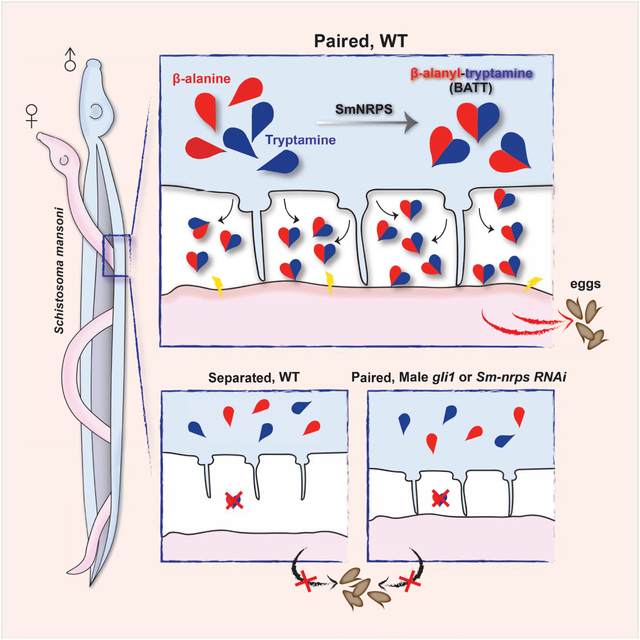
SUMMARY
Schistosomes cause morbidity and death throughout the developing world due to the massive numbers of eggs female worms deposit into the blood of their host. Studies dating back to the 1920s show that female schistosomes rely on constant physical contact with a male worm both to become and remain sexually mature, yet the molecular details governing this process remain elusive. Here, we uncover a non-ribosomal peptide synthetase that is induced in male worms upon pairing with a female and find that it is essential for the ability of male worms to stimulate female development. We demonstrate that this enzyme generates β-alanyl-tryptamine that is released by paired male worms. Furthermore, synthetic β-alanyl-tryptamine can replace male worms to stimulate female sexual development and egg-laying. These data reveal that peptide-based pheromone signaling controls female schistosome sexual maturation, suggesting avenues for therapeutic intervention and uncovering a role for non-ribosomal peptides as metazoan signaling molecules.
Keywords: schistosomes, reproductive biology, non-ribosomal peptide, NRPS
Graphical Abstract
In Brief:
A non-ribosomal peptide produced by male schistosomes when paired with a female is responsible for inducing female sexual development and egg-laying.
INTRODUCTION
Schistosomes are parasitic flatworms that infect 220 million of the world’s poorest people (Steinmann et al, 2006; WHO, 2018). These parasites claim more than 250,000 lives every year and cause chronic, debilitating symptoms in millions more (King, 2010; King & Dangerfield-Cha, 2008; van der Werf et al, 2003). The morbidity associated with schistosome infection is driven almost entirely by the parasite’s massive egg output (Grzych et al, 1991). Indeed, these parasites can produce an egg every one to five minutes over the decades they survive in the blood (Cheever et al, 1994; Harris et al, 1984). Although the parasite’s goal is to pass the eggs to the environment, many become lodged in host organs, resulting in inflammation and fibrosis that drive disease (Pearce & MacDonald, 2002). Since parasites incapable of egg production generate little pathology (Langenberg et al, 2020), strategies that blunt egg production could represent additional therapies.
In contrast to other flatworms that reproduce as hermaphrodites, schistosomes are dioecious, possessing separate male and female sexes (Cort, 1921). This sexual dimorphism was such a distinguishing characteristic that early parasitologists created the genus Schistosoma to refer to the split (schistos) body (soma) of these worms (Weinland & Wyman, 1858). The male worms are large and muscular, with a specialized groove along their ventral surface, the gynecophoral canal, that the male uses to clasp the female to allow sperm transfer. Unlike the male, the female schistosome is long and slender with much of her body mass committed to egg production. Egg production in female worms relies on two large organ systems (Figure 1A, mature): the ovary, which produces oocytes, and the vitellaria, which produce vitellocytes (yolk cells) that provide egg shell constituents and nutrition for the developing embryo.
Figure 1. gli1 is required in male worms for female sexual development.
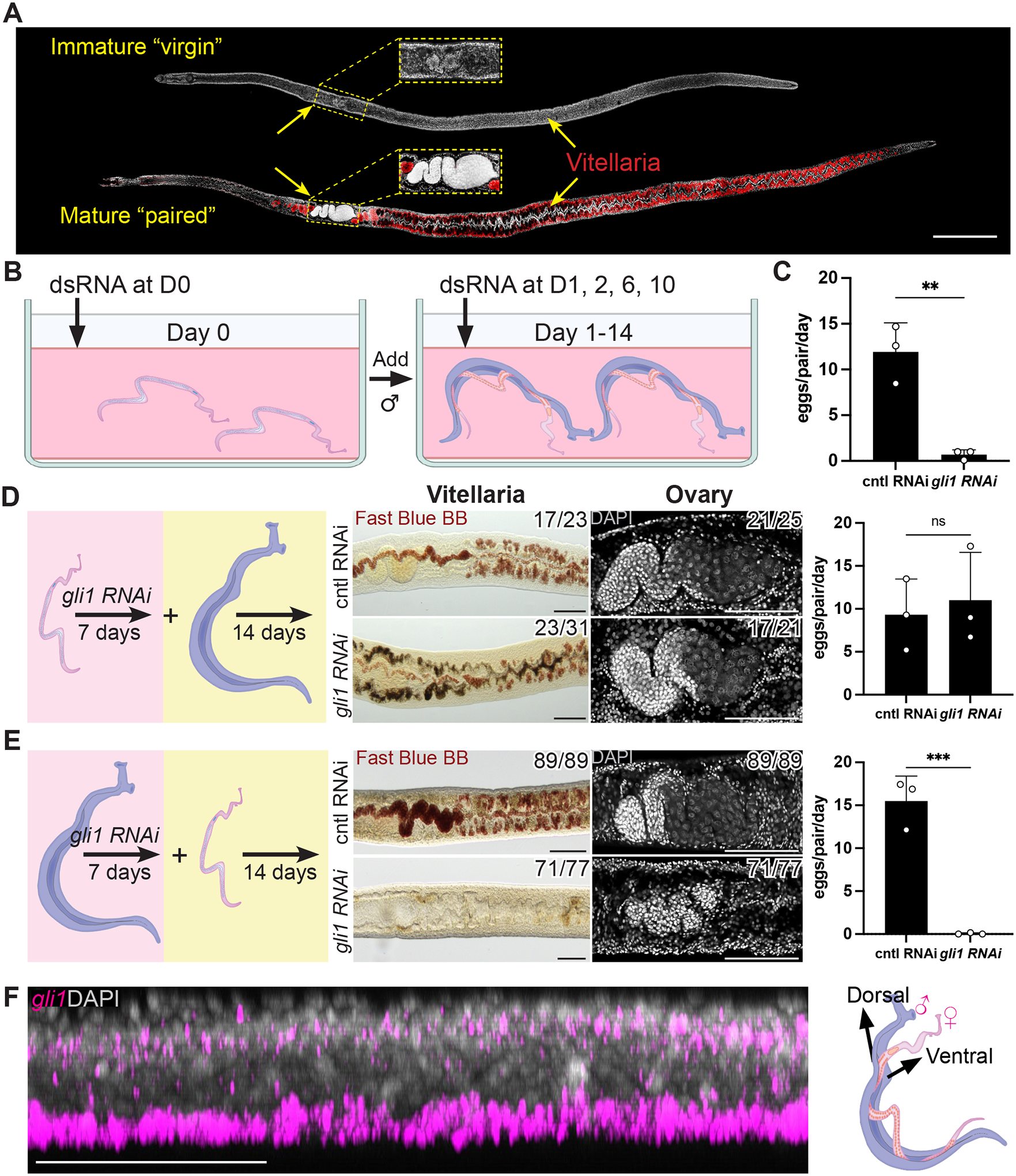
(A) DAPI (grey) and Fast Blue BB (red) labeling of sexually immature adult “virgin” and sexually mature “paired” adult females. Fast Blue BB labels mature vitellocytes. (B) Schematic of the RNAi screening regime. (C) Number of eggs laid per day per female parasite between D12–14 after pairing following RNAi. n > 23 worm pairs for each treatment group, n = 3 biological replicates. (D, E) Evaluation of sexual development (center) or egg production (right) following RNAi of gli1 in (D) female worms or (E) male worms, followed by pairing with non-treated worms of the opposite sex. Fast Blue BB (red) and DAPI (grey) staining showing the vitellaria (left) and ovary (right) from female parasites on day 14 after pairing. n > 21 parasites for each treatment, n = 3 biological replicates. Egg production rates were examined with n > 23 worms for each treatment group. (F) Sagittal confocal section of FISH showing gli1 (magenta) mRNA enriched in the ventral part of a male worm (left) where the male-female interaction occurs (right). n > 10 parasites across 3 experiments. ** p < 0.01; *** p < 0.001; ns p > 0.05. Parametric t test. Error bars represent SD. Each data point represents an individual experiment. Numbers in corner indicate fraction of worms that similar to those presented/total number of worms examined. Scale bars, A, 500 μm; D, E, F, 100 μm.
Presumably as a consequence of hailing from hermaphroditic ancestors, male and female schistosomes have evolved several unique interactions. The most striking of these, first noted nearly a century ago, is that female sexual development is entirely governed by pairing with a male schistosome (Severinghaus, 1928). Indeed, the ovaries and vitellaria of females grown in the absence of males are present as primordia that generate no differentiated oocytes or vitellocytes (Figure 1A, immature “virgin”). As a consequence, unpaired females generate no eggs. Not only do females require males to induce their sexual development, they require perpetual coupling with a male to sustain their mature reproductive status (Popiel et al, 1984). Curiously, amputated segments of male worms that lack reproductive organs can pair with virgin females and induce localized development of the female vitellaria or ovaries (Popiel & Basch, 1984; Wang et al, 2019). Thus, localized physical contact with a male, and not sperm transfer, is critical for stimulating the development of the female sexual organs. Despite progress identifying molecular pathways involved in sexual development (Grevelding et al, 2018; LoVerde et al, 2009), the molecular mechanisms by which male:female pairing induces female maturation remains one of the largest outstanding mysteries in the field.
Here, we report that the expression of the transcription factor, GLI1, is critical for male schistosomes to induce female sexual development. Transcriptional profiling and biochemical studies revealed that GLI1 is essential for the induction of a non-ribosomal peptide synthetase in male worms following pairing and that this enzyme produces a dipeptide (β-alanyl-tryptamine) exclusively in paired male worms. We find paired male worms secrete β-alanyl-tryptamine to the environment and that treatment of virgin females with this dipeptide can induce their development in the absence of a male worm. Together, these studies uncover a mechanism by which male schistosomes control female development.
RESULTS
gli1 is required in male worms for female sexual maturation
In order to uncover genes essential for male-induced female reproductive development, we performed an RNAi screen targeting components of the major signaling pathways conserved among metazoa (Table S1). Given the broad roles for these pathways in controlling metazoan development (Perrimon et al, 2012), we reasoned that targeting one or more of these pathways could uncover molecules essential for male-stimulated female development. For this screen we performed RNAi on virgin female worms, then on male:female worm pairs, and then measured the rate of female sexual development by monitoring egg production at D14 (Figure 1B).
From this screen we uncovered a single gene, gli1, that was essential for female worms to commence egg production (Figure 1C). Since both male and female worms received gli1 dsRNA, this effect could be due to gli1 depletion in male and/or female worms. To distinguish between these possibilities, we performed gli1 RNAi-treatment on either virgin female parasites or male parasites for seven days and then paired these RNAi-treated worms with untreated parasites of the opposite sex. These studies revealed that RNAi of gli1 in female worms had no effect on the development of the vitellaria or egg production (Figure 1D). However, female worms coupled with gli1 RNAi male worms displayed little vitellaria development, scant ovary differentiation, and were incapable of egg production post-pairing (Figure 1E, Figure S1A). Thus, gli1 appears to act in male worms to control one or more processes that are critical for the stimulation of female development and egg production.
Sm-nrps expression is induced in ventrally-located ciliated neurons in a gli1-dependent manner
gli1 is predicted to encode a zinc finger transcription factor related to the GLI family of proteins that are mediators of hedgehog-dependent and -independent signaling pathways (Pietrobono et al, 2019). Given its potential role in transcriptional regulation, we reasoned gli1 may control transcriptional changes in male worms that are necessary for the males to initiate processes key to induce female development. To explore this in more detail we performed whole-mount in situ hybridization (WISH) to examine the distribution of gli1 mRNA in male worms. We observed gli1 expression in cells distributed along the ventral surface of both paired (Figure 1F) and unpaired male worms (Figure S1B–C, Movie S1). By fluorescence in situ hybridization (FISH) we found that these ventrally positioned gli1+ cells expressed markers of neurons (7b2, Figure S1D) and the tegument (skin) (calpain (calp), Figure S1E) in both paired and virgin males. Since the male worm contacts the female along his ventral surface, it appears that GLI1 is expressed in cells in proximity to female parasites upon pairing. Furthermore, it appears that the distribution of gli1 mRNA is not substantially altered in male worms following pairing.
We also examined the expression of gli1 in virgin and mature female worms. We noted gli1 mRNA expressed bilaterally along the edges of the body in both mature and virgin females; by FISH we found these cells expressed the neural marker 7b2 (Figure S1G). Interestingly, we noted by WISH that mature female schistosomes expressed more gli1 throughout their bodies (Figure S1F). By FISH we observed many gli1+ cells in mature female schistosomes expressed the tegumental marker calp (Figure S1H). However, few calp/gli1 double positive cells were observed in virgin females (Figure S1H). Thus, it appears that the distribution of gli1 expression is changed in females during sexual maturation. However, since RNAi of gli1 in females resulted in no obvious phenotypes (Figure 1D), it is challenging to speculate the significance of these observations.
To explore which genes GLI1 may control in male parasites upon pairing, we performed RNAseq (Figure 2A). For these studies, virgin male parasites were treated with gli1 dsRNA and either paired with virgin female worms or co-cultured in wells with female worms but prohibited from pairing by a mesh cell-strainer. Following three days of coculture, RNA from male (Figure 2B) and female worms (Figure S2A) were processed for RNAseq. Similar to previous studies examining gene expression changes in male worms upon pairing (Lu et al, 2016; Wang et al, 2017), we noted only a small group of genes (16) that were induced > 2-fold in male worms post-pairing (p-adj. < 0.0001, Figure 2B, Table S2). Many of these genes were not induced to the same level in the absence of gli1 (Figure 2B). The most notable difference was in the expression of Smp_158480, which was induced > 20-fold in paired vs unpaired control males but only induced about 2-fold when gli1 was depleted (Figure 2B). Interestingly, this gene was down-regulated about 10-fold in the females after pairing, and this down-regulation was also dependent on male gli1 mRNA (Figure S2A, Table S3). These observations parallel previous studies that examined Smp_158480 expression in paired and unpaired worms (Haeberlein et al, 2019; Lu et al, 2019). Based on BLAST, Smp_158480 appeared to encode a non-ribosomal peptide synthetase similar to Ebony from Drosophila (Richardt et al, 2003). From here on, we will refer to Smp_158480 as Schistosoma mansoni non-ribosomal peptide synthetase (Sm-nrps).
Figure 2. RNA-seq revealed Sm-nrps as a gli1-dependent and pairing-dependent gene in male parasites.
