Abstract
Adult endochondral bones are prefigured in the embryo as cellular condensations within fields of more loosely distributed skeletal progenitors. How these early condensations are initiated and shaped has remained enigmatic, despite the wealth of research on later stages of cartilage differentiation and endochondral ossification. Using the simple larval zebrafish facial skeleton as a model, we reevaluate the involvement of the master cartilage regulator Sox9 in shaping facial condensations and find it to be largely dispensable. We then use new lineage-tracing tools to definitively show that precartilaginous condensations originate from neighboring clusters of cells termed mesenchymal condensations. These cartilage-generating mesenchymal condensations express a cohort of transcription factors that are also expressed in odontogenic mesenchyme in mammals, including barx1, lhx6a/8a, and pax9. We hypothesized that the position of each mesenchymal condensation determines the axis of growth of its corresponding precartilaginous condensation, thus influencing its final shape. Consistent with this idea, we find that positive Fgf and inhibitory Jagged-Notch signals intersect to precisely position a mesenchymal condensation in the dorsal half of the second pharyngeal arch, with loss of pathway function leading to predictable shape changes in the resulting cartilage element. Deciphering the full array of signals that control the spatial distribution of mesenchymal condensations and regulate their maturation into precartilaginous condensations thus offers a promising approach for understanding the origins of skeletal form.
Keywords: cartilage, condensation, barx1, sox9, zebrafish, pharyngeal arches
Graphical Abstract
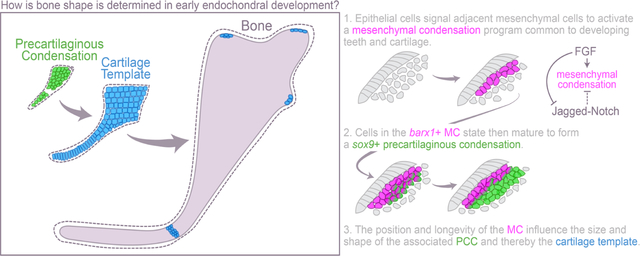
Introduction
Most of the bones in the vertebrate skeleton originate as cartilage templates that are later replaced by bone in a process called endochondral ossification. The first steps involve mesenchymal skeletal progenitor cells aggregating into precartilaginous condensations (PCCs) and then maturing into bona fide chondrocytes (Giffin et al., 2019). These cartilaginous structures later undergo hypertrophy and are invaded by blood vessels and osteoblasts that eventually convert the cartilage template into bone (Hall and Miyake, 2000). Adult endochondral bones resemble much larger versions of their initial cartilage templates, which in turn mimic the shape of their generative PCCs (Fig. 1A–C) (Karsenty, 2003; Kimmel et al., 1998; Mariani and Martin, 2003). How PCCs are shaped prior to their near-isometric growth is therefore fundamental to understanding the basis of skeletal form.
Fig. 1. Ontogeny of cartilage condensations in the pharyngeal arches.
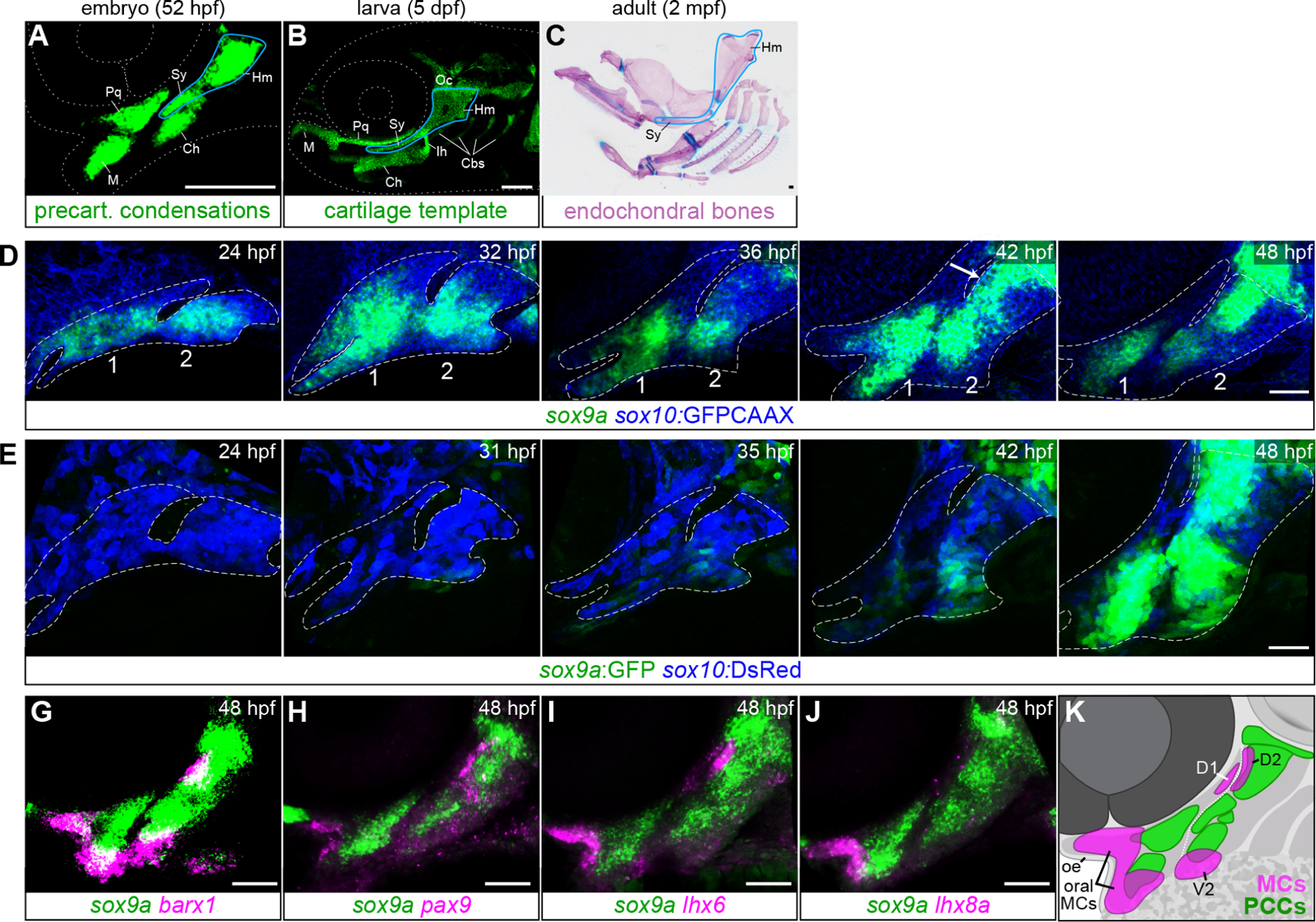
A-C, Mature endochondral bones (C) are prefigured by their cartilage templates (B), which in turn resemble their respective precartilaginous condensations (A). As an example, the hyosymplectic (a fusion of the hyomandibula (Hm) and symplectic (Sy) elements) is outlined in blue at each stage. Ch, ceratohyal; Ih, interhyal; M, Meckel’s cartilage; Oc, otic cartilage; Pq, palatoquadrate cartilage. A, Precartilaginous condensations in the embryonic face revealed by sox9a in situ hybridization. B, Facial cartilages at 5 dpf in a transgenic sox10:DsRed larva. C, Dissected endochondral bones of the adult zebrafish facial skeleton stained with Alizarin red and Alcian blue. Some bones removed for clarity. Scale bars in A-C = 100 μm. D, sox9a mRNA expression in pharyngeal arches 1 and 2 becomes gradually restricted into the PCC pattern between 24 and 48 hpf. sox10:GFPCAAX (blue) labels all CNCCs. White arrow points to de novo sox9a expression in the dorsal second arch. E, The sox9a:GFP transgene (green) is not active in early skeletal progenitors but is turned on robustly in PCCs by 48 hpf. sox10:DsRed (blue) marks all CNCCs. Dashed lines in D and E show arch boundaries. G-J, In situ hybridizations showing offset expression of mesenchymal condensation markers barx1, pax9, lhx6, and lhx8a (magenta) relative to sox9a (green) at 48 hpf. K, Schematic illustrating offset MC positions relative to PCCs at 48 hpf. D1, first arch dorsal posterior MC; D2, second arch dorsal anterior MC; V2, second arch ventral MC. Dotted white line delineates the boundary between arches 1 and 2. Confocal images are maximum intensity projections. Scale bars in D-J = 50 μm.
PCCs are defined as regions of elevated cell density within a field of mesenchymal progenitors (Fell, 1925) that must reach a critical size before initiating differentiation into cartilage (Hall and Miyake, 2000). Mesenchymal progenitors that do not condense but instead fill the spaces between the condensations have a variety of fates: some undergo apoptosis, some remain on the periphery of the cartilage as perichondral progenitor cells, while others give rise to other connective tissue cell types, including intramembranous bone, tendon, or ligament. Hall (Hall, 2015; Hall and Miyake, 2000) proposed several morphogenetic mechanisms by which condensations might form and grow, including differential adhesion, proliferation, and migration. However, how any such process is spatially deployed to produce the characteristic distribution of condensations in different parts of the body is not clearly understood. In the pharyngeal arches, which give rise to the facial skeleton, many genes are known to play some role in regulating skeletal form, but they are generally expressed in stripes or patches that bear little to no resemblance to the shapes or distribution of facial PCCs (e.g., Talbot et al., 2010).
Chick and mouse limb PCCs are enriched for cell adhesion molecules NCAM1 (encoding Neural Cell Adhesion Molecule 1), CDH2 (N-Cadherin), fibronectin, and tenascin-syndecan (Hall and Miyake, 2000) and react with the lectin peanut agglutinin (Miyake et al., 1996). PCCs also express the transcription factor SOX9 (Ng et al., 1997). SOX9 initiates chondrocyte differentiation in PCCs by activating transcription of archetypal cartilage extracellular matrix genes COL2A1 (Bell et al., 1997), ACAN (Aggrecan) (Sekiya et al., 2000), and MATN1 (Matrilin1) (Nagy et al., 2011; Rentsendorj et al., 2005), among others, and drives the condensation to mature into one or more separate cartilages. FOXC1 and other forkhead family transcription factors work in parallel with SOX9 to promote the chromatin accessibility of cartilage-associated enhancers in PCCs, facilitating SOX9-mediated transactivation of target gene expression (Xu et al., 2018; Xu et al., 2021).
