Abstract
The marine microbial natural product salinosporamide A (Marizomib) is a potent proteasome inhibitor currently in clinical trials for the treatment of brain cancer. Salinosporamide A is characterized by a complex and densely functionalized -lactam--lactone bicyclic warhead, the assembly of which has long remained a biosynthetic mystery. Here, we report an enzymatic route to the salinosporamide core catalyzed by a standalone ketosynthase, SalC. Chemoenzymatic synthesis of carrier protein-tethered substrates, as well as intact proteomics, allowed us to probe the reactivity of SalC and understand its role as an intramolecular aldolase/β-lactone synthase with roles in both transacylation and bond forming reactions. Additionally, we present the 2.85 Å SalC crystal structure that, combined with site-directed mutagenesis, allowed us to propose a bicyclization reaction mechanism. This work challenges our current understanding of the role of ketosynthase enzymes and establishes a basis for future efforts towards streamlined production of a clinically relevant chemotherapeutic.
INTRODUCTION
Salinosporamide A (1), also known as Marizomib, is a potent 20S proteasome inhibitor originally isolated from the obligate marine actinomycete Salinispora tropica and presently in phase III clinical trials for the treatment of glioblastoma, an aggressive form of brain cancer with a poor prognosis and few therapeutic options (Fig. 1a).1,2 Despite the discovery of numerous natural analogs of 1,3 as well as extensive efforts to generate derivatives via chemical synthesis4 and mutasynthesis,5,6 it is the originally discovered natural product itself that entered clinical trials. Salinosporamide A’s compact yet densely functionalized -lactam--lactone pharmacophore is distinct amongst proteasome inhibitors, including the FDA-approved bortezomib, carfilzomib, and ixazomib. The electrophilic β-lactone warhead of 1 serves as the covalent attachment site for the N-terminal catalytic threonine residue found in all three proteasome β-subunits (β1, β2, β5), resulting in pan-proteasome irreversible inhibition with nanomolar potency (Supplementary Fig. 1).7 Unlike other proteasome inhibitors, salinosporamide crosses the blood-brain barrier, leading to its advancement through glioblastoma clinical trials.
Fig. 1: Microbial natural products assembled by terminal cyclization reactions.
a, salinosporamide biosynthetic gene cluster (sal BGC) and proposed bicyclization of the SalB-PCP-tethered linear intermediate to assemble the salinosporamide A (1) pharmacophore. Bonds formed by unknown cyclization enzyme(s) are highlighted in gray. b, Representative additional examples of microbial natural products assembled via terminal cyclization reactions: obafluorin (2), fumiquinazoline F (3), vibralactone (4), nocardicin A (5), tenuazonic acid (6). Bond(s) formed in cyclization reaction and catalyzing enzyme class shown in red.
Stable isotope feeding8 and gene inactivation experiments9 revealed salinosporamide A is naturally assembled from three distinct metabolic building blocks (Fig. 1a): acetate (orange), cyclohexenylalanine (blue), and chloroethylmalonate (red). These molecules are processed by a hybrid polyketide synthase (PKS)/nonribosomal peptide synthetase (NRPS) enzymatic assembly line to produce a protein-bound linear intermediate that undergoes cyclization and offloading to yield 1 (Fig. 1a). While the pathway to the unprecedented PKS extender unit chloroethylmalonyl-CoA9 and the origins of the unusual nonproteinogenic amino acid cyclohexenylalanine are established,10 the biosynthetic reactions and enzyme(s) responsible for the assembly of the -lactam--lactone pharmacophore are unknown.
Terminal cyclization reactions in microbial biosynthesis are varied and numerous, leading to a diversity of products (Fig. 1b). However, none resemble the suspected bicyclization reaction to 1 that would invoke a two-step intramolecular aldol reaction to install the γ-lactam ring, followed by an offloading β-lactonization reaction. The chemical foundation for this proposed enzymatic reaction was achieved by an elegant biomimetic synthesis of salinosporamide.11,12 Classically, terminal thioesterase (TE) domains in microbial assembly line megasynth(et)ases are responsible for offloading via macrocyclization.13 Importantly, the first cyclizing TE capable of -lactone formation, ObiF, was recently identified in obafluorin (2) biosynthesis.14 While there is a TE encoded within the sal biosynthetic gene cluster, it is hypothesized to be an editing type II TE responsible for removing misprimed molecules from the assembly line,15 and it does not harbor the conserved GxCxG motif from ObiF required for β-lactone synthesis.14 Microbial biosynthetic pathways have also found alternative routes outside of the TE to catalyze cyclization reactions, including fungal terminal condensation domains (CT), as seen in fumiquinazoline F (3) biosynthesis;16 the hydrolase required for formation of the fused bicyclic ring system in vibralactone (4) biosynthesis;17 and the condensation domain responsible for β-lactam formation in the biosynthesis of the antibiotic nocardicin A (5).18 Recently, a terminal ketosynthase (KS) domain in the tenuazonic acid (6) PKS was shown to catalyze the Dieckmann cyclization of an amino acid β-keto thioester to yield a tetramic acid product.19–21 While these examples demonstrate a wide variety of non-canonical terminal enzymes that could be involved in a cyclization reaction, none of these previously reported examples resemble a biosynthetic precedent for the proposed bicyclization in salinosporamide A biosynthesis.
Here, we report the identification, biochemical investigation, and structural characterization of the ketosynthase homolog SalC, which catalyzes the unprecedented tandem aldol-lactonization bicyclization reaction and offloading to assemble the -lactam--lactone salinosporamide A pharmacophore. This discovery establishes the biosynthetic logic of this brain penetrant drug candidate and provides a clear roadmap for the generation of new KS-based biocatalysts in medicinal chemistry.
RESULTS
SalC, a KS, is required for salinosporamide biosynthesis
To determine the enzyme responsible for the formation of the bicyclic salinosporamide core, gene inactivation experiments were used to sequentially disrupt candidate genes in the sal BGC as described previously (Fig. 2a).9 Disruption of salO, a putative cyclase, had no effect on salinosporamide production. As expected, disruption of salA, encoding for the mixed PKS/NRPS assembly line, abolished production of salinosporamides A (1) and B (7), as did disruption of salD, which encodes a P450 enzyme. Instead, the △salD strain accumulated 8, the deoxy analog salinosporamide J. A ketosynthase, for which no other role in the pathway could be attributed, caught our attention when disruption of its encoding gene, salC, completely abolished production of 1 and 7 (Fig. 2a). Importantly, salC is conserved in all salinosporamide and cinnabaramide22 producing strains (Supplementary Fig. 2). BLAST23 and NaPDoS24 analysis of salC revealed that its gene product is a standalone ketosynthase (KS) (Supplementary Fig. 3 and 4). However, SalC lacks the canonical Cys-His-His catalytic triad associated with KSs,25 and instead has an asparagine in place of the first histidine, a variation often associated with “non-elongating” or “condensation-incompetent” KSs (KS0s) (Supplementary Fig. 5).26,27 Recent work on tenuazonic acid biosynthesis suggests KSs are capable of catalyzing cyclization reactions, and type III KSs have long been known to catalyze cyclization reactions via aldol condensations on coenzyme A-bound substrates,28 which led to our hypothesis that SalC might function as a bicyclase capable of forming the -lactam--lactone salinosporamide core.
Fig. 2: SalC is involved in late-stage salinosporamide A biosynthesis.
LCMS chromatograms of salinosporamide A standard and extracts from Salinispora cultures: wild type Salinispora tropica CNB440; S. tropica CNB440△salO; S. tropica CNB440△salA; S. tropica CNB440△salD; S. tropica CNB440△salC. 7 = salinosporamide B, 8 = salinosporamide J.
Characterizing late-stage cyclization and offloading reactions in assembly line biosynthetic pathways presents a unique set of challenges. The substrates required to probe these reactions are fully elongated and functionalized as they prepare for offloading, and thus are challenging to access synthetically. To circumvent the need for building these synthetically demanding linear substrates, previous studies of TE domains that perform macrolactonization reactions have capitalized on the reversibility of enzymatic reactions and utilized cyclized compounds as substrates for the TE to perform the reverse hydrolysis reaction.29 Therefore, to determine if salinosporamide A is the product of SalC, we chose to first probe the reaction in the reverse direction. To do so, we heterologously prepared SalC in Streptomyces (Supplementary Fig. 6 and 8), as all attempts to obtain soluble protein in E. coli failed, and incubated the recombinant enzyme with salinosporamide A. In the presence of SalC we observed a modest decrease in hydrolysis of the β-lactone (compound 9) compared to the no enzyme control, as well as decreased formation of buffer adduct products in which tris and glycerol open the β-lactone of 1 (Extended Data Fig. 1). Incubation with SalC consistently resulted in the formation of unique, non-chlorinated products indicative of the intramolecular formation of a stable tetrahydrofuran ring (10, 11) by displacement of the chloride, as judged by LCMS. Notably, this THF ring formation is the same reaction that occurs in the proteasome (Supplementary Fig. 1).7 While 9 could ultimately react to yield 10 and 11, the presence of SalC clearly increased this reaction rate. The formation of these new products as well as fewer buffer and hydrolytic cleavage products suggested that, in the presence of SalC, the hydrolysis of 1 occurs in a low-hydration environment, such as within the SalC active site. SalC’s ability to hydrolyze the β-lactone of 1 through apparent acylation further implicated SalC in the offloading bicyclization reaction.