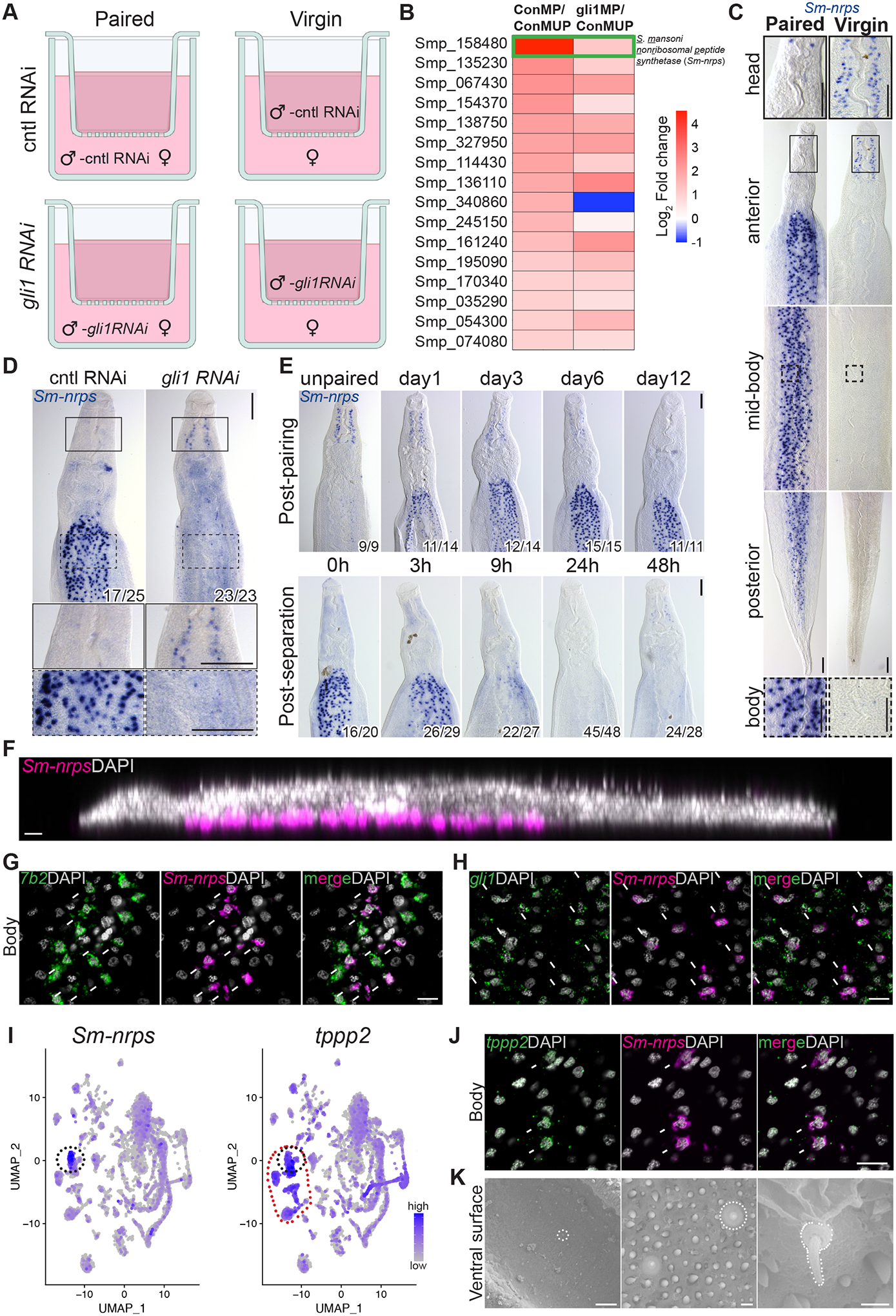
(A) Scheme to identify gli1-dependent changes in gene expression following pairing. Immature females were either paired with or separated from control (cntl) or gli1 RNAi males using a cell strainer. At three days of co-cultivation, worms were prepared for RNAseq. (B) Heatmap showing gli1-dependent transcriptional changes in paired male worms. ConMP, paired cntl male; ConMUP, unpaired cntl male; gli1MP, paired gli1 RNAi male. Green rectangle highlights the S. mansoni nonribosomal peptide synthetase (Sm-nrps) gene (Smp_158480). Genes limited to those with 2-fold change and adjusted p value < 0.0001 in the comparison of ConMP to ConMUP. (C and D) WISH showing the expression of (C) Sm-nrps mRNA in male parasites from mixed or single sex infections and (D) Sm-nrps following RNAi of gli1 in paired male parasites. n > 10 parasites across 3 experiments. Insets show expression in head or body from regions enclosed in solid or dashed rectangles, respectively. (E) WISH showing the expression of Sm-nrps at various time points post-pairing (top) or following separation from female worms (bottom). n = 3 experiments with n > 9 worms. For panels D and E, numbers indicate fraction of worms that similar to those presented/total number examined. (F) Sagittal confocal section of Sm-nrps FISH (magenta). (G and H) FISH showing expression of Sm-nrps with (G) 7b2 or (H) gli1 in the body of paired male worms. (I) Single-cell sequencing UMAP plots from adult worms showing the expression of Sm-nrps (left) and tppp2 (right). The black dotted circle highlights Sm-nrps+ cells, whereas the red dotted circle highlights tppp2+ ciliated neurons. (J) FISH for tppp2 and Sm-nrps in the body of paired male worms. In G, H and J, arrows indicate co-expression. (K) Scanning electron micrographs showing the ventral surface of a male. White dotted lines indicate sensory cilium-like structure. Representative images from 3 parasites. In panels F-H, J nuclei are gray. Scale bars, C, D, E, 100 μm; F, G, H, J, 10 μm; K, left, 20 μm, middle, 1 μm, right, 0.5 μm.
Given the robust gli1-dependent induction of Sm-nrps expression following pairing, we conducted detailed analyses of Sm-nrps expression. As suggested by our RNAseq data, WISH analysis found that Sm-nrps mRNA was abundantly expressed in paired male worms recovered from mixed sex infections, but not in virgin male worms recovered from male-only infections (Figure 2C). Sm-nrps expression in paired male worms was found in distinct cells throughout the body, but with expression limited to a few cells in the head (Figure 2C, left; Figure S2B). Conversely, virgin male parasites expressed little Sm-nrps in their bodies but possessed bilateral “lines” of Sm-nrps+ cells in their head. Interestingly, although paired gli1 RNAi parasites had little expression of Sm-nrps in their bodies, they expressed Sm-nrps in their head in a manner similar to that of virgin male worms (Figure 2C–D). This latter result suggests that gli1 does not simply regulate global Sm-nrps levels and may be required for controlling a “switch” between paired and unpaired transcriptional programs. Consistent with Sm-nrps expression being tightly controlled by pairing status, we observed that Sm-nrps mRNA levels were induced within 24 hours of pairing and were rapidly depleted in male worms within 12 hours of becoming divorced from their female mates (Figure 2E).
We next explored the molecular identity of the Sm-nrps+ pairing-responsive cells in the bodies of male worms. As was observed for gli1, Sm-nrps+ cells were almost entirely found on the ventral side of male worms (Figure 2F). Examination of Sm-nrps expression using a single cell RNAseq atlas of adult schistosomes (Wendt et al, 2020) found that this gene was most highly expressed in a sub-population of neurons (Figure 2I, left). Indeed, FISH analyses found that all Sm-nrps+ body cells (59/59 cells examined) expressed the neural marker 7b2 (Figure 2G). As anticipated by our RNAseq studies, these Sm-nrps+ neurons in the body also expressed gli1 (Figure 2H, 37/40 cells). Conversely, the small number of Sm-nrps+ cells in the head of virgin males did not express gli1+ (Figure S2C) and instead expressed the muscle marker tpm2 (Figure S2D). A closer examination of the Sm-nrps+ neuron cluster on our single cell RNASeq atlas and subsequent FISH studies found that these cells express tppp2 (Figure 2I–J, 34/38 cells examined), a marker of ciliated cell types (Wendt et al, 2020). Since ciliated cells are often associated with sensory functions (Singla & Reiter, 2006), we reasoned these Sm-nrps+ neurons on the male ventral surface could represent sensory cells involved in male:female communication. The morphology of the ventral surface where the male contacts the female worm, known as the gynecophoral canal, has received little attention due to its folded structure prohibiting detailed morphological studies. Using an optimized relaxation and fixation procedure we performed Scanning Electron Microscopy (SEM) studies of the male ventral surface. Importantly, we observed cilium-like protrusions, roughly 1 μm in length and morphologically similar to previously described schistosome sensory cilia (Collins et al, 2011), present throughout the gynecophoral canal in both paired (Figure 2K) and unpaired male worms (Figure S2E). These data suggest that Sm-nrps+ cells along the ventral surface are ciliated neurons potentially exposed to the environment within the gynecophoral canal.
We also analyzed the expression of Sm-nrps in the females by in situ hybridization. Consistent with our RNAseq studies, Sm-nrps mRNA was most highly expressed in virgin females in cells throughout the body and only weakly detected in sexually mature females (Figure S2F). Interestingly, the two lines of Sm-nrps+ head cells observed in virgin males were also present in virgin female worms (Figure S2F). By FISH, we found that Sm-nrps+ cells in both the head and body expressed the muscle marker tpm2 but not the neural marker 7b2 (Figure S2G–H). Unlike in paired males, Sm-nrps was expressed on the dorsal and lateral edges of the virgin female body (Figure S2I).
Sm-nrps is essential in male worms for female sexual maturation.
Motivated by our gene expression studies, we performed RNAi to evaluate the role of male Sm-nrps in controlling female development. For these studies, unpaired male parasites were treated with Sm-nrps or control dsRNA for a week and then paired with virgin females for 2 weeks (Figure 3A, Figure S2J). Similar to what was observed with gli1 RNAi, we found that female worms paired with Sm-nrps RNAi males laid ~90% fewer eggs compared to controls (Figure 3B). Female worms paired with Sm-nrps RNAi males also displayed reduced Fast Blue BB staining in their vitellaria and possessed poorly developed ovaries (Figure 3C). These data support the model that gli1-dependent induction of Sm-nrps levels in male worms is critical for normal female sexual development. Unlike gli1 RNAi males whose post-pairing Sm-nrps expression pattern was similar to virgin parasites (Figure 2D), Sm-nrps RNA was not detectable in heads or bodies of Sm-nrps RNAi males after pairing (Figure 3D), as expected.
Figure 3. Sm-nrps is essential in male worms for female sexual maturation.
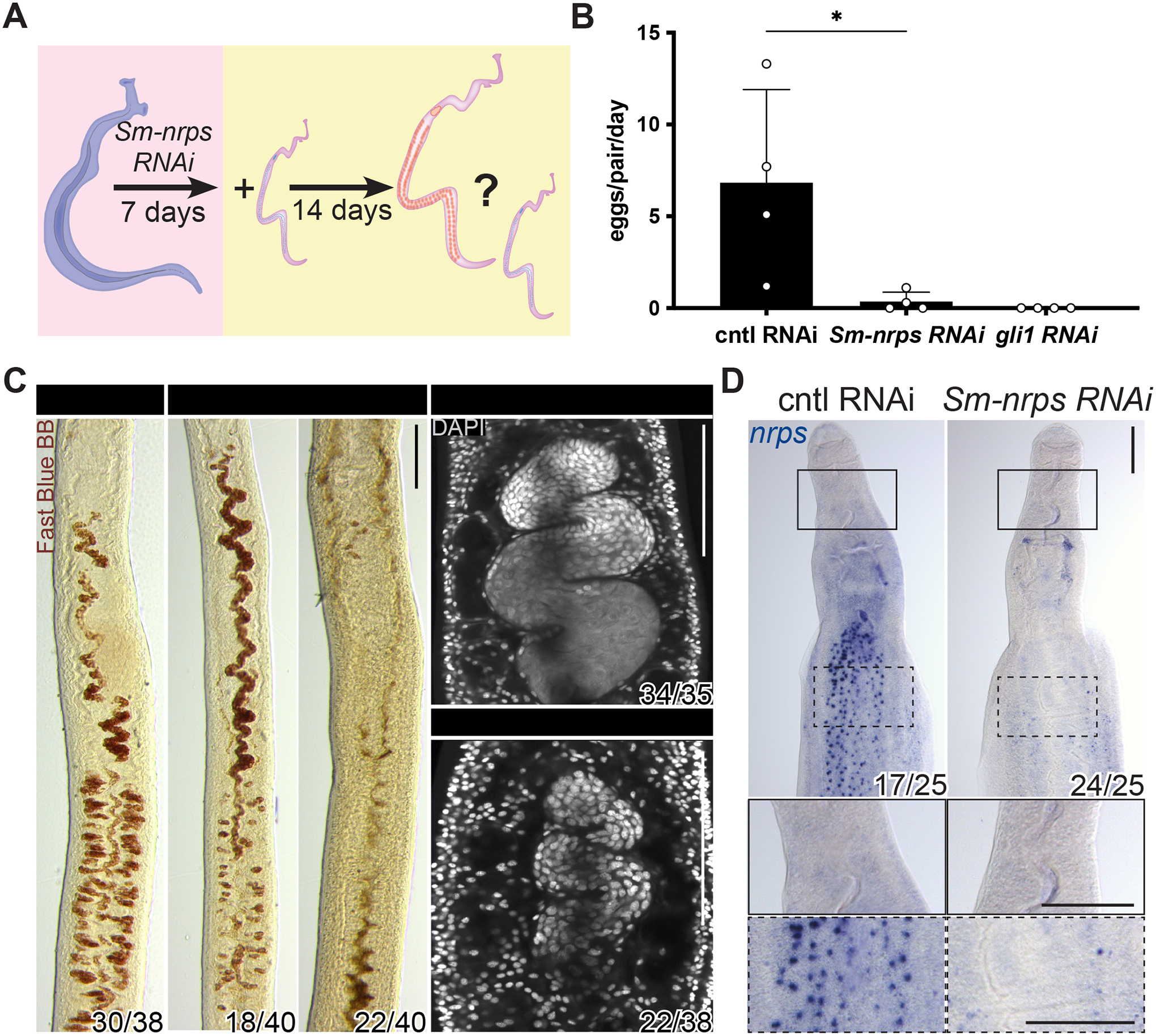
(A) Schematic of experiments to evaluate male Sm-nrps in female sexual maturation. (B) Eggs laid per day per female parasite between days 12–14 post pairing in Sm-nrps or gli1 RNAi male worms. Each data point represents an individual experiment. n > 35 parasites for each treatment group. * p < 0.05. Parametric t test. Error bars represent SD. (C) Fast Blue BB (red) and DAPI (grey) staining showing the vitellaria (left) and ovary (right) from female parasites D14 after pairing with Sm-nrps RNAi male worms. For females paired with Sm-nrps RNAi males, vitellaria development was either reduced (left) or absent (right). n > 3 experiments with n > 35 worms for each group. (D) WISH for Sm-nrps in paired male worms following RNAi. n = 3 experiments with n > 24 worms. Numbers in corner indicate fraction of worms similar to those presented/total number examined. Scale bars, C, 50 μm; D, 100 μm.
SmNRPS is a β-alanyl-bioamine nonribosomal peptide synthetase
Nonribosomal peptide synthetases (NRPSs) produce physiologically and clinically important secondary metabolites, including antibiotics (Süssmuth & Mainz, 2017), from standard and non-standard amino acids. Although these enzymes are widely found in bacteria and fungi, few have been characterized outside of these taxa (Torres & Schmidt, 2019). One exception is the D. melanogaster Ebony protein. Ebony is composed of three domains: an N-terminal amino acid adenylation domain (A domain), a peptidyl carrier protein (PCP) or thiolation domain (T domain), and a C-terminal “amine selective” domain (Izoré et al, 2019; Richardt et al, 2003). By protein alignment with Ebony, we were able to discern both an A and a T domain in the schistosome NRPS protein sequence, suggesting this protein is potentially a true metazoan NRPS enzyme (Figure 4A, Figure S3A).
Figure 4. SmNRPS functions as a β-alanyl-bioamine nonribosomal dipeptide synthetase in vitro.