Because of its essential role in chondrogenesis, understanding how SOX9 expression is spatially regulated in skeletal progenitor fields might provide an explanation for how PCCs are shaped. SOX9 is positively regulated by the Hedgehog, fibroblast growth factor (FGF), transforming growth factor-β (TGF-β), and bone morphogenetic protein (BMP) signaling pathways in the context of cartilage development (e.g., Lefebvre et al., 2019; Song and Park, 2020; Zinck et al., 2021). However, these associations are stage- and site-dependent, and no unifying pathway for upregulation of SOX9 in skeletogenic mesenchyme has been confirmed. Furthermore, whether SOX9 is even required for the formation of all PCCs is controversial. Mice with Prrx1:Cre-mediated conditional inactivation of Sox9 in limb mesenchyme did not develop discernable PNA-positive condensations in their limb buds (Akiyama et al., 2002). However, sox9a and sox9a; sox9b mutant zebrafish were reported to have histologically evident PCCs in the pharyngeal arches before cartilage differentiation halted (Yan et al., 2002; Yan et al., 2005). This discrepancy between models might be attributable to the different detection methods, or even to the scale or location of the structures assessed: the larval zebrafish facial skeleton is much smaller than the mouse limb and built from correspondingly small condensations. Alternatively, the zebrafish sox9a mutant alleles may be genetically compensated to some degree, as the mRNAs for both mutants used in the original study (Yan et al., 2002) are improperly spliced and subjected to nonsense-mediated decay, which can trigger transcriptional adaptation by related genes (El-Brolosy et al., 2019).
Another approach to understanding how PCCs are shaped may be rooted in a different class of genes that mark mesenchymal condensations (MCs; also known as precondensations (Giffin et al., 2019)). This is a general term referring to the condensations that appear early in the development of many different organs, when mesenchymal cells first come together and activate the expression of transcription factors related to their eventual cell fate (Giffin et al., 2019; Mammoto et al., 2011). One of the best-studied types of MC are those that form teeth. Here, secreted signals from the oral epithelium such as FGF8, BMP4, and SHH induce underlying neural crest-derived mesenchymal cells of the first pharyngeal arch to cluster and turn on odontogenic transcription factors such as Barx1, Lhx6, Lhx8, Msx1, Pitx1, and Pax9 (Zhang et al., 2005). These genes positively reinforce each other’s expression (Cesario et al., 2021), and mutations result in absent or malformed teeth (Denaxa et al., 2009; Miletich et al., 2011; Mitsiadis and Drouin, 2008; Peters et al., 1998). Interestingly, however, this gene signature also appears in other parts of the pharyngeal arches that do not give rise to teeth. In fact, in zebrafish, because teeth only form on the seventh pharyngeal arch (i.e., not on the first arch-derived jaws), most fish facial MC expression domains are not associated with tooth development. Whether these non-dental MCs instead give rise to cartilage appears likely but has not been directly tested. Cartilage anomalies are apparent in some mouse and zebrafish models with MC gene loss-of-function (Lanctot et al., 1999; Nichols et al., 2013; Peters et al., 1998; Sperber and Dawid, 2008; Swartz et al., 2011; Szeto et al., 1999), though none show a complete blockade of cartilage differentiation as occurs with loss of Sox9/sox9a. The mouse Dlx1/2 mutant phenotype supports related developmental pathways for tooth and cartilage, as disrupted pharyngeal arch patterning in these mice causes the upper molars to transform into nodules of cartilage (Qiu et al., 1997; Thomas et al., 1997).
As in developing teeth, expression of MC genes in non-dental populations of facial mesenchyme is activated by FGFs secreted from neighboring ectodermal or endodermal epithelia (Mandler and Neubuser, 2001) and further refined by input from other local signals like Endothelin-1 and Jagged-Notch (Barske et al., 2016; Walker et al., 2006). We thus have some understanding of how non-dental MC domains are positioned within the pharyngeal arches. This is particularly true in zebrafish where the relatively small number of cells involved facilitates high-resolution analyses of skeletal patterning and development (Mork and Crump, 2015). However, because we have not yet verified that these non-dental MCs mature into PCCs, the degree to which MC spatial patterning informs PCC, cartilage, and ultimately endochondral bone shape is unclear. Here, using a new knockin zebrafish line that tracks barx1-expressing cells, we perform a series of lineage-tracing studies to show that MCs do contribute to PCCs and eventually the cartilage template of the facial skeleton. We then explore in detail how a particular MC is positioned by a complex interplay of pro-condensation epithelial Fgf signals and anti-condensation Jagged-Notch signals from within the field of skeletal progenitors itself. Understanding how MCs form where they do and how they mature into PCCs may thus provide answers to fundamental questions about skeletal form.
Material and Methods
The Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center (Nos. 2018-0076 and 2021-0048) approved all the animal procedures carried out in this study.
Zebrafish lines and genotyping
Zebrafish (Danio rerio) embryos were raised in embryo medium at 28.5°C following standard procedures (Westerfield, 2007) and staged as described (Kimmel et al., 1995). Existing mutant and transgenic lines used in this study were maintained as heterozygotes and include barx1fh331 (Nichols et al., 2013), jag1bb1105 (Zuniga et al., 2010), notch2el515 (Barske et al., 2016), notch3fh332 (Alunni et al., 2013), sox9ahi1134Tg (Amsterdam et al., 1999; Yan et al., 2002), sox9azc81Tg (aka sox9a:GFP) (Bonkowsky and Chien, 2005; Eames et al., 2013), Tg(sox10:Gal4VP16)el159, Tg(sox10:DsRedExpress)el10, Tg(UAS:DnFgfr1a)el28, and Tg(UAS:kikGR)el377 (Das and Crump, 2012); Tg(nkx2.3:Gal4VP16)el93 (Choe et al., 2013); Tg(fli1a:Gal4VP16)el360 (Xu et al., 2018); Tg(col2a1a-R2:Gal4VP16)el647 (Barske et al., 2018); Tg(UAS:nlsGFP)el609, Tg(RUNX2:mCherry) (Barske et al., 2020); Tg(sox10:GFPCAAX) (Askary et al., 2015), Tg(scxa:mCherry)fb301 (McGurk et al., 2017); Tg(dusp6:d2GFP)pt6 (Molina et al., 2007); and Tg(UAS:DnMAML)el481, and Tg(UAS:mCherryCAAX)el597 (kind gifts from G. Crump at USC). To maintain transgenic lines, larvae expressing fluorescent reporter or marker proteins were selected under a fluorescent stereomicroscope at 5 dpf and raised to adulthood. To identify mutant carriers, the caudal fin was biopsied under tricaine anesthesia (Western Chemicals) at two weeks or three months post-fertilization, and the sample was lysed and digested with proteinase K. The barx1fh331, jag1bb1105, notch2el515, and notch3fh332 lines were genotyped as described (Alunni et al., 2013; Barske et al., 2016; Nichols et al., 2013; Zuniga et al., 2010). The sox9ahi1134Tg mutant line was genotyped with the following primers: sox9a_Hi1134_3E04_F: 5’-ACCATCAGATGTTTCCAGGGTG-3’, sox9a_Hi1134_3E03_R: 5′-AAGGGACGCTTTTCCACCTC-3′ and sox9a_t2a_wtF: 5′-GGCACTGAGAGTTTTCTGCATCTG-3′, which yield a 226-bp product for the wild-type allele and a 315-bp product for the insertion allele. The sox10:Gal4VP16 line lacks a selectable marker and was genotyped with Gal4 primers (F: 5′-CTCCCAAAACCAAAAGGTCTCC-3′; R: 5′-TGAAGCCAATCTATCTGTGACGG-3′).
Generation of new zebrafish lines
The barx1Gal4ff-ci3030 targeted knockin line (Fig. S1A) was made using a CRISPR/Cas9-non-homology-based protocol (Kimura et al., 2014). Briefly, two sgRNAs targeting sequences in the barx1 5’UTR (5’-GCTTTCATCAGGCTACCAGG-3’ and 5’-GGTGCGGTAAGAACAGAAAC-3’) were co-injected at 100 ng/μl into Tg(UAS:nlsGFP) embryos together with Cas9 RNA (100 ng/μl), a circular hsp70l:Gal4ff:pA construct, and a third gRNA targeting a bait sequence within the construct to linearize the plasmid in vivo (Kimura et al., 2014). Founders were identified by crossing to Tg(UAS:nlsGFP) at maturity and screening F1 offspring for GFP patterns that recapitulated the endogenous barx1 expression pattern.
Drug treatments and heat shocks
hsp70l:Gal4; UAS:DnFgfr1a and control siblings were heat-shocked for 2–3 hours between 20 and 24 hpf, as indicated, in a 40°C incubator, returned to 28.5°C, then fixed at 36 hpf. For combined Fgf and Notch inhibition, Gal4 expression was first induced in hsp70l:Gal4; UAS:DnFgfr1a and control siblings by heat-shocking between 20:30–23:30 or 21–24 hpf in a 40°C incubator. Embryos were then dechorionated, returned to 28.5°C, and incubated in 6.67 or 10 μM dibenzazepine (DBZ; Tocris #4489) diluted in embryo medium until fixation at 36 hpf as described (Barske et al., 2016). DMSO-only controls were performed in parallel. This experiment was repeated twice.
In situ hybridizations, immunostaining, and skeletal staining
Single- or dual-color fluorescent in situ hybridizations were carried out as previously detailed (Barske et al., 2018). Published probes used in this study include barx1 (Barske et al., 2016), dlx2a (Akimenko et al., 1994), jag1b (Zuniga et al., 2010), and sox9a (Yan et al., 2002). The fgfr2 probe was a gift from S. Paul (UCLA). Partial cDNAs for cdh2, lhx6, lhx8a, ncam1a, and pax9 were amplified and cloned into pCRBlunt-II-TOPO (Life Technologies), then sequence-verified, linearized, and used as templates for in vitro transcription with Sp6 or T7 polymerase (Roche) (Table S1). Immunostaining was performed separately or following in situ hybridization, using chick anti-GFP (Abcam ab13970, 1:300), anti-phospho-Histone H3 (Ser10) (Sigma 06–570, 1:500), or anti-Alcama (DSHB Zn8, 1:2000) with Alexa dye-conjugated secondary antibodies (1:300, Thermofisher). Alcian Blue and Alizarin Red staining of larvae and adult facial skeletons was performed as described (Ullmann, 2011; Walker and Kimmel, 2007). For all mutant/transgenic analyses, a minimum of n = 2 or 3 individuals with the genotype in question were imaged and evaluated.