SalC is a -lactone bicyclase
As our initial experiments suggested a role for SalC in the salinosporamide bicyclization reaction, we sought to develop an assay to directly explore this activity. To do so we first had to generate an appropriate substrate. The organization of the mixed PKS/NRPS modules in the sal pathway indicated that the bimodular PKS (SalA) and the NRPS didomain (SalB) work together to generate the SalB peptidyl carrier protein (PCP)-tethered β-ketoamide intermediate (Fig. 1a) prior to the bicyclization reaction. Therefore, we expressed and purified the excised SalB-PCP domain (Supplementary Fig. 7) in its apo form, confirmed by intact proteomics (Supplementary Fig. 8) as well as trypsin digestion and peptide mass fingerprinting (Supplementary Fig. 9), with the goal of modifying the serine residue with a variety of CoA-activated linear precursors.
Salinosporamide A is composed of reactive sidechain functional groups that, while essential for the potent bioactivity of this compound, complicate both chemical synthesis and in vitro enzymatic assays. To circumvent possible analytical complications,11,12 we designed a simplified analog of the linear salinosporamide chain elongation intermediate. Once cyclized, this linear precursor would yield 5-deoxy-7,8-dihydro-salinosporamide B, which we refer to as “simplisporamide”. This substrate was inspired by salinosporamide analogs that have been isolated from the bacterium itself, including the deschloro compound salinosporamide B (7), the deoxy version salinosporamide J (8), and antiprotealide (12) which contains leucine in lieu of the cyclohexenylalanine residue (Extended Data Fig. 2). As all three of these molecules are isolated from Salinispora cultures,3,30,31 we hypothesized that a substrate lacking these moieties would be recognized by the putative cyclization enzyme.
The synthesis of the linear pantetheine-activated simplisporamide precursor (13) is based on the wealth of literature precedent for the total synthesis of the salinosporamides (Supplementary Note).11,12,32 Simplisporamide linear precursors were synthesized as a mixture of diastereomers and preparative HPLC was used to separate them as confirmed by NMR. However, upon addition to buffer, these pure diastereomers rapidly equilibrated to a ~1:1 mixture (Supplementary Note), which was used for all subsequent experiments and is referred to as 13.
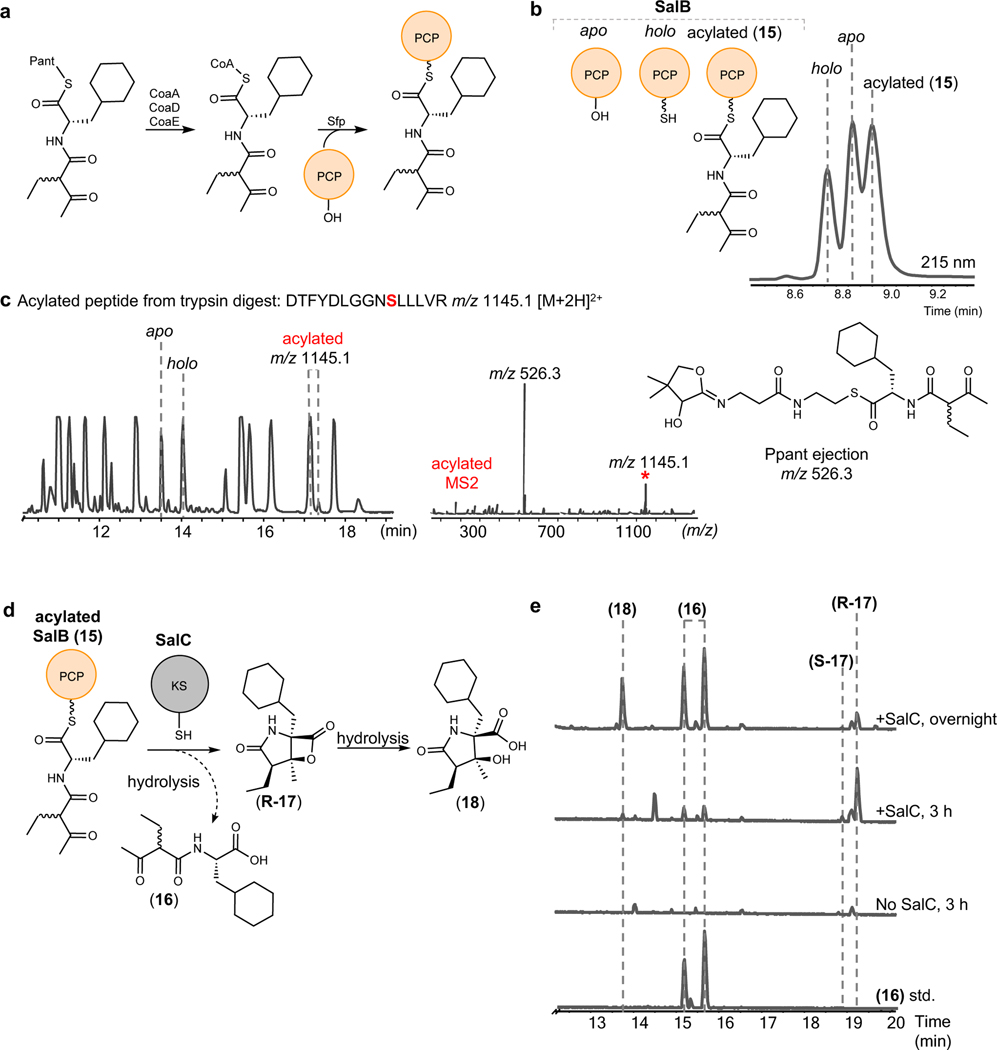
To generate the PCP-tethered linear substrate we utilized a one-pot biocatalytic synthesis beginning with the pantetheine-activated substrate (13) and proceeding through the CoA-activated substrate (14), as described previously (Fig. 3a, Extended Data Fig. 3, Supplementary Fig. 10).33 Carrier protein acylation was confirmed by both intact protein liquid chromatography mass spectrometry (LCMS) (Fig. 3b and Supplementary Fig. 11) as well as trypsin digestion of SalB and high-resolution LCMS analysis of the resulting peptide fragments (Fig. 3c). The MS-based phosphopanthetheine (PPant) ejection assay was additionally used to verify the formation of the linear precursor-acylated-SalB substrate (15) (Fig. 3c).34
Fig. 3: SalC is a γ-lactam-β-lactone bicyclase.
a, Reaction scheme for the chemoenzymatic synthesis of PCP-tethered linear substrate (15) from pantetheine-activated simplisporamide precursor (13) via in vitro CoA biosynthesis and Sfp-mediated loading. b, Intact protein LCMS chromatogram of SalB-PCP acylation assay (UV, 215 nm). See Supplementary Fig. 11 for intact proteomics HRMS data. c, LCMS chromatogram of SalB-PCP tryptic digest post acylation assay and MS2 Ppant ejection assay. d, Reaction scheme for SalC activity assay. e, LCMS chromatograms of linear hydrolysis product (16) standard, SalC activity assay with no SalC (3 h), SalC activity assay (3 h), and SalC activity assay (overnight). See Supplementary Fig. 12 for HRMS data for R-17 and Supplementary Note for full characterization.
Upon incubation of SalC (20 μM) with in situ generated 15, we observed two products with m/z 284.19 [M+H]+ at retention time (rt) 15.2 and 15.6 minutes (Fig. 3d and 3e). Co-elution with a synthetic standard revealed them to be the hydrolyzed linear β-keto amino acid (16). However, the major product of the reaction (m/z 266.17 [M+H]+, rt 19.2 min), was indicative of the formation of simplisporamide (R-17) (Fig. 3d and 3e, Supplementary Fig. 12). We noted the appearance of a minor peak with the same molecular ion at 18.9 minutes (S-17) (Fig. 3e). Critically, neither of these compounds were evident in control reactions without SalC. In a time course experiment, we visualized the depletion of these compounds accompanied by formation of a new product (rt 13.7 min) consistent with the mass (m/z 284.19 [M+H]+) of the hydrolyzed β-lactone (18) (Fig. 3e) due to degradative hydrolysis over time, which was also observed with salinosporamide A (Extended Data Fig. 1).
After demonstrating the formation of an enzymatic reaction product indicative of the bicyclic product (R-17), we sought to perform the SalC assay at a preparative scale suitable for purification of the product and NMR characterization. However, the reliance on a single turnover reaction with respect to a carrier protein presented a substantial challenge to scaling up the SalC bicyclization reaction, and as such we sought to utilize diffusible substrates to circumvent the need for stoichiometric SalB-PCP. Given the literature precedent for KSs utilizing CoA-mimics as activating groups,35 we tested SalC activity with pantetheine-, and CoA-activated linear substrates, as well as the products of the CoaA and CoaD reactions en route to the CoA activated substrate (19 and 20, respectively). When SalC was incubated with these diffusible substrates, minimal putative bicyclized product was formed (Extended Data Fig. 3), and the primary product of the reaction appeared to be 16. Therefore, we proceeded with carrier protein dependent reactions to scale up the SalC activity assay.