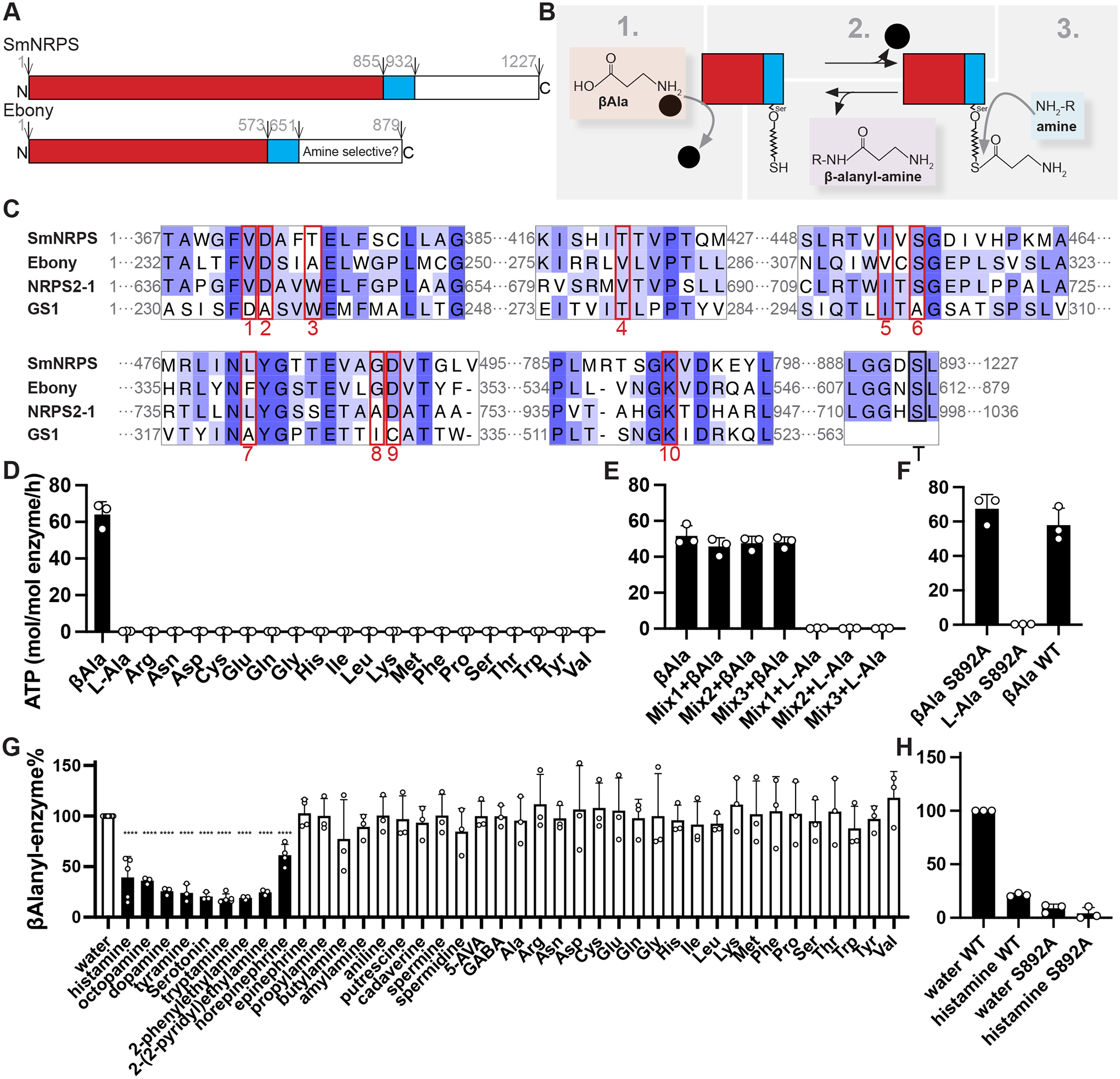
(A) Diagram showing predicted domains of SmNRPS and Ebony. (B) Schematic showing Ebony enzymology. Details see Figure S3B. (C) MUSCLE protein alignment of A and T domain regions from SmNRPS and two related enzymes. 10 amino acids boxed in red represent residues involved in βAla selectivity in Ebony and the first A domain of NRPS2–1 from Streptomyces verticillus (UniProt: Q9FB18) and Phe selectivity in Gramicidin synthetase 1 (GS1) (PDB: 1AMU) from Brevibacillus brevis. The conserved serine thiolation site in the T domain is shown by a black rectangle. (D) ATP-PPi exchange assay showing the activity of SmNRPS with βAla and the standard 20 amino acids. (E) ATP-PPi exchange assay showing the activity of SmNRPS with pools of amino acids +/− βAla. Mix1: A, R, N, C, Q, G, H, I; Mix2, L, K, M, F, P, S, T, V; Mix3, D, E, W, Y. (F) ATP-PPi exchange assay with thiolation-deficient mutant SmNRPS(S892A) with βAla as a substrate. Panels D-F share the same Y axis title. (G) Unloading assay with 3H-βAla evaluating various amines as secondary substrates. Monoamine neurotransmitters and norepinephrine (black bars) drove significant β-alanyl unloading from SmNRPS. ****p<0.0001. Parametric t test. (H) Unloading assay with thiolation-deficient SmNRPS(S892A) mutant compared to wild type enzyme. Panels G and H share the same Y axis title. All values graphed in the unloading assays were normalized to a wild-type water control within the same experiment. Each data point is the average of two technical replicates from an individual experiment. Error bars represent SD.
Biochemical studies have determined that Ebony is able to conjugate β-alanine (βAla) to a variety of biogenic amines, including serotonin, dopamine, and histamine (Hartwig et al, 2014; Richardt et al, 2003). This enzymatic process involves three steps (Figure 4B, Figure S3B): (1) adenylation of βAla catalyzed by the A domain, (2) covalent attachment of βAla to a phosphopantetheinyl group on a conserved serine (ser611) within T domain, and (3) binding of an appropriate amine in the amine selective domain to facilitate nucleophilic attack of the βAla group resulting in a dipeptide product. The specificity of A domain towards the first amino acid substrate can be predicted by the sequence of approximately ten residues within the substrate binding pocket of the A domain (Stachelhaus et al, 1999). Protein alignments of A domains that use βAla (Ebony and the first A domain of NRPS2–1, a bacterial enzyme) and the standard reference A domain from Gramicidin synthetase 1 (GS1), which uses Phe (Conti et al, 1997), found extensive similarities between SmNRPS and its homologs that use βAla (Figure 4C).
To explore the ability of the schistosome enzyme to utilize βAla, we purified full-length schistosome SmNRPS from insect cells (Figure S3C) and measured its amino aciddependent ATP-PPi exchange activity (Hartwig et al, 2014; Hoagland, 1955; Richardt et al, 2003). This assay measures the ability of the enzyme to catalyze the incorporation of 32P from [32P]PPi into ATP in the presence of an amino acid substrate (Figure S3D). Similar to Ebony, we found that SmNRPS was only active in the presence of βAla (Figure 4D–E), suggesting that the SmNRPS A domain is selective for βAla over other amino acids. Although SmNRPS had clear selectivity for βAla, its adenylation rate in the presence of βAla was roughly 8 times slower than A domain of Ebony (Figure S3E). As with Ebony (Richardt et al, 2003), the ability of SmNRPS to convert PPi to ATP was not dependent on the conserved thiolation site (Ser892) (Figure 4C, “T”) in the T domain (Figure 4F).
Next, we tested the specificity for the second amine using an “unloading assay” (Richardt et al, 2003). In this assay (Figure S3F), [3H]β-alanine is reacted with SmNRPS to generate a [3H]β-alanine-NRPS intermediate by the adenylation and covalent conjugation in the T domain. Then various amines were added to the enzyme to determine whether they can displace the tritiated moiety from the enzyme. We observed that all 20 common amino acids, various monoamines/polyamines, and epinephrine possessed little activity in this assay. However, several monoamine neurotransmitters including histamine, serotonin, dopamine, and octopamine were all capable of displacing >50% of [3H]β-alanyl moiety from the loaded enzyme (Figure 4G). Similar to Ebony (Hartwig et al, 2014; Richardt et al, 2003), mutation of the conserved thiolation site (S892) in the T domain to alanine (S892A) reduced [3H]β-alanine conjugation to the enzyme by ~90% (Figure 4H). These data confirm that SmNRPS is a non-ribosomal peptide synthetase and indicate this enzyme possesses β-alanyl-bioamine synthase activity in vitro.
SmNRPS generates β-alanyl-tryptamine that is secreted from paired male schistosomes.
Our biochemical studies suggested that SmNRPS may conjugate βAla to one or more natural monoamines in vivo. To determine the identity of the in vivo SmNRPS product, we treated paired worms with either natural βAla or 13C-labeled βAla (Figure 5A). We then performed liquid chromatography-mass spectrometry (LC-MS) analysis on male extracts, searching for metabolites that possessed a +3-Dalton mass shift in extracts from the [13C3]βAla-labeled worms. These studies identified a metabolite (Figure 5B) corresponding to the molecular weight of β-alanyl-tryptamine (BATT) (Figure 5C) that was shifted +3 Da in extracts from [13C3]βAla labeled worms. LC-MS analysis of an unloading assay mixture with tryptamine confirmed that this ion from schistosome extracts was BATT (Figure S4A). Importantly, we did not find any evidence of [13C3]βAla incorporation into molecules with masses corresponding to other β-alanyl-bioamines (e.g., β-alanyldopamine, -histamine, -serotonin, etc.) that SmNRPS generates in vitro (Figure 4G). The incorporation of [13C3]βAla into BATT was only observed in male parasites (Figure 5D) and this ion was not observed in extracts from paired female worms (Figure S4B). To determine if the production of BATT in male worms was dependent on Sm-nrps expression, we performed RNAi studies along with LC-MS analyses (Figure 5E). We found that RNAi of either gli1 or Sm-nrps blocked the ability of worms to incorporate [13C3]βAla into BATT by >90% (Figure 5F) and abolished total BATT levels (Figure S4C). Taken together, our data suggest BATT to be the physiologically relevant product of SmNRPS in male worms.
Figure 5. SmNRPS produces β-alanyl-tryptamine (BATT) in paired male parasites in a gli1-dependent fashion.
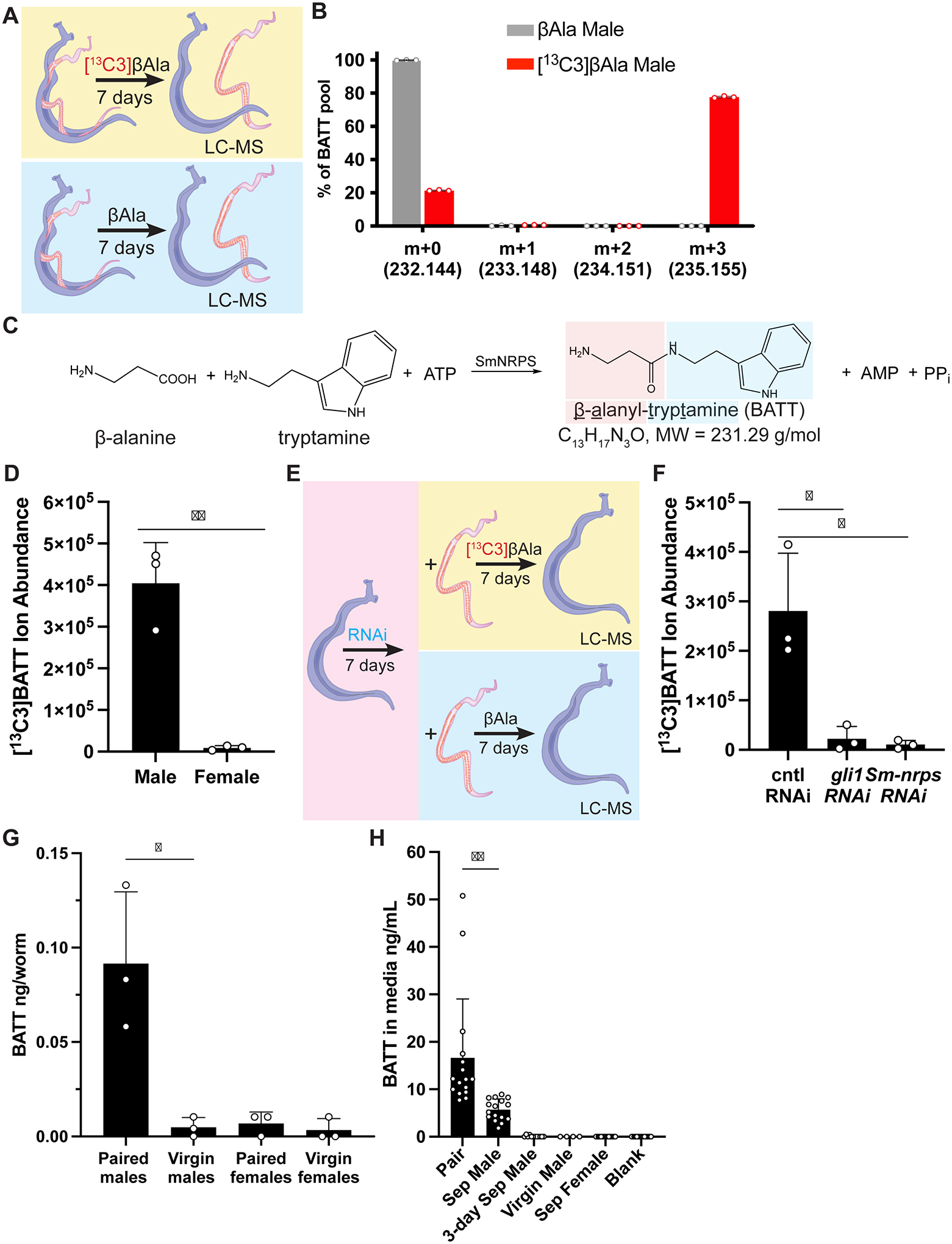
(A) A schematic diagram showing strategy to uncover SmNRPS products utilizing [13C3]βAla. (B) Plot showing relative abundance of ions in paired male parasites treated with standard (gray) or [13C3] (red) βAla. n = 3 biological replicates. (C) Chemical structure and formula of β-alanyl-tryptamine (BATT) and its potential synthesis by SmNRPS. (D) Abundance of [13C3]BATT in paired males and female parasites 7 days after [13C3]βAla treatment. (E) Regiment for evaluating effects of RNAi on [13C3]BATT production. (F) Abundance of [13C3]BATT in gli1 or Sm-nrps RNAi paired male worms following 7D of [13C3]βAla treatment. (G) Measurement of BATT in extracts from virgin or 14-day paired virgin (paired) male/female parasites. For panels C, F, G, each dot is an individual biological replicate derived from extracts from 30 parasites. (H) BATT quantity in media conditioned by parasites for two days. “Sep Male” and “Sep Female” represents media from parasites unpaired at the beginning of the experiment. “3-day Sep Male”, represents media from males separated from females 3D prior to experiment. Each dot represents the BATT concentration in media conditioned from an individual worm or worm pair. ** p < 0.01, * p < 0.05. Parametric t test. Error bars are SD.
Using chemically synthesized BATT as a standard, we next quantified naturally occurring BATT levels in paired and unpaired male worms. As suggested by the expression of Sm-nrps (Figure 2C), we only detected high levels of BATT in samples from paired male worms (Figure 5G). In principle, BATT could act either as an endogenous factor in males required for them to promote female development or BATT could be released from male worms to act directly on females. To explore these possibilities, we cultured paired male worms or isolated male or female worms and quantified BATT levels in media after 48 hours of culture. BATT was not detected in media conditioned by virgin male, virgin female, or male worms that had been separated from females three days prior to the experiment (Figure 5H). In contrast, media conditioned by males continuously paired with female worms contained an average of 16.6 ng/ml (71 nM) BATT (Figure 5H). Interestingly, male worms separated from females on day 0 of the experiment released ~70% less BATT into the media than males that remained continuously paired (Figure 5H), indicating that, like Sm-nrps expression (Figure 2E), BATT production drops rapidly in the absence of female worms. In summary, our data suggest that SmNRPS induction upon pairing results in the production of BATT and this molecule is released from the male worm into the environment.