Imaging
Larval skeletons were imaged with a Zeiss AxioImager.Z1 compound microscope, and adults with a Zeiss StereoDiscovery V8. Transgenic or fluorescently stained embryos and larvae were imaged with a Nikon A1R inverted confocal and are presented as single optical sections or maximum intensity projections, as indicated. Live embryos and larvae were anesthetized in Tricaine in low-percentage agarose prior to imaging. Brightness and contrast were modified evenly across samples using Adobe Photoshop CC2019. Time-lapse imaging of barx1Gal4ff; UAS:kikGR; sox10:DsRed embryos was initiated at 48 hpf and continued for approximately 42 hours, using a 20x objective to capture a 144 μm μm z-stack every 12 minutes. barx1Gal4ff; UAS:mCherryCAAX; sox9a:GFP embryos were imaged for 23 hours starting at 36 hpf, with a 20x objective capturing a 56 μm z-stack every 12 minutes.
Data analysis
Colocalization of barx1Gal4ff; UAS:nlsGFP with sox10:DsRed expression in the hyomandibula, ceratohyal, and Meckel’s cartilages at 6 dpf was determined using the Spot Colocalization extension in Imaris (Bitplane). Four to six replicates were counted for each element. pHH3+GFP+ cells in the dorsal hyoid domain of barx1Gal4ff; UAS:nlsGFP or sox9a:GFP embryos (n = 9 and 7, respectively) were counted in NIS Elements software and compared using unpaired two-tailed t tests in GraphPad (Prism), with p < 0.05 deemed significant.
Results
Ontogeny of sox9a+ precartilaginous condensations in the developing zebrafish face
To study how precartilaginous condensations are positioned within the zebrafish pharyngeal arches, we first closely examined the spatiotemporal expression of the most robust PCC marker known in fish, sox9a. At least three waves of sox9a expression occur in the zebrafish cranial neural crest (CNC) lineage that gives rise to the facial skeleton. Expression is present in premigratory crest (wave 1; ~11.5 hpf) (Zhao et al., 2014), absent during migration (~14–16 hpf) (McKeown et al., 2005; Yan et al., 2005), then reactivated in the subset of cells that reach the pharyngeal region and adopt a skeletogenic fate (wave 2; ~20 hpf) (Fig. S2A–B). Broad expression throughout the arches is gradually concentrated by 36 hpf into limited intermediate and ventral arch regions (Fig. 1D). Foci within these regions intensify in expression, and a new domain in the dorsal second arch appears by 42 hpf (wave 3). This rough approximation of the future cartilage pattern is refined over the next ten hours to yield the recognizable pattern of PCCs prefiguring both dorsal and ventral facial cartilages (Fig. 1A,D). This third wave of expression is recapitulated by the sox9azc81tg enhancer trap line (hereafter sox9a:GFP), which inserted approximately 120 kb upstream of the sox9a transcription start site (Bonkowsky and Chien, 2005; Eames et al., 2013). The earliest reported expression of this line in the CNC is at 34.5 hpf (Talbot et al., 2012). We observed GFP expression in the otic placode by 16 hpf but no credible GFP in premigratory (11.5 hpf) or migratory (16 hpf) crest or skeletogenic precursors (20 hpf; Fig. S2C). GFP expression was first detected in intermediate/ventral regions at approximately 36 hpf (Fig. 1E) and mimics endogenous sox9a by 48 hpf. In line with their early enrichment of sox9a expression, intermediate- and ventrally-located PCCs are the first to differentiate into chondrocytes at approximately 56 hpf, when they begin to upregulate col2a1a and sox10 and produce Alcian blue-reactive sulfated glycosaminoglycans (Barske et al., 2016; Schilling and Kimmel, 1997). The third wave of PCC-associated sox9a expression appears to be the most critical for facial development, as sox9a mutants were reported to be indistinguishable from sibling controls until chondrocyte differentiation stalls after 54 hpf (Yan et al., 2002).
We next compared sox9a expression at 48 hpf to that of the putative MC genes barx1, lhx6, lhx8a, and pax9. Though all four genes are expressed in the mesenchyme of the first pharyngeal arch immediately under the oral epithelium (‘oral MCs’ in Fig. 1K; also see Askary et al. (2017) for pitx1), only barx1, lhx6 and pax9 are also present in the dorsal anterior second arch (‘D2’), just barx1 and lhx6 are expressed in the ventral second arch MC (‘V2’), and only lhx6 is present in the dorsal posterior first arch (‘D1’)(Fig. 1G–J). These MC foci are offset from sox9a+ PCCs (Fig. 1G–K) (also see Barske et al., 2016), again raising the question of how and whether the two cell populations are related.
Facial condensation markers are not overtly affected in sox9a mutants
In an effort to substantiate the report that histologically defined PCCs still form in the absence of sox9a in zebrafish (Yan et al., 2002), we examined the expression of fish homologs of canonical amniote limb PCC markers SOX9, CDH2, NCAM1, and FGFR2 in the sox9ahi1134Tg mutant line (Amsterdam et al., 1999). sox9a itself was still present at 48 hpf (Fig. 2A), albeit at reduced intensity, as previously noted at other stages (Yan et al., 2002). This reduction might be attributable to failed autoregulation, nonsense-mediated decay, or reduced probe affinity for the mutant transcript. cdh2 and ncam1a proved less informative, as they were not enriched in wild-type zebrafish PCCs at 48 hpf (Fig. S3A–B). fgfr2 was only partially co-expressed with sox9a at 48 hpf (Fig. S3C) but more closely matched the sox9a+ PCC pattern by 56 hpf (Fig. 2B). We noted absent or weakened fgfr2 expression in the Hm and Ch PCCs of sox9a mutants at this stage (Fig. 2B), but this may reflect the block in chondrocyte differentiation rather than defects in PCC formation. Nevertheless, the fact that sox9a is still expressed in its usual PCC pattern in sox9a mutants indicates that Sox9a is not itself required for setting up the pattern: however, determining whether mutant PCCs molecularly resemble wild-type PCCs will require development of a new tool for purifying mutant cells, as the sox9a:GFP transgene is linked to the wild-type sox9a locus.
Fig. 2. Facial condensation markers are not appreciably affected in sox9a mutants.
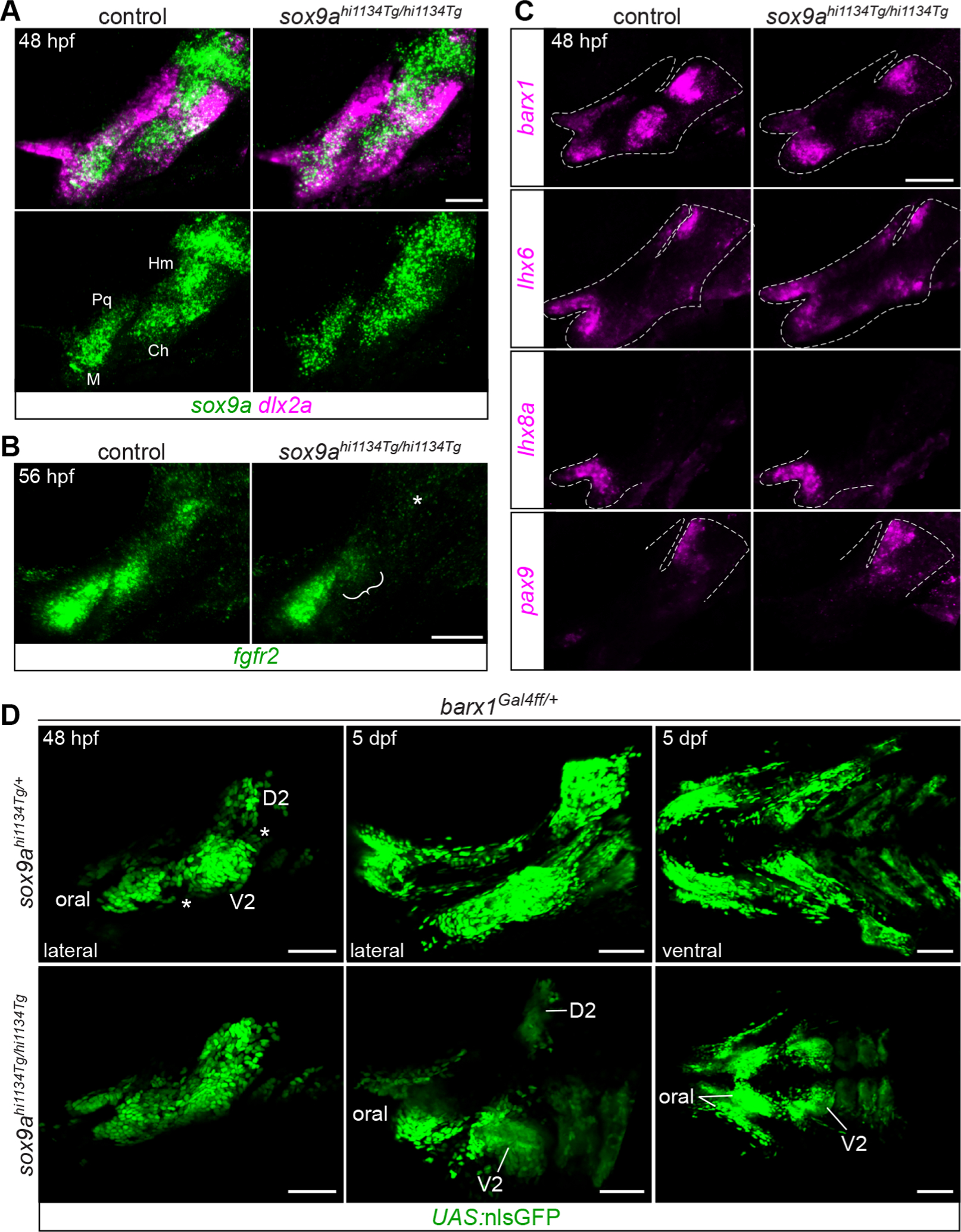
A, sox9a is expressed in its typical PCC pattern in sox9a mutants, though more diffusely (n = 3/3 mutants examined). dlx2a marks undifferentiated skeletal progenitors. PCCs labeled as in Fig. 1. B, fgfr2 expression is weaker in the ceratohyal PCC (bracket) and absent in the hyomandibula PCC (asterisk) in sox9a mutants at 56 hpf (n = 2/2). C, MC markers barx1, lhx6, lhx8a, and pax9 are expressed normally in sox9a mutants at 48 hpf (n = 3/3, 2/2, 3/3, 2/2, respectively). Dashed lines indicate approximate arch boundaries. D, Live imaging of GFP-expressing barx1-lineage cells in sox9a mutants revealed less well-separated MCs at 48 hpf (gaps in control marked by asterisks) and a massive reduction in GFP+ cells by 6 dpf. Both lateral and views are presented. MC labels as in Fig. 1. Images are maximum intensity projections. Scale bars A-C = 50 μm, D = 100 μm.