The inherent instability of the simplisporamide β-lactone presented an additional challenge for a larger scale reaction in an aqueous environment.36 During fermentative production of 1, degradation via hydrolysis of the β-lactone is mitigated by the addition of the absorbent resin XAD-7 to the culture media, which stabilizes and captures 1 for subsequent purification.37 Inspired by this strategy, we included an absorbent resin in a large scale SalC assay to protect the cyclized product as it is offloaded from the protein. Without resin, the product degraded rapidly to yield 18. The major product (R-17) was captured by the resin (Supplementary Fig. 13), purified, and confirmed by NMR (for complete tables of NMR assignments, spectra, and key correlations, see Supplementary Note).
The strained β-lactone of 1 is the key pharmacophore of the molecule, serving as the electrophilic trap for proteasome inhibition, and so we aimed to directly link SalC to the formation of this molecular warhead and therefore the bioactivity of the molecule. We performed the SalC activity assay with resin to trap the simplisporamide product, as well as control assays without SalC or 13. The extracts of these resins (Supplementary Fig. 14) were directly tested for in vitro inhibitory activity against the human 20S proteasome chymotrypsin-like protease activity (β5 subunit) (Extended Data Fig. 4). Importantly, this fluorescence-based assay is robust, commercially available, and performed in a 96-well plate format which makes it amenable for high-throughput work. The bioassay showed that SalC was required for proteasome inhibition, and that the proteasome was not inhibited when SalC or substrate were excluded from the assay. This result further confirmed that SalC is responsible for the formation of the bioactive β-lactone moiety.
SalC is acylated by SalB-PCP
The proposed bicyclization reaction performed by SalC is intramolecular, in contrast to the intermolecular nature of canonical KS biosynthesis. As such, SalC-mediated cyclization could occur on the carrier protein, or the substrate could be transferred onto SalC itself for cyclization to occur (Supplementary Fig. 15a). To distinguish between these two possibilities, we synthesized a mechanistic probe lacking the β-ketone group (21) that would be capable of acylating SalB and SalC, but would be incompetent for cyclization and offloading. Therefore, this substrate would remain trapped on the protein, potentially allowing for detection via intact protein mass spectrometry (Fig. 4a). Using 21, we performed the previously described chemoenzymatic acylation assay with apo-SalB-PCP to yield holo-acylated-SalB-PCP (22). 22 was then purified away from residual CoA proteins and small molecule intermediates to ensure all transacylation originated from the carrier protein. Column purified 22 was incubated with SalC (m/z 66613 [M+H]+) and then subjected to intact protein LC-HRMS analysis, revealing a new peak (m/z 66864 [M+H]+) (Fig. 4b) indicative of chain transfer of the mechanistic probe from the SalB-PCP domain to SalC (Supplementary Fig. 16 and 17). We also performed this transacylation assay with diffusible substrates: the pantetheine-activated linear probe (21), the products of the CoaA and CoaD reactions with 21 (23 and 24, respectively), as well as the CoA-activated linear probe (25) (Extended Data Fig. 5, and Supplementary Figs. 18–23). While SalC was capable of being acylated by diffusible substrates, the largest amount of acylated SalC was observed with carrier protein-activated substrate. Finally, small molecule LCMS revealed that once acylated with the mechanistic probe, SalC was able to hydrolyze off 21 over time as well (Supplementary Fig. 15b).
Fig. 4: Transacylation of SalC by acylated-SalB.
a, Reaction scheme for the chemoenzymatic synthesis of 22 and subsequent transacylation assay with SalC. b, UV chromatograms (215 nm) of intact protein LCMS of apo-SalB-PCP and SalC (bottom trace; small amount of holo-SalB-PCP present likely due to native E. coli PPTase modification), SalB-PCP acylated with the mechanistic probe (21) and purified by size exclusion chromatography to yield 22 (middle trace), and the complete transacylation assay using 22 and SalC (top trace). See Supplementary Figs. 16 and 17 for intact proteomics HRMS data.
Structure-guided mutagenesis investigated bicyclization
To elucidate the molecular basis for the observed bicyclase activity of this unusual ketosynthase, we solved a crystal structure of SalC to 2.85 Å resolution (PDB: 7S2X, Supplementary Tables 3 and 4). Crystallography revealed that the oligomeric state of SalC is a homotetramer (Fig. 5a), verified via size exclusion chromatography (Supplementary Fig. 24).38 Overall, SalC possesses high structural homology39 to trans-AT KSs (Extended Data Fig. 6), although they only share less than 40% sequence identity. Each monomeric subunit of SalC is composed of two domains, the N-terminal KS domain, which forms tetrameric interfaces, and the C-terminal flanking domain, which does not contact domains from other subunits. The KS domain of SalC resembles a canonical ketosynthase with the classic αβαβα thiolase fold40 (Extended Data Fig. 7); whereas the C-terminal flanking subdomain resembles those seen in trans-AT KSs (Extended Data Fig. 6), a ~100 residue structure homologous to the KS-AT adapters in cis-AT PKS systems but with an unknown function.41,42
Fig. 5: SalC structure and active site mutagenesis.
a, SalC overall tetrameric structure with each monomeric domain colored differently (KS domains in green and brown, flanking subdomain in teal and yellow). b, SalC active site with proposed catalytic residues labeled. c, UV (215 nm) chromatograms from intact protein LCMS transacylation assays using column purified 22 and SalC mutants. d, LCMS chromatograms of SalC activity assays (3 h) with WT SalC and SalC active site variants using 15. See Supplementary Figs. 27–30 for intact proteomics HRMS data.
The active site of SalC lacks the established Cys-His-His catalytic triad found in canonical KS domains25 and instead contains a Cys180-Asn316-His353 triad similar to non-elongating KSs (Supplementary Fig. 5, Supplementary Fig. 25). The SalC active site (Fig. 5b) also contains a conserved lysine (Lys348) found in both PKS KSs and fatty acid synthase (FAS) β-keto-acyl-ACP synthases. Examination of the active site also revealed the presence of a tyrosine residue (Tyr284) located 3.6 Å from Cys180 and positioned on a loop that protrudes into the active site. Tyr284 is conserved in all SalC homologs from salinosporamide producing Salinispora strains and the cinnabaramide SalC homolog CinC (Extended Data Figs. 7 and 8). Superimposition of SalC with a KS from the bacillaene PKS pathway (bae) (Extended Data Fig. 9) bound to its native chain elongation intermediate (PDB ID: 4NA2)42 revealed the SalC Tyr284 residue is oriented towards the thioester α-carbonyl of the bae intermediate, suggesting it may play a key role in salinosporamide bicyclization. To investigate the role these residues play in SalC bicyclization, we performed site-directed mutagenesis experiments of active site residues implicated in catalysis (Supplementary Fig. 26). Resulting SalC variants were subjected to SalB-to-SalC transacylation assays with 22 (Fig. 5c, Supplementary Fig. 27–30), as well as SalC cyclization activity assays with 15 (Fig. 5d).
As SalC is acylated by the SalB-PCP domain to form a linear acyl-enzyme intermediate prior to cyclization, it stands to reason that mutating the active site Cys180Ala both abrogated chain transfer and abolished production of the cyclized product (Fig. 5c and 5d, Supplementary Fig. 27). Furthermore, the Cys180Ser mutation had the same effect (Supplementary Fig. 28).43 The Asn316Ala mutant did not retain full transacylation activity in the chain transfer assay, and showed reduced production of the bicyclized product to less than 4% of wild type SalC (Fig. 5c and 5d, Supplementary Fig. 29). The His353Asn mutation was selected as type III KSs, known to perform cyclization reactions, typically have an asparagine in place of the final histidine in the catalytic triad. In the case of SalC, this mutant retained roughly 40% of the wild type SalC cyclization activity, but, notably, it largely appeared to convert SalC from a bicyclase to a hydrolase (Fig. 5c, Supplementary Fig. 30). This mutant produced 43-times the amount of 16 compared to wild type SalC, which may be due to a potential role of His353 in deprotonating the thioester α-proton. Without this deprotonation, the intramolecular aldol reaction cannot occur and the stuck linear intermediate is hydrolyzed off SalC as 16. The Tyr284Phe mutation, unfortunately, resulted in unstable protein and was thus unable to be assessed.