BATT is sufficient to induce egg laying in virgin female parasites.
The secretion of BATT from male worms suggested that BATT may act to directly stimulate female development. To examine this possibility, we first evaluated the effects of low concentration BATT on female gene expression after short-term treatment. Our previous RNAseq studies (Figure S2, Table S3) identified genes up- and down-regulated in female worms post-pairing whose regulation was dependent on gli1 function in male worms. We selected two such genes for analysis: a rotund (Smp_169260) homolog that was induced in females only when paired with gli1+ males and Sm-nrps that was down-regulated in females post-pairing with gli1+ males (Figure 6A). Treatment of separated females for 24 h with 0.5 μM synthetic BATT induced both a significant decrease in Sm-nrps mRNA and an increase in rotund mRNA (Figure 6B) when compared to controls treated with βAla alone. The magnitude of the transcriptional changes of Sm-nrps and rotund following BATT treatment were comparable in magnitude to those resulting from pairing with male worms (Figure 6A–B).
Figure 6. Synthetic β-alanyl-tryptamine (BATT) is sufficient to induce egg-laying in virgin female parasites.
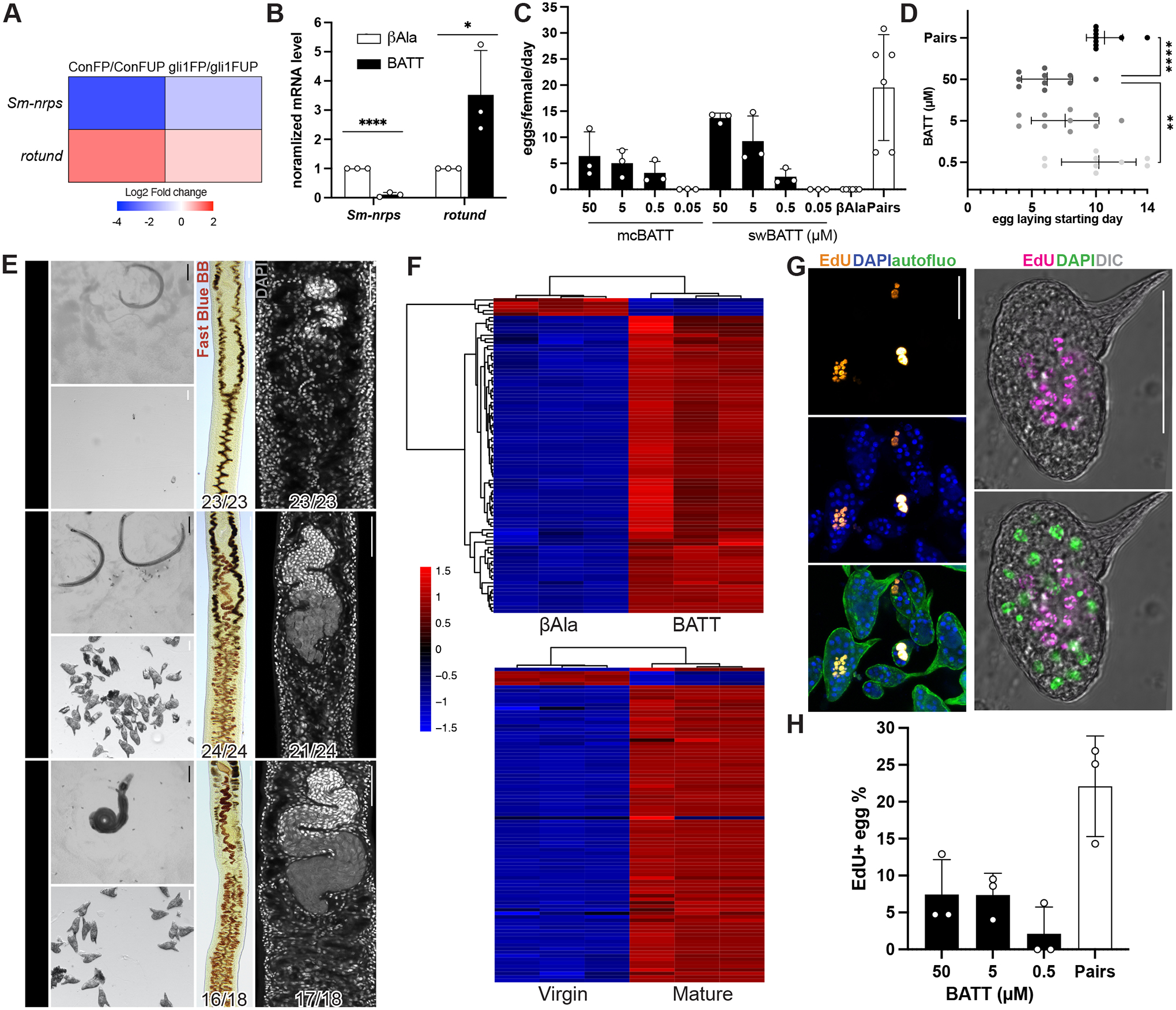
(A) Heatmap expression of Sm-nrps and rotund mRNA levels in female parasites following pairing with control or gli1 RNAi parasites described in Figure 2A. (B) qRT-PCR showing relative expression of Sm-nrps and rotund following treatment of unpaired female worms with 0.5 μM BATT for 1 day. **** p < 0.0001, * p < 0.05. t test. n = 3 biological replicates. (C) Eggs laid per day per female parasite between days 12–14 following treatment with two sources of synthetic BATT (mcBATT or swBATT) or βAla treatment or following pairing with male worms. n > 45 females for each treatment group; n > 10 for the paired group. (D) Time course of egg production following treatment with three increasing concentrations of BATT (Y axis) or following pairing. **** p < 0.0001, ** p < 0.01. Parametric t test. (E) Images of worms (top left), eggs (bottom left), Fast Blue BB (middle) and DAPI (right) staining 14 days after exposure of virgin females with to βAla, 50 μM BATT, or to pairing. Representative images from 3 experiments with n > 23 worms for chemical treatment group and n > 10 for paired group. Numbers in corner indicate fraction of worms similar to those presented/total number of worms examined. (F) Top, clustered heatmap of 89 differentially expressed genes following 8D of 5 μM BATT treatment. Bottom, heatmap showing expression of same 89 genes in virgin vs sexually mature female S. mansoni. (G, left) EdU-labeling (yellow) of proliferative embryonic cells within the eggs laid by virgin females treated with 50 μM BATT. Autofluorescence (autofluo, green) and DAPI (blue) shows the eggshell and nuclei, respectively. (G, right) Confocal image of EdU-labeled (magenta) egg laid by a virgin female treated with 50 μM BATT. DAPI, green. DIC, Differential Interference Contrast. (H) Percentages of EdU+ eggs laid by virgin female parasites between day 12–14 after BATT treatment or pairing. n > 250 eggs. In plots, each dot represents an individual experiment and error bars represent SD. Black scale bars, 500 μm; white scale bars, 50 μm.
Encouraged by the gene expression changes induced by BATT, we evaluated its effects on virgin female development. Strikingly, virgin female schistosomes commenced egg production with ≥ 0.5 μM BATT as early as 4 days after initial treatment (Figure 6C–D). No egg production was observed with βAla treatment or 50nM BATT (Figure 6C). This latter result is consistent with the observation that conditioned media that contained ~70 nM BATT also failed to induce sexual development (Figure S4D). Examination of virgin females treated with ≥ 0.5 μM BATT found they produced mature Fast-Blue BB+ vitellocytes and possessed ovaries that contained mature oocytes (Figure 6E, Figure S5A–B). Indeed, examination of the expression of vitellocyte-specific marker p48 and the oocyte-specific marker bmpg confirmed that BATT treatment induces maturation of the female reproductive machinery (Figure S5C). We observed similar female stimulatory activity using two batches of synthetic BATT (mcBATT and swBATT), confirming the specificity of this effect (Figure 6C, S5A–B). The rate of egg production following BATT treatment was lower than that induced by male:female pairing (Figure 6C), and only about 16% of BATT treated females contained bmpg+ ovaries despite visible ovary differentiation (Figure S5C). These differences may indicate either that some feature of pairing is required for the complete egg laying phenomenon in addition to BATT or, alternatively, that simple addition of BATT to the medium is not as effective as localized delivery.
To further evaluate the effects of BATT on female development, we performed RNAseq analyses comparing virgin females at D8 following treatment with 5 μm BATT or βAla (Table S4). We then selected the 89 most differentially regulated genes (log2 fold change +/− 2, adjusted p < 0.05) following BATT treatment and evaluated how these genes were regulated in virgin vs sexually mature worms using a previously collected dataset (see Key resources table). Remarkably, of the 87 of 89 genes that were up- or down-regulated following BATT treatment were similarly regulated in sexually mature vs virgin worms (Figure 6F, Figure S6A). We next queried where these 89 BATT-regulated genes were expressed on our single cell RNAseq atlas (Wendt et al, 2020). Consistent with BATT stimulating development of the sexual organs, roughly 60% of BATT-regulated genes were most highly expressed in cell clusters associated with late stages of development of the ovary and vitellaria (“Late Female Germ Cells” and “Differentiated Vitellaria”, Figure S6B). Taken together, these data highlight that BATT induces molecular changes associated with female sexual development.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| For WISH: Anti-Digoxigenin-AP | Sigma-Aldrich | Cat# 11093274910 RRID:AB_2734716 |
| For FISH: Anti-Digoxigenin-horseradish peroxidase | Roche | Cat# 11207733910 RRID:AB_514500 |
| For FISH: Anti-Fluorescein-horseradish peroxidase | Sigma-Aldrich | Cat# 11426346910 |
| For FISH: Anti-DNP-horseradish peroxidase | Vector Laboratories | Cat# MB-0603-5 |
| Bacterial and virus strains | ||
| MAX Efficiency™ DH10Bac™ Competent Cells for bacto-bac cloning | Thermo Fisher Scientific | Cat# 10361012 |
| Biological samples | ||
| N/A | ||
| Chemicals, peptides, and recombinant proteins | ||
| ABC169 media supplement: bovine washed red blood cells | Lampire Biological Products | Cat# 7240808 |
| ABC169 media supplement: porcine cholesterol concentrate | Rocky Mountain Biologicals | Cat# PCC-BDG-VCG |
| ABC169 media supplement: L-ascorbic acid | Sigma-Aldrich | Cat# A5960 |
| tricaine | Sigma-Aldrich | Cat# A5040 |
| Fast Blue BB | Sigma-Aldrich | Cat# F3378 |
| EdU (5-ethynyl-2’-deoxyuridine) | Invitrogen | Cat# E10187 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| FAST SYBR Green Master Mix | Bio-rad | Cat# 1725122 |
| cOmplete protease inhibitor | Sigma-Aldrich | Cat# 11873580001 |
| Ni-NTA agarose | Qiagen | Cat# 4561 |
| Strep-Tactin Sepharose resin | Fisher Scientific | Cat# NC9891255 |
| [32P]PPi | PerkinElmer | Cat# NEX019001MC |
| [3H]βAlanine | American Radiolabeled Chemicals | Cat# ART 0205 |
| [13C3]βAlanine | Cambridge Isotope Laboratories | Cat# clm-8755-PK |
| βAlanine | Sigma-Aldrich | Cat# 146064 |
| mcBATT | Synthesized by MuseChem, owned by this paper | Batch# M21X01198 |
| swBATT | This paper | Order# MCC-RC-106 |
| Critical commercial assays | ||
| Direct-zol RNA MiniPrep | Genesee Scientific | Cat# 11-330 |
| Illumina TruSeq stranded mRNA library kit | Illumina | Cat# 20020595 |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat# 1708891 |
| Deposited data | ||
| gli1 RNAi RNAseq | GEO | GSE184849 |
| BATT treatment female RNAseq | GEO | GSE191062 |
| Single sex female RNAseq 1 | WormBase ParaSite | ERR1328130_1, ERR1328130_2 |
| Single sex female RNAseq 2 | WormBase ParaSite | ERR1328191_1, ERR1328191_2 |
| Single sex female RNAseq 3 | WormBase ParaSite | ERR1328215_1, ERR1328215_2 |
| Mixed sex female RNAseq 1 | WormBase ParaSite | ERR1328194_1, ERR1328194_2 |
| Mixed sex female RNAseq 2 | WormBase ParaSite | ERR1328218_1, ERR1328218_2 |
| Mixed sex female RNAseq 3 | WormBase ParaSite | ERR1328156_1, ERR1328156_2 |
| Experimental models: Cell lines | ||
| Sf9 | ATCC | CRL-1711 |
| Experimental models: Organisms/strains | ||
| Parasites: Schistosoma mansoni | NIAID Schistosomiasis Resource Center of the Biomedical Research Institute | NMRI |
| Infected snails: Biomphalaria glabrata | NIAID Schistosomiasis Resource Center of the Biomedical Research Institute | NMRI |
| Infected female mice | NIAID Schistosomiasis Resource Center of the Biomedical Research Institute | Swiss-Webster |
| Oligonucleotides | ||
| Forward and reverse primers for riboprobe and dsRNA synthesis | This paper, Table S5 | N/A |
| Forward and reverse primers for qPCR detection | This paper, Table S5 | N/A |
| Recombinant DNA | ||
| pJC53.2-gli1 for RNAi and in situ hybridization | This paper | Plasmid# pJW186, Smp_266960 |
| pJC53.2-Sm-nrps for RNAi and in situ hybridization | This paper | Plasmid# pRC90, Smp_158480 |
| pJC53.2-7b2 for in situ hybridization | This paper | Plasmid# pLZ180, Smp_073270 |
| pJC53.2-tppp2 for in situ hybridization | This paper | Plasmid# pLZ266, Smp_097490 |
| pJC53.2-calp for in situ hybridization | This paper | Plasmid# pGW17, Smp_214190 |
| pJC53.2-tpm2 for in situ hybridization | This paper | Plasmid# pLZ93, Smp_031770 |
| pJC53.2-p48 for in situ hybridization | This paper | Plasmid# pLZ31, Smp_241610 |
| pJC53.2-bmpg for in situ hybridization | This paper | Plasmid# pLZ262, Smp_078720 |
| Protein expression construct: pFastBac1-His6-StrepII-TEV-SmNRPS, codon optimized to insect cells | This paper | Plasmid# pRC135_OP |
| Protein expression construct: pFastBac1-His6-StrepII-TEV-SmNRPS_S892A, codon optimized to insect cells | This paper | Plasmid# pRC135.S892A_OP |
| Protein expression construct: pFastBac1-His6-StrepII-TEV-Ebony | This paper | Plasmid# pRC136, protein NCBI: NM_079707.4 |
| Software and algorithms | ||
| MassHunter Profinder software | Agilent Technologies | Version B.08.00 SP3 |
| T-Coffee sequence alignment | Snapgene 4.3.4 | http://tcoffee.crg.cat/ |
| MUSCLE sequence alignment | Snapgene 4.3.4 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| STAR | Dobin et al., 2013 [1] | N/A |
| DESseq2 | Love et al., 2014 [2] | N/A |
| Other | ||
| 40 μm cell strainer for female/male worm separation | Thermo Fisher Scientific | Cat# 22-363-547 |
| speed vacuum | Thermo Fisher Scientific | Cat# SPD1010-115 |
| 6550 iFunnel Q-TOF mass spectrometer | Agilent Technologies | Cat# G6550BA |
| Acquity UPLC® HSS T3 column | Waters | Cat# 186003539 |
| AB Sciex 6500+ QTRAP® mass spectrometer | Framingham | N/A |
| Nexera X2 UHPLC/HPLC | Shimadzu and Columbia | N/A |
Dobin, A., et al., STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 2013. 29(1): p. 15–21.
Love, M.I., W. Huber, and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550.