Expression of barx1, lhx6, lhx8a, and pax9 was unaffected in sox9a mutants at 48 hpf (Fig. 2C). This result supports the model that their expression is independent of Sox9a function and marking an earlier stage of pre-skeletal condensation and differentiation, similar to results for the earlier skeletal progenitor markers Twist1 and Prrx1 in Sox9 mutant mouse limbs (Liu et al., 2018).
Fate of barx1+ cells in the zebrafish facial skeleton
As a means of determining the fate of cells expressing MC genes, we elected to make a Gal4 driver line for barx1, which was the most robustly expressed of the MC genes assayed. Several previous studies had proposed that barx1+ or barx2+ cells become cartilage (Barlow et al., 1999; Lorda-Diez et al., 2011; Meech et al., 2005; Meulemans and Bronner-Fraser, 2007; Sperber and Dawid, 2008), but definitive evidence was lacking. We used a non-homology-based plasmid knockin method (Kimura et al., 2014) to insert the Gal4ff sequence into the barx1 5’UTR, creating the barx1ci3030 allele (hereafter barx1Gal4ff; Fig. S1A). This driver recapitulates endogenous barx1 expression at 36 and 48 hpf when crossed to a UAS:nlsGFP reporter (Fig. S1B–C). The knockin is a hypomorphic allele with a less pronounced craniofacial phenotype when homozygosed than the barx1fh331 TILLING mutant (Fig. S1D) (Nichols et al., 2013). Though less definitive than a Cre-based lineage-tracing system, this Gal4-based method exploits the long half-life of GFP to identify cells that have recently passed through a barx1+ state.
To test the hypothesis that barx1-expressing MC cells give rise to facial cartilages, we performed static and live time-lapse imaging of barx1Gal4ff/+embryos carrying green fluorescent UAS:nlsGFP or UAS:kikGR reporters and the sox10:DsRed transgene between 48 hpf and 6 dpf (Fig. 3A–B, S4; Movies S1–S4). The sox10:DsRed transgene is first expressed in migrating neural crest then robustly re-activated in differentiating chondrocytes starting at 56 hpf (Barske et al., 2016). This analysis revealed two populations of sox10:DsRed+ chondrocytes: one that clearly emerged from within clusters of green fluorescent cells (simplified to GFP hereafter) and one that seemed to arise de novo from GFP− precursors. The DsRed+GFP+ population included most of the lower jaw cartilage (Meckel’s; Movie S2) and part of the palatoquadrate (Pq) in the first arch, most of the ceratohyal (Movie S3) and hyomandibula (Movie S4) in the second arch, and the ceratobranchial (Cb) cartilages in the posterior arches. In contrast, DsRed+GFP− chondrocytes were observed in the jaw joint region between Meckel’s and Pq, the symplectic cartilage (Sy, which fuses with the hyomandibula to form the hyosymplectic), the interhyal joint between the hyomandibula and ceratohyal, the ventroposterior edge of Pq, the otic cartilage, the midline ventral cartilages, and the medial tips of Meckel’s and the ceratohyal. Colocalization between nlsGFP and DsRed expression in the hyomandibula, ceratohyal, and Meckel’s cartilages at 6 dpf was estimated at 33, 42, and 32%, respectively (Fig. 3C), reflecting the uneven distribution of barx1-lineage cells in these cartilages.
Fig. 3. barx1-lineage cells give rise to facial cartilage.
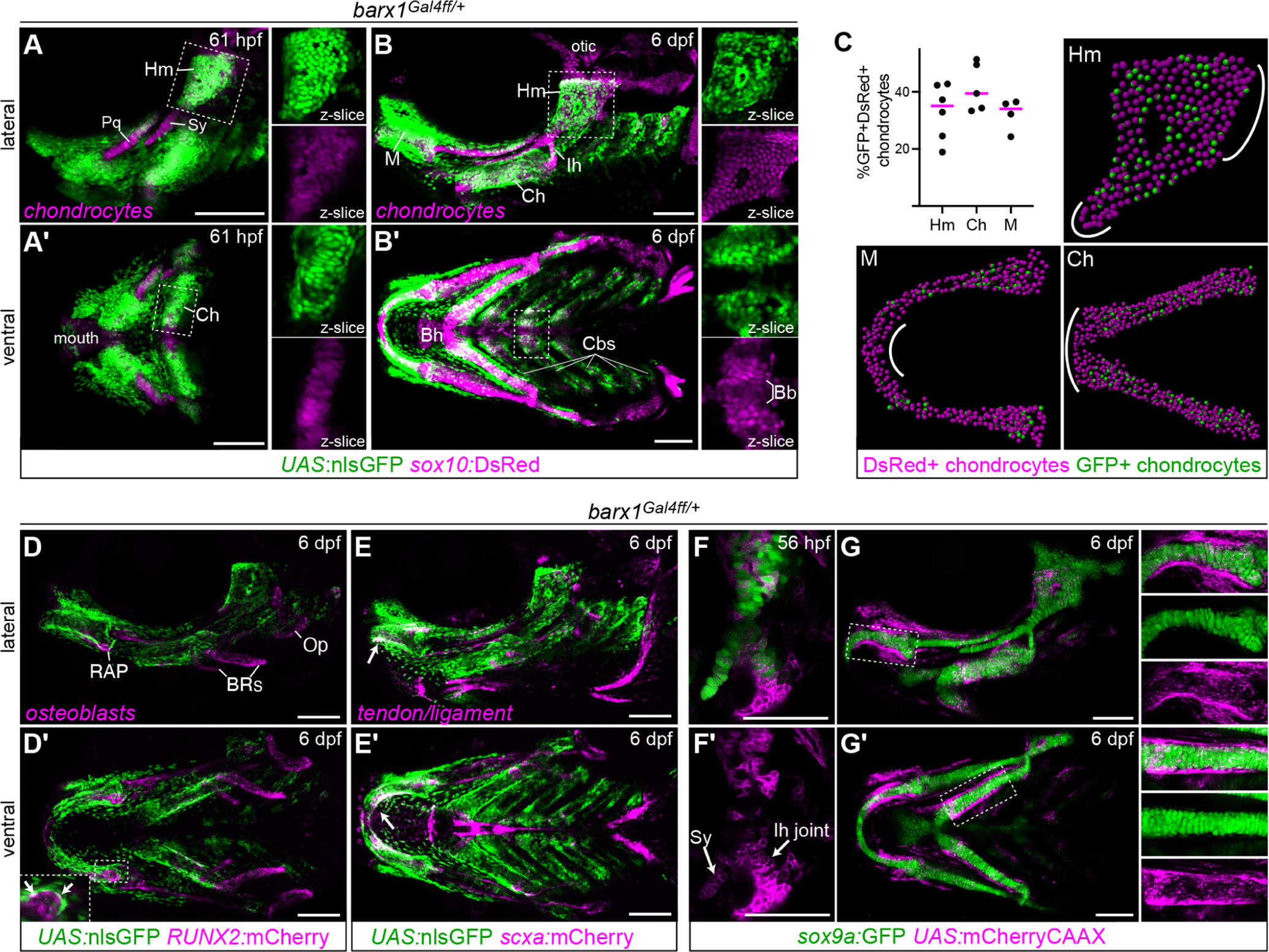
A-B, Live imaging of barx1Gal4ff; UAS:nlsGFP; sox10:DsRed embryos at 61 hpf and 6 dpf reveals colocalization of barx1-lineage GFP+ cells with DsRed+ chondrocytes in most facial cartilages. Enlarged single-channel z-slice views of the boxed regions are shown to the right of each main panel, illustrating the high degree of GFP/DsRed colocalization in the Hm and Ch cartilages and the absence of GFP in the midline basibranchial cartilage (Bb). GFP expression is also present in perichondral cells and other unidentified mesenchyme. Bh, basihyal; other labels as in the Fig. 1 legend. C, Colocalization analysis between barx1Gal4ff; UAS:nlsGFP and sox10:DsRed at 6 dpf. Graph shows the percentage of double-positive chondrocytes among all chondrocytes counted in the given element. Four to six biological replicates were counted for each element using Imaris. Representative spot-transformed images are shown, with areas depleted for GFP+ cells indicated with white lines. D, Minimal colocalization between GFP and RUNX2:mCherry indicates that most RUNX2+ intramembranous bone osteoblasts derive from barx1-negative precursors or turned barx1 off very early in arch development. Double-positive cells were occasionally observed in the perichondrium (arrows in inset) and likely correspond to perichondral osteoblasts engaged in the early stages of perichondral ossification. Op, opercle bone; BRs, branchiostegal ray bones; RAP, retroarticular process. E, Minimal colocalization between GFP and scxa:mCherry indicates that most tendon/ligament cells are also not recently derived from barx1lineage cells. The major exception was the GFP+ tenocytes lining the posterior rim of Meckel’s cartilage (white arrows). Lateral views shown in top panels; ventral views in A’-D’. F, In barx1Gal4ff; UAS:mCherryCAAX embryos, mCherry expression is detectable in the Sy and Ih joint regions at the onset of definitive chondrogenesis at 56 hpf. G, At 6 dpf, barx1-driven mCherry expression is largely excluded from differentiated sox9a:GFP+ chondrocytes but apparent in the perichondrium. Enlarged single-channel projection views of the boxed regions are shown to the right of each main panel, showing minimal mCherry expression within GFP+ chondrocytes. Images are maximum intensity projections (mips) unless otherwise noted. Scale bars = 100 μm.