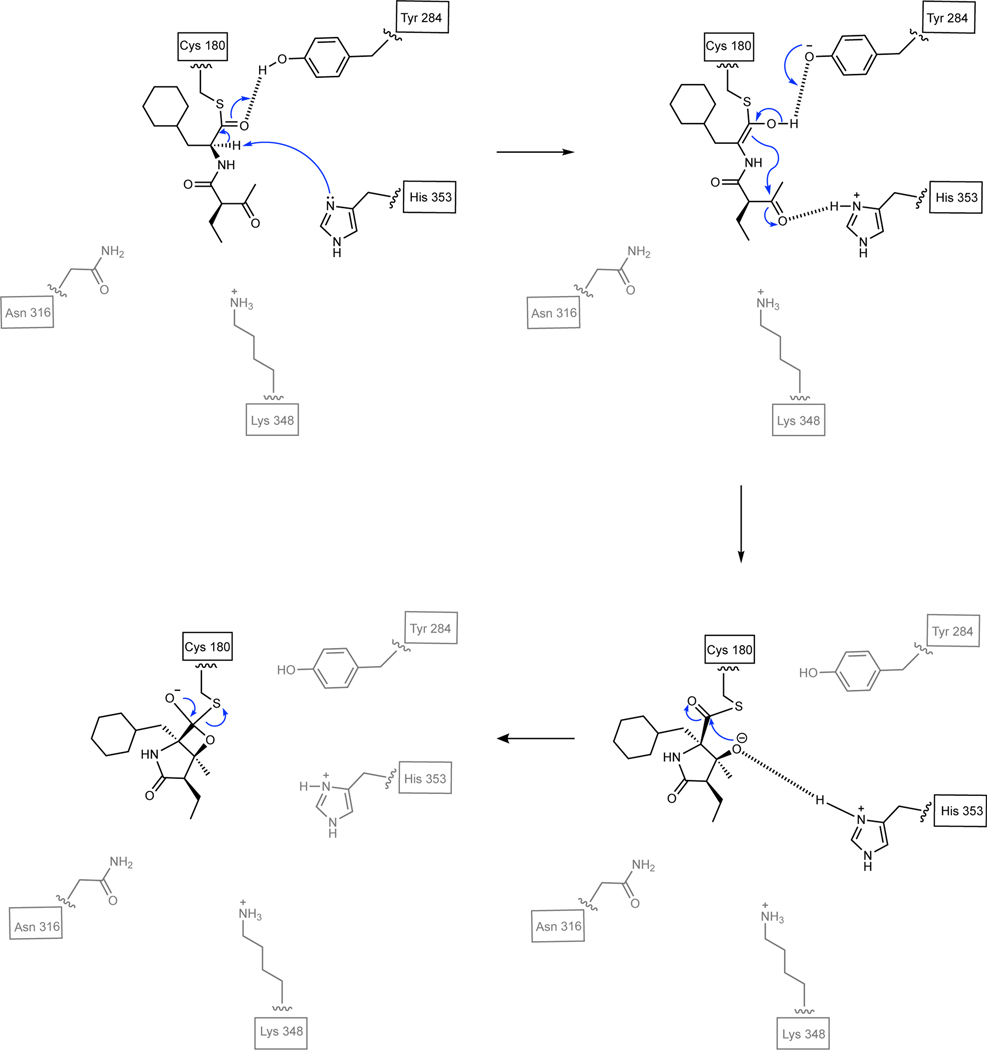
Based on the structure of SalC and results from our mutagenesis campaign, we propose a possible mechanism (Fig. 6, Extended Data Fig. 10) initiated by deprotonation of Cys180 by His353, followed by acylation of the resulting thiolate with the SalB-PCP-tethered substrate (15).25 Lys348 may serve as a general base capable of deprotonating His353, although there are other basic residues near the active site that could perform this deprotonation. Asn316 may play a stabilizing role in this reaction as this residue appears critical for transacylation. Hydrogen bonding of the thioamide carbonyl to the Tyr284 phenol facilitates deprotonation of the thioester α-carbon by His353, thereby generating an enol intermediate that could react via an intramolecular aldol reaction to form the γ-lactam. The resulting oxyanion is presumably stabilized by dipole interactions with backbone amides, as is hypothesized for KSs.25 Subsequent β-lactonization through a tetrahedral intermediate releases the bicyclic simplisporamide product from SalC. Finally, Cys180 is reprotonated by His353 to restart the catalytic cycle as previously proposed for KSs from type I PKS systems.25
Fig. 6: Abbreviated key bicyclization steps of the proposed SalC mechanism with 15.
Once substrate undergoes transacylation from acylated SalB (15) to Cys180 of SalC (not shown), hydrogen bonding by Tyr284 facilitates deprotonation of the thioester α-proton by His353. An intramolecular aldol reaction forms the γ-lactam and the resulting oxyanion is presumably stabilized by dipole interactions with backbone amides (not shown).25 Subsequent β-lactonization through a tetrahedral intermediate leads to release of simplisporamide (R-17) from SalC. See Extended Data Fig. 10 for further details.
DISCUSSION
The proteasome is a well-established target for cancer therapeutics due to its essential role in intracellular protein degradation, a process that is upregulated in rapidly proliferating cancer cells.44 Three constitutive proteasome inhibitors (PIs) are currently FDA approved as cancer therapeutics. Marizomib, however, is fundamentally different from these approved PIs as it is the only non-peptidic compound in advanced clinical trials, and targets all three catalytic subunits of the human 20S proteasome with single-digit nanomolar inhibitory activity against the β5 subunit.7 This pan-proteasome inhibitory activity allows 1 to retain clinical activity in patients resistant to other PIs as resistance is typically due to mutations in a single proteasome subunit.45 Resistance is a major obstacle towards using PIs in the clinic, which underscores the importance of the continued development of 1 and its analogs. Finally, salinosporamide A’s ability to cross the blood-brain barrier facilitates a new route to treat neurological diseases, which often require invasive surgery for drug delivery to the brain. While salinosporamide A is still in Phase III clinical trials, 1’s potent activity and ability to enter the brain could provide hope for patients with glioblastoma.
The work described here establishes a ketosynthase homolog, SalC, acts as a bicyclase and installs the salinosporamide pharmacophore responsible for 1’s clinically relevant bioactivity. This discovery not only solves a long-standing biosynthetic enigma, but also expands the repertoire of known reactivities for the ketosynthase family of enzymes. Rather than a decarboxylative Claisen condensation to extend a nascent polyketide chain, the KS SalC plays an unexpected dual role in the sal pathway as an intramolecular aldolase/-lactone synthase, a reaction for which there is no biosynthetic precedent. Furthermore, by using a proteasome inhibition assay to screen for formation of this reactive feature, we were able to directly link the biosynthetic activity of this enzyme to the bioactivity of the molecule.
This ability to use a fluorescence-based assay to validate SalC’s biosynthetic ability is especially intriguing as it opens the door for high-throughput screening efforts to generate SalC variants with desirable qualities for biocatalysis. The unusual reactivity of SalC has the potential to be harnessed for chemoenzymatic routes to β-lactone synthesis, as well as complement existing syntheses of 1 that perform bicyclization as the terminal step.12 Several features of SalC make the enzyme well suited for this.
First, SalC is notable for its ability to perform a complex biosynthetic reaction on a variety of substrates. Typically, the scaffold of assembly line-based natural products is constructed by a PKS and/or NRPS and then functionalized post-release from the terminal module via tailoring reactions such as oxidations or glycosylations which are essential for the bioactivity of these compounds. Instead, the sal pathway expends metabolic energy upfront to generate complex precursors that are then assembled by SalC, a standalone enzyme that zips together complex pieces in a single step. While TEs are known to be highly selective for their native substrate,46 SalC is able to perform a complex cyclization reaction on a variety of precursors to form the fused bicyclic core that is the constant feature amid the wealth of salinosporamide biosynthetic diversity.
Beyond its flexible substrate preference, SalC also appears capable of resolving a diastereomeric substrate mixture, with a strong preference for forming simplisporamide with the R stereocenter at the ethyl side chain. The minor epimer is produced as well (S-17), but in an approximately 1:10 ratio. This preference is borne out in the native bacterium as well; naturally produced salinosporamide F is the C2 epimer of salinosporamide A but is produced in very low yield compared to 1.30 Furthermore, this work also lays the foundation to engineer or design SalC biocatalysts capable of working on small molecule diffusible substrates (Extended Data Fig. 3).
Finally, SalC is potentially useful as a biocatalyst because it is capable of installing a β-lactone, a chemical moiety known for its reactivity. While the assembly of β-lactam natural products has been studied for decades,18 the study of β-lactone biosynthesis has largely been hindered by the instability of these molecules. Only in the last five years have routes to β-lactone biosynthesis been discovered, with just three examples prior to this work. Furthermore, SalC catalyzes an entirely new reaction type from the previously characterized enzymes which include adenylate-forming β-lactone synthetases47,48, thioesterases,14 and α/β hydrolases.17 SalC is also different in that it must first perform an aldol reaction to generate the moieties required for β-lactonization; the nucleophilic alcohol is not already installed on the molecule as in the obafluorin and olefinic hydrocarbon pathways. The discovery of SalC as a standalone KS capable of forming a β-lactone suggests the possibility of this type of reactivity in other pathways, particularly where terminal standalone C or KS domains are evident. In fact, NaPDoS analysis reveals that SalC and its homologs, including the cinnabaramide homolog CinC, belong to their own unique clade among known KSs (Supplementary Fig. 4), demonstrating the unique functionality of this enzyme and the potential for discovery of other bicyclized natural products.