BATT treatment elicited egg production in virgin female worms several days faster than pairing with virgin male worms (Figure 6D), suggesting that it may take several days before virgin male worms can produce sufficient quantities of BATT to stimulate female development. Previous studies have shown that in vitro pairing with castrated male worm segments can induce virgin females to lay eggs that contain parthenogenic embryos capable of limited development (Wang et al, 2019). Examination of the eggs produced by BATT-treated virgin females found that these eggs (Figure 6G) possessed a lateral spine characteristic of S. mansoni eggs (Figure S5D) and that small fraction of these eggs (~5%) contained clusters of proliferative EdU+ embryonic cells (Figure 6H). Together, these data support the model that BATT can replace a male partner to trigger egg production.
DISCUSSION
The data presented here indicate that male:female pairing induces the GLI1-dependent expression of Sm-nrps in male worms, resulting in the production of BATT. BATT is then released from the male to act as a pheromone that stimulates female sexual development (Figure 7). This implies that males first “sense” the presence of the female to commence BATT production and that females possess the necessary signaling machinery to interpret the presence of BATT.
Figure 7. Model for BATT-mediated regulation of female development from male pheromone secretion.
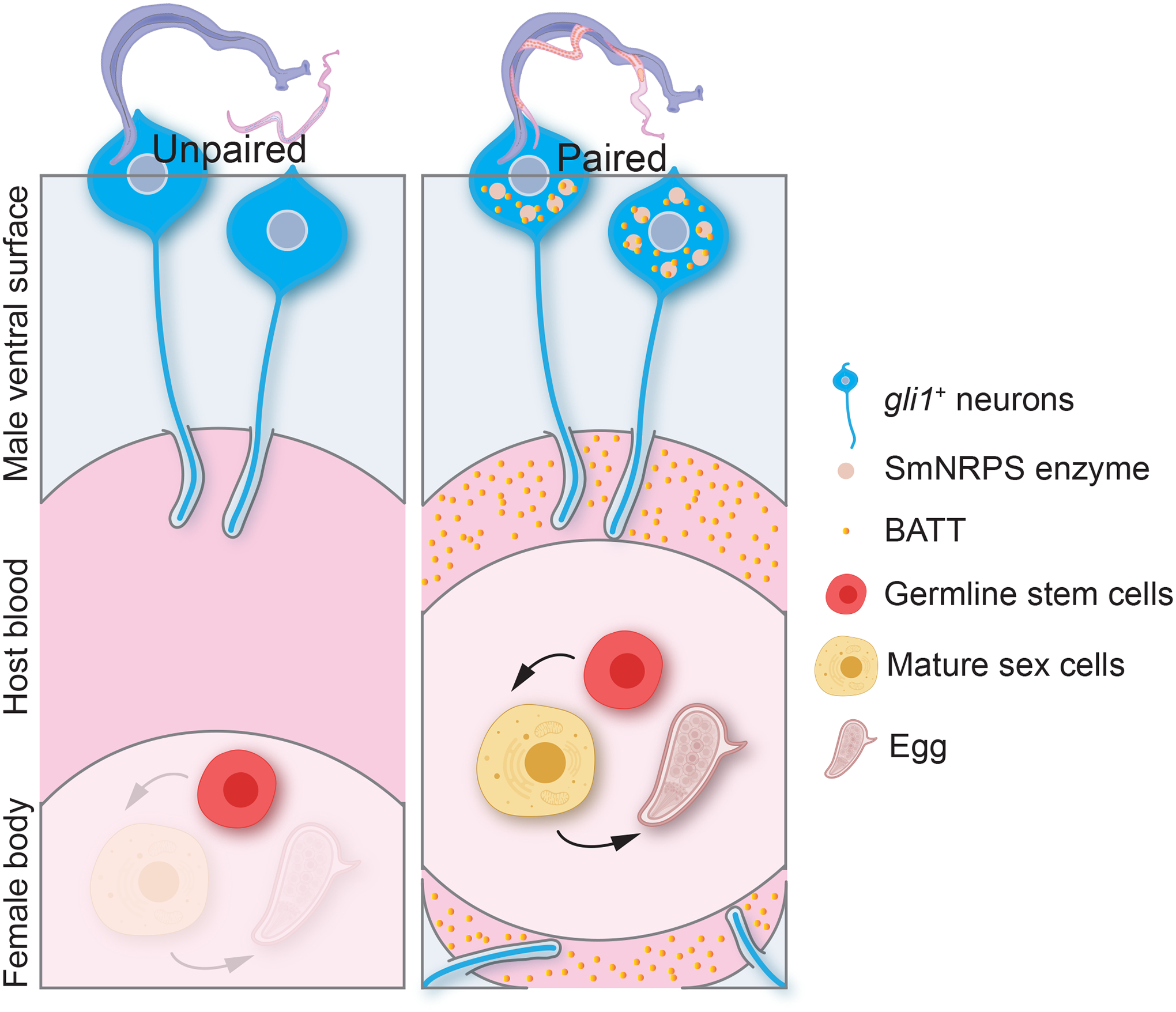
In the absence of the female, gli1+ ciliated neurons protrude into the gynecophoral canal express little SmNRPS and BATT is not produced. When females are present, males sense the presence of the female, perhaps via gli1+ ciliated neurons, inducing the expression of SmNRPS and BATT synthesis. Synthesized BATT is then released and concentrations sufficient to induce female sexual development accumulate in the gynecophoral canal.
Pairing with amputated male worm segments induces development of female reproductive organs only in regions in direct contract with the male worm (Popiel & Basch, 1984; Wang et al, 2019). So, if BATT is diffusible into the medium (Figure 5H), why does it not stimulate the development of regions not in contact with male worms? We find that single paired male worms can condition media with BATT to a concentration of ~70 nM over 48 hours and that this conditioned media cannot induce virgin female sexually maturity (Figure S4D). Similar concentrations of synthetic BATT are also not sufficient to stimulate virgin female development (Figure 6C, Figure S5A–B). Since the volume of the gynecophoral canal, where males probably release BATT, is much smaller than the total culture volumes for these experiments, the local BATT concentrations female experience while paired are probably in the micromolar range in which BATT induces development.
Ciliated neurons play numerous roles in sensory perception in animals (Singla & Reiter, 2006). Thus, the expression of Sm-nrps in ciliated neurons on the male’s ventral surface suggests the possibility that these cells are critical for detecting the presence of the female and in turn stimulating BATT production. Interestingly, studies in C. elegans have demonstrated that ciliated sensory neurons can release material into the environment enclosed in exosome-like vesicles (Wang et al, 2014b). This observation, along with the fact adult schistosomes produce exosome-like vesicles (Sotillo et al, 2016) and that vesicles have been observed in adult schistosome sensory cilia (Hockley, 1973; Morris & Threadgold, 1967), suggests that Sm-nrps+ neurons could be responsible not only for the perception of the female but the release of BATT into the environment.
NRPS enzymes are widely found in bacteria and fungi where they produce secondary metabolites that can act as toxins, pigments, siderophores, and antibiotics (Süssmuth & Mainz, 2017). However, relatively few NRPS enzymes have been characterized in animals (Shou et al, 2016; Torres & Schmidt, 2019; Wang et al, 2014a), and thus their potential roles in metazoan biology remains largely unexplored. Perhaps the best characterized metazoan NRPS enzyme is Ebony, which is critical for Drosophila cuticle pigmentation (Wright, 1987) and neurotransmission (Borycz et al, 2002). Unlike NRPS enzymes in bacteria and fungi, Ebony is not thought to produce any biologically active molecules, but rather inactivate them. For instance, histamine is a key neurotransmitter in the visual system and is deactivated and recycled by its conversion to β-alanyl-histamine (carcinine) by Ebony in glial cells (Borycz et al, 2012; Stenesen et al, 2015; Ziegler et al, 2013). In contrast to flies, our data suggest that the SmNRPS product BATT directly stimulates signaling in the female, culminating in female development. Since BATT has not previously been recognized as a signaling molecule, the nature of the signaling pathway(s) that mediates its effect, and whether they are related to pathways characterized in other metazoa, remains unclear.
Interestingly, genes for SmNRPS-like proteins with A domains potentially selective for β-alanine are found not only in insects and in all schistosome species that infect humans, but also in the genomes of free-living and parasitic flatworms, rotifers, annelids, cnidarians, and at least one hemichordate (Figure S7, Table S5). Thus, BATT-like molecules may participate in cellular signaling in diverse invertebrate organisms to regulate diverse processes. Because schistosomes are the only major group of dioecious flatworms, the presence of SmNRPS in hermaphroditic flatworms is intriguing. Should SmNRPS enzymes in hermaphroditic flatworms produce BATT-like molecules necessary for development of the female reproductive system, it would suggest that, during the transition from hermaphroditism to dioecy, SmNRPS product formation was “outsourced” to the male where it could act as a pheromone, rather than an endogenous hormone, to dictate the timing of female development. Clearly exploring SmNRPS function, not just in flatworms, but a variety of metazoa may uncover more paradigms for cell-to-cell, or even animal-to-animal, communication.
BATT is capable of inducing egg production in virgin females days before females paired with virgin males (Figure 6D) and inducing transcriptional changes associated with sexual development (Figure 6F). Yet the rate of egg production, the quality of eggs and the sexual development induced by BATT (especially with the ovary) were inferior to those induced by intact male worms (Figure 6C, Figure S5A–B). Thus, BATT may act in parallel with other factors to support robust female development. One possible explanation is that BATT is a key signal to initiate development, yet the male worms provide additional factors key for optimal development. Indeed, radiolabeling studies have suggested that male worms transfer molecules such as glucose (Conford & Huot, 1981) and cholesterol (Haseeb et al, 1985; Popiel & Basch, 1986) to female worms. Also, since immune signals are important for optimal female development (Davies et al, 2001), there may be host factors that potentiate the response to BATT. Alternatively, females may perceive pairing with a male and alter their behavior or metabolism in a BATT-independent manner. Yet another possibility is that BATT is packaged (for instance, in vesicles or with a co-factor) and delivered from males in a manner optimal for female BATT recognition or absorption. Clearly, more detailed assessments of the similarities between paired females and those treated with BATT will provide additional insights into the mechanisms that ensure robust female development.
Nearly a century ago, Aura Severinghaus detailed the role of the male schistosome in female development and offered that “it is conceivable that hormones produced by the male may in some way be connected to the phenomenon “(Severinghaus, 1928). Our studies identify BATT as a male-derived pheromone that stimulates sexual development in female schistosomes. These observations provide key insights into one of the longest-standing mysteries in schistosome biology and suggest that therapeutically blocking BATT signaling could present an opportunity to blunt both schistosome transmission and egg-induced pathology. SmNRPS is a clear target, but the identification of BATT also permits study of its receptor and downstream signaling in females. Studies of this pathway both in schistosomes and other metazoa may elucidate additional cellular signaling mechanisms as well as targets for therapeutic intervention.
LIMITATIONS OF THE STUDY
BATT is a pheromone released from males that induces female S. mansoni sexual development. Yet, important questions remain. First, it is unclear how BATT is released from male worms and how and where it acts on female worms. Our data suggest that SmNRPS is expressed in ciliated neurons present on the ventral side of male, and that sensory cilia are present on the male ventral surface, but technical limitations have prohibited our ability to definitively show these ventral sensory cells are the source of BATT for release. Second, our work has only focused on a single species of schistosome, so it is not clear how generalizable our results are to other species. However, studies in S. japonicum have shown that NRPS is induced in males upon coupling with a female (Lu et al, 2019; Wang et al, 2017). Also, studies have shown that males of one schistosome species can induce the sexual development of females of another (Armstrong, 1965). Taken together, it is reasonable to assume that the effects of BATT are generalizable. However, given the promiscuity of SmNRPS for various monoamines (Figure 4G), it remains possible that some schistosome species, of which there are hundreds that infect mammals and birds, generate species-specific BATT-like molecules. This could be a strategy in regions endemic to multiple species to ensure sexual reproduction only occurs with mates capable of producing viable offspring.
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James J. Collins III (jamesj.collins@utsouthwestern.edu).
Materials availability
This study did not generate new or unique reagents. Synthetic BATT can be shared upon request.
Data and code availability
Two of the RNA sequencing data sets collected by this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The existing, publicly available data for single sex and mixed sex female RNAseq were obtained from the accession numbers listed in the key resources table. Microcopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
EXPERIMENTAL MODEL AND SUBJECT DETAILS
S. mansoni parasites
S. mansoni is the Naval Medical Research Institute (NMRI) strain provided by the NIAID Schistosomiasis Resource Center at the Biomedical Research Institute (BRI) (Rockville, MD). Infected Biomphalaria glabrata snails (NMRI) and infected mice (Swiss-Webster female mice) were received on a regular basis for parasite recovery and in-house life cycle maintenance. Adult S. mansoni were perfused from the hepatic portal vein of mice 6–7 weeks after infection. 37 °C DMEM (Fisher Scientific MT10014CV, NH, USA) with 5% horse serum (Sigma-Aldrich H1138, MO, USA) and heparin was used as perfusion solution. Recovered parasites were washed several times with perfusion solution before culture in BM169 media (Basch, 1981) supplemented with 1 × Antibiotic-Antimycotic (Sigma-Aldrich A5955, MO, USA). Worm pairs were harvested from mice infected with pooled cercaria shed by several B. glabrata snails each parasitized with more than 5 miracidia, ensuring a mixed pool of the two sexes. Parasites derived from such infections are referred to as “Mixed Sex”. “Virgin” parasites were harvested from mice infected with cercaria shed by a single snail parasitized by a single miracidium. Adult males which were unpaired for more than 3 days or virgins from single miracidia infections were considered “unpaired”. For regular maintenance, parasites were cultivated in cell culture petri dishes/plates with ~0.5 mL media per worm (pair) at 37°C in 5% CO2. Due to the slow metabolism rate of virgin females, ~0.15 mL media/worm was used. Media was changed every other day unless mentioned otherwise. For experiments monitoring reproduction and sexual development, parasites were cultured in ABC169 media (Wang et al, 2019). dsRNA treatment was performed in BM169 unless described otherwise. To separate worm pairs, worms were suspended in BM169 with 0.25% tricaine (Sigma-Aldrich A5040, MO, USA) and agitated for 5 min. Unpaired worms were processed further or washed 2X in BM169 and returned to culture.
S. mansoni infected mice
6–8 week old female Swiss-Webster mice (Mus musculus) of were infected with cercaria by tail exposure and perfused 6–7 weeks later. Mouse infections were performed either in-house or by BRI. Experiments with and care of mice were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of UT Southwestern Medical Center (approval APN: 2017–102092).
B. glabrata snails
B. glabrata snails of NMRI strain were maintained in artificial pond water (0.125 mg/L FeCl3•6H2O, 32 mg/L CaCl2•2H2O, 25 mg/mL MgSO4•7H2O in pH 7.2 phosphate buffer) and fed with 16% layer chicken feed. For snail infections, livers harvested from “Mixed sex” infected mice were blended and resuspended artificial pond water to liberate miracidia from eggs. Released miracidia were incubated with snails for either “Mixed sex” or “Virgin” infections for 2h and then cultured in artificial pond water.
METHOD DETAILS
RNA interference (RNAi)
dsRNA production and RNAi treatment were essentially performed as previously described (Collins et al, 2010; Collins et al, 2013). Oligo sequences used to generate dsRNA templates are listed in Table S6. A dsRNA derived from two bacterial genes was used as a negative control for all RNAi experiments (Collins et al, 2010). D0 represents the first day of the experiment.
For RNAi screening, virgin females were treated with 30 μg/mL dsRNA on D0 in BM169 and then paired with male parasites from D1 onwards in ABC169. Media was supplemented with an additional 30 μg/mL dsRNA on D1/2/6/10 until D14 where phenotypes were monitored. For RNAi treatment of either virgin male or female worms, the unpaired worms were treated with 30 μg/mL dsRNA in BM169 media for one week. After a week, opposite sex worms were added in fresh BM169. The following day, BM169 was replaced by ABC169 for the duration of the experiment. For RNAi experiments in Figure 1D–E, Figure 2A–B (RNAseq), Figure 3D, Figure 5F and Figure S4C, negative control, gli1 or Sm-nrps dsRNA was added on D0/1/2/3 with fresh media and followed by media change every other day until pairing; for experiments in Figure 3B–C, dsRNAs were added every day with media change before pairing. In cases where egg output was quantified, existing eggs were removed from the culture during the last media change (D12 post pairing). Two days later, egg number and the number of female parasites was counted to calculate egg/female/day.