We next investigated whether barx1-lineage cells also contribute to any of the other major skeletal cell types present at 6 dpf. GFP+ cells were abundant in the perichondrium of GFP+ cartilages, the nascent gill filaments, and scattered mesenchymal cells between skeletal elements (Fig. 3B, D–E). To specifically test for contributions to bone and ligament/tendon lineages, we separately crossed barx1Gal4ff; UAS:nlsGFP fish to RUNX2:mCherry and scxa:mCherry transgenic fish and imaged their offspring at 6 dpf. Intramembranous bone osteoblasts expressing RUNX2:mCherry were consistently GFP− (Fig. 3D, S4). RUNX2:mCherry+ cells within the perichondrium were occasionally GFP+, indicating that some osteoblasts mediating perichondral ossification pass through a barx1+ state. Connective tissues marked by scxa:mCherry were nearly all GFP− (Fig. 3E, S4), with the exception of the tenocytes that line the inner edge of Meckel’s cartilage and connect the lower jaw to the intermandibularis muscles (Chen and Galloway, 2014). These analyses demonstrate that barx1-lineage cells extensively contribute to the cartilaginous facial skeleton and structures that form within the perichondrium, but less overtly to other skeletal derivatives. The caveat to the negative results is that we cannot readily distinguish GFP− cells that never expressed barx1 from those that expressed it early but have since degraded all their GFP protein.
barx1+ cells activate sox9a expression as they mature into precartilaginous condensations
To more closely examine how barx1+ MCs transition into PCCs before becoming cartilage, we next created barx1Gal4ff; UAS:mCherryCAAX; sox9a:GFP embryos. Expression from this GFP enhancer trap line initiates earlier than sox10:DsRed (>42 hpf vs. >56 hpf), in PCCs rather than differentiating chondrocytes. Live imaging starting at ~36 hpf showed sox9a:GFP expression initiating in mCherry+ cells (Movie S5) consistent with barx1+ MCs maturing into sox9a+ PCCs. We also observed that sox9a:GFP+ cells in the PCCs corresponding to the future palatoquadrate (Pq), symplectic (Sy), and interhyal joint cartilages were mCherry+ at 56 hpf (Fig. 3F). This contradicted the barx1Gal4ff; UAS:nlsGFP; sox10:DsRed results, where Pq, Sy and interhyal DsRed+ chondrocytes emerged from nlsGFP− domains. One possible explanation is that because Pq and Sy chondrocytes are among the first in the face to fully differentiate (Schilling and Kimmel, 1997), their progenitors may have expressed barx1 at early stages but shut the gene off and turned over their GFP protein by the time sox10:DsRed begins to turn on in differentiating chondrocytes. Interhyal joint progenitors may likewise shut down barx1 expression early to facilitate joint formation. By 6 dpf, barx1-driven mCherry expression is limited to perichondral cells and excluded from mature chondrocytes (Fig. 3G). This indicates that mCherryCAAX turns over more rapidly than nlsGFP, and that the nlsGFP observed in chondrocytes at this stage in barx1Gal4ff; UAS:nlsGFP larvae (Fig. 3B) largely derives from protein perdurance rather than continued protein production.
To substantiate this finding, we created barx1Gal4ff; UAS:kikGR embryos and photoconverted all the kikGR protein in the first two arches using a UV laser at 50 or 72 hpf (Fig. S5). They were then reimaged at 6 dpf. Green fluorescence at this stage represents new kikGR protein synthesized after 50 or 72 hpf, indicative of continued Gal4 transactivation of the kikGR transgene. Green fluorescence was strongest in perichondral and mesenchymal cells (see single-channel z-slice images of Meckel’s cartilage in Fig. S5), though some green chondrocytes were observed in the Hm cartilage, particularly in the larvae exposed to UV at 50 hpf. This supports that Gal4 activity declines after 50 hpf as cells differentiate, but persists longer in cartilages like the Hm that differentiate later (Barske et al., 2016). Conversely, red but no green protein was observed in the Ih joint region in the embryos converted at 50 hpf, and even minimal red in the embryos converted at 72 hpf. This finding aligns with the conclusion that cells in the Ih region are among the first to turn off barx1 expression.
To determine what happens to barx1+ MCs when chondrocyte differentiation is blocked, we generated sox9a−/−; barx1Gal4ff; UAS:nlsGFP embryos and imaged at 48 hpf (before the mutant phenotype is apparent) and 5 dpf (Fig. 2D). Compared with sibling controls, GFP+ cells were more evenly distributed across the mutant arches at 48 hpf, with the typical GFP−gaps between MC foci less discernible. At 5 dpf, though no cartilage is present, GFP+ cells were detected at those sites enriched for GFP+ perichondrium in controls, specifically the anterior dorsal second arch D2 domain and in the oral region around the upper and lower jaws. A large GFP+ aggregate in the ventral second arch domain was also consistently observed (n = 8/8 mutants); cells in this region are for unknown reasons less susceptible to loss of sox9a and differentiate into small balls of cartilage on either side of the midline (Yan et al., 2002). Otherwise, the MC cells that would have normally differentiated into cartilage have disappeared by this stage, in contrast to the abundant GFP+ chondrocytes in controls.
Proliferation rates are low in both precartilaginous and mesenchymal condensations
Kimmel et al. (1998) observed minimal proliferation in cartilage-forming regions of the pharyngeal arches from 48–84 hpf (Kimmel et al., 1998), when PCCs are forming and maturing into chondrocytes. This finding, taken together with our data showing that barx1-expressing cells contribute to PCCs, indicates that PCCs might grow by incorporating MC cells rather than by intrinsic proliferation. To assess general proliferation trends in the two types of condensations, we quantified proliferation in the dorsal hyoid arch by phospho-Histone H3 immunostaining of barx1Gal4ff; UAS:nlsGFP and sox9a:GFP embryos at 48 hpf. Challenges in accurate detection of cell boundaries in the sox9a:GFP embryos precluded counting the total number of GFP+ cells; we therefore only determined the total number of proliferating GFP+ cells. Few pHH3+GFP+ double positive cells were observed in either line (mean ± std. error = 3.44 ± 0.77 and 3.86 ± 0.51, respectively, data not shown), indicating that neither type of cellular condensation is robustly proliferating at this transitional stage.
Positioning of mesenchymal condensations within the arches by Fgf signaling
We next returned to the question of how MCs are induced to form at precise spatial coordinates within the arches (Fig. 1K). Numerous signaling pathways regulate MC marker expression in facial skeletogenic cells. Given our findings that MCs give rise to PCCs and then to cartilage, we hypothesized that integration of these patterning signals determines the spatial distribution of MCs in the arches and in turn the ultimate pattern of PCCs and cartilage in the face. To dissect where these signals need to be received to achieve skeletal pattern, we used transgenic tools to block two of these pathways – Fgf and Notch – in specific facial cell populations and then evaluated cartilage patterning and MC gene expression.
In the mouse, Fgfs from the first arch epithelium activate Barx1, Lhx6/8, Pitx1, and Pax9 expression in the underlying odontogenic mesenchyme (Grigoriou et al., 1998; Mandler and Neubuser, 2001; St Amand et al., 2000; Tucker et al., 1999; Tucker et al., 1998). Previous studies similarly found that treatment of zebrafish embryos with the Fgf inhibitor su5402 blocked facial barx1 and lhx6a/8a expression (Jackman et al., 2004; Sperber and Dawid, 2008). We confirmed the effect on MC genes using the heat-shock inducible hsp70l:Gal4 driver to activate UAS:DnFgfr1a, which dominantly interferes with Fgf receptor activation when present at high levels (Amaya et al., 1991; Das and Crump, 2012). hsp70l:Gal4; UAS:DnFgfr1a embryos heat-shocked from 21–24 hpf showed normal expression of sox9a but reduced expression of barx1, lhx6, and pax9 at 36 hpf, with the anterior D2 domain that gives rise to the Hm cartilage appearing the most sensitive (Fig. 1K, 4A–D). This MC domain also specifically expresses the Fgf reporter dusp6:GFP starting at approximately 30 hpf (Fig. 4F), supporting that it may directly receive Fgf signaling.
Fig. 4. Intersection of Fgf and Notch signaling positions dorsal mesenchymal condensations.
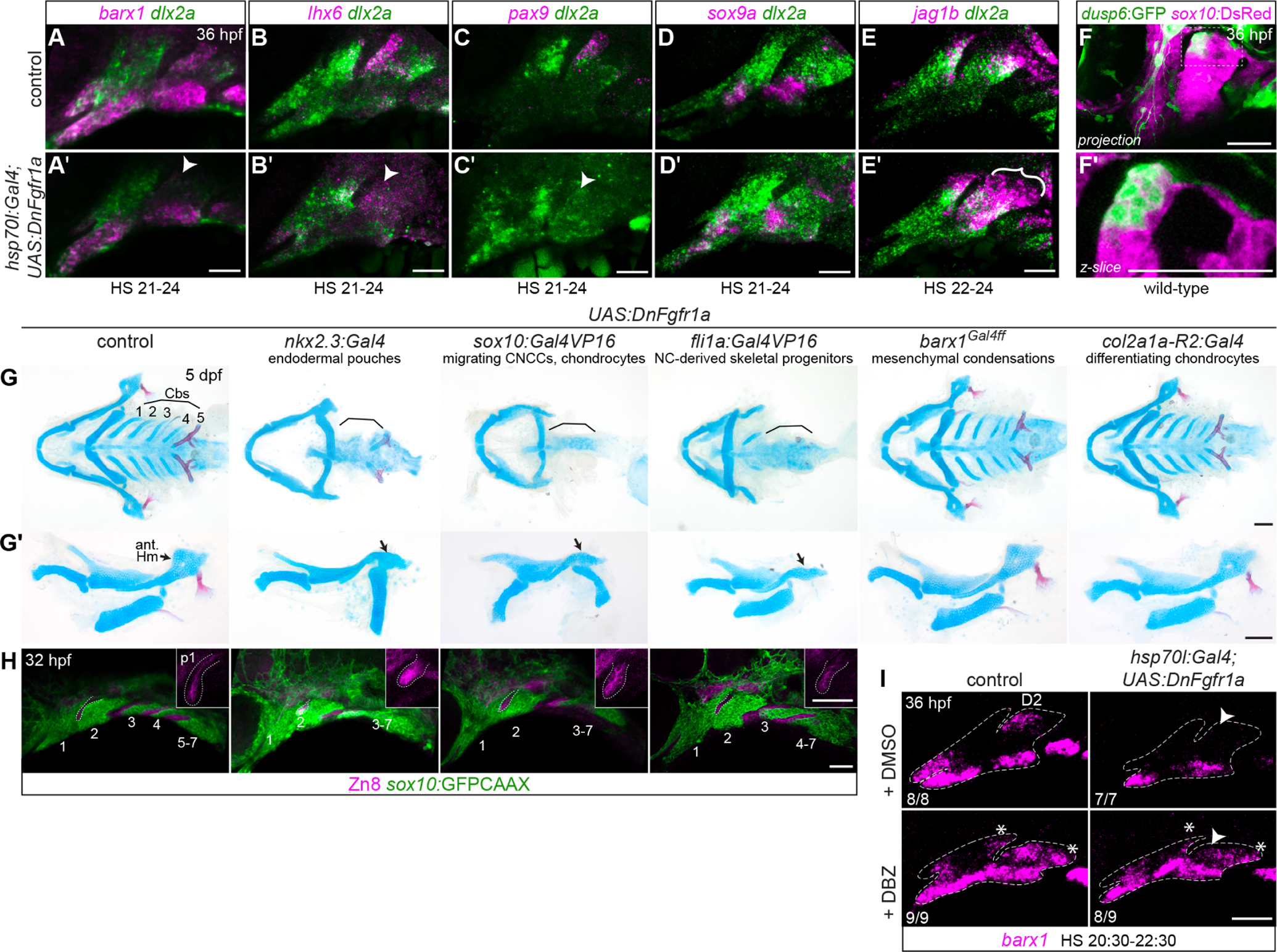
A-E, Effects of transient systemic inhibition of Fgf signaling on condensation-related gene expression. Transgenic hsp70l:Gal4; UAS:DnFgfr1a embryos were heat-shocked for the indicated durations and then fixed at 36 hpf. dlx2a (green) labels all arch NCCs. A-C, Expression of MC genes barx1, lhx6, and pax9 was consistently weakened in Fgf-inhibited embryos. Diminished expression in the D2 MC is noted with arrowheads. D, Pre-PCC sox9a expression is not appreciably affected by Fgf inhibition. E, jag1b expression expands ventrally and across the anterior-posterior axis of the dorsal second arch in Fgf-inhibited embryos. F, Enrichment of dusp6:GFP reporter expression in the anterior dorsal second arch. sox10:DsRed marks all CNC-derived cells at this stage. G, Expression of dominant-negative Fgfr1a under different cell-type specific Gal4 drivers affects facial cartilage development. Full viscerocrania are shown as ventral preparations in G; dissected mandibular and hyoid cartilages are shown from the lateral perspective in G’. Brackets highlight the missing Cbs; black arrows point to the anterior hyomandibula cartilage. H, Segmentation of the posterior arches is disrupted in DnFgfr1a transgenics that lose posterior arch cartilages. sox10:GFPCAAX marks NC-derived cells, and Zn8 immunostaining marks pharyngeal epithelia. Arches are numbered, and dotted lines show the position of the first pharyngeal pouch. Insets are enlarged single optical sections of the first pouch (p1) stained with Zn8. I, Epistatic effects of Fgf and Notch signaling on dorsal barx1 expression. When Fgf signaling is inhibited, the barx1 D2 domain is lost (arrowhead, top right panel); when Notch is inhibited with the DBZ inhibitor, barx1 is ectopically expressed in dorsal posterior cells (asterisks, bottom left); when both pathways are inhibited, the D2 domain does not recover, but ectopic dorsal posterior expression is present (bottom right). Ratios refer to the number of embryos with the presented phenotype in each group, pooled from two independent iterations of this experiment. Dashed lines indicate approximate arch boundaries. Scale bars in A-E, G, H = 50 μm; in F = 100 μm.