Almost 18 years after its initial discovery, and four years after entering phase III clinical trials, a key mystery in the biosynthesis of salinosporamide A has finally been revealed. Using biochemical and structural data, we identified and characterized an unusual ketosynthase responsible for performing previously unprecedented biochemistry. The salinosporamide bicycle is the most synthetically challenging portion of the molecule and, as such, salinosporamide derivatives are prepared by slow and low-yielding fermentative approaches. To this day, salinosporamide A is produced by fermentation for clinical trials, a process which requires the use of specialized reactors to handle the saltwater media necessary for Salinispora spp. growth. However, the identification of SalC as a bicyclase capable of generating the γ-lactam-β-lactone pharmacophore completes the minimum genetic architecture required for assembly of the salinosporamide scaffold and has the potential to fundamentally alter how salinosporamides are produced. As our clinical understanding of alternative proteasomes grows,44 and we begin to understand the role these cellular machines play not just in cancer but in autoimmune and parasitic diseases as well, a hybrid synthetic-biocatalytic blueprint for producing brain-penetrant salinosporamide molecules is now feasible. Understanding the unique reactivity of the SalC bicyclase is the first step towards harnessing that activity towards the efficient, straightforward production of new-to-nature bioactive salinosporamides.
METHODS
Software and tools used
The SalC protein sequence was analyzed via BLAST,23 and all alignments were made using Clustal Omega (1.2.4) software. Geneious Prime 2019.2.3 software was used for plasmid maps and primer design. The NaPDoS platform24 was used to examine SalC in relation to other KSs, cblaster (1.2.9)49 was used to examine SalC homologs in salinosporamide producing strains, and clinker (v0.0.21) was used for visualization.50
Bacterial strains and growth conditions
E. coli strains were grown in LB broth or agar at 37 °C with appropriate antibiotics for selection (apramycin 50 μg/mL, chloramphenicol 25 μg/mL, kanamycin 50 μg/mL, nalidixic acid 25 μg/mL). For conjugation purposes, E. coli was grown using 2TY media (1.6% w/v tryptone, 1% w/v yeast extract, 0.5% w/v NaCl) with appropriate antibiotic selection. For protein production, TB media (1.2% w/v tryptone, 2.4% w/v yeast extract, 0.4% (v/v) glycerol, 2.31% w/v KH2PO4, 12.54% w/v K2HPO) was used. All liquid cultures were shaken at 200 rpm
Streptomyces strains were grown on SFM agar plates (2% w/v D-mannitol, 2% w/v soya flour, 2% w/v agar) for conjugation and strain maintenance at 30 °C, or in TSBY liquid media (3% w/v tryptic soy broth, 10.3% w/v sucrose, 0.5% w/v yeast extract) at 30 °C with shaking at 220 rpm.
For protein production in S. coelicolor CH999, a TSBY preculture (100 μL) was used to inoculate the 30 mL ‘primary culture’ of Super YEME media (0.3% w/v yeast extract, 0.5% w/v peptone, 1% w/v glucose, 0.3% w/v malt extract, 34% w/v sucrose, 0.5% w/v glycine, 0.235% v/v MgCl2∙6H2O (2.5 M), 7.5×10−3% w/v L-proline, 7.5×10−3 w/v % L-arginine, 7.5×10−3 % w/v L-cysteine, 0.01% L-histidine, 1.5×10−3 % w/v uracil, pH 7.2) with 50 mg/mL apramycin in a 250 mL flask with bottom spring. This culture was grown at 30 °C with shaking at 220 rpm. After 4 days, 4 mL of primary culture was used to inoculate 40 mL of Super-YEME supplemented with apramycin to generate the ‘secondary culture’. This culture was grown at 30 °C for 2 days. Finally, 600 mL of ‘expression culture’ was inoculated using 30 mL of secondary culture supplemented with apramycin (50 mg/mL) in 2.8 L flasks with metal springs. After 2 days, thiostrepton was added to a final concentration of 10 ug/mL to induce protein expression.
Salinispora strains were grown in sea water based A1 media (1% w/v soluble starch, 0.4% w/v yeast extract, 0.2% w/v peptone, 0.1% w/v CaCO3) at 30 °C with shaking at 220 rpm, or A1 agar at 30 °C supplemented with apramycin (50 mg/mL) and naldixic acid (25 mg/mL) for mutant strains. For production of salinosporamide, liquid A1 was supplemented with 2% v/v KBr (20 g/L) and 0.8% v/v Fe2(SO4)3, (8 g/L) and XAD7 resin was added to the cultures after 24 hrs of growth.
DNA isolation and manipulation
Genomic DNA (gDNA) was isolated following standard procedures from Practical Streptomyces Genetics.51 Phusion High-Fidelity DNA Polymerase (NEB) with GC buffer was used to amplify genes from the sal cluster using gDNA as a template.
PCR reactions were carried out in a BioRad MyCycler with gradient option. PCR products were purified using Qiagen QIAquick PCR and Gel Cleanup Kit according to the manufacturer’s instructors. Vectors (pET28, pET28-MBP, and pCJW93) were linearized with restriction enzymes for cloning purposes. For cloning, Gibson assembly (NEB HiFi DNA Assembly Master Mix) was used to combine linearized vector and purified PCR products before subsequent transformation into chemically competent E. coli DH10B cells. Plasmid DNA was isolated using the QIAPrep Spin Miniprep Kit (Qiagen) and cloning was verified by Sanger sequencing (Genewiz).
For protein expression and production of SalC in Streptomyces, pCJW93-SalC was transformed into E. coli ET1256752 and then conjugated into S. coelicolor CH999 via triparental intergeneric mating facilitated by E. coli ET12567/pUB30753 following established procedures.54 Exconjugates were grown on SFM media containing 10 mM MgCl2 for exactly 18 hours at which time plates were overlaid with 0.5 mg/mL naldixic acid followed by 1 mg/mL apramycin. Plates were incubated at 30 °C until the appearance of exconjugants, which were subsequently replated for further rounds of selection.
Salinispora tropica CNB440 mutants were made by inactivating each gene by replacement with an apramycin resistance cassette (aac(3)IV) using λ-Red recombination, as described previously.9
Salinosporamide identification and purification
Wild type and knockout S. tropica CNB440 strains were grown for five days after addition of XAD7 absorbent resin. Resin was harvested and extracted with ethyl acetate. The organic phase was evaporated and samples were reconstituted in acetonitrile and filtered through a 0.2 μm filter for subsequent LCMS analysis.
Salinosporamides were identified by LCMS analysis using an Agilent 1260 Infinity LC system coupled to an Agilent 6530 Accurate-Mass Q-TOF. A solvent system of acetonitrile and water both containing 0.1% formic acid (v/v) was used. A 10 μL aliquot was injected on a Phenomenex Kinetex C18 reversed-phase HPLC column (5 μm, 250 mm × 4.6 mm) and eluted over a 40 minute method with a gradient from 20% to 100% over 25 minutes. 100% acetonitrile was held for 5 minutes before acetonitrile percentage was dropped to 20%. Flow rate was 0.75 mL/min. Eluent was detected using electrospray ionization-mass spectrometry (ESI-MS) monitoring m/z 70–3,200 in positive mode with a speed of 32,500 m/z /s and 30 kV collision energy. MS data were analyzed using Agilent MassHunter Qualitative Analysis B.05.01 software. Salinosporamide A was purified via methods described previously.5
SalC expression and purification from Streptomyces
S. coelicolor CH999-pCJW9354-SalC were grown as described above and harvested at 4 °C by centrifuging at 16,000 x g for 20 min. The cell pellet was resuspended in harvest buffer (100 mM Tris, 300 mM NaCl, 0.8 mM TCEP, 10% glycerol) and lysed on ice with a QSonica sonicator (6 mm tip at 60% amplitude for 15 cycles of 15 seconds pulse on followed by 45 seconds pulse off). The lysate was then centrifuged for 60 min at 4 °C, 44,000 x g and subjected to column chromatography.
Protein purification was performed on an ÄKTApurifier instrument (GE Healthcare) with the modules Box-900, UPC-900, R-900 and Frac-900 with all buffers filtered through a nylon membrane 0.2 μm GDWP (Merck) prior to use. FPLC data was analyzed with UNICORN 5.31 (Built 743) software. SalC was initially purified by Ni2+-affinity chromatography using 5 mL HisTrap FF (GE Healthcare) columns pre-equilibrated in buffer A (300 M NaCl, 100 mM Tris, 25 mM imidazole, 0.8 mM TCEP, pH 8.0). Lysate was loaded onto column at 1.5 mL/min after which the column was washed with 8 column volumes (40 mL) of buffer A. Protein was eluted by a linear gradient of 0 – 100% buffer B (300 M NaCl, 100 mM Tris, 250 imidazole, 0.8 mM TCEP, pH 8.0) over 30 min at a flow rate of 2.0 mL/min. Protein-containing fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in the presence of reducing agents (SDS-PAGE, 12% acrylamide) and then combined and concentrated to a volume of ~3 mL using Amicon Ultra centrifugal filters with 50 kDa molecular weight cut-off (EMD Millipore). Concentrated protein was then loaded onto a HiLoad Superdex 200 prep grade size exclusion (SEC) column (16 cm x 60 cm, GE Healthcare) equilibrated in 50 mM Tris, 150 mM NaCl, 0.8 mM TCEP, 10% glycerol, pH 8.0 buffer and eluted at a constant flow rate of 1.0 mL/min. Fractions containing the target protein were pooled, concentrated, aliquoted, and flash frozen for storage at −80 °C. Protein concentration was determined by Bradford assay.