Parasite/egg staining and imaging
Whole-mount in situ hybridization (WISH), fluorescent in situ hybridization (FISH) (Collins et al, 2013), Fast Blue BB, DAPI, and EdU labeling (Wang et al, 2019) were performed as previously described. Riboprobes were synthesized from templates generated using primers listed in Table S6. Non-quantitative phenotypes were evaluated by reporting the fraction of worms with a given phenotype/total number of worms examined.
Gene expression analyses
For RNAseq analysis of gli1 RNAi-treated worms, dsRNA treatment was performed on male worms for a week at 30 μg/mL, and these male worms were either co-cultured with virgin female parasites separated by 40 μm cell strainers (Fisher Scientific 22–363-547, NH, USA) or allowed to pair for three days in ABC169. Each of the three biological replicates included worms pooled from three technical replicates. Following three days of pairing, worms were separated with 0.25% tricaine in BM169 and collected in Trizol. RNA was extracted using Zymo Direct-zol kit (Genesee Scientific 11–331, CA, USA). The samples were then prepared by Illumina TruSeq stranded mRNA library kit. All 24 samples were sequenced with one flow cell on Illumina NextSeq 550 sequencer with 78bp read lengths. Reads were mapped with STAR (Dobin et al, 2013) and data were analyzed using DESeq2 (Love et al, 2014). Raw and processed data have been deposited in NCBI (GSE184849).
For RNAseq analysis of BATT-treated virgin females, virgin female parasites in six-well plates were separated from equal number of virgin males by 40 μm cell strainer. 5 μM BATT or βAla was refreshed every other day with fresh ABC169 media from day0. On day8, females were processed for RNAseq as above. Each of the three biological replicates included worms pooled from two technical replicates. RNA was extracted same as described above. The samples were then prepared using the Illumina TruSeq stranded mRNA library kit. All samples were sequenced on one flow cell on an Illumina NextSeq 550 sequencer with 78bp read lengths. Raw and processed data have been deposited in NCBI (GSE191062). Heatmaps were generated with variance stabilized transformed counts using heatmap in R. The post-pairing transcriptome change analyses were performed from publicly-available pair-ended RNAseq data from virgin (single sex) and sexually mature (mixed sex) females (WormBase ParaSite). The accession numbers of the three single sex female biological replicates and the three mixed sex female biological replicates are in the key resources table.
For qPCR analyses, worms were collected in 150 μL Trizol, homogenized, and RNA was extracted using Zymo Direct-zol kit and reverse transcribed with iScript (Bio-rad 1708891, CA, USA). Real time quantitative PCR was performed using FAST SYBR Green Master Mix (Bio-rad 1725122, CA, USA) on a QuantStudio3 real-time PCR system (Fisher Scientific, NH, USA). cytochrome c (Smp_900000) was used as an endogenous control for normalization. The data were analyzed by Applied biosystems on the Thermo Fisher Cloud (Thermo Fisher Scientific, MA, USA).
Scanning electron microscopy (SEM)
Freshly perfused male worms were rocked gently in BM169 containing 0.25% tricaine for 5 min and beheaded under a light microscope. To relax the male trunks such that the ventral surface was exposed for viewing by SEM, the trunk pieces were then rocked in a gradient of MgCl2 diluted in BM169 containing 0.25% tricaine as follows: 0.12 M MgCl2 for 30 min, 0.18 M MgCl2 for 20 min, 0.3 M MgCl2 for 5 min and 0.6 M MgCl2 for 1 min). After fixation in 4% formaldehyde in PBS for 30 min, worms were further fixed with 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4 °C. After three rinses in 0.1 M sodium cacodylate buffer, they were post-fixed with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer for 2 hours. Worms were rinsed with water and dehydrated with increasing concentration of ethanol, followed by increasing concentrations of hexamethyldisilazane in ethanol. Worms were air dried under the hood, mounted on SEM stubs with the ventral surface facing up and sputter coated with gold/palladium in a Cressington 108 auto sputter coater. Images were acquired with a Field-Emission Scanning Electron Microscope (Zeiss Sigma, Jena, Germany) at an accelerating voltage of 10kV.
Protein expression and purification
SmNRPS or Ebony were expressed in Sf9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen, MA, USA). Codon-optimized DNA of SmNRPS (Genescript, NJ, USA) or original cDNA of Ebony was cloned into pFastBac1 with a His6 tag, a StrepII tag and a TEV site before the N terminus. Third pass virus (40 mL) was used to infect 1L of Sf9 cells at 2–2.5 × 106 cells/mL. After 72h, cells were centrifuged at 1000 g for 10 min, frozen in liquid N2 and stored at −80 °C. Cell pellets were thawed in a 37 °C water bath and lysed with 60 ml of lysis buffer (20 mM Tris-HCl (pH 8.0), 250 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.024 mg/mL DNAse, 3 mM 2-mercaptoethanol, 0.2 mM PMSF, Sigma complete protease inhibitor (2 tablet/100 ml) and 10% glycerol for 1 h shaking at 4 °C. The following steps were performed on ice or at 4 °C. Lysates were homogenized using a Dounce homogenizer and centrifuged at 45,000 rpm for 50 min. The supernatant was supplemented with 10 mM imidazole (pH 8.0) and incubated with 3 mL equilibrated Qiagen Ni-NTA agarose (50% suspension in lysis buffer) with rotation for at least 1.5 h. The resin was centrifuged at 500g for 10 min, resuspended with 10 mL supernatant and loaded to a glass chromatography column. The resin was washed with Buffer 1 (20 mM Tris-HCl (pH 8.0), 250 mM NaCl, 5 mM MgCl2, 0.05% Triton X-100, 3 mM 2-mercaptoethanol, 10 mM imidazole (pH 8.0), 1 μg/mL aprotinin, 2 μg/mL leupeptin, 0.2 mM PMSF and 10% glycerol until A280 in the eluate returned to baseline, followed by Buffer 1 supplemented with 450 mM NaCl and 20mM imidazole. Protein was eluted with Buffer 1 containing 250 mM imidazole. Fractions containing SmNRPS were pooled and applied to a column of 0.9 mL Strep-Tactin Sepharose resin (Fisher Scientific NC9891255, NH, USA) that had been pre-equilibrated with the Ni-NTA elution buffer. The column was washed with 8 mL of 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 3 mM MgCl2, 0.5 mM DTT and 10% glycerol. SmNRPS was eluted in the same buffer but with only 150 mM NaCl and with 10 mM desthiobiotin. The first 1.8 mL elution was performed immediately. The column was left overnight in 1.8 mL elution buffer followed by elution with 4 more 1.8 mL fractions. Most of the SmNRPS was concentrated in the second fraction. Fractions containing SmNRPS or Ebony according to Coomassie Blue-stained SDS-PAGE were dialyzed against 20 mM Tris-HCl, 150 mM NaCl, 3 mM MgCl2 and 3 mM DTT, and 10% glycerol) and stored at −80 °C. Protein concentration was estimated by comparison with BSA standards on the same gel.
Biochemical analysis of SmNRPS
The ATP/PPi exchange and βAla unloading assays were adapted from previously reported methods (Hartwig et al, 2014; Richardt et al, 2003). For the exchange assay, each reaction contained 100 μL of 50 mM sodium phosphate buffer (pH 7.0), 25 mM MgCl2, 1 mM ATP, 50 μM [32P]PPi (0.11 cpm/fmol), 0.1 mg/mL BSA, 0.1 mM EDTA, 0.25 μM enzyme and 0.2 mM of the stated amino acid(s). Reactions were incubated at 37 °C for 15 min and quenched with a 1 mL ice cold 1.7% activated charcoal slurry (Millipore Sigma 53663, MA, USA) in 0.1 M Na4PPi and 0.56 M HClO4. Samples were centrifuged at 1000 g for 5 min and the charcoal pellet was washed twice with 1 mL of 5 μM Na4PPi. Charcoal pellets were resuspended with 0.5 mL deionized water and radioactivity was monitored by scintillation counting.
The unloading assay involves two reaction steps, loading and unloading. In the loading step, 0.25 μM enzyme was first incubated for 30 min at 37 °C in a loading cocktail of 1 mM ATP, 50 mM sodium phosphate (pH 7.0), 25 mM MgCl2, 0.1 mg/mL BSA, 0.1 mM EDTA, 5 μM βAla and 10 μCi/mL [3H]βAla (American Radiolabeled Chemicals ART 0205, MO, USA). This mixture was chilled on ice. The unloading reaction was initiated by adding 2.22 μL of 100 μM amine to 20 μL of the loading reaction incubation at 37 °C for 10 min. The reaction was then placed on ice, 15 μL of 2.5% (w/v) BSA was added, and protein was precipitated on ice with 10% (w/v) chilled trichloroacetic acid. After 30 min, the solution was transferred to a 1.5 mL conical tube, and centrifuged for 15 min at ~12,000 rpm. The pellet was washed twice with 1 mL of 10% (w/v) chilled trichloroacetic acid, resuspended in 200 μL formic acid, and subjected to scintillation counting. Note that only 2–10% of the enzyme could be loaded with βAla in the initial loading step. This may reflect either inefficient covalent addition of the phosphopantetheinate cofactor in the Sf9 cells (Hartwig et al., 2014), partial denaturation of the enzyme or erroneous estimation of enzyme concentration based on Coomassie Blue staining. Regardless, amounts of loaded enzyme at time zero was normalized to the water unloading control within the same experiment (Figure 4G–H).
Metabolomic analysis
For stable heavy isotope tracing, [13C3]βAla (Cambridge Isotope Laboratories clm-8755-PK, MA, USA) or normal βAla (Sigma-Aldrich 146064, MO, USA) were added to ABC169 media at 1 mM final concentration and refreshed every other day during media changes for 7 days. Samples of 30 male or female worms were rinsed twice in PBS, flash frozen in liquid N2, and homogenized in 1 mL ice-cold 80% methanol in water (vol/vol) using an ultrasonicator (30 s × 3 times at the highest setting). Homogenates were vortexed and 200 μL was diluted in 800 μL ice-cold 80% methanol. This mixture was vortexed again (1 min), centrifuged at maximum speed for 15 min at 4 °C, the supernatant was transferred to new tube, dried overnight using a SpeedVac, and stored at −80 °C. The in vitro reaction mix used for the retention time reference of BATT contained 100 μL of 50 mM sodium phosphate buffer pH 7.0, 25 mM MgCl2, 0.1 mg/mL BSA, 0.1 mM EDTA, 1 mM βAla, 0.25 mM tryptamine, 5 μM SmNRPS protein and 1 mM ATP. The mixture was incubated at 37°C for 2.5 h, then added with 1 mL ice-cold 80% methanol, vortexed for 1 min, centrifuged at maximum speed for 15 min at 4 °C. The supernatant was dried to a pellet with a SpeedVac and resuspended in 100 μL deionized H2O.
Metabolite analysis data acquisition was performed by reverse-phase chromatography on a 1290 UHPLC liquid chromatography (LC) system interfaced to a high-resolution mass spectrometry (HRMS) 6550 iFunnel Q-TOF mass spectrometer (MS) (Agilent Technologies, CA, USA). The MS was operated in both positive and negative (ESI+ and ESI−) modes. Analytes were separated on an Acquity UPLC® HSS T3 column (1.8 μm, 2.1 × 150 mm, Waters, MA, USA). The column was kept at room temperature. Mobile phase A composition was 0.1% formic acid in water and mobile phase B composition was 0.1% formic acid in 100% ACN. The LC gradient was 0 min: 1% B; 5 min: 5% B; 15 min: 99% B; 23 min: 99% B; 24 min: 1% B; 25 min: 1% B. The flow rate was 250 μL/min. The sample injection volume was 5 μL. ESI source conditions were set as follows: dry gas temperature 225 °C and flow 18 L/min, fragmentor voltage 175 V, sheath gas temperature 350 °C and flow 12 L/min, nozzle voltage 500 V, and capillary voltage +3500 V in positive mode and −3500 V in negative. The instrument was set to acquire over the full m/z range of 40–1700 in both modes, with the MS acquisition rate of 1 spectrum s−1 in profile format. When quantification was required, both samples and BATT concentration standards were spiked with same amount of tryptophan-d5 as the internal standard.
Raw data files (.d) were processed using Profinder B.08.00 SP3 software (Agilent Technologies, CA, USA) with an in-house database containing retention time and accurate mass information on 600 standards from Mass Spectrometry Metabolite Library (IROA Technologies, MA, USA) which was created under the same analysis conditions. The in-house database matching parameters were: mass tolerance 10 ppm; retention time tolerance 0.5 min; coelution coefficient 0.5. Peak integration result was manually curated in Profinder for improved consistency and exported as a spreadsheet (.csv).
BATT detection in conditioned media
One worm or worm pair were placed in one well of a 96 well plate with 150 μL ABC169 for two days. Worm pairs were either freshly perfused, freshly separated or separated for 3 days prior to experiment set up. Negative control contained only medium ABC169. After 48 hours, 100 μL media from each well were sampled and mixed with 100 μL methanol containing 0.2% formic acid. The mixtures were vortexed for 15 s, incubated for 10 min at RT and centrifuged at ~18000g for 5 min. 150 μL supernatant was removed to a clean tube and stored at −20 °C. Before analysis, tryptophan-d5 was spiked into the samples as an internal standard. The negative control contained only ABC169 medium. In some experiments, media conditioned by paired was collected to evaluate virgin female development. For these experiments, ~10 virgin females were cultured in 1 mL of 48 h pair conditioned media. Every other day, conditioned media was refreshed with media freshly conditioned by worm pairs of the previous 48 hours. Such worms were cultured alongside BATT (5 μM) or mock treated virgin females and harvested at day 14.
The BATT concentration was measured in conditioned media by LC-MS/MS using an AB Sciex (Framingham, MA, USA) 6500+ QTRAP® mass spectrometer coupled to a Shimadzu (Columbia, MD, USA) Prominence LC. BATT was detected in positive MRM (multiple reaction monitoring) mode by following the precursor to fragment ion transitions of 231.8 to 185.2 (qualifier ion). An Agilent C18 XDB column (5 μm, 50 × 4.6 mm) was used for chromatography with the following conditions: Buffer A: H20 + 0.1% formic acid; Buffer B: MeOH + 0.1% formic acid; 0–1.5 min 3% B (97% A); 1.5–3.5 min gradient to 100% B (0% A); 3.5–4.5 min 3% B (97% A). Tryptophan-d5 (transition 210.206 to 192.400) was used as internal standard. Concentrations of BATT were determined by comparison to a standard curve prepared by spiking synthetic BATT in DMSO into ABC169 followed by standard sample processing. A limit of detection was defined as a level three times that observed in blank media and the limit of quantitation (LOQ) as the lowest point on the standard curve that gave an analyte signal above the LOD and within 20% of nominal upon back-calculation. The LOQ for BATT was 0.5 ng/mL.
BATT synthesis and treatment
One batch of β-alanyl-tryptamine (N-(2-(1H-indol-3-yl)ethyl)-3-aminopropanamide) was synthesized by MuseChem (Batch M21X01198, NJ, USA). We refer to this compound as mcBATT. The certificate of analysis showed the purity as 99.88% by HPLC and an ESI-MS confirmed molecular weight of 231.2 g/mol. However, the product was a green, sticky, hygroscopic solid.