General Fgf inhibition also impairs formation of the facial cartilages (Crump et al., 2004a; David et al., 2002; Larbuisson et al., 2013). However, these phenotypes reflect the sum of multiple requirements for Fgf signaling in facial development: in addition to regulating MC formation, Fgfs are required earlier for facial skeletal progenitors to be correctly specified (Blentic et al., 2008; Das and Crump, 2012) and for pouching of the pharyngeal endoderm (Choe and Crump, 2014; Lovely et al., 2016). Formation of the first pouch requires Fgf signaling between 10–14 hpf; when the pathway is inhibited at later stages, the first pouch is still apparent but posterior pouches do not form, the third CNC stream does not subdivide into arches 3–7, and the gill Cb cartilages do not form (Crump et al., 2004a). To narrow down the key cell types that require Fgf reception for condensation and cartilage formation, we crossed the UAS:DnFgfr1a line to cell-type specific drivers nkx2.3:Gal4VP16 (endodermal pouches) (Choe et al., 2013), sox10:Gal4VP16 (migrating neural crest and differentiating chondrocytes) (Das and Crump, 2012), fli1a:Gal4VP16 (NC-derived skeletal progenitors) (Xu et al., 2018), barx1Gal4ff (mesenchymal condensations), and col2a1a:Gal4VP16 (differentiating chondrocytes).
Larvae carrying UAS:DnFgfr1a with the nkx2.3, sox10, or fli1a driver each lost the anterior Hm and Cb cartilages (with the occasional exception of ceratobranchial 5) (Fig. 4G–G´). Zn8 staining of pharyngeal epithelia revealed that pouching of posterior endoderm was disrupted (Fig. 4H), resulting in incomplete segmentation of the third CNC stream and failure of Cb formation. A more severe version of this phenotype was reported for fgfr1a; fgfr1b mutants, which lose the Cbs as well as the entire Hm cartilage (Leerberg et al., 2019). Loss of the anterior Hm was also observed in integrin-α5 mutants, which lack the first endodermal pouch (Crump et al., 2004b). However, the first pouch had formed in each of the UAS:DnFgfr1a transgenics, including those with the nkx2.3:Gal4 endodermal driver. As endogenous nkx2.3 expression does not turn on until ≥12 hpf (Li et al., 2019), by the time Gal4/DnFgfr1a protein levels have accumulated, the cells forming the first pouch may no longer require Fgf signaling. Instead, loss of the anterior Hm might be caused by earlier disruption of the D2 MC, which feeds cells to the Hm PCC/cartilage from the anterior border (Movie S1). Reception of Fgf signaling is thus cell-autonomously required in both endoderm and neural crest-derived cells for pouching and anterior Hm formation.
Larvae carrying UAS:DnFgfr1a with the barx1 or col2a1a drivers were indistinguishable from controls. This suggests that once cells have differentiated far enough to express these markers, they become insensitive to loss of Fgf reception. Alternatively, these drivers may not activate sufficiently high levels of DnFgfr1a to interfere with endogenous signaling.
Notch reception is required within neural crest-derived cells to pattern facial cartilage
We had previously observed that the same anterior D2 MC is also patterned by Notch signaling. Jag1-Notch2/3 signaling in the posterior dorsal first and second arches inhibits barx1 expression posteriorly, thereby confining it to more anterior domains of each arch (Barske et al., 2016). jag1b and notch2; notch3 mutants show ectopic barx1 in the dorsal posterior cells of the first and second arches and later develop abnormal bulges on the posterior palatoquadrate and hyomandibula cartilages. These bulges were rescued in jag1b; barx1 double mutants (Barske et al., 2016), indicating that the ectopic barx1 expression specifically contributed to their formation. This Notch-dependent restriction of MC gene expression may be limited to barx1. pax9 expression is not altered in jag1b mutants, whereas lhx6 is already weakly expressed in posterior cells in wild-type embryos, indicating that it is not actively repressed by endogenous Notch activity (Fig. 5C). Transplant experiments also previously demonstrated that jag1b is required in the NC to pattern the facial cartilages (Zuniga et al., 2010). However, it remained unclear whether the reception of this signal by Notch receptors also occurs in NC cells, as Notch receptors are also expressed in arch epithelia (Barske et al., 2016; Zhang et al., 2017). To determine which cell type requires Notch reception, we used the same battery of Gal4 drivers in combination with a UAS:DnMAML line carrying a dominant-negative version of the Notch effector Mastermind-like. Only larvae carrying the NCC driver sox10:Gal4VP16 with UAS:DnMAML showed a skeletal phenotype similar to the jag1b and notch2; notch3 mutants (Fig. 5A–B), confirming that Notch acts within NC-derived cells to pattern cartilage.
Fig. 5. Tissue-specific blockade of Notch reception in migrating neural crest disrupts facial skeletal patterning.
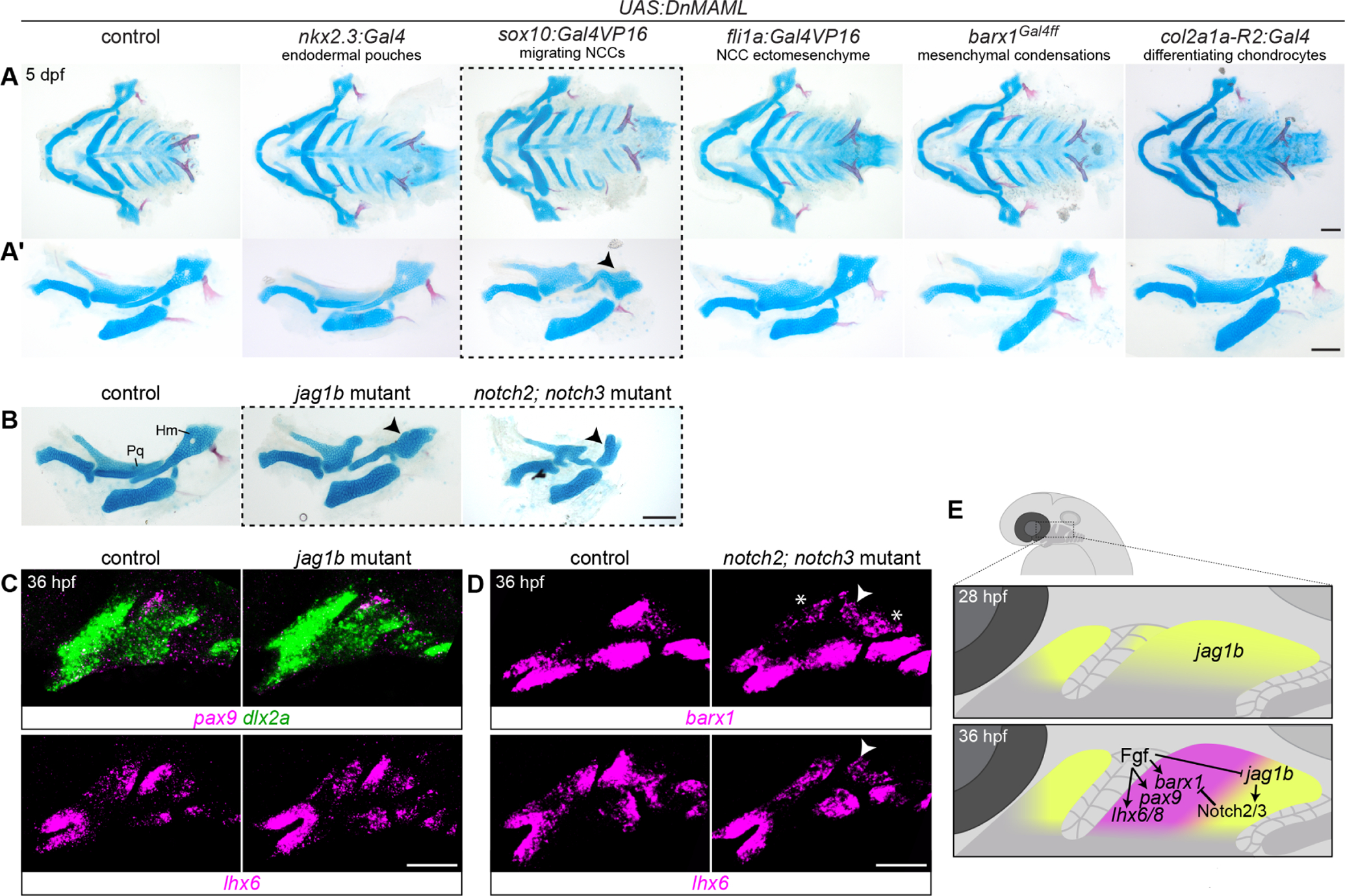
A, Five tissue-specific Gal4 drivers were crossed to UAS:DnMAML transgenic fish, and effects on facial skeletal development were evaluated by Alcian blue and Alizarin red staining at 5 dpf. Only the sox10:Gal4VP16 neural crest driver altered the dorsal palatoquadrate (Pq) and hyomandibula (Hm) cartilages in a manner reminiscent of jag1b and notch2; notch3 mutants (dotted lines; compare to mutants shown in B). Ventral mounts of the viscerocrania are shown in A, while dissected mandibular and hyoid skeletons are presented in A’. Black arrowheads indicate reduction in the anterior hyomandibula. C, Expression of other MC genes pax9 and lhx6 is not altered in jag1b mutants. dlx2a marks arch CNC-derived cells. D, barx1 and lhx6 expression is reduced in the dorsal anterior second arch (white arrowheads) in the more severe notch2; notch3 mutants at 36 hpf. barx1 is also ectopically expression in the posterior first and second arches (asterisks). E, Summary model of Fgf and Notch regulation of MC genes in the developing dorsal hyoid arch between 28 and 36 hpf, prior to PCC formation. Scale bars in A-B = 100 μm; in C-D = 50 μm.