Protein expression and purification from E. coli
E. coli BL21Gold(DE3) transformed with expression plasmids for CoaA, CoaD, CoaE, Sfp, and SalB-PCP were inoculated in 10 mL LB containing kanamycin. After overnight incubation, this 10 mL culture was used to inoculate 1 L of TB media in a 2.8 L baffled flask supplemented with kanamycin. Flasks were incubated at 37 °C, 220 rpm until OD600 reached 0.6–0.8 at which time flasks were cooled for 1 hr at 18 °C before induction with 1 M isopropyl--D-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM. Flasks were incubated overnight at 18 °C, 220 rpm, and harvested by centrifugation at 4 °C, 5,000 × g for 10 min. From this point on all subsequent steps were performed at 4 °C or on ice. The cell pellet was subsequently resuspended in lysis buffer (500 mM NaCl, 20 mM Tris, 10% glycerol, pH 8.0) and lysed by sonication with a Qsonica sonicator (6 mm tip at 40% amplitude for 10 cycles of 15 seconds pulse on followed by 45 seconds pulse off). Lysate was then centrifuged at 14,000 × g for 45 min at 4 °C. Protein purification was performed as described in the previous section using buffer A (1 M NaCl, 20 mM Tris, 25 mM imidazole, pH 8.0) and buffer B (1 M NaCl, 20 mM Tris, 250 imidazole, pH 8.0).
CoaA, CoaD, CoaE, and Sfp proteins were buffer exchanged using PD-10 desalting columns (GE Healthcare) to storage buffer (20 mM Tris, 10% glycerol, pH 8.0) as described previously.33 Protein concentration was determined by Bradford assay before the samples were aliquoted, flash frozen, and stored at −80 °C.
SalB-PCP was further purified by SEC using a HiLoad Superdex 75 prep grade gel filtration column (16 cm x 60 cm, GE Healthcare) preequilibrated with storage buffer (20 mM Tris, 200 mM NaCl, 10% glycerol, pH 8.0). Fractions with the target protein were pooled and concentrated with centrifugal filters again. Protein concentration was determined using the Bradford assay and then flash frozen and stored at −80 °C.
Tryptic fingerprinting SalB protein
To confirm the identity of the purified SalB protein as well as monitor conversion from apo to the holo and acylated-holo forms, the protein was digested using the Trypsin Single Proteomics Grade Kit (Sigma Aldrich) and analyzed by High Resolution-LCMS analysis. A 10 μL aliquot was injected onto a Phenomenex Aeris WIDEPORE XB-C18 200 Å, LC column(3.6 μM, 250 mm x 4.6 mm) and analyzed with an Agilent 1260 Infinity LC system coupled to an Agilent 6530 Accurate-Mass Q-TOF. A solvent system of acetonitrile and water both containing 0.1% formic acid (v/v) was used. Peptide fragments were eluted over a 43-min method with a gradient from 5 to 30% acetonitrile over 4 min, 30 to 65% acetonitrile over the next 15 min, and then to 100% over 5 min. 100% acetonitrile was held for 6 minutes before concentration was dropped to 5%. Flow rate was 0.75 mL/min. Eluent was detected using electrospray ionization-mass spectrometry (ESI-MS) monitoring m/z 70–3,200 in positive mode with a speed of 32,500 m/z /s and 30 kV collision energy. MS data were analyzed using Agilent MassHunter Qualitative Analysis B.05.01 software. Peptide fragments were compared to predicted trypsin digest fragments using the ExPASy PeptideMass tool.
SalC hydrolysis assay
To probe the SalC reaction in the reverse direction, 50 μL assays containing purified SalC (20 μM), SalB-PCP (50 μM), and 1 mM salinosporamide A were incubated in SalB buffer (20 mM Tris, 200 mM NaCl, 0.8 mM TCEP, 10% glycerol pH 8.0). After three hours the assays were quenched with three volumes of acetonitrile and centrifuged at 21,000 × g for 30 minutes to precipitate protein. Samples were filtered and then analyzed by LCMS with the method for salinosporamide identification described earlier.
Chemical synthesis
The syntheses and accompanying spectral data of the pantetheine-tethered substrates 13 and 21 are described in the Supplementary Notes file.
SalC activity assays
Linear pantetheine activated substrates (13 or 21, 250 μM) were incubated with purified CoaA (7 μM), CoaD (10 μM), CoaE (10 μM), Sfp (10 μM) and SalB-PCP (250 μM) in reaction buffer (50 mM phosphate, 100 mM NaCl, 0.8 mM TCEP, 10 mM MgCl2, pH 7.5) in a 50 μL reaction to allow carrier protein loading to occur. The reaction was initiated with the addition of 5 mM ATP. After 3 h, 25 μL of acylated SalB-PCP was removed and used for trypsin fingerprinting or intact MS. SalC (20 μM) was added to the remaining 25 μL for activity assays. Upon testing different reaction conditions (buffer, pH, concentrations of all proteins, time), these conditions were found to be optimal.
Assays with diffusible substrates omitted required CoA or Sfp proteins. After the addition of SalC, reactions were incubated for an additional 3 h at 30 °C. Reactions were quenched by the addition of three volumes of acetonitrile and centrifuged at 21,000 × g for 30 min to precipitate protein. Finally, samples were analyzed via HR-LCMS using the method for salinosporamide identification described earlier.
Preparative scale SalC activity assay
For purification of the SalC assay product, reactions were set up as described above at a 50 mL scale in an Erlenmeyer flask. SalB-PCP loading was allowed to proceed for 12 hours, at which point approximately 1 gram of XAD7 resin and SalC (20 μM) were added to the reaction mixture and assays were allowed to proceed overnight. In the morning, resin was extracted (3×) with ethyl acetate. Organic extracts were combined and evaporated to dryness. Samples were resuspended in acetonitrile and the peak corresponding to R-17 was purified by preparative HPLC using a Phenomenex Luna C18 column (5 μm, 100 mm, 2mm i.d.), along with an Agilent Technologies system composed of a PrepStar pump, a ProStar 410 autosampler, and a ProStar UV detect (Agilent Technologies, Inc, Santa Clara, USA). The sample were eluted by a gradient from 20–100% acetonitrile over 40 min at a flow rate of 10 mL/min. The peak corresponding to R-17 was collected and dried by rotary evaporation and lyophilization. Finally, R-17 was purified using a small-scale silica column with a dichloromethane/acetonitrile solvent system. All stages of purification were monitored by LCMS analysis.
Structural characterization of R-17
All NMR data were collected at the UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences NMR Facility on a 600 MHz Varian NMR spectrometer (Topspin 2.1.6 software, Bruker) with a 1.7 mm cryoprobe. Deuterated chloroform containing TMS standard was used as a solvent. All spectra for purified R-17 can be found in the Supplementary Notes.
Proteasome inhibition assay
SalC activity assays (+/− SalC and +/− 13) were run on a 1 mL scale with the addition of resin, as described in the Preparative scale SalC activity assay section. Resin extracts were resuspended in 50 μL buffer and used for proteasome inhibition assays and LCMS analysis.
Proteasome inhibition assays were performed using the QUANTIZYME® Assay System (Enzo Life Sciences), designed to measure chymotrypsin-like protease activity (β5 subunit) of the purified 20S human proteasome using the fluorogenic substrate Suc-LLVY-AMC. Inhibitor (0.5 μM epoxomicin, 0.5 μM salinosporamide A, or 10 μL of resuspended SalC activity assay extracts) was added to 0.2 μg proteasome in kit buffer (50 mM Tris-HCl, pH 7.5, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.03% SDS) and incubated for 15 minutes. Substrate was added to a final concentration of 75 μM. Proteasome activity was measured by reading fluorescence of the cleaved substrate at 355 nm (excitation) and 460 nm (emission) using Spectra max M2 (Molecular Devices) every 5 minutes for 30 minutes. All assays performed in duplicate.
Chain transfer assays and intact protein MS
SalB-PCP was acylated with the panthetheine-activated mechanistic probe 21 using the same methods described earlier. This holo-acyl-SalB-PCP was purified away from residual CoA proteins and small molecules using a Superdex 75 prep grade gel filtration column (16 cm x 60 cm, GE Healthcare) preequilibrated with buffer (50 mM Tris, 150 mM NaCl, 5% glycerol, pH 8.0). Purified 22 or diffusible substrates (21–25) were incubated with SalC or a SalC variant (20 μM) for 3 h, and then subjected to LC-HRMS analysis. All intact protein LC-HRMS was performed by the UC San Diego Molecular Mass Spectrometry Facility on an Agilent 1260 Infinity Binary LC coupled with a 6230 Accurate-Mass TOFMS.
Crystallization of SalC
SalC was crystallized by hanging drop crystallization at room temperature. 1.0 μL 1.7–3.4 mg/mL SalC in gel filtration buffer was mixed with 1.0 μL well solution (0.10 M HEPES pH 8.5, 30 % (w/v) PEG 3350, and 0.30 M KCl) to make a 2 μL hanging drop in a sealed well with 400 μL well solution. Transparent plate crystals grew in 24–48 h. The crystals were then transferred to a drop of LV Cryo Oil (MitGen) and flash-cooled in liquid nitrogen.