We then synthesized a second batch locally (swBATT). The synthesis was a two-step sequence. Boc protected BATT (Boc-BATT, tert-Butyl (3-((2-(1H-indol-3-yl)ethyl)amino)-3-oxopropyl)carbamate) was first synthesized as follows: i-Pr2NEt (1.4 mL, 8.0 mmol) was added to a solution of Boc-β-Ala-OH (1.0 g, 5.29 mmol), tryptamine (851.9 mg, 5.32 mmol) and HATU (2.42 g, 6.4 mmol) in DMF (24 mL) in a 50 mL round bottom flask. The reaction was stirred overnight and the reaction went to completion as judged by HPLC/MS analysis. Water was added to the reaction mixture, which was then extracted with EtOAc. The organic layer was washed several times with water and then with brine, dried over Na2SO4, filtered and condensed. The crude mixture was purified via ISCO flash chromatography in 1–10% MeOH/DCM to give 1.55 g of the Boc-BATT in 88% yield. For the second step, TFA (1.0 mL) was added dropwise to a solution of Boc-BATT (497.5 mg, 1.5 mmol) in 10 mL DCM in a scintillation vial. The reaction went to completion within 2 hours and was thereafter condensed under reduced pressure. The crude mixture was purified by reverse phase ISCO flash chromatography in 0–40% H2O/CH3CN. Fractions containing the desired product were combined and condensed on a speed vacuum (Thermo Fisher Scientific SPD1010–115, MA, USA) to give 505.4 mg of the TFA salt (98% yield). Proton NMR was performed on both Boc-BATT and the final product, swBATT, for confirmation. This product was a brown, extremely hygroscopic, oil-like compound.
Concentrated stock solutions of mcBATT or swBATT were prepared in sterile filtered deionized water and stored at < −20 °C. Because of the hygroscopic nature of both preparations and the unexpected colors, we estimated the concentration of stock solutions by UV absorbance in comparison to solutions of tryptamine after extensive dilution. Concentrated aqueous solutions of BATT displayed broad absorbance maxima above 300 nm, consistent with their appearance. Upon further dilution, the absorbance maxima sharpened and shifted to the UV such that < 1 mM BATT from either batch displayed an absorbance spectrum with a 240–300 nm spectrum essentially identical to that of tryptamine. The colors of the semi-solid and concentrated solutions thus appear to result from stacking of the indole rings. For treatment of worms, the compound was directly added into ABC169 media to achieve desired concentration. Compound was refreshed every other day with fresh media.
QUANTIFICATION AND STATISTICAL ANALYSIS
For RNAseq experiments, base calling was performed by Illumina cassava 1.9 software. Sequenced reads were trimmed for adaptor sequence, and masked for low-complexity or low-quality sequence, then mapped to the S. mansoni genome (v7) using STAR (Dobin et al, 2013). Pairwise comparisons of differential gene expression were performed with DESseq2 (Love et al, 2014). For statistical analysis of non-sequencing data, GraphPad Prism software processed and presented the data as the mean with SD. Statistical significance was calculated by unpaired two-tailed parametric t test. Unpaired ordinary one-way ANOVA was also performed in Figure 4G. P values < 0.05 are considered significant (NS, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Supplementary Material
(A) qPCR showing the relative mRNA level of gli1 in cntl RNAi or gli1 RNAi males at the end of two-weeks of pairing. *** p < 0.001. Parametric t test. Data from three biological replicates. (B) Whole-mount colorimetric in situ hybridization showing expression of the gli1 gene in a paired male worm (top) and a virgin male worm (bottom). (C) FISH showing the expression of gli1 (magenta) mRNA in the ventral region of paired and virgin males. (D) Two-color FISH for 7b2 (green) and gli1 (magenta) mRNAs in paired (top) or virgin (bottom) male worms. (E) Two-color FISH for calp (green) and gli1 (magenta) mRNAs in paired (top) or virgin (bottom) male worms. (F) Whole-mount colorimetric in situ hybridization showing expression of the gli1 gene in a sexually mature female worm (top) and a virgin female worm (bottom). Arrows indicate gli1 mRNA expression in virgin female. (G) Two-color FISH for 7b2 (green) and gli1 (magenta) mRNAs in sexually mature (top) or virgin (bottom) female worms. (H) Two-color FISH for calp (green) and gli1 (magenta) mRNAs in sexually mature (top) or virgin (bottom) female worms. For (E, G-H), arrows indicate co-expression. All images are representative of 3 experiments with n > 10. DAPI, gray. Scale bars, B, F, 100 μm; C, D, E, G, H, 20 μm.
(A) Heatmap of up-regulated and down-regulated genes in females after pairing with cntl RNAi males or gli1 RNAi males. Con_FP, virgin females paired with cntl RNAi males; Con_FUP, virgin females left unpaired and co-cultured cntl RNAi males; gli1_FP, virgin females paired with gli1 RNAi males; gli1_FUP, virgin females left unpaired and co-cultured with gli1 RNAi males. Genes limited to those with 2-fold change and adjusted p value < 0.0001 of Con_FP/Con_FUP. (B) FISH showing head and body expression of Sm-nrps mRNA (magenta) in the male parasites in paired and virgin worms. (C and D) Two-color FISH for (C) gli1 or (D) the muscle marker tpm2 with Sm-nrps in the head of virgin male parasites. Arrows indicate cells co-expression. (E) Scanning electron microscopy showing the ventral surface of a paired male parasite and an unpaired male parasite. White dotted lines enclose sensory cilium-like structure. Representative images from 3 parasites. (F) Whole-mount colorimetric in situ hybridization showing expression of the gli1 gene in a sexually mature female worm (top) and a virgin female worm (bottom). (G, H) Two-color FISH for tpm2 or 7b2 with Sm-nrps mRNAs in the (G) head or (H) body of a virgin female parasite. (I) Confocal sagittal section of Sm-nrps FISH (magenta) showing mRNA enriched in the dorsal surface and the body edges of a virgin female worm. (J) qPCR showing relative Sm-nrps levels following RNAi treatment in male worms two-weeks post-pairing. *** p < 0.001. Parametric t test. Data from three biological replicates. All FISH and WISH images are representatives of data from 3 experiments with n > 10 parasites. DAPI, gray. Scale bars, B-D, G-I, 20 μm; E, 0.5 μm; F, 100 μm.
(A) MUSCLE Alignment of NRPS enzymes. SmNRPS and Ebony represent full-length protein sequences. NRPS2–1 includes the first A and the following T domain encoded from the blmIV gene. GS1 is the A domain from gramicidin synthetase 1. The region within the red arrows represents the A domain whereas the region in the blue arrows is the T domain. (B) Enzymatic reactions performed by Ebony and SmNRPS. (C) SDS-PAGE gel images of fractions from SmNRPS protein purification. The Elution2 fraction from Strep-Tactin purification was dialyzed against the storage buffer and used for assays. Arrows point to where SmNRPS bands migrate (~ 150 kDa). (D) Reaction for ATP-PPi exchange assay. The assay measures the reverse reaction by the production of [32P] ATP from [32P]PPi. (E) ATP-PPi exchange assay with Drosophila Ebony and SmNRPS. **** p < 0.0001. Parametric t test. Error bars are SD. (F) Reaction for unloading assay. Bioamine reacts with the covalently [3H]βAla-labeled enzyme to release free enzyme.
(A) LC/MS extracted-ion chromatogram (EIC) of m/z at 232.144 showing retention time of BATT from paired male extracts (bottom) and an in vitro reaction mixture (top) containing SmNRPS, βAla, tryptamine, ATP and magnesium. Paired male data shows a sample extracted from paired worms exposed to control RNAi. Y axis is ion abundance and X axis is retention time from 1.5min to 8.5min. Time is shown for all peaks. (B) LC-MS showing abundance of BATT in paired male and female parasites. (C) LC-MS showing BATT abundance following RNAi treatment in paired male worms. (D) Images of Fast Blue BB (left) and DAPI (right) staining 14 days after exposure of virgin females to male:female conditioned media (same in Figure 5H), 5 μM BATT and no treatment. Representative images from 3 experiments with n > 22 worms. Numbers in corner indicate fraction of worms similar to those presented/total number of worms examined. Scale bar, 100 μm.
(A) DAPI staining of virgin females after 14 days of 0.05 μM-50 μM mcBATT or swBATT treatment; 100 μM βAla treatment was used a control. (B) Fast Blue BB (red) staining of virgin females after 14 days of 0.05 μM-50 μM mcBATT or swBATT treatment, 100 μM βAla treatment was used a control. Panels A and B are from the same experiments as data shown in Figure 6E. (C) FISH for p48 (magenta) in the vitellaria and bmgp (green) of in the ovary of virgin females after 14 days of 50 μM mcBATT or βAla treatment. (D, left) EdU staining (gold) of eggs laid by virgin female worms 14 days after pairing with a male. Autofluorescence (autofluo, green) shows egg shape. (D, right) Confocal micrograph of EdU labeling and Differential Interference Contrast (DIC) of an egg with laid by a virgin female worm 14 days after pairing. A, B, representative images from 3 experiments with n > 18. C, representative images from 3 experiments with n > 14 worms. Numbers in corner indicate fraction of worms similar to those presented/total number of worms examined. Scale bars, 100 μm.
(A) Left, hierarchal clustered heatmap of 89 genes differentially-regulated following BATT treatment (log2 fold change > +/− 2, adjusted p < 0.05, base mean > 50). The expression of these same 89 genes were evaluated in publicly-available RNAseq data from virgin and sexually mature female parasites (Right). Arrows indicate two instances where gene expression trend was different between two datasets. Accession numbers of virgin and sexually mature female data can be found in key resources table. (B) Dot plot showing expression of the 89 BATT-regulated genes across scRNAseq clusters from adult parasites (Wendt et al, 2020). Each row represents a cluster type and each row a gene. Size of circles and color reflect percentage of cells expressing marker and the level of expression, respectively. For ease of visualization, in some cases clusters of related cell types were collapsed into a single cluster (e.g., 31 clusters of neurons and 8 clusters of muscles were reduced to single cluster.)
Multiple sequence alignment of A domains from NRPS enzymes. Residues aligned to those involved in amino acid specificity in the reference Phe-specific A domain of GS1 are marked in red rectangles. Fruit fly, Drosophila melanogaster; S. mansoni, Schistosoma mansoni; S. haematobium, Schistosoma haematobium; S. japonicum, Schistosoma japonicum; planarian, Schmidtea mediterranea; tapeworm, Hymenolepis diminuta; annelid, Capitella teleta; rotifer, Rotaria magnacalcarata; cnidarian, Hydra vulgaris; hemichordate, Saccoglossus kowalevskii. Alignment was analyzed using T-Coffee. Aligned amino acids were colored by percentage identity (dark blue, 100%; white 0%).
Movie S1. Confocal stacks from dorsal to ventral tissue of FISH for gli1 mRNA (magenta) and DAPI (gray) staining in paired and virgin male worms, related to Figure 1.
Table S1. List of signaling pathway factors screened by RNAi, related to Figure 1.
Table S2 RNAseq data from paired and unpaired male worms treated with gli1(RNAi), related to Figure 2.
Table S3. RNAseq data from female worms paired with gli1 or control (RNAi) male worms, related to Figure 2.
Table S4. RNAseq data from virgin females following 8D of BATT treatment, related to Figure 6.
Table S5. A-domain specificity code from SmNRPS-like enzymes found in various metazoans, related to Figure 7.
Table S6. Sequences of oligonucleotides used for RNAi, in situ hybridization and qPCR, related to the STAR Methods.
Highlights.
gli1 is essential for male schistosomes to induce female sexual development.
Male:female pairing induces male Sm-nrps expression in a gli1 dependent manner.
Sm-nrps is essential for male worms to induce female sexual development.
SmNRPS makes β-alanyl-tryptamine, a pheromone that stimulates female development.
ACKNOWLEDGEMENTS
We thank members from Collins lab, Phillip A. Newmark and Melanie H. Cobb for comments on the manuscript; the NIAID Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) for providing infected snails and infected mice through NIH-NIAID Contract HHSN272201700014I for distribution through BEI Resources; Helmut Krämer for providing Ebony cDNA; the UT Southwestern Electron Microscopy and Genomics Core Facilities; and Francisco Ortiz from Preclinical Pharmacology Core Lab. Biorender was used for some schematic material. This work was funded by NIH grants R01AI121037 (J.J.C.), R01AI150776 (J.J.C.), and R35CA22044901 (R.J.D); Welch Foundation grant I-1948–20180324 (J.J.C.) and the Burroughs Wellcome Fund (J.J.C.). R.J.D. is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
R.J.D. is on the SABs for Agios Pharmaceuticals, Vida Ventures and Nirogyone Therapeutics, and is a founder/advisor for Atavistik Biosciences.
