S.M., Khan M.S., Chowdhury M.E.H. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021;132:104319. doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Durand T., Mordan T., Thome N., Cord M. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. WILDCAT: Weakly Supervised Learning Of Deep ConvNets For Image Classification, Pointwise Localization And Segmentation; pp. 5957–5966. [Google Scholar]
- 67.Zhang H., Wu J., Shi H., Jiang Z., Ji D., Yuan T., Li G. Multidimensional Extra Evidence Mining for Image Sentiment Analysis. IEEE Access. 2020;8:103619–103634. [Google Scholar]
- 68.Inoue N., Goto K. 2020 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC) 2020. Semi-Supervised Contrastive Learning With Generalized Contrastive Loss And Its Application To Speaker Recognition; pp. 1641–1646. [Google Scholar]
- 69.Ronneberger O., Fischer P., Brox T. U-Net: Convolutional Networks For Biomedical Image Segmentation. MICCAI. 2015 [Google Scholar]
- 70.Dosovitskiy, Alexey, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit and Neil Houlsby. “An Image Is Worth 16x16 Words: Transformers For Image Recognition At Scale.” ArXiv abs/2010.11929 (2021): n. pag.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.