Data collection and processing
A data set of SalC was collected at Advanced Light Source (Berkeley, California, USA) on beamline 8.2.2 using a ADSC Q315R detector at a temperature of 100 K. Reflections could be observed to 2.85 Å. Data were indexed and integrated using XDS,55 with high resolution cut-off to 2.85 Å, based on CC1/2 = ~ 0.7. Data statistics are listed in Supplementary Table 3.
Structure determination and refinement
The structure of SalC was determined to 2.85-Å resolution by molecular replacement (MR) using Phaser56 implemented in CCP4. The ketosynthase domain of DEBS1 from Saccharopolyspora erythraea (PDB ID: 2HG4)57, which shared 33.5% identity with SalC, was used to generate a homology model of SalC by I-TASSER58 as the MR search model. The MR results in one SalC tetramer per asymmetric unit (ASU). Resulting LLG, Rval and TFZ were 8178.1, 44.8 and 66.5, respectively. The MR solution was used as an input for AutoBuild implemented in Phenix59 for initial model building. After the AutoBuild run, the atomic coordinates and B-factors were iteratively refined in Phenix Refine with model building and manual adjustment of the model in Coot.60 Water molecules were added manually throughout real space refinements using Fo-Fc electron density contoured to 3.0 σ as criteria. Four-fold non-crystallographic symmetry (NCS) restraints were used throughout refinement. Final cycles of refinements include Translation-Libration-Screw-rotation (TLS) parameterizations with two TLS group per SalC monomer. The division is assigned based on the two domains of SalC. A composite-omit electron density map calculated by Phenix Composite_omit_map was used to verify the model. The refinement statistics are in Supplementary Table 3, and the final model of the SalC structure contains residues listed in Supplementary Table 4. All structure figures were rendered in PyMOL.
SalC mutagenesis
SalC mutagenesis was performed by generating two PCR products using the salC forward primer and a mutagenic reverse primer and a mutagenetic forward primer and the salC reverse primer. Three-piece Gibson assembly was used to assemble these two amplicons into linearized pCJW93. pCJW93-SalC mutants were subsequently conjugated into S. coelicolor CH999. Following expression and purification, SalC variants were subjected to SalC activity assays and chain transfer assays as described above.
Data availability
Strains and plasmids used in this study are described in Supplementary Table 1. All oligonucleotides (Integrated DNA Technology) are shown in Supplementary Table 2. The salinosporamide (sal) biosynthetic gene cluster from S. tropica CNB440 is available in the MIBiG database (accession BGC0001041). Other salinosporamide BGCs and the cinnabaramide BGC used for alignments are available in the IMG JGI database. Functional KS and non-elongating KS protein sequences used for alignments can be found in the Supplementary material and through MIBiG. Atomic coordinates and structure factors for the reported crystal structures in this work have been deposited to the Protein Data Bank under accession number 7S2X (native SalC). Additionally, the following PBD datasets were used for SalC structural comparison: 2HG4, 4WKY, 2QO3, 4NA2. Source data for the proteasome inhibition assay (Extended Data Fig. 4) is provided with this paper. Other relevant data supporting the findings of this study are available in this published article or its Supplementary files.
Extended Data
Extended Data Fig. 1. Salinosporamide A hydrolysis and subsequent THF ring formation is accelerated by the presence of SalC.
a, LCMS chromatograms of salinosporamide A (1) hydrolysis assay with and without SalC. * indicates compound retains chloride and no THF ring formation has occurred as evidenced by characteristic isotope pattern. b, Structures of compounds putatively identified by LCMS in a.
Extended Data Fig. 2. Naturally produced analogs of salinosporamide A that served as inspiration for simplisporamide.
Extended Data Fig. 3. SalC activity assay with diffusible substrates.
a, Reaction scheme depicting chemoenzymatic synthesis and subsequent SalB-PCP acylation assay to generate (15). b, LCMS chromatograms (BPCs and EICs m/z 266.18) of SalC activity assay with diffusible substrates shown in part (a) including pantetheine-activated (13), CoaA product-activated (19), CoaD product-activated (20), CoA activated (14), and carrier protein-activated substrate (15). Substrates were activated via in vitro CoA enzyme biosynthesis. All SalC activity assays contained apo-SalB-PCP as well.
Extended Data Fig. 4. Proteasome inhibitory activity of SalC assay product using purified human 20S proteasome.
a, Proteasome activity determined by reading fluorescence (AFUs) of the cleaved substrate (Suc-LLVY-AMC) at 355 nm (excitation) and 460 nm (emission) every five minutes after substrate was added for 30 min in the presence of various inhibitors. Blank (no proteasome added) = black, control (no inhibitor) = gray, epoxomicin (0.5 μM) = green, SalC reaction product = red, no SalC control = dark blue, no substrate control = orange, salinosporamide A (0.5 μM) = light blue. Samples run in duplicate, all data points shown. b, Magnification of y-axis of plot from a to examine successful inhibition of the 20S proteasome by epoxomicin, SalC reaction product, and salinosporamide A. c, Percent proteasome inhibition at 30 min, relative to control (no inhibitor, 0% inhibition). See Supplementary Fig. 14 for corresponding LCMS traces of extracts used in these assays.
Extended Data Fig. 5. Acylation of SalC with diffusible substrates.
a, Reaction scheme depicting chemoenzymatic synthesis to generate (22) b, UV chromatograms (215 nm) of intact protein LCMS for transacylation assay with diffusible substrates. Transacylation assay utilized linear mechanistic probe (21) activated in different ways (CoA-precursor-activated (23, 24), CoA-activated (25), and SalB-PCP-tethered, all generated in situ) and SalC. Transacylation assay with column purified 22 shown for comparison.
Extended Data Fig. 6. SalC overlay with a trans-AT KS.
SalC structure aligned with closest Dali server homolog, the trans-AT KS OzmN KS2 (PDB ID: 4WKY) from the hybrid NRPS/PKS oxazolomycin pathway, RMSD 0.842 . Overall structure of SalC dimer (colors shown as previous, KS monomers in brown and green, flanking subdomains in yellow and teal) with OzmN KS2 (gray).
Extended Data Fig. 7. SalC overlay with functional type I KS.
SalC KS aligned with DEBS KS3 (PDB: 2QO3), RMSD 1.225 . a, Overall structure of SalC dimer (colors shown as previous, KS monomers in brown and green, flanking subdomains in yellow and teal) with DEBS KS3 (gray). b, Active site overlay of SalC (green) and DEBS KS3 (gray).
Extended Data Fig. 8. Tyr284 is conserved in SalC homologs.

Condensed alignment showing conservation of Tyr284 in all SalC homologs from Salinispora and Streptomyces cinnabarigriseus JS360 (CinC) but not in canonical elongating KSs. All SalC homologs from Salinispora strains found in JBI IMG database. For functional KS sequences refer to Supplementary Figure 5.
Extended Data Fig. 9. SalC structure overlaid with bacillaene PKS (bae) KS2 bound to its natural intermediate.
SalC is shown in green and BaeKS2 is shown in grey, bae intermediate in gold. PDB ID: 4NA2, RMSD 1.071 .
Extended Data Fig. 10. Proposed active site mechanism of SalC.