Intersection of Jagged-Notch and Fgf signaling in facial development
barx1 is activated in the anterior D2 MC domain at approximately 32 hpf, well after it turns on ventrally (< 26 hpf) (Barske et al., 2016). This activation is coincident with the gradual restriction of jag1b to the posterior half of the second arch. jag1b is initially expressed in arch epithelia and in NC-derived cells across the full dorsal extent of each arch at 28 hpf (Zuniga et al., 2010). The epithelial domain then fades, and jag1b expression becomes confined to the dorsal-posterior corners of each arch by 32 hpf. This retraction of jag1b expression does not depend on Barx1, as barx1 mutants show normal jag1b expression (Barske et al., 2016). We therefore hypothesized that the same signals (i.e., Fgf) that activate barx1 in the anterior domain may simultaneously inhibit jag1b. We tested this idea by blocking Fgf signaling after anterior pouch formation has completed, using hsp70l:Gal4; UAS:DnFgfr1a embryos heat-shocked between 22–24 hpf. jag1b was robustly expressed across the entire dorsal domain at 36 hpf in doubly transgenic larvae, appearing expanded relative to control siblings showing the posterior restriction typical for this stage (n = 4/4 affected; Fig. 4E).
To test whether activation of barx1 in the D2 domain by Fgf occurs indirectly via this repression of jag1b, we simultaneously inhibited both Fgf and Notch signaling in hsp70l:Gal4; UAS:DnFgfr1a embryos heat-shocked from 20:30–22:30 hpf and then treated with the gamma-secretase inhibitor DBZ from 24–36 hpf. barx1 expression was restored in this case, albeit only in the ectopic posterior domains that form when Notch is inhibited, not in the typical anterior D2 domain (Fig. 4I). These results suggest that Fgf signaling likely directly activates barx1 in the anterior hyoid arch, not just indirectly through its inhibition of jag1b expression.
A reanalysis of barx1 and lhx6 expression in notch2; notch3 mutants revealed a similar pattern to these doubly inhibited embryos: expression was reduced in the anterior dorsal hyoid arch and elevated in the posterior (Fig. 5D, (Barske et al., 2016)). This phenotype prefigures the defects in the anterior Hm cartilage seen in severe Notch pathway mutants (Fig. 5A–B), which were exacerbated rather than rescued when both jag1b and barx1 were mutated (Barske et al., 2016). One interpretation of this phenotype is that early Notch signaling events may enhance Fgf receptivity in the anterior dorsal second arch, thereby initiating a feedback loop that eventually leads to the retraction of jag1b expression to the posterior domain.
Discussion
This study confirms that non-odontogenic mesenchymal condensations in the pharyngeal arches give rise to precartilaginous condensations and thus inform the shapes of facial cartilages. MCs form at specific coordinates in the arches, adjacent to endodermal or ectodermal arch epithelia. Using a new barx1 Gal4ff knockin line, we show for the first time that most – but not all – PCCs and cartilage in the face clearly pass through a barx1+ MC state. We propose that epithelial signals induce underlying mesenchyme to condense (as in developing teeth); then, as daughter cells are pushed further away from the source of the signal, they activate sox9a expression and advance to the PCC stage of differentiation. Whether these zebrafish MCs are true condensations with elevated cell density or unique adhesive properties remains to be determined. However, where MCs are located determines where PCCs form and the axis along which they grow. To illustrate this, we show that positive Fgf and inhibitory Notch signals intersect during early arch development to place the dorsal hyoid MC in its anterior position (Fig. 5E). This has the effect of positioning the hyomandibular cartilage, which evolves from this MC, at the right site to connect to the otic cartilage above and the intermediate Sy cartilage below, thereby buttressing the jaw skeleton to the neurocranium.
Some cartilages did not express the barx1 lineage marker (Fig. 3, Movies S1–S4). One possible interpretation is that some cartilages, like the otic cartilage surrounding the ear, do not require passage through the MC state. Alternatively, they may traverse the MC state quickly and shut down MC markers earlier than other cartilages – this appears to be the case for the symplectic and interhyal joint cartilage (Fig. 3F). Exclusion of barx1 expression from joint-forming regions is consistent with Nichols et al. (2013), who found that Barx1 function represses formation of ectopic joints within the ventral cartilages in zebrafish and is sufficient to block joint formation when misexpressed broadly throughout the arches (Nichols et al., 2013). Joint chondrocytes may require the absence of Barx1 in order to maintain their immature, Col2a1a-negative state (Askary et al., 2015). Most of the seemingly barx1-negative cartilages are elongated rod shapes rather than large and flat or thick rods, suggesting a possible correlation between cartilage/PCC shape and the duration of MC contribution. A final possibility is that their associated MCs simply did not express barx1 – not all MCs express all recognized MC transcription factors (Fig. 1G–J). However, none of the other assayed MC markers were expressed, for example in the barx1-negative otic cartilage.
The question of how exactly MCs progress into the PCC state is still open. We note that in zebrafish, facial MCs consistently form directly under epithelia, whereas PCCs are positioned more centrally. MC cell divisions that increase the distance from a signal source may thus contribute to maturation. Careful pseudo-time analyses of single-cell RNAseq data from skeletal progenitors collected before and during PCC emergence should reveal the transcriptional changes that accompany intermediate stages in this MC-PCC progression. Further, the degree of similarity between cartilage- and tooth-fated MCs warrants further investigation, as does the identity of factors or conditions that push them in either direction. Clues may come via comparing first arch MCs from zebrafish to those of related species like cavefish that have retained teeth on their oral jaws (Hammer et al., 2016). It remains to be seen how similar these fine-scale patterning mechanisms in the zebrafish embryo are to the analogous processes operating in the limb and axial skeletons as well as in the faces of much larger vertebrate embryos. It nevertheless seems likely that shifts in the distribution and size of mesenchymal condensations contributed to the innumerable changes in skeletal form that occurred throughout vertebrate evolution.
Supplementary Material
Highlights.
Endochondral bone shape is prefigured in early precartilaginous condensations.
Precartilaginous condensations arise from precursor mesenchymal condensations.
Mesenchymal condensations in the face are positioned by Fgf and Notch signaling.