REFERENCE
- Armstrong JC (1965) Mating behavior and development of schistosomes in the mouse. J Parasitol, 51, 605–16. [PubMed] [Google Scholar]
- Basch PF (1981) Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J Parasitol, 67(2), 179–85. [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Edwards TN, Boulianne GL & Meinertzhagen IA (2012) The metabolism of histamine in the Drosophila optic lobe involves an ommatidial pathway: β-alanine recycles through the retina. J Exp Biol, 215(Pt 8), 1399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Loubani M & Meinertzhagen IA (2002) tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. J Neurosci, 22(24), 10549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever AW, Macedonia JG, Mosimann JE & Cheever EA (1994) Kinetics of egg production and egg excretion by Schistosoma mansoni and S. japonicum in mice infected with a single pair of worms. Am J Trop Med Hyg, 50(3), 281–95. [DOI] [PubMed] [Google Scholar]
- Collins JJ, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, Sweedler JV & Newmark PA (2010) Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol, 8(10), e1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ III, King RS, Cogswell A, Williams DL & Newmark PA (2011) An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl Trop Dis, 5(3), e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, Wang B, Lambrus BG, Tharp ME, Iyer H & Newmark PA (2013) Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature, 494(7438), 476–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conford EM & Huot ME (1981) Glucose transfer from male to female schistosomes. Science, 213(4513), 1269–71. [DOI] [PubMed] [Google Scholar]
- Conti E, Stachelhaus T, Marahiel MA & Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J, 16(14), 4174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cort WW (1921) Sex in the Trematode Family Schistosomidae. Science, 53(1367), 226–8. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Grogan JL, Blank RB, Lim KC, Locksley RM & McKerrow JH (2001) Modulation of blood fluke development in the liver by hepatic CD4+ lymphocytes. Science, 294(5545), 1358–61. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevelding CG, Langner S & Dissous C (2018) Kinases: Molecular Stage Directors for Schistosome Development and Differentiation. Trends Parasitol, 34(3), 246–260. [DOI] [PubMed] [Google Scholar]
- Grzych JM, Pearce E, Cheever A, Caulada ZA, Caspar P, Heiny S, Lewis F & Sher A (1991) Egg deposition is the major stimulus for the production of Th2 cytokines in murine schistosomiasis mansoni. J Immunol, 146(4), 1322–7. [PubMed] [Google Scholar]
- Haeberlein S, Angrisano A, Quack T, Lu Z, Kellershohn J, Blohm A, Grevelding CG & Hahnel SR (2019) Identification of a new panel of reference genes to study pairing-dependent gene expression in Schistosoma mansoni. Int J Parasitol, 49(8), 615–624. [DOI] [PubMed] [Google Scholar]
- Harris AR, Russell RJ & Charters AD (1984) A review of schistosomiasis in immigrants in Western Australia, demonstrating the unusual longevity of Schistosoma mansoni. Trans R Soc Trop Med Hyg, 78(3), 385–8. [DOI] [PubMed] [Google Scholar]
- Hartwig S, Dovengerds C, Herrmann C & Hovemann BT (2014) Drosophila Ebony: a novel type of nonribosomal peptide synthetase related enzyme with unusually fast peptide bond formation kinetics. FEBS J, 281(22), 5147–58. [DOI] [PubMed] [Google Scholar]
- Haseeb MA, Eveland LK & Fried B (1985) The uptake, localization and transfer of [4–14C]cholesterol in Schistosoma mansoni males and females maintained in vitro. Comp Biochem Physiol A Comp Physiol, 82(2), 421–3. [DOI] [PubMed] [Google Scholar]
- Hoagland MB (1955) An enzymic mechanism for amino acid activation in animal tissues. Biochim Biophys Acta, 16(2), 288–9. [DOI] [PubMed] [Google Scholar]
- Hockley DJ (1973) Ultrastructure of the tegument of Schistosoma. Adv Parasitol, 11(0), 233–305. [DOI] [PubMed] [Google Scholar]
- Izoré T, Tailhades J, Hansen MH, Kaczmarski JA, Jackson CJ & Cryle MJ (2019) nonribosomal peptide synthetase Ebony encodes an atypical condensation domain. Proc Natl Acad Sci U S A, 116(8), 2913–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CH (2010) Parasites and poverty: the case of schistosomiasis. Acta Trop, 113(2), 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CH & Dangerfield-Cha M (2008) The unacknowledged impact of chronic schistosomiasis. Chronic Illn, 4(1), 65–79. [DOI] [PubMed] [Google Scholar]
- Langenberg MCC, Hoogerwerf MA, Koopman JPR, Janse JJ, Kos-van Oosterhoud J, Feijt C, Jochems SP, de Dood CJ, van Schuijlenburg R, Ozir-Fazalalikhan A, Manurung MD, Sartono E, van der Beek MT, Winkel BMF, Verbeek-Menken PH, Stam KA, van Leeuwen FWB, Meij P, van Diepen A, van Lieshout L, van Dam GJ, Corstjens PLAM, Hokke CH, Yazdanbakhsh M, Visser LG & Roestenberg M (2020) A controlled human Schistosoma mansoni infection model to advance novel drugs, vaccines and diagnostics. Nat Med, 26(3), 326–332. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W & Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12), 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVerde PT, Andrade LF & Oliveira G (2009) Signal transduction regulates schistosome reproductive biology. Curr Opin Microbiol, 12(4), 422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Sessler F, Holroyd N, Hahnel S, Quack T, Berriman M & Grevelding CG (2016) Schistosome sex matters: a deep view into gonad-specific and pairing-dependent transcriptomes reveals a complex gender interplay. Sci Rep, 6, 31150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Spänig S, Weth O & Grevelding CG (2019) Males, the Wrongly Neglected Partners of the Biologically Unprecedented Male-Female Interaction of Schistosomes. Front Genet, 10, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP & Threadgold LT (1967) A presumed sensory structure associated with the tegument of Schistosoma mansoni. J Parasitol, 53(3), 537–9. [PubMed] [Google Scholar]
- Pearce EJ & MacDonald AS (2002) The immunobiology of schistosomiasis. Nat Rev Immunol, 2(7), 499–511. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Pitsouli C & Shilo BZ (2012) Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol, 4(8), a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrobono S, Gagliardi S & Stecca B (2019) Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front Genet, 10, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popiel I & Basch PF (1984) Reproductive development of female Schistosoma mansoni (Digenea: Schistosomatidae) following bisexual pairing of worms and worm segments. J Exp Zool, 232(1), 141–50. [DOI] [PubMed] [Google Scholar]
- Popiel I & Basch PF (1986) Schistosoma mansoni: cholesterol uptake by paired and unpaired worms. Exp Parasitol, 61(3), 343–7. [DOI] [PubMed] [Google Scholar]
- Popiel I, Cioli D & Erasmus DA (1984) The morphology and reproductive status of female Schistosoma mansoni following separation from male worms. Int J Parasitol, 14(2), 183–90. [DOI] [PubMed] [Google Scholar]
- Richardt A, Kemme T, Wagner S, Schwarzer D, Marahiel MA & Hovemann BT (2003) Ebony, a novel nonribosomal peptide synthetase for beta-alanine conjugation with biogenic amines in Drosophila. J Biol Chem, 278(42), 41160–6. [DOI] [PubMed] [Google Scholar]
- Severinghaus AE (1928) Sex Studies on Schistosoma japonicum. Quarterly Journal of Microscopical Science, 71, 653–702. [Google Scholar]
- Shou Q, Feng L, Long Y, Han J, Nunnery JK, Powell DH & Butcher RA (2016) A hybrid polyketide-nonribosomal peptide in nematodes that promotes larval survival. Nat Chem Biol, 12(10), 770–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V & Reiter JF (2006) The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science, 313(5787), 629–33. [DOI] [PubMed] [Google Scholar]
- Sotillo J, Pearson M, Potriquet J, Becker L, Pickering D, Mulvenna J & Loukas A (2016) Extracellular vesicles secreted by Schistosoma mansoni contain protein vaccine candidates. Int J Parasitol, 46(1), 1–5. [DOI] [PubMed] [Google Scholar]
- Stachelhaus T, Mootz HD & Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol, 6(8), 493–505. [DOI] [PubMed] [Google Scholar]
- Steinmann P, Keiser J, Bos R, Tanner M & Utzinger J (2006) Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis, 6(7), 411–25. [DOI] [PubMed] [Google Scholar]
- Stenesen D, Moehlman AT & Krämer H (2015) The carcinine transporter CarT is required in Drosophila photoreceptor neurons to sustain histamine recycling. Elife, 4, e10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süssmuth RD & Mainz A (2017) Nonribosomal Peptide Synthesis-Principles and Prospects. Angew Chem Int Ed Engl, 56(14), 3770–3821. [DOI] [PubMed] [Google Scholar]
- Torres JP & Schmidt EW (2019) The biosynthetic diversity of the animal world. J Biol Chem, 294(46), 17684–17692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf MJ, de Vlas SJ, Brooker S, Looman CW, Nagelkerke NJ, Habbema JD & Engels D (2003) Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop, 86(2–3), 125–39. [DOI] [PubMed] [Google Scholar]
- Wang H, Fewer DP, Holm L, Rouhiainen L & Sivonen K (2014a) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A, 111(25), 9259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen R & Collins JJ (2019) Systematically improved in vitro culture conditions reveal new insights into the reproductive biology of the human parasite Schistosoma mansoni. PLoS Biol, 17(5), e3000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH & Barr MM (2014b) C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol, 24(5), 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu Y, Shen H, Qing T, Zheng Y, Li Q, Mo X, Wang S, Li N, Chai R, Xu B, Liu M, Brindley PJ, McManus DP, Feng Z, Shi L & Hu W (2017) Dynamic transcriptomes identify biogenic amines and insect-like hormonal regulation for mediating reproduction in Schistosoma japonicum. Nat Commun, 8, 14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinland DF & Wyman J (1858) Human cestoides : an essay on the tapeworms of man : giving a full account of their nature, organization, and embryonic development; the pathological symptoms they produce, and the remedies which have proved successful in modern practice. Cambridge: Metcalf and Company. [Google Scholar]
- Wendt G, Zhao L, Chen R, Liu C, O’Donoghue AJ, Caffrey CR, Reese ML & Collins JJ (2020) A single-cell RNA-seq atlas of Schistosoma mansoni identifies a key regulator of blood feeding. Science, 369(6511), 1644–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, W. H. O. (2018) Schistosomiasis and soil-transmitted infections: number of people treated in 2017.: Weekly Epidemiological Record.
- Wright TR (1987) The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv Genet, 24, 127–222. [PubMed] [Google Scholar]
- Ziegler AB, Brüsselbach F & Hovemann BT (2013) Activity and coexpression of Drosophila black with ebony in fly optic lobes reveals putative cooperative tasks in vision that evade electroretinographic detection. J Comp Neurol, 521(6), 1207–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) qPCR showing the relative mRNA level of gli1 in cntl RNAi or gli1 RNAi males at the end of two-weeks of pairing. *** p < 0.001. Parametric t test. Data from three biological replicates. (B) Whole-mount colorimetric in situ hybridization showing expression of the gli1 gene in a paired male worm (top) and a virgin male worm (bottom). (C) FISH showing the expression of gli1 (magenta) mRNA in the ventral region of paired and virgin males. (D) Two-color FISH for 7b2 (green) and gli1 (magenta) mRNAs in paired (top) or virgin (bottom) male worms. (E) Two-color FISH for calp (green) and gli1 (magenta) mRNAs in paired (top) or virgin (bottom) male worms. (F) Whole-mount colorimetric in situ hybridization showing expression of the gli1 gene in a sexually mature female worm (top) and a virgin female worm (bottom). Arrows indicate gli1 mRNA expression in virgin female. (G) Two-color FISH for 7b2 (green) and gli1 (magenta) mRNAs in sexually mature (top) or virgin (bottom) female worms. (H) Two-color FISH for calp (green) and gli1 (magenta) mRNAs in sexually mature (top) or virgin (bottom) female worms. For (E, G-H), arrows indicate co-expression. All images are representative of 3 experiments with n > 10. DAPI, gray. Scale bars, B, F, 100 μm; C, D, E, G, H, 20 μm.
(A) Heatmap of up-regulated and down-regulated genes in females after pairing with cntl RNAi males or gli1 RNAi males. Con_FP, virgin females paired with cntl RNAi males; Con_FUP, virgin females left unpaired and co-cultured cntl RNAi males; gli1_FP, virgin females paired with gli1 RNAi males; gli1_FUP, virgin females left unpaired and co-cultured with gli1 RNAi males. Genes limited to those with 2-fold change and adjusted p value < 0.0001 of Con_FP/Con_FUP. (B) FISH showing head and body expression of Sm-nrps mRNA (magenta) in the male parasites in paired and virgin worms. (C and D) Two-color FISH for (C) gli1 or (D) the muscle marker tpm2 with Sm-nrps in the head of virgin male parasites. Arrows indicate cells co-expression. (E) Scanning electron microscopy showing the ventral surface of a paired male parasite and an unpaired male parasite. White dotted lines enclose sensory cilium-like structure. Representative images from 3 parasites. (F) Whole-mount colorimetric in situ hybridization showing expression of the gli1 gene in a sexually mature female worm (top) and a virgin female worm (bottom). (G, H) Two-color FISH for tpm2 or 7b2 with Sm-nrps mRNAs in the (G) head or (H) body of a virgin female parasite. (I) Confocal sagittal section of Sm-nrps FISH (magenta) showing mRNA enriched in the dorsal surface and the body edges of a virgin female worm. (J) qPCR showing relative Sm-nrps levels following RNAi treatment in male worms two-weeks post-pairing. *** p < 0.001. Parametric t test. Data from three biological replicates. All FISH and WISH images are representatives of data from 3 experiments with n > 10 parasites. DAPI, gray. Scale bars, B-D, G-I, 20 μm; E, 0.5 μm; F, 100 μm.
(A) MUSCLE Alignment of NRPS enzymes. SmNRPS and Ebony represent full-length protein sequences. NRPS2–1 includes the first A and the following T domain encoded from the blmIV gene. GS1 is the A domain from gramicidin synthetase 1. The region within the red arrows represents the A domain whereas the region in the blue arrows is the T domain. (B) Enzymatic reactions performed by Ebony and SmNRPS. (C) SDS-PAGE gel images of fractions from SmNRPS protein purification. The Elution2 fraction from Strep-Tactin purification was dialyzed against the storage buffer and used for assays. Arrows point to where SmNRPS bands migrate (~ 150 kDa). (D) Reaction for ATP-PPi exchange assay. The assay measures the reverse reaction by the production of [32P] ATP from [32P]PPi. (E) ATP-PPi exchange assay with Drosophila Ebony and SmNRPS. **** p < 0.0001. Parametric t test. Error bars are SD. (F) Reaction for unloading assay. Bioamine reacts with the covalently [3H]βAla-labeled enzyme to release free enzyme.
(A) LC/MS extracted-ion chromatogram (EIC) of m/z at 232.144 showing retention time of BATT from paired male extracts (bottom) and an in vitro reaction mixture (top) containing SmNRPS, βAla, tryptamine, ATP and magnesium. Paired male data shows a sample extracted from paired worms exposed to control RNAi. Y axis is ion abundance and X axis is retention time from 1.5min to 8.5min. Time is shown for all peaks. (B) LC-MS showing abundance of BATT in paired male and female parasites. (C) LC-MS showing BATT abundance following RNAi treatment in paired male worms. (D) Images of Fast Blue BB (left) and DAPI (right) staining 14 days after exposure of virgin females to male:female conditioned media (same in Figure 5H), 5 μM BATT and no treatment. Representative images from 3 experiments with n > 22 worms. Numbers in corner indicate fraction of worms similar to those presented/total number of worms examined. Scale bar, 100 μm.
(A) DAPI staining of virgin females after 14 days of 0.05 μM-50 μM mcBATT or swBATT treatment; 100 μM βAla treatment was used a control. (B) Fast Blue BB (red) staining of virgin females after 14 days of 0.05 μM-50 μM mcBATT or swBATT treatment, 100 μM βAla treatment was used a control. Panels A and B are from the same experiments as data shown in Figure 6E. (C) FISH for p48 (magenta) in the vitellaria and bmgp (green) of in the ovary of virgin females after 14 days of 50 μM mcBATT or βAla treatment. (D, left) EdU staining (gold) of eggs laid by virgin female worms 14 days after pairing with a male. Autofluorescence (autofluo, green) shows egg shape. (D, right) Confocal micrograph of EdU labeling and Differential Interference Contrast (DIC) of an egg with laid by a virgin female worm 14 days after pairing. A, B, representative images from 3 experiments with n > 18. C, representative images from 3 experiments with n > 14 worms. Numbers in corner indicate fraction of worms similar to those presented/total number of worms examined. Scale bars, 100 μm.
(A) Left, hierarchal clustered heatmap of 89 genes differentially-regulated following BATT treatment (log2 fold change > +/− 2, adjusted p < 0.05, base mean > 50). The expression of these same 89 genes were evaluated in publicly-available RNAseq data from virgin and sexually mature female parasites (Right). Arrows indicate two instances where gene expression trend was different between two datasets. Accession numbers of virgin and sexually mature female data can be found in key resources table. (B) Dot plot showing expression of the 89 BATT-regulated genes across scRNAseq clusters from adult parasites (Wendt et al, 2020). Each row represents a cluster type and each row a gene. Size of circles and color reflect percentage of cells expressing marker and the level of expression, respectively. For ease of visualization, in some cases clusters of related cell types were collapsed into a single cluster (e.g., 31 clusters of neurons and 8 clusters of muscles were reduced to single cluster.)
Multiple sequence alignment of A domains from NRPS enzymes. Residues aligned to those involved in amino acid specificity in the reference Phe-specific A domain of GS1 are marked in red rectangles. Fruit fly, Drosophila melanogaster; S. mansoni, Schistosoma mansoni; S. haematobium, Schistosoma haematobium; S. japonicum, Schistosoma japonicum; planarian, Schmidtea mediterranea; tapeworm, Hymenolepis diminuta; annelid, Capitella teleta; rotifer, Rotaria magnacalcarata; cnidarian, Hydra vulgaris; hemichordate, Saccoglossus kowalevskii. Alignment was analyzed using T-Coffee. Aligned amino acids were colored by percentage identity (dark blue, 100%; white 0%).
Movie S1. Confocal stacks from dorsal to ventral tissue of FISH for gli1 mRNA (magenta) and DAPI (gray) staining in paired and virgin male worms, related to Figure 1.
Table S1. List of signaling pathway factors screened by RNAi, related to Figure 1.
Table S2 RNAseq data from paired and unpaired male worms treated with gli1(RNAi), related to Figure 2.
Table S3. RNAseq data from female worms paired with gli1 or control (RNAi) male worms, related to Figure 2.
Table S4. RNAseq data from virgin females following 8D of BATT treatment, related to Figure 6.
Table S5. A-domain specificity code from SmNRPS-like enzymes found in various metazoans, related to Figure 7.
Table S6. Sequences of oligonucleotides used for RNAi, in situ hybridization and qPCR, related to the STAR Methods.
Data Availability Statement
Two of the RNA sequencing data sets collected by this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The existing, publicly available data for single sex and mixed sex female RNAseq were obtained from the accession numbers listed in the key resources table. Microcopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request