Catalysis is initiated by deprotonation of Cys180 followed by transacylation of the SalB-PCP tethered substrate through a tetrahedral intermediate (not shown). Lys348 deprotonates His353, and hydrogen bonding of the thioamide carbonyl to Tyr284 facilitates deprotonation of the thioester α-proton by His353. An intramolecular aldol reaction forms the γ-lactam; the oxyanion is presumably stabilized by dipole interactions with backbone amides, as is hypothesized for KSs.25 Subsequent β-lactonization through a tetrahedral intermediate leads to release of simplisporamide from SalC. Finally, Cys180 is reprotonated by His353.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to P.R. Jensen (Scripps Institution of Oceanography) for the Salinispora tropica CNB440 strain, B.M. Dungan (UC San Diego) for assistance with NMR, K. E. Creamer (UC San Diego) for the NaPDoS2.0 analysis and helpful bioinformatics discussions, Dr. Y. Su (UC San Diego Molecular Mass Spectrometry Facility) for intact proteomics experiments, and J.P. Noel and G. Louie (Salk Institute for Biological Studies) for beamline coordination. Beamline 8.2.2 of the Advanced Light Source, a U.S. DOE Office of Science User Facility under Contract No. DE-AC02-05CH11231, are supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30-GM124169-01. This work was supported by NIH grants R01CA127622 (B.S.M.), R01AI047818 (B.S.M.), and F31HD101307 (K.D.B.), R35GM134910 (D.R.), the Robert A. Welch Foundation grant number AA-1280 (D.R.) and the São Paulo Research Foundation (FAPESP) grant number 2011/21358-5 (D.B.B.T).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Roth P et al. A phase III trial of marizomib in combination with temozolomide-based radiochemotherapy versus temozolomide-based radiochemotherapy alone in patients with newly diagnosed glioblastoma. Am. J. Clin. Oncol 39, 2004–2004 (2021). [Google Scholar]
- 2.Feling RH et al. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed 42, 355–357 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Williams PG et al. New cytotoxic salinosporamides from the marine Actinomycete Salinispora tropica. J. Org. Chem 70, 6196–6203 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Macherla VR et al. Structure-activity relationship studies of salinosporamide A (NPI-0052), a novel marine derived proteasome inhibitor. J. Med. Chem 48, 3684–3687 (2005). [DOI] [PubMed] [Google Scholar]
- 5.McGlinchey RP et al. Engineered biosynthesis of antiprotealide and other unnatural salinosporamide proteasome inhibitors. J. Am. Chem. Soc 130, 7822 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nett M, Gulder TAM, Kale AJ, Hughes CC & Moore BS Function-oriented biosynthesis of β-lactone proteasome inhibitors in Salinispora tropica. J. Med. Chem 52, 6163–6167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groll M, Huber R & Potts BCM Crystal structures of salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of β-lactone ring opening and a mechanism for irreversible binding. J. Am. Chem. Soc 128, 5136–5141 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Beer LL & Moore BS Biosynthetic convergence of salinosporamides A and B in the marine actinomycete Salinispora tropica. Org. Lett 9, 845–848 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Eustáquio AS et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-l-methionine. Proc. Natl. Acad. Sci. U.S.A 106, 12295–12300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahlstedt S, Fielding EN, Moore BS & Walsh CT Prephenate decarboxylases: a new prephenate-utilizing enzyme family that performs non-aromatizing decarboxylation en route to diverse secondary metabolites. Biochemistry 49, 9021–9023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma G, Nguyen H & Romo D Concise total synthesis of (+/−)-salinosporamide A, (+/−)-cinnabaramide A, and derivatives via a bis-cyclization process: implications for a biosynthetic pathway? Org. Lett 9, 2143–2146 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen H, Ma G, Gladysheva T, Fremgen T & Romo D Bioinspired total synthesis and human proteasome inhibitory activity of (−)-salinosporamide A, (−)-homosalinosporamide A, and derivatives obtained via organonucleophile promoted bis-cyclizations. J. Org. Chem 76, 2–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischbach MA & Walsh CT Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev 106, 3468–3496 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Schaffer JE, Reck MR, Prasad NK & Wencewicz TA β-Lactone formation during product release from a nonribosomal peptide synthetase. Nat. Chem. Biol 13, 737–744 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Gulder TAM & Moore BS Salinosporamide natural products: potent 20S proteasome inhibitors as promising cancer chemotherapeutics. Angew. Chem. Int. Ed 49, 9346–9367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X et al. Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Bio l 8, 823–830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng K-N et al. A hydrolase-catalyzed cyclization forms the fused bicyclic β-lactone in vibralactone. Angew. Chem. Int. Ed 59, 7209–7213 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Gaudelli NM, Long DH & Townsend CA β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature 520, 383–387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun C-S et al. Unique features of the ketosynthase domain in a non-ribosomal peptide synthetase–polyketide synthase hybrid enzyme, tenuazonic acid synthetase 1. J. Biol. Chem.( 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun C-S, Motoyama T & Osada H Biosynthesis of the mycotoxin tenuazonic acid by a fungal NRPS–PKS hybrid enzyme. Nat. Commun 6, 8758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo X & Gulder TAM Biosynthetic strategies for tetramic acid formation. Nat. Prod. Rep (2021). [DOI] [PubMed] [Google Scholar]
- 22.Rachid S et al. Mining the cinnabaramide biosynthetic pathway to generate novel proteasome inhibitors. ChemBioChem 12, 922–931 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Ziemert N et al. The Natural Product Domain Seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7, e34064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins T, Kapilivsky J, Cane DE & Khosla C Roles of conserved active site residues in the ketosynthase domain of an assembly line polyketide synthase. Biochemistry 55, 4476–4484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helfrich EJN & Piel J Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep 33, 231–316 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Masschelein J et al. A dual transacylation mechanism for polyketide synthase chain release in enacyloxin antibiotic biosynthesis. Nat. Chem 11, 906–912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satou R et al. Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases. J. Biol. Chem 288, 34146–34157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boddy CN, Schneider TL, Hotta K, Walsh CT & Khosla C Epothilone C macrolactonization and hydrolysis are catalyzed by the isolated thioesterase domain of epothilone polyketide synthase. J. Am. Chem. Soc 125, 3428–3429 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Reed KA et al. Salinosporamides D-J from the marine actinomycete Salinispora tropica, bromosalinosporamide, and thioester derivatives are potent inhibitors of the 20S proteasome. J. Nat. Prod 70, 269–276 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Manam RR et al. Antiprotealide is a natural product. J. Nat. Prod 72, 295–297 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Ling T, Macherla VR, Manam RR, McArthur KA & Potts BCM Enantioselective total synthesis of (−)-salinosporamide A (NPI-0052). Org. Lett 9, 2289–2292 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Agarwal V et al. Chemoenzymatic synthesis of acyl coenzyme A substrates enables in situ labeling of small molecules and proteins. Org. Lett 17, 4452–4455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorrestein PC et al. Facile detection of acyl and peptidyl intermediates on thiotemplate carrier domains via phosphopantetheinyl elimination reactions during tandem mass spectrometry. Biochemistry 45, 12756–12766 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khosla C, Tang Y, Chen AY, Schnarr NA & Cane DE Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem 76, 195–221 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Denora N, Potts BCM & Stella VJ A mechanistic and kinetic study of the beta-lactone hydrolysis of salinosporamide A (NPI-0052), a novel proteasome inhibitor. J. Pharm. Sci 96, 2037–2047 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Tsueng G & Lam KS Stabilization effect of resin on the production of potent proteasome inhibitor NPI-0052 during submerged fermentation of Salinispora tropica. J. Antibiot 60, 469–472 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Dutta S et al. Structure of a modular polyketide synthase. Nature 510, 512–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm L Using Dali for protein structure comparison. Methods Mol. Biol 2112, 29–42 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Tang Y, Chen AY, Kim C-Y, Cane DE & Khosla C Structural and mechanistic analysis of protein interactions in module 3 of the 6-Deoxyerythronolide B synthase. Chem. Biol 14, 931–943 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohman JR et al. Structural and evolutionary relationships of “AT-less” type I polyketide synthase ketosynthases. Proc. Natl. Acad. Sci. U.S.A 112, 12693–12698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gay DC et al. A close look at a ketosynthase from a trans-acyltransferase modular polyketide synthase. Structure 22, 444–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies C, Heath RJ, White SW & Rock CO The 1.8 Å crystal structure and active-site architecture of β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure 8, 185–195 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Cromm PM & Crews CM The proteasome in modern drug discovery: second life of a hHighly valuable drug target. ACS Cent. Sci 3, 830–838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niewerth D et al. Antileukemic activity and mechanism of drug resistance to the marine Salinispora tropica proteasome inhibitor salinosporamide A (Marizomib). Mol. Pharmacol 86, 12–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen DA, Koch AA & Sherman DH Identification of a thioesterase bottleneck in the pikromycin pathway through full-module processing of unnatural pentaketides. J. Am. Chem. Soc 139, 13450–13455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christenson JK et al. β-lactone synthetase found in the olefin biosynthesis pathway. Biochemistry 56, 348–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson SL et al. Global analysis of adenylate-forming enzymes reveals β-lactone biosynthesis pathway in pathogenic Nocardia. J. Biol. Chem 295, 14826–14839 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 49.Gilchrist CLM et al. cblaster: a remote search tool for rapid identification and visualisation of homologous gene clusters. Bioinformatics Adv. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilchrist CLM & Chooi Y-H clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics (2021). [DOI] [PubMed] [Google Scholar]
- 51.Kieser T Practical Streptomyces Genetics. (John Innes Foundation, 2000). [Google Scholar]
- 52.MacNeil DJ et al. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Flett F, Mersinias V & Smith CP High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting Streptomycetes. FEMS Microbiol. Lett 155, 223–229 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson CJ et al. Increasing the efficiency of heterologous promoters in actinomycetes. J. Mol. Microbiol. Biotechnol 4, 417–426 (2002). [PubMed] [Google Scholar]
- 55.Kabsch W XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy AJ et al. Phaser crystallographic software. J Appl Cryst 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Y, Kim C-Y, Mathews II, Cane DE & Khosla C The 2.7-Å crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U.S.A 103, 11124–11129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy A, Kucukural A & Zhang Y I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids used in this study are described in Supplementary Table 1. All oligonucleotides (Integrated DNA Technology) are shown in Supplementary Table 2. The salinosporamide (sal) biosynthetic gene cluster from S. tropica CNB440 is available in the MIBiG database (accession BGC0001041). Other salinosporamide BGCs and the cinnabaramide BGC used for alignments are available in the IMG JGI database. Functional KS and non-elongating KS protein sequences used for alignments can be found in the Supplementary material and through MIBiG. Atomic coordinates and structure factors for the reported crystal structures in this work have been deposited to the Protein Data Bank under accession number 7S2X (native SalC). Additionally, the following PBD datasets were used for SalC structural comparison: 2HG4, 4WKY, 2QO3, 4NA2. Source data for the proteasome inhibition assay (Extended Data Fig. 4) is provided with this paper. Other relevant data supporting the findings of this study are available in this published article or its Supplementary files.