Acknowledgements
We thank the Zebrafish International Resource Center for supplying the sox9ahi1134Tg fish line. We are also grateful to Danielle Fritsch and Eric Alley of the CCHMC Division of Veterinary Services for fish care and to Matthew Kofron at CCHMC Confocal Imaging Core for imaging assistance. This work was supported by the National Institute of Dental and Craniofacial Research (R00 DE026239 to L.B.) and the Cincinnati Children’s Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
Nothing to declare.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M, 1994. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci 14, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B, 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16, 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunni A, Krecsmarik M, Bosco A, Galant S, Pan L, Moens CB, Bally-Cuif L, 2013. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development 140, 3335–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya E, Musci TJ, Kirschner MW, 1991. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 66, 257–270. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N, 1999. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev 13, 2713–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A, Mork L, Paul S, He X, Izuhara AK, Gopalakrishnan S, Ichida JK, McMahon AP, Dabizljevic S, Dale R, Mariani FV, Crump JG, 2015. Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev Cell 35, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AJ, Bogardi JP, Ladher R, Francis-West PH, 1999. Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev Dyn 214, 291–302. [DOI] [PubMed] [Google Scholar]
- Barske L, Askary A, Zuniga E, Balczerski B, Bump P, Nichols JT, Crump JG, 2016. Competition between Jagged-Notch and Endothelin1 Signaling Selectively Restricts Cartilage Formation in the Zebrafish Upper Face. PLoS Genet 12, e1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L, Fabian P, Hirschberger C, Jandzik D, Square T, Xu P, Nelson N, Yu HV, Medeiros DM, Gillis JA, Crump JG, 2020. Evolution of vertebrate gill covers via shifts in an ancient Pou3f3 enhancer. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L, Rataud P, Behizad K, Del Rio L, Cox SG, Crump JG, 2018. Essential Role of Nr2f Nuclear Receptors in Patterning the Vertebrate Upper Jaw. Dev Cell 44, 337–347 e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS, 1997. SOX9 directly regulates the type-II collagen gene. Nat Genet 16, 174–178. [DOI] [PubMed] [Google Scholar]
- Blentic A, Tandon P, Payton S, Walshe J, Carney T, Kelsh RN, Mason I, Graham A, 2008. The emergence of ectomesenchyme. Dev Dyn 237, 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky JL, Chien CB, 2005. Molecular cloning and developmental expression of foxP2 in zebrafish. Dev Dyn 234, 740–746. [DOI] [PubMed] [Google Scholar]
- Cesario J, Ha S, Kim J, Kataria N, Jeong J, 2021. Candidate positive targets of LHX6 and LHX8 transcription factors in the developing upper jaw. Gene Expr Patterns 43, 119227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JW, Galloway JL, 2014. The development of zebrafish tendon and ligament progenitors. Development 141, 2035–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Collazo A, Trinh le A, Pan L, Moens CB, Crump JG, 2013. Wnt-dependent epithelial transitions drive pharyngeal pouch formation. Dev Cell 24, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Crump JG, 2014. Tbx1 controls the morphogenesis of pharyngeal pouch epithelia through mesodermal Wnt11r and Fgf8a. Development 141, 3583–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB, 2004a. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development 131, 5703–5716. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Kimmel CB, 2004b. An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol 2, E244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Crump JG, 2012. Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet 8, e1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David NB, Saint-Etienne L, Tsang M, Schilling TF, Rosa FM, 2002. Requirement for endoderm and FGF3 in ventral head skeleton formation. Development 129, 4457–4468. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Sharpe PT, Pachnis V, 2009. The LIM homeodomain transcription factors Lhx6 and Lhx7 are key regulators of mammalian dentition. Dev Biol 333, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames BF, Delaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, McFadden M, Sasaki MM, Kimmel CB, 2013. FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution. BMC Dev Biol 13, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR, 2019. Genetic compensation triggered by mutant mRNA degradation. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell HB, 1925. The histogenesis of cartilage and bone in the long bones of the embryonic fowl. J Morphol 40, 417–459. [Google Scholar]
- Giffin JL, Gaitor D, Franz-Odendaal TA, 2019. The Forgotten Skeletogenic Condensations: A Comparison of Early Skeletal Development Amongst Vertebrates. J Dev Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V, 1998. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 125, 2063–2074. [DOI] [PubMed] [Google Scholar]
- Hall BK, 2015. Bones and cartilage : developmental and evolutionary skeletal biology, Second edition. ed. Elsevier/AP, Academic Press is an imprint of Elsevier, Amsterdam. [Google Scholar]
- Hall BK, Miyake T, 2000. All for one and one for all: condensations and the initiation of skeletal development. Bioessays 22, 138–147. [DOI] [PubMed] [Google Scholar]
- Hammer CL, Atukorala ADS, Franz-Odendaal TA, 2016. What shapes the oral jaws? Accommodation of complex dentition correlates with premaxillary but not mandibular shape. Mech Dev 141, 100–108. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW, 2004. Fgf signaling is required for zebrafish tooth development. Dev Biol 274, 139–157. [DOI] [PubMed] [Google Scholar]
- Karsenty G, 2003. The complexities of skeletal biology. Nature 423, 316–318. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF, 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC, 1998. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol 203, 245–263. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S, 2014. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4, 6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J, 1999. Hindlimb patterning and mandible development require the Ptx1 gene. Development 126, 1805–1810. [DOI] [PubMed] [Google Scholar]
- Larbuisson A, Dalcq J, Martial JA, Muller M, 2013. Fgf receptors Fgfr1a and Fgfr2 control the function of pharyngeal endoderm in late cranial cartilage development. Differentiation 86, 192–206. [DOI] [PubMed] [Google Scholar]
- Leerberg DM, Hopton RE, Draper BW, 2019. Fibroblast Growth Factor Receptors Function Redundantly During Zebrafish Embryonic Development. Genetics 212, 1301–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Angelozzi M, Haseeb A, 2019. SOX9 in cartilage development and disease. Curr Opin Cell Biol 61, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ning G, Yang S, Yan Y, Cao Y, Wang Q, 2019. BMP signaling is required for nkx2.3-positive pharyngeal pouch progenitor specification in zebrafish. PLoS Genet 15, e1007996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CF, Angelozzi M, Haseeb A, Lefebvre V, 2018. SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorda-Diez CI, Montero JA, Diaz-Mendoza MJ, Garcia-Porrero JA, Hurle JM, 2011. Defining the earliest transcriptional steps of chondrogenic progenitor specification during the formation of the digits in the embryonic limb. PLoS One 6, e24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovely CB, Swartz ME, McCarthy N, Norrie JL, Eberhart JK, 2016. Bmp signaling mediates endoderm pouch morphogenesis by regulating Fgf signaling in zebrafish. Development 143, 2000–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, Mammoto A, Torisawa YS, Tat T, Gibbs A, Derda R, Mannix R, de Bruijn M, Yung CW, Huh D, Ingber DE, 2011. Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev Cell 21, 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandler M, Neubuser A, 2001. FGF signaling is necessary for the specification of the odontogenic mesenchyme. Dev Biol 240, 548–559. [DOI] [PubMed] [Google Scholar]
- Mariani FV, Martin GR, 2003. Deciphering skeletal patterning: clues from the limb. Nature 423, 319–325. [DOI] [PubMed] [Google Scholar]
- McGurk PD, Swartz ME, Chen JW, Galloway JL, Eberhart JK, 2017. In vivo zebrafish morphogenesis shows Cyp26b1 promotes tendon condensation and musculoskeletal patterning in the embryonic jaw. PLoS Genet 13, e1007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown SJ, Lee VM, Bronner-Fraser M, Newgreen DF, Farlie PG, 2005. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev Dyn 233, 430–444. [DOI] [PubMed] [Google Scholar]
- Meech R, Edelman DB, Jones FS, Makarenkova HP, 2005. The homeobox transcription factor Barx2 regulates chondrogenesis during limb development. Development 132, 2135–2146. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M, 2007. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One 2, e787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletich I, Yu WY, Zhang R, Yang K, Caixeta de Andrade S, Pereira SF, Ohazama A, Mock OB, Buchner G, Sealby J, Webster Z, Zhao M, Bei M, Sharpe PT, 2011. Developmental stalling and organ-autonomous regulation of morphogenesis. Proc Natl Acad Sci U S A 108, 19270–19275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Drouin J, 2008. Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev Biol 313, 887–896. [DOI] [PubMed] [Google Scholar]
- Miyake T, Cameron AM, Hall BK, 1996. Stage-specific onset of condensation and matrix deposition for Meckel’s and other first arch cartilages in inbred C57BL/6 mice. J Craniofac Genet Dev Biol 16, 32–47. [PubMed] [Google Scholar]
- Molina GA, Watkins SC, Tsang M, 2007. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork L, Crump G, 2015. Zebrafish Craniofacial Development: A Window into Early Patterning. Curr Top Dev Biol 115, 235–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Kenesi E, Rentsendorj O, Molnar A, Szenasi T, Sinko I, Zvara A, Oommen ST, Barta E, Puskas LG, Lefebvre V, Kiss I, 2011. Evolutionarily conserved, growth plate zone-specific regulation of the matrilin-1 promoter: L-Sox5/Sox6 and Nfi factors bound near TATA finely tune activation by Sox9. Mol Cell Biol 31, 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P, 1997. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol 183, 108–121. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Pan L, Moens CB, Kimmel CB, 2013. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development 140, 2765–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R, 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev 12, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL, 1997. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and −2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol 185, 165–184. [DOI] [PubMed] [Google Scholar]
- Rentsendorj O, Nagy A, Sinko I, Daraba A, Barta E, Kiss I, 2005. Highly conserved proximal promoter element harbouring paired Sox9-binding sites contributes to the tissue- and developmental stage-specific activity of the matrilin-1 gene. Biochem J 389, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB, 1997. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 124, 2945–2960. [DOI] [PubMed] [Google Scholar]
- Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M, 2000. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem 275, 10738–10744. [DOI] [PubMed] [Google Scholar]
- Song H, Park KH, 2020. Regulation and function of SOX9 during cartilage development and regeneration. Semin Cancer Biol 67, 12–23. [DOI] [PubMed] [Google Scholar]
- Sperber SM, Dawid IB, 2008. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Dev Biol 321, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Amand TR, Zhang Y, Semina EV, Zhao X, Hu Y, Nguyen L, Murray JC, Chen Y, 2000. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev Biol 217, 323–332. [DOI] [PubMed] [Google Scholar]
- Swartz ME, Sheehan-Rooney K, Dixon MJ, Eberhart JK, 2011. Examination of a palatogenic gene program in zebrafish. Developmental dynamics 240, 2204–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto DP, Rodriguez-Esteban C, Ryan AK, O’Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisua-Belmonte JC, Rosenfeld MG, 1999. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev 13, 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB, 2010. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Walker MB, Carney TJ, Huycke TR, Yan YL, BreMiller RA, Gai L, Delaurier A, Postlethwait JH, Hammerschmidt M, Kimmel CB, 2012. fras1 shapes endodermal pouch 1 and stabilizes zebrafish pharyngeal skeletal development. Development 139, 2804–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT, 1997. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development 124, 4811–4818. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Ferguson CA, Bach I, Rosenfeld MG, Sharpe PT, 1999. Conserved regulation of mesenchymal gene expression by Fgf-8 in face and limb development. Development 126, 221–228. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT, 1998. Transformation of tooth type induced by inhibition of BMP signaling. Science 282, 1136–1138. [DOI] [PubMed] [Google Scholar]
- Ullmann B, 2011. Juvenile No Acid Bone & Cartilage Stain, ZFIN Protocol Wiki. University of Oregon, Zebrafish Information Network (ZFIN). [Google Scholar]
- Walker MB, Kimmel CB, 2007. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem 82, 23–28. [DOI] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Coffin Talbot J, Stock DW, Kimmel CB, 2006. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol 295, 194–205. [DOI] [PubMed] [Google Scholar]
- Westerfield M, 2007. THE ZEBRAFISH BOOK: A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- Xu P, Balczerski B, Ciozda A, Louie K, Oralova V, Huysseune A, Crump JG, 2018. Fox proteins are modular competency factors for facial cartilage and tooth specification. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Yu HV, Tseng KC, Flath M, Fabian P, Segil N, Crump JG, 2021. Foxc1 establishes enhancer accessibility for craniofacial cartilage differentiation. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH, 2002. A zebrafish sox9 gene required for cartilage morphogenesis. Development 129, 5065–5079. [DOI] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH, 2005. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069–1083. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang L, Wong EYM, Tsang SL, Xu PX, Lendahl U, Sham MH, 2017. An Eya1-Notch axis specifies bipotential epibranchial differentiation in mammalian craniofacial morphogenesis. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YD, Chen Z, Song YQ, Liu C, Chen YP, 2005. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res 15, 301–316. [DOI] [PubMed] [Google Scholar]
- Zhao C, Andreeva V, Gibert Y, LaBonty M, Lattanzi V, Prabhudesai S, Zhou Y, Zon L, McCann KL, Baserga S, Yelick PC, 2014. Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS Genet 10, e1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinck NW, Jeradi S, Franz-Odendaal TA, 2021. Elucidating the early signaling cues involved in zebrafish chondrogenesis and cartilage morphology. J Exp Zool B Mol Dev Evol 336, 18–31. [DOI] [PubMed] [Google Scholar]
- Zuniga E, Stellabotte F, Crump JG, 2010. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development 137, 1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


